Significance
Tissue resident macrophages are long-lived, self-replenishing myeloid cells. They harbor and support HIV-1 replication, but unlike CD4+ T cells, do not succumb to virus-induced cell death. Here, we have screened a panel of 90 long noncoding RNAs (lncRNA) and identified a lncRNA, SAF, that plays a critical role in the resistance of HIV-1–infected macrophages to activation of apoptotic caspases. We have further shown that down-regulation of SAF expression with siRNA treatment can activate effector caspase-3/7 specifically in virus-infected macrophages without affecting the uninfected and bystander cells. Overall, our study describes the approach of modulating the lncRNA SAF for targeted elimination of HIV-1–infected macrophages that can lead to reduction and potential clearance of these viral reservoir cells.
Keywords: HIV, macrophage, apoptosis, lncRNA, AIDS
Abstract
Long noncoding RNAs (lncRNAs) impart significant regulatory functions in a diverse array of biological pathways and manipulation of these RNAs provides an important avenue to modulate such pathways, particularly in disease. Our knowledge about lncRNAs’ role in determination of cellular fate during HIV-1 infection remains sparse. Here, we have identified the impact of the lncRNA SAF in regulating apoptotic effector caspases in macrophages, a long-lived cellular reservoir of HIV-1, that are largely immune to virus-induced cell death. Expression of SAF is significantly up-regulated in HIV-1–infected human monocyte-derived macrophages (MDM) compared with bystander and virus-nonexposed cells. A similar enhancement in SAF RNA expression is also detected in the HIV-1–infected airway macrophages obtained by bronchoalveolar lavage of HIV-1–infected individuals. Down-regulation of SAF with siRNA treatment increases caspase-3/7 activity levels in virus-infected MDMs. This induction of apoptotic caspases occurs exclusively in HIV-1–infected macrophages and not in bystander cells, leading to a significant reduction in HIV-1 replication and overall viral burden in the macrophage culture. This study identifies targeting of the lncRNA SAF as a potential means to specifically induce cell death in HIV-1–infected macrophages.
Cell death is a prominent feature of HIV-1 pathogenesis that is characterized by severe loss of T lymphocytes and other immune cells during the disease progression (1–6). Infection of activated CD4+ T cells, the primary target of HIV-1, results in efficient and productive virus replication but rapidly leads to a virus-induced cytopathic death of the infected cells (7). Another cellular target of HIV-1 infection are the different macrophage lineages (8). Tissue macrophages are long-lived cells that are maintained in their local tissue environment through self-renewal (9–12). These myeloid cells reside in various tissues and vary in their phenotype and function depending upon their location (13). Several tissue macrophages such as brain microglia, liver Kupffer cells, and alveolar macrophages have long been recognized to harbor and support HIV-1/Simian immunodeficiency virus (SIV) infection at different stages of the disease (14–21). In fact, in recent studies, using humanized myeloid-only mice and CD4+ T cell-depleted rhesus macaques, it has been shown that tissue macrophages can support HIV-1/SIV replication and maintain prolonged plasma viremia even in the absence of T cells (22–24). Although the half-life of infected macrophages estimated on the basis of the viral decay kinetics in these macaques was determined to be relatively short, the high viral burden, rapid rate of disease progression, and aberrant immune activation in the CD4+ T cell-depleted milieu may all impact the macrophage half-life. Nonetheless, these studies emphasize the contribution of tissue macrophages to HIV-1/SIV viral persistence. Moreover, several other in vivo and in vitro studies indicate that in comparison with CD4+ T cells, macrophages support relatively lower levels of HIV-1 replication and are relatively resistant to virus-induced cell death (25–28). This unique ability of macrophages to sustain viral replication while retaining extended cell survival makes them an ideal candidate for viral persistence even during long-term antiretroviral therapy, which poses a major hurdle in the HIV-1 cure efforts. A complex host-pathogen interaction must be in play to maintain such a delicate balance between productive HIV-1 infection and macrophage survival. A comprehensive understanding of how HIV-1 modulates cell death and/or survival pathways in infected macrophages is likely to be a requirement for the effective elimination of the infection.
Apoptosis is the most commonly reported form of virus-induced programmed cell death, and several studies have examined the mechanisms involved in induction or evasion of apoptosis by HIV-1 (29–31). These studies are mainly focused on the role of various protein modulators, either viral (HIV-1 Nef, Vpr, Tat, and Env) or cellular (Fas, TNF-α, Bcl-2, Bax, FLICE, p53) factors (32–34). However, there is only limited information available on how HIV-1 infection affects nonprotein-coding regulatory elements such as the long noncoding RNAs (lncRNA). lncRNAs are RNA transcripts that are larger than 200 nucleotides in length yet lack a protein-coding ORF. They are now known to play a significant role in regulating diverse cellular pathways both in health and disease, including viral infections such as HIV-1 (35, 36). The lncRNAs NEAT1 and NRON have been shown to inhibit HIV replication by regulating nuclear localization of viral transcripts (37, 38). Several lncRNAs modulate cell apoptotic pathways at both extrinsic and intrinsic levels. For instance, SAF (FAS-AS1) and HOXA-AS2 are antiapoptotic lncRNAs that regulate death receptor functions in the extrinsic pathway, while the lncRNAs Malat1, GAS5, and MEG3 act on the intracellular regulator p53 to induce apoptosis (39–43). The possible roles of such lncRNAs during HIV-1 infection have yet to be explored.
In the current study, we determined that the lncRNA expression profile in HIV-1–infected human macrophages was distinct from that of their noninfected counterparts. We utilized an in vitro HIV-1 infection model of human primary monocyte-derived-macrophage (MDM) culture to quantify and compare expression of 90 well-characterized lncRNAs in three MDM cell populations: HIV-1–infected, HIV-1–exposed but uninfected bystander, and virus-nonexposed control MDMs. Across the panel of 90 lncRNAs, we observed significant up-regulation in expression of the lncRNA SAF (FAS-AS1) in HIV-1–infected MDMs in comparison with both bystander and nonexposed cells. Moreover, we also found that SAF expression was up-regulated in HIV-1–infected airway macrophages recovered by bronchoalveolar lavage (BAL) from HIV-1-positive donors from Malawi. SAF is an antiapoptotic lncRNA that protects against Fas-mediated programmed cell death (43–45). We further demonstrated that siRNA-mediated down-regulation of the lncRNA SAF in MDMs led to activation of apoptotic effector caspase-3/7 specifically in HIV-1–infected cells but not in bystander cells. The increase in cell death lead to a reduction in viral burden in the MDM culture, indicating a potential opportunity to reduce or eliminate HIV-1–infected macrophage reservoirs by modulation of lncRNA-mediated cell survival.
Results
HIV-1 Infection of Human MDMs Does Not Activate Apoptotic Effector Caspases or Lead to Cell Death.
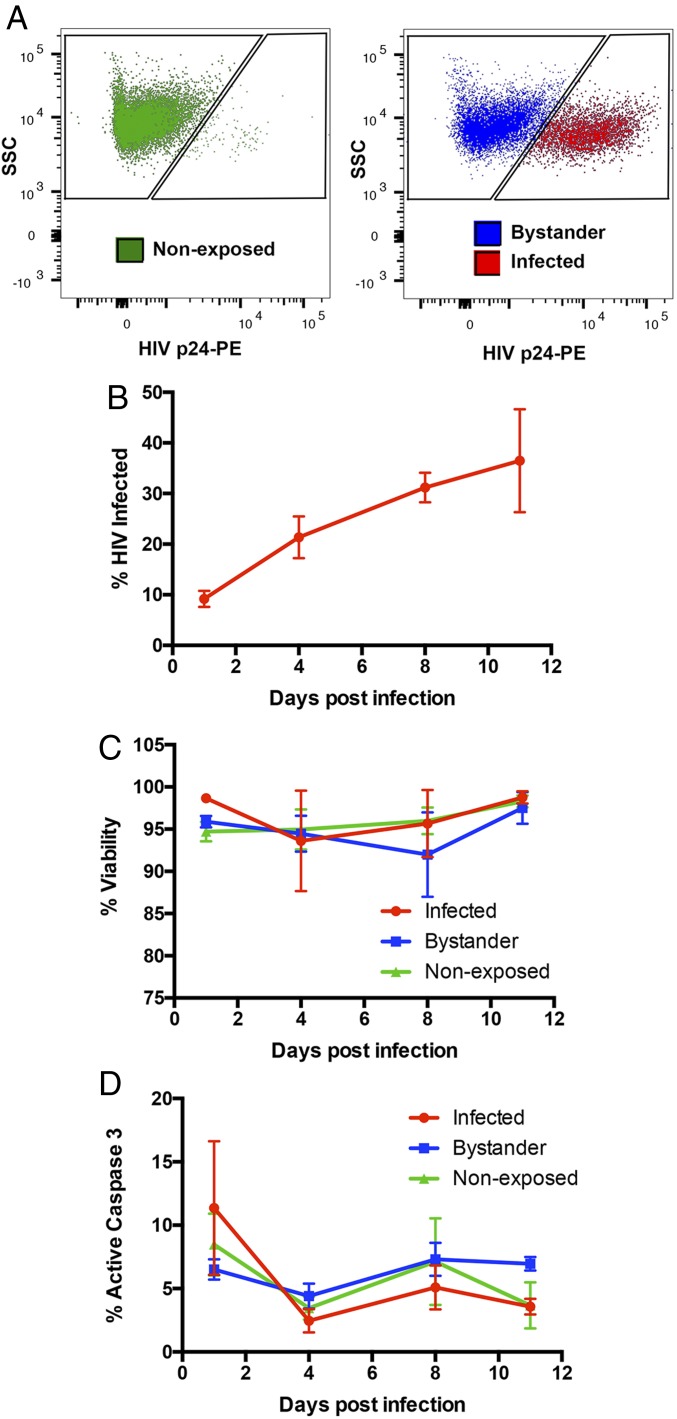
To understand the dynamics of viral replication and activation of apoptosis in HIV-1–infected primary macrophages, human MDMs were infected with a replication-competent vesicular stomatitus virus-G (VSV-G) pseudotyped glycoprotein pseudotyped HIV-1 (isolate HXB3) virus (HXB3) virus expressing a R5-tropic BaL envelope (HXB3/BaL) and monitored up to 11 d postinfection (dpi) for viral infection and induction of apoptosis. As illustrated in Fig. 1A, HIV-1–infected macrophages were distinguished from uninfected bystanders by flow cytometric detection of intracellular expression of the viral capsid (p24) protein. The proportion of p24-positive HIV-1–infected cells within the virus-infected culture increased progressively from 9.2% on 1 dpi to 36.5% on 11 dpi when the experiment was terminated (Fig. 1B). This demonstrates that HIV-1 establishes a productive and spreading viral infection in the primary macrophage culture. However, this productive infection did not increase cell death in the infected culture. Throughout the 11 d of infection, cell viability remained high in infected (mean, 97 ± 1.7%), and in bystander (95 ± 1.4%) or nonexposed (96 ± 0.8%) cells (Fig. 1C). Level of active caspase-3/7, which are effector proteases of apoptosis, remained low in infected cells (2.5–11.4%), at levels of expression comparable to those of bystander (4.4–7.3%) and nonexposed cells (3.5–8.5%) throughout the infection (Fig. 1D). Together, these data indicate that primary human macrophages support productive HIV-1 infection but resist the activation of apoptotic effector caspases and cell death that is usually associated with HIV-1 infection of lymphocytes.
Fig. 1.
HIV-1 infection and apoptosis in MDMs. (A) Representative flow cytometry plots showing HIV-1 p24 staining of MDMs and demarcation of virus-nonexposed (Green), virus-exposed but uninfected bystander (Blue), and virus-infected (Red) cells. Cells were gated for singlets (FSC-H vs. FSC-A) and MDM (SSC-A vs. FSC-A) before analysis of HIV-1 p24 expression. (B) The percentage (mean ± SEM) of HIV-1–infected MDMs was determined by flow cytometry staining with PE-conjugated anti-p24 antibody on days 1, 4, 8, and 11 postinfection (n = 4). (C and D) The percentage (mean ± SEM) of viable (C) and active caspase 3-positive (D) MDMs was determined by flow cytometry staining with fixable viability dye eFluor506 and CellEvent caspase-3/7 green detection reagent, respectively, within virus-nonexposed (green), bystander (blue), and infected (red) cells at indicated days postinfection (n = 4).
Expression of lncRNA SAF Is Up-Regulated in HIV-1–Infected MDMs.
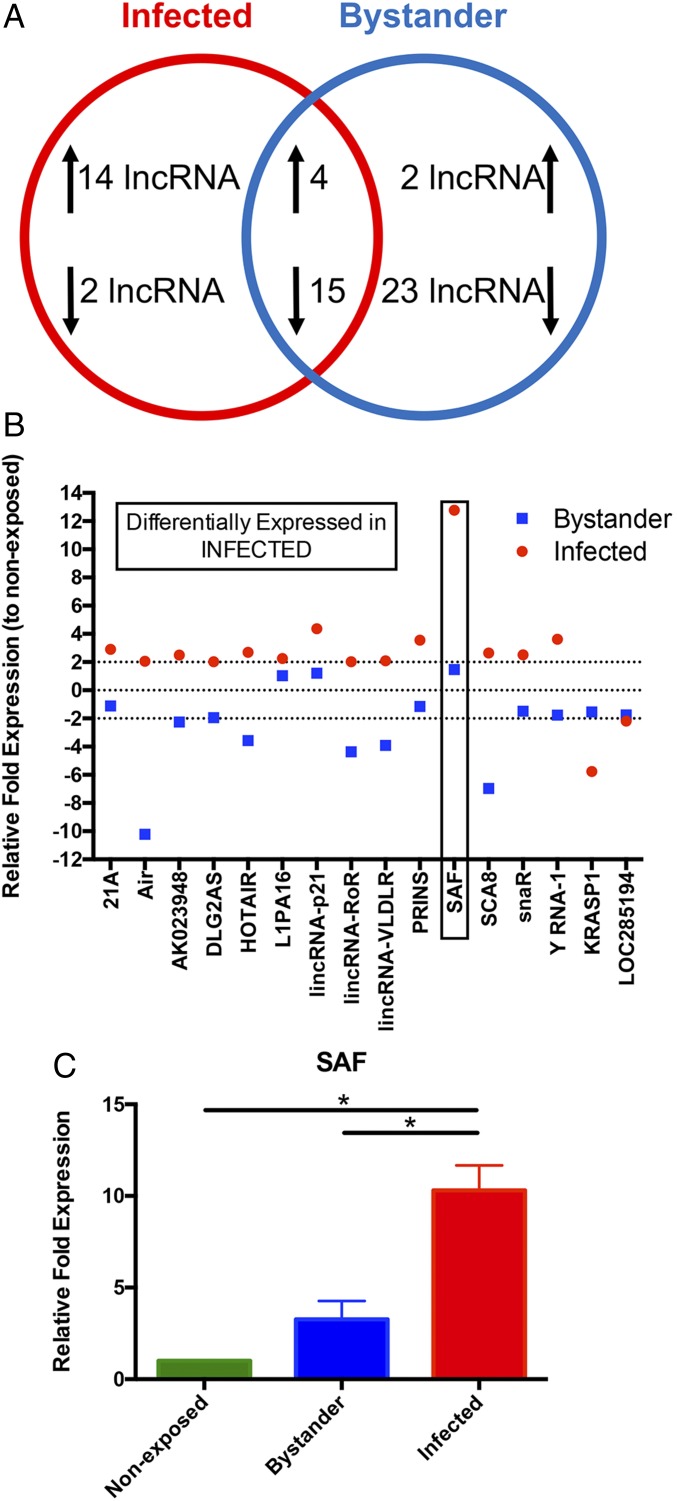
There is increasing evidence that lncRNAs play important roles in HIV-1 infection. However, most reports have focused on identifying those lncRNAs that impact viral replication (37, 38). The differential expression of lncRNAs in HIV-1–infected and exposed but uninfected macrophages has not been explored. To identify the lncRNAs that are differentially expressed in HIV-1–infected versus bystander cells, we infected MDMs with a replication-competent mCherry-reporter HIV-1 virus for 7 d and sorted HIV-1–infected (mCherry-positive) and bystander (mCherry-negative) MDMs. Uninfected, nonexposed MDMs were also processed through the cell sorter as control cells. The expression levels of 90 well-characterized lncRNAs, including a number of lncRNAs implicated in apoptosis, were determined using a quantitative real-time PCR (qRT-PCR) based array. Out of the 90, expression of 71 lncRNAs was detected in all three groups and therefore was used for further analysis (SI Appendix, Fig. S1). Comparison of the lncRNA expression profile among nonexposed, bystander, and virus-infected MDMs revealed that 18 lncRNAs were up-regulated (≥twofold) and 17 were down-regulated (≤twofold) in HIV-1–infected MDMs, whereas in bystander cells only six were up-regulated. A considerable number of lncRNAs (36 out of 71) were down-regulated in the bystander MDMs (Fig. 2A). Changes in 19 lncRNAs followed a similar pattern in both virus-infected and bystander cells. A further examination of the 16 lncRNAs that were differentially expressed in HIV-1–infected MDMs (Fig. 2B) revealed that the change was most pronounced in the lncRNA SAF. This antiapoptotic lncRNA had the highest increase in expression in HIV-1–infected cells but exhibited minimal up-regulation in the bystander MDMs, indicating that this variation was likely to be HIV-1 infection-specific. To further validate this lncRNA profiler array data, we measured SAF expression in HIV-1–infected, bystander, and nonexposed cells using a different set of published qRT-PCR primers (43) in four independent MDM cultures. Consistent with the lncRNA profiler data, the independent qRT-PCR results (Fig. 2C) confirmed that SAF expression is significantly elevated in HIV-1–infected cells compared with bystander or virus-nonexposed control MDMs.
Fig. 2.
lncRNA SAF expression in HIV-1–infected MDMs. (A) Venn diagram summarizing changes in lncRNA expression profile in HIV-1–infected and bystander cells compared with virus-nonexposed MDMs. At least twofold up- or down-regulation compared with nonexposed MDMs was considered as a change in expression. (B) A summary of the relative expressions of the 16 lncRNAs that were differentially expressed in HIV-1–infected (red dot) MDMs compared with bystander (blue box) cells. The lncRNA SAF is identified with a box. (C) Fold changes (mean ± SEM) in expression of the lncRNA SAF were determined by qRT-PCR in nonexposed, bystander, and virus-infected MDMs (n = 4). Significance of difference among groups determined by one-way ANOVA is indicated above the groups, *P < 0.05.
Expression of lncRNA SAF Is Enhanced in HIV-1–Infected Human BAL-Derived Airway Macrophages.
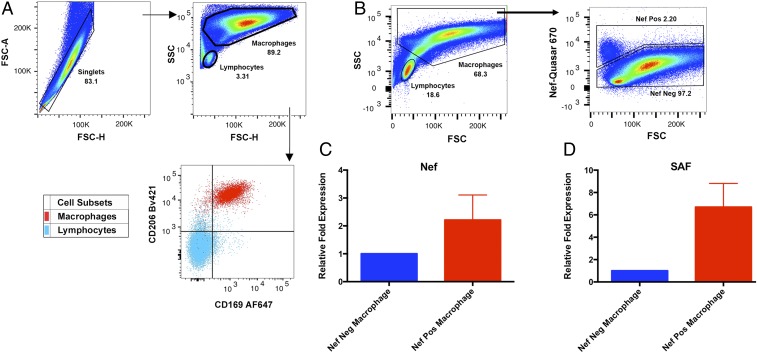
To determine if the increase in SAF expression observed in MDMs in vitro also occurs in vivo during HIV-1 infection, we measured SAF expression in bronchoalveolar lavage (BAL)-derived airway macrophages from HIV-1–infected individuals. Airway macrophages in HIV-1–infected individuals have previously been shown to harbor the virus, and viral RNA has been detected by fluorescent in situ hybridization (FISH) and PCR-based assays (14, 46). We obtained airway macrophages from three HIV-1–infected, antiretroviral therapy naïve individuals by BAL. Following a previously published gating strategy for human BAL cells (14, 47), the two major targets of HIV-1, macrophages and lymphocytes, can be differentiated on the basis of their relative size and granularity. Surface marker analysis of the cells gated as macrophages demonstrates that they uniformly express the macrophage surface markers CD206 and CD169 (47), while the smaller cells gated as lymphocytes do not (Fig. 3A). HIV-1–infected cells within this airway macrophage population were detected and flow sorted on the basis of FISH staining with fluorescent (Quasar 670)-labeled HIV-1 Nef probes (Fig. 3B). Analysis of the Nef-positive cell population by qRT-PCR demonstrated a two to threefold higher level of Nef RNA transcripts in those cells (Fig. 3C), indicating an enrichment of infected cells through flow cytometry sorting. When we compared the levels of expression of the lncRNA SAF, we found that SAF RNA transcripts were approximately sixfold more abundant in Nef-positive cells in comparison with the Nef-negative cells in two of the three individuals (Fig. 3D). Expression of SAF could not be compared in the third individual as it was undetectable by PCR amplification in the Nef-positive cells. These data indicate that during natural HIV-1 infection, the virus induces enhancement of SAF expression levels in human airway macrophages comparable to those observed in MDMs in vitro.
Fig. 3.
Expression of lncRNA SAF in BAL-derived airway macrophages from HIV-1–infected individuals. (A) Representative flow cytometry plots showing the gating strategy and surface expression of CD206 and CD169 in macrophages and lymphocytes, performed on study participant HITUB1096Z. (B) Representative flow cytometry plots showing gating strategy for sorting Nef-positive and Nef-negative BAL-derived airway macrophages. BAL cells were first gated for singlets (FSC-H vs. FSC-A) and then macrophages (SSC-A vs. FSC-A) before analysis of HIV Nef-Quasar670 probe staining. (C) Fold changes (mean ± SEM) in expression of the HIV-1 Nef RNA was determined by qRT-PCR in Nef-negative and Nef-positive airway macrophages (n = 3). (D) Fold changes (mean ± SEM) in expression of the lncRNA SAF was determined by qRT-PCR in Nef-negative and Nef-positive airway macrophages (n = 2). This analysis was performed on study participants AMAC282, AMAC283, and AMAC292.
Inhibition of lncRNA SAF Activates Apoptotic Effector Caspase-3/7 in HIV-1–Infected MDMs.
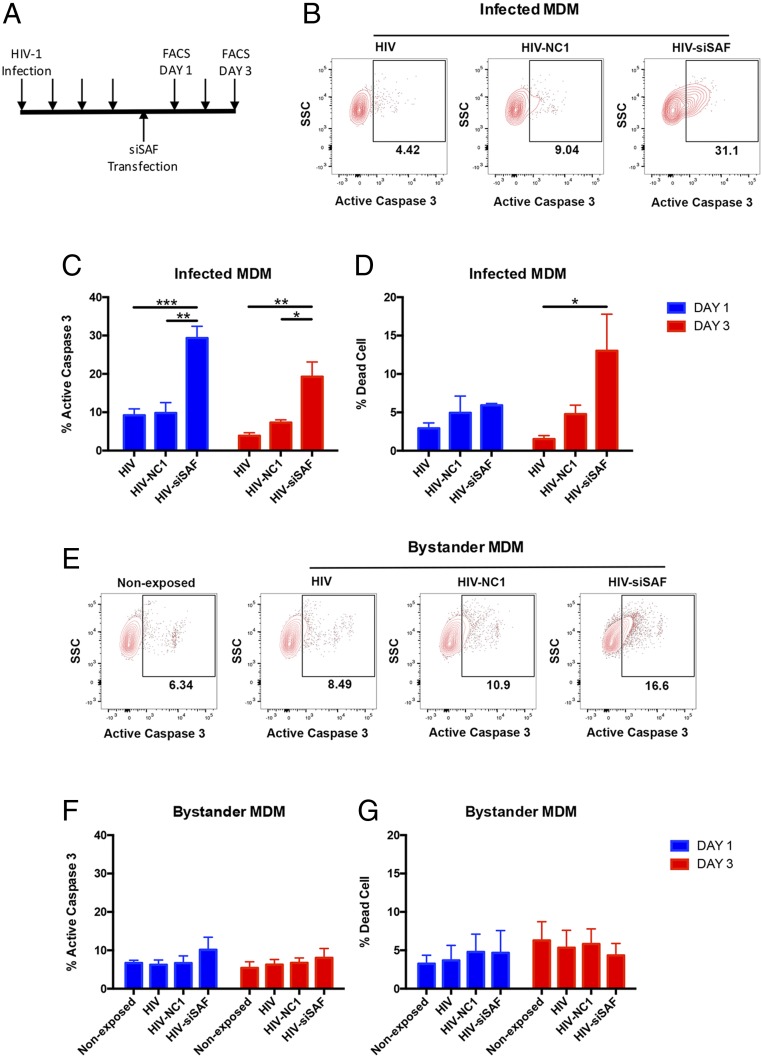
The lncRNA SAF has been shown to protect cells from induction of apoptosis (44, 45). Since expression of SAF was up-regulated in HIV-1–infected macrophages both in vivo and in vitro, we tested if the lncRNA SAF is directly involved in the protection of HIV-1–infected MDMs against virus-induced cell death. We used siRNA to reduce SAF expression levels in MDMs. Using Viromer Blue transfection reagent and a Cy3-labeled siRNA, we achieved an average transfection efficiency of about 70% in MDMs (SI Appendix, Fig. S2 A and B). Transfection of MDMs with siSAF resulted in a threefold reduction in SAF lncRNA level in comparison with cells treated with negative-control siRNA (NC1), demonstrating that the SAF lncRNA level in MDM can be effectively manipulated with siRNAs (SI Appendix, Fig. S2C). Next, to assess how down-regulation of SAF expression in HIV-1–infected MDMs impacts activation of caspase-3/7, we infected MDMs with VSV-G pseudotyped HIV-1 (HXB3/BaL) virus, allowed the infection to establish for 4 d, and then treated the MDMs with either siSAF or control NC1 siRNA (Fig. 4A). As shown in Fig. 4 B and C, control NC1 siRNA treatment of MDMs did not have a substantial effect on active caspase-3/7 levels, but siSAF treatment led to a significant induction in active caspase-3/7 in HIV-1–infected MDMs compared with untreated or NC1 control siRNA-treated MDMs. After 1 d of transfection, there were approximately threefold more cells expressing active caspase-3/7 within HIV-1–infected MDMs in siSAF-treated culture than control NC1-treated or untreated MDMs (Fig. 4C). On day 3 posttransfection, although the number of active caspase-3/7–positive cells was still significantly higher among HIV-1–infected MDMs in siSAF-treated culture, the effect of siSAF appeared to wane (Fig. 4C). In contrast to HIV-1–infected MDMs, active caspase-3/7 levels in neither the nonexposed nor the bystander cells were significantly affected by the siSAF treatment (Fig. 4 E and F). Although siSAF treatment led to a rapid induction of active caspase-3/7 on day 1, significant loss of cell viability in HIV-1–infected MDMs was only observed on day 3 posttransfection, which is consistent with the progressive nature of a programmed cell death pathway (Fig. 4D). Virus-nonexposed and bystander cells again did not show any increase in cell death due to siSAF treatment (Fig. 4F). These data strongly indicate that siSAF treatment can induce activation of apoptotic effector caspases and cell death specifically in HIV-1–infected macrophages while the uninfected bystander cells remain largely unaffected.
Fig. 4.
Effect of siSAF treatment on apoptosis of HIV-1–infected MDMs. (A) Schematic presentation of the experiment timeline. MDMs were transfected with negative-control siRNA (NC1) or siSAF 4 d after infection with HIV-1 virus and analyzed for induction of apoptosis and cell death on day 1 and day 3 posttransfection. Each arrow indicates a day. (B and E) Representative flow cytometry plots showing active caspase 3 staining of HIV-1–infected (B) or nonexposed and bystander (E) MDMs with or without siRNA treatment, as indicated above the panel. Infected and bystander cells were determined based on HIV-1 p24 staining of cells with preceding gating for singlets (FSC-H vs. FSC-A) and MDM (SSC-A vs. FSC-A). (C and F) The percentage of active caspase three positive cells (mean ± SEM) was determined by flow cytometry staining with CellEvent caspase-3/7 green detection reagent in virus-infected (C) or nonexposed and bystander (F) MDMs on day 1 (blue bar) and day 3 (red bar) post siRNA transfection (n = 4). (D and G) The percentage of dead cells (mean ± SEM) was determined by flow cytometry with fixable viability dye eFluor506 in virus-infected (D) or nonexposed and bystander (G) MDMs on day 1 (blue bar) and day 3 (red bar) post siRNA transfection (n = 4). HIV, virus-infected and untreated; HIV-NC1, virus-infected and negative-control siRNA NC1-treated; HIV-siSAF, virus-infected and siSAF-treated. Significance of difference among groups determined by one-way ANOVA is indicated above the groups, *P < 0.05, **P < 0.01, and ***P < 0.001.
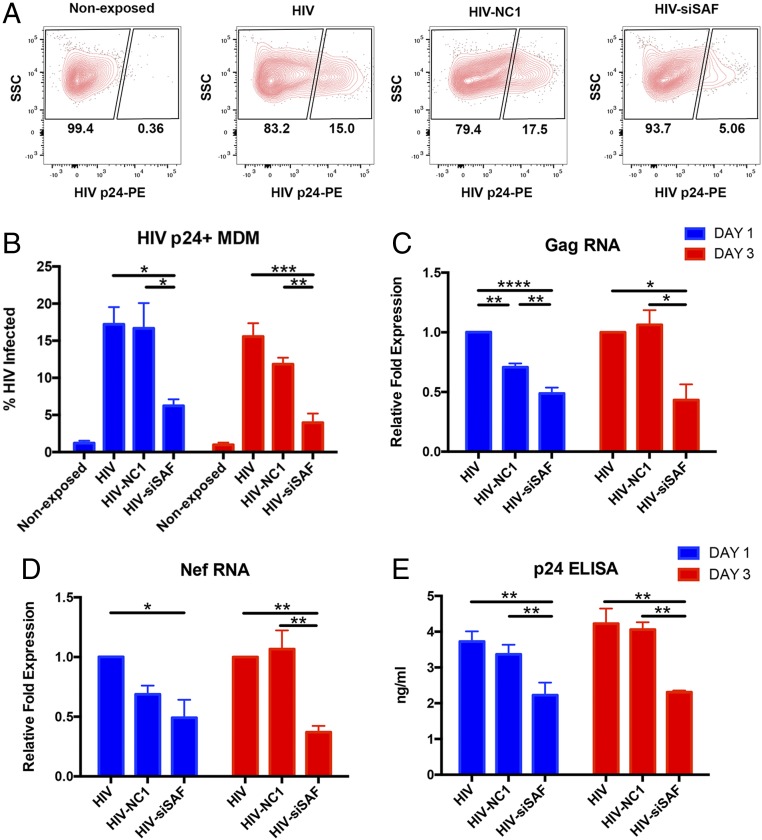
Inhibition of lncRNA SAF Reduces HIV-1 Infection Burden in MDMs.
As siSAF treatment rendered HIV-1–infected MDMs significantly more prone to activation of apoptotic effector caspases, we examined how this affected total HIV-1 viral burden in the MDM culture. We assessed this first by identification of HIV-1 p24-positive cells within the culture and observed that the proportion of virus-infected cells was reduced significantly on day 1 post siSAF treatment (Fig. 5 A and B). The effect was even further pronounced by day 3 posttransfection when the proportion of HIV-1–infected cells in siSAF-treated culture was about 67–75% lower than control NC1-treated or untreated cultures, respectively (Fig. 5B). We then quantified HIV-1 viral RNA transcript levels in the cells to compare ongoing viral replication in the MDM cultures. As shown in Fig. 5 C and D, levels of both Gag and Nef viral RNA were significantly reduced in siSAF-treated MDMs as early as day 1 posttransfection and remained low for 3 d. We noted that the control NC1 siRNA treatment caused a slight reduction in the viral RNA levels on day 1, but the levels recovered to those of untreated cells by day 3 posttransfection. Finally, we assessed the effect of siSAF treatment on virus production and spreading by measuring virion-associated p24 protein levels in the culture supernatants. Consistent with a reduced viral RNA transcript level, the amount of viral p24 protein was significantly decreased in the culture supernatant by day 1 and remained so 3 d posttransfection (Fig. 5E). These results determine that siSAF treatment leads to a marked reduction in HIV-1 replication and total viral burden in the MDM culture.
Fig. 5.
Effect of siSAF treatment on HIV-1 infection in MDMs. (A) Representative flow cytometry plots showing p24-positive HIV-1–infected cells in virus-nonexposed and virus-infected MDMs with or without siRNA treatment, as indicated above the panel. Cells were gated for singlets (FSC-H vs. FSC-A) and MDM (SSC-A vs. FSC-A) before analysis of HIV-1 p24 expression. (B) The percentage of viral p24-positive (mean ± SEM) HIV-1–infected cells was determined by flow cytometry staining with PE-conjugated anti-p24 antibody in untreated, negative-control siRNA NC1-treated and siSAF-treated MDMs on day 1 (blue bar) and day 3 (red bar) post siRNA-transfection (n = 4). (C and D) Fold changes (mean ± SEM) in expression of the viral Gag (C) and Nef (D) RNA was determined by qRT-PCR in untreated, negative-control siRNA NC1-treated and siSAF-treated MDMs on day 1 (blue bar) and day 3 (red bar) post siRNA-transfection (n = 3). Expression levels of housekeeping genes GAPDH, U6, and 18S rRNA was used to normalize data. (E) The amount of viral p24 protein (mean ± SEM) was determined by ELISA in the supernatant of HIV-1–infected cells in untreated, negative-control siRNA NC1-treated and siSAF-treated MDMs on day 1 (blue bar) and day 3 (red bar) post siRNA-transfection (n = 3). HIV, virus-infected and untreated; HIV-NC1, virus-infected and negative-control siRNA NC1-treated; HIV-siSAF, virus-infected and siSAF-treated. Significance of difference among groups determined by one-way ANOVA is indicated above the groups, *P < 0.05, **P < 0.01 and ***P < 0.001, ****P < 0.0001.
Discussion
It has become increasingly evident that lncRNAs play an important role in the virus–host interaction and pathogenesis. In the context of HIV-1 infection, studies on lncRNA expression and their role in viral pathogenesis have mostly been limited to viral infection of lymphoid or monocytic cell lines (35–38). In addition, most of these studies compared data from HIV-1–infected cultures as a whole to that of uninfected cultures without assessing potential changes to uninfected bystander cells that were nonetheless exposed to virus. Since only a fraction of the virus exposed cells become productively infected with HIV-1, a scenario that is particularly true of macrophages, this approach has a reduced capacity to discriminate between alterations in HIV-1–infected versus bystander cells within the same culture. In the current study, we describe the impact of HIV-1 infection on lncRNA expression in a primary human cell of myeloid origin specifically delineating the differences between virus-infected and bystander cells. These two cell subsets showed dramatic differences in lncRNA expression levels upon exposure to HIV-1 virus. Only 19 lncRNAs shared a similar pattern of expression in both infected and bystander MDMs compared with control, nonexposed MDMs. In contrast, most of the differentially expressed lncRNAs were up-regulated in HIV-1–infected MDMs, while exposed but uninfected bystander cells showed a pattern of down-regulation in expression of those specific lncRNA species. Although we focused our study on SAF, which was the most up-regulated lncRNA, the second most up-regulated lncRNA on the list was lincRNA-p21 that has recently been shown to play an important antiapoptotic role during HIV-1 infection in tissue culture (48). These findings demonstrate how HIV-1 cellular infection status differentially affects lncRNA expression levels and emphasizes the importance of distinguishing between the infected and bystander cell subsets during studies of virus-induced alterations in cellular pathways.
The lncRNA that was most markedly affected by HIV-1 infection in MDMs was SAF (FAS-AS1). It is a 1.5-kb antisense lncRNA that is transcribed from the intron 1 region of the Fas gene (43). Expression of this lncRNA has been shown to prevent apoptosis by inducing alternative splicing of the Fas gene and thereby increasing production of soluble FAS from the cells (45). Our data revealed that expression of this antiapoptotic lncRNA SAF was significantly up-regulated in HIV-1–infected MDMs in comparison with bystander and nonexposed cells. Significantly, a similar increase in SAF expression was also observed in HIV-1–infected airway macrophages from HIV-1–infected human volunteers. Interestingly, in contrast to our findings in MDMs and airway macrophages, expression of SAF lncRNA is reportedly decreased in HIV-1–infected T cells that are susceptible to virus-induced cell death (38). This raises the possibility that the antiapoptotic lncRNA SAF is differentially regulated in macrophages and T lymphocytes during HIV-1 infection. This observation is consistent with the respective fate of these two cell types following HIV-1 infection as delayed activation of caspase-3 and apoptosis has also been associated with the impaired killing of HIV-1–infected macrophages relative to CD4+ T cells by cytotoxic T cells (49). A similar observation was also reported in macaques where virus-specific CD8+ T cells failed to kill SIV-infected macrophages and limit viral replication in these myeloid cells (50, 51). Moreover, bystander MDMs did not show a statistically significant increase in SAF expression in comparison with virus-nonexposed control MDMs. The data suggest that active viral infection and/or replication, rather than mere exposure to the HIV-1 virion, is required for induction of SAF lncRNA-dependent cell survival in macrophages. Although a number of HIV-1 viral proteins have been implicated in either inducing or inhibiting cellular apoptotic pathways, further investigation would be needed to ascertain any precise roles of viral proteins in modulation of this prosurvival lncRNA.
Our data demonstrate that HIV-1–mediated evasion of induction of apoptotic effector caspases in macrophages can be effectively negated or subverted by siRNA-mediated down-regulation of SAF expression. Most importantly, such genetic modulation was highly specific because siRNA treatment induced activation of caspase-3/7 and loss of cell viability in the HIV-1–infected macrophages alone, leaving the bystander and nonexposed cells unaffected. Our results also showed that this selective targeting of the infected cells can result in a significant decline in viral reproduction and burden. Indeed, transfection of MDMs with siSAF reduced HIV-1 replication and virus production by nearly twofold within 1 d of treatment. On day 1 posttransfection, an increase in caspase-3/7 activity was associated with a corresponding inhibition in virus replication even without a discernable decrease in cell viability. Although active caspase-3/7 is an important effector in the induction of apoptotic pathway, recent publications suggest that effector-triggered immunity or mitochondrial outer membrane permeabilization mediated activation of caspase-3/7 might also induce other parallel pathways such as inflammasome and IL-1β maturation (52, 53). The downstream pathway(s) of siSAF-mediated activation of caspase-3/7 and subsequent cell death remain somewhat enigmatic. However, with a siRNA-based treatment approach, which would be transient, a single round of transfection was sufficient to sustain reduced levels of virus infection for at least 3 d. Such an anti-viral effect could potentially be enhanced through multiple rounds of treatment. The in vivo significance of these results is supported by the demonstration that comparable up-regulation of the lncRNA SAF is observed in HIV-1–infected macrophages from the airways of HIV-1–positive individuals. Previous analysis of BAL-derived macrophages in Malawi indicated that HIV-1 was present predominantly in a subset of small alveolar macrophages and that alveolar macrophages account for ∼80% of the myeloid cells recoverable by BAL (14, 47). Interestingly, alveolar macrophages are now known to originate from fetal stem cells early in embryonic development and are capable of extended life span measurable in years. Such long-lived tissue resident macrophages would be perfect candidates as reservoirs for HIV-1 persistence, especially if stabilized by cell survival programs such as driven by the lncRNA SAF. Overall, this study emphasizes the role of lncRNAs as an important regulator in the cellular response to HIV-1 infection and pathogenesis as well as highlights the potential of targeting long noncoding RNAs such as SAF as a future therapeutic intervention specifically aimed toward HIV-1–infected long-lived reservoirs.
Materials and Methods
Virus Production and Infection.
The HIV-1 infectious molecular clone, pWT/BaL (HXB3/BaL, Catalog 11414), was obtained from the NIH AIDS Reagent Program. The plasmid encoding HIV-1 molecular clone NL43-IRES-mCherry with R5-tropic env (BaL) was generated by replacing the EGFP sequence with that of mCherry in the original plasmid pBR43IeG-nef+ R5env (a kind gift from Thorsten R. Mempel, Massachusetts General Hospital, Boston). Replication-competent VSV-G pseudotyped HIV-1 virus was prepared by cotransfecting 293FT cells with the respective HIV-1 molecular clone and VSV-G expression plasmid using Lipofectamine 3000 reagent (Life Technologies). Transfection media was replaced with fresh antibiotic free DMEM media 8 h posttransfection. After 72 h, cell culture supernatant containing HIV-1 virus was harvested, centrifuged to remove cell debris, passed through 0.45-µm filter, and stored at −80 °C in aliquots. Titer of the virus stock was determined by p24 ELISA (RETROtek HIV-1 p24 antigen ELISA kit; ZeptoMetrix), and 50 ng/mL of virus was used to infect MDMs.
MDM Differentiation and siRNA Transfection.
Human monocytes were obtained from peripheral blood mononuclear cells of healthy individuals by counter current centrifugal elutriation with an average purity of >97% (Elutriation Core Facility, University of Nebraska Medical Center). Monocytes were maintained in DMEM media supplemented with 10% human serum, 100 U/mL penicillin, and 100 µg/mL streptomycin (Invitrogen). Cells were cultured at 37 °C with 6% CO2 for 7–8 d to fully differentiate into macrophages (MDMs).
For siRNA treatment, 1.5 × 106 MDMs were transfected with either 10 nM each of three Dicer-substrate short interfering RNAs (DsiRNAs) targeted to SAF (hs.Ri.FAS-AS1.13; IDT) or a nonspecific control, NC1 (negative control DsiRNA; IDT) using Viromer Blue transfection reagent (Lipocalyx) according to the manufacturer’s instructions. Transfection efficiency was monitored with a Cy3-labeled DsiRNA (IDT) control.
lncRNA Profiling and qRT-PCR.
For comparative lncRNA expression analysis of infected and bystander cells, fully differentiated MDMs were infected with the replication-competent VSV-G pseudotyped NL43–IRES-mCherry-Bal virus. On day 7 postinfection, cells were harvested by gentle scraping following a 10-min incubation in cold PBS, washed once, and processed with a Bio-Rad S3 cell sorter to separate mCherry-positive HIV-1–infected and mCherry-negative bystander cells. Similar to the HIV-1–infected culture, virus-nonexposed MDMs were also passed through the cell sorter and recovered as mCherry negative as an experimental control. Total RNA was extracted from sorted cells using TRIzol reagent (Invitrogen) according to manufacturer’s instructions, followed by DNase treatment (Turbo DNA-free kit; Invitrogen) to remove genomic DNA. The same amount of RNA was used for cDNA synthesis and subsequent quantification of lncRNA expression using the Human LncRNA Profiler qPCR Array kit (System Bioscience) and ABI 7500 Fast Real-time PCR system (Applied Bioscience) in accordance to the manufacturer’s protocol. Expression of SAF lncRNA was further independently verified with previously published (43) primer sets (Forward primer: CAT CTC AGC CTC TTG GTG TAA and Reverse primer: ATG GGA GAT ATG GGA TTG AAC) and iTaq Universal SYBR Green Supermix (Bio-Rad). HIV-1 viral transcripts were quantified by qRT-PCR of Gag (Forward primer: AAG CAC TGG GAC CAG GAG C and Reverse primer: TGG TAG CTG GAT TTG TTA CTT GGC) and Nef (Forward primer: TAG TGT GAT TGG ATG GCC TGC and Reverse primer: ACA AGC ATT GTT AGC TGC TG), following a similar procedure. Expression of the housekeeping genes: GAPDH (Forward primer: GAC AAG CTT CCC GTT CTC AG and Reverse primer: GAG TCA ACG GAT TTG GTC GT), U6 (Forward primer: CTC GCT TTG GCA CA and Reverse primer: AAC GCT TCA CGA ATT TGC GT), and 18S rRNA (Forward primer: GGC CCT GTA ATT GGA ATG AGT C and Reverse primer: CCA AGA TCC AAC TAC GAG CTT) was used for normalization of qRT-PCR expression data.
Flow-Cytometry for HIV-1 p24 and Detection of Active Caspase-3/7.
MDMs were harvested by gentle scraping following a 10-min incubation in cold PBS. Cells were stained for intracellular active caspase-3/7 enzyme using the CellEvent Caspase-3/7 Green Detection reagent (Invitrogen) according to the manufacturer’s instruction with some modifications. Briefly, cells were resuspended in 1 mL PBS to which 1 µl CellEvent caspase-3/7 reagent was added and then incubated at 37 °C for 30 min. During the last 10 min of incubation, 1 µL of Fixable Viability Dye eFluor 506 (eBioscience) was added to the cells. Thereafter, the cells were washed with PBS and fixed and permeabilized using 1× Cytofix/cytoperm solution (BD Bioscience) for 30 min. For detection of HIV-1–infected MDMs, the cells were then stained with an antibody against the HIV-1 core protein p24 (KC57-RD1; Beckman Coulter) for 30 min at room temperature. Finally, the cells were fixed in 1% paraformaldehyde in PBS and analyzed with a BD LSRII flow cytometer. Flow cytometry data were analyzed using FlowJo software. Cells were first gated for singlets [forward scatter height (FSC-H) vs. forward scatter area (FSC-A)] and macrophage [side scatter area (SSC-A) vs. FSC-A]. The macrophage gate was further analyzed for expression of HIV-1 p24 protein and gated into bystander [p24-phycoerythrin (PE) negative] and infected (p24-PE positive) MDMs. From these gated populations, cell viability and apoptosis of bystander and infected MDMs were subsequently determined based on staining for viability dye eFluor 506 and active caspase-3/7, respectively.
Study Subjects and Bronchoalveolar Lavage.
Bronchoalveolar lavage (BAL) was obtained from four HIV-positive, antiretroviral treatment naive individuals (aged ≥18 y) at the Queen Elizabeth Central Hospital in Blantyre, Malawi. Age, gender, plasma viral load, and CD4 T cell counts of the study participants at the time of bronchoscopy are listed in SI Appendix, Table S1. The study received ethical approval from the research ethics committees of the College of Medicine, Malawi (Research protocol P.10/08/708), the Liverpool School of Tropical Medicine, UK (Research protocol 08.54), and Cornell University (Research protocol 908000698). All study participants provided written informed consent.
FISH Staining and Flow Cytometry Sorting of BAL-Derived Airway Macrophages.
Cells isolated from whole BAL fluid were stained with predetermined optimal concentrations of fluorochrome-conjugated monoclonal antibodies against human cell surface proteins as detailed (47). Briefly, BAL cells (1 × 106 cells) were incubated with anti-CD206 Brilliant Violet 421 and anti-CD169 Alexa Fluor 647 antibodies for 15 min in the dark at room temperature. The cells were washed, resuspended in PBS, and acquired on a BD LSRFortessa flow cytometer (Becton Dickinson). Data were analyzed using FlowJo v10.5.0 (FlowJo LLC). FISH staining for HIV-1 Nef RNA in BAL-derived airway macrophages were carried out as previously described (14, 54). FISH-stained BAL cells were first gated for singlets (FSC-H vs. FSC-A) and macrophages (SSC-A vs. FSC-A) and then sorted into Nef-positive and Nef-negative cells based on staining for HIV Nef-Quasar670 probes (LGC Biosearch Technologies) using a BD FACS Aria cell sorter.
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism software. Unless otherwise indicated, one-way ANOVA with Tukey’s multiple comparison test was used for all statistical analysis and a P value below 0.5 was considered significant.
Supplementary Material
Acknowledgments
We thank all the members of the D.G.R. laboratory for helpful discussions and technical support. This work was supported by NIH Grants AI118582 and AI136097 (to D.G.R.) and Wellcome Trust Grants 088696/Z/09/Z (to H.C.M.) and 105831/Z/14/Z (to K.C.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818662116/-/DCSupplemental.
References
- 1.Fahey JL, et al. The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N Engl J Med. 1990;322:166–172. doi: 10.1056/NEJM199001183220305. [DOI] [PubMed] [Google Scholar]
- 2.Margolick JB, Donnenberg AD. T-cell homeostasis in HIV-1 infection. Semin Immunol. 1997;9:381–388. doi: 10.1006/smim.1997.0096. [DOI] [PubMed] [Google Scholar]
- 3.Moir S, et al. B cells in early and chronic HIV infection: Evidence for preservation of immune function associated with early initiation of antiretroviral therapy. Blood. 2010;116:5571–5579. doi: 10.1182/blood-2010-05-285528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alter G, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106:3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 5.Donaghy H, et al. Loss of blood CD11c(+) myeloid and CD11c(-) plasmacytoid dendritic cells in patients with HIV-1 infection correlates with HIV-1 RNA virus load. Blood. 2001;98:2574–2576. doi: 10.1182/blood.v98.8.2574. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley JM, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho DD, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 8.Verani A, Gras G, Pancino G. Macrophages and HIV-1: Dangerous liaisons. Mol Immunol. 2005;42:195–212. doi: 10.1016/j.molimm.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Sieweke MH, Allen JE. Beyond stem cells: Self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 14.Jambo KC, et al. Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol. 2014;7:1116–1126. doi: 10.1038/mi.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan CA, Watkins BA, Kufta C, Dubois-Dalcq M. Infection of brain microglial cells by human immunodeficiency virus type 1 is CD4 dependent. J Virol. 1991;65:736–742. doi: 10.1128/jvi.65.2.736-742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol. 2015;21:276–289. doi: 10.1007/s13365-014-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Housset C, et al. Immunohistochemical evidence for human immunodeficiency virus-1 infection of liver Kupffer cells. Hum Pathol. 1990;21:404–408. doi: 10.1016/0046-8177(90)90202-g. [DOI] [PubMed] [Google Scholar]
- 18.DiNapoli SR, et al. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naive, SIV-infected Asian macaques. JCI Insight. 2017;2:e91214. doi: 10.1172/jci.insight.91214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koenig S, et al. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 20.Avalos CR, et al. Quantitation of productively infected monocytes and macrophages of simian immunodeficiency virus-infected macaques. J Virol. 2016;90:5643–5656. doi: 10.1128/JVI.00290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gama L, et al. SIV latency in macrophages in the CNS. Curr Top Microbiol Immunol. May 17, 2018 doi: 10.1007/82_2018_89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honeycutt JB, et al. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest. 2016;126:1353–1366. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honeycutt JB, et al. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med. 2017;23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Micci L, et al. CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog. 2014;10:e1004467. doi: 10.1371/journal.ppat.1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 26.Chugh P, et al. Infection of human immunodeficiency virus and intracellular viral Tat protein exert a pro-survival effect in a human microglial cell line. J Mol Biol. 2007;366:67–81. doi: 10.1016/j.jmb.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giri MS, Nebozhyn M, Showe L, Montaner LJ. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J Leukoc Biol. 2006;80:1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 28.Igarashi T, et al. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci USA. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyaard L, et al. Programmed death of T cells in HIV-1 infection. Science. 1992;257:217–219. doi: 10.1126/science.1352911. [DOI] [PubMed] [Google Scholar]
- 30.Terai C, Kornbluth RS, Pauza CD, Richman DD, Carson DA. Apoptosis as a mechanism of cell death in cultured T lymphoblasts acutely infected with HIV-1. J Clin Invest. 1991;87:1710–1715. doi: 10.1172/JCI115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 32.Swingler S, Mann AM, Zhou J, Swingler C, Stevenson M. Apoptotic killing of HIV-1-infected macrophages is subverted by the viral envelope glycoprotein. PLoS Pathog. 2007;3:1281–1290. doi: 10.1371/journal.ppat.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummins NW, Badley AD. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis. 2010;1:e99. doi: 10.1038/cddis.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badley AD, Sainski A, Wightman F, Lewin SR. Altering cell death pathways as an approach to cure HIV infection. Cell Death Dis. 2013;4:e718. doi: 10.1038/cddis.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair M, Sagar V, Pilakka-Kanthikeel S. Gene-expression reversal of lncRNAs and associated mRNAs expression in active vs latent HIV infection. Sci Rep. 2016;6:34862. doi: 10.1038/srep34862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas S, et al. Differentially expressed host long intergenic noncoding RNA and mRNA in HIV-1 and HIV-2 infection. Sci Rep. 2018;8:2546. doi: 10.1038/s41598-018-20791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep. 2015;5:8639. doi: 10.1038/srep08639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596-12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Y, et al. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao H, Zhang X, Frazão JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem. 2013;114:2375–2383. doi: 10.1002/jcb.24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 42.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Yan MD, et al. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 44.Villamizar O, et al. Fas-antisense long noncoding RNA is differentially expressed during maturation of human erythrocytes and confers resistance to Fas-mediated cell death. Blood Cells Mol Dis. 2016;58:57–66. doi: 10.1016/j.bcmd.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Villamizar O, Chambers CB, Riberdy JM, Persons DA, Wilber A. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget. 2016;7:13810–13826. doi: 10.18632/oncotarget.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cribbs SK, Lennox J, Caliendo AM, Brown LA, Guidot DM. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses. 2015;31:64–70. doi: 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mwale A, et al. B cell, CD8+ T cell and gamma delta T cell infiltration alters alveolar immune cell homeostasis in HIV-infected Malawian adults. Wellcome Open Res. 2018;2:105. doi: 10.12688/wellcomeopenres.12869.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barichievy S, et al. Viral apoptosis evasion via the MAPK pathway by use of a host long noncoding RNA. Front Cell Infect Microbiol. 2018;8:263. doi: 10.3389/fcimb.2018.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clayton KL, et al. Resistance of HIV-infected macrophages to CD8+ T lymphocyte-mediated killing drives activation of the immune system. Nat Immunol. 2018;19:475–486. doi: 10.1038/s41590-018-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vojnov L, et al. The majority of freshly sorted simian immunodeficiency virus (SIV)-specific CD8(+) T cells cannot suppress viral replication in SIV-infected macrophages. J Virol. 2012;86:4682–4687. doi: 10.1128/JVI.06324-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rainho JN, et al. Nef is dispensable for resistance of simian immunodeficiency virus-infected macrophages to CD8+ T cell killing. J Virol. 2015;89:10625–10636. doi: 10.1128/JVI.01699-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vince JE, et al. The mitochondrial apoptotic effectors BAX/BAK activate caspase-3 and -7 to trigger NLRP3 inflammasome and caspase-8 driven IL-1β activation. Cell Rep. 2018;25:2339–2353.e4. doi: 10.1016/j.celrep.2018.10.103. [DOI] [PubMed] [Google Scholar]
- 53.Chauhan D, et al. BAX/BAK-induced apoptosis results in caspase-8-dependent IL-1β maturation in macrophages. Cell Rep. 2018;25:2354–2368.e5. doi: 10.1016/j.celrep.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 54.Wilburn KM, et al. Heterogeneous loss of HIV transcription and proviral DNA from 8E5/LAV lymphoblastic leukemia cells revealed by RNA FISH:FLOW analyses. Retrovirology. 2016;13:55. doi: 10.1186/s12977-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.