Abstract
The majority of proteins excreted by human cells and borne at the cell surface are modified with carbohydrates. Glycoproteins mediate a wide range of processes and adopt fundamental roles in many diseases. The carbohydrates covalently attached to proteins during maturation in the cell directly impact protein structure and function as integral and indispensable components. However, the ability to study the structure of glycoproteins to high resolution was historically limited by technical barriers including a limited availability of appropriate recombinant protein expression platforms, limited methods to generate compositional homogeneity and difficulties analyzing glycoprotein composition. Furthermore, glycoproteins and in particular the glycan moieties themselves often exhibit a high degree of conformational heterogeneity. Solution NMR spectroscopy is a powerful tool to study biological macromolecules that is capable of characterizing mobile elements of molecules with atomic-level resolution. Methods to express glycoproteins, incorporate stable isotope labels and analyze glycoproteins have recently opened new avenues to prepare and investigate glycoproteins. These methods are accessible to many laboratories with experience expressing and purifying proteins from prokaryotic expression hosts.
Keywords: N-glycoprotein, O-glycoprotein, HEK293, glycoprotein expression, glycan remodeling, glycosyltransferase, glycosylhydrolase
1. Introduction
Glycoproteins represent a staggeringly small fraction of proteins analyzed by high-resolution techniques despite high concentrations at the cell surface and critical roles in many human diseases. Though one half or more of all human proteins are predicted to contain a carbohydrate chain linked through an asparagine residue (N-linked glycan; (Apweiler, Hermjakob, & Sharon, 1999)), only ~2% of all human protein structures in the Protein Data Bank show at least the first three residues of what is likely a much larger N-glycan (14% of these are the crystallizable fragment of immunoglobulin G (IgG Fc), as of May 2018). Many reasons account for the dramatic under-representation in the structural database and in structure/function studies. These factors include: challenges associated with preparing glycoproteins, difficulties crystallizing glycoproteins or resolving disordered glycans, and the requirement of specialized techniques to characterize glycoprotein composition. Solution NMR spectroscopy is particularly well suited to explore the structure and function of glycoproteins. Furthermore, recent advances in protein expression and glycoprotein analyses surmount these barriers. The goal of this article is to describe practical considerations to prepare human glycoproteins with appropriate glycans for analysis by multinuclear solution NMR spectroscopy.
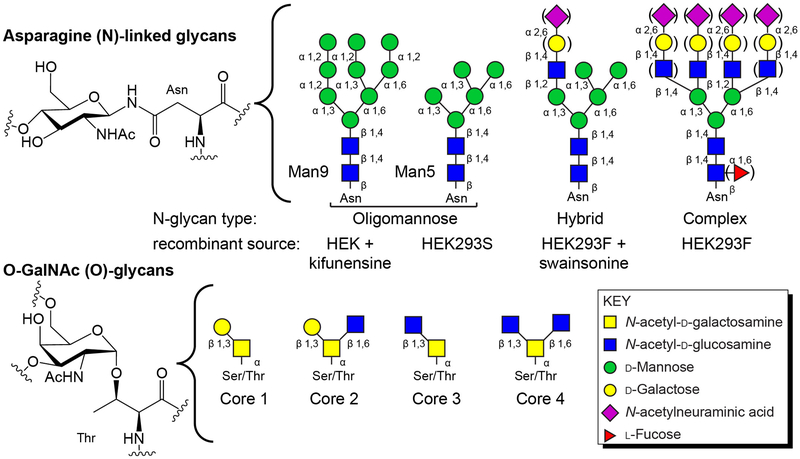
Mammals target many proteins to the secretory pathway with a ~20 amino acid signal peptide at the N-terminus that localizes the translating ribosome to an endoplasmic reticulum (ER) transmembrane pore complex (Moremen, Tiemeyer, & Nairn, 2012). The nascent polypeptide chain is imported into the ER lumen during which time the oligosaccharyltransferase activity associated with the pore complex recognizes a three amino acid sequence of [Asn / any non-proline residue / serine or threonine] and transfers a large 14-residue carbohydrate chain from a dolichol diphosphate donor to the Asn sidechain (Figure 1). Modified polypeptides then fold in the lumen of the ER where the N-glycan remodeling continues. Most glycoproteins then proceed to the Golgi where those designated for secretion are exported to the surface of the cell. During this transport, other types of modification may occur, including O-linked N-acetylgalactosamine (O-GalNAc) modifications to Ser or Thr residues in flexible regions (Figure 1). N- and O-glycans are further modified during transport and the types and extent of remodeling is due in large part to the enzymes expressed in the Golgi. As a result, most glycoproteins exhibit significant compositional heterogeneity.
Figure 1. N- and O-linked carbohydrates are two common modifications found on cell surface, secreted, Golgi and ER proteins.
Above, the structure of a glucosamine residue linked through an Asn residue to the protein and common N-glycan forms. Below, an O-GalNAc glycan linked through a Thr residue. Core structures, shown in cartoon forms, are often modified beyond the composition shown here.
Glycans are integral to protein function and harbor epitopes for specific receptors, provide stability, assist folding, impact oligomerization, and change the structure and function of proteins (reviewed extensively: (A. Varki, 2017)). Furthermore, blocking N-glycosylation is fatal to human cells in culture. Given the essential role glycans adopt in protein function, it is crucial to develop methods to simplify glycoprotein production and analysis. Furthermore, the appropriate investigation of disease-related factors often requires proteins in a form as similar to the native form as possible, including relevant modifications (Baum & Crocker, 2009). The development of multiple powerful expression and analytical tools provides new opportunities to probe glycoproteins using solution NMR spectroscopy. Here we describe the techniques utilized by our lab and the various considerations of targeted glycoprotein studies. Here we present strategies to prepare and analyze glycoproteins by solution NMR spectroscopy that utilize advances in glycoprotein expression using cultured human cells, strategies to produce homogenous glycoproteins with stable isotope labels, advances in mass spectrometry to analyze glycan composition, and specific NMR experiments to probe glycans. As such, the references cited note details of the individual techniques (with references to videos, where available).
The timing of this review should also mark a transition in glycoprotein studies. The recent development of simplified expression and analysis techniques imparts capabilities on the experimentalist that were previously accessible to only a few labs in the world. As a result, studies of glycoproteins are poised to reward the ambitious experimentalists who recognize a new opportunity to characterize previously inaccessible targets. These techniques are poised to promote a revolution in the structural biology of appropriately-processed human proteins that will form the foundation of relevant models for disease processes.
2. Glycoprotein Production
2.1. Suitable expression hosts
The vast majority of isotopically-enriched proteins analyzed by solution NMR spectroscopy are expressed using Escherichia coli. This system, however, fails for glycoprotein expression in multiple key ways. E. coli does not contain the mammalian glycosylation machinery and thus does not synthesize glycoproteins. Mammalian chaperones assist protein folding but are missing in prokaryotic hosts. Lastly, the prokaryotic cytosol is reductive and incompatible with disulfide bond formation that occurs in the oxidative endoplasmic reticulum of eukaryotes. Currently, no holistic prokaryotic expression strategy to produce uniformly labeled mammalian glycoproteins exists, though multiple groups are engineering E. coli to surmount this limitation (Lee, Nam, Nuhn, Wang, Schneider, & Ge, 2017; Schein et al., 1992; Valderrama-Rincon et al., 2012; Wang & Amin, 2014). Eukaryotic microbes, namely the yeasts, have an analogous oxidative secretory system with glycosylation machinery. Unfortunately, yeasts synthesize glycans that are very different from mammalian glycans though efforts are ongoing to correct this deficiency (Amano et al., 2008; Ajit Varki et al., 2017; Wang & Lomino, 2012). Thus, at present, mammalian protein expression to obtain appropriate glycoforms is most effectively performed using a mammalian host.
2.2. Human embryonic kidney (HEK)293 cells
HEK293 cells provide appropriate glycosylation machinery and support high yield expression. Though mammalian cells grow in considerably more complex media than E. coli, the development of rapid transient transfection methods for suspension cultures greatly reduces preparation times compared to stable transfections of adherent cells (Backliwal, Hildinger, Chenuet, Wulhfard, De Jesus, & Wurm, 2008; Wurm & Bernard, 1999). Cells for suspension cultures are commercially available, including HEK293F, and work with commercial vectors (ThermoFisher). Recent advances by Kelley Moremen and coworkers at the University of Georgia greatly expanded the system for secretory pathway proteins (Barb, Meng, Gao, Johnson, Moremen, & Prestegard, 2012; Moremen et al., 2018). A complete protocol for this expression system is available with an accompanying video (Subedi, Johnson, Moniz, Moremen, & Barb, 2015). The equipment required to culture these cells includes a humidified, heated CO2 shaker, a sterile BSL-2 biological safety cabinet, a centrifuge to pellet cultured cells, and a microscope or other device to count cells. HEK293F cells grow much slower than E. coli and divide only every 20-24h. Despite the significant differences in handling E. coli and HEK293F cells, the latter are robust, forgiving, and can even be vortexed. It is important to note that handling HEK293 cells requires a higher standard for safety and sterility compared to handling E. coli. Not only must the cultures be protected from contamination, the scientist must be protected because human cells can harbor human diseases. Enhanced safety training is strongly advised, including training for handling human tissues and fluids plus other certifications required by the sponsoring institute.
2.3. Transient transfection of HEK293
Transient transfection allows a faster route from plasmid preparation to protein expression because the time-consuming task of selecting stable transfectants is avoided. Plasmid DNA is combined with a transfection reagent like the cationic polymer polyethyleneimine (PEI) to precipitate DNA on the cell surface (Longo, Kavran, Kim, & Leahy, 2013). These plasmids contain an E. coli replication sequence plus appropriate selectable markers, thus the DNA for transfection is easily prepared using standard plasmid purification procedures. One unusual aspect of the transfection, in comparison to E. coli transformation, is the use of relatively high concentrations of plasmid DNA (~2.5 μg DNA / mL transfection) with a three-fold mass excess of PEI. The use of actively dividing cells with high viability (>95%) increases protein yield. Following addition of the DNA, cells are incubated for 24 h, at which point the culture is diluted with an equal volume of medium containing 4.4 mM valproic acid (a histone deacetylase inhibitor that prevents loss of the transfected DNA) (Backliwal, Hildinger, Kuettel, Delegrange, Hacker, & Wurm, 2008). Protein expression usually follows the dilution step and cultures maintain a high degree of viability for 4-6 days. It is advisable to harvest the medium containing the secreted protein once cell viability drops below 50% (as judged by trypan blue staining). One limiting aspect of this expression system is proteolysis by enzymes released from apoptotic or lysed cells. Sensitive proteins may require culture harvest at an earlier time point. We have observed limited proteolysis of a few unstructured residues at the IgG1 Fc C-terminus and the degradation of N- and C-terminal poly-His tags (data not shown). The pGen2 vector used in our lab contains an N-terminal GFP tag that allows for easy protein expression monitoring (Subedi, Johnson, et al., 2015). With highly expressing proteins like GFP-hCD16a (~200 mg/L), the culture medium becomes visibly green on the day following culture dilution. Furthermore, some expressed proteins may not be released by the cells. This scenario is evident when the cell pellet, but not the medium, is green. In the latter example it is advisable to screen different protein constructs to identify one that will secrete soluble protein.
2.4. Stable isotope labeling in HEK293
HEK293F cells will grow in a chemically defined medium that contains multiple sources of carbon and nitrogen atoms, unlike many mammalian cell lines that require serum, a complex mixture of chemicals, growth factors and proteins. The use of a chemically-defined medium allows for substitution with isotope-labeled sugars or amino acids that are incorporated into the expressed glycoprotein (Dutta, Saxena, Schwalbe, & Klein-Seetharaman, 2012). Labeling can be achieved in one of two ways. It is possible to add a labeled amino acid or carbohydrate directly to the standard expression medium. This works well for [13CU]-glucose, though the percentage of label at any site will be directly proportional to the amount of label in the medium. For example, we routinely add 3 g/L [13CU]-glucose to a commercial medium that contains 4 g/L glucose for a final [13C]-glycan content of roughly 40%. The primary advantage of this labeling strategy is the ease of medium preparation; no further adjustment to the commercial medium is required. One disadvantage of this strategy is that mass spectrometry-based analysis of glycan composition, as discussed in the next section, will be complicated by a mixture of isotopologues. We have scant experience with adding labeled amino acids to a complete medium, and it is unclear how much or if label scrambling will occur.
Complete substitution of unlabeled amino acid(s) or glucose in the medium is achieved by purchasing or preparing a specially formulated medium lacking the specific labeling target. It is possible to purchase medium completely devoid of amino acids and carbohydrates to prepare any custom labeling scheme. In this case, we reconstitute the medium with 100 mg/L of each amino acid except Gln which is added at 1 g/L as well as 3 g/L glucose. Next, the medium is adjusted to an osmolarity of 300 mOsm/L with sodium chloride then sterilized before use. This strategy has proven effective with selective or uniform 13C, 15N labeling of amino acids plus glucose and often results in higher protein expression when compared to the commercial medium, likely due to a higher concentration of many amino acids. In our lab, yields depend on the protein expressed, ranging from 10 to 200 mg/L.
3. Strategies to obtain glycoprotein compositional homogeneity
Glycoproteins are modified by glycosyltransferases and glycosylhydrolases in the ER and Golgi, however, the degree of remodeling for each glycan on each protein is not explicitly defined by a template (Moremen et al., 2012). This template-independent synthesis leads to potentially enormous compositional variability that is evident as a smear in an SDS-PAGE gel instead of a single, tight band. This section contains a description of multiple methods available to obtain homogeneity that manipulate N-glycans during protein expression, post purification, or a combination of both. These methods have advanced glycoprotein studies, however, it is not always clear what types of glycans are found on glycoproteins in the human body, and proteins expressed by HEK293F cells do not necessarily contain the correct types of N-glycans, thus, caution is suggested (Patel, Roberts, Subedi, & Barb, 2018). Another aspect that is not addressed by any of these techniques is the accessibility of the N-glycosylation site to the oligosaccharyltransferase. Asparagine residues in the extreme N-terminus or within 50 residues of the C-terminus may not be efficiently modified (Kelleher & Gilmore, 2006).
3.1. The lec1−/− HEK293 cell line
One variation of the HEK293F cell line, HEK293S, contains the deletion of a key enzyme that catalyzes a branchpoint in N-glycan biosynthesis, Gnt1/Lec1 (Reeves, Callewaert, Contreras, & Khorana, 2002; Stanley, Narasimhan, Siminovitch, & Schachter, 1975) (for a complete review of N-glycan biosynthesis see (Ajit Varki et al., 2017)). The result is the expression of proteins with predominantly Man5 oligomannose N-glycans, though a small percentage of Man6, Man7, Man8 and Man9 forms should be expected (Figure 1).
3.2. Inhibitors of glycosylation steps
Small molecules that inhibit specific N-glycan processing steps likewise restrict the manifold of N-glycan compositions. HEK293F cells, with a full complement of processing enzymes, can be treated with kifunensine to produce predominantly Man9 oligomannose N-glycans (Figure 1). Swainsonine addition inhibits another mannosidase, Golgi mannosidase 2, and results in the production of glycans with predominantly hybrid type N-glycans (Figure 1). Both of these inhibitors are relatively inexpensive, potent (5-10 μM), and well-tolerated. Likewise, fucosylation can be effectively reduced by 90-95% by adding 100-250 μM 2-deoxy-2-fluoro-fucose to the culture medium.
3.3. Glycan remodeling following purification
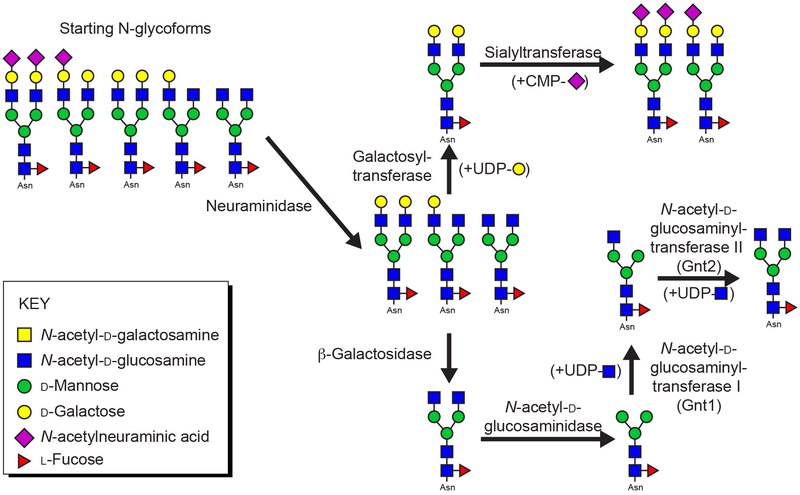
Once expressed, glycans are often accessible to glycan modifying enzymes in vitro. This approach provides two advantages: glycan homogeneity as well as a mechanism to introduce selective carbohydrate labels with labeled sugar nucleotides. Multiple methods to prepare labeled sugar nucleotides are published, including UDP-[13C]-galactose, [13C, 15N]-CMP-N-acetylneuraminic acid and [13C, 15N]-UDP-N-acetylglucosamine (Azurmendi, Vionnet, Wrightson, Trinh, Shiloach, & Freedberg, 2007; Barb, 2015; Barb & Prestegard, 2011; Macnaughtan et al., 2008; Yamaguchi et al., 1998). Many of these methods were developed to remodel glycoproteins with predominantly complex-type biantennary glycans with a varying level of galactose and N-acetylneuraminic acid residues following purification from human serum or HEK293F expression (Figure 2). In principle, these techniques are applicable to many other N-glycoprotein N-glycans but have not been evaluated using complex-type N-glycans with more than two branches.
Figure 2. N-glycan homogeneity can be achieved through post purification remodeling.
The predominant N-glycoforms found on hIgG1 Fc expressed from HEK293F cells can be remodeled with specific enzymes and substrates. The glycans are predominantly a complex-type biantennary N-glycan with varying levels of galactose and sialic acid incorporation (shown on the left side of the figure). NMR active isotopes can be incorporated using the glycosyltransferase enzymes noted above. Also note the paucimannose structure in the bottom right of the figure with two N-acetylglucosamine and three mannose residues (in this example the paucimannose N-glycan contains a core fucose residue).
The first in vitro remodeling step is conducted with a commercially available neuraminidase to efficiently remove N-acetylneuraminic acids residues from the non-reducing termini. Neuraminidase-treated glycans generally remain inhomogeneous, but can be further treated in a galactosylation reaction catalyzed by commercially available bovine lactose synthase or trimmed further with a β-galactosidase (Figure 2). The galactosylation reaction is rapid, easily completed, and will produce a homogenous N-glycan, depending on variability in the starting material. However, the β-galactosidase reaction has proven more challenging in our lab. Through β-galactosidase enzymes are commercially available, we screen multiple enzyme preparations to identify a single supply that will efficiently remove galactose residues from IgG1 Fc. As sources and suppliers change, we have repeated this exercise many times in the past decade.
Galactosylated N-glycans are suitable substrates for ST6Gal-I catalyzed sialylation (Macnaughtan et al., 2008). ST6Gal-I expressed to high yield in HEK293F cells with an N-terminal GFP fusion (Barb, Meng, et al., 2012). It is well known that ST6Gal-I sialylates the galactose residue on the Manα1-3Manβ branch of a complex-type biantennary N-glycan with 10-fold greater activity than the galactose residue on the Manα1-6Manβ branch (Barb, Meng, et al., 2012; Paulson, Rearick, & Hill, 1977). We were able to achieve high levels of IgG1 Fc with two sialic acids on each glycan only by using high amounts of ST6Gal-I (15 moles substrate : 1 mole enzyme). The reaction is incubated for multiple days and washed with fresh CMP-N-acetylneuraminic acid each day to remove the CMP hydrolysis product that also inhibits the reaction. This is the most challenging glycosyltransferase reaction we have yet to encounter with IgG1 Fc as a substrate.
Shorter N-glycans can be formed by further digestion with a commercially available N-acetylhexosaminidase to generate paucimannose N-glycans (which may or may not contain a core fucose residue; defined in Figure 2). These glycans can be extended by one N-acetylglucosamine residue attached to the Manα1-3Manβ residue using Gnt1, expressed from HEK293F cells (Barb, 2015). Though the Man5 oligomannose N-glycan is the native substrate for this reaction, Gnt1 efficiently modifies the paucimannose N-glycan (containing two N-acetylglucosamine and three mannose residues). A second N-acetylglucosamine branch can be added to the Manα1-6Manβ residue with Gnt2.
The Man5 N-glycans from HEK293S expression (Figure 1) are effective acceptor substrates for Gnt1 to create a Man5 N-glycan with an N-acetylglucosamine residue on the Manα1-3Manβ residue (Barb, 2015). We have not identified a suitable mannosidase that expresses in sufficient quantities to convert the Man5-GlcNAc N-glycan in vitro into the cognate Gnt2 substrate, a Man3-GlcNAc N-glycan.
4. Compositional Analysis of Glycoproteins
One critical aspect of glycan remodeling is assessing compositional heterogeneity arising from incomplete remodeling reactions in vivo and in vitro. Though many techniques are available, we use predominantly mass spectrometry-based methods due to rapidity and precision. In addition to confirming sample homogeneity, N-glycan processing during recombinant protein expression is a useful reporter of glycan accessibility. Analyses of N-glycan composition before enzymatic remodeling has the potential to directly support observations made by NMR spectroscopy (Subedi, Hanson, & Barb, 2014). For example, N-glycans that stabilize protein structure often form van Der Waals contacts with amino acids at the protein surface. These interactions restrict N-glycan processing and restrict enzymatic N-glycan remodeling in vivo and in vitro.
4.1. Electrospray ionization mass spectrometry (ESI-MS) of intact glycoproteins
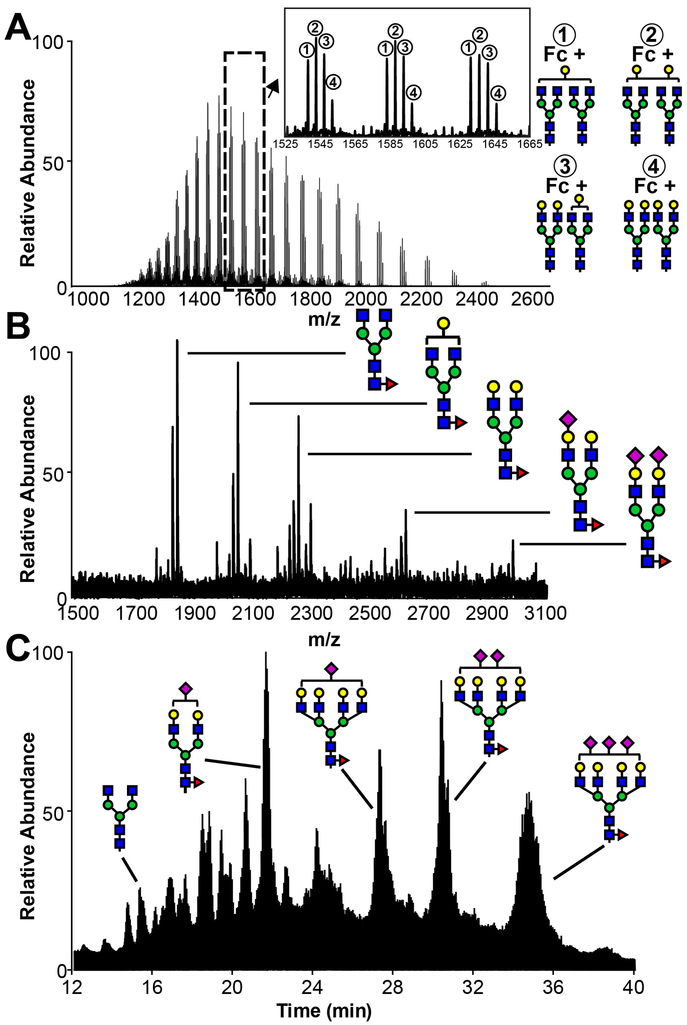
One advantage of ESI-MS is the ability to analyze intact glycoproteins rapidly with minimal sample preparation. Generally, a small amount of glycoprotein (1 μg) is loaded onto a C4 or C8 reversed phase column and eluted with an acetonitrile gradient into a mass spectrometer. Chromatography eliminates sample preparation steps including desalting, and the instrument time is often brief (15-20 min per sample). Intact mass spectrometry generates an envelope of peaks with different mass-to-charge ratios (m/z) for each protein as shown in Figure 3A. Each different protein mass will generate a unique envelope, thus, the number of distinct glycoprotein masses that can be resolved in a single experiment is somewhat limited. As a result, this approach is the most effective when there are only a small number of predominant forms as observed for IgG Fc. A spectrum can be deconvoluted manually or with multiple software platforms to provide the intact mass of each species with potentially very high resolution (±1 Da at ~50 kDa). One drawback to this technique is that all modifications to a single protein contribute to the intact mass; oxidation, alkylation, proteolysis, etc, introduce challenges to match the observed mass with a calculated composition. ESI-MS of intact glycoproteins works very well when assessing the efficacy of an in vitro glycan remodeling reaction, e.g. where ±162 Da (a hexose) is easily confirmed. This approach is of limited applicability with isotope-labeled proteins that are more challenging to deconvolute, particularly in the case of label incorporation at lower percentages (<95%).
Figure 3. N-Glycan composition as probed with mass spectrometry.
A. Total ion current from an ESI-MS analysis of intact hIgG1 Fc, a homodimer containing two N-glycans. B. Total ion current from MALDI-MS of permethylated hIgG1 Fc F243S N-glycans. C. Total ion current from an ESI-MS analysis of procainamide-derivatized N-glycans from recombinant CD16a.
4.2. Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS)
N-glycans are directly observable using MALDI-MS. First, 25-200 μg of glycoprotein is proteolyzed then digested with PNGaseF, a commercially available enzyme that specifically cleaves N-glycans from Asn side chains. Peptides are removed by passing the mixture over a C18 column to collect the flow-through which contains N-glycans. Next, released N-glycans are permethylated in a chemical reaction using anhydrous sodium hydroxide in dimethyl sulfoxide with iodomethane (Anumula & Taylor, 1992). A video protocol using related methods is available (Zaare, Aguilar, Hu, Ferdosi, & Borges, 2016). Following a two-phase extraction with methylene chloride and water, N-glycans are spotted on a MALDI plate with dihydroxybenzoic acid as the matrix. Permethylation replaces all labile hydrogens attached to nitrogen and oxygen residues with a methyl group, eliminating charges on the N-glycan. MALDI generates sodium or potassium adducts that are detectable as singly-charged species with a TOF mass analyzer (Figure 3B). These data are relatively simple to analyze as the predominant species only contain carbohydrate residues with a single metal ion and a single charge, though the methyl modifications must be accounted for. Permethylated N-glycans can be detected using multiple platforms, but we find MALDI-MS rapid and sufficiently sensitive. Another advantage of this approach is that even very basic instruments will produce reliable data. One disadvantage of the approach is the relatively lengthy preparation, which may take 2-3 days though many samples can be analyzed in parallel. MALDI-MS of permethylated N-glycans is not as sensitive as the analysis of procainamide derivatized N-glycans by ESI-MS, as described below. It should also be noted that modification with sialic acids, including N-acetylneuraminic acid, is labile in weak acids including acetic acid. Thus, it is important to avoid low pH as much as possible to accurately assess sialylation.
4.3. Analysis of N-glycans by hydrophilic interaction chromatography (HILIC)-ESI-MS/MS
HILIC-ESI-MS/MS is the standard technique used by our laboratory to characterize the compositional heterogeneity of recombinant proteins due to high sensitivity and precision (Figure 3C). This technique requires a only small amount of glycoprotein (1-5 μg). In lieu of permethylation, N-glycans are derivatized at the reducing end by reacting with procainamide to form a Schiff base that is reduced with sodium cyanoborohydride (Patel et al., 2018). The reaction mixture is next directly injected onto a HILIC column (Waters) in 75% acetonitrile, 25% H2O, 0.1% formic acid and 0.01% trifluoroacetic acid, and eluted with an increasing linear water gradient. HILIC separates glycans by size, with larger glycans eluting later in the profile. Raw data are analyzed with byonic (Protein Metrics) and validated manually by matching the monoisotopic peak with the calculated mass. Retention time provides an additional validation and correlates linearly with N-glycan mass (Patel et al., 2018). Analysis of N-glycans using a Q-Exactive Orbitrap mass spectrometer (Thermo Scientific) provided mass accuracy greater than 0.01 Da. Fragmentation in the second MS dimension can confirm the composition and provide insight regarding the configuration of individual N-glycan species (core vs. branch fucosylation, etc). These methods are straightforward and easily validated.
Studies of proteins with multiple N-glycans per polypeptide chain may require added sophistication to analyze each N-glycan separately. The analysis of glycopeptides provides more information than the analysis of released N-glycans, including what type of N-glycans are found at specific sites in the protein. Glycoproteomics approaches are widely applied to analyze glycopeptides (Kolarich, Jensen, Altmann, & Packer, 2012; Plomp, Bondt, de Haan, Rombouts, & Wuhrer, 2016). Though linking glycans and peptides introduces complexity into the analysis as the critical data is found in MS2 spectra, multiple programs are available to guide the analysis including byonic and pGlyco2.0 (Liu et al., 2017). Another potentially challenging aspect of glycopeptide analysis involves identifying appropriate proteases and reaction conditions to separate the individual N-glycans. Once digested, the mixture of peptides and glycopeptides can be analyzed using the HILIC separation described above. Peptides separate based on N-glycan composition with aglycosylated peptides eluting very early from the column. C18 reversed-phase columns may also be used, but this approach often requires glycopeptide purification prior to chromatography. A simple clean-up procedure reduces the ion-suppressing effects of non-glycosylated peptides which are predominant and often ionize more efficiently than glycopeptides.
5. Solution NMR of labeled glycoproteins
NMR spectra of isotope labeled glycoproteins are collected with the same pulse sequences applied to most [13C, 15N]-labeled proteins. Here we describe considerations for NMR analyses of glycoproteins with labeled backbone residues and labeled carbohydrates.
5.1. Analysis of glycoproteins with backbone labels
N-glycans promote folding and stability in secreted glycoproteins, therefore many glycoproteins are stable at high temperatures. IgG1 Fc, for example, is stable for weeks to months at 50 ºC. N-glycans, however, can restrict protein tumbling in solution, leading to broader linewidths and reduced signal in 3d experiments requiring multiple coherence transfer steps. We often observe a benefit to using TROSY-based experiments (Pervushin, Riek, Wider, & Wuthrich, 1997) even in moderately sized proteins without benefit of perdeuteration. Unfortunately, deuterium labeling to reduce spoiling of the TROSY-effect due to dipolar interactions with remote 1H has not yet been demonstrated using mammalian cells. It is assumed mammalian cells are unlikely to survive in high percentages of 2H2O. Supplementing expression medium with [2H, 13C, 15N]-labeled amino acids is very expensive, tolerated by insect cells and potentially applicable to protein expression in HEK293 (Franke et al., 2018). We have observed improvements in spectral quality by removing N-glycans that do not contribute or contribute minimally to protein structure and retaining those that are essential for protein structure and function. Non-essential N-glycans are removed by mutating the Asn residue to Gln. In our experience, as discussed below, removing non-essential N-glycans does reduce protein stability to a moderate degree for CD16a.
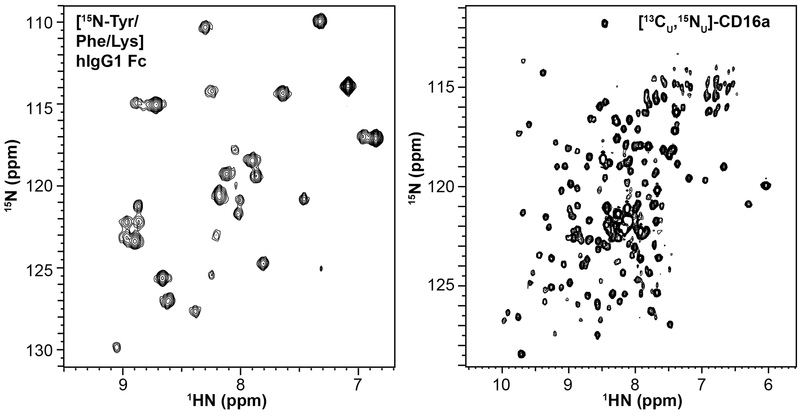
Example spectra from isotope-labeled glycoproteins expressed in HEK293F cells are shown in Figure 4. IgG1 Fc is a ~50 kDa protein composed of two identical polypeptide chains, each with an N-glycan attached at residue 297. The IgG1 Fc N-glycan does not contribute substantially to the protein rotational correlation time (Subedi & Barb, 2015). [15Namide-Tyr/Phe/Lys]-IgG1 Fc produced from HEK293F cells provides a clear fingerprint of the protein in an 15N-HSQC-TROSY spectrum, with minimal evidence for metabolic scrambling of the labeled amino acids. The 37 Tyr, Phe or Lys residues in this construct provide 29 clear peaks in a 15N-HSQC spectrum. Kato and coworkers assigned the protein backbone using traditional triple resonance experiments (Yagi et al., 2015).
Figure 4. Glycoproteins expressed at high yield with high percentages of 15N and 13C incorporated into the polypeptide backbne.
(left) Selective labeling to incorporate 15N into the amides of Tyr, Phe and Lys residues of glycosylated IgG1 Fc, a ~50 kDa homodimer with one N-glycan per chain, observed in a 2d HSQC-TROSY collected at 50 °C. (right) Complete 15N and 13C labeling of a CD16a variant with two N-glycans at N45 and N162 observed in a 2d projection of a 3d HNCA-TROSY experiment collected at 30 °C. The CD16a variant is a ~23 kDa monomer. These spectra were collected 16.4T.
The extracellular antibody-binding domain of CD16a / Fc γ receptor 3a is a smaller monomeric protein of ~23 kDa with as many as five N-glycans. Mutating three asparagine residues to prevent N-glycosylation at N38, N74 and N169 does not reduce antibody binding affinity but does reduce the stability of the protein. The result is a soluble CD16a variant with two N-glycans at N45 and N162 that provides well dispersed NMR spectra (Figure 4B)(Ferrara, Stuart, Sondermann, Brunker, & Umana, 2006). An HNCA spectrum of [13C, 15N]-CD16a also shows clear evidence for efficient resonance transfer and high levels of label incorporation using the uniform labeling method described in section 2.4, above (Figure 4B). Thus, these strategies to label amino acids are suitable for targeted studies with selective labeling or global studies of structure and motion using standard solution NMR pulse sequences.
Glycan truncation is another approach to limit the negative impact of large N-glycans on protein tumbling without mutating asparagine residues. Glycan truncation is achievable by expressing protein with the HEK293S cell line that produces only oligomannose N-glycans that can be trimmed by endoglycosidase F to a single asparagine-linked N-acetylglucosamine. The preservation of stability upon truncation is likely due to the predominant stabilizing effect of the first N-acetylglucosamine residue; Hanson et al. demonstrated that the first monosaccharide residue contributed roughly 2/3 of the energy towards stabilizing CD2 when compared to the stabilization provided by the full-length N-glycan (Hanson, Culyba, Hsu, Wong, Kelly, & Powers, 2009). Thus, in some cases, truncating the N-glycan may allow sufficient sensitivity for successful backbone assignment through traditional triple-resonance experiments.
5.2. Analysis of glycoproteins with uniformly-labeled carbohydrates
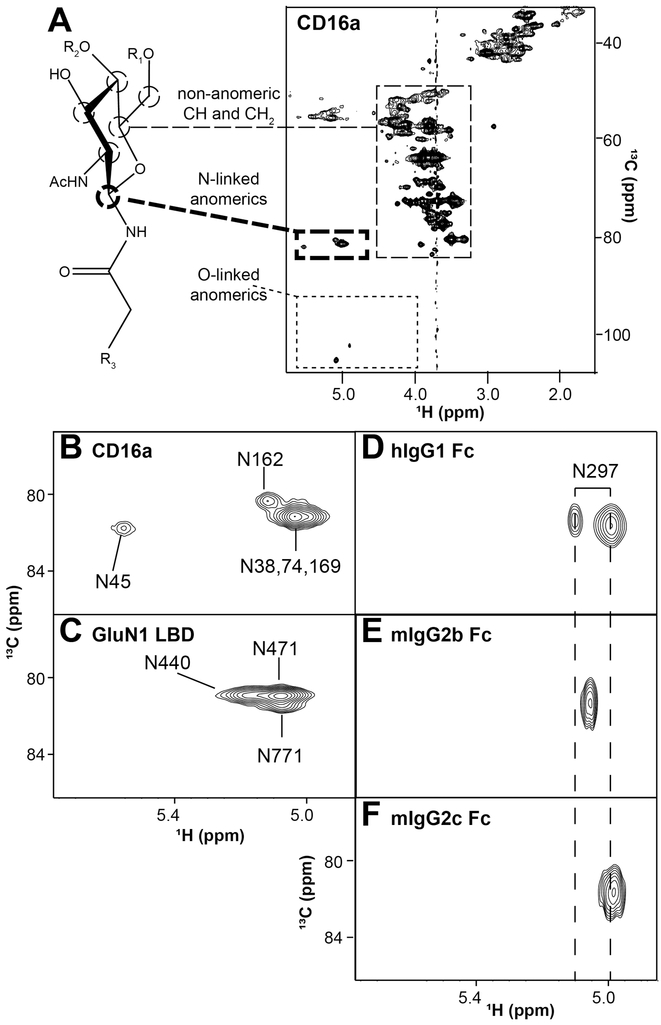
Expressing glycoproteins in standard culture medium (containing [12C]-glucose) supplemented with [13C]-glucose provides an effective and relatively simple strategy to rapidly assess N-glycans using NMR (Subedi & Barb, 2015; Subedi, Falconer, & Barb, 2017). This strategy predominantly incorporates [13C] into the glycans with little evidence for scrambling into amino acids (Figure 5A). It is likely the high concentration of amino acids in the medium prevents metabolic scrambling. The advantage of this approach arises from the unique spectral signature of each N-glycan: the anomeric carbon (C1 for aldohexoses like glucose, galactose and mannose) resonates at a greater frequency (~100 ppm) compared to other carbohydrate carbons at (50-80 ppm). However, the anomeric carbon of the N-acetylglucosamine residue, attached through a nitrogen atom to the Asn sidechain, resonates at roughly 80 ppm with a 1H1 chemical shift around 5.0 ppm. Though there are many anomeric protons and carbons in a glycan, there is only one N-linked anomeric carbon per N-glycan. Furthermore, the location proximal to the polypeptide provides a single site for each N-glycan that is positioned to report differences arising from interactions with the amino acid residues and variations in amino acid type. These Asn-linked correlations can be easily distinguished in a 2d 1H-13C HSQC spectrum of CD16a (Figure 5A). Furthermore, the signals of interest appear in a region of the spectrum with few signals from amino acids. This glycoprotein labeling and pulse sequence strategy provides an N-glycan fingerprint for each glycoprotein, similar to a 2d 1H-15N HSQC for backbone amide correlations in proteins.
Figure 5. Proteins with 13C-labeled N-glycans provide a single, unique signal for each N-glycan.
A. A 13C-HSQC of [13C-glycan] CD16a with 5 N-glycans shows predominantly signal corresponding to carbohydrate and potentially limited scrambling to amino acids. Of the three distinct regions noted in the spectrum, crosspeaks from the anomeric asparagine-linked H1-C1 atoms are separated from the other signals, with one coherence for each N-glycan residue (expanded in B). C-F Similar analyses of four glycoproteins yields comparable results. These spectra were collected 16.4T.
Significant differences in peak positions arising from Asn-linked 1H1-13C1 correlations have proven valuable to identify specific N-glycans in proteins that relate to protein function. CD16a contains as many as five N-glycans that show three distinct N-glycan peaks (Figure 5B). Mutating Asn residues to Gln revealed which peaks belonged to which N-glycans (Subedi et al., 2017). As mentioned above, the two N-glycans that are important for antibody binding affinity, N45 and N162, show distinct peaks that are separate from peaks arising from the N38, N74 and N169 glycans. The unique environment for each peak was destroyed by proteolysis causing the peaks to collapse upon the large intense peaks formed by N38, N74 and N169. Thus, the chemical environment of the Asn-linked 1H1 and 13C1 atoms of the N45 and N162-glycans are likely influenced by surrounded polypeptide residues. A similar analysis of the GluN1 ligand binding domain from the N-methyl-D-aspartate receptor (NMDAR) with three N-glycans revealed similar differences that could be destroyed through proteolysis (G. Subedi and A. Barb, in press).
Analysis of IgG Fc provided further insight into the chemical environment surrounding the glycan at the point of attachment. IgG1 Fc from human binds tightly to CD16a and shows two clear signals of differing intensities and linewidths originating from the Asn-linked N-acetylglucosamine residue (Fig 5D) (Subedi & Barb, 2015). The appearance of two peaks for a single H1-C1 correlation likely results from slow conformational exchange. Interestingly, two IgG1 Fc forms that cannot bind CD16a, D265A and proteolyzed IgG1 Fc, showed only a single peak that matched the position of the minor peak observed for intact IgG1 Fc. This result indicates the conformation contributing to the more intense peak on the right hand side of the intact IgG1 Fc spectrum is consistent with a conformation that binds receptor. The same analytical approach was applied to mouse IgG2b Fc and IgG2c Fc. Unlike human Fc, the mouse proteins showed only one peak for the Asn-linked N-acetylglucosamine residue; however, the peak from IgG2c Fc was found further to the right and mIgG2c bound mouse Fc γ receptor IV with ~5-fold greater affinity that mIgG2b Fc, consistent with the stabilized conformation observed in hIgG1 Fc (Falconer & Barb, 2018).
These results demonstrate that peaks corresponding to the Asn-linked N-acetylglucosamine residue provide a wealth of information regarding individual N-glycan behavior in many different glycoproteins.
5.3. Analysis of glycoproteins with specifically-labeled carbohydrates
Labeling glycoprotein glycans with [13C, 15N]-labeled carbohydrate residues following purification greatly simplifies spectral complexity and assists peak assignment. Our laboratory has used this approach extensively to study the IgG1 Fc N-glycans (Barb, 2015; Barb, Ho, Flanagan-Steet, & Prestegard, 2012; Barb, Meng, et al., 2012; Barb et al., 2011; Subedi et al., 2014). A wealth of labeling strategies and nuclei are accessible though labeled sugar nucleotides are time consuming or expensive to obtain. Furthermore, some glycosyltransferase reactions are challenging to optimize to acheive complete protein labeling. One example of post-purification labeling is the addition of [13C2]-galactose to the IgG1 Fc N-glycans. Spin relaxation and relaxation-dispersion measurements demonstrated that the galactose residues were mobile, in contrast to previous interpretations of structures determined by X-ray crystallography (Barb et al., 2011). Furthermore, adding N-acetylglucosamine, galactose and N-acetylneuraminic acid residues to the Fc N-glycan branches reduced carbohydrate motion after each addition (Barb, 2015; Barb, Meng, et al., 2012). Glycan labeling after purification represents an orthogonal approach to study the structure, function and interactions of glycoprotein glycans. This strategy often benefits from the greater motion, and thus narrower lines, experienced by residues distal to the reducing end point of attachment in large proteins. This property is in contrast to backbone atoms in structured regions or select carbohydrate residues located closer to the glycan reducing end.
6. Summary
Glycoproteins should no longer represent a forbidden territory to structural biologists and NMR spectroscopists due to the extraordinary advances in glycoprotein expression platforms, MS-based analytic methods and NMR strategies. The goal to accurately recapitulate native disease processes will push laboratories to manufacture proteins that mimic glycan composition as well as behavior as closely as possible. Future steps towards understanding these complex molecules in human and animal health will require no less.
Acknowledgement/Funding
This material is based upon work supported by the National Institutes of Health under Award No. R01 GM115489 (NIGMS) and the Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology at Iowa State University. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
Works Cited
- Amano K, Chiba Y, Kasahara Y, Kato Y, Kaneko MK, Kuno A, … Narimatsu H (2008). Engineering of mucin-type human glycoproteins in yeast cells. Proc Natl Acad Sci U S A, 105(9), 3232–3237. doi: 10.1073/pnas.0710412105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anumula KR, & Taylor PB (1992). A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem, 203(1), 101–108. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, & Sharon N (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta, 1473(1), 4–8. [DOI] [PubMed] [Google Scholar]
- Azurmendi HF, Vionnet J, Wrightson L, Trinh LB, Shiloach J, & Freedberg DI (2007). Extracellular structure of polysialic acid explored by on cell solution NMR. Proc Natl Acad Sci U S A, 104(28), 11557–11561. doi: 10.1073/pnas.0704404104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backliwal G, Hildinger M, Chenuet S, Wulhfard S, De Jesus M, & Wurm FM (2008). Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res, 36(15), e96. doi: 10.1093/nar/gkn423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backliwal G, Hildinger M, Kuettel I, Delegrange F, Hacker DL, & Wurm FM (2008). Valproic acid: a viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnol Bioeng, 101(1), 182–189. doi: 10.1002/bit.21882 [DOI] [PubMed] [Google Scholar]
- Barb AW (2015). Intramolecular N-glycan/polypeptide interactions observed at multiple N-glycan remodeling steps through [(13)C,(15)N]-N-acetylglucosamine labeling of immunoglobulin G1. Biochemistry, 54(2), 313–322. doi: 10.1021/bi501380t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, Ho TG, Flanagan-Steet H, & Prestegard JH (2012). Lanthanide binding and IgG affinity construct: potential applications in solution NMR, MRI, and luminescence microscopy. Protein Sci, 21(10), 1456–1466. doi: 10.1002/pro.2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, & Prestegard JH (2012). NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry, 51(22), 4618–4626. doi: 10.1021/bi300319q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb AW, & Prestegard JH (2011). NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol, 7(3), 147–153. doi: 10.1038/nchembio.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum LG, & Crocker PR (2009). Glycoimmunology: ignore at your peril! Immunol Rev, 230(1), 5–8. doi: 10.1111/j.1600-065X.2009.00800.x [DOI] [PubMed] [Google Scholar]
- Dutta A, Saxena K, Schwalbe H, & Klein-Seetharaman J (2012). Isotope labeling in mammalian cells. Methods Mol Biol, 831, 55–69. doi: 10.1007/978-1-61779-480-3_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DJ, & Barb AW (2018). Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1. PLoS One, 13(2), e0192123. doi: 10.1371/journal.pone.0192123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara C, Stuart F, Sondermann P, Brunker P, & Umana P (2006). The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J Biol Chem, 281(8), 5032–5036. doi: 10.1074/jbc.M510171200 [DOI] [PubMed] [Google Scholar]
- Franke B, Opitz C, Isogai S, Grahl A, Delgado L, Gossert AD, & Grzesiek S (2018). Production of isotope-labeled proteins in insect cells for NMR. J Biomol NMR. doi: 10.1007/s10858-018-0172-7 [DOI] [PubMed] [Google Scholar]
- Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, & Powers ET (2009). The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A, 106(9), 3131–3136. doi: 10.1073/pnas.0810318105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, & Gilmore R (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology, 16(4), 47R–62R. doi: 10.1093/glycob/cwj066 [DOI] [PubMed] [Google Scholar]
- Kolarich D, Jensen PH, Altmann F, & Packer NH (2012). Determination of site-specific glycan heterogeneity on glycoproteins. Nat Protoc, 7(7), 1285–1298. doi: 10.1038/nprot.2012.062 [DOI] [PubMed] [Google Scholar]
- Lee KB, Nam DH, Nuhn JAM, Wang J, Schneider IC, & Ge X (2017). Direct expression of active human tissue inhibitors of metalloproteinases by periplasmic secretion in Escherichia coli. Microb Cell Fact, 16(1), 73. doi: 10.1186/s12934-017-0686-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MQ, Zeng WF, Fang P, Cao WQ, Liu C, Yan GQ, … Yang PY (2017). pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat Commun, 8(1), 438. doi: 10.1038/s41467-017-00535-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo PA, Kavran JM, Kim MS, & Leahy DJ (2013). Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol, 529, 227–240. doi: 10.1016/B978-0-12-418687-3.00018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughtan MA, Tian F, Liu S, Meng L, Park S, Azadi P, … Prestegard JH (2008). 13C-sialic acid labeling of glycans on glycoproteins using ST6Gal-I. J Am Chem Soc, 130(36), 11864–11865. doi: 10.1021/ja804614w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, … Jarvis DL (2018). Expression system for structural and functional studies of human glycosylation enzymes. Nat Chem Biol, 14(2), 156–162. doi: 10.1038/nchembio.2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, & Nairn AV (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol, 13(7), 448–462. doi: 10.1038/nrm3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KR, Roberts JT, Subedi GP, & Barb AW (2018). Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function. J Biol Chem, 293(10), 3477–3489. doi: 10.1074/jbc.RA117.001207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC, Rearick JI, & Hill RL (1977). Enzymatic properties of beta-D-galactoside alpha2 leads to 6 sialytransferase from bovine colostrum. J Biol Chem, 252(7), 2363–2371. [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, & Wuthrich K (1997). Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A, 94(23), 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomp R, Bondt A, de Haan N, Rombouts Y, & Wuhrer M (2016). Recent Advances in Clinical Glycoproteomics of Immunoglobulins (Igs). Mol Cell Proteomics, 15(7), 2217–2228. doi: 10.1074/mcp.O116.058503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, & Khorana HG (2002). Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A, 99(21), 13419–13424. doi: 10.1073/pnas.212519299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein CH, Boix E, Haugg M, Holliger KP, Hemmi S, Frank G, & Schwalbe H (1992). Secretion of mammalian ribonucleases from Escherichia coli using the signal sequence of murine spleen ribonuclease. Biochem J, 283 (Pt 1), 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Narasimhan S, Siminovitch L, & Schachter H (1975). Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine--glycoprotein N-acetylglucosaminyltransferase activity. Proc Natl Acad Sci U S A, 72(9), 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi GP, & Barb AW (2015). The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure, 23(9), 1573–1583. doi: 10.1016/j.str.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi GP, Falconer DJ, & Barb AW (2017). Carbohydrate-Polypeptide Contacts in the Antibody Receptor CD16A Identified through Solution NMR Spectroscopy. Biochemistry, 56(25), 3174–3177. doi: 10.1021/acs.biochem.7b00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi GP, Hanson QM, & Barb AW (2014). Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure, 22(10), 1478–1488. doi: 10.1016/j.str.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi GP, Johnson RW, Moniz HA, Moremen KW, & Barb A (2015). High Yield Expression of Recombinant Human Proteins with the Transient Transfection of HEK293 Cells in Suspension. J Vis Exp(106), e53568. doi: 10.3791/53568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, … DeLisa MP (2012). An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol, 8(5), 434–436. doi: 10.1038/nchembio.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A (2017). Biological roles of glycans. Glycobiology, 27(1), 3–49. doi: 10.1093/glycob/cww086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, … Seeberger PH (2017). Essentials of glycobiology (Third edition ed.). Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Wang LX, & Amin MN (2014). Chemical and chemoenzymatic synthesis of glycoproteins for deciphering functions. Chem Biol, 21(1), 51–66. doi: 10.1016/j.chembiol.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LX, & Lomino JV (2012). Emerging technologies for making glycan-defined glycoproteins. ACS Chem Biol, 7(1), 110–122. doi: 10.1021/cb200429n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm F, & Bernard A (1999). Large-scale transient expression in mammalian cells for recombinant protein production. Curr Opin Biotechnol, 10(2), 156–159. [DOI] [PubMed] [Google Scholar]
- Yagi H, Zhang Y, Yagi-Utsumi M, Yamaguchi T, Iida S, Yamaguchi Y, & Kato K (2015). Backbone (1)H, (13)C, and (15)N resonance assignments of the Fc fragment of human immunoglobulin G glycoprotein. Biomol NMR Assign, 9(2), 257–260. doi: 10.1007/s12104-014-9586-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Kato K, Shindo M, Aoki S, Furusho K, Koga K, … Shimada I (1998). Dynamics of the carbohydrate chains attached to the Fc portion of immunoglobulin G as studied by NMR spectroscopy assisted by selective 13C labeling of the glycans. J Biomol NMR, 12(3), 385–394. [DOI] [PubMed] [Google Scholar]
- Zaare S, Aguilar JS, Hu Y, Ferdosi S, & Borges CR (2016). Glycan Node Analysis: A Bottom-up Approach to Glycomics. J Vis Exp(111). doi: 10.3791/53961 [DOI] [PMC free article] [PubMed] [Google Scholar]