Abstract
Toll-like receptor 9 (TLR9) recognizes DNA in endosomes and activates distinct signaling pathways to stimulate the production of proinflammatory cytokines and type I interferons (IFN). The assembly of signaling platforms on microtubule-associated proteins 1A/1B light chain 3B (LC3)-decorated endosomal vesicles is required to transduce TLR9 signals that stimulate the production of IFN, but not interleukin-12 p40 (IL-12p40). LC3-associated phagocytosis (LAP), a form of noncanonical autophagy, is critical for the activation of interferon regulatory factor 7 (IRF7) and for IFN synthesis. Here we showed that, after the stimulation of TLR9 by CpG oligonucleotides, the autophagy protein LC3 and the kinase IKKα were recruited to endosomes that contained TLR9. The recruitment of IKKα and LC3 to such signaling endosomes was not stimulated by catalysts of classical autophagosome formation, but involved LAP formation, which required ATG5 but not FIP200. Additionally, we found that the LC3-IKKα complex further associated with both TRAF3 and IRF7. We identified three putative LC3-interacting regions (LIRs) in IKKα, and mutagenesis suggested that two of these were critical for direct binding to LC3. Moreover, mutation of the same LIR sequences failed to rescue type I IFN production in IKKα-deficient dendritic cells upon reconstitution. Together, these data suggest a direct link between LAP formation and IKKα recruitment downstream of TLR9 activation that is necessary to facilitate type I IFN production.
One sentence summary:
Autophagy protein LC3 directly recruits kinase IKKα to endosomal platforms necessary for type I IFN production after TLR9 stimulation
Editor’s summary:
LC3 recruits IKKα after TLR9 activation
Toll like receptor 9 (TLR9) is a pattern recognition receptor involved in innate sensing of extracellular DNA within the endosomes of phagocytes. Hayashi, et al found that IKKα kinase associated with the autophagy protein LC3 deposited on endosomes after TLR9 stimulation of macrophages. Formation of this LC3-IKKα signaling platform through non-canonical autophagy also recruited other effectors involved in type I interferon signaling. Mutagenesis of LC3 interacting regions in IKKα suggested these effects were mediated by a direct interaction of the kinase with LC3. Together, these data identify IKKα as a critical component of LC3-associated phagocytosis-mediated endosome-based signaling platforms necessary for stimulating type I interferon production after TLR9 engagement.
Introduction
Innate immune cells are equipped with an array of pattern recognition receptors (PRRs) that recognize evolutionarily conserved pathogen-associated molecular patterns and activate the production of antimicrobial defense effector molecules. Toll-like receptor 9 (TLR9) is a PRR expressed in the endosomes of immune cells that recognizes DNA (1). Upon ligand engagement, TLR9 activates two distinct downstream signaling pathways to stimulate the production of proinflammatory cytokines and type I interferons (IFNs), including IFN-α and IFN-β (2). DNA-dependent dimerization of TLR9 stimulates the recruitment of MyD88 (or myeloid differentiation primary response 88), and the following signaling molecules: interleukin-1 receptor-associated kinase 4 (IRAK4), IRAK2, TNF receptor associated factor 6 (TRAF6), transforming growth factor beta-activated kinase 1 (TAK1), and its regulatory components TAK1-binding protein 2/3 (TAB2/3). This signaling complex ultimately triggers the transcription of pro-inflammatory cytokines through activation of nuclear factor-kappa B (NF-κB), interferon regulatory factor 5 (IRF5), and mitogen-activated protein kinases (MAPKs) (2). Activation of TLR9 also stimulates a second MyD88-dependent pathway that requires IRAK1 (3), TRAF3 (4, 5), IκB kinase alpha (IKKα) (6), osteopontin (7), dedicator of cytokinesis 2 (DOCK2) (8), a plasmacytoid dendritic cell (PDC) -specific receptor (PDC-TREM) (9), viperin (10), and adaptor protein 3 (AP-3) (11). This pathway results in IRF7 phosphorylation and subsequent type I IFN production.
In macrophages stimulated with the TLR9 ligand–like DNA-containing immune complexes or CpG, microtubule-associated protein 1A/1B-light chain 3 (LC3), an autophagy-associated protein, is recruited to the phagosome containing TLR9 independently of canonical autophagy through a process known as LC3-associated phagocytosis (LAP) (12). LAP facilitates the fusion between zymosan-containing phagosomes and lysosomes to mediate rapid acidification of the cargo-containing phagosomes in a TLR2-dependent manner (13). This process is associated with the recruitment of LC3, Beclin 1, and phosphatidylinositol 3 (PI(3)) kinase, as well as the components of the ubiquitin-like conjugation system, autophagy related 5 and 7 (ATG5 and 7). Similarly, autophagy protein-dependent vacuole formation is also observed in macrophages treated with other TLR ligands such as lipopolysaccharide (LPS; TLR4 ligand), Porphyromonas gingivalis lipopolysaccharide and lipoteichoic acid (PAM3CSK4; a TLR½ ligand), and DNA-immune complex and CpG oligodeoxynucleotides (TLR9 ligands) (12, 13). Notably, LAP is required for TLR9-mediated IRF7 activation and production of type I IFN, but not for IL-12p40 secretion (12).
Three distinct features of LAP distinguish it from canonical autophagosomes. First, unlike the double membrane structures characteristic of canonical autophagosomes, LAP generates single membrane enveloped vesicles coated with LC3 on the cytosolic side of the phagosomal membrane. Secondly, LAP initiation occurs independently of proteins associated with the autophagy pre-initiation complex, namely Unc-51 like autophagy activating kinase ½ (ULK½), RB1-inducible coiled-coil protein 1 (RB1CC1 or FIP200), or ATG13, which are required for initiation of canonical autophagy. Finally, the class III PI(3)K-associated protein, Rubicon, is required for LAP (14). It stabilizes phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3 or VPS34) activity to sustain PtdIns(3)P presence on LAP-engaged phagosome (LAPosome) and stabilize the NOX2 complex for ROS production, both of which are critical for progression of LAP. Other autophagy-related genes identified to be involved in LAP formation are other components of class III PI(3)K-associated proteins, Beclin-1, Vps34, Atg14, UVRAG (UV radiation resistance associated or Vps38) (14), and components of ubiquitin-like conjugation system (Atg5, Atg7) (14).
Here, we interrogated the relationship between IKKα and LAP formation after TLR9 stimulation. We found that LC3 was recruited to TLR9-containing LAPosomes and provided an anchor for IKKα recruitment. We identified two LC3-binding motifs within IKKα that were critical for both its interaction with LC3 and the stimulation of type I IFN production downstream of TLR9. Our findings provide a missing mechanistic link between IKKα and LAP-dependent signaling downstream of TLR9.
Results
IKKα is recruited to TLR9-containing endosomes and associates with LC3 after stimulation with CpG-A
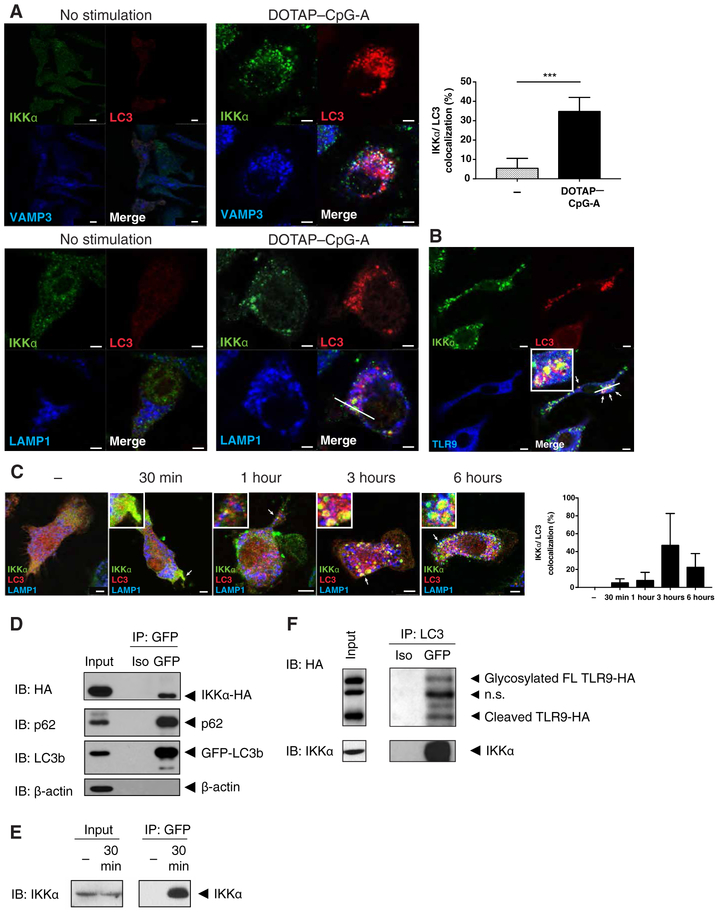
Because LAPs and IKKα are required for TLR9 signaling and IFN-α production, we examined the localization of IKKα and LC3 after transfection of macrophages with cationic lipid vesicles (DOTAP or 1,2-di-(9Z-octadecenoyl)-3-trimethylammonium-propane (chloride salt)) containing the TLR9 ligand CpG-A. In macrophages, this approach delivers TLR9 ligands to the appropriate cellular compartment to stimulate robust IFN-α production downstream of TLR9 (15). We found that IKKα and LC3 colocalized only after DOTAP-CpG-A stimulation in mouse fetal-liver macrophages and RAW264.7 cells (Figure 1A and fig. S1A). Throughout this study, LC3 refers to the LC3b isoform. After the activation of TLR9 in macrophages, both early (vesicle-associated membrane protein 3; VAMP3+) and late (lysosome-associated membrane protein 1; LAMP1+) endosomes recruited IKKα and LC3 (Figure 1A upper and lower panels, respectively). IKKα and LC3 also colocalized with TLR9-containing endosomes after stimulation of fetal-liver macrophages or RAW264.7 cells stably expressing HA-tagged TLR9 (TLR9-HA) and mCherry-tagged LC3 (Cherry-LC3) (Figure 1B and fig. S1B). A time course analysis revealed that IKKα and LC3 began to colocalize as early as 30 min after CpG-A stimulation and decorated large LAMP1+ endosomes by 3 to 6 hours after stimulation (Figure 1C). Immunoprecipitation analysis of lysates of HEK293T cells transfected with IKKα-HA and GFP-LC3 showed that IKKα and LC3 physically interacted with each other (Figure 1D). This interaction was confirmed in primary bone marrow–derived macrophages (BMMs) and was found within 30 min of stimulation with DOTAP-CpG-A (Figure 1E). Furthermore, IKKα-LC3 complexes also contained TLR9 (Figure 1F). These data indicate that IKKα was mobilized upon DOTAP-CpG-A stimulation to bind to LC3 on TLR9-containing endosomes.
Figure 1. IKKα binds to LC3 after DOTAP-CpG stimulation.
(A) Confocal microscopy images from Ikkα KO fetal-liver macrophages that were retrovirally co-transduced with wild-type IKKα-HA, TLR9-GFP, and Cherry-LC3 plasmids then stimulated with DOTAP-CpG-A 3μM. Images are representative of 3 or more independent experiments. Scale bars: 2.5um (B) Confocal microscopy images from fetal-liver macrophages heterozygous for IKKα that were retrovirally co-transduced with wild-type IKKα-HA, TLR9-GFP, and Cherry-LC3 plasmids then stimulated with DOTAP-CpG-A. Images are representative of 3 or more independent experiments. Scale bars: 2.5 um (C) Confocal microscopy images from Ikkα KO fetal-liver macrophages that were retrovirally co-transduced with wild-type IKKα-HA, TLR9-GFP, and Cherry-LC3 plasmids then stimulated with DOTAP-CpG-A for the indicated times. Images are representative of 3 or more independent experiments. Scale bars: 2.5um (D) HEK293T cells were co-transfected with wild-type IKKα-HA and GFP-LC3, then cell lysates were immunoprecipitated for GFP and blotted for the indicated protein. Blots are representative of 3 or more independent experiments. (E) Day 7 BMMs were stimulated with DOTAP-CpG-A for 30 min, then the pre-nuclear cell lysates were immunoprecipitated for GFP-LC3 and blotted for the indicated proteins. Blots are representative of 3 or more independent experiments. (F) RAW264.7 cells stably expressing TLR9-HA and Cherry-LC3 were stimulated with DOTAP-CpG-A for 6 hours, then cell lysates were immunoprecipitated for LC3 and blotted for the indicated proteins. Blots are representative of 3 or more independent experiments.
*** denotes P < 0.001 by t test.
Canonical autophagy induction is not sufficient to recruit IKKα to LC3
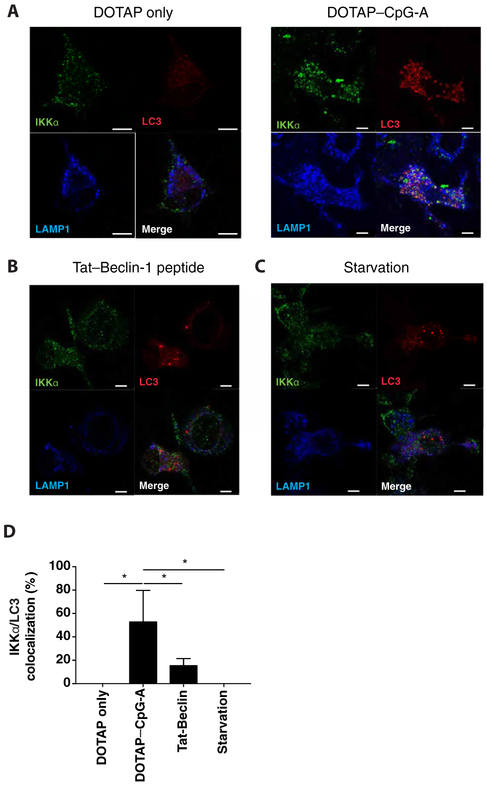
To determine whether LC3-IKKα interactions occurred as a consequence of the activation of canonical autophagy, we stimulated autophagy in BMMs and examined the colocalization of IKKα and LC3. Autophagy can be induced in various ways, such as serum starvation or treatment with a Tat-Beclin-1 peptide (16). Although confocal analysis showed that autophagy was induced in response to both stimuli, as expected, as measured by the formation of LC3 puncta (Figure 2A-D), colocalization between IKKα and LC3 was not observed under these conditions. Similar results were obtained when we analyzed BMMs for IKKα and LC3 colocalization after treatment with DOTAP alone (Figure 2A). These data indicate that neither canonical autophagy induction alone nor exposure to cationic lipid was sufficient to trigger IKKα-LC3 complex formation. Furthermore, these results suggest that the interaction between IKKα and LC3 on endosomes required CpG stimulation.
Figure 2. Autophagy induction is not sufficient to trigger LC3-IKKα interaction.
(A-C) Wild-type hematopoietic stem cells were retrovirally transduced with retrovirus expressing Cherry-LC3 then differentiated into BMMs that were either treated with DOTAP alone (A), DOTAP-CpG 3μM (A), Tat-Beclin 1 peptide 1μM (B), or starved in serum-deprived media (C) for 6 hours. After treatment, cells were stained for the indicated proteins and analyzed by confocal microscopy. Images are representative of 3 or more independent experiments. Scale bars: 5um (D) Percent IKKα-LC3 co-localization from confocal analysis BMMs expressing Cherry-LC3 (A-C). Data are means ± SEM from 3 or more independent experiments.
IKKα binding to LC3 requires LAP but not canonical autophagy
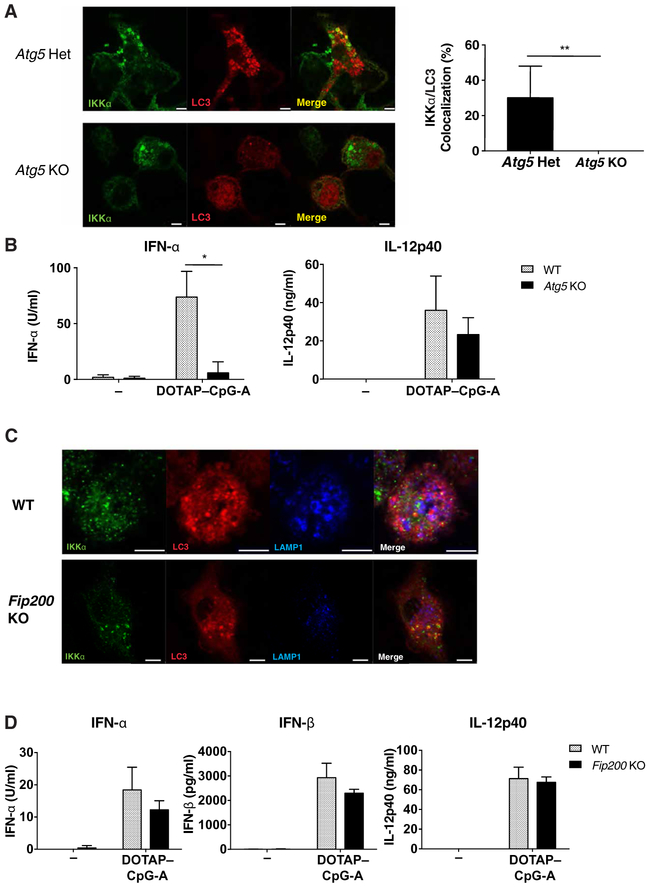
Because noncanonical autophagy is required for LAP formation in response to TLR9 stimulation (12), we tested the involvement of this pathway in TLR9-mediated IKKα-LC3 colocalization. ATG5 is critical for both canonical autophagy (17) and for LAP formation (18). When fetal-liver macrophages from Atg5−/− mice were stimulated with DOTAP-CpG-A, no colocalization between IKKα and LC3 was found (Figure 3A). However, ATG5 did not interact with IKKα or LC3 by immunoprecipitation studies (Figure S2), suggesting that the requirement for ATG5 for IKKα-LC3 complex formation is likely upstream of complex formation. As shown previously (19), Atg5-deficient macrophage cells were unable to secrete IFN-α, but secreted similar amounts of pro-inflammatory cytokine IL-12 to wild-type cells (Figure 3B). These data suggested that although IRF7-mediated IFN-α production was impaired in the absence of ATG5, proinflammatory cytokine production downstream of NFκB activation was not. In contrast to ATG5, activation of the autophagy pre-initiation complex that includes FIP200 is required for the induction of autophagy, but not LAP formation (12). Note that we found that IKKα still colocalized with LC3 in Fip200−/− macrophages stimulated with DOTAP-CpG-A (Figure. 3C). Similarly, when Fip200-deficient bone marrow-derived dendritic cells (BMDCs) were stimulated through TLR9 with DOTAP-CpG, we found no defect in the production of IFN-α, IFN-β, or IL-12p40 (Figure 3D). Together, these data indicate that LAP formation, but not canonical autophagy, was required for the interaction between LC3 and IKKα upon TLR9 stimulation and for the signaling required for type I IFN production.
Figure 3. LC3-IKKα interaction depends on LAP, but not canonical autophagy.
(A) Atg5+/− or Atg5−/− fetal-liver hematopoietic stem cells (HSCs) were retrovirally transduced with wild-type IKKα-HA and Cherry-LC3, then differentiated into macrophages and stimulated with DOTAP-CpG-A 1μM for 6h. After stimulation, cells were stained for the indicated proteins and analyzed by confocal microscopy. Images are representative of 3 or more independent experiments. Scale bars: 5μm (B) Wild-type or Atg5−/− fetal-liver macrophages were stimulated with DOTAP-CpG for 24h and secretion of IFN-α and IL-12 was measured by ELISA. Data are means ± SEM from 3-5 independent experiments. (C) Wild-type and Fip200fl/flxLysM-Cre HSCs were retrovirally transduced with Cherry-LC3, then differentiated to BMMs and stimulated with DOTAP-CpG for 6 hours. After treatment, cells were stained and analyzed by confocal microscopy. Images are representative of 3 or more independent experiments. Scale bars: 5um. (D) Wild-type and Fip200fl/flxLysM-Cre BMDCs were stimulated with DOTAP-CpG 1μM for 24 hours and the cytokine accumulation in the supernatants was assayed by ELISA. Data are means ± SEM from 3 or more independent experiments. * denotes P < 0.05 by t test.
The LC3-IKKα complex recruits TRAF3 and IRF7
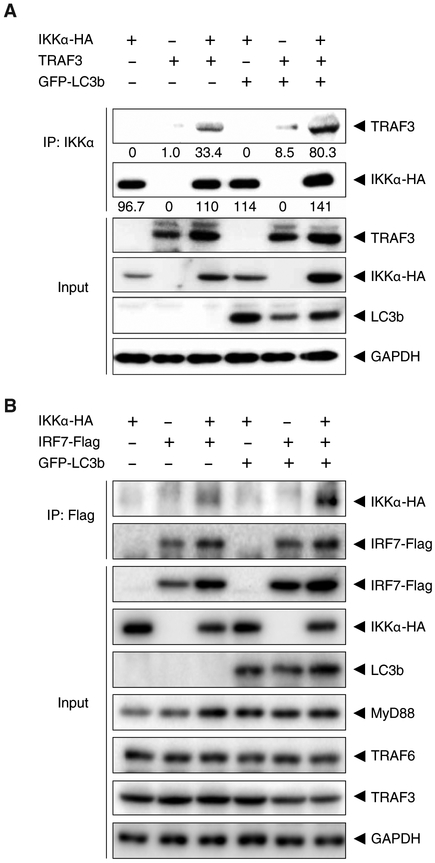
To determine whether LC3-IKKα complexes recruited other key molecules involved in type I IFN production, HEK293T cells were co-transfected with plasmids encoding IKKα–HA, GFP-LC3b, and TRAF3. When cell lysates were subjected to immunoprecipitation of IKKα, we found that IKKα associated with TRAF3, and that this association was enhanced in the presence of LC3 (Figure 4A). We also examined the recruitment to the LC3-IKKα complex of IRF7, the key transcription factor for the expression of type I IFN genes downstream of TLR9. Using a similar approach, we co-transfected HEK293T cells with plasmids encoding IKKα–HA, GFP-LC3b, and IRF7-Flag, as well as plasmids encoding MyD88, TRAF6, and TRAF3, to better mimic TLR9 signaling (6). Cell lysates were subjected to immunoprecipitation of the Flag-tagged proteins and Western blotting results suggested that the association between IKKα and IRF7 was enhanced in the presence of LC3 (Figure 4B). Thus, these data suggest that LC3 recruits IKKα, and that this LC3-IKKα complex may further assemble a larger LAP-dependent signalosome (LAPosome) involving TRAF3 and IRF7.
Figure 4. IKKα-LC3 complex recruits TRAF3 and IRF7.
(A) HEK293T cells were transfected with plasmids encoding wild-type IKKα-HA, GFP-LC3, and TRAF3. Cell lysates were immunoprecipitated IKKα and blotted for the indicated proteins. Blots are representative of 3 independent experiments. (B) HEK293T cells were transfected with plasmids encoding wild-type IKKα-HA, GFP-LC3b, MyD88, TRAF3, TRAF6, and IRF7-Flag. Cell lysates were subjected to immunoprecipitated for the Flag tag and blotted for the indicated proteins. Blots are representative of 3 independent experiments.
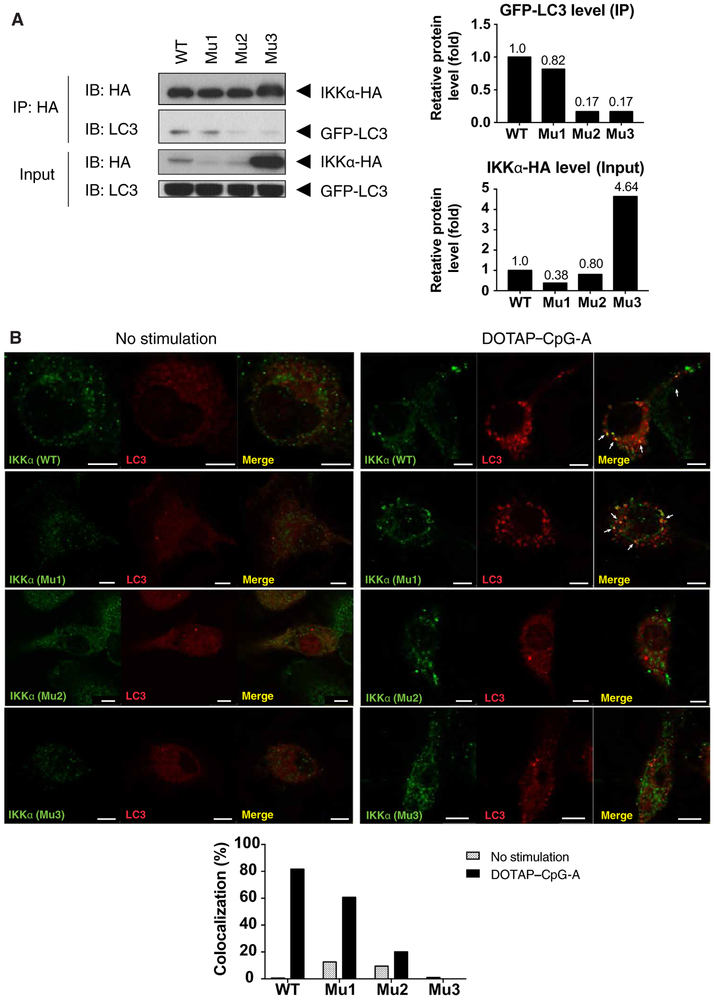
Interactions between IKKα and LC3 depend on the LC3-interacting regions in IKKα
The LC3-interacting region (LIR), which is defined by a conserved hydrophobic motif of WXXL where X is any amino acid, is used by various proteins to interact with LC3 (20, 21). Sequence analysis of IKKα uncovered three putative LIRs, which we designated as LIR-1 (residues 275 to 278), LIR-2 (residues 651 to 654), and LIR-3 (residues 740 to 743) (Figure S3A). We mutated each LIR sequence by replacing the tryptophan residue in the motif with an alanine residue to examine their role in LC3-IKKα complex formation. After co-transfection of HEK293T cells with plasmids encoding each individual LIR3-mutant IKKα-HA and LC3-GFP, Western blotting analysis showed an increase in the abundance of IKKα protein (Figure 5A). These data suggest that mutation of the LIR-3 sequence of IKKα may have affected protein stability. Nevertheless, IKKα-HA was immunoprecipitated from all cell lysates in equivalent abundance. Western blotting analysis of immunoprecipitates of LC3 showed that mutation of the LIR-2 and LIR-3 sequences, but not the LIR-1 sequence, abolished the interaction between IKKα and LC3. These data suggested that LIR2 and LIR3 might be involved in the binding of IKKα to LC3 (Figure 5A). We confirmed this observation by confocal microscopy, which showed that wild-type IKKα and the LIR-1 mutant IKKα, but not LIR-2 and LIR2 mutant IKKα, colocalized with LC3 (Figure 5B). Together, these results suggest that LIRs near the C terminus of IKKα are required for binding to LC3 and recruitment to the LAPosomes after TLR9 stimulation.
Figure 5. Interaction between IKKα and LC3 depends on LC3 interacting region in IKKα.
(A) HEK293T cells were transfected with different LIR mutants, immunoprecipitated for LC3, and blotted for the indicated proteins. Blots are representative of 3 or more independent experiments (B) IKKα-deficient bone marrow cells were retrovirally reconstituted with IKKα-HA (wild-type, LIR-1, LIR-2, or LIR-3 mutant) and Cherry-LC3, then differentiated into BMDCs and stimulated with 0.5μM DOTAP-CpG-A for 6 hours. After treatment, cells were analyzed by confocal microscopy. Images are representative of 3 independent experiments. Scale bars: 5um.
Mutations in the LIR motifs do not affect the kinase activity of IKKα
The kinase activity of IKKα is required for noncanonical NF-κB signaling by mediating the ubiquitin-dependent degradation of NF-κB p100 and its processing into p52 (22). A subset of TNF receptor superfamily members, such as lymphotoxin beta receptor (LTβR), B-cell-activating factor of the tumor-necrosis-factor family (BAFF) and CD40, induce noncanonical NF-κB activation (23). To examine the possibility that LIR mutations interfered with the kinase activity of IKKα, we transduced IKKα-deficient mouse embryonic fibroblasts (MEFs) with retroviruses expressing wild-type IKKα or IKKα-LIR mutants and analyzed p52 nuclear translocation after LTβR stimulation. The mutant IKKα constructs were expressed; however, more IKKα protein accumulated in cells expressing the LIR-3 mutant than in cells expressing WT IKKα (Figure S3). Agonistic antibody crosslinking of LTβR induced p52 nuclear translocation in MEFs expressing either wild-type or LIR mutant IKKα, but not in control, IKKα-deficient MEFs (Figure S3B). These data suggest that LIR mutant IKKα constructs retain kinase activity.
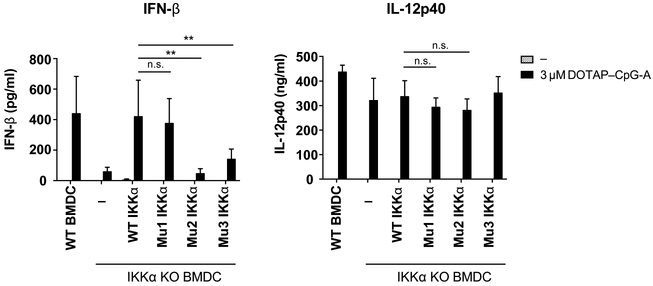
Interaction between IKKα and LC3 is required for type I IFN production
Finally, we sought to identify the physiological importance of the interaction between IKKα and LC3. IKKα is specifically required for type I IFN production in response to TLR9 activation, because IKKα deficiency in both plasmacytoid dendritic cells (pDCs) and conventional dendritic cells (cDCs) results in impaired IFN production, but intact IL-12p40 production (6, 24). Therefore, we reconstituted Ikkα−/− BMDCs with wild-type or LIR-mutant IKKα to examine whether LIR sequence mutations had any effect on type I IFN production after TLR9 stimulation. Consistent with previous results (6), Ikkα−/− pDCs showed significantly reduced IFN-α production upon TLR9 stimulation, but had no defect in IL-12p40 production (Figure S4). Similarly, when BMDCs from Ikkα−/− fetal liver chimeras were stimulated through TLR9, they produced substantially less IFN-β than did wild-type cells, but secreted normal amounts of IL-12p40 (Figure 6). Upon reconstitution of Ikkα−/− BMDCs with wild-type or mutant IKKα constructs, only those cells expressing wild-type or LIR-1 mutant IKKα exhibited restored IFN-β production (Figure 6). Furthermore, in HEK293T cells overexpressing LC3, TRAF3, TRAF6, MyD88, and IRF7, only those expressing either WT IKKα or the LIR-1 mutant IKKα, but not LIR-2 or LIR-3 mutant IKKα, exhibited phosphorylation of IRF7 (Figure S5). Computational modeling using SWISS-MODEL suggests that LIR-2 could be located on the surface of the protein, whereas it appears that LIR-1 may be buried close to the kinase core (Figure S6). Thus, the LIR-2 domain, but not the LIR-1 domain, of IKKα may be accessible for binding to LC3. Together, our results suggest that IKKα recruitment and interaction with LC3 and TLR9 on LAPosomes is critical for stimulating type I IFN production.
Figure 6. Interaction between LC3 and IKKα is crucial for type I IFN production in GM-CSF-differentiated BMDCs.
Ikkα−/− bone marrow cells were retrovirally reconstituted with IKKα-HA (wild-type, LIR-1, LIR-2, or LIR-3 mutant), then differentiated into BMDCs and stimulated with DOTAP-CpG-A. After 24 hours the supernatants were harvested and cytokine production was assayed by ELISA. Data are means ± SEM from 3 independent experiments. ** denotes P < 0.01 by t test.
Discussion
In this study, we provide evidence for a link between two previously identified pathways required for the TLR9 signaling that stimulates type I IFN production. We showed that, shortly after stimulation of macrophage and dendritic cells with CpG-A, IKKα was recruited to the LC3 embedded in TLR9-positive endosomes. This process did not occur after the induction of canonical autophagy. In addition, macrophages that lacked FIP200, a critical component of autophagosome formation, were still capable of exhibiting IKKα recruitment to LC3-positive endosomes and signaling for type I IFN production downstream of TLR9. These data suggest that noncanonical autophagy, namely, LAP, is important for the IKKα recruitment and TLR9 signaling. To gain a better understanding of the IKKα-LC3 interaction, we identified and examined three putative LIRs in IKKα. Alanine replacement of the critical tryptophan motif of the LIR (WXXL) suggested that mutations in the LIR-2 and LIR-3 domains reduced the binding of IKKα to LC3. Furthermore, reconstitution of Ikkα−/− BMDCs with these mutants failed to restore type I IFN production. In contrast, wild-type IKKα and the LIR-1 mutant IKKα bound to LC3 and restored the TLR signaling required for type I IFN production in Ikkα−/− BMDCs. Collectively, our study highlights a link between two known pathways for IFN induction by TLR9, namely, LAP and IKKα.
Although this type of experiment cannot rule out the possibility that a loss of binding may be secondary to protein structural distortion, the IKKα mutants exhibited no defects in the activation of noncanonical NF-κB signaling (fig S3B). Specifically, all of our IKKα mutants were capable of p52 activation downstream of LTβR signaling mediated by NF-κB-inducing kinase (NIK) (fig S3B). Because NIK is required for the phosphorylation of IKKα (25), these data suggest that none of the LIR mutations that we generated interfered with phosphorylation by NIK. However, we did not directly examine this possibility in our current study.
IKKα is required for canonical autophagy induced by starvation or inflammatory signals. IKKα and IKKβ are phosphorylated upon starvation, and cells deficient in these genes have impaired autophagy induction (26)(27). Downstream of TNF receptor signaling, activated IKKα also phosphorylates and stabilizes Atg16L1 and prevents its degradation (28). In our current study, canonical autophagy induced by starvation or Tat-Beclin1 peptide treatment did not result in IKKα recruitment to the LC3-coated LAPosomes (Fig. 2), suggesting that IKKα phospohyrylation induced by these signals is not sufficient for IKKα-LC3 interaction. In addition, IKKα-deficient BMDC reconstituted with LIR mutants 2 and 3 had only slightly fewer LC3 puncta per cell (Fig. 5B). Thus, it is possible that IKKα also participates in LAP formation, and that these mutations may have interfered with that process. Nevertheless, there was little association of IKKα LIR mutants with LC3 puncta (Fig. 5C), which suggests that these LIR regions in IKKα control their recruitment to LAPosomes.
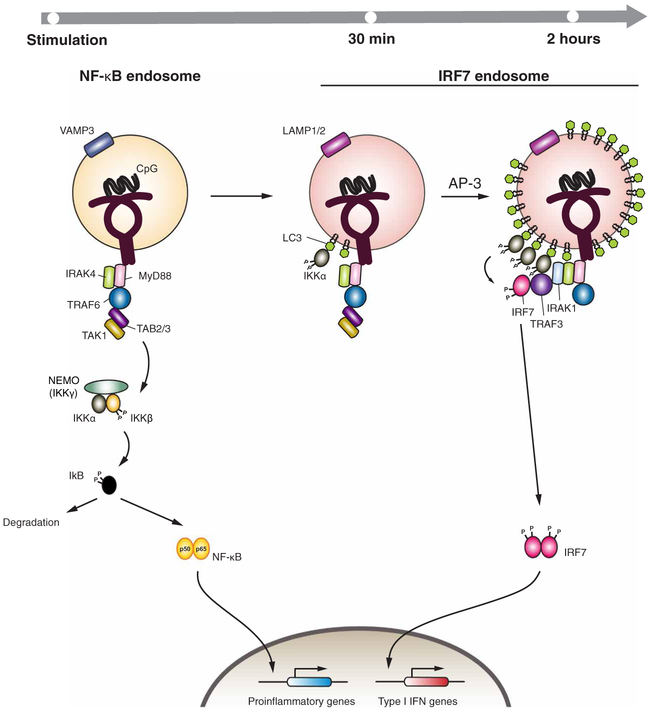
Based on our current study, we propose a revised model for TLR9 signaling (Figure 6). After stimulation, ligand-bound TLR9 dimerizes and recruits the signaling adaptor MyD88 and downstream signaling molecules, including IRAK4 and the E3 ubiquitin ligase TRAF6. Autoubiquitylation of TRAF6 mediates recruitment of the kinase TAK1 and its regulatory proteins TAB2 and TAB3, which phosphorylates and activates IKKβ. This induces the NF-κB–mediated transcription of proinflammatory genes (NF-κB endosome, Figure 7). Activation of TLR9 also stimulates the ATG5-dependent recruitment of LC3 to the endosomal membrane through the process of LAP (12). Our data suggest that IKKα is recruited to the LC3 through the LIR-2 and LIR-3 domains. This likely brings IKKα into proximity with IRF7, which it may phosphorylate to activate the transcription of genes encoding type I IFNs.
Figure 7. Schematic of IKKα complex recruitment to LC3-containing LAPosomes necessary for TLR9 signaling and IFN production.
Following stimulation, ligand-bound TLR9 dimerizes and recruits signaling adaptor MyD88 and downstream signaling molecules including IRAK4 and the E3-ligase, TRAF6. Autoubiquitination of TRAF6 mediates recruitment of TAK1 and its regulatory proteins TAB2/3, which phosphorylates and activates IKKβ. This induces NF-κB mediated transcription of proinflammatory genes (NF-κB endosome). Activation of TLR9 also triggers ATG5-dependent recruitment of LC3 to the endosomal membrane through the process of LAP. IKKα is recruited to the LC3 through LIR-2 and LIR-3 domains. AP-3 is required for the formation of lysosome-related organelle decorated with LAMP2. IKKα recruitment to the membrane bound LC3 brings this kinase to the proximity of IRF7, where it phosphorylates IRF7 and activates type I IFN gene transcription.
These data provide a link between IKKα and LAP for type I IFN production. We suggest that the direct recruitment of IKKα is LAP-dependent, because the recruitment was lost in Atg5−/− BMMs, but not in Fip200−/− BMMs. Our data suggest that LC3 acts as an anchor for IKKα on TLR9-positive endosomes and is necessary for the production of type I IFN (Figure 7). Although Atg8 family proteins (including LC3) recruit various cargoes into autophagosomes by interacting with adaptor proteins (21), our work suggests that IKKα may be directly recruited to LAPosomes through its interaction with LC3. Additionally, as LC3 decorates the outer membrane of TLR9-containing endosomes, this enables IKKα to be recruited to a signaling platform on the cytosolic face of the membrane. This suggests that LC3 may also mediate the recruitment of other molecules to the outer membrane of autophagosomes or the LAPosome for various cellular processes.
Second, our results contribute to the expanding the list of potential roles of LAP in regulating molecular and cellular processes. The central component of LAP is LC3, which interacts with various proteins through its LIRs. The role of LIRs in the selective recruitment of autophagic cargoes have been extensively studied, because LIRs provide docking sites for adaptor proteins, such as p62, an ubiquitin-binding protein, neighbor of BRCA1 gene 1 (NBR1), nuclear domain 10 protein 52 (NDP52), Nip-like protein X (NIX), and ATG19 (21). A proteomics analysis showed that the ATG8 family, which includes the LC3 and GABARAP subfamilies, has 67 high-confidence interactions with other cellular proteins (29). In addition, proteomic analysis identified Rubicon and UVRAG-containing Class III PI(3)K complex proteins as being associated with the TLR2-induced LAPosome (14). Our study identifies IKKα as another LC3-binding, LAPosome--associated protein, and suggests that the LIR-LC3 interaction may be involved in kinase recruitment to the signaling platform on the endosomal membrane.
Our results suggest that LIR-2 and LIR-3 may both be important for the ability of IKKα to interact with LC3 after TLR9 stimulation. Although the crystal structure of IKKα has not been solved yet, its likely structure can be extrapolated from that of IKKβ due to similarity between two molecules (52% identical in amino acid sequence) (30). Our prediction of IKKα structure using SWISS-MODEL suggests that LIR-2 could be located near the surface of the protein, which has higher accessibility for other proteins interactions (Figure S6). Unfortunately, SWISS-MODEL could not predict the likely structure of LIR-3, because no corresponding region exists in IKKβ. Whereas yeast and other fungi have only one Atg8-encoding gene, humans have multiple Atg8 subfamily genes, including a single GATE-16 gene, two GABARAP genes, and four LC3 genes with splice variants (31). Whereas all Atg8 subfamily proteins are recruited to autophagosomes, it remains unknown if they play redundant roles in mediating TLR9 signaling. It is possible that only a subset of Atg8 proteins form specific interactions with IKKα.
In autoimmune disease, recognition of self-RNA and DNA by a variety of PRRs may trigger excessive amounts of type I IFN to be produced. The activation of TLR7 and TLR9 is linked to the exacerbation of autoimmune diseases such as systemic lupus erythematosus (SLE) (32) and psoriasis (33). Similarly, a universal deficiency in LAP components impaired clearance of dying cells and exacerbated SLE pathogenesis in mice (34). Based on our current study, inhibiting LAPosome signaling in pDCs is expected to ameliorate SLE symptoms by reducing the amount of type I IFN produced without impacting phagocytic clearance. Specific inhibition of IKKα-LC3 interaction may provide a selective means to reduce the amount of type I IFN produced by pDCs in patients with SLE or psoriasis, while leaving the LAP-dependent phagosomal degradation intact. Further investigation is still needed to determine the cell type differences in LAP usage as a potential target of intervention in SLE pathogenesis.
Materials and Methods
Mice
Age- and sex-matched C57BL/6 (wild-type) mice from the National Cancer Institute (Frederick, MD) were used for isolating bone marrow. CD45.1 congenic C57BL/6 mice were purchased from Charles River for fetal liver chimera generation. All procedures used in this study complied with federal guidelines and the institutional policies of the Yale Animal Care and Use Committee. Ikkα−/− mice were a kind gift from Dr. Michael Karin at University of California, San Diego.Atg5+/− mice (35) were bred in Yale University animal facility.
Fetal liver chimera generation
Ikkα and Atg5 fetal liver chimeric mice were generated by standard protocols, as described previously (19). Briefly, 2 to 3 ×106 liver cells from E15.5 to E18.5 embryos born to Ikkα+/− or Atg5+/− parents were screened for the Ikkα−/− or Atg5−/− genotype and injected into recipient C57BL/6 mice through tail vein. Before the transfer of liver cells, the recipient mice were lethally irradiated with 4.75Gy of irradiation twice, 3 hours apart. This procedure led to complete chimerism of hematopoietic cells by 8 weeks after transplantation.
Cell lines and primary cell culture
RAW267.4 cells were purchased from ATCC and cultured in RPMI supplemented with 10% FCS, penicillin/streptomycin, 10 mM HEPES (Invitrogen), and 2-mercaptoethanol (Sigma). TLR9-HA– and Cherry-LC3b–coexpressing cells were prepared by transduction with MSCV2.2 retrovirus and the cells were selected with puromycin (2 μg/ml). Bone-marrow-derived pDCs (36), bone-marrow derived DCs (37), bone-marrow derived macrophages (11) were prepared as previously described. Hematopoietic stem cells (HSCs) were transduced with retroviruses expressing the gene of interest at Day 0 and Day 1 by spinoculation at 1500 xg for 2 hours. After spinoculation, viral media was removed and cells were resuspended with fresh media to be plated for further cell differentiation. Cell culture media was replaced at Day 3, Day 5 throughout 7-day BMM and BMDC differentiation. Bones from Fip200fl/fl × LysM-Cre mice were a kind gift from H. W. Virgin (Washington University at St. Louis). IKKα-deficient mouse embryonic fibroblasts (MEFs) were generated from E14.5 fetuses as previously described (38).
Reagents and antibodies
LPS was purchased from List biological laboratories; CpG 2216 (CpG type-A) was purchased from TriLink BioTechnologies; and both poly(I:C) and R848 were purchased from Invivogen. DOTAP was obtained from Roche. The Tat-Beclin 1 peptide was purchased from Sigma-Aldrich. Mouse anti-HA tag (H3663), mouse anti-Flag-tag M2 (F1804), mouse anti-β-actin (A1978) antibodies were from Sigma-Aldrich. Mouse anti-GAPDH (GTX627408) antibody was from GeneTex. Rabbit anti-LC3b (ab48394), rabbit anti-Myd88 (ab2064), and mouse anti-GFP (ab1218) were from Abcam. Rabbit anti-IKKα (2682), rabbit anti-phosphorylated-IKKα/β (2697), rabbit anti-p100/p52 (4882), rabbit anti-TRAF3 (4729), rabbit anti-TRAF6 (8028), and rabbit anti-Flag-tag HRP(2044) antibodies were ontained from Cell Signaling Technology. HRP-conjugated goat antibody against mouse IgG and HRP-conjugated antibody against rabbit IgG were purchased from Jackson ImmunoResearch and Cell Signaling Technology, respectively. TrueBlot ULTRA HRP anti-mouse (13-8817-80) and TrueBlot HRP anti-rabbit (18-8816-31) antibodies were obtained from Rockland. For flow cytometry, anti-phosphorylated IRF7 antibody (558630) was from BD Biosciences. Agonistic LT-βR antibody [5G11] was purchased from Abcam.
Plasmids
TRAF3-HA was constructed as described previously (11). TLR9-HA and the MyD88 plasmids were a gift from R. Medzhitov. Plasmid encoding GFP-LC3b was a gift from N. Mizushima. Plasmid encoding the mIRF7-Flag was a gift from K. Hoshino and T. Kaisho. The plasmids pcDNA3-IKKα-HA (#23296), pMIG-II (#52107), TRAF6 (#21624), and c-Flag pcDNA3 (#20011) were purchased from Addgene. Human IRF7 plasmid (LV798857) was purchased from Applied Biological Materials Inc. The ATG5-Flag was constructed by subcloning the gene from our previously generated ATG5-IRES-EGFP into the c-Flag pcDNA3 vector. Cherry-LC3b and the mutant IKKα (Mutant 1-3, AA mutant) were subcloned by standard PCR protocols. Retrovirus vectors containing TLR9-HA and Cherry-LC3b were generated by subcloning the genes into pMSCV2.2 and pMSCVpuro, respectively. Retrovirus vectors containing mutant IKKα were generated by subcloning the genes into pMIG-II vector.
Immunoprecipitation and Western blotting
Cells were lysed with lysis buffer containing 20 mM Tris-HCl (pH 7.5), 1% NP-40, 150 mM NaCl, proteinase inhibitor cocktail (Roche), phosphatase inhibitor (PhosSTOP tablets, Roche) for 30 minutes (HEK 293T cells) or overnight (RAW264.7 and BMMs). Lysates were harvested and centrifuged at 21,130 xg for 10 min to obtain a post-nuclear supernatant. The supernatant was then subjected to immunoprecipitation assay with antibodies and Dynabeads Protein G (Thermo Fisher Scientific). Beads were then washed with wash buffers, followed by Western blotting analysis, which was peformed as previously described (11).
Immunofluorescence microscopy
Transduced RAW264.7 and BMMs were stimulated on day 7 of differentiation with 3 μM DOTAP-CpG or 1 μM Tat-Beclin 1 peptide for the times indicated in the figure legends, and then stained for immunofluorescence microscopy as previously described (39). The following antibodies were used for immunofluorescence microscopy analysis: mouse anti-IKKα from Santa Cruz Biotechnology; rabbit anti-HA, mouse anti-GFP, and rabbit anti-LAMP1 were from Abcam; rabbit anti-VAMP3 was from Synaptic Systems; rat anti-RFP [5F8] was obtained from chromotek; Alexa Fluor 488 anti-mouse antibody was from Invitrogen, biotin-anti mouse, streptavidin-A488, streptavidin-Cy5, Cy3 anti-rabbit, Cy3 anti-rat, Cy5 anti-rabbit, and A647 anti-rabbit antibodies were from Jackson ImmunoResearch. After staining, coverslips were mounted on slides with ProLong Gold (Invitrogen) and imaged with a Leica TCS SP8 confocal microscope using a 64× objective lens. A minimum of 15t to 20 cells were examined per slide.
ELISA
The amounts of IFN-α protein present in the cell culture medium and serum were measured by ELISA as previously described (40). The amounts of IFN-β protein present in the cell culture medium and serum were measured with a Verikine mouse IFN-β ELISA (PBL Assay Science). The amounts of IL-12p40 protein present in the cell culture medium and serum were measured according to manufacturer’s instructions with specific antibodies (BioLegend).
Flow cytometry
HEK293T cells were transfected with plasmids encoding WT or mutant IKKα constructs, human IRF7, Myd88, TRAF3, TRAF6, and GFP-LC3b. The cells were then fixed, permeabilized in 200 μl of BD Cytofix/Cytoperm (BD Biosciences) for 20 min at 4°C, and stained with antibodies Alexa Fluor 647–conjugated anti-pIRF7 antibody (pS477/pS479, BD Biosciences) for 30 min in BD Perm/Wash buffer at 4°C. Mean fluorescent Intensity (MFI) of phosphorylated IRF7 in each sample was analyzed by Flowjo (Treestar).
Statistical analysis
Normally distributed continuous variable comparisons were performed with the Student’s t-test using Prism software (GraphPad). Data are presented as means ± SEM. P < 0.05 was considered to be statistically significant.
Supplementary Material
Figure S1. LC3 binds to TLR9 and IKKα after DOTAP-CpG-A stimulation.
Figure S2. ATG5 does not interact with LC3 or IKKα.
Figure S3. LIR mutants of IKKα maintain intact kinase activity.
Figure S4. Flt3L-derived Ikkα-deficient pDCs show impaired IFNα production but intact IL-12p40 after CpG-A stimulation.
Figure S5. WT and LIR-1, but not LIR-2 or LIR-3, mutant IKKα induces IRF7 phosphorylation.
Figure S6. LIR prediction based on IKKα structure by SWISS-MODEL.
Acknowledgments:
We thank M. Karin and H. W. Virgin for kindly providing IKKα heterozygous mice and bones from LysM-Cre FIP200fl/fl mice, respectively. We also thank M. Linehan, H. Dong, Y. Kumamoto, P. Wong, and X. Liu for technical support.
Funding: K.H. is a recipient of a Nakajima Foundation Predoctoral Fellowship. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health (NIH) grants R01AI064705, R01AI081884, R01AI054359, and R56AI125504 to A.I.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S, A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, Matsuda M, Coban C, Ishii KJ, Kawai T, Takeuchi O, Akira S, Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med 201, 915–923 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, Mann M, Karin M, Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G, Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T, IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature 440, 949–953 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, Glimcher LH, Cantor H, Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol 7, 498–506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh K, Tanaka Y, Nishikimi A, Nakamura R, Yamada H, Maeda N, Ishikawa T, Hoshino K, Uruno T, Cao Q, Higashi S, Kawaguchi Y, Enjoji M, Takayanagi R, Kaisho T, Yoshikai Y, Fukui Y, Selective control of type I IFN induction by the Rac activator DOCK2 during TLR-mediated plasmacytoid dendritic cell activation. J Exp Med 207, 721–730 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watarai H, Sekine E, Inoue S, Nakagawa R, Kaisho T, Taniguchi M, PDC-TREM, a plasmacytoid dendritic cell-specific receptor, is responsible for augmented production of type I interferon. Proc Natl Acad Sci U S A 105, 2993–2998 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S, Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Sasai M, Linehan MM, Iwasaki A, Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, Coyle AJ, Kolbeck R, Green DR, Sanjuan MA, Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37, 986–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR, Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR, Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T, Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B, Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weidberg H, Shvets E, Elazar Z, Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80, 125–156 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Shibutani ST, Saitoh T, Nowag H, Munz C, Yoshimori T, Autophagy and autophagy-related proteins in the immune system. Nat Immunol 16, 1014–1024 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A, Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315, 1398–1401 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F, Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Birgisdottir AB, Lamark T, Johansen T, The LIR motif - crucial for selective autophagy. J Cell Sci 126, 3237–3247 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Sun SC, Non-canonical NF-kappaB signaling pathway. Cell Res 21, 71–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jost PJ, Ruland J, Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood 109, 2700–2707 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Hoshino K, Sasaki I, Sugiyama T, Yano T, Yamazaki C, Yasui T, Kikutani H, Kaisho T, Critical role of IkappaB Kinase alpha in TLR7/9-induced type I IFN production by conventional dendritic cells. J Immunol 184, 3341–3345 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Wang RP, Zhang M, Li Y, Diao FC, Chen D, Zhai Z, Shu HB, Differential regulation of IKK alpha-mediated activation of IRF3/7 by NIK. Mol Immunol 45, 1926–1934 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, Tailler M, Delahaye N, Tesniere A, De Stefano D, Younes AB, Harper F, Pierron G, Lavandero S, Zitvogel L, Israel A, Baud V, Kroemer G, The IKK complex contributes to the induction of autophagy. EMBO J 29, 619–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comb WC, Cogswell P, Sitcheran R, Baldwin AS, IKK-dependent NF-kappaB-independent control of autophagic gene expression. Oncogene 30, 1727–1732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamanti MA, Gupta J, Bennecke M, De Oliveira T, Ramakrishnan M, Braczynski AK, Richter B, Beli P, Hu Y, Saleh M, Mittelbronn M, Dikic I, Greten FR, IKKalpha controls ATG16L1 degradation to prevent ER stress during inflammation. J Exp Med 214, 423–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behrends C, Sowa ME, Gygi SP, Harper JW, Network organization of the human autophagy system. Nature 466, 68–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karin M, How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18, 6867–6874 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z, Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 12, 226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banchereau J, Pascual V, Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25, 383–392 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, Gilliet M, Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med 202, 135–143 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, Linkermann A, Virgin HW, Green DR, Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N, The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 36.D’Amico A, Wu L, The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med 198, 293–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato A, Linehan MM, Iwasaki A, Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells. Proc Natl Acad Sci U S A 103, 17343–17348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S, Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med 202, 1333–1339 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi K, Sasai M, Iwasaki A, Toll-like receptor 9 trafficking and signaling for type I interferons requires PIKfyve activity. Int Immunol 27, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A, Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med 198, 513–520 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. LC3 binds to TLR9 and IKKα after DOTAP-CpG-A stimulation.
Figure S2. ATG5 does not interact with LC3 or IKKα.
Figure S3. LIR mutants of IKKα maintain intact kinase activity.
Figure S4. Flt3L-derived Ikkα-deficient pDCs show impaired IFNα production but intact IL-12p40 after CpG-A stimulation.
Figure S5. WT and LIR-1, but not LIR-2 or LIR-3, mutant IKKα induces IRF7 phosphorylation.
Figure S6. LIR prediction based on IKKα structure by SWISS-MODEL.