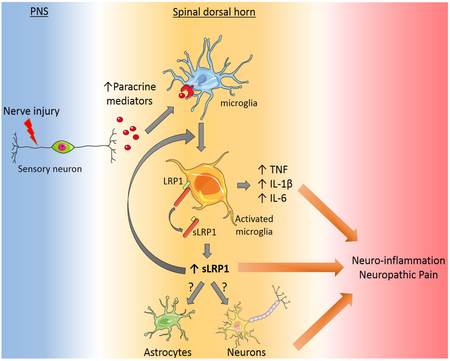
Abstract
Following injury to the peripheral nervous system (PNS), microglia in the spinal dorsal horn (SDH) become activated and contribute to the development of local neuro-inflammation, which may regulate neuropathic pain processing. The molecular mechanisms that control microglial activation and its effects on neuropathic pain remain incompletely understood. We deleted the gene encoding the plasma membrane receptor, LDL Receptor-related Protein-1 (LRP1), conditionally in microglia using two distinct promoter-Cre recombinase systems in mice. LRP1 deletion in microglia blocked development of tactile allodynia, a neuropathic pain-related behavior, after partial sciatic nerve ligation (PNL). LRP1 deletion also substantially attenuated microglial activation and pro-inflammatory cytokine expression in the SDH following PNL. Because LRP1 shedding from microglial plasma membranes generates a highly pro-inflammatory soluble product, we demonstrated that factors which activate spinal cord microglia, including lipopolysaccharide (LPS) and colony-stimulating factor-1, promote LRP1 shedding. Proteinases known to mediate LRP1 shedding, including ADAM10 and ADAM17, were expressed at increased levels in the SDH after PNL. Furthermore, LRP1-deficient microglia in cell culture expressed significantly decreased levels of interleukin-1β and interleukin-6 when treated with LPS. We conclude that in the SDH, microglial LRP1 plays an important role in establishing and/or amplifying local neuro-inflammation and neuropathic pain following PNS injury. The responsible mechanism most likely involves proteolytic release of LRP1 from the plasma membrane to generate a soluble product that functions similarly to pro-inflammatory cytokines in mediating crosstalk between cells in the SDH and in regulating neuropathic pain.
Keywords: LRP1, microglia, neuropathic pain, central sensitization, membrane protein shedding
Graphical Abstract:
1 │. INTRODUCTION
Neuropathic pain is a debilitating condition caused by injury to the somatosensory nervous system (Dworkin et al., 2003). Development and maintenance of neuropathic pain may be regulated by peripheral sensitization, which is characterized by hyper-excitability of injured primary afferent neurons, and central sensitization, which is defined by the International Association of the Study of Pain as increased responsiveness of nociceptive neurons in the CNS to normal or sub-threshold afferent input (Woolf, 2014; Gilron et al., 2015). Glia and immune cells in the spinal dorsal horn (SDH) may contribute to the pathogenesis of neuropathic pain (Marchand et al., 2005; Scholz and Woolf, 2007; Grace et al., 2014); however, the responsible mechanisms remain incompletely understood. Identifying novel pathways that contribute to neuropathic pain is an important goal for drug development (Cohen and Mao, 2014; Bouhassira and Attal, 2018).
After peripheral nerve injury, factors produced by injured sensory neurons, such as colony-stimulating factor-1 (CSF-1), serve as triggers for glial activation in the SDH (Guan et al., 2016). Activated microglia and astrocytes then secrete diverse extracellular mediators to establish paracrine pathways that may exacerbate neuropathic pain (Scholz and Woolf, 2007; Ren and Dubner, 2008; Schomberg and Olson, 2012). Many cytokines expressed by activated glia, including TNFα and interleukin-1β (IL-1β), are pro-inflammatory. For this reason, preclinical and clinical trials studies have been performed to test the efficacy of anti-inflammatory agents in treating pain (Sommer et al., 2001; Nguyen et al., 2015; Vanelderen et al., 2015).
LDL Receptor-related protein-1 (LRP1) is an endocytic receptor that binds structurally and functionally diverse ligands (Herz and Strickland, 2001; Gonias and Campana, 2014). An important property of LRP1 is its ability to couple endocytosis with activation of cell-signaling by functioning in conjunction with receptors such as the N-methyl-D-aspartate Receptor, Trk Receptor, and p75NTR (Shi et al., 2009; Mantuano et al., 2013; Stiles et al., 2013). In bone marrow-derived macrophages (BMDMs), membrane-anchored LRP1 is anti-inflammatory and its deletion increases expression of pro-inflammatory cytokines (May et al., 2013; Staudt et al., 2013; Mantuano et al., 2016). Mice with LRP1-deficient macro-phages demonstrate exacerbated responses to lipopolysaccharide (LPS) (Mantuano et al., 2016). However, the activity of LRP1 in inflammation is not straightforward because membrane-anchored LRP1 may be proteolytically released from the cell-surface by the proteinases, ADAM10 and ADAM17 (Liu et al., 2009; Gorovoy et al., 2010; Shackleton et al., 2016), generating a soluble product, shed LRP1 (sLRP1), that is robustly pro-inflammatory (Gorovoy et al., 2010; Brifault et al., 2017). sLRP1 is present in normal human plasma (Quinn et al., 1999) and at increased levels in plasma from patients with rheumatoid arthritis and systemic lupus erythematosus (Gorovoy et al., 2010). sLRP1 is present in osteoarthritic cartilage (Yamamoto et al., 2017), broncho-alveolar lavage fluid from patients with respiratory distress syndrome (Wygrecka et al., 2011), and in CSF, where its concentration increases with aging (Liu et al., 2009).
LRP1 is expressed by microglia, especially when these cells are activated (Marzolo et al., 2000; Chuang et al., 2016). Thus, microglia may serve as a source of sLRP1 and as a target for sLRP1 once it is generated. However, in microglia, like in macrophages, membrane-anchored LRP1 expresses anti-inflammatory activity (Pocivavsek et al., 2009b, 2009a; Yang et al., 2016). Conditional deletion of LRP1 in microglia exacerbates experimental autoimmune encephalomyelitis in mice (Chuang et al., 2016), probably reflecting loss of the membrane-anchored form of the receptor. Theoretically, LRP1 deletion in macrophages or microglia also may attenuate inflammation in a specific mouse model system if the activity of sLRP1 predominates: however, such results have not been previously reported.
Herein, we show that conditional deletion of LRP1 in microglia blocks development of neuropathic pain-related behavior in mice subjected to partial sciatic nerve ligation (PNL). LRP1 deletion also inhibits microglial activation and pro-inflammatory cytokine expression in the SDH. Agents that activate spinal cord microglia, including CSF-1, induced LRP1 shedding. Furthermore, ADAM10 and ADAM17 were expressed at increased levels in the SDH following PNL. These results demonstrate that microglial LRP1 regulates neuro-inflammation and neuropathic pain following PNS injury. We propose that the responsible pathway involves LRP1 shedding to generate a pro-inflammatory “cytokine-like” soluble product in the SDH.
2 │. MATERIALS AND METHODS
2.1 │. Proteins and reagents
Receptor-associated protein (RAP) was expressed as a GST fusion protein in bacteria and purified as described (Herz et al., 1991). LPS, serotype 055.B5, and Tamoxifen (TAM) were from Sigma-Aldrich. Recombinant mouse CSF-1 and Quantikine ELISA kits were purchased from R&D systems. All primers and probes for RT-qPCR experiments were from Applied Biosystems.
2.2 │. Mice
Wild type C57BL/6J mice were from the Jackson Laboratory. Mice with LoxP sites partially flanking the LRP1 gene (Rohlmann et al., 1998) were crossed with LysM-Cre mice (Clausen et al., 1999) to generate LysM-Cre-positive-LRP1fl/fl mice. In experiments, LysM-Cre-positive-LRP1fl/fl mice were compared with littermate control mice that carried two loxP-flanked LRP1 genes but were LysM-Cre-negative (LysM-Cre-negative-LRP1fl/fl mice). LRP1fl/fl mice also were bred with Cx3cr1-CreERT mice (Goldmann et al., 2013) to generate Cx3cr1-CreERT-LRP1fl/fl mice. In most experiments, Cx3cr1-CreERT-LRP1fl/fl mice were compared with littermate control mice that also were Cx3cr1-CreERT-positive but expressed two wild-type LRP1 genes (Cx3cr1-CreERT-LRP1 mice). To activate CreERT in cells in which it is expressed, Cx3cr1-CreERT-LRP1fl/fl mice and Cx3cr1-CreERT-LRP1 mice were treated with TAM (150 mg/kg, IP) twice, 1 week apart before conducting experiments, four weeks later. In control experiments, Cx3cr1-CreERT-LRP1 and Cx3cr1-CreERT-LRP1fl/fl mice were treated with TAM vehicle (5% ethanol and corn oil) instead of TAM. All experiments were conducted using 2 to 3 months-old male mice. All animal experiments were approved by the Institutional Animal Care and Use Committee at University of California San Diego.
2.3 │. Surgery
PNL (Seltzer model) studies were performed as previously described (Seltzer et al., 1990) and modified for mice (Orita et al., 2013). Male mice (2-3 months old) were anesthetized with isoflurane (5% for induction and 2% for maintenance). An incision was made and the left sciatic nerve was exposed at mid-thigh level. A 9-0 nylon suture (Ethicon) was inserted into the nerve and ligated so that the dorsal one-third to one-half of the nerve was included. The wound was closed with surgical skin staples. In sham-operated animals, the nerve was exposed at mid-thigh level but not ligated.
2.4 │. Behavioral testing
Tactile allodynia was tested by applying 0.04 to 4 g von Frey filaments (Stoelting) to the plantar surface of the ipsilateral hind paw. All mice were baseline tested for three days prior to surgery and then tested again on days 2, 3, 7, 10, 14, 17 and 21 following PNL or sham operation. Mice were allowed to acclimate to the room for at least 30 minutes and placed on the mesh stand for at least 45 min before testing. Von Frey filaments were presented in a consecutive fashion either ascending or descending using the up-down method as previously described (Dixon, 1980; Chaplan et al., 1994). The filament that caused paw withdrawal 50% of the time (the 50% PWT) was determined.
Motor testing was performed using an accelerating Rotarod (Ugo Basile). The Rotarod speed was increased from 4 to 40 rotations per min over a 120 s time period. Mice received two training trials on separate days before the surgery. The latency time to failure was measured in three different trials on days 2, 3, 7, 10, 14, 17 and 21 after surgery. All behavioral testing was done by a blinded investigator.
2.5 │. Tissue harvest, labeling, and image analysis
Spinal cords centered at L3-L4 and sciatic nerves were harvested 3 days after PNL or sham operation. Mice were deeply anesthetized with isoflurane and subjected to intra-cardiac perfusion with fresh PBS followed by 4% paraformaldehyde. All tissue was paraffin-embedded. Tissue sections (4 µm) were prepared (at least three from each harvested tissue). For immunofluorescence (IF) microscopy, sections were incubated with primary antibodies directed against the microglial cell specific marker, Iba1 (019-19741; Wako), and LRP1 β-chain (L2170; Sigma). The secondary antibodies were Alexa 594-conjugated-donkey anti-rabbit IgG (A21207; Invitrogen) and Alexa 488-conjugated-donkey anti-goat IgG (A21206; Invitrogen). Sections were mounted and viewed using a confocal laser scanning microscope (LSM 880 with Airyscan; Zeiss) and subjected to images analysis using IMARIS software (Bitplane). Co-localization of LRP1 immunostain with Iba1 immunostain was determined using the IMARIS Coloc® module. Each cohort consisted of four mice. Three sections from each mouse were analyzed.
For immunohistochemistry (IHC), tissue sections were incubated with 10% nonfat milk and then with primary antibodies against CD11b (ab133357; Abcam) or Iba1 for 1 h. Next, sections were incubated with anti-mouse antibodies conjugated to horseradish peroxidase and developed with 3’3 diaminobenzidine (DAB, Jackson ImmunoResearch). Control sections were treated with secondary antibody only. Light microscopy was performed using a Leica DFC420 microscope with Leica Imaging Software 2.8.1 (Leica Biosystems).
Activation of microglia in the SDH was determined by Iba1 immunostaining and image analysis using ImageJ software (NIH). The area immunostained by Iba1-specific antibody was determined in the ipsilateral SDH as a percentage of the total area examined. Two regions of interest, corresponding to the Laminae I-II and III-IV, were delineated and Iba1 staining density was determined in each of these regions in three separate microscopic sections per animal. At least four separate animals were analyzed in each cohort.
To quantitate migration of CD11b-positive cells into crush-injured sciatic nerves, sections of distal nerve were examined by a blinded investigator. The number of CD11b-positive cells within the endoneurium was determined. Three sections of nerve were examined per animal. Four mice were studied per condition.
2.6 │. Isolation of microglia and cell culture
Brain and spinal cords were collected from mice and homogenized using the Neural Tissue Dissociation kit (Miltenyi Biotec). For studies in which cells were analyzed without culturing, homogenates were subjected to centrifugation on 30% Percoll gradients, as previously described (Nikodemova and Watters, 2012). The microglia were then isolated by magnetic cell sorting using CD11b-Microbeads (Miltenyi Biotec). Cultures of brain-derived microglia were established as previously described (Brifault et al., 2017). Cultures of microglia from adult mouse spinal cords were established as described by Yip et al. (Yip et al., 2009). Briefly, spinal cord tissue was digested in papain (1 mg/mL) and triturated for mechanical dissociation. Cells were plated in complete medium consisting of Dulbecco’s Modified Eagle’s Medium/F-12 (DMEM/F-12), supplemented with GlutaMAX™, 15% fetal bovine serum (FBS), and 100 units/mL Antibiotic-antimycotic (all reagents from ThermoFisher/ Gibco). After 4 hours, the medium was replaced to clear debris and non-adherent cells. Primary microglial cell cultures were studied when the cells were approximately 80% confluent within 4 days. Primary cultures were examined for homogeneity by IF microscopy for GFAP (astrocytes), Iba1 (microglia), βIII tubulin (neurons), and Olig1 (oligodendrocytes). All reported experiments were performed using cultures that included greater than 95% microglia.
For gene expression and immunoblot or RAP ligand blot analysis, microglia were transferred to low-serum medium containing 0.5% FBS for 30 min and then treated with 100 ng/mL LPS or increasing concentrations of recombinant mouse CSF-1 (10, 25 or 50 ng/mL). Control cells were treated with vehicle (20 mM sodium phosphate, 150 mM NaCl, pH 7.4, PBS) (1 µL/mL).
2.7 │. Immunoblot analysis and RAP ligand blotting
Cells were extracted in RIPA buffer (20 mM sodium phosphate, 150 mM NaCl, pH 7.4, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Complete Protease Inhibitor Cocktail and phosphatase inhibitor (Roche Diagnostics). Equal amounts of cellular protein (20 µg) were loaded onto Protean TGX gels (Bio-Rad) and electrotransferred to PVDF membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in 10 mM Tris-HCl, 150 mM NaCl, pH 7.4, and 0.1% Tween 20 (TBS-T buffer) and incubated with the following primary antibodies: anti-LRP1 β-chain; anti-GAPDH (clone GT239; GTX627408; GeneTex); and anti-β-actin (3700; Cell Signaling Technology). Primary antibodies were detected with HRP-conjugated species-specific secondary antibody (Cell Signaling Technology).
RAP ligand blotting was performed to detect sLRP1 in conditioned medium (CM). Samples of CM were analyzed without concentrating as previously described (Brifault et al., 2017). CM was subjected to SDS-PAGE and electrotransferred to PVDF membranes, which were blocked and then incubated with 100 nM GST-RAP in 5% nonfat milk for 1 h at 22 °C. The membranes were then washed three times and incubated with GST-specific antibody coupled to horseradish peroxidase (84-814; Genesee Scientific). Conjugated antibody was detected with ECL reagent Prosignal™ (Prometheus) and the Azure C300 imaging system. As a control, the equivalent samples were subjected to immunoblot analysis to detect an intracellular epitope in the LRP1 β-chain (absent in sLRP1).
2.8 │. Analysis of conditioned medium by ELISA
Microglia were allowed to condition medium that contained 0.5% FBS. TNFα and IL-6 in CM were quantified using mouse quantikine ELISA kits (R&D systems).
2.9 │. Real-time qPCR
Mice were subjected to PNL or sham operation. Three days later, tissue was harvested from the ipsilateral and contralateral SDHs (centered at L3-L4) and the sciatic nerve 3 mm distal from the injury site. Total RNA was isolated and purified using the NucleoSpin® RNA kit (Macherey-Nagel). cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was performed using TaqMan® gene expression products on an AB Step one Plus Real-Time PCR System (Applied Biosystems). The relative change in gene expression was calculated using the 2ΔΔCt method and GAPDH mRNA was used as a standard. The primer-probe sets used included: TNFα (Mm00443258_m1); IL-6 (Mm00446190_m1); IL1-β (Mm00434228_m1); CD11b/ITGAM (Mm00434455_m1); LRP1 (Mm00464608-m1); and GAPDH (Mm99999915_g1).
2.10 │. Statistics
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software Inc.). All results are expressed as mean ± SEM. Comparisons between two groups were performed using two-tailed unpaired t-test with a confidence interval set at 95%. Multiple comparisons were investigated by one-way ANOVA followed by Tukey’s or Dunnett’s multiple comparison tests to detect pair-wise between-group differences. Behavioral data for each group were analyzed by repeated measure ANOVA followed by Bonferroni’s post hoc test. p < 0.05 was considered statistically significant.
3 │. RESULTS
3.1 │. LRP1 promotes microglial activation in response to LPS
Purified sLRP1 is a potent activator of brain-derived microglia in culture, which also induces neuro-inflammation when injected directly into the SDH (Brifault et al., 2017). By contrast, membrane-anchored microglial LRP1 is reported to express anti-inflammatory activity (Pocivavsek et al., 2009a, 2009b; Yang et al., 2016). To study the activity of membrane-anchored and shed LRP1 collectively in microglia, we harvested microglia from the brains of mice that are homozygous for the floxed LRP1 gene (LRP1fl/fl) and LysM-Cre-positive or -negative. LysM drives expression of Cre in monocytes, macrophages, neutrophils, and microglia, although the level of Cre recombinase expressed in microglia may depend on whether the cells are activated or in culture (Cho et al., 2008; Goldmann et al., 2013). When prepared according to the method used here, brain-derived microglia from LysM-Cre-positive-LRP1fl/fl mice in culture are 85% LRP1-deficient at the protein level (Brifault et al., 2017).
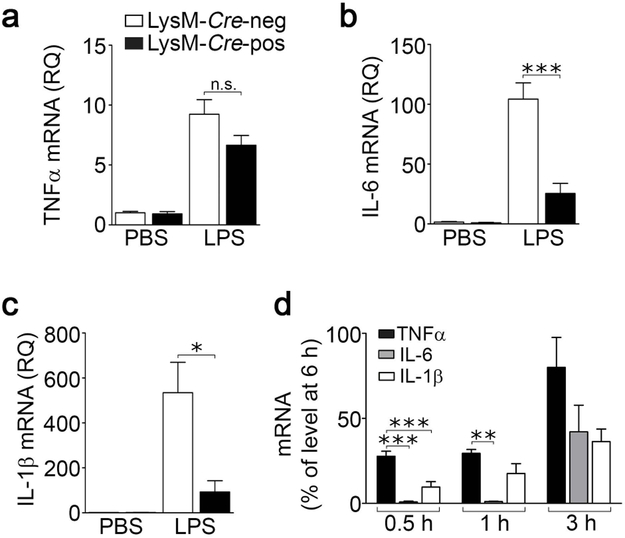
LRP1-expresing and –deficient microglia were treated with LPS (100 ng/mL) or vehicle for 24 h. LPS activates innate immune system pathways in brain-derived microglia and also induces LRP1 shedding (Olson and Miller, 2004; Brifault et al., 2017). In LRP1-expressing cells, LPS increased expression of the mRNAs encoding TNFα (Fig. 1a), IL-6 (Fig. 1b), and IL-1β (Fig. 1c). A similar increase in TNFα mRNA was observed in LRP1-deficient cells. By contrast, the effects of LPS on expression of IL-6 and IL-1β were significantly attenuated in LRP1-deficient microglia (***p < 0.001, *p < 0.05). Given the known pro-inflammatory activity of sLRP1, one explanation for these data is decreased availability of LRP1 for shedding in microglia isolated from LysM-Cre-positive-LRP1fl/fl mice.
FIGURE 1.
LRP1 regulates the microglial response to LPS. Brain-derived microglia were isolated from LysM-Cre-positive-LRP1fl/fl and LysM-Cre-negative-LRP1fl/fl mice and cultured in low-serum (0.5% FBS) medium for 30 minutes. The cells were treated with LPS (100 ng/mL) or vehicle (PBS, 1 µL/mL). After 24 h, RNA was harvested and RT-qPCR was performed to quantify mRNA for (a) TNFα, (b) IL-6 and (c) IL-1β. Cells from LysM-Cre-negative-LRP1fl/fl mice are shown with open bars; cells from LysM-Cre-positive-LRP1fl/fl mice are shown with closed bars (mean ± s.e.m.; n = 3; n.s., not significant, *p < 0.05, ***p < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). (d) Primary microglia was treated for up to 6 hours with LPS (100 ng/mL). Expression of TNFα, IL-6 and IL-1β mRNA was determined by RT-qPCR at the indicated times. mRNA levels for each cytokine were expressed as a percentage of the level of that same cytokine present at 6 h (mean ± s.e.m.; n = 3; **p < 0.01, ***p < 0.001, one-way ANOVA followed by Tukey’s post-hoc test).
Next, we performed studies to examine why LRP1 deficiency may selectively affect expression of IL-1β and IL-6 but not TNFα in LPS-treated microglia in culture. In mouse monocytes and glia, LPS rapidly activates TNFα gene transcription; TNFα protein is detected in conditioned medium within 1 h (Shakhov et al., 1990; Minogue et al., 2012). By contrast, IL-1β and IL-6 are produced in a secondary wave, which may be modulated by factors produced earlier in the LPS response (Ghezzi et al., 2000; Beurel and Jope, 2009; Minogue et al., 2012). We hypothesized that a factor or factors produced selectively by LRP1-expressing microglia early in the course of the response to LPS, including possibly sLRP1, contribute to the observed increases in expression of IL-1β and IL-6. In Fig. 1d, we examined expression of TNFα, IL-1β and IL-6 as a function of time in LPS-treated brain-derived microglia. Cytokine expression at early time points was plotted as a percentage of that detected at 6 h. TNFα mRNA was increased at 0.5 and 1 h. IL-1β mRNA was increased to a significantly lesser extent at 0.5 h, compared with TNFα, and IL-6 mRNA was not detected at 0.5 or 1.0 h. These results confirm, in our cell culture model system, that the mRNAs encoding IL-1β and IL-6 are expressed at later times compared with TNFα and thus, subject to regulatory signals generated earlier in the LPS response, including possibly sLRP1.
3.2 │. LPS and CSF-1 promote LRP1 shedding from spinal cord microglia
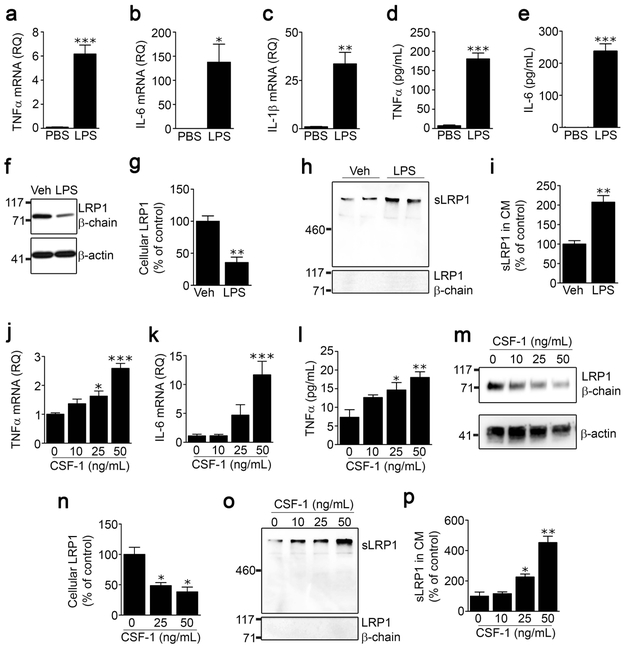
Microglia from different regions of the CNS vary phenotypically (Lai et al., 2012; Baskar Jesudasan et al., 2014). Because of our interest in the role of microglia in the response to PNS injury, we isolated microglia from mouse spinal cords and studied LRP1 shedding. To begin, we examined the response to LPS. Figs. 2a-c show that LPS (100 ng/mL) induced expression of the mRNAs encoding TNFα, IL-6, and IL-1β in spinal cord microglia, as anticipated. Analysis of protein in CM by ELISA confirmed that TNFα and IL-6 were increased in response to LPS (Fig. 2d, e).
FIGURE 2.
LPS and CSF-1 induce LRP1 shedding from spinal cord microglia. Microglia were isolated from spinal cords of adult C57BL/6J wild-type mice and cultured in low-serum medium. The cells were treated with LPS (100 ng/mL) or vehicle for 24 h. Expression of (a) TNFα, (b) IL-6 and (c) IL-1β was determined by RT-qPCR. (d, e) TNFα protein and IL-6 protein in CM were measured by ELISA (mean ± s.e.m. of 4 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t-test). (f) Cell extracts were subjected to immunoblot analysis to detect LRP1 β-chain (upper panel) and β-actin as a loading control (lower panel). (g) Densitometry was performed to compare cellular LRP1 in microglia before and after LPS treatment (mean ± s.e.m; n = 3 independent immunoblotting experiments; **p < 0.01, two-tailed unpaired t-test) (h) Conditioned medium was collected and subjected to RAP ligand-blotting to detect sLRP1 (upper panel). The same samples were subjected to immunoblot analysis using an antibody that detects an epitope in the LRP1 β-chain, absent in sLRP1 (lower panel). These immunoblots were performed concurrently with those shown in panel “f’ of this figure, providing a positive control. (i) Densitometry was performed to quantify sLRP1 in CM (mean ± s.e.m; n = 4 independent experiments; **p < 0.01, two-tailed unpaired t-test). (j, k) Microglia were treated with increasing concentrations of mouse CSF-1. RNA was isolated 24 h later and RT-qPCR was performed to determine expression of TNFα and IL-6. (l) CM was recovered. TNFα protein was determined by ELISA (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Dunnett’s post-hoc test). (m, n) Microglia were treated with the indicated concentrations of CSF-1. Cellular LRP1 was determined by immunoblot analysis using anti LRP1 β-chain antibody and quantified by densitometry (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, one-way ANOVA followed by Dunnett’s post-hoc test). (o) sLRP1 released into CM was detected by RAP ligand-blotting. The same samples were subjected to immunoblot analysis using LRP1 β-chain antibody. (p) Densitometry was performed to quantify sLRP1 in CM (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, **p < 0.01, one-way ANOVA followed by Dunnett’s post-hoc test).
Next, we examined LRP1 shedding and the effects of LPS on this process in spinal cord microglia. Treating cells with LPS for 24 h decreased the total abundance of cellular LRP1 in cell extracts, as determined by immunoblot analysis using an antibody that recognizes an intra-cytoplasmic epitope present in the LRP1 β-chain of intact membrane-anchored LRP1 but absent in sLRP1 (Fig. 2f). Densitometry analysis of three separate studies showed that cellular LRP1 was decreased by 64 ± 8% in LPS-treated cells (p < 0.01) (Fig. 2g). To detect LRP1 shedding from spinal cord microglia, CM from the same LPS-treated and control cells was examined by RAP ligand-blotting. RAP, which is expressed as a GST-fusion protein, binds directly to the 515-kDa LRP1 α-chain, which is intact in sLRP1 (Quinn et al., 1999) and is then detected using GST-specific antibody (Brifault et al., 2017). Fig. 2h shows that LRP1 α-chain was detected in CM from spinal cord microglia by RAP ligand-blotting and increased in CM recovered from LPS-treated cells. To confirm that the LRP1 α-chain, detected by RAP ligand-blotting, was derived from sLRP1 and not full-length cellular LRP1 in cell fragments, the equivalent CM preparations were subjected to immunoblot analysis using the LRP1 β-chain antibody. No signal was detected (Fig. 2h, lower panel). Densitometry analysis of four independent experiments showed that LPS increased sLRP1 in CM by 2.1 ± 0.2-fold (Fig. 2i). In control experiments, we determined by MTT assay that LPS does not affect microglial cell viability. Overall, these results show that LPS decreases the total abundance of cellular LRP1 in microglia, confirming the report by Marzolo et al (2000). Our results further show that the decrease in cellular LRP1 may be at least partially attributed to LRP1 shedding.
CSF-1 is a known activator of spinal cord microglia in vivo following peripheral nerve injury (Guan et al., 2016). We therefore tested whether CSF-1 induces LRP1 shedding from cultured spinal cord microglia. To confirm that CSF-1 activates spinal cord microglia, first we tested the effects of CSF-1 on expression of pro-inflammatory cytokines. Fig. 2j, k show that CSF-1 dose-dependently increased expression of the mRNAs encoding TNFα and IL-6. CSF-1 also increased accumulation of TNFα protein in CM (Fig. 2l). Immunoblot analysis of cell extracts using LRP1 β-chain antibody showed that CSF-1 decreased the abundance of cellular LRP1 in spinal cord microglia (Fig. 2m, n). This result was not anticipated because CSF-1 has been reported to increase LRP1 expression in peripheral macrophages (Hussaini et al., 1990). Analysis of CM from the same cell cultures showed that CSF-1 dose-dependently increased LRP1 shedding from spinal cord microglia (Fig. 2o, p). The effects of CSF-1 on LRP1 shedding from spinal cord microglia may explain its effects on the total abundance of cellular LRP1 protein.
3.3 │. LRP1 and proteinases that induce LRP1 shedding are up-regulated in the SDH following sciatic nerve injury.
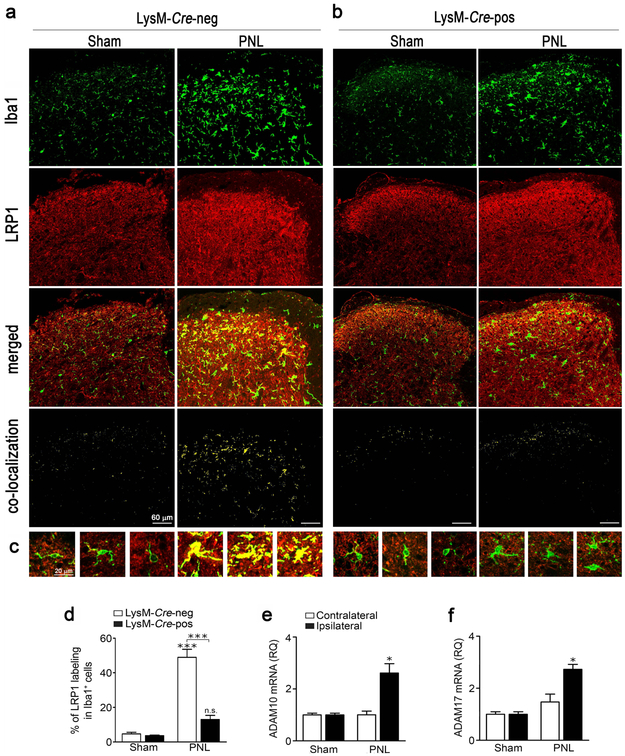
To study LRP1 expression in SDH microglia in situ, LysM-Cre-negative-LRP1fl/fl and LysM-Cre-positive-LRP1fl/fl mice were subjected to PNL or sham operation. Spinal cords were harvested 3 days later and cross sections of SDH tissue centered at L4 were immunostained for the microglial marker, Iba1 (green), and for LRP1 (red). In sham-operated LysM-Cre-negative-LRP1fl/fl mice, LRP1 immunopositivity in the SDH was diffuse (Fig. 3a, middle panels), as anticipated because diverse cell types in the CNS, including neurons, astrocytes, and oligodendrocytes, express LRP1 (Wolf et al., 1992; Bu et al., 1994; Gaultier et al., 2009; Auderset et al., 2016) . However, Iba1-immunopositive microglia were mostly LRP1-negative (see panel labeled “merged”). Trace co-localization of LRP1 with Iba-1-immunopositive cells was detected using IMARIS Coloc® (see panel labeled “co-localization”).
FIGURE 3.
Sciatic nerve PNL induces expression of microglial LRP1, ADAM10, and ADAM17 in the SDH. (a, b) LysM-Cre-positive-LRP1fl/fl or LysM-Cre-negative-LRP1fl/fl mice were subjected to PNL or sham-operation. Three days later, spinal cords were collected. Sections of the SDH at the L3-L4 level were immunostained to detect Iba1 (green) and LRP1 (red). Representative IF confocal microscopy images are presented for tissue harvested from (a) LysM-Cre-negative-LRP1fl/fl mice and (b) LysM-Cre-positive-LRP1fl/fl mice (n = 4/group). The merged images show cells that label for Iba1 and LRP1. Using IMARIS software, the degree of co-localization between LRP1 and Iba1 was calculated and a separate co-localization channel was built and shown here in yellow. (c) Representative images of single cell projections for the 4 experimental groups are shown. (d) The degree of co-localization of LRP1 with Iba1 is expressed as a percentage of the area that is Iba1-immunopositive which also was LRP1-immunopositive (mean ± s.e.m.; n = 4 animals/group; ***p < 0.001, signs directly over the bars compare PNL and sham operation in the same genotype; one-way ANOVA followed by Tukey’s post-hoc test). (e, f) C57BL6/J mice were subjected to PNL or sham-operation. Three days later, ipsilateral and contralateral SDH tissue (L3-L4 lumbar segments) was collected. RT-qPCR was performed to quantify expression of ADAM10 and ADAM17 mRNAs (mean ± s.e.m.; n = 3 for sham-operated animals and n = 6 for animals subjected to PNL; *p < 0.05 PNL vs Sham in the ipsilateral SDH, one-way ANOVA followed by Tukey’s post-hoc test).
When LysM-Cre-negative-LRP1fl/fl mice were subjected to PNL, Iba-1-immunopositivity was substantially increased, which may reflect an increase in the number of microglia, changes in microglial morphology such as hypertrophied cell bodies, and/or increased Iba1 expression (Sasaki et al., 2001). LRP1-immunopositivity appeared diffusely increased throughout the SDH following PNL and importantly, many microglia became LRP1-immunopositive, as indicated by the emergence of yellow cells in the “merged” and “co-localization” images (Fig. 3a, lower panels). Thus, in LysM-Cre-negative-LRP1fl/fl mice, LRP1 expression in SDH microglia appears to be induced by peripheral nerve injury, concomitant with microglial activation.
In sham-operated LysM-Cre-positive-LRP1fl/fl mice, Iba1 immunoreactivity appeared similar to that detected in LysM-Cre-negative-LRP1fl/fl animals (Fig. 3b). Minimal co-localization of LRP1 with Iba1 was observed. Following PNL, Iba1-immunopositivity increased, although perhaps to a lesser extent than in wild-type mice. PNL also caused a diffuse increase in LRP1 immunoreactivity throughout the SDH, as was observed in LysM-Cre-negative-LRP1fl/fl mice; however, in LysM-Cre-positive-LRP1fl/fl animals, Iba1-positive cells remained largely LRP1 immunonegative, as seen in the “co-localization” and “merged” images. Fig. 3c shows single cell projections from each of the four conditions. Co-localization of LRP1 with Iba1 is clearly present only in cells from LysM-Cre-negative-LRP1fl/fl mice subjected to PNL.
Image analysis was conducted to quantify LRP1 immunopositivity in Iba1-positive cells of each slide. In LysM-Cre-negative-LRP1fl/fl mice subjected to PNL, 49 ± 5% of the area occupied by Iba1 immunostain also stained positively for LRP1 (Fig. 3d). In LysM-Cre-positive-LRP1fl/fl mice subjected to PNL, only 13 ± 2% of the area occupied by Iba1 immunostain also stained positively for LRP1. These results demonstrate that in LysM-Cre-positive-LRP1fl/fl mice, LRP1 is largely deleted in SDH microglia; this deletion event may occur prior to or concomitant with microglial activation in response to PNL.
ADAM10 and ADAM17 are the major proteinases responsible for LRP1 shedding in the CNS (Liu et al., 2009; Shackleton et al., 2016). We therefore examined expression of ADAM10 and ADAM17, at the mRNA level, in SDH tissue, 3 days after PNL or sham-operation in wild-type mice. Both proteinases were significantly up-regulated by PNL on the ipsilateral side of the SDH (*p < 0.05) (Fig. 3e, f), compared with sham-operated animals. No change in expression of ADAM10 or ADAM17 was observed in contralateral SDH tissue.
3.4 │. LRP1 deletion in myeloid cells attenuates microglial activation in the SDH after PNL
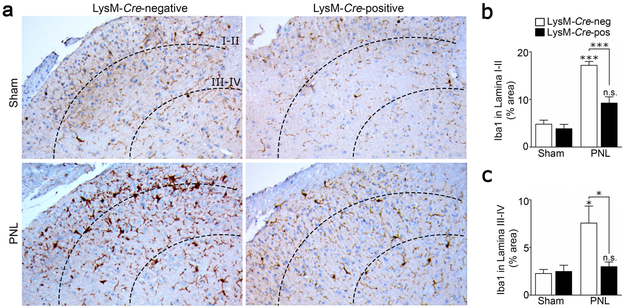
Upon activation, microglial cells acquire a hypertrophied or amoeboid morphology, proliferate, and express increased levels of Iba1 (Sasaki et al., 2001; Gu et al., 2016; Xu et al., 2016). Collectively, these processes lead to an increase in the density of the microglial marker Iba1 in the ipsilateral SDH following PNS injury, as determined by IHC. To test the hypothesis that LRP1 gene deletion in microglia in LysM-Cre-positive-LRP1fl/fl mice attenuates microglial activation, spinal cord tissue was harvested from LysM-Cre-positive-LRP1fl/fl and LysM-Cre-negative-LRP1fl/fl mice, 3 days after PNL or sham operation. IHC and image analysis were performed to quantitate the two-dimensional area occupied by Iba1 immunostaining in Laminae I-II and III-IV of the ipsilateral SDH, as previously described (Wirenfeldt et al., 2009).
Representative Iba1 IHC images are shown in Fig. 4a. In sham-operated LysM-Cre-negative-LRP1fl/fl and LysM-Cre-positive-LRP1fl/fl mice, Iba1 immunostaining in the SDH appeared approximately the same. After PNL, a substantial increase in the density of Iba1 immunostaining was observed selectively in LysM-Cre-negative-LRP1fl/fl mice. Quantitative analysis of Iba1-immunostaining confirmed this observation. Iba1 staining density was significantly increased in Laminae I-II of LysM-Cre-negative-LRP1fl/fl mice in response to PNL (Fig. 4b). Although a modest increase in Iba staining in Laminae I-II also was observed following PNL in LysM-Cre-positive-LRP1fl/fl mice, the increase did not attain statistical significance. Most importantly, Iba1 immunopositivity was significantly lower in LysM-Cre-positive-LRP1fl/fl mice subjected to PNL compared with LysM-Cre-negative-LRP1fl/fl mice subjected to PNL (***p < 0.001). The equivalent result was obtained when we analyzed Iba1 immunopositivity in Laminae III-IV (Fig. 4c, *p < 0.05). We conclude that following PNL, microglial activation in vivo in the SDH is attenuated when LRP1 is deleted under the control of the LysM promoter.
FIGURE 4.
LRP1 deletion attenuates microglial activation in the SDH following PNS injury. (a) Representative images showing Iba1 IHC on the ipsilateral SDH (L3-L4) 3 days after sciatic nerve PNL or sham operation in LysM-Cre-positive-LRP1fl/fl and LysM-Cre-negative-LRP1fl/fl mice. Laminae I-II and Laminae III-IV are delineated by dotted lines. (b, c) The percentage of tissue immunostained with Iba1 in Lamina I-II and Lamina III-IV following PNL or sham operation was determined for both mouse genotypes by image analysis (mean ± s.e.m.; n = 4 sham-operated animals, n = 6 LysM-Cre-negative-LRP1fl/fl mice subjected to PNL and n = 9 LysM-Cre-positive-LRP1fl/fl animals subjected to PNL; *p < 0.05, ***p < 0.001; one-way ANOVA followed by Tukey’s post-hoc analysis).
3.5 │. TAM-induced LRP1 deficiency in microglia in adult mice attenuates microglial activation following PNL
To further study the activity of microglial LRP1 following PNL, we established a second model system in which LRP1fl/fl mice express CreERT under the control of the Cx3cr1 promoter (Goldmann et al., 2013; Chuang et al., 2016). To activate Cre recombinase, adult mice were treated with TAM (150 mg/kg, IP) twice, 1 week apart. Because Cx3cr1 is expressed in cells of myeloid lineage, CreERT is activated and LRP1 is initially deleted in monocytes and macrophages, as well as microglia. However, the pool of circulating monocytes is constantly renewed while long-lived microglia are not replenished. Thus, 4 weeks after TAM administration, LRP1 expression in microglia should still be neutralized whereas LRP1 expression in monocytes and macrophages should be restored (Goldmann et al., 2013). For this reason, the Cx3cr1-CreERT model system is thought to confer greater specificity for studying microglia compared with the LysM-Cre model system. Moreover, because gene deletion occurs in adult mice, this model system precludes genetic compensation that may occur when gene deletion occurs during development. In our experiments, control mice carried the Cx3cr1-CreERT transgene, but had wild-type (non-floxed) LRP1. Control Cx3cr1-CreERT-LRP1 mice were treated with TAM, equivalently to Cx3cr1-CreERT-LRP1fl/fl mice.
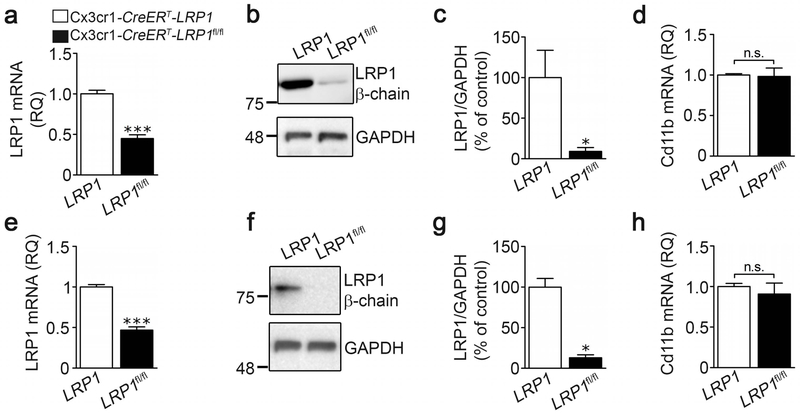
To characterize this second model system, microglia were isolated from the brains of Cx3cr1-CreERT-LRP1fl/fl mice (LRP1fl/fl) and control Cx3cr1-CreERT-LRP1 (LRP1) mice, four weeks after TAM treatment. LRP1 mRNA expression was determined without establishing the cells in culture. Brain-derived microglia from Cx3cr1-CreERT-LRP1fl/fl mice demonstrated a 65 ± 5% decrease in LRP1 mRNA, compared with microglia from Cx3cr1-CreERT-LRP1 mice (Fig. 5a). LRP1 protein was decreased by 91 ± 5%, as determined by immunoblot analysis (Fig. 5b) and densitometry analysis of four replicates (Fig. 5c). Fig. 5d shows that CD11b mRNA expression was not changed in microglia from Cx3cr1-CreERT-LRP1fl/fl mice. In control experiments, we determined that LRP1 expression is equivalent when we compared brain-derived microglia isolated from Cx3cr1-CreERT-LRP1fl/fl mice and Cx3cr1-CreERT-LRP1 mice treated with vehicle (5% EtOH and corn oil) instead of TAM (Supplementary Figure S1a, b). Thus, Cx3cr1-CreERT is silent in the absence of TAM. LRP1 expression also was equivalent when we compared cells isolated from control Cx3cr1-CreERT-LRP1 mice treated with TAM versus vehicle (results not shown), demonstrating that TAM does not independently regulate LRP1 expression in microglia.
FIGURE 5.
Characterization of microglia isolated from the brains and spinal cords of Cx3cr1-CreERT-LRP1fl/fl mice. (a) Microglia were isolated from brains of TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice (black bars, LRP1fl/fl) and Cx3cr1-CreERT-LRP1 mice (open bars, LRP1). RT-qPCR was performed to determine the relative abundance of LRP1 mRNA (mean ± s.e.m.; n = 4 independent experiments; ***p < 0.001, unpaired two-tailed t-test). (b) Immunoblot analysis was performed to determine cellular LRP1 protein using β-chain-specific antibody. Membranes were re-probed for GAPDH as a loading control. (c) Densitometry was performed to determine the relative level of cellular LRP1 protein, standardized against the loading control (mean ± s.e.m.; n = 4; *p < 0.05, unpaired t-test). (d) RNA was isolated from brain microglia. RT-qPCR was performed to determine the relative abundance of CD11b mRNA (mean ± s.e.m.; n = 4; p = 0.88, unpaired two-tailed t-test). (e) Microglia were isolated from spinal cords of TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice (black bars, LRP1fl/fl) and Cx3cr1-CreERT-LRP1 mice (open bars, LRP1). RT-qPCR was performed to determine the relative abundance of LRP1 mRNA (mean ± s.e.m.; n = 4; ***p < 0.001, unpaired two-tailed t-test). (f) Protein extracts from spinal cord microglia were immunoblotted to detect LRP1 β-chain and GAPDH, as a loading control. (g) Densitometry was performed to determine the relative level of LRP1 protein, standardized against the loading control (mean ± s.e.m.; n = 4; *p < 0.05, unpaired t-test). (h) RNA was isolated from spinal microglia. RT-qPCR was performed to determine CD11b mRNA (no statistical difference was observed using one-way ANOVA: p = 0.53).
Next, we examined microglia harvested from spinal cords. In cells from TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice, LRP1 mRNA was decreased by 53 ± 4% (Fig. 5e). LRP1 protein was decreased by 87 ± 4%, as determined by immunoblot analysis (Fig. 5f) and densitometry (Fig. 5g). cd11b mRNA was unchanged in spinal cord microglia from Cx3cr1-CreERT-LRP1fl/fl mice compared with Cx3cr1-CreERT-LRP1 mice (Fig. 5h).
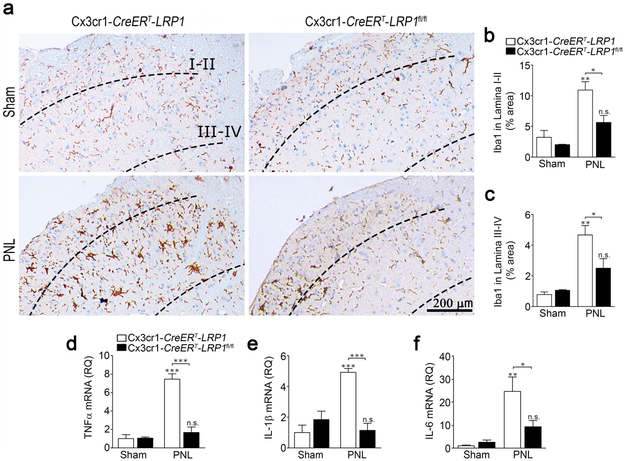
Having confirmed that the Cx3cr1-CreERT model system is effective, Cx3cr1-CreERT-LRP1fl/fl and control Cx3cr1-CreERT-LRP1 mice were subjected to PNL or sham operation 4 weeks after TAM treatment. Fig. 6a shows that once again, in this model system, following sham operation, Iba1 immunostaining in the SDH was similar in LRP1-deficient and control animals. After PNL, a significant increase in Iba1 density was apparent in the ipsilateral SDH of control Cx3cr1-CreERT-LRP1 mice in both Laminae I-II (Fig. 6b) and Laminae III-IV (Fig. 6c). In Cx3cr1-CreERT-LRP1fl/fl mice, although a trend towards increased Iba1 immunostaining was observed following PNL, the increase did not attain statistical significance. Most importantly, once again in this model system, Iba1 density was significantly lower in Cx3cr1-CreERT-LRP1fl/fl mice subjected to PNL compared with Cx3cr1-CreERT-LRP1 mice subjected to PNL in both Laminae I-II and Laminae III-IV. These results suggest that in two distinct mouse model systems, LRP1 deficiency in microglia attenuates microglial activation in response to peripheral nerve injury.
FIGURE 6.
LRP1 deletion in microglia in adult mice attenuates microglial activation in the SDH following PNL. (a) TAM-treated Cx3cr1-CreERT-LRP1fl/fl (LPR1fl/fl) mice and Cx3cr1-CreERT-LRP1 (LRP1) mice were subjected to PNL or sham-operation. SDH tissue was isolated 3 days later. Representative IHC images for Iba1 are shown. (b, c) Densitometry analysis was performed to assess the percentage of tissue area occupied by Iba1 immunostaining in Laminae I-II and in Laminae III-IV (mean ± s.e.m.; n = 3 for sham-operated animals and n = 4 for animals subjected to PNL;*p < 0.05, **p < 0.01, n.s. not significant, the signs directly over the bars compare PNL with sham-operation in the same genotype; one-way ANOVA followed by Tukey’s post-hoc analysis). (d-f) RNA was harvested from the ipsilateral SDH. RT-qPCR was performed to quantify mRNAs encoding (d) TNFα, (e) IL-1β, and (f) IL-6 (mean ± s.e.m.; n = 4/group; *p < 0.05, **p < 0.01, ***p < 0.001, the stars directly over the bars compare PNL with sham operation in the same genotype; one-way ANOVA followed by Tukey’s post-hoc analysis).
3.6 │. Spinal cord inflammation is decreased following PNL in mice with LRP1-deficient microglia
We subjected TAM-treated Cx3cr1-CreERT-LRP1fl/fl and control Cx3cr1-CreERT-LRP1 mice to PNL or sham operation and harvested ipsilateral SDH three days later. RNA was isolated and the mRNAs encoding various pro-inflammatory mediators were determined. Fig. 6d shows that PNL significantly increased TNFα mRNA expression in Cx3cr1-CreERT-LRP1 mice but not in Cx3cr1-CreERT-LRP1fl/fl mice. Similar results were obtained when we examined IL-1β mRNA expression (Fig. 6e) and IL-6 mRNA expression (Fig. 6f). For all three cytokines, the mRNA level following PNL was significantly higher in Cx3cr1-CreERT-LRP1 mice compared with Cx3cr1-CreERT-LRP1fl/fl mice. When Cx3cr1-CreERT-LRP1fl/fl and control Cx3cr1-CreERT-LRP1 mice were pre-treated with vehicle instead of TAM, so microglial LRP1 expression was unaltered, IL-6 mRNA expression was equivalent in the two mouse strains, before and after PNL (Supplementary Fig. S1c). As a further control, we examined cytokine expression on the contralateral side of TAM-treated Cx3cr1-CreERT-LRP1fl/fl and Cx3cr1-CreERT-LRP1 mice. Significant changes in expression of TNFα and IL-6 were not observed (Supplementary Fig. S2). The design of our experiments precluded determining the cellular source of the pro-inflammatory cytokine mRNAs. However, these results demonstrate that microglial LRP1 amplifies the overall pro-inflammatory response observed in the SDH following PNL.
3.7 │. Macrophage infiltration into injured sciatic nerves is not altered in TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice
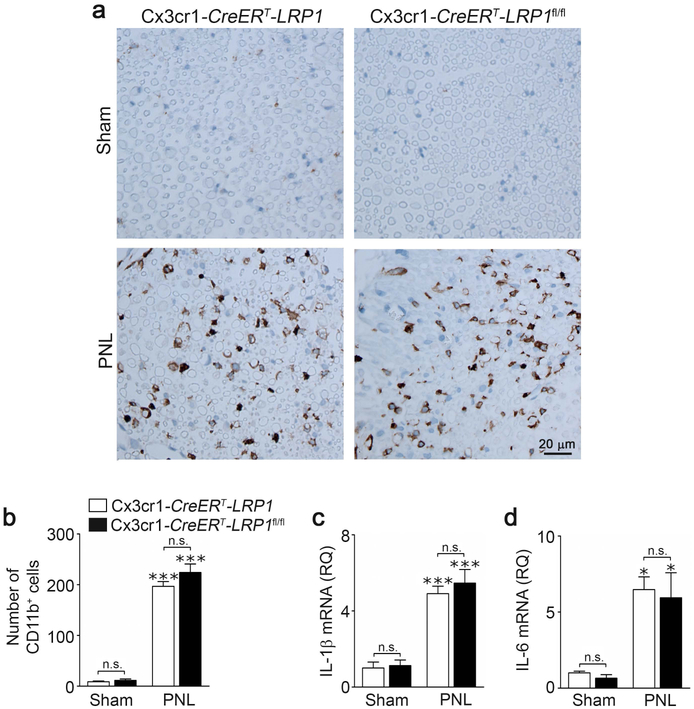
LRP1-deficient peripheral monocytes and macrophages accumulate at sites of inflammation in greater abundance (Overton et al., 2007; Staudt et al., 2013). Although the Cx3cr1-CreERT model system is designed to restore gene expression in monocytes four weeks after TAM treatment (Goldmann et al., 2013; Chuang et al., 2016), we performed control experiments to confirm that macrophage infiltration and inflammatory mediator expression were not altered in injured sciatic nerves in Cx3cr1-CreERT-LRP1fl/fl mice, under the conditions of our experiments.
Fig. 7a shows that very few CD11b-positive cells were detected in sciatic nerves in sham-operated Cx3cr1-CreERT-LRP1 or Cx3cr1-CreERT-LRP1fl/fl mice. When Cx3cr1-CreERT-LRP1 or Cx3cr1-CreERT-LRP1fl/fl mice were subjected to PNL and studied 3 days later, nerve tissue 3 mm distal from the injury site was infiltrated with CD11b-positive cells equivalently (Fig. 7a, b). Expression of the pro-inflammatory cytokines, IL-1β (Fig. 7c) and IL-6 (Fig. 7d), also was equivalent in distal sciatic nerve tissue from Cx3cr1-CreERT-LRP1 and Cx3cr1-CreERT-LRP1fl/fl mice harvested 3 days after PNL. These results suggest that the changes in microglial activation and SDH cytokine expression, observed in Cx3cr1-CreERT-LRP1fl/fl mice, are not secondary to changes occurring in the injured sciatic nerve.
FIGURE 7.
TAM-induced LRP1 deficiency in microglia does not alter myeloid cell infiltration or inflammation in injured sciatic nerves. (a) TAM-treated Cx3cr1-CreERT-LRP1fl/fl (LRP1fl/fl) mice and Cx3cr1-CreERT-LRP1 (LRP1) mice were subjected to PNL or sham-operation. Sciatic nerve tissue distal to the injury site was harvested 3 days later. The area of the nerve directly downstream of the ligation was immunostained to detect the myeloid cell marker CD11b. Images represent n = 3 sham-operated animals and 4 mice subjected to PNL for each genotype. (b) The number of CD11b-positive cells present in the nerves was counted. Although PNL significantly increased the number of cells, there was no change with mouse genotype (mean ± s.e.m.; n = 3 sham-operated animals and n = 4 mice subjected to PNL; ***p < 0.001, n.s., not significant, the stars directly over the bars refer to a comparison of PNL with sham operation in the same genotype, one-way ANOVA followed by Tukey’s post-hoc analysis). (c, d) RNA was harvested from distal sciatic nerve tissue. RT-qPCR was performed to determine expression of IL-1β and IL-6 (mean ± s.e.m.; n = 3 sham-operated animals and n = 4 mice subjected to PNL; *p < 0.05, ***p < 0.001, n.s., not significant using one-way ANOVA followed by Tukey’s post-hoc test).
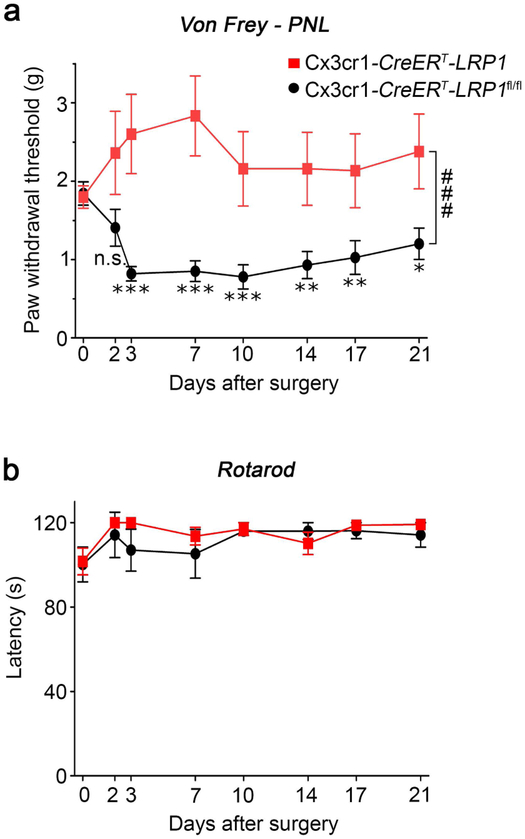
3.8 │. LRP1 deficiency in microglia blocks development of allodynia
To determine whether LRP1 deficiency in microglia alters pain-related behavior following PNS injury, cohorts of nine TAM-treated Cx3cr1-CreERT-LRP1 and Cx3cr1-CreERT-LRP1fl/fl mice were subjected to PNL. Tactile allodynia was assessed using von Frey filaments. The 50% paw withdrawal threshold (PWT) was determined (Chaplan et al., 1994). Prior to injury, sensitivity to punctate stimulation of the ipsilateral hind paw was similar in Cx3cr1-CreERT-LRP1 and Cx3cr1-CreERT-LRP1fl/fl mice (Fig. 8a). Three days after PNL, control Cx3cr1-CreERT-LRP1 mice developed significantly reduced PWTs, indicating development of tactile allodynia (Vogel et al., 2006). The PWTs remained significantly different from those recorded prior to surgery throughout the 3 weeks study. By contrast, tactile allodynia did not develop in Cx3cr1-CreERT-LRP1fl/fl mice. Comparison of the two curves by two-way repeated measures ANOVA demonstrated a highly significant difference in tactile allodynia in Cx3cr1-CreERT-LRP1fl/fl mice following PNL, compared with control animals (###p < 0.0001).
FIGURE 8.
LRP1 deletion in microglia prevents development of allodynia after sciatic nerve PNL. (a) TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice (red curve) and Cx3cr1-CreERT-LRP1 mice (black curve) were baseline tested with von Frey hairs for three days and then subjected to PNL. The mean baseline value for each group is presented at Day 0. PWTs were determined on the indicated days throughout 3 weeks (mean ± s.e.m; n = 9/group; *p < 0.05, **p < 0.01, ***p < 0.001 as compared with the corresponding basal PWTs; ###p < 0.0001 as compared with control mice; repeated measures ANOVA with post-hoc Bonferroni’s test). (b) TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice (red curve) and Cx3cr1-CreERT-LRP1 mice (black curve) were baseline tested and then subjected to Rotarod-testing (mean ± s.e.m.; n = 5; p = 0.5, repeated measures ANOVA). Significant differences between the two genotypes were not observed.
As a control, groups of 4 TAM-treated Cx3cr1-CreERT-LRP1 and Cx3cr1-CreERT-LRP1fl/fl mice were subjected to sham operation. Through the duration of the study, significant changes in the PWT were not detected in either cohort (Supplementary Fig. S3a). In a separate control study, we compared Cx3cr1-CreERT-LRP1 mice that were treated with TAM or vehicle and then subjected to PNL. In these mice with wild-type LRP1, TAM treatment did not affect PWTs (Supplementary Fig. S3b). Motor function was not disturbed by PNL in TAM-treated Cx3cr1-CreERT-LRP1fl/fl or Cx3cr1-CreERT-LRP1 mice, as determined by Rotarod testing (Fig. 8b). Collectively, these results demonstrate that the changes in microglial activation and neuro-inflammation, observed when mice with LRP1-deficient microglia are subjected to sciatic nerve injury, are associated with a block in the development of tactile allodynia.
4 │. DISCUSSION
Following injury to the PNS, extracellular mediators produced by cells in the SDH, including many pro-inflammatory mediators, establish crosstalk that may lead to the development of, maintain, and/or amplify neuropathic pain (Scholz and Woolf, 2007). However, therapeutic trials in which anti-inflammatory strategies have been applied to treat clinical pain have not yet been successful (Nguyen et al., 2015; Vanelderen et al., 2015). Therefore, identifying novel molecular pathways that regulate neuropathic pain is an important objective for future drug discovery. Herein, we studied mice in which a single gene, LRP1, was deleted conditionally in microglia. The gene deletion event resulted in a decrease in the ability of microglia to mount a complete response to a pro-inflammatory challenge (LPS) when the cells were cultured in vitro, suggesting an essential role of LRP1 in microglia activation. When mice with LRP1-deficient microglia were subjected to sciatic nerve PNL, microglial activation and neuro-inflammation in the ipsilateral SDH were substantially attenuated. Strikingly, development of tactile allodynia was blocked. Although membrane-anchored LRP1 is considered anti-inflammatory (Pocivavsek et al., 2009b, 2009a; Staudt et al., 2013; Chuang et al., 2016; Mantuano et al., 2016), LRP1 shedding converts LRP1 into a potently pro-inflammatory soluble product that activates microglia (Brifault et al., 2017). Because our results demonstrate that LRP1 is shed by spinal cord microglia, most likely as a consequence of microglial activation, sLRP1 may represent a promising new molecular target for treating neuropathic pain.
The effects of LRP1 deficiency on spinal cord microglial activation were similar in two gene deletion model systems. In the LysM-Cre model system, gene deletion occurs in cells of myeloid origin during development (Clausen et al., 1999) although, in microglia, genes may be deleted when the cells become activated, such as after PNL (Coyle, 1998). In the Cx3cr1-CreERT model system, gene deletion occurs only in adults that are treated with TAM (Goldmann et al., 2013). Thus, the likelihood that other gene regulatory events compensated for LRP1 deletion was decreased in Cx3cr1-CreERT-LRP1fl/fl mice, compared with LysM-Cre-positive-LRP1fl/fl mice.
Our observation that LRP1 deficiency in microglia inhibits the development of tactile allodynia following PNL was made using the Cx3cr1-CreERT model system. In control experiments, we ruled out changes in macrophage function outside the CNS as contributing to the observed PWTs. This was an anticipated result because, in the Cx3cr1-CreERT model system, four weeks after TAM treatment, LRP1 deficiency should be limited to microglia and not peripheral monocytes that infiltrate injured nerves (Goldmann et al., 2013; Chuang et al., 2016). From our results, we draw the following important conclusions. First and foremost, our results confirm that microglia do indeed regulate pain processing. Second, microglial activation and development of neuro-inflammation in the SDH are correlated with development of neuropathic pain-related behavior. Third, a single gene deletion event in microglia is sufficient to prevent development of allodynia in response to PNS injury.
Because, in cells of myeloid origin, membrane-anchored LRP1 expresses anti-inflammatory activity (Overton et al., 2007; Pocivavsek et al., 2009b, 2009a; Gorovoy et al., 2010; May et al., 2013; Staudt et al., 2013; Mantuano et al., 2016; Yang et al., 2016; Brifault et al., 2017), proteolytic shedding of LRP1 serves as a key reaction that converts the activity of LRP1 from anti-inflammatory to pro-inflammatory (Gorovoy et al., 2010; Brifault et al., 2017). However, to date, in in vivo model systems, only the anti-inflammatory activity of LRP1 has been observed (Overton et al., 2007; Staudt et al., 2013; Chuang et al., 2016; Mantuano et al., 2016). The results presented here demonstrate effects of microglial LRP1 that may be characterized as “pro-inflammatory” in vivo and the preponderance of our evidence suggests that the observed effect of LRP1 on microglial activation and SDH neuro-inflammation are due to sLRP1. Agents known to activate spinal cord microglia, including LPS and CSF-1, induced LRP1 shedding from spinal cord microglia. Furthermore, in response to PNL, proteinases known to cause LRP1 shedding in the CNS were expressed at increased levels exclusively on the ipsilateral side of the SDH. We previously demonstrated that purified sLRP1 induces expression of multiple pro-inflammatory cytokines, including TNFα, IL-6 and IL-1β when injected into the spinal cord, mimicking the activity of sLRP1 in cell culture (Brifault et al., 2017). We now show that expression of the equivalent cytokines is decreased in the SDH following PNL in mice with LRP1-deficient microglia. TNFα, IL-6 and IL-1β are known to modulate spinal cord synaptic transmission and play an important role in central sensitization (Obreja et al., 2002; Kawasaki et al., 2008; Liu et al., 2017). Thus, the absence of demonstrable tactile allodynia in TAM-treated Cx3cr1-CreERT-LRP1fl/fl mice may reflect either direct interaction of sLRP1 with SDH neurons or changes in the local concentration of other factors, such as TNFα, IL-6, and IL-1β, secondary to sLRP1-induced gene regulatory events. Because sLRP1 is a soluble protein, it may function in an autocrine or paracrine manner. Whether astrocytes and neurons respond to sLRP1 directly remains to be tested.
Some factors known to promote LRP1 shedding from macrophages and microglia, such as LPS and interferon-γ, also down-regulate LRP1 expression at the transcriptional level (LaMarre et al., 1993; Hussaini et al., 1996; Marzolo et al., 2000). LRP1 shedding and transcriptional down-regulation both decrease the abundance of membrane-anchored LRP1, which has anti-inflammatory properties, and thus may be viewed as having the common purpose of sustaining pro-inflammatory mediator production. At the same time, decreased LRP1 protein expression may decrease sLRP1 production. Other factors such as CSF-1, which we now understand promotes LRP1 shedding, may increase LRP1 expression (Hussaini et al., 1990). Under conditions in which LRP1 shedding is activated, this pattern may be more pro-inflammatory due to replenishing of the substrate for sLRP1 generation.
Identifying novel therapeutic approaches for treating chronic pain in patients is an important medical goal. The proteinases implicated in LRP1 shedding target other membrane proteins as substrates as well. In particular, ADAM17 cleaves pro-TNF and releases from the plasma membrane the well-described pro-inflammatory cytokine, TNFα (Black et al., 1997; Lunn et al., 1997). sLRP1 may therefore be a member of a pro-inflammatory protein network, released from membrane-anchored precursors when proteinases such as ADAM10 and ADAM17 are expressed and activated. We suggest that membrane protein shedding may be targeted for pain therapeutics development. Our results implicating microglial LRP1 in the pathogenesis of neuropathic pain justify future work to further characterize reactions that lead to LRP1 shedding, the full continuum of membrane proteins that may be shed simultaneously, and approaches for limiting this process.
Supplementary Material
Main Points:
LRP1 deletion in microglia attenuates microglial activation, spinal inflammation, and neuropathic pain after sciatic nerve injury in mice.
The pro-inflammatory activity of microglial LRP1 is explained by shed LRP1, a soluble “cytokine-like” factor.
Acknowledgements:
We are grateful to Dr. Don Pizzo for assistance with the immunohistochemistry studies and Mr. Michael Banki for his technical help.
Funding:
Grants R01 HL136395 and R01 NS097590 from the National Institutes of Health and Grant 1I01RX002484 from the Veterans Administration.
Footnotes
Conflict of interest
The authors declare no competing financial interests or conflicts of interest.
References
- Auderset L, Cullen CL, Young KM (2016) Low Density Lipoprotein-Receptor Related Protein 1 Is Differentially Expressed by Neuronal and Glial Populations in the Developing and Mature Mouse Central Nervous System. PLoS One 11:e0155878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar Jesudasan SJ, Todd KG, Winship IR (2014) Reduced Inflammatory Phenotype in Microglia Derived from Neonatal Rat Spinal Cord versus Brain Stangel M, ed. PLoS One 9:e99443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Jope RS (2009) Lipopolysaccharide-induced interleukin-6 production is controlled by glycogen synthase kinase-3 and STAT3 in the brain. J Neuroinflammation 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RA et al. (1997) A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385:729–733. [DOI] [PubMed] [Google Scholar]
- Bouhassira D, Attal N (2018) Emerging therapies for neuropathic pain: new molecules or new indications for old treatments? Pain 159:576–582. [DOI] [PubMed] [Google Scholar]
- Brifault C, Gilder AS, Laudati E, Banki M, Gonias SL (2017) Shedding of membrane-associated LDL receptor-related protein-1 from microglia amplifies and sustains neuroinflammation. J Biol Chem 292:18699–18712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G, Maksymovitch E a, Nerbonne JM, Schwartz a L (1994) Expression and function of the low density lipoprotein receptor-related protein (LRP) in mammalian central neurons. J Biol Chem 269:18521–18528. [PubMed] [Google Scholar]
- Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL (1994) Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 53:55–63. [DOI] [PubMed] [Google Scholar]
- Cho I-H, Hong J, Suh EC, Kim JH, Lee H, Lee JE, Lee S, Kim C-H, Kim DW, Jo E-K, Lee KE, Karin M, Lee SJ (2008) Role of microglial IKKbeta in kainic acid-induced hippocampal neuronal cell death. Brain 131:3019–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T-Y, Guo Y, Seki SM, Rosen AM, Johanson DM, Mandell JW, Lucchinetti CF, Gaultier A (2016) LRP1 expression in microglia is protective during CNS autoimmunity. Acta Neuropathol Commun 4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. [DOI] [PubMed] [Google Scholar]
- Cohen SP, Mao J (2014) Neuropathic pain: mechanisms and their clinical implications. BMJ 348:f7656. [DOI] [PubMed] [Google Scholar]
- Coyle DE (1998) Partial peripheral nerve injury leads to activation of astroglia and microglia which parallels the development of allodynic behavior. Glia 23:75–83. [PubMed] [Google Scholar]
- Dixon WJ (1980) Efficient Analysis of Experimental Observations. Annu Rev Pharmacol Toxicol 20:441–462. [DOI] [PubMed] [Google Scholar]
- Dworkin RH et al. (2003) Advances in Neuropathic Pain. Arch Neurol 60:1524. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Wu X, Le Moan N, Takimoto S, Mukandala G, Akassoglou K, Campana WM, Gonias SL (2009) Low-density lipoprotein receptor-related protein 1 is an essential receptor for myelin phagocytosis. J Cell Sci 122:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P, Sacco S, Agnello D, Marullo A, Caselli G, Bertini R (2000) LPS induces IL-6 in the brain and in serum largely through TNF production. Cytokine 12:1205–1210. [DOI] [PubMed] [Google Scholar]
- Gilron I, Baron R, Jensen T (2015) Neuropathic Pain: Principles of Diagnosis and Treatment. Mayo Clin Proc 90:532–545. [DOI] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Müller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, Luedde T, Heikenwalder M, Jung S, Prinz M (2013) A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16:1618–1626. [DOI] [PubMed] [Google Scholar]
- Gonias SL, Campana WM (2014) LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am J Pathol 184:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonias SL, Gaultier A, Jo M (2011) Regulation of the Urokinase Receptor (uPAR) by LDL Receptor-related Protein-1 (LRP1). Curr Pharm Des 17:1962–1969. [DOI] [PubMed] [Google Scholar]
- Gorovoy M, Gaultier A, Campana WM, Firestein GS, Gonias SL (2010) Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J Leukoc Biol 88:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace PM, Hutchinson MR, Maier SF, Watkins LR (2014) Pathological pain and the neuroimmune interface. Nat Rev Immunol 14:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu N, Peng J, Murugan M, Wang X, Eyo UB, Sun D, Ren Y, DiCicco-Bloom E, Young W, Dong H, Wu L-J (2016) Spinal Microgliosis Due to Resident Microglial Proliferation Is Required for Pain Hypersensitivity after Peripheral Nerve Injury. Cell Rep 16:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Kuhn JA, Wang X, Colquitt B, Solorzano C, Vaman S, Guan AK, Evans-Reinsch Z, Braz J, Devor M, Abboud-Werner SL, Lanier LL, Lomvardas S, Basbaum AI (2016) Injured sensory neuron–derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat Neurosci 19:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Goldstein JL, Strickland DK, Ho YK, Brown MS (1991) 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. J Biol Chem 266:21232–21238. [PubMed] [Google Scholar]
- Herz J, Strickland DK (2001) LRP: a multifunctional scavenger and signaling receptor. J Clin Invest 108:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussaini IM, LaMarre J, Lysiak JJ, Karns LR, VandenBerg SR, Gonias SL (1996) Transcriptional regulation of LDL receptor-related protein by IFN-gamma and the antagonistic activity of TGF-beta(1) in the RAW 264.7 macrophage-like cell line. J Leukoc Biol 59:733–739. [DOI] [PubMed] [Google Scholar]
- Hussaini IM, Srikumar K, Quesenberry PJ, Gonias SL (1990) Colony-stimulating factor-1 modulates alpha 2-macroglobulin receptor expression in murine bone marrow macrophages. J Biol Chem 265:19441–19446. [PubMed] [Google Scholar]
- Kawasaki Y, Zhang L, Cheng J-K, Ji R-R (2008) Cytokine mechanisms of central sensitization: distinct and overlapping role of interleukin-1beta, interleukin-6, and tumor necrosis factor-alpha in regulating synaptic and neuronal activity in the superficial spinal cord. J Neurosci 28:5189–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Dhami KS, Dibal CD, Todd KG (2012) Neonatal rat microglia derived from different brain regions have distinct activation responses. Neuron Glia Biol 7:5–16. [DOI] [PubMed] [Google Scholar]
- LaMarre J, Wolf BB, Kittler EL, Quesenberry PJ, Gonias SL (1993) Regulation of macrophage alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein by lipopolysaccharide and interferon-gamma. J Clin Invest 91:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang J, Tran H, Verbeek MM, Reiss K, Estus S, Bu G (2009) LRP1 shedding in human brain: roles of ADAM10 and ADAM17. Mol Neurodegener 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou L-J, Wang J, Li D, Ren W-J, Peng J, Wei X, Xu T, Xin W-J, Pang R-P, Li Y-Y, Qin Z-H, Murugan M, Mattson MP, Wu L-J, Liu X-G (2017) TNF-α Differentially Regulates Synaptic Plasticity in the Hippocampus and Spinal Cord by Microglia-Dependent Mechanisms after Peripheral Nerve Injury. J Neurosci 37:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn CA, Fan X, Dalie B, Miller K, Zavodny PJ, Narula SK, Lundell D (1997) Purification of ADAM 10 from bovine spleen as a TNFalpha convertase. FEBS Lett 400:333–335. [DOI] [PubMed] [Google Scholar]
- Mantuano E, Brifault C, Lam MS, Azmoon P, Gilder AS, Gonias SL (2016) LDL receptor-related protein-1 regulates NFκB and microRNA-155 in macrophages to control the inflammatory response. Proc Natl Acad Sci U S A 113:1369–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantuano E, Lam MS, Gonias SL (2013) LRP1 assembles unique co-receptor systems to initiate cell signaling in response to tissue-type plasminogen activator and myelin-associated glycoprotein. J Biol Chem 288:34009–34018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand F, Perretti M, McMahon SB (2005) Role of the Immune system in chronic pain. Nat Rev Neurosci 6:521–532. [DOI] [PubMed] [Google Scholar]
- Marzolo MP, von Bernhardi R, Bu G, Inestrosa NC (2000) Expression of alpha(2)-macroglobulin receptor/low density lipoprotein receptor-related protein (LRP) in rat microglial cells. J Neurosci Res 60:401–411. [DOI] [PubMed] [Google Scholar]
- May P, Bock HH, Nofer J-R (2013) Low density receptor-related protein 1 (LRP1) promotes anti-inflammatory phenotype in murine macrophages. Cell Tissue Res 354:887–889. [DOI] [PubMed] [Google Scholar]
- Minogue AM, Barrett JP, Lynch MA (2012) LPS-induced release of IL-6 from glia modulates production of IL-1β in a JAK2-dependent manner. J Neuroinflammation 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C, Palazzo C, Grabar S, Feydy A, Sanchez K, Zee N, Quinquis L, Ben Boutieb M, Revel M, Lefèvre-Colau M-M, Poiraudeau S, Rannou F (2015) Tumor necrosis factor-α blockade in recurrent and disabling chronic sciatica associated with post-operative peridural lumbar fibrosis: results of a double-blind, placebo randomized controlled study. Arthritis Res Ther 17:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodemova M, Watters JJ (2012) Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J Neuroinflammation 9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obreja O, Rathee PK, Lips KS, Distler C, Kress M (2002) IL-1β potentiates heat-activated currents in rat sensory neurons: involvement of IL-1RI, tyrosine kinase, and protein kinase C. FASEB J 16:1497–1503. [DOI] [PubMed] [Google Scholar]
- Orita S, Henry K, Mantuano E, Yamauchi K, De Corato A, Ishikawa T, Feltri ML, Wrabetz L, Gaultier A, Pollack M, Ellisman M, Takahashi K, Gonias SL, Campana WM (2013) Schwann cell LRP1 regulates remak bundle ultrastructure and axonal interactions to prevent neuropathic pain. J Neurosci 33:5590–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton CD, Yancey PG, Major AS, Linton MF, Fazio S (2007) Deletion of Macrophage LDL Receptor-Related Protein Increases Atherogenesis in the Mouse. Circ Res 100:670–677. [DOI] [PubMed] [Google Scholar]
- Pocivavsek A, Burns MP, Rebeck GW (2009a) Low-density lipoprotein receptors regulate microglial inflammation through c-Jun N-terminal kinase. Glia 57:444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocivavsek A, Mikhailenko I, Strickland DK, Rebeck GW (2009b) Microglial low-density lipoprotein receptor-related protein 1 modulates c-Jun N-terminal kinase activation. J Neuroimmunol 214:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KA, Pye VJ, Dai YP, Chesterman CN, Owensby DA (1999) Characterization of the soluble form of the low density lipoprotein receptor-related protein (LRP). Exp Cell Res 251:433–441. [DOI] [PubMed] [Google Scholar]
- Ren K, Dubner R (2008) Neuron-glia crosstalk gets serious: role in pain hypersensitivity. Curr Opin Anaesthesiol 21:570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Hammer RE, Herz J (1998) Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J Clin Invest 101:689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Ohsawa K, Kanazawa H, Kohsaka S, Imai Y (2001) Iba1 Is an Actin-Cross-Linking Protein in Macrophages/Microglia. Biochem Biophys Res Commun 286:292–297. [DOI] [PubMed] [Google Scholar]
- Scholz J, Woolf CJ (2007) The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci 10:1361–1368. [DOI] [PubMed] [Google Scholar]
- Schomberg D, Olson JK (2012) Immune responses of microglia in the spinal cord: Contribution to pain states. Exp Neurol 234:262–270. [DOI] [PubMed] [Google Scholar]
- Seltzer Z, Dubner R, Shir Y (1990) A novel behavioral model of neuropathic pain disorders produced in rats by partial sciatic nerve injury. Pain 43:205–218. [DOI] [PubMed] [Google Scholar]
- Shackleton B, Crawford F, Bachmeier C (2016) Inhibition of ADAM10 promotes the clearance of Aβ across the BBB by reducing LRP1 ectodomain shedding. Fluids Barriers CNS 13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhov AN, Collart MA, Vassalli P, Nedospasov SA, Jongeneel CV (1990) Kappa B-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor alpha gene in primary macrophages. J Exp Med 171:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mantuano E, Inoue G, Campana WM, Gonias SL (2009) Ligand binding to LRP1 transactivates Trk receptors by a Src family kinase-dependent pathway. Sci Signal 2:ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C, Lindenlaub T, Teuteberg P, Schäfers M, Hartung T, Toyka KV (2001) Anti-TNF-neutralizing antibodies reduce pain-related behavior in two different mouse models of painful mononeuropathy. Brain Res 913:86–89. [DOI] [PubMed] [Google Scholar]
- Staudt ND, Jo M, Hu J, Bristow JM, Pizzo DP, Gaultier A, VandenBerg SR, Gonias SL (2013) Myeloid cell receptor LRP1/CD91 regulates monocyte recruitment and angiogenesis in tumors. Cancer Res 73:3902–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles TL, Dickendesher TL, Gaultier A, Fernandez-Castaneda A, Mantuano E, Giger RJ, Gonias SL (2013) LDL receptor-related protein-1 is a sialic-acid-independent receptor for myelin-associated glycoprotein that functions in neurite outgrowth inhibition by MAG and CNS myelin. J Cell Sci 126:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gool B, Dedieu S, Emonard H, Roebroek AJM (2015) The Matricellular Receptor LRP1 Forms an Interface for Signaling and Endocytosis in Modulation of the Extracellular Tumor Environment. Front Pharmacol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanelderen P, Van Zundert J, Kozicz T, Puylaert M, De Vooght P, Mestrum R, Heylen R, Roubos E, Vissers K (2015) Effect of Minocycline on Lumbar Radicular Neuropathic Pain. Anesthesiology 122:399–406. [DOI] [PubMed] [Google Scholar]
- Vogel C, Stallforth S, Sommer C (2006) Altered pain behavior and regeneration after nerve injury in TNF receptor deficient mice. J Peripher Nerv Syst 11:294–303. [DOI] [PubMed] [Google Scholar]
- Wirenfeldt M, Clare R, Tung S, Bottini A, Mathern GW, Vinters HV (2009) Increased activation of Iba1+ microglia in pediatric epilepsy patients with Rasmussen’s encephalitis compared with cortical dysplasia and tuberous sclerosis complex. Neurobiol Dis 34:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf BB, Lopes MB, VandenBerg SR, Gonias SL (1992) Characterization and immunohistochemical localization of alpha 2-macroglobulin receptor (low-density lipoprotein receptor-related protein) in human brain. Am J Pathol 141:37–42. [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ (2014) What to call the amplification of nociceptive signals in the central nervous system that contribute to widespread pain? Pain 155:1911–1912. [DOI] [PubMed] [Google Scholar]
- Wygrecka M, Wilhelm J, Jablonska E, Zakrzewicz D, Preissner KT, Seeger W, Guenther A, Markart P (2011) Shedding of low-density lipoprotein receptor–related protein-1 in acute respiratory distress syndrome. Am J Respir Crit Care Med 184:438–448. [DOI] [PubMed] [Google Scholar]
- Xu F, Huang J, He Z, Chen J, Tang X, Song Z, Guo Q, Huang C (2016) Microglial polarization dynamics in dorsal spinal cord in the early stages following chronic sciatic nerve damage. Neurosci Lett 617:6–13. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Santamaria S, Botkjaer KA, Dudhia J, Troeberg L, Itoh Y, Murphy G, Nagase H (2017) Inhibition of Shedding of Low-Density Lipoprotein Receptor-Related Protein 1 Reverses Cartilage Matrix Degradation in Osteoarthritis. Arthritis Rheumatol (Hoboken, NJ) 69:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu C-C, Zheng H, Kanekiyo T, Atagi Y, Jia L, Wang D, N’songo A, Can D, Xu H, Chen X-F, Bu G (2016) LRP1 modulates the microglial immune response via regulation of JNK and NF-κB signaling pathways. J Neuroinflammation 13:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip PK, Kaan TKY, Fenesan D, Malcangio M (2009) Rapid isolation and culture of primary microglia from adult mouse spinal cord. J Neurosci Methods 183:223–237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.