Summary
The recovery and persistence of rare and endangered species are often threatened by genetic factors, such as the accumulation of deleterious mutations, loss of adaptive potential, and inbreeding depression [1]. Island foxes (Urocyon littoralis), the dwarfed descendants of mainland gray foxes (U. cinereoargenteus), have inhabited California’s Channel Islands for >9,000 years [2-4]. Previous genomic analyses revealed island foxes have exceptionally low levels of diversity and elevated levels of putatively deleterious variation [5]. Nonetheless, all six populations have persisted for thousands of generations, and several populations rebounded rapidly following recent severe bottlenecks [6, 7]. Here, we combine morphological and genomic data with population genetic simulations to determine the mechanism underlying the enigmatic persistence of these foxes. First, through analysis of genomes from 1929-2009, we show that island foxes have remained at small population sizes with low diversity for many generations. Second, we present morphological data indicating an absence of inbreeding depression in island foxes, confirming that they are not afflicted with congenital defects common to other small and inbred populations. Lastly, our population genetic simulations suggest long-term small population size results in a reduced burden of strongly deleterious recessive alleles, providing a mechanism for the absence of inbreeding depression in island foxes. Importantly, the island fox illustrates a scenario in which genetic restoration through human-assisted gene flow could be a counterproductive or even harmful conservation strategy. Our study sheds light on the puzzle of island fox persistence, a unique success story that provides a model for the preservation of small populations.
Keywords: deleterious variation, genetic diversity, inbreeding depression, small populations, skeletal morphology, island evolution, genomics, island fox, gray fox, Channel Islands
Results
Minimal impact of recent demography on island fox genomes
Previous analysis of whole genome sequences from island foxes sampled in 1988 revealed dramatically reduced levels of diversity and increased levels of putatively deleterious alleles relative to the mainland gray fox [5]. On the smallest island, San Nicolas, the population is nearly monomorphic across its entire genome [5]. Beginning in the 1990s, four populations declined >90% due to novel predators (San Miguel, Santa Rosa, Santa Cruz) and disease (Santa Catalina), but subsequently rebounded under human management in the fastest recovery of any mammal under the Endangered Species Act to date [6, 7]. We sequenced whole genomes of foxes sampled in 2000-2009 from each island to determine whether recent extreme bottlenecks reduced variation, or whether very low genetic variation is a persistent feature of island fox genomes. Additionally, we sequenced DNA isolated from bone fragments from a 1929 San Nicolas island fox, and DNA from a Northern California gray fox. Combined with existing sequences, our dataset includes 16 genomes with 13-23X coverage (Table S1). All sequences were aligned to the domestic dog (Canis familiaris) reference genome, canFam3.1. Phylogenetic analyses show that island fox genomes cluster by population, and southern and northern island populations define distinct clusters, consistent with prior studies (Figures 1A, B, S1) [4, 5, 8, 9].
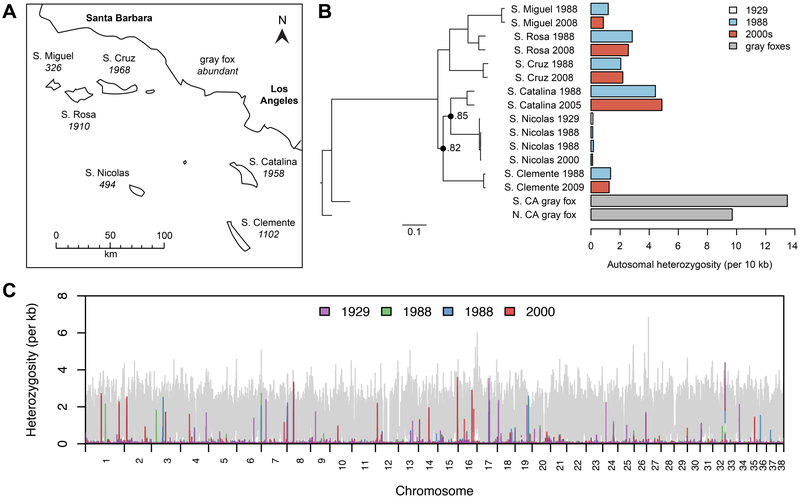
Figure 1. Genetic relationships and heterozygosity of island fox genomes.
(A) Map of the Channel Islands with estimated island fox census sizes [8]. (B) Left: maximum-likelihood tree based on >12,000 SNPs. Nodes have 100% bootstrap support except where noted. Right: mean per-site heterozygosity across the autosomal genome. See also Figure S1, Table S1. (C) Heterozygosity in 100 kb windows with a 10 kb step size across the autosomal genome shows the genomic flatlining punctuated by occasional peaks of heterozygosity in San Nicolas foxes. Heterozygosity in the Southern California gray fox is shown in grey. X-axis coordinates correspond to the reference dog genome. See also Tables S2, S3.
Overall, we did not find significant changes in genome-wide heterozygosity between island foxes sampled from the same population at different times (Figure 1B). Among populations that bottlenecked in the 1990s, two genomes showed slightly higher heterozygosity after 2000 than in 1988 (Santa Catalina, Santa Cruz), whereas two showed slightly lower heterozygosity (San Miguel, Santa Rosa). Similarly, in San Nicolas and San Clemente, genome-wide heterozygosity was essentially unchanged from 1988 to after 2000. These minor differences between genomes over time are not statistically significant (Wilcoxon signed rank test, p=1), and may be expected due to small inter-individual differences in heterozygosity. Robinson et al. [5] hypothesized that the genomic flatlining of San Nicolas foxes may have resulted, in part, from a suspected bottleneck in the 1970s [10]. However, the 1929 genome bears exceptionally low diversity (1.33 × 10−5 heterozygotes/bp), similar to genomes from 1988 and 2000 (mean=1.35 × 10−5 heterozygotes/bp) (Figure 1 B, C). The genomic flatlining of San Nicolas foxes is therefore an enduring characteristic of this population, underscoring the mystery concerning its long-term persistence.
Previous analysis of remnant peaks of heterozygosity in San Nicolas genomes revealed strong enrichment for olfactory function [5]. We revisited the question of whether patterns of heterozygosity in San Nicolas foxes are consistent with neutral evolution, or the result of selection maintaining diversity at functionally important loci. Among peaks of heterozygosity from all four San Nicolas genomes, only 43.9% were present in two or more individuals. The 1929 genome was the most divergent, with only 41.5% of its peaks shared with at least one other individual, suggesting peaks of heterozygosity are not strongly conserved over time. We conducted coalescent simulations [11] under plausible models of San Nicolas demographic history [5] to determine whether variation in the sequenced genomes is predicted by these neutral demographic models. We found that the empirical numbers of peaks, average peak widths, and proportions of shared peaks all fell within the middle 95% of values obtained through simulation (Table S2), suggesting that demographic processes can account for empirical patterns of variation in peaks of heterozygosity within and between San Nicolas fox genomes.
Despite the lack of evidence that peaks of heterozygosity are constrained by selection, peaks in the 1929 and 2000 San Nicolas genomes are strongly enriched for olfactory receptor (OR) genes (p≤3.57 × 10−16), as they are in the genomes from 1988 (p≤2.78 × 10−15) (Table S3, KEGG:04740). Strikingly, 39.3-50.7% of peaks in San Nicolas genomes overlap OR genes. However, these peaks vary between individuals; 37.2% of peaks containing OR genes are found in only one San Nicolas genome, despite consistency in coverage and peak thresholds (1.67-1.72 × 10−4 heterozygotes/bp) across all four individuals. Peaks of heterozygosity in mainland gray fox genomes are also enriched for OR genes (p≤4.98 × 10−3). Enrichment of OR genes in gray fox peaks is unsurprising, since OR genes are highly variable in mammals generally, suggesting they have an elevated rate of evolution [12]. Given high initial variation, OR genes may be among the last to lose diversity as drift erodes heterozygosity in island genomes over time. Alternatively, high variation in OR gene regions may be due to alignment errors, since OR gene repertoires evolve rapidly through gene duplication, pseudogenization, and loss [12]. Nonetheless, even if peaks of heterozygosity containing OR genes result from read misalignment, the fact that peaks are not consistently observed across San Nicolas foxes implies some variability, either copy number variation or true heterozygosity, persists in these regions.
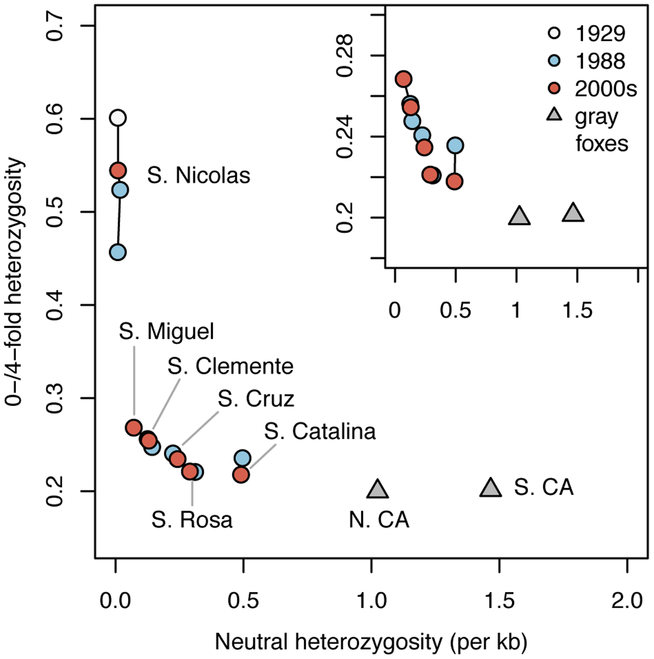
We found that levels of non-neutral variation are largely similar between island fox genomes sampled at different times. Comparison of heterozygosity at zero-fold and four-fold degenerate sites within protein-coding regions (i.e. sites where all or none of the possible nucleotide changes alter the encoded amino acid, respectively) showed that island fox genomes have a high ratio of amino acid-changing to non-amino acid-changing variants relative to gray foxes (Figure 2). We calculated heterozygosity within a set of putatively neutral loci across the genome as a proxy for effective population size. Together, the elevated proportions of putatively damaging mutations at zero-fold degenerate sites and the reduction in neutral heterozygosity demonstrate the genomic consequences of enhanced drift relative to selection on the islands. Neutral diversity and ratios of zero-fold to four-fold heterozygosity were essentially unchanged between genomes from 1988 and after 2000. All San Nicolas genomes showed a strongly elevated ratio of heterozygosity at zero-fold relative to four-fold degenerate sites, although the small number of heterozygous sites produces high variance of this statistic [5].
Figure 2. Island fox genomes contain high proportions of amino acid-changing mutations and low neutral diversity.
A negative relationship exists between neutral genetic diversity (proxy for effective population size) and the ratio of heterozygosity at zero-fold relative to four-fold degenerate sites (proxy for efficacy of selection). Solid black lines connect individuals from the same population. Inset: The relationship holds even when removing San Nicolas. See also Figure S2, Table S4.
We further examined predicted loss-of-function (LOF) and putatively deleterious missense mutations (Figure S2, Table S4). Across all populations, island foxes had elevated homozygosity of derived alleles, with more than double the number of homozygous LOF and deleterious missense alleles compared to gray foxes (p<1.75 × 10−10). Further, island genomes contained an excess total number of putatively deleterious alleles per genome. Relative to gray foxes, island foxes from 1988 had 2.3% more deleterious missense alleles (p=1.11 × 10−10) and 4.9% more LOF alleles (p=2.43 × 10−3) per genome, and island foxes from after 2000 contained 1.7% more deleterious missense alleles (p=4.68 × 10−9) and 3.3% more LOF alleles per genome (p=0.0560). Thus, regardless of whether deleterious mutations are additive or recessive, island foxes are predicted to have reduced fitness compared to gray foxes due to elevated homozygosity and an increased total number of deleterious alleles.
Congenital skeletal defects are rare in island and gray foxes
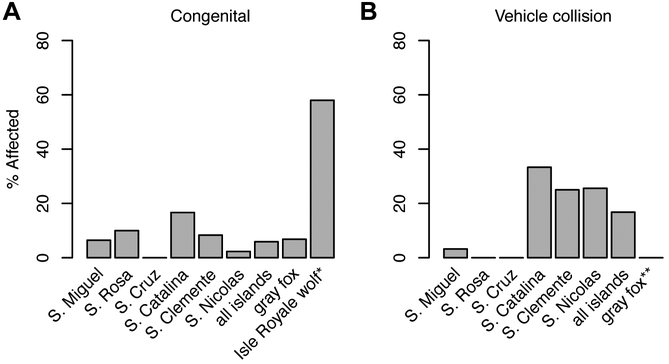
Previous studies of inbred carnivoran skeletons have found a suite of pathologies linked to low genetic diversity, particularly in the skull and vertebral column [13-15]. For example, a study of gray wolves (Canis lupus) on Isle Royale, Michigan found that 58% of individuals exhibited vertebral anomalies within ten generations of the population’s founding due to extreme inbreeding [15]. We inspected skulls (n=141) and complete skeletons (n=163) of adult foxes sampled in 1929-2013 from all six islands and the mainland to identify anomalies that could indicate inbreeding depression. Congenital deformities were present but rare in island foxes; among 119 specimens, only seven possessed developmental pathologies. Observed congenital defects included extra lumbar vertebrae (n=1), lumbosacral transitional vertebrae (LSTV; n=1), sacrocaudal transitional vertebrae (SCTV; n=3), and maloccluded teeth (n=2) (Figure 3A, Table S5). Among 43 San Nicolas specimens, only one congenital defect (maloccluded incisors) was observed. In contrast, traumatic pathologies indicating prior injuries, primarily from suspected vehicular collisions, were common in island foxes, as evidenced by specimens with bone fractures and indications of infection (Figure 3B, Table S5). Unlike the rare congenital malformations we observed, vehicular impacts likely compromise mobility and contribute to early mortality.
Figure 3. Prevalence of skeletal pathologies in island and gray foxes.
Sample sizes: S. Miguel, n=31; S. Rosa, n=10; S. Cruz, n=5; S. Catalina, n=6; S. Clemente, n=24; S. Nicolas, n=43; all islands, n=119; gray foxes, n=44; Isle Royale wolves, n=36. See also Table S5. (A) Incidence of congenital defects in gray and island foxes (this study), compared to Isle Royale wolves *[15]. (B) Incidence of traffic-related pathologies in gray and island foxes. **Absence of traffic-related pathologies in gray foxes possibly due to ascertainment bias against retrieval of mainland fox road kill.
We note that although island foxes have low levels of congenital skeletal defects, this finding does not preclude the presence of soft tissue traits commonly associated with inbreeding, such as syndactyly and reproductive anomalies [16, 17]. Further study is needed to rule out the presence of such traits in island foxes, but they have not been previously reported despite over a decade of intensive management, including several years of captive breeding [6], ejaculate quality assays [18], and necropsy of road killed individuals [19].
Among 44 gray fox specimens, few pathologies were observed. Congenital defects in gray foxes included LSTV (n=1), SCTV (n=1), and achondroplastic dwarfism (n=1). In contrast to island foxes, traumatic pathologies were uncommon in gray foxes. Healed fractures consistent with vehicular impacts were not observed, though we note that this may be due to ascertainment bias if road killed mainland foxes were less likely to be included in museum collections.
Using Fisher’s exact test, we determined that there was no significant difference (p=0.663) in the proportion of congenital defects between island foxes and mainland gray foxes (Figure 3A). However, there was a significant difference between the islands in the proportion of probable collision injuries (p=3.90 × 10−3) (Figure 3B). We calculated relative risk scores and performed a permutation test to assess significant differences in collision injuries between populations (Table S5). We found that foxes on the three islands with paved roads and substantial human habitation (Santa Catalina) or naval bases (San Clemente, San Nicolas) face significantly higher risks of vehicle collisions. No probable collisions were recorded in the Santa Cruz, Santa Rosa or gray fox samples. Overall, our morphological assessment supports the hypothesis that island foxes do not exhibit canonical signs of inbreeding depression, though some populations are adversely affected by vehicular traffic.
Predicted genetic variation in large mainland versus small island populations
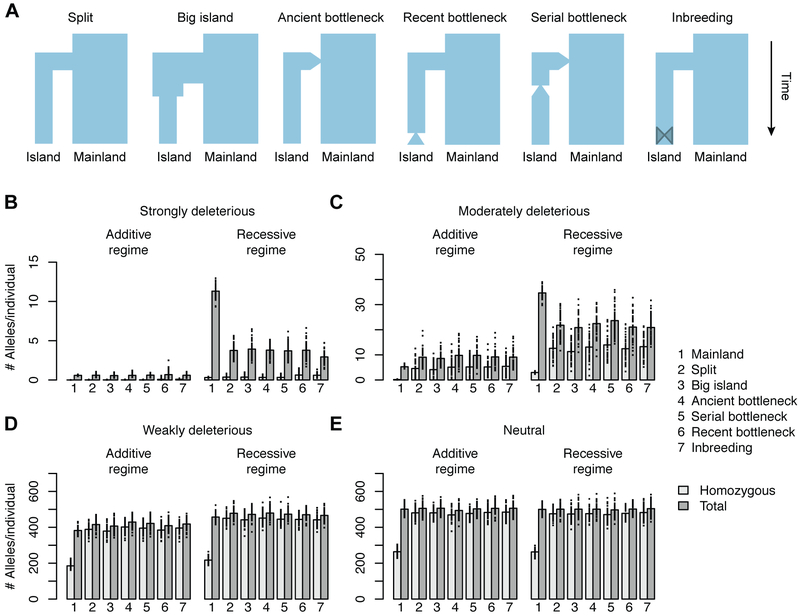
We hypothesized that the absence of inbreeding depression in island foxes may be attributed to purging of strongly deleterious recessive mutations in island populations relative to the mainland, despite the overall accumulation of deleterious variants observed in island genomes. To test this hypothesis, we conducted forward-in-time simulations [20] under a range of demographic models (Figure 4A), each consisting of a mainland population of 10,000 diploids giving rise to an island population with a final size of 1,000 diploids. After 10,000 generations, we counted the number of strongly deleterious (s<−0.01; Figure 4B), moderately deleterious (−0.01≤s<−0.001; Figure 4C), weakly deleterious (−0.001≤s<0; Figure 4D), and neutral (s=0; Figure 4E) mutations per individual. To explore the impact of dominance, we performed one set of simulations with recessive mutations (h=0) and one set of simulations with additive mutations (h=0.5).
Figure 4. The number of strongly deleterious recessive alleles per individual is reduced in island populations.
(A) Depictions of demographic models used in simulations (not to scale). At the end of the simulations, Nmainland=10,000 diploids, Nisland=1,000 diploids. (B-E) Charts showing the total number of derived alleles and the number of homozygous derived alleles per individual by dominance and selection strength. Dots represent the mean per individual within a single replicate; bar height represents the mean across all fifty replicates. Selection strengths: (B) strongly deleterious, −1≤s<−0.01; (C) moderately deleterious, −0.01≤s<−0.001; (D) weakly deleterious, −0.001≤s< 0; (E) neutral, s=0. Simulations include a mixture of neutral and deleterious alleles in which deleterious alleles are either entirely additive (Additive Regime) or entirely recessive (Recessive Regime). See also Table S6.
Results were strikingly consistent across all six demographic scenarios, indicating that the predominant factor driving levels of variation is long-term small population size on the islands, rather than transient effects. Except in the case of strongly deleterious mutations, homozygosity was always higher in island genomes relative to mainland genomes (p<10−7). The total number of deleterious alleles per individual on the islands relative to the mainland varied according to selection and dominance coefficients (Figure 4B-E). In simulations where mutations were additive, island genomes contained, on average, 9.97% more deleterious alleles per individual overall (p≤3.09 × 10−5), primarily due to the accumulation of weakly and moderately deleterious alleles. However, numbers of strongly deleterious additive alleles were equivalent between the islands and the mainland, which is expected since these mutations sharply reduce fitness in both homozygotes and heterozygotes, and are thus readily removed by selection in small and large populations.
In contrast, rare recessive mutations are exposed to selection more frequently in small populations due to elevated homozygosity, and are therefore removed more efficiently than in larger populations. Although the total number of recessive deleterious alleles was not significantly different between individuals from the islands and the mainland, moderately and strongly deleterious recessive alleles were greatly depleted. Island genomes contained, on average, 37.1% fewer moderately deleterious recessive alleles (p<10−7) and 67.7% fewer strongly deleterious recessive alleles (p<10−7) compared to the mainland. Furthermore, there were no simulations in which the average number of strongly deleterious recessive alleles was higher in island genomes compared to the mainland (Figure 4B).
We also performed simulations under a range of smaller island population sizes (N=500, 200, 100, 50) to determine whether purging of strongly deleterious mutations still occurred. We expected that population size would modulate selection against deleterious alleles, as predicted by the nearly neutral theory, whereby mutations with ∣s∣<1/2N behave as if they are neutral despite their effects on fitness [21]. Overall, we found that purging of recessive deleterious alleles became weaker and the accumulation of additive deleterious alleles became more severe as population size declined (Table S6). However, exceptionally deleterious recessive alleles (s<−0.1) were always depleted in the smaller island populations (p<10−7). In sum, these findings suggest that individuals derived from historically small populations carry a reduced burden of strongly deleterious recessive alleles relative to individuals from a large outbred population, reducing the former’s risk of inbreeding depression.
Discussion
Our genomic analyses show that recent catastrophic bottlenecks had limited impact on island fox genomes, indicating that exceptionally low heterozygosity and higher levels of putatively deleterious alleles are caused by historically small population size. We confirm the apparent lack of inbreeding depression in island foxes through detailed morphological assessment, and, through demographic simulations, demonstrate that this lack of inbreeding depression may be explained by purging of strongly deleterious alleles in persistently small populations. Potentially, the purging of strongly deleterious alleles in island foxes may have enabled their long-term persistence, as well as rapid recovery from recent severe bottlenecks.
Sluggish recovery from bottlenecks is a characteristic of severely inbred populations [16, 22]. For example, the wolf population of Isle Royale National Park, founded less than a hundred years ago, is intensely inbred with evidence of inbreeding depression (Figure 1), has not increased in size despite an abundance of prey, and is destined for extinction [15, 23]. Large outbred populations, such as the mainland source population for Isle Royale wolves, likely carry a greater hidden load of recessive deleterious mutations, increasing the risk of inbreeding depression following population decline. Experiments with houseflies (Musca domestica) have found that purging with population persistence is more likely when the rate of inbreeding is slow, as might occur in a chronically small population, rather than fast, as in a sudden extreme bottleneck [24]. A previous history of purging of strongly deleterious recessive variants due to long-term small population size likely pre-adapted island foxes to rapid recovery from recent bottlenecks.
Few examples of intentional inbreeding to facilitate purging of strongly deleterious alleles are known [25], though purging as a result of long-term reduced population sizes may conceivably be a factor in other cases of persistence with low genome-wide diversity [26, 27]. In contrast, examples of inbreeding depression are frequent in both wild and captive environments, and minimizing inbreeding is commonly a paramount concern of captive management efforts [28, 29]. Under long-term small population size, weakly deleterious mutations may accumulate, slightly beneficial mutations may be lost, and depletion of genetic variation could compromise adaptability [1]. Rapid recovery of island fox populations following recent bottlenecks suggests weakly deleterious mutations have not seriously compromised their fitness, at least for several thousand generations, though the progressive fixation and accumulation of such mutations over time is predicted to ultimately lead to mutational meltdown and extinction [30]. Our results suggest this process may take place gradually over thousands of generations, if it occurs at all before other non-genetic factors cause extinction.
One final consideration in the persistence of island foxes is that historically, island foxes may have benefitted from a relatively benign environment. Recently, however, island fox populations have been threatened by anthropogenic changes in their environment (e.g. traffic collisions, novel diseases and predators) [6]. A recent decline in the San Nicolas population due to unknown causes has resurrected the idea that genetic rescue, or human-assisted migration, may be necessary to preserve island fox populations [3, 9]. If the recent decline is not the result of inbreeding depression, but is rather due to environmental stressors, genetic rescue may not materially enhance the likelihood of population persistence and would compromise the genetic distinction of the population. Genetic rescue should not be used as a conservation technique for historically small populations, as on many islands, without compelling evidence of inbreeding depression. A systematic survey to detect inbreeding depression (assays of sperm quality, reproductive success, survivorship, etc.) is necessary before considering genetic rescue as a conservation strategy for island foxes. We advocate for continued close monitoring of island fox populations, “watchful waiting”, as in slowly developing cancers, to ensure a swift response in the event of a catastrophic decline.
In conclusion, the small population paradigm emphasizing an “extinction vortex,” partly driven by genetic factors [31], may not be an appropriate model when purging of strongly deleterious alleles has occurred through historically small population size. Of course, the fate of many small populations may be extinction before purging can occur. However, the independent long-term persistence of all six island fox populations for thousands of years suggests small populations on the order of a few hundred individuals can persist over evolutionary timescales, and provides a model for the preservation of small fragmented populations.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jacqueline Robinson (jacqueline.robinson@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
DNA samples from island foxes sampled in 2000-2009 from each population and from a Northern California gray fox were obtained from the archive of Dr. Robert K. Wayne for whole genome sequencing on Illumina HiSeq machines at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley. Additionally, we sequenced DNA isolated from miscellaneous bone fragments from a San Nicolas island fox specimen collected in 1929 (#15477, Donald R. Dickey Collection, University of California, Los Angeles). Finally, we generated new higher quality sequence data from a 1988 Santa Rosa island fox, as the Santa Rosa genome from Robinson et al. [5] was of relatively low quality compared to the other sequences. Further sample details are provided in Table S1. We incorporated the genomes from Robinson et al. [5] yielding a total of 16 genomes at 13-23X coverage.
METHOD DETAILS
Read alignment and processing followed the methods outlined in Robinson et al. [5]. Briefly, reads were aligned to the domestic dog reference genome, canFam3.1, with BWA MEM [32], followed by removal of duplicate and low quality reads (reads with mapping quality Phred score <30 and reads not mapped in proper pairs), base quality score recalibration with the Genome Analysis Toolkit [33], joint genotyping with FreeBayes [34], and finally variant and genotype filtering. Variants that were not biallelic SNPs with Phred score ≥30, variant sites with more than 4 missing genotypes, and variant sites with more than 75% of genotypes called as heterozygous were filtered out. Heterozygous genotypes at sites with skewed allele balance (<0.2 or >0.8) were also excluded. Sites in CpG islands, repetitive regions, or with excess depth (>99th percentile total depth) were masked. At least two observations of the alternate allele on each of the forward and reverse strands were required as evidence of a variant site. Further, genotypes with fewer than 6 supporting reads, with Phred score less than 20, or with depth greater than the 99th percentile for a given individual were filtered out.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of variation in island and gray fox genomes
We evaluated the genetic distance between individuals with a maximum-likelihood phylogenetic tree and principal components analysis (PCA), both based on a set of 12,249 SNPs pruned for linkage disequilibrium (maximum=0.2) with a minimum minor allele frequency of 0.1 and no missing data. The pruning and PCA were conducted with SNPRelate [35]. The tree was generated in SNPhylo from 1,000 bootstrap replicates [36].
We assessed genetic diversity by calculating heterozygosity, here defined as the number of heterozygous genotypes divided by the number of called sites within a single individual. Heterozygosity was calculated for the entire autosomal genome and in 100 kb sliding windows with a 10 kb step size. Windows with more than 20% of sites failing quality filters, or with fewer than 20 kb of confidently called sequence were excluded. Peaks of heterozygosity within a genome were defined as windows with heterozygosity greater than two standard deviations above the mean, based on the genome-wide distribution of per-window heterozygosity. Overlapping windows of high heterozygosity were merged using BEDTools [37]. Peaks were assessed for putative functional relevance by conducting a gene ontology enrichment analysis in gProfileR [38]. The g:SCS correction method within gProfileR was used to assess significance.
Since analysis of olfactory receptor genes may be complicated by their elevated rate of evolution, which could produce technical artifacts due to improper read alignment, we employed several stringent filters (described above) that should reduce the inclusion of likely gene paralogs in our analyses. These filters excluded regions of excessive depth (indicating possible gene copy number differences) and low sequence complexity (indicating repetitive or non-unique sequence), reads with low mapping quality (indicating non-unique alignment or excessive mismatches to the reference), variants with skewed allele balance in heterozygotes or excess heterozygosity, and windows with a large fraction of sites failing filters. Manual inspection of a subset of San Nicolas heterozygous peak regions revealed few that were obviously the result of alignment errors, although this possibility cannot be ruled out with the existing data. We confirmed that technical differences were not driving variation in peaks containing OR genes between San Nicolas genomes by determining that >98% of peaks overlapping OR genes were confidently called across all four individuals.
Coordinates of the set of 13,647 putatively neutral 1-kb loci in the dog reference genome were those employed by Robinson et al. ([5], originally from [39]). These regions were designed to be distant from functional regions (≥10 kb from coding sequence, ≥100 bp from conserved non-coding sequence) while excluding regions likely to have problematic alignment due to repetitive content or low mappability, and were spaced ≥30 kb apart to reduce the effects of non-independence due to linkage. Coordinates of zero-fold degenerate sites (where all mutations are amino acid-changing) and four-fold degenerate sites (where all mutations result in the same amino acid) within protein-coding regions were those employed by Robinson et al. ([5], originally from [40]). Levels of deleterious variation were evaluated by calculating the proportion of derived alleles per genome at synonymous and non-synonymous sites in coding regions as follows.
Variant annotation was conducted with Ensembl’s Variant Effect Predictor (VEP; [41]) and the Sorting Intolerant From Tolerant (SIFT; [42]) tool. SIFT classifies non-synonymous mutations at each site as likely to be deleterious or tolerated on the basis of amino acid conservation across taxa. Following Robinson et al. [5], likelihood ratio tests were used to evaluate whether the proportion of homozygotes and the proportion of derived alleles differed significantly between gray and island foxes at synonymous, tolerated, deleterious, and LOF variant sites. The test compared the likelihoods under two models; under the null model, there is a single mean value (q) for all gray and island foxes, and under the alternative model, the island fox mean (qi) may differ from the gray fox mean (qg). Log-likelihood values for the null and alternative models were used to calculate the likelihood ratio test statistic, Λ=−2(log-likelihoodnull – log-likelihoodalternative). Asymptotically, Λ is χ2-distributed with one degree of freedom. This distribution was used to calculate p-values. For allele tests, the null model assumes the ratio of qi to qg found at synonymous SNPs. This is a conservative test of whether the difference in the proportion of derived alleles between island and mainland foxes is greater than that seen at synonymous SNPs, which should be equivalent under neutrality, but was found to differ slightly, reflecting possible technical biases such as the under-calling of heterozygotes.
Coalescent simulations under San Nicolas demographic models
We performed neutral coalescent simulations in msprime [11] under plausible models of San Nicolas demographic history to evaluate empirical patterns of genome-wide diversity in this population. The parameters of demographic models inferred in Robinson et al. [5] were used. Briefly, these parameters were obtained through approximate Bayesian computation, and were designed to match observed levels of heterozygosity within 13,647 putatively neutral 1-kb loci (described above) in the 1988 San Nicolas genome. Parameter values from the 100 models with the best fit to the empirical data constituted the posterior distribution. Here, we sampled parameters with replacement from the joint posterior distribution 1,000 times to simulate complete San Nicolas genomes under neutrality. The simulated genomes consisted of 220 "chromosomes" of 10 Mb in length to emulate the size of the autosomal dog reference genome. We used a mutation rate of 2 × 10−8 per site per generation, and assumed a generation time of one year. A single per-site recombination rate was randomly sampled for each chromosome, drawn from the recombination rates inferred in a study in dogs [43]. The genome architecture, mutation rate, and recombination rates were chosen to be consistent with Robinson et al. [5].
Within each simulation, we randomly sampled one individual in the generation corresponding to the year 1929, two individuals in 1988, and one individual in 2000, and obtained the coordinates of peaks of heterozygosity within these genomes as we did in our analysis of the empirical data. Specifically, we calculated heterozygosity in 100 kb windows with a 10 kb step size for each individual. To take into account the variance in missing data rates across windows in the sequenced genomes, we randomly selected a subset of sites within each simulated window to match the number of sites observed empirically in each San Nicolas genome. Overlapping windows were merged prior to calculating the number of peaks of heterozygosity, the mean width of peaks of heterozygosity, and the proportion of unique peaks within each individual. To determine whether the empirical values we obtained from the sequenced genomes were expected based on the simulated results, we calculated the percentile rankings of where the empirical values fell within the distributions obtained from simulation. In all cases, the empirical values fell within the middle 95% interval of the simulated statistics, implying that the empirical results are expected under neutrality in San Nicolas demographic models.
Morphological assay of gray and island fox museum specimens
Skulls (n=141) and complete skeletons (n=163) of adult gray and island foxes sampled between 1929 and 2013 were obtained from collections in the Los Angeles County Museum of Natural History, the Santa Barbara Museum of Natural History, and the Donald Ryder Dickey Bird and Mammal Collection at the University of California, Los Angeles. Specimens were placed in anatomical position and surveyed for skeletal abnormalities including irregular development, healed fracture, and bony growths (from infection, muscle traction or other causes). Vertebrae were identified and counted with the exception of distal caudal vertebrae, which were missing from many specimens, and are also variable between islands. Specimen information and morphological descriptions are available in a table archived at https://doi.org/10.5281/zenodo.1345794.
We defined “congenital” abnormalities as pathologies of the axial skeleton not related to trauma or injury. Observed congenital defects included extra vertebrae, transitional vertebrae (LSTV: malformed final lumbar vertebra with characteristics of a sacral vertebra, SCTV: malformed final sacral vertebra with characteristics of a caudal vertebra), and maloccluded teeth. LSTV is implicated in function impairment but SCTV has no clinical significance [15, 44]. Malocclusion has been found to have a significant genetic component in small canids generally [45, 46] and in reproductively isolated fox (Vulpes vulpes) populations specifically [47]. A genetic basis has not been established for extra vertebrae or transitional vertebrae, but the high incidence of congenital vertebral anomalies in highly inbred wolf populations on Isle Royale and in Scandinavia implies that there is a heritable component in canids [14, 15]. Vertebral abnormalities are absent or rare in outbred wolf and coyote (Canis latrans) populations that have been surveyed [14, 15, 48].
We binned traumatic pathologies into a broad category of “trauma” and a more conservative category of trauma resulting from collision with a vehicle. Injury pathologies were broadly defined to include fracture, infection, osteoarthritis, osteophytes and evidence of abnormal muscle use (e.g. traction, roughened insertion). Only healed injuries were recorded, thus all foxes obtained as traffic fatalities were counted only if they had previously survived a collision. Thus, the frequency of vehicle collision rates is an underestimate. To identify probable vehicle collisions, we expanded the diagnosis of Harris [49] in which multiple bones from the same side of the body were fractured, particularly in the hindlimb and ribs. In our analyses, evidence of muscular strain on the same side as multiple fractured elements were included as evidence of trauma to that side of the body, since we surveyed animals that survived collisions but Harris necropsied fresh traffic fatalities.
Fisher's exact test was used to test for significant differences in the prevalence of congenital and traumatic pathologies between island and mainland foxes. Additionally, relative risk scores for congenital and traumatic pathologies were calculated for all population pairs. The relative risk score is the ratio of the incidence of pathology between two populations, where incidence is defined as the number of individuals with the pathology divided by the number of individuals examined. For example, if the incidence of pathology in one population is 1/10, and the incidence in a second population is 2/50, the relative risk of pathology in the first population relative to the second is 0.1/0.04=2.5; likewise the relative risk in the second population relative to the first is 0.04/0.1=0.4. Statistical significance was determined by calculating relative risk scores in 1,000 permutations where population labels were assigned to specimens at random. Empirical relative risk scores greater than the 95th percentile of scores obtained through permutation were deemed significant.
Forward simulations of genetic variation in island versus mainland populations
We simulated neutral and deleterious variation under six different demographic models, each involving the establishment of a small island population (N=1,000, 500, 200, 100, or 50 individuals) from a large mainland population (N=10,000 individuals) to evaluate the possibility of purging in island populations. Simulations were conducted with SLiM [20]. In the most basic model (Split Model), an island population of is established by sampling individuals from the mainland population with replacement, such that the mainland population is unaffected by the formation of the island population. The mainland population was kept at constant size for 10,000 generations. A range of island population sizes was chosen because census sizes of island fox populations vary from a few hundred to just under 2,000 individuals per island [8]. Recent estimates suggest an initial colonization of the islands by foxes at ~9,200 years BP [4], and we assumed a one-year generation time. The purpose of the basic split model was to compare a small population against a larger one, without any additional complexities (e.g. inbreeding).
The other five models are variations on the split model, reflecting plausible elements of island fox history, specifically: 1) an ancient bottleneck to simulate a small founding population (Ancient Bottleneck Model); 2) a recent bottleneck 30 generations ago, such as may have occurred in San Nicolas during the 1970s [10] (Recent Bottleneck Model); 3) a serial bottleneck (at 10,000 and 2,000 generations ago) to mimic the possibility of serial colonization of the Northern and then the Southern Channel Islands (Serial Bottleneck Model); 4) a model in which the island population is initially large (N=2,000 for ~2,000 generations), as may have occurred if the foxes initially the Northern Channel Islands when they constituted a single landmass (Santarosae) when sea levels were lower >10,000 years ago [50] (Big Island Model); and 5) a model with recent strong inbreeding, where individuals are twice as likely to mate with close relatives than non-kin (Inbreeding Model). All bottlenecks consist of a starting population of ten individuals that doubles in size each generation until the final population size (1,000, 500, 200, 100, or 50 individuals) is reached.
Each simulated individual consists of a diploid 2 Mb genome, consisting of 2,000 “genes” carried on 38 chromosomes proportional to chromosome lengths in the dog genome. Each gene was represented by a contiguous 1 kb sequence that accumulated mutations at a rate of 1 × 10−8 per nucleotide per generation. 30% of these mutations were selectively neutral, and the remaining 70% were deleterious, with selection coefficients drawn from a gamma distribution of fitness effects [51]. Recombination was incorporated by including a single base pair in between each gene that did not accumulate mutations, but where crossovers occurred at a rate of 1 × 10−3 per site per generation. This was done to mimic intergenic regions 100 kb in length with a recombination rate of 1 × 10−8 per site per generation without simulating extraneous non-coding sequence. Each model was run for 10,000 generations following a 100,000-generation burn-in period.
The average number of alleles and the average number of homozygous alleles carried by each individual were calculated for deleterious (s<0) and neutral mutations (s=0). We grouped deleterious mutations arbitrarily as strongly (s<−0.01), moderately (−0.01≤s<−0.001), and weakly deleterious (−0.001≤s<0). We categorized deleterious mutations by selection coefficient (s), rather than population-rescaled coefficients (Ns), specifically to explore the effects of different population sizes on the accumulation/depletion of mutations with non-zero selection coefficients. Within a simulation, deleterious mutations were either entirely additive (h=0.5) or entirely recessive (h=0.0). Fifty replicates were performed for each dominance value and each island size (N=1,000, 500, 200, 100, or 50) under each of the six models (3,000 simulations total). One-way ANOVA and Tukey HSD post hoc tests were used to evaluate significant differences in the number of total alleles and the number of homozygous alleles between different models.
DATA AND SOFTWARE AVAILABILITY
All newly generated raw whole genome sequence reads have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA478450. Previously sequenced reads from Robinson et al. [5] are available under BioProject PRJNA312115. A table containing specimen information and phenotypic descriptions used for the morphological assessment is available at https://doi.org/10.5281/zenodo.1345794. Scripts for coalescent and forward simulations are available at https://github.com/jarobin/islandfox2018, and archived at https://doi.org/10.5281/zenodo.1345812.
Supplementary Material
Acknowledgements
We thank the National Park Service, the Catalina Island Conservancy, Dennis Hedgecock, and William Andelt for sharing samples used for genomic sequencing. We thank Paul Collins for advice and access to specimens, and the Los Angeles County Museum of Natural History, the Santa Barbara Museum of Natural History, and the Donald Ryder Dickey Bird and Mammal Collection at the University of California, Los Angeles for the use of their collections. This work was supported with funding from a UC President’s Catalyst Award to RKW and National Institutes of Health grant R35GM119856 to KEL. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Frankham R (2005). Genetics and extinction. Biol. Cons 126, 131–140. [Google Scholar]
- 2.Collins PW (1991). Interaction between Island Foxes (Urocyon littoralis) and Indians on Islands off the Coast of Southern California: I. Morphologic and Archaeological Evidence of Human Assisted Dispersal. J. Ethnobiol 11, 51–81. [Google Scholar]
- 3.Vellanoweth RL (1998). Earliest island fox remains on the southern Channel Islands: evidence from San Nicolas Island, California. J. Calif. Great Basin Anthropol 20, 100–108. [Google Scholar]
- 4.Hofman CA, Rick TC, Hawkins MT, Funk WC, Ralls K, Boser CL, Collins PW, Coonan T, King JL, Morrison SA et al. (2015). Mitochondrial genomes suggest rapid evolution of dwarf California Channel Islands foxes (Urocyon littoralis). PLoS ONE 10, e0118240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, Marsden CD, Lohmueller KE and Wayne RK (2016). Genomic flatlining in the endangered island fox. Curr. Biol 26, 1183–1189. [DOI] [PubMed] [Google Scholar]
- 6.Coonan TJ, Schwemm CA and Garcelon DK (2010). Decline and recovery of the island fox: a case study for population recovery (New York: Cambridge University Press; ). [Google Scholar]
- 7.US Fish and Wildlife Service (2016). Removing the San Miguel Island Fox, Santa Rosa Island Fox, and Santa Cruz Island Fox From the Federal List of Endangered and Threatened Wildlife, and Reclassifying the Santa Catalina Island Fox From Endangered to Threatened; final rule. Fed. Reg 81, 53315–53333. [Google Scholar]
- 8.Wayne RK, George SB, Gilbert D, Collins PW, Kovach SD, Girman D and Lehman N (1991). A morphologic and genetic study of the island fox, Urocyon littoralis. Evolution 45, 1849–1868. [DOI] [PubMed] [Google Scholar]
- 9.Funk WC, Lovich RE, Hohenlohe PA, Hofman CA, Morrison SA, Sillett TS, Ghalambor CK, Maldonado JE, Rick TC, Day MD et al. (2016). Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis). Mol. Ecol 25, 2176–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laughrin LL (1980). Populations and status of the island fox In The California islands: proceedings of a Multi-disciplinary symposium, Power DM, ed. (Santa Barbara, CA: Santa Barbara Museum of Natural History; ), pp. 745–749. [Google Scholar]
- 11.Kelleher J, Etheridge AM and McVean G (2016). Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Comp. Biol 12, e1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niimura Y and Nei M (2007). Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marker LL and Dickman AJ (2004). Dental Anomalies and Incidence of Palatal Erosion in Namibian Cheetahs (Acinonyx jubatus jubatus). J. Mammal 85, 19–24. [Google Scholar]
- 14.Räikkönen J, Bignert A, Mortensen P and Fernholm B (2006). Congenital defects in a highly inbred wild wolf population (Canis lupus). Mamm. Biol 71, 65–73. [Google Scholar]
- 15.Räikkönen J, Vucetich JA, Peterson RO and Nelson MP (2009). Congenital bone deformities and the inbred wolves (Canis lupus) of Isle Royale. Biol. Cons 142, 1025–1031. [Google Scholar]
- 16.Roelke ME, Martenson JS and O'Brien SJ (1993). The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Curr. Biol 3, 340–350. [DOI] [PubMed] [Google Scholar]
- 17.Wildt DE, Bush M, Howard JG, O’Brien SJ, Meltzer D, Van Dyk A, Ebedes H and Brand DJ (1983). Unique seminal quality in the South African cheetah and a comparative evaluation in the domestic cat. Biol. Reprod 29, 1019–1025. [DOI] [PubMed] [Google Scholar]
- 18.Asa CS (2010). The importance of reproductive management and monitoring in canid husbandry and endangered species recovery. Int. Zoo Yearb 44, 102–108. [Google Scholar]
- 19.Snow NP, Andelt WF and Gould NP (2011). Characteristics of road-kill locations of San Clemente Island foxes. Wildl. Soc. Bull 35, 32–39. [Google Scholar]
- 20.Haller BC and Messer PW (2016). SLiM 2: flexible, interactive forward genetic simulations. Mol. Biol. Evol 34, 230–240. [DOI] [PubMed] [Google Scholar]
- 21.Ohta T (1992). The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst 23, 263–286. [Google Scholar]
- 22.Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, Bouzat JL and Paige KN (1998). Tracking the long-term decline and recovery of an isolated population. Science, 282, 1695–1698. [DOI] [PubMed] [Google Scholar]
- 23.Hedrick PW, Peterson RO, Vucetich LM, Adams JR and Vucetich JA (2014). Genetic rescue in Isle Royale wolves: genetic analysis and the collapse of the population. Conserv. Genet 15, 1111–1121. [Google Scholar]
- 24.Day SB, Bryant EH and Meffert LM (2003). The influence of variable rates of inbreeding on fitness, environmental responsiveness, and evolutionary potential. Evolution 57, 1314–1324. [DOI] [PubMed] [Google Scholar]
- 25.Crnokrak P and Barrett SC (2002). Perspective: purging the genetic load: a review of the experimental evidence. Evolution 56, 2347–2358. [DOI] [PubMed] [Google Scholar]
- 26.Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN et al. (2015). Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benazzo A, Trucchi E, Cahill JA, Delser PM, Mona S, Fumagalli M, Bunnefeld L, Cornetti L, Ghirotto S, Girardi M et al. (2017). Survival and divergence in a small group: The extraordinary genomic history of the endangered Apennine brown bear stragglers. Proc. Natl. Acad. Sci. USA 114, E9589–E9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralls K and Ballou J (1983). Extinction: lessons from zoos In Genetics and Conservation, Schonewald-Cox C, Chambers S, MacBryde B, Thomas L eds. (Menlo Park, CA: Benjamin/Cummings; ), pp. 164–184. [Google Scholar]
- 29.Crnokrak P and Roff DA (1999). Inbreeding depression in the wild. Heredity 83, 260–270. [DOI] [PubMed] [Google Scholar]
- 30.Lynch M, Conery J and Burger R (1995). Mutational meltdowns in sexual populations. Evolution 49,1067–1080. [DOI] [PubMed] [Google Scholar]
- 31.Gilpin ME and Soulé ME (1986). Minimum Viable Populations: Processes of Species Extinction In Conservation Biology: The Science of Scarcity and Diversity, Soulé ME ed. (Sunderland, MA: Sinauer; ), pp. 19–34. [Google Scholar]
- 32.Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA MEM. arXiv:1303.3997v2 [Google Scholar]
- 33.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrison E and Marth G (2012). Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [Google Scholar]
- 35.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C and Weir BS (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee TH, Guo H, Wang X, Kim C and Paterson AH (2014). SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan AR and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reimand J, Arak T and Vilo J (2011). g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 35, W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B et al. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10, e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vila C, Marques-Bonet T, Schnabel RD, Wayne RK and Lohmueller KE (2016). Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. USA 113, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P and Cunningham F (2010). Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar P, Henikoff S and Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 43.Wong AK, Ruhe AL, Dumont BL, Robertson KR, Guerrero G, Shull SM, Ziegle JS, Millon LV, Broman KW, Payseur BA et al. (2010). A comprehensive linkage map of the dog genome. Genetics 184, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thrall DE and Robertson ID (2015). Atlas of Normal Radiographic Anatomy and Anatomic Variants in the Dog and Cat, First Edition. (St. Louis, MO: Elsevier Saunders; ) [Google Scholar]
- 45.Pengilly D (1984). Developmental versus functional explanations for patterns of variability and correlation in the dentitions of foxes. J. Mammal 65, 34–43. [Google Scholar]
- 46.Trut LN, Oskina IN and Kharlamova AV (2001). Experimental studies of early canid domestication In The Genetics of the Dog, Second Edition, Ostrander EA, ed. (Cambridge, MA: CAB International; ), pp. 12–37. [Google Scholar]
- 47.Bouwmeester J, Mulder JL and Bree PV (1989). High incidence of malocclusion in an isolated population of the red fox (Vulpes vulpes) in the Netherlands. J. Zool 219, 123–136. [Google Scholar]
- 48.Wobeser G (1992): Traumatic degenerative and developmental lesions in wolves and coyotes from Saskatchewan. J. Wildl. Dis 28, 268–275. [DOI] [PubMed] [Google Scholar]
- 49.Harris S (1977). Spinal arthritis (spondylosis deformans) in the red fox, Vulpes vulpes, with some methodology of relevance to zooarchaeology. J. Archaeol. Sci 4, 183–195. [Google Scholar]
- 50.Porcasi P, Porcasi JF and O’Neill C (1999). Early Holocene coastlines of the California Bight: the Channel Islands as first visited by humans. Pacific Coast Archaeological Society Quarterly 35, 1–24. [Google Scholar]
- 51.Kim BY, Huber CD and Lohmueller KE (2017). Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly generated raw whole genome sequence reads have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA478450. Previously sequenced reads from Robinson et al. [5] are available under BioProject PRJNA312115. A table containing specimen information and phenotypic descriptions used for the morphological assessment is available at https://doi.org/10.5281/zenodo.1345794. Scripts for coalescent and forward simulations are available at https://github.com/jarobin/islandfox2018, and archived at https://doi.org/10.5281/zenodo.1345812.