Abstract
Abnormal pH is a common feature of malignant tumours and has been associated clinically with sub-optimal outcomes. Amide proton transfer magnetic resonance imaging (APT MRI) holds promise as a means to non-invasively measure tumour pH, yet multiple factors collectively make quantification of tumour pH from APT MRI data challenging. The purpose of this study was to improve our understanding of the biophysical sources of altered APT MRI signals in tumours. Combining in vivo APT MRI measurements with ex vivo histological measurements of protein concentration in a rat model of brain metastasis, we determined that the proportion of APT MRI signal originating from changes in protein concentration was approximately 66%, with the remaining 34% originating from changes in tumour pH. In a mouse model of hypopharyngeal squamous cell carcinoma (FaDu), APT MRI showed that a reduction in tumour hypoxia was associated with a shift in tumour pH. The results of this study extend our understanding of APT MRI data and may enable the use of APT MRI to infer the pH of individual patients' tumours as either a biomarker for therapy stratification or as a measure of therapeutic response in clinical settings.
Keywords: amide proton transfer, magnetic resonance imaging, pH, tumour, validation
Introduction
The metabolic phenotype of tumour cells is such that they reverse the pH gradient across the cell membrane with respect to normal cells, with a slightly alkaline intracellular pH (pHi) and an acidic extracellular pH (pHe) (1). The severity of the acidosis of pHe has been shown to correlate with resistance to chemo- and radiotherapy treatments; tumours with a very acidic pHe are more resistant to radiotherapy (2), and ion-trapping of weakly basic chemotherapy agents prevents their entry to the cell (3). An alkalotic pHi also makes tumour cells less susceptible to cell death via apoptotic pathways (4) and promotes tumour proliferation (5). Determination of pHe or pHi in a non-invasive, reliable manner may, therefore, aid in the stratification of patients into personalised therapeutic strategies, or inform on treatment response. Thus, there is a clear need for non-invasive, reliable quantification of tumour pH in the clinic.
In addition to pH changes, tumours often have regions of acute and chronic hypoxia as a result of both an increased oxygen consumption rate of tumour cells compared to normal cells, and an abnormal vasculature reducing delivery of oxygen (6,7). Tumours with larger fractions of hypoxia are more resistant to chemotherapy as a result of this abnormal vasculature, and are more resistant to radiotherapy because of a reduced oxygen enhancement effect of radiotherapy. Consequently, measurement of tumour hypoxia may also aid in the stratification of patients into optimal therapies, and reduction of tumour hypoxia is a common target for therapies. Importantly, a close link between tumour hypoxia and pH has been demonstrated (8). If tumour hypoxia is reduced then a shift in the metabolic phenotype of tumour cells that were previously hypoxic would be expected, with a resultant change in their pHi and pHe.
Chemical exchange saturation transfer (CEST) is a magnetic resonance imaging (MRI) contrast mechanism that utilises the chemical exchange of labile protons in biomolecules with solvent water protons (9). Amide proton transfer (APT) is a variant of CEST MRI that is sensitive to the exchange of amide protons resident on the backbone and sidechains of proteins. APT MRI has been used previously in the imaging of tumours for non-invasive staging (10,11), differentiation of radiation necrosis from recurrent tumour (12,13) and definition of infiltrating tumour rim tissue (14). Additionally, since the APT signal is dependent on the exchange rate of amide protons with solvent water protons, and this exchange rate is base catalysed (15), APT is sensitive to the pH of tissue. Previous studies have measured the pH of tissue in ischaemic stroke lesions using calibrated APT signals (16,17). Quantification of tumour pH from APT MRI data, however, remains a significant challenge.
Preclinical and clinical APT MRI studies have usually attributed the altered APT signal in tumours to an increase in cytosolic protein concentration because the cells are rapidly proliferating (10,18). This interpretation is contested, however, by work suggesting that APT signal changes in tumours are a result of alterations in water T1 relaxation time in these tissues (19). Despite the metabolic phenotype of tumour cells altering the intra- and extracellular pH of their microenvironment, this factor tends to be ignored when interpreting the APT signal change in tumours. The reason for this apparent oversight is that APT MRI is weighted towards measuring the intracellular compartment (18) and the intracellular pH change in tumours is less marked than the extracellular pH change (20,21). The combination of these factors has limited, to date, the use of APT MRI for quantitation of biophysical parameters such as protein concentration or pH in tumours, and this limitation will remain until the complex interplay between these factors is deconvolved.
The aim of this study, therefore, was to determine the proportion of APT signal change between normal appearing and tumour tissue that is caused by protein concentration and pH changes, respectively, using a preclinical model of brain metastasis. Subsequently, we tested the sensitivity of our approach to pharmacological modulation of tumour pH following alleviation of tumour hypoxia in subcutaneous tumours. By understanding the biophysical sources of altered APT signals in tumours, our goal was to improve the interpretation of APT signal changes in tumours.
Methods and Materials
In vivo brain tumour models
All animal experiments were approved by the UK Home Office (Animals [Scientific Procedures] Act 1986) and conducted in accordance with the Guidelines for the Welfare and Use of Animals in Cancer Research (22). For the brain metastasis model, female Berlin Druckrey IX (BDIX) rats (180 – 340 g; n = 15; Charles River, UK) were focally microinjected with 1000 ENU1564 cells (kind gift from Prof. G. Stoica, Texas A&M University) in 1 µL phosphate buffered saline into the left striatum (co-ordinates 1 mm anterior, 3 mm lateral from bregma, 3.5 mm depth), as described previously (23). MRI experiments were performed four weeks post-injection. This animal model has been previously shown to exhibit similar histological characteristics to human brain metastatic growth, with an infiltrating tumour rim and necrotic tumour core, allowing observation of these two distinct areas (23).
Subcutaneous model of hypopharyngeal squamous carcinoma
Female BALB/c nude mice (age 55–70d; n = 18; Charles River, UK) were implanted with subcutaneous tumours on their right flank. Tumours were induced by injection of 1x106 FaDu hypopharyngeal carcinoma cells in Matrigel (Corning, USA). Once tumours reached a volume of 100 mm3 measured by callipers, mice were randomly split into Atovaquone-treated or control groups. Atovaquone is an anti-malarial drug which has recently been shown to alter the oxygen consumption rate of cancer cells in vitro and in vivo, reducing tumour hypoxia (24). Atovaquone was administered in drinking water (50 mg kg−1 d−1) with 2% dimethyl sulfoxide (DMSO) and 0.1% carboxymethylcellulose (CMC). Control mice were treated with DMSO and CMC only. After 7 days of treatment, MRI was performed on each mouse.
MRI experiments
All MRI experiments were performed using a 9.4 T Varian Inova spectrometer (Agilent Technologies, Santa Clara, CA, USA). Animals were anaesthetised with 2 – 3 % isoflurane in a mixture of 30 % oxygen and 70 % nitrogen. Respiration and rectal temperature were monitored and maintained at 40 – 60 breaths/min and 37 °C, respectively. For imaging of rats with brain tumours, a 72 mm diameter volume transmit coil and 4-channel surface receive array (Rapid Biomedical) were used, with the rat head immobilised using a custom cradle. Prior to placement of the rats in the MRI scanner, a tail vein was cannulated to allow for injection of contrast agents during imaging. For a subset of the rats (n = 10), 60 mg/kg pimonidazole (Hypoxyprobe, USA) was injected intraperitoneally prior to imaging. For imaging of mice with subcutaneous FaDu tumours, a 26 mm diameter volume transmit-receive coil was used, with mice positioned supine. Insulation was placed around the mice to prevent excessive heat loss and provide a small amount of immobilisation.
The CEST MRI pulse sequence used for rat imaging comprised a pulsed saturation scheme of 50 saturation pulses, with each pulse comprising a 20 ms Gaussian radiofrequency (RF) pulse with flip angle 184° followed by a 20 ms crusher gradient, for a total saturation duration of 2 s with equivalent continuous wave RF power of 0.55 µT. This saturation scheme was preferentially sensitive to the exchange of amide protons at pH 7.02 in vivo The use of a pulsed saturation scheme was motivated by matching saturation parameters of previous human APT MRI studies at lower field strengths, which are more restricted in terms of hardware and specific absorption rate considerations (15,25). Following saturation, a spin-echo echo planar imaging (SE-EPI) readout with field of view = 32 mm x 32 mm measured the Z-magnetisation. Other sequence parameters were TR = 5 s, TE = 27 ms, 1 average, 10 slices, slice thickness = 1 mm, in-plane resolution 500 μm x 500 μm. A full Z-spectrum was measured following saturation at 49 saturation frequencies unevenly sampled between -4.1 and 5.0 ppm, with a further two measurements following saturation at ±300 ppm for normalisation.
CEST MRI data were acquired from mice using a multi-slice gradient echo sequence (TR = 195 ms, TE = 1.4 ms) with constant TR respiration gating using a SPLICER acquisition scheme (26). Briefly, within each TR a CEST saturation pulse (Gaussian shape, duration 20 ms, flip angle 180°) was applied followed by a 1 ms crusher gradient and gradient echo readout of one k-space line. Data acquisition that was corrupted by a breath was reacquired. Data were acquired linearly through k-space to ensure that CEST saturation was in the steady state when acquiring the centre of k-space (27). Other sequence parameters were slice thickness = 2 mm, in-plane resolution 469 µm x 469 µm. A full Z-spectrum was measured following saturation at 35 saturation frequencies evenly sampled between –5.1 and 5.1 ppm, with a further two measurements at ±300 ppm for normalisation.
Quantitative maps of the T1 and T2 relaxation times were acquired to correct for the concomitant change in T1 and T2 in tumours when analysing CEST MRI data (see MRI Data Analysis). For rats, T1 and T2 relaxation times were determined using inversion recovery (TR = 10 s, TE = 8.22 ms, inversion time (TI) varied in 9 steps from 13.14 – 8000 ms, signals fitted to MZ = M0(1-2exp(-TI/T1))) and spin echo (TR = 10 s, TE varied in 10 steps from 30 – 160 ms, signals fitted to MZ = M0exp(-TE/T2)) experiments, respectively. In both cases the same SE-EPI readout used for CEST imaging was used to acquire images. The slice plan for T1 and T2 mapping was identical to CEST MRI to enable co-registration of the images. T1 and T2 times were quantified post-mortem immediately after in vivo CEST MRI in a subset of mice used for the hypoxia alleviation experiment (n = 5 Atovaquone treated, n = 5 control) using the same method. Mice were euthanised with an overdose of pentobarbital and re-placed in the MRI, with their temperature continually maintained at 37 °C to minimise variations in relaxation times owing to reduced thermal regulation post-mortem. The relaxation times were measured post-mortem because no respiration-gated relaxation time mapping sequence was available on the spectrometer used in this study.
For rats, T1-weighted gradient echo anatomical imaging was performed pre- and post-injection of gadolinium contrast agent (Omniscan, GE Healthcare, USA) to elucidate the extent of blood-brain barrier breakdown (TR = 500 ms, TE = 20 ms, same slice plan as CEST MRI but with in-plane resolution 125 μm x 125 μm).
MRI Data Analysis
The APT effect from the measured CEST MRI data was quantified using the APTR* metric as described previously (25,28–30). Briefly, data were fit to the Bloch-McConell equations using BayCEST in the FMRIB Software Library (FSL; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/baycest) assuming a 3-pool exchange model comprising water, amide protons at 3.5 ppm, and a combined NOE+MT pool at -2.41 ppm. Correction for water T1 and T2 effects was implemented by allowing BayCEST to fit water T1 and T2 from the data, with prior values set based on the average T1 and T2 time within the whole rat brain or whole tumour for mice, yielding an APTR* value that is sensitive only to amide proton concentration and exchange rate (which is itself proportional to 10pH). A previous study (30) has shown that APTR* quantified using this analysis approach is specific to changes in protein concentration and pH in biologically relevant phantoms. The exchange rate and concentration estimated by BayCEST were used to generate an idealised two-pool Z-spectrum, and APTR* calculated using equation 1. The calculation compares the signal at the amide proton frequency from this two-pool Z-spectrum (Sw+a(3.5ppm)) to the signal from an idealised one-pool Z-spectrum (Sw(3.5ppm)), normalised by the unsaturated signal (Mw0). The T1 and T2 relaxation times in the idealised simulations were set as 1.8s and 50ms for water, and as 1.8s and 1ms for amide protons.
| (1) |
Regions of interest (ROI) were defined to measure the mean ± standard deviation APTR* from areas of biologically similar tissue. For rats, an initial abnormality volume was defined manually based on the T1-weighted post-Gd anatomical images, and down-sampled to the resolution of the CEST images. Since the model of brain metastasis used here is known to reflect the heterogeneity present in human disease (23), with a characteristic rim of infiltrating tumour cells surrounding a core of necrotic tissue and cellular debris, the water T1 time was used within the abnormality region as a tissue classifier: voxels with T1 < 1.6 s were classified as normal tissue, voxels with 1.6 s < T1 < 2.2 s as tumour, and tissue with T1 > 2.2s as necrotic (see Supplementary Figure S1 for example T1 and T2 maps). The automatic segmentation was implemented using MATLAB (MathWorks Inc., Natick, MA, USA) and adjusted manually to ensure an accurate ‘rim-core’ pattern was present in the full tumour volume. An ROI of approximately equal size to the tumour volume was defined in an equivalent anatomical location in the contralateral hemisphere to represent normal tissue, ensuring that the contralateral ROI contained the same composition of tissue (i.e. striatal) as the region encompassing the tumour ROI.
Regions of interest for infiltrating tumour rim, necrotic tumour core, and contralateral hemisphere were then used to calculate the relative APTR* as rAPTR* = APTR*(Tumour) / APTR*(Normal). The relative APTR* metric calculated this way normalises differences in raw APTR* values between animals, thereby allowing groupwise statistics to be calculated. rAPTR* measurements from all animals and all ROIs were combined using a random effects model to generate a weighted mean that accounts for the size of the ROI in each animal as well as the variability of APTR* within each ROI (see Supplementary Methods for further details). Groupwise rAPTR* values are reported as a mean ± 95 % CI, and were compared to a hypothetical mean of 1 (which would suggest no difference from normal tissue) using a one-sample t-test; statistical significance was defined as P < 0.05.
For analysis of the subcutaneous tumours in mice, the whole tumour region was manually segmented. Since no normal control tissue was available to permit calculation of rAPTR*, the absolute APTR* values for all voxels in tumours from both Atovaquone-treated and control groups were calculated and the cumulative frequency of APTR* values compared using the Kolmogorov-Smirnov test, with statistical significance defined as P < 0.05.
Ex vivo protein concentration measurements and histology
Immediately following MRI experiments, rats were split into two groups. One group (n = 10) were sacrificed under terminal anaesthesia by transcardial perfusion-fixation with 0.9% heparinised saline, followed by periodate-lysine-paraformaldehyde (PLP) containing 0.025% glutaraldehyde. The brains of these animals were collected for histology. The remaining rats (n = 5) were sacrificed under terminal anaesthesia by transcardial perfusion with 0.9% heparinised saline. The brains of these rats were extracted and sliced into 2 mm slices using an ice-cold rat brain matrix (Braintree Scientific, USA). Tissue biopsies of the tumour and contralateral brain tissue were taken for subcellular fractionation and protein concentration quantification. The biopsies were snap-frozen in liquid nitrogen and stored at −80°C until further processing.
Mice in the hypoxia alleviation experiment were also split into two groups following MRI. One group (n = 5 Atovaquone-treated animals and n = 6 control animals) were injected intraperitoneally with 0.01 ml g−1 EF5 (2-(2-Nitro-1H-imidazol-1-yl)-N-(2,2,3,3,3-pentafluoropropyl) acetamide) and sacrificed under terminal anaesthesia by transcardial perfusion-fixation with 0.9% heparinised saline, followed by PLP containing 0.025% glutaraldehyde. The tumours of these animals were excised for histology. The remaining animals were sacrificed following MRI under terminal anaesthesia without perfusion, and their tumours excised and stored at −80°C until further processing.
The snap-frozen biopsies from both mice and rats were homogenised in Cytoplasmic Extraction Buffer (from the Subcellular Protein Fractionation Kit for Tissues, Thermo Scientific, UK), sedimented by centrifugation (500 x g for 5 min at 4°C), and the supernatant recovered; this sample contained the cytoplasmic protein from the tissue biopsy. The pellet was resuspended in PBS and mixed until homogenous; this sample contained the remaining (non-cytoplasmic) protein from the biopsy. The protein concentration of these fractions was quantified by bicinchoninic acid (BCA) assay (Pierce BCA Assay Kit, Thermo Scientific, UK). Mean and standard deviation protein concentrations were taken from triplicate measurements, and were combined across animals and cellular locations to give group-wise protein concentration estimates. The total protein concentration was derived as the sum of the two fractions. The protein concentrations were compared using an unpaired two-tailed t-test, with statistical significance defined as P < 0.05.
Tissue from both rats and mice that had been perfusion-fixed was sectioned at 10 µm thickness. Tissue sections were stained non-specifically for all proteins using 1.25 % w/v Coomassie Brilliant Blue G-250 dye (Thermo Scientific, UK) in 0.9 % NaCl with 0.5 % Tween for 30 min. The addition of Tween to the staining solution ensures that cellular membranes are permeated and that Coomassie also stains intracellular proteins. Subsequently, sections were washed in PBS for 3 x 15 min or until the destain solution ran clear, and imaged at 200 x magnification using a Leica Biosystem ScanScope CS2 scanner (Aperio, ePathology Solutions, Milton Keynes, UK). To convert the blue stain from Coomassie into protein concentration measurements, homogenised samples of naïve rat brain tissue were processed in a similar way to experimental tissue and serially diluted, as described previously (31). The protein concentration of each dilution was measured by both BCA assay and Coomassie staining to produce a standard curve (see Supplementary Figure S2 for more details). For rat tissue, tissue sections adjacent to those stained for Coomassie Blue were stained for hypoxia (pimonidazole; Hypoxyprobe, USA) and blood vessel (CD31; AF3628, R&D Systems, UK) markers. For mouse tissue, adjacent tissue sections to those stained with Coomassie Blue were stained for the hypoxia marker EF5 and cellular nucleus marker DAPI, visualised using immunofluorescence, to evaluate the effect of Atovaquone treatment on tumour hypoxia.
Estimation of contribution of protein concentration and pH to APT signals using isoAPTR*
An isoAPTR* analysis (32) was performed to determine the contribution of protein concentration to measured APTR* differences between tumour and normal tissue. Briefly, a library of over 500,000 theoretical APTR* values was generated for a range of amide proton concentrations (relative concentration to water 0 – 4 x 10-3, n = 501) and pH values (5.00 – 7.65, n = 1001, pH dependence of exchange rate defined by kamide = 5.57 × 10pH−6.4) using a two-pool model of the Bloch-McConnell equations. The T1 and T2 relaxation times for water were set as 1.8 s and 50 ms, respectively, and for amide protons as 1.8 s and 1 ms, respectively. Lines of constant APTR* (isoAPTR* lines) were defined in pH-amide concentration space corresponding to those measured from tumour and normal appearing tissue in vivo. The biophysical source of the difference in APTR* between these two tissue areas can then be determined by (1) assuming a normal tissue pH value and (2) inferring a change in amide proton concentration from some independent measurement. Using a normal brain pH of 7.1 (15) and the APTR* measured in normal appearing brain in vivo, the normal brain amide proton concentration was estimated. Subsequently, the change in amide proton concentration between tumour and normal tissue was assumed to be the same as the protein concentration change measured by Coomassie staining. Combining the tumour amide proton concentration estimate with the tumour measured APTR*, an estimate of the tumour pH may be found. By comparing the measured tumour rim APTR* with the APTR* expected to be measured if there was no pH change in the tumour, the proportion of APTR* signal change attributable to pH and protein concentration changes, respectively, was determined.
Since the tumour pH estimated using isoAPTR* depends on the initial value of the normal tissue pH assumed, the steps of the isoAPTR* method were repeated but using different assumptions of normal tissue pH within a realistic physiological range (7.0 – 7.3) and the final tumour pH change taken as the average of all of these repeated calculations.
Results
APTR* is elevated in the tumour rim and tumour core of brain metastases
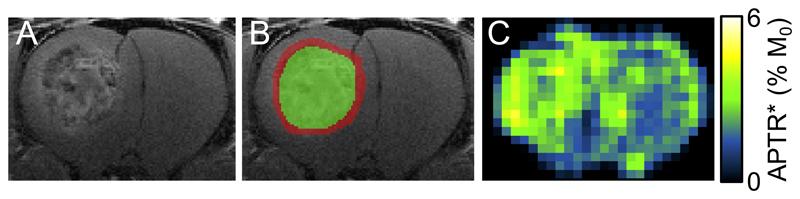
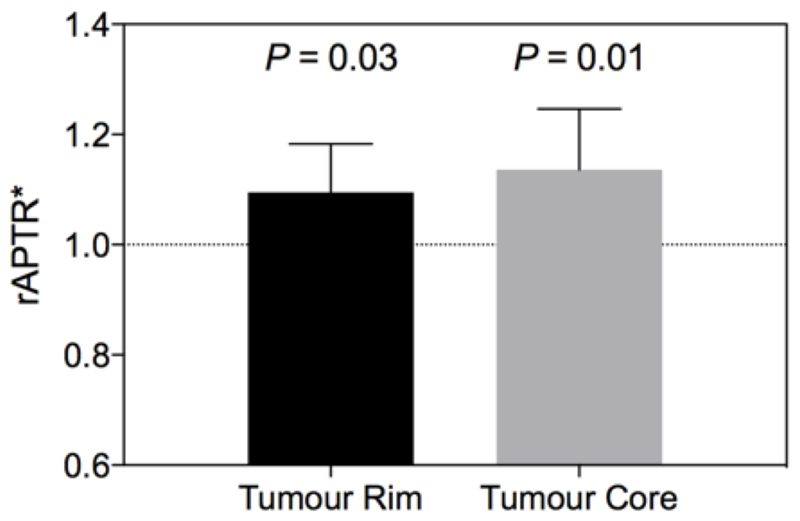
Post-Gd T1-weighted imaging showed the heterogeneity typical of human disease, with a contrast-enhancing rim and hypo- to isointense central area; taken to reflect an infiltrating rim and necrotic core, respectively (Figure 1A,B). APTR* was visually hyperintense in tumour regions (Figure 1C). Conventional MTRasym measurements were elevated in the tumour, and concomitant changes in the T1 and T2 relaxation times were also evident (see Supplementary Figures S2, S3 and Supplementary Table S1). rAPTR* in both rim and core ROIs were significantly greater than 1, indicating increased APTR* values in tumour areas (Figure 2, rAPTR*(Rim) = 1.10 ± 0.09, rAPTR*(Core) = 1.14 ± 0.01, mean ± 95 % CI).
Figure 1.
Representative post-gadolinium image (A) with tumour rim (red) and core (green) regions of interest overlaid (B), and APTR* map (C) from preclinical model of brain metastasis. The contrast-enhancing tumour rim and hypo- to isointense tumour core resemble images obtained from human metastatic foci, and illustrate the intratumoural heterogeneity of an infiltrating rim region and necrotic core. Enhancement in the APTR* map in the region of the tumour is evident. No tumour burden in the contralateral hemisphere is apparent.
Figure 2.
APTR* is elevated in both tumour rim and tumour core. P values show significance level of a one-sample t-test comparing to a hypothetical mean of rAPTR* = 1 (which would indicate no difference from normal tissue). Error bars are 95% CI. The number of voxels for each tissue type is 1347 for tumour rim, and 1475 for tumour core.
Protein concentration is elevated in tumour rim assessed histologically
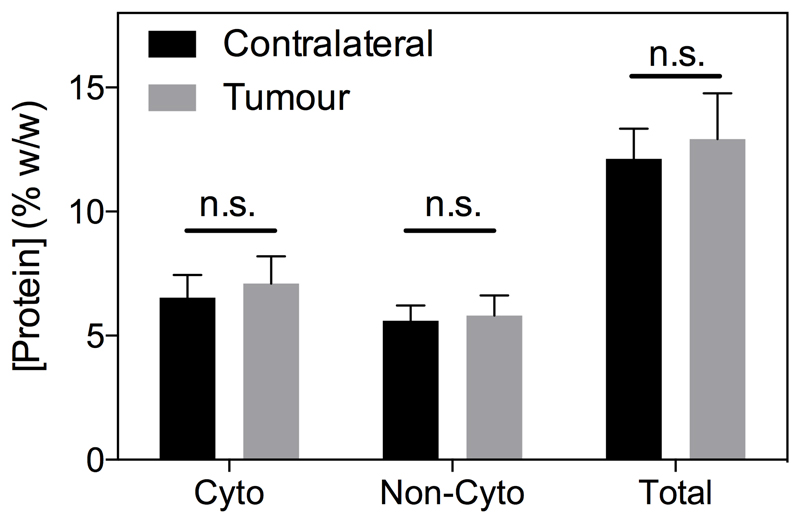
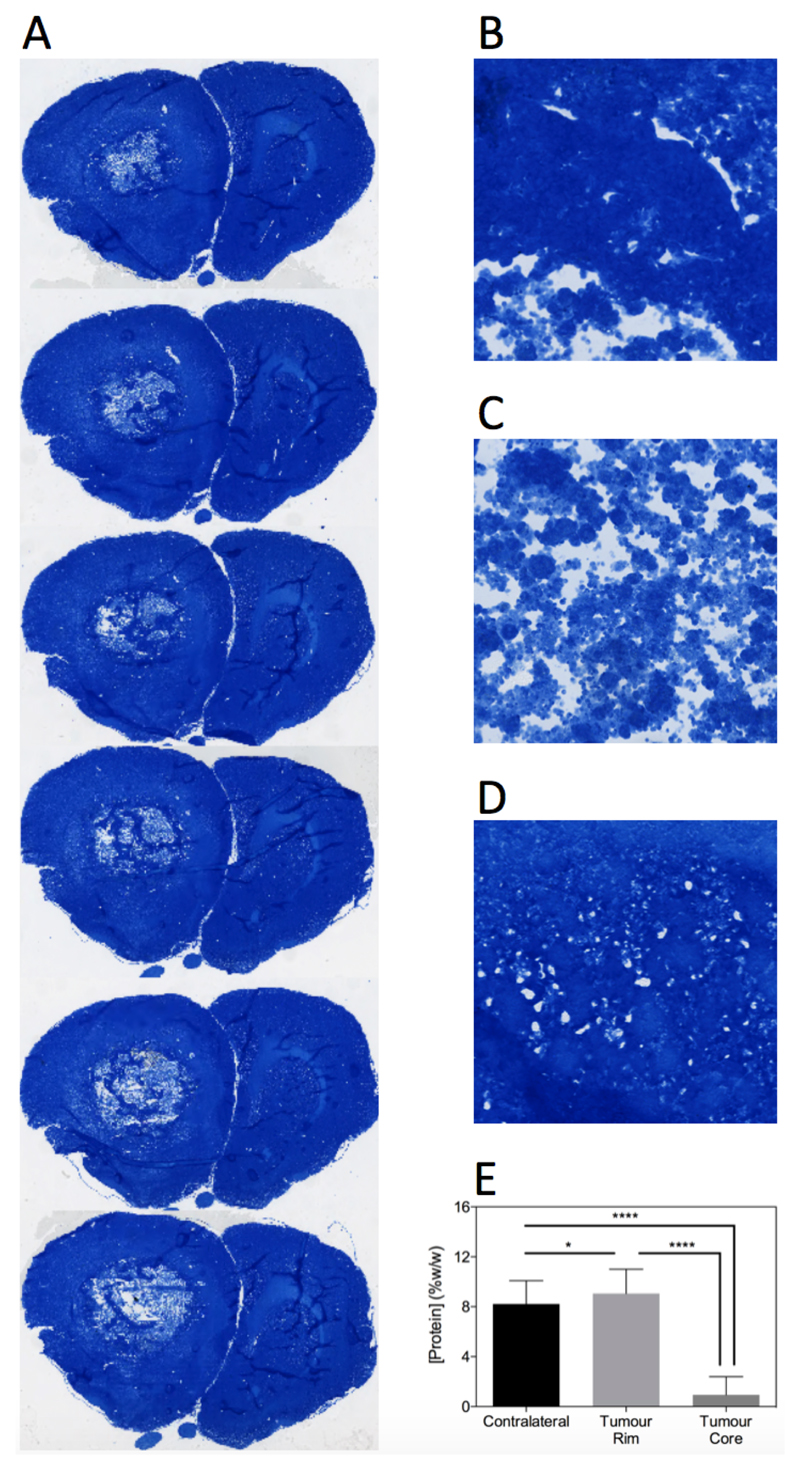
No significant difference in cytoplasmic, non-cytoplasmic or total protein concentration was evident between the biopsied tumour and contralateral brain tissue (Figure 3). However, the protein concentration measurements are likely biased owing to tumour heterogeneity, meaning mixed tissue types (necrotic tumour core and infiltrating tumour rim) were likely assayed in the same sample. Consequently, quantitative protein concentration measurements were made using Coomassie staining of tissue sections in a second cohort of animals, yielding spatial information on protein concentration heterogeneity. Example sections through a tumour volume of a representative rat are shown in Figure 4A, as well as zoomed regions of the tumour rim, necrotic core and contralateral hemisphere from a single section (Figure 4B-D). These sections show a clear pattern of lighter blue staining (corresponding to lower protein concentration) in the core areas, darker blue staining in the tumour rim areas, and an intermediate level of blue staining in the contralateral hemisphere. The same spatial distribution was maintained over all sections and all animals (18 sections per animal) with a small but significant increase in protein concentration in the tumour rim evident (normal 8 ± 2 % w/w, tumour 9 ± 2 % w/w, mean ± s.d., P < 0.05, Figure 4E). Pimonidazole and CD31 vessel staining confirmed that core regions were largely necrotic, whilst the rim regions maintained vessel structure but also showed a degree of hypoxia (Supplementary Figure S4).
Figure 3.
Cytoplasmic, non-cytoplasmic and total protein concentration measured by BCA assay of tissue biopsies (n = 5) is not significantly different between tumour (grey) and normal (black) tissue. Statistical comparison was unpaired t-test comparing contralateral and tumour protein concentration in each fraction.
Figure 4.
Coomassie staining reveals the spatial heterogeneity of protein concentration in tumours. A. Examples of Coomassie stained tissue sections through a tumour of a representative rat. B-D. Magnified (200x) regions of tumour rim, tumour core and contralateral tissue, respectively. Group-wise protein concentration measurements (E) show that tumour rim has a significantly higher protein concentration than contralateral tissue, whereas tumour core has significantly lower protein concentration than contralateral tissue. **** = P < 0.0001, * = P < 0.05 (one-way repeated measures ANOVA, followed by Tukey’s multiple comparison test).
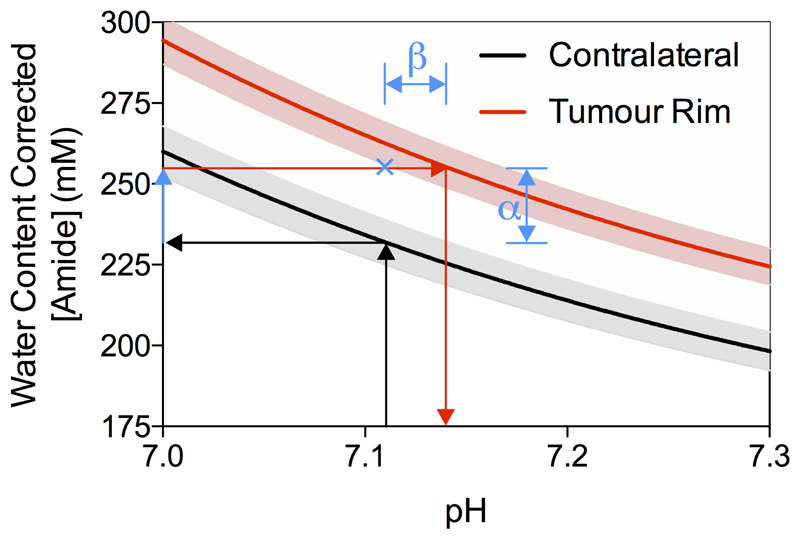
Protein concentration changes account for 66 % of measured APTR* signal change
Using the isoAPTR* method with the normal and tumour tissue measured APTR* (normal 3.43 ± 0.10 % M0, tumour 3.77 ± 0.09 % M0, mean ± 95 % CI) and the measured protein concentration increase in the tumour rim (from Coomassie measurements), it was found that approximately 66 % of the APTR* change was caused by protein concentration (α in Figure 5). Thus, the remaining 34% signal change (β in Figure 5) reflects an increase in tumour pH to 7.14 ± 0.01. The contralateral hemisphere was used as a measurement of normal tissue since no evidence of tumour burden was observed using the post-Gd T1-weighted imaging (Figure 1A). Additionally, previous studies have confirmed histologically that there is no tumour present in the contralateral hemisphere in this model (23).
Figure 5.
Demonstration of the isoAPTR* method to measure the pH of ENU tumours. Black and red lines indicate the isoAPTR* lines for the APTR* values measured in the contralateral and tumour rim ROIs, respectively, with 95 % CI shown in grey for contralateral and pink for tumour rim. Arrows show the isoAPTR* methodology, where the contralateral tissue pH is assumed to be 7.11 and used with the measured APTR* to estimate the amide proton concentration. Using the relative increase in protein concentration in the tumour rim tissue measured by Coomassie staining, a tumour pH of 7.14 ± 0.01 is measured. The blue cross shows the APTR* value that would be expected with no pH change in the tumour, indicating that approximately 66% of the observed APTR* change reflects protein concentration changes (α), with the remaining 34% a result of pH changes in the tumour (β).
Atovaquone treatment alleviates tumour hypoxia with no concomitant alteration of protein concentration
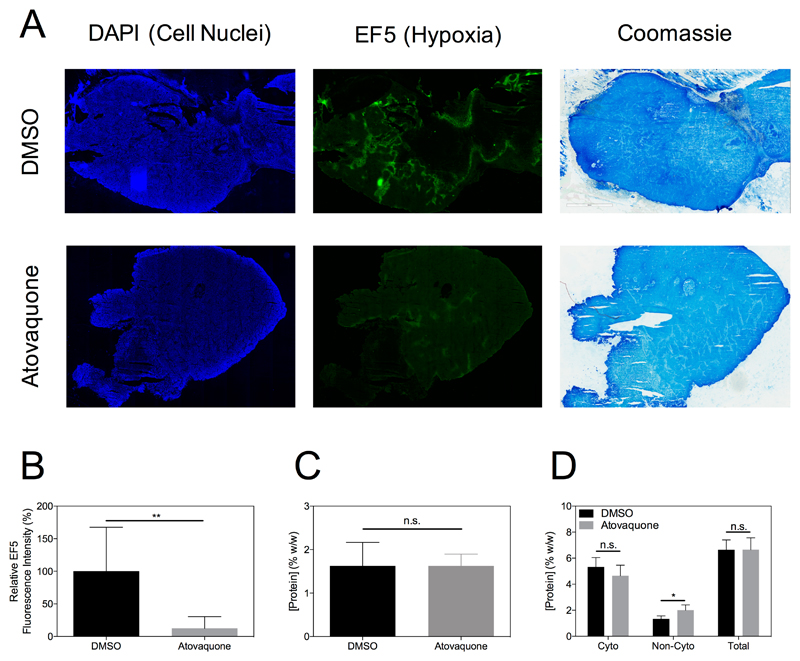
Representative sections of tumours from mice in the control (DMSO) or Atovaquone treated groups are shown in Figure 6A stained for DAPI (cell nuclei), EF5 (hypoxia) and Coomassie (protein concentration). The alleviation of tumour hypoxia in the Atovaquone treated tumour is evident as a reduction in the intensity of the fluorescent signal in the image. The relative fluorescence intensity over all animals showed the significant effect of Atovaquone in reducing tumour hypoxia (Figure 6B). Importantly, no concomitant alteration in cytoplasmic protein concentration was observed, as shown by similar intensities in the Coomassie stained images in Figure 6A, the non-significant difference between protein concentration quantified from these images in Figure 6C, and the quantitation of protein concentration in various subcellular fractions by BCA assay in Figure 6D.
Figure 6.
Representative tissue sections from tumours in the DMSO or Atovaquone treatment groups stained for DAPI, EF5 and Coomassie show reduction in tumour hypoxia due to Atovaquone with no concomitant alteration of protein concentration (A). The reduction in tumour hypoxia is evident as a significant reduction in EF5 fluorescence intensity across all animals (B, P < 0.01, unpaired t-test). Cytoplasmic protein concentration as measured by Coomassie staining (C) or BCA assay (D) was not significantly different between Atovaquone and DMSO groups (P > 0.05, unpaired t-test).
APTR* measures pH change associated with alleviation of hypoxia due to Atovaquone treatment
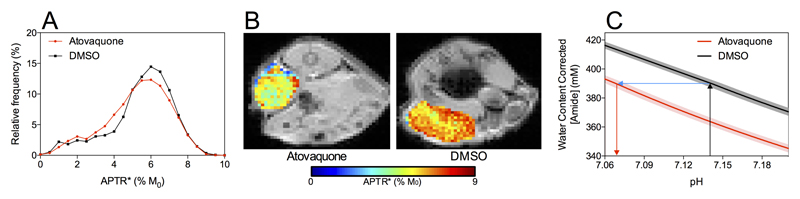
The histogram of APTR* measurements from Atovaquone treated tumours was significantly different from the histogram of APTR* measured from control animals, with the median APTR* being lower in Atovaquone treated animals (Figure 7A). The spatial heterogeneity of the APTR* maps in tumours, necessitating the measurement of effect size by cumulative frequency distributions, is shown in Figure 7B for two representative animals, clearly showing a higher APTR* in control (DMSO) treated animals compared to Atovaquone treated. Crucially, since no significant change in protein concentration was measured between the two groups using BCA assay and Coomassie staining techniques, it is likely that this APTR* difference is a result of a change in tumour pH associated with the reduction in tumour hypoxia. The size of this pH change was estimated as –0.07 pH units by isoAPTR* (Figure 7C), reducing tumour pH from 7.14 ± 0.01 to 7.07 ± 0.01. The tumour pH of 7.14 ± 0.01 in the DMSO group was assumed from the previous estimate of tumour pH in the rat model of brain metastasis.
Figure 7.
Histogram of APTR* values measured from tumours on mice treated with Atovaquone to decrease tumour hypoxia or with DMSO as control (A). The median APTR* was significantly lower in Atovaquone-treated animals (P < 0.001, Mann-Whitney test). The heterogeneity of APTR* values within single tumours is shown in representative animals from each group (B) to demonstrate the necessity of the histogram analysis. isoAPTR* analysis measured a tumour pH reduction of 0.07 pH units (C), consistent with the decrease in tumour pH expected with a reduction of tumour hypoxia.
Discussion
In this study, we sought to better understand the contribution of protein concentration and pH changes in tumours to APT signals. We performed in vivo APT MRI and ex vivo protein concentration measurements, and combined information from both modalities to determine that approximately 66% of the measured APT signal change was explained by a protein concentration increase. The remaining 34% was assumed to be a result of an alkalosis of tumour intracellular pH (compared to normal tissue) to 7.14 ± 0.01, in agreement with previous studies showing that the intracellular pH in tumour cells is slightly alkalotic compared to normal cells (4,21). Elucidation of the contribution of pH effects to altered APT signals in tumours leads to improved understanding of APT measurements in the clinic, and may enable inferences regarding the pH of individual patients’ tumours in clinical settings. Additionally, we used the anti-malarial drug Atovaquone to reduce tumour hypoxia and measured the associated alteration in tumour pH using APT MRI, which opens the possibility of using APT MRI for assessing therapeutic response. The observed shift in intracellular pH of Atovaquone-treated tumours, to values typically seen in normal cells, is consistent with expectations based on the link between hypoxia and tumour pH (6,33).
This study represents the first application of the APTR* method (29) to tumour imaging, with all previous in vivo studies investigating ischaemic stroke. The measured increase in APTR* in the tumour rim and core in this study is in agreement with previous studies that have reported an increased APT signal in viable tumour and necrotic tissue using other non-quantitative APT MRI analysis methods. Although the BCA-derived protein assay showed no differences in protein concentration between normal brain and tumour tissue, the Coomassie histology revealed a reduced protein concentration in the necrotic core region compared to normal brain tissue. Tissue detachment during the sectioning and staining process is a common problem in acellular tissue areas, which could bias the Coomassie protein concentration measurements in the necrotic regions. For this reason, only tumour rim areas were used in the subsequent isoAPTR* analysis. Additional potential limitations to using the Coomassie stain as a protein concentration measurement technique include inaccurate measurements as a result of poor sectioning technique and difficulties with co-registration of the APT MRI data with histology resolution images. Nevertheless, Coomassie staining retains more information about tumour heterogeneity than simple biopsies of tumour tissue. The agreement between the BCA-derived protein concentration measurements in this study and previous measurements from a 9L glioma model (19), suggests that those studies may not have fully captured tumour protein heterogeneity and that future studies using other tumour models may benefit from protein concentration measurements made using the Coomassie staining procedure.
The invasive nature of histology limits the clinical translatability of the methods used in this preclinical study. The histological measurement of protein concentration, and the associated estimation of pH by isoAPTR*, is not necessarily meant for clinical translation, however, but rather to aid in the interpretation of APT MRI. Whereas prior clinical studies have interpreted the APT signal in tumours in terms of changes in intracellular protein content alone, the results of this study suggest that the APT signal change in tumours also reflects changes in pH. Additionally, it may not be necessary to use ex vivo histological measurements of protein concentration to estimate tumour pH using isoAPTR* if tumour pH can be selectively modulated, as was done using Atovaquone in this study. Alternative endogenous CEST MRI methods such as AACID (34) have been used in this way to investigate the tumour pH change associated with administration of dichloroacetate (35), topiramate (36) and lonidamine (37) in preclinical experiments. Additionally, exogenous diaCEST (38–41) and paraCEST (42,43) contrast agents have been developed that successfully measure tumour extracellular pH, though these methods are limited by contrast agent availability to the tumour, which may be limited in areas of particularly poor perfusion (44) or in the brain where the blood-brain barrier may be partially intact in the early stages of tumour growth.
The isoAPTR* method used in this study assumes that the measurement of protein concentration can be used to infer a change in the amide proton concentration, which is the true biophysical origin of the APT signal. This simplification requires further assumptions, for instance that the number of amide protons available for exchange per protein molecule does not change. Protein conformation changes, proteolysis, or differences in protein size between the two tissues may also affect the validity of this assumption. Although it is unlikely that protein structure would be sufficiently different between tumour and normal tissue to affect the conclusions of this work, further studies to investigate the effect of differences in protein structure and size on the APT MRI signal in tumours are warranted.
This study combined in vivo APT MRI measurements with ex vivo histological measurements of protein concentration in a model of brain metastasis to determine that the proportion of APT signal change originating from changes in protein concentration is ca. 66%, with the remaining 34% originating from changes in tumour pH. Furthermore, a significant change in tumour pH associated with a pharmacologically-induced reduction in tumour hypoxia was measured using APT MRI in a subcutaneous tumour model. This study extended our understanding of APT MRI, and may enable the use of APT MRI to infer the pH of individual patients’ tumours as a biomarker either for therapy stratification or of therapeutic response in clinical settings.
Supplementary Material
Statement of Significance.
These findings advance our understanding of amide proton transfer magnetic resonance imaging (APT MRI) of tumours and may improve the interpretation of APT MRI in clinical settings.
Acknowledgements
The authors would like to thank Karla Watson, Jade Harris and Jessica Law for assistance with animal husbandry.
Grant Support: This work was supported by Cancer Research UK (C5255/A15935), the CRUK/EPSRC Cancer Imaging Centre in Oxford (grant number C5255/A16466), and the Medical Research Council (MC_ST_U13080, MR/K501256/1). GH was supported by Cancer Research UK through their Clinician Scientist Awards programme (C34326/A19590 & C34326/A13092). SS was supported by Cancer Research UK grants C5255/A12678, C2522/A10339, EPSRC grant C2522/A10339, and the MRC Unit Grant for the Oxford Institute for Radiation Oncology.
Footnotes
Potential Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Hanahan D, Weinberg RAA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Tannock IF, Rotin D. Acid pH in Tumors and Its Potential for Therapeutic Exploitation. Cancer Res. 1989;49:4373–84. [PubMed] [Google Scholar]
- 3.Wojtkowiak JW, Verduzco D, Schramm KJ, Gillies RJ. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol Pharm. 2011;8:2032–8. doi: 10.1021/mp200292c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–7. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 5.Flinck M, Kramer SH, Pedersen SF. Roles of pH in control of cell proliferation. Acta Physiol. 2018;223:e13068. doi: 10.1111/apha.13068. [DOI] [PubMed] [Google Scholar]
- 6.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 7.Eales KL, Hollinshead KER, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis. 2016;5:e190. doi: 10.1038/oncsis.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiche J, Brahimi-Horn MC, Pouysségur J. Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med. 2010;14:771–94. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Zijl PCM, Lam WW, Xu J, Knutsson L, Stanisz GJ. Magnetization Transfer Contrast and Chemical Exchange Saturation Transfer MRI. Features and analysis of the field-dependent saturation spectrum. Neuroimage. 2017;168:222–41. doi: 10.1016/j.neuroimage.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50:1120–6. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Zhu H, Lim M, Blair L, Quinones-Hinojosa A, Messina SA, et al. Three-Dimensional Amide Proton Transfer MR Imaging of Gliomas: Initial Experience and Comparison With Gadolinium Enhancement. J Magn Reson Imaging. 2013;38:1119–28. doi: 10.1002/jmri.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nat Med. 2011;17:130–4. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrabian H, Desmond KL, Soliman H, Sahgal A, Stanisz GJ. Differentiation between Radiation Necrosis and Tumor Progression Using Chemical Exchange Saturation Transfer. Clin Cancer Res. 2017;23:3667–75. doi: 10.1158/1078-0432.CCR-16-2265. [DOI] [PubMed] [Google Scholar]
- 14.Zaiss M, Windschuh J, Paech D, Meissner J-E, Burth S, Schmitt B, et al. Relaxation-compensated CEST-MRI of the human brain at 7T: Unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage. 2015;112:180–8. doi: 10.1016/j.neuroimage.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9:1085–90. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 16.Tee YK, Harston GWJ, Blockley N, Okell TW, Levman J, Sheerin F, et al. Comparing different analysis methods for quantifying the MRI amide proton transfer (APT) effect in hyperacute stroke patients. NMR Biomed. 2014;27:1019–29. doi: 10.1002/nbm.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun PZ, Wang E, Cheung JS. Imaging acute ischemic tissue acidosis with pH-sensitive endogenous amide proton transfer (APT) MRI--correction of tissue relaxation and concomitant RF irradiation effects toward mapping quantitative cerebral tissue pH. Neuroimage. 2012;60:1–6. doi: 10.1016/j.neuroimage.2011.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan K, Fu Z, Yang C, Zhang K, Jiang S, Lee D-H, et al. Assessing Amide Proton Transfer (APT) MRI Contrast Origins in 9 L Gliosarcoma in the Rat Brain Using Proteomic Analysis. Mol Imaging Biol. 2015;17:479–87. doi: 10.1007/s11307-015-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Zaiss M, Zu Z, Li H, Xie J, Gochberg DF, et al. On the origins of chemical exchange saturation transfer (CEST) contrast in tumors at 9.4 T. NMR Biomed. 2014;27:406–16. doi: 10.1002/nbm.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Lin Y, Gillies RJ. Tumor pH and its measurement. J Nucl Med. 2010;51:1167–70. doi: 10.2967/jnumed.109.068981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhujwalla ZM, Aboagye EO, Gillies RJ, Chacko VP, Mendola CE, Backer JM. Nm23-transfected MDA-MB-435 human breast carcinoma cells form tumors with altered phospholipid metabolism and pH: a 31P nuclear magnetic resonance study in vivo and in vitro. Magn Reson Med. 1999;41:897–903. doi: 10.1002/(sici)1522-2594(199905)41:5<897::aid-mrm7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 22.Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, et al. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–77. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serres S, Martin CJ, Sarmiento Soto M, Bristow C, O’Brien ER, Connell JJ, et al. Structural and functional effects of metastases in rat brain determined by multimodal MRI. Int J Cancer. 2014;134:885–96. doi: 10.1002/ijc.28406. [DOI] [PubMed] [Google Scholar]
- 24.Ashton TM, Fokas E, Kunz-Schughart LA, Folkes LK, Anbalagan S, Huether M, et al. The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat Commun. 2016;7 doi: 10.1038/ncomms12308. 12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harston GWJ, Tee YK, Blockley N, Okell TW, Thandeswaran S, Shaya G, et al. Identifying the ischaemic penumbra using pH-weighted magnetic resonance imaging. Brain. 2015;138:36–42. doi: 10.1093/brain/awu374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinchesh P, Allen PD, Beech JS, Fokas E, Gilchrist S, Kersemans V, et al. Dynamic Reacquisition for Respiratory Gated, Constant TR 2D multi-slice MRI. Proc 23rd Sci Meet Int Soc Magn Reson Med; 2015. p. 2574. [Google Scholar]
- 27.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, et al. In vivo imaging of glucose uptake and metabolism in tumors. Nat Med. 2013;19:1067–72. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chappell MA, Groves AR, Whitcher B, Woolrich MW. Variational Bayesian Inference for a Nonlinear Forward Model. IEEE Trans Signal Process. 2009;57:223–36. [Google Scholar]
- 29.Chappell MA, Donahue MJ, Tee YK, Khrapitchev AA, Sibson NR, Jezzard P, et al. Quantitative Bayesian model-based analysis of amide proton transfer MRI. Magn Reson Med. 2013;70:556–67. doi: 10.1002/mrm.24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray KJ, Larkin JR, Tee YK, Khrapitchev AA, Karunanithy G, Barber M, et al. Determination of an optimally sensitive and specific chemical exchange saturation transfer MRI quantification metric in relevant biological phantoms. NMR Biomed. 2016;29:1624–33. doi: 10.1002/nbm.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JA, Curella P, Zahniser NR. A new densitometric procedure to measure protein levels in tissue slices used in quantitative autoradiography. Brain Res. 1988;447:60–6. doi: 10.1016/0006-8993(88)90965-1. [DOI] [PubMed] [Google Scholar]
- 32.Ray KJ, Larkin JR, Chappell MA, Sibson NR. isoAPTR*: A novel method to measure tumour pHi using CEST MRI. Proc 25th Sci Meet Int Soc Magn Reson Med; 2017. p. 1972. [Google Scholar]
- 33.Stubbs M, McSheehy PM, Griffiths JR, Bashford CL. Causes and consequences of tumour acidity and implications for treatment. Mol Med Today. 2000;6:15–9. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 34.McVicar N, Li AX, Gonçalves DF, Bellyou M, Meakin SO, Prado MA, et al. Quantitative tissue pH measurement during cerebral ischemia using amine and amide concentration-independent detection (AACID) with MRI. J Cereb Blood Flow Metab. 2014;34:690–8. doi: 10.1038/jcbfm.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albatany M, Li A, Meakin S, Bartha R. Dichloroacetate induced intracellular acidification in glioblastoma: in vivo detection using AACID-CEST MRI at 9.4 Tesla. J Neurooncol. 2018;136:255–62. doi: 10.1007/s11060-017-2664-9. [DOI] [PubMed] [Google Scholar]
- 36.Marathe K, McVicar N, Li A, Bellyou M, Meakin S, Bartha R. Topiramate induces acute intracellular acidification in glioblastoma. J Neurooncol. 2016;130:465–72. doi: 10.1007/s11060-016-2258-y. [DOI] [PubMed] [Google Scholar]
- 37.Mcvicar N, Li AX, Meakin SO, Bartha R. Imaging chemical exchange saturation transfer (CEST) effects following tumor-selective acidification using lonidamine. NMR Biomed. 2015;28:566–75. doi: 10.1002/nbm.3287. [DOI] [PubMed] [Google Scholar]
- 38.Chen LQ, Howison CM, Jeffery JJ, Robey IF, Kuo PH, Pagel MD. Evaluations of extracellular PH within in vivo tumors using acidocest MRI. Magn Reson Med. 2014;72:1408–17. doi: 10.1002/mrm.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M, Chen C, Shen Z, Zhang X, Chen Y, Lin F, et al. Extracellular pH is a biomarker enabling detection of breast cancer and liver cancer using CEST MRI. Oncotarget. 2017;8:45759–67. doi: 10.18632/oncotarget.17404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longo DL, Sun PZ, Consolino L, Michelotti FC, Uggeri F, Aime S. A General MRI-CEST Ratiometric Approach for pH Imaging: Demonstration of in Vivo pH Mapping with Iobitridol. J Am Chem Soc. 2014;136:14333–6. doi: 10.1021/ja5059313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon BF, Jones KM, Chen LQ, Liu P, Randtke EA, Howison CM, et al. A comparison of iopromide and iopamidol, two acidoCEST MRI contrast media that measure tumor extracellular pH. Contrast Media Mol Imaging. 2015;10:446–55. doi: 10.1002/cmmi.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li AX, Suchy M, Li C, Gati JS, Meakin S, Hudson RHE, et al. Magn Reson Med. Vol. 66. Wiley-Blackwell; 2011. In vivo detection of MRI-PARACEST agents in mouse brain tumors at 9.4 T; pp. 67–72. [DOI] [PubMed] [Google Scholar]
- 43.Ferrauto G, Di Gregorio E, Auboiroux V, Petit M, Berger F, Aime S, et al. CEST-MRI for glioma pH quantification in mouse model: Validation by immunohistochemistry. NMR Biomed. 2018:e4005. doi: 10.1002/nbm.4005. [DOI] [PubMed] [Google Scholar]
- 44.Jones KM, Randtke EA, Yoshimaru ES, Howison CM, Chalasani P, Klein RR, et al. Clinical Translation of Tumor Acidosis Measurements with AcidoCEST MRI. Mol Imaging Biol. 2017;19:617–25. doi: 10.1007/s11307-016-1029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.