Abstract
Infectious diseases are caused by pathogenic microorganisms and can be transmitted between individuals and populations thus threatening the general public health and potentially the economy. Efficient diagnostic tools are needed to provide accurate and timely guidance for case identification, transmission disruption and appropriate treatment administration. Point of care (POC) tests provide actionable results near the patient and thereby serve as a personal “radar”. In this review, we review clinical needs for POC testing for several major pathogens, including malaria parasites, human immunodeficiency virus (HIV), human papillomavirus (HPV), dengue, Ebola and Zika viruses and Mycobacterium tuberculosis (TB). We compare different molecular approaches, including pathogen nucleic acid and protein, circulating microRNA and antibodies, used in the POC tests. Finally, we review recent advances in novel POC technologies focusing on microfluidic and plasmonic-based approaches.
Introduction
Most infectious diseases are caused by pathogenic microorganisms including viruses, bacteria, parasites and fungi. Compared with other diseases, infectious diseases can be exponentially transmitted among populations in a relatively short period of time thus threatening the general public health and potentially the economy. It is estimated that over half of the world population are at risk for infectious diseases, making them one of the most dangerous threats to humanity[1].
“Without diagnostics, medicine is blind.”[2] Adequate and prompt treatment to illnesses cannot be made properly without diagnosis in the first place. Sensitive, specific and rapid diagnostic testing not only paves the way toward effective treatment but also plays a critical role in preventing the transmission of infectious diseases. While central clinical laboratories offer sensitive and specific assays, such as blood culture, high-throughput immunoassays, polymerase chain reaction (PCR) and mass spectrometry (MS) tests, they are often time and labor intensive, costly, and dependent on sophisticated instruments and well trained operators. On the other hand, point-of-care (POC) tests provide rapid ‘on-site’ results at the site of care delivery, and in resource-limited settings, supporting timely and proper treatment [3]. According to the World Health Organization (WHO), POC tests that address infectious disease control needs, especially for the developing countries, should follow “ASSURED” criteria: (1) affordable, (2) sensitive, (3) specific, (4) user-friendly, (5) rapid and robust, (6) equipment-free and (7) deliverable to end-users [4].
Here in, we review literature published in the past decade and indexed in Pubmed, on the development of POC tests for infectious diseases. Based on the number of published studies in this field, we have chosen to focus on several major infectious disease-causing microorganisms, including malaria parasites, human immunodeficiency virus (HIV), human papillomavirus (HPV), dengue, Ebola and Zika viruses, and Mycobacterium tuberculosis (TB) bacteria. We first review the pathological processes, impact on public health, and POC needs for the detection of these microorganisms, then focus on several key biomarkers used in developed POC tests, including pathogen nucleic acids and proteins, circulating microRNAs and antibodies, comparing their roles during the entire process of disease management. Finally, we review advancements in microfluidics and plasmonics, two technologies that we have seen significant innovations in the past decade in developing POC tests for infectious diseases. These technologies, together with others in the “POCT Toolbox” (Figure 1), act as personal radar in the fight against infectious diseases, towards the goal of patient-centralized diagnosis and treatment, as shown in Figure 1.
Figure 1.
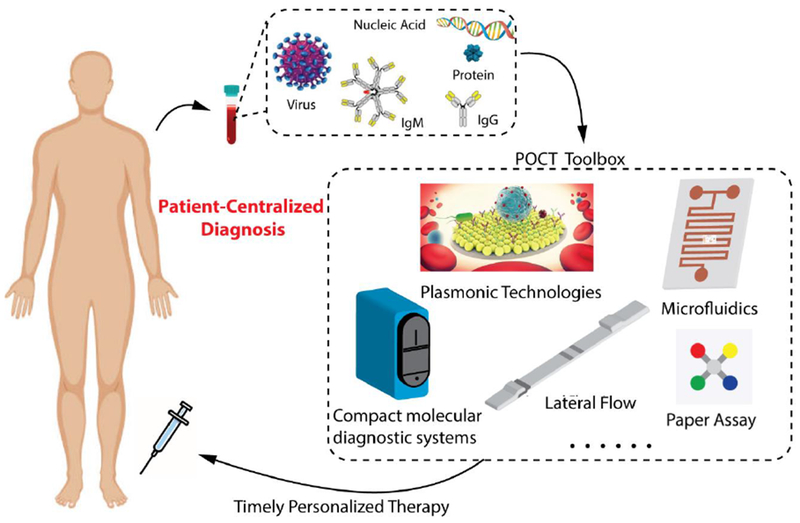
Point of Care Tests (POCT) (such as com pact molecular diagnostic systems, lateral flow assays, microfluidics, plasmonic technologies and paper-based assays etc.) detect a variety of infectious diseases-related biomarkers, including virus particles, nucleic acids, proteins and antibodies. They serve as the foundation of “patient centralized” diagnosis and treatment of infectious diseases. Partly adapted from [5] with permission.
Pathogen Detection Needs at the POC
Malaria Parasites
Over 300 million patients every year in tropical areas (such as sub-Saharan Africa) suffer from malaria [6, 7]. According to the “Malaria case management: operations manual” recommended by the World Health Organization (WHO), effective malaria management relies heavily on early diagnosis and prompt artemisinin-combined therapy (ACT) [8]. The first proof of malaria parasites in human blood was observed under microscope in the 1880s [9]. Since then, microscopy examination of Giemsa-stained blood film has been established as the gold standard in malaria diagnosis, which requires highly qualified and well-trained operators and reliable equipment [10], both are often in low supply in the areas where malaria is most prevalent [11]. To address this issue, the last decade has seen significant development of malaria rapid diagnostic tests (RDTs), with the goal to enable fast and reliable testing in remote settings where clinical diagnostics resources are not routinely available [11–14]. For example, the RDT lateral flow strips developed in a study can detect proteins derived from malaria parasites in blood, generating a series of clearly visible lines [9]. Rathod et al. used microfluidic channels to successfully mimic the capillary environment for more accurate in-field malaria diagnosis [15].
HIV
Over 40 million people are affected by HIV worldwide. About 85% of them are living in developing countries, where clinical diagnostics and antiretroviral therapy (ART) monitoring platforms are limited [4]. HIV infection causes a variety of immune system dysfunctions [16]. CD4+ T-lymphocytes are reported as the host cells for HIV viruses[17]. The gp120 envelope glycoprotein of HIV virus binds to the CD4 receptor, initiating infection and cell damage. At the early stage of HIV infection, although no obvious signs may appear, the number of CD4+T cells in the patient body declines, undermining the immune system and eventually making the patient succumb to opportunistic infections (e.g. pneumonia) [18]. No established cure is currently available for late-stage AIDS, with several anti-retrovirus drugs reported effective in suppressing symptom onset [19]. These drugs are reported to be more effective in the earlier stages of HIV infection [20]. Early HIV infection detection may also prevent unknowing transmissions, underscoring the importance of early HIV diagnosis. Fourth-generation p24 antigen (Ag)/Ab combination (combo) enzyme immunoassays (EIA) detecting HIV p24 Ag and antibodies (followed by HIV-1/2 differentiation and rt-PCR confirmation), have significantly narrowed the diagnostic window to within 2 weeks from the time of transmission [21]. Food and Drug Administration (FDA) approved fourth-generation HIV-Ag/Ab assays include ARCHITECT HIV Ag/Ab EIA (Abbott Laboratories), GS HIV combo Ag/Ab EIA (Bio-Rad Laboratories and Walter Reed Army Institute of Research), Vitros HIV combo assay (Ortho Clinical Diagnostic), BioPlex 2200 HIV Ag-Ab assay (Bio-Rad Laboratories), and ADVIA Centaur HIV combo (Siemens Healthcare Diagnostics) [22, 23]. Commercially available HIV Rapid Diagnostic Tests (RDTs) such as Multispot HIV-1/HIV-2 Rapid Test (Bio-Rad Laboratories), HIV 1/2/O rapid test device (ABON), Determine HIV 1/2 (Alere), OraQuick Rapid HIV-1/2 Antibody Test (OraSure Technologies) and DPP HIV 1/2 (Chembio) can detect and sometimes differentiate between antibodies to HIV-1/2 in the POC setting [24]. For therapy monitoring, since the number of CD4+ T-lymphocytes declines with HIV infection and rebounds with effective ART [25], the enumeration of CD4+T-lymphocytes and quantitation of HIV viral load can be used to monitor HIV infection [26, 27]. However, these conventional methods used for CD4+T-lymphocytes and HIV viral load quantitation, including flow cytometry EIAs and quantitative RT-PCR, are limited by long turn-around-time, the need for sophisticated instruments and well-trained operators, and associated high costs [28]. There is an urgent demand for POC devices that can accurately detect and monitor HIV/AIDS in resource-limited settings, as emphasized by the World Health Organization (WHO) [29]. These POC devices for HIV detection should be accurate, inexpensive, easy to use and disposable to enable detection of HIV infection, and quantitation of CD4+ T-lymphocytes and HIV viral load in resource-limited settings[30, 31]. To meet the clinical needs, the lower detection limit of the devices needs to be at least 200 CD4+ cells per μL and 400 copies of HIV per mL of whole blood [9].
HPV
Over 50,000 women die from cervical cancer every year in Africa. In the United States, the incidence of cervical cancer was 40.1 in whites and 73.1 in nonwhites per 100,000 females prior to the wide use of Papanicolaou (Pap)-based screening, in selected areas in 1947-1948. Thanks to Pap-based screening directed precancerous lesions treatment, the incidence dropped to 7.7 per 100,000 women in 2012, underscoring the critical role of screening and early detection in cervical cancer prevention [32, 33]. Cervical intra-epithelial neoplasia (CIN), caused by persistent infection with one or more oncogenic types of HPV [34], is the target for cervical cancer screening. HPV DNA testing and/or Pap cytology, followed by colposcopy and biopsy are currently the gold standard in cervical cancer screening in developed countries. As reported by Sankaranarayanan R et al., in a large randomized trial in India, one round of HPV screening based on DNA testing in women over age 30 significantly reduced advanced cervical cancer incidence and mortality by 50% [35]. However, these tests require expensive laboratory settings and depend on a reliable recall system, making it not suitable to be deployed in large scale in resource-limited settings such as in developing countries[36]. In fact, 85% of the global cervical cancer burden is from the developing countries [37]. An alternative test to Pap-based cervical cancer screening is visual inspection with acetic acid (VIA), as recommended by the WHO for areas where resources are limited [37], which provides immediate results at a low cost. But visual inspection with VIA is not sensitive, more operator dependent, and lacks objective means of quality assurance, which may lead to over-or under-treatment [38, 39]. Simple, affordable, POC test platforms for HPV virus with both high sensitivity and high specificity are urgently needed to improve cervical cancer prevention in developing countries.
Dengue Virus and Ebola Virus
It is estimated that approximately 3 billion people from over 120 countries are at risk for Dengue virus (DENV, 4 serotypes: DENV1–DENV4) infection [40]. DENV belongs to the genus Flavivirus in the family Flaviviridae [41], with a single-stranded, positive-sense RNA genome. DENV spreads via mosquitoes and is concentrated in tropics and subtropics areas in Latin America and Asia [42]. It is the leading mosquito-borne viral infection and disease in humans, with an estimation of 390 million new infected people every year [40, 43]. In south China, multiple dengue fever outbreaks have taken place in the past ten years [44]. DENV infections can cause a range of syndromes, from dengue fever, to the potentially life-threatening severe dengue shock syndrome [43]. According to the 2009 WHO revised case definition, three forms of DENV infection caused diseases are 1) dengue, 2) dengue with warming signs, and 3) severe dengue [45]. Moreover, there are no FDA-approved protective vaccines or specific antiviral therapies to treat dengue. One dengue vaccine, Dengvaxia, is available for only patients with past DENV infections but not dengue-naive individuals [46]. Accurate and rapid detection of DENV infection is important to ensure timely management of severe dengue diseases, while avoiding over-treatment of cases with similar clinical presentations but no DENV infections. Current diagnostic strategies in central clinical laboratories for DENV infection includes virus isolation, nonstructural protein 1 (NS1) antigen immunoassays, reverse transcription-PCR (RT-PCR), and serological detection of DENV specific antibodies such as IgM and IgG [46]. Among them, RT-PCR is the method with optimal sensitivity and specificity, commonly being used as a gold standard for DENV detection [47]. However, these laboratory-based diagnostic strategies require expensive instruments and licensed operators, limiting their use in remote resource-limited regions. Simple, rapid, accurate, and affordable POCTs for DENV detection with timely on-site confirmation of suspected cases is in high demand.
Syndromes of dengue fever resemble those of other viral hemorrhagic fevers, such as those caused by Ebola virus (EBOV) [48]. EBOV is an enveloped, nonsegmented, negative single-strand RNA virus [49], first discovered in 1976 [48]. Five species of EBOV have been discovered so far: Bundibugyo, Sudan, Reston, Tai Forest and Zaire, the last of which caused over 11,000 fatalities during a recent outbreak from 2014 to 2016 in West Africa [49, 50]. EBOV detection during outbreak was primarily carried out with reverse transcription polymerase chain reaction (RT-PCR) assay [51]. Although RT-PCR can be used to successfully diagnose Ebola infections with high sensitivity and specificity, it requires laboratory-based instrument and professional training to obtain accurate results, which are usually limited in the outbreak areas. Confirmed Ebola diagnosis was made in less than 60% of the cases during the 2014-16 outbreak, due to limited availability of diagnostic tests [52]. This emphasizes the need for POC diagnostic tools during Ebola outbreak [53]. Broadhurst et al. compared the in-field performance of the ReEBOV Antigen Rapid Test kit with a benchmark RT-PCR assay for the detection of EBOV. The rapid diagnostic test demonstrated a sensitivity of 100% [95% CI 87·7–100]) [54]. Brangel et al. developed a lateral flow based POC test to detect Sudan virus with a customized smart phone application to collect both test results and geographical information. Compared with standard ELISA, this POC test detected glycoprotein monoplex with 100% sensitivity and 98% specificity[55]. Sebba et al. developed another quick POC test using surface-enhanced Raman spectroscopy nanoparticle tags (SERS nanotags) to differentiate Ebola from other endemic febrile diseases, including Lassa and malaria. This POC test can be completed in two hands-on steps and <30 min with 90.0% sensitivity and 97.9% specificity for Ebola [51].
Due to the highly contagious nature of pathogens such as Ebola, and the usually rapidly developed critical conditions of infected patients, POC testing in or close to containment facilities is also needed in well-resourced countries [56]. Guidelines are available from the Centers for Disease Control and Prevention (CDC) for infection prevention and control during sample collection, transportation, testing and disposal [57]. Real-world laboratory testing experiences have also been reported from different U.S. institutions [58, 59]. These practical aspects are important considerations when choosing POC technologies and implementing them in patient care workflows.
Mycobacterium tuberculosis
Approximately 10.4 million new cases of Tuberculosis (TB) was estimated in 2016 according to WHO, with less than 64% cases diagnosed [2], preventing timely therapeutic interventions [60]. Therefore, even though TB has now become a largely treatable disease, it remains the worldwide leading infectious cause of death [2], claiming around 1.3 million deaths every year[4]. It would be impossible to achieve the goal of the End TB strategy, with 90% reduction in incidence and 95% reduction in mortality by 2035 [61], without improved TB diagnostic tools to deliver timely therapeutic interventions. Currently standard diagnostic tools for TB include QuantiFERON-TB, liquid culture and smear microscopy [62], many of which require costly instruments, well trained individuals and large volume of samples [63]. Accurate and rapid POC diagnostics will be the key to achieve the End TB strategy. Recent years have seen impressive progresses in the field of TB POC diagnostics. In December 2010, WHO endorsed the POC Xpert® MTB/RIF assay, developed by Alland et al., in TB endemic countries [64]. Xpert® MTB/RIF assay uses a cartridge-based integrated miniature PCR system with minimal technical expertise requirement, obtaining test results from unprocessed sputum samples within 90 minutes [65]. WHO-endorsed assay tools such as urine lateral flow lipoarabinomannan (LF-LAM) and loop-mediated isothermal amplification (TB-LAMP) have also been developed, obviating the need for complicated instruments (such as thermal cycle controlling systems) [2].
Zika Virus
Zika virus (ZIKV), an Aedes mosquito-borne flavivirus was first reported in Brazil in 2015 and has rapidly spread throughout the tropical and subtropical areas of America since then [66–68]. ZIKV has been reported to lead to congenital microcephaly, Guillain- Barré syndrome (GBS)[69], and other severe neurological defects in newborns whose mothers have been infected by ZIKV during pregnancy [70]. According to an economics model by Lee et al. [71], the estimated total costs (including direct medical costs and productivity losses) will range from 0.5 to 2 billion US dollars if the ZIKV emergency occurred across six US states. ZIKV is mainly propagated via mosquitoes, with other routes coexisting such as sexual and perinatal transmission and blood transfusions [72]. Because ZIKV-infection caused symptoms such as fever and chills are similar to many other febrile diseases [68], accurate and rapid detection of ZIKV is critical for proper and timely therapeutic interventions. ZIKV detection also plays critical roles in infection spread tracking, risk management throughout pregnancy, treatment and vaccine efficacy monitoring, blood supply safety assurance and determining whether sexual partners harbor infections [72]. The Food and Drug Administration (FDA) recently authorized emergency use of the IgM Antibody Capture Enzyme-Linked Immunosorbent Assay (Zika MAC-ELISA) and Trioplex rRT-PCR laboratory test to detect ZIKV [73]. However, these assays require central lab settings including bulky instruments and well trained operators. Simple, accurate and rapid POC diagnostic tools for ZIKV detection are the key to effective treatment and prevention [74, 75].
Biomarkers in Infectious Disease POCT
The National Institutes of Health (NIH) Director’s Initiative on Biomarkers and Surrogate Endpoints define a biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention”[76]. Almost all the molecules or cells involved in the infection process of infectious diseases can be used as biomarkers, such as proteins, nucleic acids and antibodies. For example, during the HIV infection, the levels of HIV RNA genome, capsid protein p24 and different kinds of antibodies each has distinct profile signatures and can be used to assess the stages of the infection process, as shown in Figure 2. In the following section, we will review different biomarkers used in POC tests for infectious diseases, and their roles in assessing the disease stages and treatments.
Figure 2.
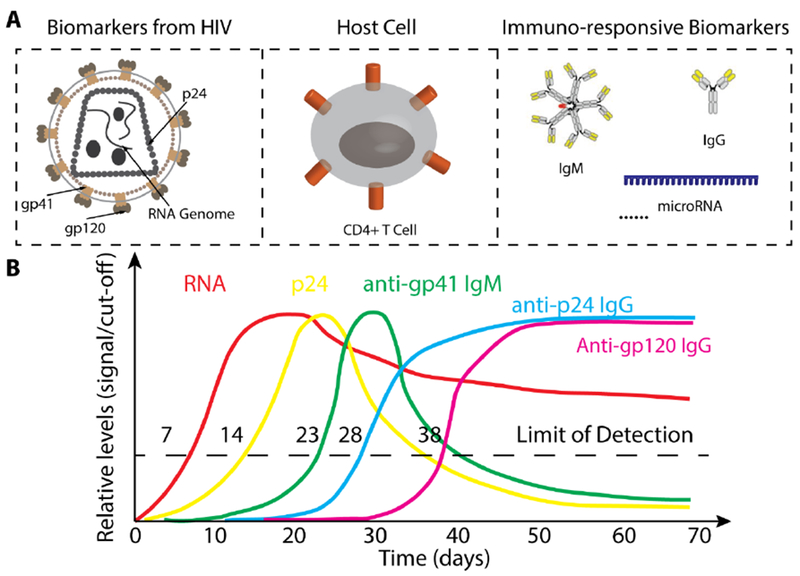
(A) Different biomarkers used for the diagnosis and monitoring of HIV infection. (B) Kinetics of different biomarkers during HIV infection. Refer to [77] for further information. Partly remade from [77].
Pathogen Nucleic Acids
Since almost all infectious diseases are caused by pathogens carrying nucleic acids (except for rare cases such as Prions), pathogen nucleic acids (RNA or DNA) can naturally serve as biomarkers for the diagnosis of infectious diseases. As a matter of fact, nucleic acid tests (NAT) for the detection of pathogen specific nucleic acid sequences have been widely used in centralized laboratories [78, 79]. The amount of pathogen genome nucleic acids also directly reflects the load of pathogen during infection. For example, quantitative RNA detection has been used to monitor HIV viral load during early infection and after treatment [77], as shown in Figure 2. A drawback using pathogen nucleic acids as biomarkers is the inability to differentiate between infection and colonization. Furthermore, traditional PCR-based NATs involve multiple sample purification/preparation steps and require costly instruments such as programable thermocycler, which renders them not suitable for POC use [80]. A lot of efforts have been made in developing accurate, simple and cost effective diagnostic tools for the detection of infectious disease-specific nucleic acids in the past decade. Multiple approaches have been exploited including: replacing PCR with isothermal amplification methods such as recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP), simplifying experimental procedures with integrated microfluidic devices, and synthetic biology approach. Maffert et al. systematically reviewed recent developments in POC nucleic acid detection for infectious diseases [80].
Antibodies
The presence of anti-pathogen antibodies can serve as biomarkers to evaluate the infectious state. During the infectious process, the immune system produces massive amount of antibodies, the level of which may be much higher than the level of pathogens. The level of antibodies may remain high during the entire infection process, while the antigen level may drop significantly at the late stage of infection. For example, at the late stage of HIV infection, anti-p24 antibody remains detectable while p24 drops down to an undetectable level [77], as shown in Figure 2. In this scenario, the antibodies are more useful for the diagnosis of infectious diseases. From the technology perspective, it is often easier to build immunoassays to detect antibodies than those to detect antigens, which require costly generation and preparation of antibodies. As an example, HIV antibody tests can easily detect antibodies up to several mg/ml with good specificity, achieving major success in performance and market share for HIV diagnosis [81, 82]. However, when the level of antibody does not corelate well with the infectious stage, antibody tests are not suitable to be used for infectious disease diagnosis. For example, infants who are not HIV virus-infected might get maternal antibody prenatally and via breastmilk and be tested antibody positive [83, 84]. People who have not yet seroconverted after HIV infection [85] or people with no or atypical antibody responses are also not suitable to be diagnosed with antibody tests [86, 87].
Pathogen Proteins
All pathogens causing infectious diseases carry proteins, such as capsid and envelope proteins. These proteins can be used as valuable biomarkers for infectious disease diagnosis. For example, the HIV virus capsid protein p24 has long been recognized as a possible substitute biomarker to HIV antibodies, which have dominated the market of HIV POC tests [77, 88]. The p24 protein is a small protein with high copy numbers, encoded by the gag gene with a molecular weight of ~24 kDa, and polymerizes to form a cup-like shell to protect the RNA genome of HIV virus [89]. Just like the genome RNA of HIV, p24 presents very early during HIV infection and can be detected before seroconversion. The Alere Determine™ HIV-1/2 AG/AB Combo rapid test, which detects both HIV-1/2 antibodies and HIV-1 p24 antigen, was approved by FDA and achieved CLIA-waived status for fingerstick whole blood [90].
Unlike nucleic acids, which can be amplified using PCR, ultra-low levels of proteins are not easily detected, which renders p24 detection later than RNA detection during HIV infection [77], as shown in Figure 2.
Circulating microRNAs
MicroRNAs (miRNAs) are non-coding RNA molecules with small sizes (~20 nt), and function to post-transcriptionally regulate gene expression [91–93]. Over 60% of mammalian mRNAs are under the regulation of corresponding miRNAs [94, 95]. MiRNAs also play critical roles in host immune response during infection [96–98], known to be routinely released into extracellular environments, especially by Immune cells [99, 100] as messengers for cell-to-cell communication [101]. It was first reported in 2008 that circulating miRNAs were detected in plasma [102] and serum samples [103, 104]. Interestingly, it has been found that extracellular miRNAs are extremely stable in body fluids including plasma, serum, urine, saliva, and semen, protected by RNA-binding proteins, high-density lipoprotein particles and lipid vesicles [105, 106]. Although the exact functions of the extracellular miRNA network are still under investigation, the potential of using circulating miRNA expression signatures as biomarkers to monitor pathological states has attracted increasing attention. For example, Fu et al. have used miRNA microarray platform (Exiqon miRCURY™ LNA) to detect 92 differentially expressed miRNAs in serum samples from patients with TB infections [107]. They found that the levels of circulating miR-93* and miR-29a were upregulated significantly in serum samples from the TB cases compared to the healthy controls. It has also been reported that two pairs of plasma miRNAs (miR-495-3p in combination with let-7b-5p, miR-151a-5p, or miR-744-5p; and miR-376a-3p in combination with miR-16-5p) can be potentially used as biomarkers for HIV-associated neurological disorders (HAND) [108].
Technology Advancements in Infectious Disease POCT
The past decade has seen significant technology advancements for the development of POCTs for infectious disease diagnosis, such as compact molecular diagnostic systems, lateral flow assays, microfluidics, plasmonic technologies and paper-based assays. Among them, microfluidics has been considered as one of the most promising solutions, offering miniaturization and integration of most of the functional modules used in central laboratory diagnostics into a portable chip [109]. Meanwhile plasmonic technologies including surface plasmon resonance (SPR), localized surface plasmon resonance (LSPR) and surface-enhanced Raman scattering (SERS), offer ideal properties as readout modules for POCTs, such as high sensitivity, label free and real time monitoring. The integration of plasmonics and microfluidic technologies can potentially serve as an ideal platform for the development of POCTs for the diagnosis of infectious diseases toward inexpensive, robust, and portable solutions [5]. In the following section, we will review recent technology advancements in microfluidics and plasmonics in the diagnosis of infectious diseases. The compact molecular diagnostic systems, typically benchtop instruments, are not reviewed here. Refer to [110, 111] for detailed reviews on these systems.
Microfluidics
Microfluidics is a technology used to manipulate very small volume of fluids (10−9 to 10−18 L) [112], offering precise, programable, spatial and temporal control of the fluids [113]. Through microfluidics technology, samples and reagents can be transported, mixed, and reacted in specific micro chambers in a precisely controlled manner [112, 114]. It is naturally an ideal platform for POC test development with many desired features such as automation, integration, and miniaturization [115, 116]. An ideal microfluidic system with “sample-to-answer” characteristic for POCT [109] is illustrated in Figure 3A. In the following section we will review several recent advances in using microfluidics technology for infectious disease POCT.
Figure 3.
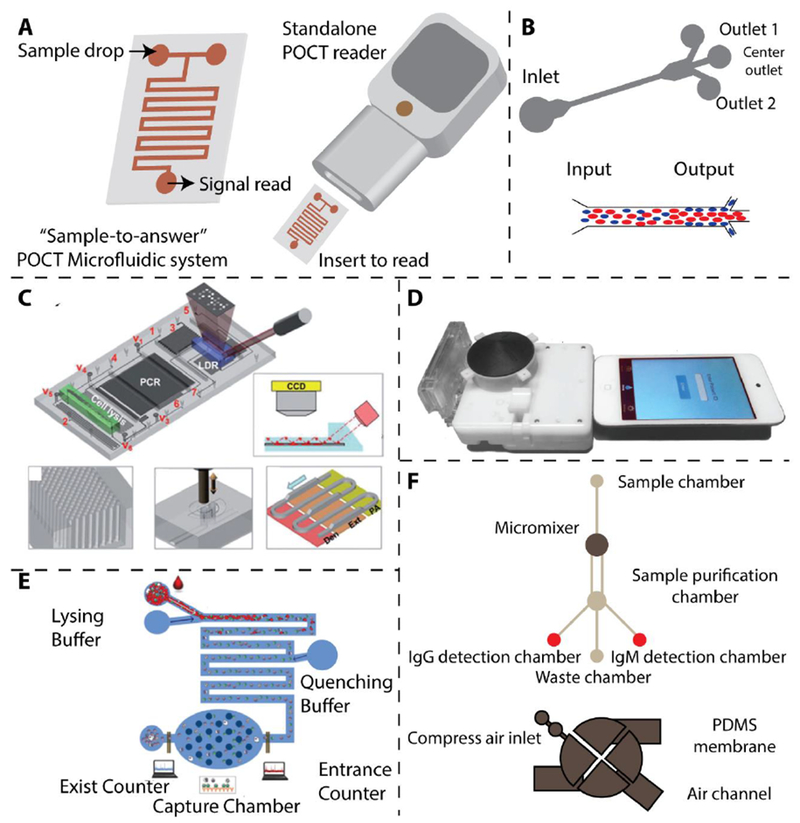
(A) Schematic of an ideal microfluidic system with: “Sample-to-answer” characteristic for POCT [109]. (B) Working principle of microfluidic device for the separation of malaria infected red blood cells (iRBC) with the concept of margination. Less deformable iRBCs are concentrated to the peripheral walls of microfluidic channel [118]. (C) An integrated microfluidic chip for sensitive detection of DNA from M. tuberculosis with on-chip PCR [120]. Reproduced with permission. (D) Microfluidic dongle for the sensitive detection of HIV [122]. Reproduced with permission. (E) Microfluidic device for the sensitive detection of HIV via electrical impedance measurement [123]. Reproduced with permission. (F) Magnetic microbeads-assisted microfluidic device for the sensitive detection of anti-dengue antibodies [124]. Refer to [4, 122] for further information.
During malaria infection, the infected red blood cells (iRBCs) progressively lose deformability as the parasites mature in the cells [117]. Based on this fact, Hou et al. designed and fabricated a microfluidic device to investigate the potential of using deformability as a biomarker to monitor the infection stages of malaria (Figure. 3B) [118, 119]. They found that the less deformable iRBCs were more likely to be displaced to the walls of the microfluidic channels. By splitting the main microchannel into side channels, they isolated more than 80% of the iRBCs (in trophozoite/schizont stages) into the side channels. However, as other diseases such as sickle cell anemia also involve RBC deformability changes, the specificity of this assay still needs to be improved. As shown in Figure 3C, Wang et al. developed an automated microfluidic device for the detection of single-base variations in multi-drug resistant forms of M. tuberculosis by integrating cell lysis, DNA isolation, PCR amplification, and signal readout into a single small cartridge [120]. They implemented micropillar array in the microchannels to increase the interaction surface for DNA adsorption to enhance the colorimetric signal for readout. The Sia group from Columbia University developed a POC microfluidic chip for the simultaneous detection of HIV and syphilis using silver enhanced immunoassays [121]. They used air bubbles to separate reagents in the microfluidic channels, and sliver reduction to enhance the colorimetric signals, enabling ELISA-like sensitivity and specificity within 20 min. Later on they have also integrated the microfluidic device into a small cartridge which can be easily readout by a mobile device such as an iPod touch (Figure 3D) [122]. Watkins et al. developed a microfluidic chip to count CD4+and CD8+ T cells for HIV infection monitoring, using differential electrical impedance measurement (Figure 3E) [123]. In their design, when the target CD4+ or CD8+ lymphocytes flow through a specific area of the microfluidic channel, a spike in impedance with specific amplitude and width is recorded. Their CD4+ and CD8+ lymphocyte counting can be completed within 20 min, with results matching well with the results via flow cytometry. Lee et. al developed an integrated microfluidic device to sensitively diagnose DENV infection by detecting specific IgG and IgM antibodies (Figure 3F) [124]. They used magnetic microbeads and micromixers to efficiently capture IgG and IgM antibodies. On-chip built magnetic coils are used to collect the purified antibodies for subsequent fluorescence readouts.
Paper-based microfluidics is considered as a low cost and user-friendly technology for infectious disease detection in POCT [125–128]. In addition, the capability of colorimetric readout also makes them useful in many resource-limited circumstances. Whiteside et al. demonstrated a microfluidic paper-based analytical device (μPAD) for the detection of antibodies to the HIV-1 envelope antigen gp41 [125]. In this design, the testing is simple and relatively fast (within 1h), and a small volume of sample (1-10 μl) is required. Other methods using microfluidics and paper-based devices for the detection of infectious diseases have been developed [126–128]. Some representative microfluidic technologies for POCT are listed In Table 1.
Table 1.
Summary of microfluidic and plasmonic POCT technologies for infectious diseases.
| POCT | Pathogen | Analyte | Detection Method | Limit of Detection | Assay time | Reference |
|---|---|---|---|---|---|---|
| Microfluidic device (non-paper-based) | Malaria | Red blood cell | Deformation | N/A | N/A | [118] |
| Malaria | Red blood cell | Deformation | N/A | N/A | [119] | |
| M. tuberculosis | DNA | Colorimetric | 50 cells/ml | N/A | [144] | |
| HIV | Antibody | Colorimetric | N/A | 20 min | [145] | |
| HIV | Antibody | Optical | N/A | 15 min | [122] | |
| HIV | CD4 and CD8 T cell | Electricity | 12 cells/μl | 20 min | [123] | |
| Dengue | IgG and IgM antibodies | Fluorescence | 21 pg | 30 min | [124] | |
| Microfluidic device (Paper-based) | HIV | HIV-1 gp41 | Colorimetric | N/A | 51 min | [125] |
| Ebola | Viral RNA | Colorimetric | 107 copy/ml | 20 min | [126] | |
| TB | TB-DNA | Colorimetric | 1.95×10−2 ng/ml | 60 min | [127] | |
| ZIKA | Viral RNA | Colorimetric | 1 copy/μl | 15 min | [128] | |
| Plasmonic Technology | HIV | A, B, C, D, E, G and panel subtypes | LSPR | 98±39 copies/ml for HIV subtype D | 1h for capture and 10 min for detection and analysis | [138] |
| Ebola | VSV glycoprotein | LSPR | 106 PFU/ml | More than 90 min | [146] | |
| TB | TB-DNA | Colorimetric | 10 μg/ml | Less than 2h | [147] | |
| ZIKA | Viral RNA | Fluorescence | 1.7 copy/ml | 3 min | [148] |
Plaque-forming units: PFU
Plasmonic Technologies
Plasmonics studies the interaction between light and the conductive electrons of metallic nanomaterials [129]. Common plasmonic metals include gold, silver and aluminum [129, 130]. Various plasmonic nanomaterials have been designed and fabricated to target POC applications owing to their label-free nature, facile optical tunability and high sensitivity to surrounding medium [131, 132]. For example, Peng et al. developed a coulometric POC test using engineered phage-induced gold nanoparticle aggregation to detect bacterial pathogens [133]. Recent progresses in highly sensitive optical transducers have further driven rapid development of plasmonic applications. Among the various optical sensing platforms, the unique surface plasmon resonance (SPR) properties of plasmonic nanomaterials make it a highly promising method for chemical and biological sensing and clinical diagnostics [134–137]. Based on the sensitivity of the SPR to the changes in the dielectric properties of the surrounding medium, and the enhancement of the electromagnetic (EM) field in proximity to noble metal nanostructures, two important classes of plasmonic sensors have evolved: localized surface plasmon resonance (LSPR) and surface-enhanced Raman scattering (SERS) sensors.
The LSPR relies on the high sensitivity of plasmonic nanomaterial to refractive index changes [129]. It has been used for label-free, fluorescence-free and repeatable HIV viral load detection using unprocessed whole blood (Figure 4A) [138]. This sensing platform is based on the binding events of biomarkers that lead to the LSPR wavelength shift. This technology enables the detection and quantification of multiple HIV subtypes with high sensitivity, specificity, and relatively short assay time (1 h for capture and 10 min for detection and analysis). Additionally, prism coupling configuration for SPR excitation has been employed for label-free clinical protein detection (Figure 4B) [139]. In this configuration, the capture of biological samples is monitored via the change in refractive index, which results in the change in reflected light.
Figure 4.
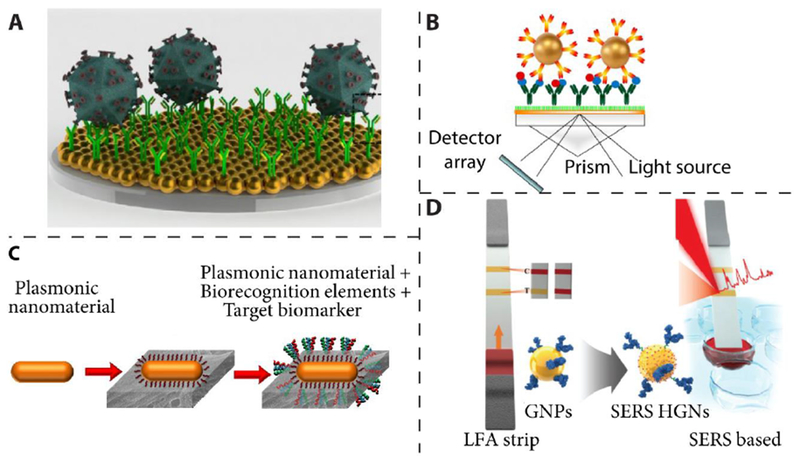
(A) Illustration of nanoplasmonic viral load platform for the detection of intact virus. Reproduced with permission from [138]. (B) Schematic representation of SPR-based protein sandwich assay. Reproduced with permission from [139]. (C) Schematic representing the LSPR-based biosensor with peptide recognition elements. Reproduced with permission from [141]. (D) Illustration of the configuration of SERS-based lateral flow assay for detection of staphylococcal enterotoxin B. Reproduced with permission from [143].
Paper-based devices offer numerous advantages such as high surface area, small sample volume requirement, portable, flexible, and low cost [140]. Plasmonic paper LSPR device has been demonstrated for the selective and sensitive detection of protein biomarkers (Figure 4C), which makes it ideal for POC diagnosis of infectious diseases in a resource-limited setting [141].
SERS involves the large amplification of the Raman scattering from analytes adsorbed on or nearby to a nanostructured metal surface [142]. Extensive efforts have been dedicated to the design and fabrication of SERS device with large signal amplification and uniform enhancement. SERS-based lateral flow assay has been developed for detection of staphylococcal enterotoxin B with ultrahigh sensitivity compared to ELISA-based detection methods (Figure 4D) [143]. Some representative plasmonic technologies for POCT are listed In Table 1.
Conclusions and Outlook
In summary, with its simplicity, short turnaround time, and wide accessibility, POC tests pave the way for prompt diagnosis of infectious diseases especially in resources-lim ited settings, which in turn enables timely and effective patientcentric treatment and management. Although a number of biomarkers have been successfully used as targets in POC tests for infectious diseases, biomarkers with better sensitivity and specificity are still needed. Systematic characterization of a set of biomarker signatures for a single infectious disease may prove to be a useful approach in future biomarker screening. Since many infectious diseases may present with similar clinical symptoms, POC tests with multiplex functionality are also highly desirable. Significant advances have been achieved in novel technology development for POC infectious disease testing in the past decade, including microfluidics and plasmonic technologies. Stringent clinical validations are still needed for these technologies to be translated from research to clinical practice. Many practical issues including infection control, testing in confined environment, information technology connectivity and optimization of clinical pathway are also important considerations for successful implementation to meet clinical challenges [149].
Highlights.
POCT plays critical roles in diagnosis, treatment and prevention of infectious diseases.
Simple, accurate, multiplex and widely accessible POC tests are needed for many major pathogens.
POCT technologies have advanced significantly in the past decade, including microfluidics and plasmonics.
Acknowledgement
We would like to acknowledge NIH RO1DA035868, Penn Center for Precision Medicine and Department of Pathology and Laboratory Medicine at University of Pennsylvania for funding support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hwang H, Hwang BY, Bueno J, Biomarkers in Infectious Diseases, Dis Markers 2018. ( 2018) 8509127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Garcia-Basteiro AL, DiNardo A, Saavedra B, Silva DR, Palmero D, Gegia M, Migliori GB, Duarte R, Mambuque E, Centis R, Cuevas LE, Izco S, Theron G, Point of care diagnostics for tuberculosis, Pulmonology 24(2) (2018) 73–85. [DOI] [PubMed] [Google Scholar]
- [3].Chen H, Hagstrom AE, Kim J, Garvey G, Paterson A, Ruiz-Ruiz F, Raja B, Strych U, Rito-Palomares M, Kourentzi K, Conrad JC, Atmar RL, Willson RC, Flotation Immunoassay: Masking the Signal from Free Reporters in Sandwich Immunoassays, Sci Rep 6 (2016) 24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tay A, Pavesi A, Yazdi SR, Lim CT, Warkiani ME, Advances in microfluidics in combating infectious diseases, Biotechnol Adv 34(4) (2016) 404–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tokel O, Inci F, Demirci U, Advances in plasmonic technologies for point of care applications, Chem Rev 114(11) (2014) 5728–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sachs J, Malaney P,The economic and social burden of malaria, Nature 415(6872) (2002) 680–5. [DOI] [PubMed] [Google Scholar]
- [7].Hay SI, Okiro EA, Gething PW, Patil AP, Tatem AJ, Guerra CA, Snow RW, Estimating the global clinical burden of Plasmodium falciparum malaria in 2007, PLoS Med 7(6) (2010) e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kim S, Nhem S, Dourng D, Menard D, Malaria rapid diagnostic test as point-of-care test: study protocol for evaluating the VIKIA Malaria Ag Pf/Pan, Malar J 14 (2015) 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee WG, Kim YG, Chung BG, Demirci U, Khademhosseini A, Nano/Microfluidics for diagnosis of infectious diseases in developing countries, Adv Drug Deliv Rev 62(4-5) (2010) 449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jorgensen P, Chanthap L, Rebueno A, Tsuyuoka R, Bell D, Malaria rapid diagnostic tests in tropical climates:the need for a cool chain, Am J Trop Med Hyg 74(5) (2006) 750–4. [PubMed] [Google Scholar]
- [11].Moody A, Rapid diagnostic tests for malaria parasites, Clin Microbiol Rev 15(1) (2002) 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bell D Wongsrichanalai C, Barnwell JW, Ensuring quality and access for malaria diagnosis: how can it be achieved?, Nat Rev Microbiol 4(9 Suppl) (2006) S7–20. [DOI] [PubMed] [Google Scholar]
- [13].Bell D Wongsrichanalai C, Barnwell JW, Ensuring quality and access for malaria diagnosis: how can it be achieved?, Nature Reviews Microbiology 4(2006) S7. [DOI] [PubMed] [Google Scholar]
- [14].Rafael ME, Taylor T, Magill A, Lim YW, Girosi F, Allan R, Reducing the burden of childhood malaria in Africa: the role of improved, Nature 444Suppl 1 (2006) 39–48. [DOI] [PubMed] [Google Scholar]
- [15].Antia M, Herricks T, Rathod PK, Microfluidic Modeling of Cell-Cell Interactions in Malaria Pathogenesis, PLOS Pathogens 3(7) (2007) e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fauci AS, HIV and AIDS: 20 years of science, Nature Medicine 9(2003) 839. [DOI] [PubMed] [Google Scholar]
- [17].Dalgleish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, Weiss RA, The Cd4 (T4) Antigen Is an Essential Component of the Receptor for the Aids Retrovirus, Nature 312(5996) (1984) 763–767. [DOI] [PubMed] [Google Scholar]
- [18].Dalgleish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, Weiss RA, The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus, Nature 312 (1984) 763. [DOI] [PubMed] [Google Scholar]
- [19].A.S.S.W.G.o.H.I.V.C. The International, Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun T-W, Churchill M, Mascio MD, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud M-C, Peterlin M, Poll G, Routy J-P, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Lint CV, Verdin E, Woolfrey A, Zaia J, Barré-Sinoussi F, Towards an HIV cure: a global scientific strategy, Nature Reviews Immunology 12 (2012) 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kitahata MM, Gange SJ, Abraham AG, Merriman B, Saag MS, Justice AC, Hogg RS, Deeks SG, Eron JJ, Brooks JT, Rourke SB, Gill MJ, Bosch RJ, Martin JN, Klein MB, Jacobson LP, Rodriguez B, Sterling TR, Kirk GD, Napravnik S, Rachlis AR, Calzavara LM, Horberg MA, Silverberg MJ, Gebo KA, Goedert JJ, Benson CA, Collier AC, Van Rompaey SE, Crane HM, McKaig RG, Lau B, Freeman AM, Moore RD, Investigators N-A, Effect of early versus deferred antiretroviral therapy for HIV on survival, N Engl J Med 360(18) (2009) 1815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stone M, Bainbridge J, Sanchez AM, Keating SM, Pappas A, Rountree W, Todd C, Bakkour S, Manak M, Peel SA, Coombs RW, Ramos EM, Shriver MK, Contestable P, Nair SV, Wilson DH, Stengelin M, Murphy G, Hewlett I, Denny TN, Busch MP, Comparison of Detection Limits of Fourth- and Fifth-Generation Combination HIV Antigen-Antibody, p24 Antigen, and Viral Load Assays on Diverse HIV Isolates, J Clin Microbiol 56(8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stone M, Bainbridge J, Sanchez AM, Keating SM, Pappas A, Rountree W, Todd C, Bakkour S, Manak M, Peel SA, Coombs RW, Ramos EM, Shriver MK, Contestable P, Nair SV, Wilson DH, Stengelin M, Murphy G, Hewlett I, Denny TN, Busch MP, Comparison of Detection Limits of Fourth- and Fifth-Generation Combination HIV Antigen-Antibody, p24 Antigen, and Viral Load Assays on Diverse HIV Isolates, Journal of Clinical Microbiology 56(8) (2018) e02045–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Qiu X, Sokoll L, Yip P, Elliott DJ, Dua R, Mohr P, Wang XY, Spencer M, Swanson P, Dawson GJ, Hackett J, Comparative evaluation of three FDA-approved HIV Ag/Ab combination tests using a genetically diverse HIV panel and diagnostic specimens, Journal of Clinical Virology 92 (2017) 62–68. [DOI] [PubMed] [Google Scholar]
- [24]. https://www.who.int/diagnostics_laboratory/evaluations/PQ_list/en/.
- [25].Simon V, Ho DD, HIV-1 dynamics in vivo: implications for therapy, Nature Reviews Microbiology 1 (2003) 181. [DOI] [PubMed] [Google Scholar]
- [26].O’Gorman MR, Gelman R, Inter- and intrainstitutional evaluation of automated volumetric capillary cytometry for the quantitation of CD4- and CD8-positive T lymphocytes in the peripheral blood of persons infected with human immunodeficiency virus. Site Investigators and the NIAID New CD4 Technologies Focus Group, Clin Diagn Lab Immunol 4(2) (1997) 173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M, Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection, Nature 373 (1995) 123. [DOI] [PubMed] [Google Scholar]
- [28].Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W, H.I.V.R.A.V.L.A.W.G. Forum for Collaborative, HIV-1 viral load assays for resource-limited settings, PLoS Med 3(10) (2006) e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Willyard C, Simpler tests for immune cells could transform AIDS care in Africa, Nature Medicine 13 (2007) 1131. [DOI] [PubMed] [Google Scholar]
- [30].Cohen J, Monitoring Treatment: At What Cost?, Science 304(5679) (2004) 1936. [DOI] [PubMed] [Google Scholar]
- [31].Linder V, Sia SK, Whitesides GM, Reagent-loaded cartridges for valveless and automated fluid delivery in microfluidic devices, Anal Chem 77(1) (2005) 64–71. [DOI] [PubMed] [Google Scholar]
- [32].Jemal A, Bray F, Forman D, O’Brien M, Ferlay J, Center M, Parkin DM, Cancer burden in Africa and opportunities for prevention, Cancer 118(18) (2012) 4372–84. [DOI] [PubMed] [Google Scholar]
- [33].Mohammed SI, Ren W, Flowers L, Rajwa B, Chibwesha CJ, Parham GP, Irudayaraj JM, Point-of-care test for cervical cancer in LMICs, Oncotarget 7(14) (2016) 18787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJF, Peto J, Meijer CJLM, Muñoz N, Human papillomavirus is a necessary cause of invasive cervical cancer worldwide, The Journal of Pathology 189(1) (1999) 12–19. [DOI] [PubMed] [Google Scholar]
- [35].Sankaranarayanan R, Nene BM, Shastri SS, Jayant K, Muwonge R, Budukh AM, Hingmire S, Malvi SG, Thorat R, Kothari A, Chinoy R, Kelkar R, Kane S, Desai S, Keskar VR, Rajeshwarkar R, Panse N, Dinshaw KA, HPV screening for cervical cancer in rural India, N Engl J Med 360(14) (2009) 1385–94. [DOI] [PubMed] [Google Scholar]
- [36].Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F, Ferlay J, Worldwide burden of cervical cancer in 2008, Ann Oncol 22(12) (2011) 2675–86. [DOI] [PubMed] [Google Scholar]
- [37].Campos NG, Tsu V, Jeronimo J, Mvundura M, Kim JJ, Estimating the value of point-of-care HPV testing in three low- and middle-income countries: a modeling study, BMC Cancer 17(1) (2017) 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mahé C, Gaffikin L, Screening test accuracy studies: how valid are our conclusions? Application to visual inspection methods for cervical screening, Cancer Causes & Control 16(6) (2005) 657–666. [DOI] [PubMed] [Google Scholar]
- [39].Denny LA, Sankaranarayanan R, De Vuyst H, Kim JJ, Adefuye PO, Alemany L, Adewole IF, Awolude OA, Parham G, de Sanjose S, Bosch FX, Recommendations for cervical cancer prevention in sub-saharan Africa, Vaccine 31 Suppl 5 (2013) F73–4. [DOI] [PubMed] [Google Scholar]
- [40].Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI, The global distribution and burden of dengue, Nature 496 (2013) 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gubler DJ, The global pandemic of dengue/dengue haemorrhagic fever: current status and prospects for the future, Ann Acad Med Singapore 27(2) (1998) 227–34. [PubMed] [Google Scholar]
- [42].Rigau-Perez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV, Dengue and dengue haemorrhagic fever, Lancet 352(9132) (1998) 971–7. [DOI] [PubMed] [Google Scholar]
- [43].Guzman MG, Harris E, Dengue, The Lancet 385(9966) (2015) 453–465. [DOI] [PubMed] [Google Scholar]
- [44].Chen B, Liu Q, Dengue fever in China, The Lancet 385(9978) (2015) 1621–1622. [DOI] [PubMed] [Google Scholar]
- [45].Wang WK, Gubler DJ, Potential Point-of-Care Testing for Dengue Virus in the Field, J Clin Microbiol 56(5) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schwartz LM, Halloran ME, Durbin AP, Longini IM, The dengue vaccine pipeline: Implications for the future of dengue control, Vaccine 33(29) (2015) 3293–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pang J, Chia PY, Lye DC, Leo YS, Progress and Challenges towards Point-of-Care Diagnostic Development for Dengue, Journal of Clinical Microbiology 55(12) (2017) 3339–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Benzine JW, Brown KM, Agans KN, Godiska R, Mire CE, Gowda K, Converse B, Geisbert TW, Mead DA, Chander Y, Molecular Diagnostic Field Test for Point-of-Care Detection of Ebola Virus Directly From Blood, J Infect Dis 214(suppl 3) (2016) S234–S242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kaushik A, Tiwari S, Dev Jayant R, Marty A, Nair M, Towards detection and diagnosis of Ebola virus disease at point-of-care, Biosensors and Bioelectronics 75 (2016) 254–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tan D-X, Korkmaz A, Reiter RJ, Manchester LC, Ebola virus disease: potential use of melatonin as a treatment, Journal of Pineal Research 57(4) (2014) 381–384. [DOI] [PubMed] [Google Scholar]
- [51].Sebba D, Lastovich AG, Kuroda M, Fallows E, Johnson J, Ahouidi A, Honko AN, Fu H, Nielson R, Carruthers E, Diedhiou C, Ahmadou D, Soropogui B, Ruedas J, Peters K, Bartkowiak M, Magassouba N, Mboup S, Amor YB, Connor JH, Weidemaier K, A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases, Sci Transl Med 10(471) (2018). [DOI] [PubMed] [Google Scholar]
- [52].Dhillon RS, Srikrishna D, Sachs J, Controlling Ebola: next steps, Lancet 384(9952) (2014) 1409–11. [DOI] [PubMed] [Google Scholar]
- [53].Clark DV, Kibuuka H, Millard M, Wakabi S, Lukwago L, Taylor A, Eller MA, Eller LA, Michael NL, Honko AN, Olinger GG Jr., Schoepp RJ, Hepburn MJ, Hensley LE, Robb ML, Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study, Lancet Infect Dis 15(8) (2015) 905–12. [DOI] [PubMed] [Google Scholar]
- [54].Broadhurst MJ, Kelly JD, Miller A, Semper A, Bailey D, Groppelli E, Simpson A, Brooks T, Hula S, Nyoni W, Sankoh AB, Kanu S, Jalloh A, Ton Q, Sarchet N, George P, Perkins MD, Wonderly B, Murray M, Pollock NR, ReEBOV Antigen Rapid Test kit for point-of-care and laboratory-based testing for Ebola virus disease: a field validation study, Lancet 386(9996) (2015) 867–74. [DOI] [PubMed] [Google Scholar]
- [55].Brangel P, Sobarzo A, Parolo C, Miller BS, Howes PD, Gelkop S, Lutwama JJ, Dye JM, McKendry RA, Lobel L, Stevens MM, A Serological Point-of-Care Test for the Detection of IgG Antibodies against Ebola Virus in Human Survivors, ACS Nano 12(1) (2018) 63–73. [DOI] [PubMed] [Google Scholar]
- [56].Jelden KC, Iwen PC, Herstein JJ, Biddinger PD, Kraft CS, Saiman L, Smith PW, Hewlett AL, Gibbs SG, Lowe JJ, Ebola US Treatment Center Clinical Laboratory Support, Journal of Clinical Microbiology 54(4) (2016) 1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].https://www.cdc.gov/vhf/ebola/laboratory-personnel/index.html.
- [58].Duncan A, Winkler AM, Kraft CS, Hill CE, Parslow TG, Burd EM, Ryan EL, Ritchie JC, Cardella JC, Laboratory Test Support for Ebola Patients Within a High-Containment Facility, Laboratory Medicine 45(3) (2014) e109–e111. [DOI] [PubMed] [Google Scholar]
- [59].Iwen PC, Pirruccello SJ, Hinrichs SH, Wisecarver JL, Lowe JJ, Gibbs SG, Sambol AR, Herrera VL, Stiles K, Salerno KJ, Garrett JL, An Integrated Approach to Laboratory Testing for Patients with Ebola Virus Disease, Laboratory Medicine 45(4) (2014) e146–e151. [DOI] [PubMed] [Google Scholar]
- [60].McNerney R, Daley P, Towards a point-of-care test for active tuberculosis: obstacles and opportunities, Nature Reviews Microbiology 9 (2011) 204. [DOI] [PubMed] [Google Scholar]
- [61].Uplekar M, Weil D, Lonnroth K, Jaramillo E, Lienhardt C, Dias HM, Falzon D, Floyd K, Gargioni G, Getahun H, Gilpin C, Glaziou P, Grzemska M, Mirzayev F, Nakatani H, Raviglione M, WHO’s new End TB Strategy, The Lancet 385(9979) (2015) 1799–1801. [DOI] [PubMed] [Google Scholar]
- [62].DHEDA K, RUHWALD M, THERON G, PETER J, YAM WC, Point-of-care diagnosis of tuberculosis: Past, present and future, Respirology 18(2) (2013) 217–232. [DOI] [PubMed] [Google Scholar]
- [63].Mani V, Wang S, Inci F, De Libero G, Singhal A, Demirci U, Emerging technologies for monitoring drug-resistant tuberculosis at the point-of-care, Advanced Drug Delivery Reviews 78 (2014) 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lawn SD, Nicol MP, Xpert® MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance, Future microbiology 6(9) (2011) 1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NTN, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D, Rapid Detection of <em>Mycobacterium tuberculosis</em> and Rifampin Resistance by Use of On-Demand, Near-Patient Technology, Journal of Clinical Microbiology 48(1) (2010) 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Benelli G, Mehlhorn H, Declining malaria, rising of dengue and Zika virus: insights for mosquito vector control, Parasitology Research 115(5) (2016) 1747–1754. [DOI] [PubMed] [Google Scholar]
- [67].Lewnard JA, Gonsalves G, Ko AI, Low Risk of International Zika Virus Spread due to the 2016 Olympics in Brazil, Annals of internal medicine 165(4) (2016) 286–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Waggoner JJ, Pinsky BA, Zika Virus: Diagnostics for an Emerging Pandemic Threat, Journal of clinical microbiology 54(4) (2016) 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F, Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study, The Lancet 387(10027) (2016) 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C, Instrument-Free Point-of-Care Molecular Detection of Zika Virus, Anal Chem 88(14) (2016) 7289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lee BY, Alfaro-Murillo JA, Parpia AS, Asti L, Wedlock PT, Hotez PJ, Galvani AP, The potential economic burden of Zika in the continental United States, PLoS Negl Trop Dis 11(4) (2017) e0005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kindhauser MK, Allen T, Frank V, Santhana RS, Dye C, Zika: the origin and spread of a mosquito-borne virus, Bull World Health Organ 94(9) (2016) 675–686C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mauk MG, Song J, Bau HH, Liu C, Point-of-Care Molecular Test for Zika Infection, Clin Lab Int 41 (2017) 25–27. [PMC free article] [PubMed] [Google Scholar]
- [74].Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawché F, Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study, Lancet (London, England) 387(10027) (2016) 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming G-L, Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth, Cell stem cell 18(5) (2016) 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].G. Biomarkers Definitions Working, Biomarkers and surrogate endpoints: preferred definitions and conceptual framework, Clin Pharmacol Ther 69(3) (2001) 89–95. [DOI] [PubMed] [Google Scholar]
- [77].Gray ER, Bain R, Varsaneux O, Peeling RW, Stevens MM, McKendry RA, p24 revisited: a landscape review of antigen detection for early HIV diagnosis, AIDS 32(15) (2018) 2089–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Niemz A, Ferguson TM, Boyle DS, Point-of-care nucleic acid testing for infectious diseases, Trends Biotechnol 29(5) (2011) 240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen H, Crum M, Chavan D, Vu B, Kourentzi K, Willson RC, Nanoparticle-Based Proximity Ligation Assay for Ultrasensitive, Quantitative Detection of Protein Biomarkers, ACS Appl Mater Interfaces 10(38) (2018) 31845–31849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Maffert P, Reverchon S, Nasser W, Rozand C, Abaibou H, New nucleic acid testing devices to diagnose infectious diseases in resource-limited settings, Eur J Clin Microbiol Infect Dis 36(10) (2017) 1717–1731. [DOI] [PubMed] [Google Scholar]
- [81].Denkinger CM, Nicolau I, Ramsay A, Chedore P, Pai M, Are peripheral microscopy centres ready for next generation molecular tuberculosis diagnostics?, Eur Respir J 42(2) (2013) 544–7. [DOI] [PubMed] [Google Scholar]
- [82].Nath N, Wunderlich C, Darr FW 2nd, Douglas DK, Dodd RY, Immunoglobulin level in donor blood reactive for antibodies to human immunodeficiency virus, J Clin Microbiol 25(2) (1987) 364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rakusan TA, Parrott RH, Sever JL, Limitations in the laboratory diagnosis of vertically acquired HIV infection, J Acquir Immune Defic Syndr 4(2) (1991) 116–21. [PubMed] [Google Scholar]
- [84].Kellerman S, Essajee S, HIV testing for children in resource-limited settings: what are we waiting for?, PLoS medicine 7(7) (2010) e1000285–e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Busch MP, Satten GA, Time course of viremia and antibody seroconversion following human immunodeficiency virus exposure, Am J Med 102(5B) (1997) 117–24; discussion 125-6. [DOI] [PubMed] [Google Scholar]
- [86].Jindai K, Kunzer B, Van TT, Striker R, Human immunodeficiency virus testing pitfalls and clinical suspicion, Am J Emerg Med 32(11) (2014) 1442 e1–2. [DOI] [PubMed] [Google Scholar]
- [87].Chin BS, Lee SH, Kim GJ, Kee MK, Suh SD, Kim SS, Early identification of seronegative human immunodeficiency virus type 1 infection with severe presentation, J Clin Microbiol 45(5) (2007) 1659–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].George E, Beauharnais CA, Brignoli E, Noel F, Bois G, De Matteis Rouzier P, Altenor M, Lauture D, Hosty M, Mehta S, Wright PF, Pape JW, Potential of a simplified p24 assay for early diagnosis of infant human immunodeficiency virus type 1 infection in Haiti, J Clin Microbiol 45(10) (2007) 3416–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P, Mature HIV-1 capsid structure by cryo-electron microscopy and allatom molecular dynamics, Nature 497(7451) (2013) 643–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Parker MM, Bennett SB, Sullivan TJ, Fordan S, Wesolowski LG, Wroblewski K, Gaynor AM, Performance of the Alere Determine™ HIV-1/2 Ag/Ab Combo Rapid Test with algorithm-defined acute HIV-1 infection specimens, Journal of Clinical Virology 104 (2018) 89–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, Shaughnessy RG, Circulating microRNAs as Potential Biomarkers of Infectious Disease, Frontiers in immunology 8(2017) 118–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lee RC, Feinbaum RL, Ambros V, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell 75(5) (1993) 843–54. [DOI] [PubMed] [Google Scholar]
- [93].Wightman B, Ha I, Ruvkun G, Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans, Cell 75(5) (1993) 855–62. [DOI] [PubMed] [Google Scholar]
- [94].O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D, Physiological and pathological roles for microRNAs in the immune system, Nat Rev Immunol 10(2) (2010) 111–22. [DOI] [PubMed] [Google Scholar]
- [95].Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD, MicroRNAs: new regulators of immune cell development and function, Nat Immunol 9(8) (2008) 839–45. [DOI] [PubMed] [Google Scholar]
- [96].Lu L-F, Liston A, MicroRNA in the immune system, microRNA as an immune system, Immunology 127(3) (2009) 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].O’Connell RM, Rao DS, Baltimore D, microRNA regulation of inflammatory responses, Annu Rev Immunol 30 (2012) 295–312. [DOI] [PubMed] [Google Scholar]
- [98].Chen CZ, Schaffert S, Fragoso R, Loh C, Regulation of immune responses and tolerance: the microRNA perspective, Immunol Rev 253(1) (2013) 112–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Robbins PD, Morelli AE, Regulation of immune responses by extracellular vesicles, Nat Rev Immunol 14(3) (2014) 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].de Candia P, De Rosa V, Casiraghi M, Matarese G, Extracellular RNAs: A Secret Arm of Immune System Regulation, J Biol Chem 291(14) (2016) 7221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY, Secreted monocytic miR-150 enhances targeted endothelial cell migration, Mol Cell 39(1) (2010) 133–44. [DOI] [PubMed] [Google Scholar]
- [102].Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM, Detection and characterization of placental microRNAs in maternal plasma, Clin Chem 54(3) (2008) 482–90. [DOI] [PubMed] [Google Scholar]
- [103].Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL, Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma, Br J Haematol 141(5) (2008) 672–5. [DOI] [PubMed] [Google Scholar]
- [104].Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M, Circulating microRNAs as stable blood-based markers for cancer detection, Proceedings of the National Academy of Sciences of the United States of America 105(30) (2008) 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K, The microRNA spectrum in 12 body fluids, Clin Chem 56(11) (2010) 1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M, Argonaute 2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma, Proc Natl Acad Sci U S A 108(12) (2011) 5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fu Y, Yi Z, Wu X, Li J, Xu F, Circulating microRNAs in patients with active pulmonary tuberculosis, J Clin Microbiol 49(12) (2011) 4246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kadri F, LaPlante A, De Luca M, Doyle L, Velasco-Gonzalez C, Patterson JR, Molina PE, Nelson S, Zea AH, Parsons CH, Peruzzi F, Defining Plasma MicroRNAs Associated With Cognitive Impairment In HIV-Infected Patients, J Cell Physiol 231(4) (2016) 829–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Jung W, Han J, Choi J-W, Ahn CH, Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies, Microelectronic Engineering 132 (2015) 46–57. [Google Scholar]
- [110].Holland CA, Kiechle FL, Point-of-care molecular diagnostic systems — past, present and future, Current Opinion in Microbiology 8(5) (2005) 504–509. [DOI] [PubMed] [Google Scholar]
- [111].Zarei M, Advances in point-of-care technologies for molecular diagnostics, Biosens Bioelectron 98 (2017) 494–506. [DOI] [PubMed] [Google Scholar]
- [112].Whitesides GM, The origins and the future of microfluidics, Nature 442 (2006) 368. [DOI] [PubMed] [Google Scholar]
- [113].Beebe DJ, Mensing GA, Walker GM, Physics and Applications of Microfluidics in Biology, Annual Review of Biomedical Engineering 4 (1) (2002) 261–286. [DOI] [PubMed] [Google Scholar]
- [114].Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Haeggstrom E, Kuritzkes DR, Demirci U, Rapid automated cell quantification on HIV microfluidic devices, Lab Chip 9(23) (2009) 3364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dittrich PS, Manz A, Lab-on-a-chip: microfluidics in drug discovery, Nat Rev Drug Discov 5(3) (2006) 210–8. [DOI] [PubMed] [Google Scholar]
- [116].West J, Becker M, Tombrink S, Manz A, Micro total analysis systems: latest achievements, Anal Chem 80(12) (2008) 4403–19. [DOI] [PubMed] [Google Scholar]
- [117].Cranston HA, Boylan CW, Carroll GL, Sutera SP, Williamson JR, Gluzman IY, Krogstad DJ, Plasmodium falciparum maturation abolishes physiologic red cell deformability, Science 223(4634) (1984) 400–403. [DOI] [PubMed] [Google Scholar]
- [118].Hou HW, Bhagat AAS, Lin Chong AG, Mao P, WeiTan KS, Han J, Lim CT, Deformability based cell margination—A simple microfluidic design for malaria-infected erythrocyte separation, Lab on a Chip 10(19) (2010) 2605–2613. [DOI] [PubMed] [Google Scholar]
- [119].Alizadehrad D, Imai Y, Nakaaki K, lshikawa T, Yamaguchi T, Quantification of red blood cell deformation at high-hematocrit blood flow in microvessels, Journal of Biomechanics 45(15) (2012) 2684–2689. [DOI] [PubMed] [Google Scholar]
- [120].Wang H, Chen H-W, Hupert ML, Chen P-C, Datta P, Pittman TL, Goettert J, Murphy MC, Williams D, Barany F, Soper SA, Fully integrated thermoplastic genosensor for the highly sensitive detection and identification of multi-drug-resistant tuberculosis, Angewandte Chemie (International ed. in English) 51(18) (2012) 4349–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK, Microfluidics-based diagnostics of infectious diseases in the developing world, Nat Med 17(8) (2011) 1015–9. [DOI] [PubMed] [Google Scholar]
- [122].Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK, A smartphone dongle for diagnosis of infectious diseases at the point of care, Science Translational Medicine 7(273) (2015) 273re1–273re1. [DOI] [PubMed] [Google Scholar]
- [123].Watkins NN, Hassan U, Damhorst G, Ni H, Vaid A, Rodriguez W, Bashir R, Microfluidic CD4+ and CD8+ T Lymphocyte Counters for Point-of-Care HIV Diagnostics Using Whole Blood, Science Translational Medicine 5(214) (2013) 214ral70–214ral70. [DOI] [PubMed] [Google Scholar]
- [124].Lee Y-F, Lien K-Y, Lei H-Y, Lee G-B, An integrated microfluidic system for rapid diagnosis of dengue virus infection, Biosensors and Bioelectronics 25(4) (2009) 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Cheng C-M, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM, Paper-Based ELISA, Angewandte Chemie International Edition 49(28) (2010) 4771–4774. [DOI] [PubMed] [Google Scholar]
- [126].Magro L, Jacquelin B, Escadafal C, Garneret P, Kwasiborski A, Manuguerra J-C, Monti F, Sakuntabhai A, Vanhomwegen J, Lafaye P, labeling P, Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics, Scientific Reports 7(1) (2017) 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tsai T-T, Huang C-Y, Chen C-A, Shen S-W, Wang M-C, Cheng C-M, Chen C-F, Diagnosis of Tuberculosis Using Colorimetric Gold Nanoparticles on a Paper-Based Analytical Device, ACS Sensors 2(9) (2017) 1345–1354. [DOI] [PubMed] [Google Scholar]
- [128].Kaarj K, Akarapipad P, Yoon J-Y, Simpler, Faster, and Sensitive Zika Virus Assay Using Smartphone Detection of Loop-mediated Isothermal Amplification on Paper Microfluidic Chips, Scientific Reports 8(1) (2018) 12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Halas NJ, Lai S, Chang W-S, Link S, Nordlander P, Plasmons in Strongly Coupled Metallic Nanostructures, Chemical Reviews 111(6) (2011) 3913–3961. [DOI] [PubMed] [Google Scholar]
- [130].Knight MW, King NS, Liu L, Everitt HO, Nordlander P, Halas NJ, Aluminum for Plasmonics, ACS Nano 8(1) (2014) 834–840. [DOI] [PubMed] [Google Scholar]
- [131].Tian L, Tadepalli S, Hyun Park S, Liu K-K, Morrissey JJ, Kharasch ED, Naik RR, Singamaneni S, Bioplasmonic calligraphy for multiplexed label-free biodetection, Biosensors and Bioelectronics 59 (2014) 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sepúlveda B, Angelomé PC, Lechuga LM, Liz-Marzán LM, LSPR-based nanobiosensors, Nano Today 4(3) (2009) 244–251. [Google Scholar]
- [133].Peng H, Chen IA, Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages, ACS Nano (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Jarvis RM, Goodacre R, Characterisation and identification of bacteria using SERS, Chemical Society Reviews 37(5) (2008) 931–936. [DOI] [PubMed] [Google Scholar]
- [135].Homola J, Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species, Chemical Reviews 108(2) (2008) 462–493. [DOI] [PubMed] [Google Scholar]
- [136].Golightly RS, Doe ring WE, Natan MJ, Surface-Enhanced Raman Spectroscopy and Homeland Security: A Perfect Match?, ACS Nano 3(10) (2009) 2859–2869. [DOI] [PubMed] [Google Scholar]
- [137].Vo-Dinh T, Wang H-N, Scaffidi J, Plasmonic nanoprobes for SERS biosensing and bioimaging, Journal of Biophotonics 3(1-2) (2010) 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Inci F, Tokel O, Wang S, Gurkan UA, Tasoglu S, Kuritzkes DR, Demirci U, Nanoplasmonic Quantitative Detection of Intact Viruses from Unprocessed Whole Blood, ACS Nano 7(6) (2013) 4733–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Uludag Y, Tothill IE, Cancer Biomarker Detection in Serum Samples Using Surface Plasmon Resonance and Quartz Crystal Microbalance Sensors with Nanoparticle Signal Amplification, Analytical Chemistry 84(14) (2012) 5898–5904. [DOI] [PubMed] [Google Scholar]
- [140].Martinez AW, Phillips ST, Butte MJ, Whitesides GM, Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays, Angewandte Chemie International Edition 46(8) (2007) 1318–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Tadepalli S, Kuang Z, Jiang Q, Liu K-K, Fisher MA, Morrissey JJ, Kharasch ED, Slocik JM, Naik RR, Singamaneni S, Peptide Functionalized Gold Nanorods for the Sensitive Detection of a Cardiac Biomarker Using Plasmonic Paper Devices, Scientific Reports 5 (2015) 16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Nie S, Emory SR, Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering, Science 275(5303) (1997) 1102–1106. [DOI] [PubMed] [Google Scholar]
- [143].Hwang J, Lee S, Choo J, Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B, Nanoscale 8(22) (2016) 11418–11425. [DOI] [PubMed] [Google Scholar]
- [144].Wang H, Chen H-W, Hupert ML, Chen P-C, Datta P, Pittman TL, Goettert J, Murphy MC, Williams D, Barany F, Soper SA, Fully Integrated Thermoplastic Genosensor for the Highly Sensitive Detection and Identification of Multi-Drug-Resistant Tuberculosis, Angewandte Chemie International Edition 51(18) (2012) 4349–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK, Microfluidics-based diagnostics of infectious diseases in the developing world, Nature Medicine 17(2011) 1015. [DOI] [PubMed] [Google Scholar]
- [146].Yanik AA, Huang M, Kamohara O, Artar A, Geisbert TW, Connor JH, Altug H, An Optofluidic Nanoplasmonic Biosensor for Direct Detection of Live Viruses from Biological Media, Nano Letters 10(12) (2010) 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Veigas B, Jacob JM, Costa MN, Santos DS, Viveiros M, Inacio J, Martins R, Barquinha P, Fortunato E, Baptista PV, Gold on paper–paper platform for Au-nanoprobe TB detection, Lab on a Chip 12(22) (2012) 4802–4808. [DOI] [PubMed] [Google Scholar]
- [148].Adegoke O, Morita M, Kato T, lto M, Suzuki T, Park EY, Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays, Biosensors and Bioelectronics 94 (2017) 513–522. [DOI] [PubMed] [Google Scholar]
- [149].Wang P, Kricka LJ, Current and Emerging Trends in Point-of-Care Technology and Strategies for Clinical Validation and Implementation, Clinical Chemistry 64(10) (2018) 1439–1452. [DOI] [PubMed] [Google Scholar]


