Abstract
Nanomedicines have been demonstrated to have passive or active tumor targeting behaviors, which are promising for cancer chemotherapy. However, most of the nanomedicines still suffer from suboptimal targeting effect and drug leakage, resulting in unsatisfactory treatment outcome. Herein, a hierarchical responsive nanomedicine (HRNM) is developed for programmed delivery of chemotherapeutics. The HRNMs are prepared via the self-assembly of cyclic RGD peptide conjugated triblock copolymer, poly(2-(hexamethyleneimino)ethyl methacrylate)-poly(oligo-(ethylene glycol) monomethyl ether methacrylate)-poly reduction-responsive camptothecin (PC7A-POEG-PssCPT). In blood circulation, RGD peptides are shielded by POEG coating; therefore, the nanosized HRNMs can achieve effective tumor accumulation through passive targeting. Once the HRNMs reach tumor site, due to the acidic tumor microenvironment induced hydrophobic-to-hydrophilic conversion of PC7A chains, the RGD peptides will be exposed for enhanced tumor retention and cellular internalization. Moreover, in response to the glutathione (GSH) inside cells, active CPT drugs will be released rapidly for chemotherapy. The in vitro and in vivo results confirmed effective tumor targeting, potent antitumor effect and reduced systemic toxicity of the HRNMs. This HRNM is promising for enhanced chemotherapeutic delivery.
Keywords: Hierarchical responsiveness, Programmed tumor targeting, Triggered drug release, Polyprodrug, Nanomedicine
A hierarchical responsive nanomedicine, which can achieve programmed targeting and triggered drug release, is developed for enhanced chemotherapeutic delivery. In physiological condition, the nanomedicine shows high stability without obvious drug leakage. Once they reach tumor microenvironment, the activated RGD peptides could achieve enhanced cellular uptake. Then active camptothecin moieties would be rapidly released in response to the intracellular glutathione.
Cancer, a kind of disease features abnormal cell proliferation, has been one of the major threats to human beings.[1] To treat cancer, chemotherapy is a commonly used approach in the clinic; however, the conventional chemotherapy suffers from low bioavailability and adverse effects to normal tissues, which is mainly due to the nonspecific biodistribution of therapeutic drugs.[2] Nanomedicines have shown promising passive tumor targeting behavior through the enhanced permeability and retention (EPR) effect.[3] Furthermore, active targeting could be achieved by conjugating nanomedicines with ligands that specifically bind to overexpressed receptors on the cancer cells.[4]
To date, however, most of the nanomedicines still face many challenges to achieve satisfactory treatment outcome. For example, conventional nanomedicines use nanocarriers for physical loading of drugs, exhibiting uncontrolled drug release behaviors such as burst release and premature drug leakage. By exploiting the stimuli-responsiveness of the smart nanomaterials, controlled drug release can be achieved, permitting improved bioavailability and reduced side effects.[5] One strategy to achieve the on-demand drug release is the use of stimuli-responsive nanocarriers for encapsulation of drugs;[6] however, most of them still suffer from low drug loading content and drug leakage during blood circulation.[7] The use of stimuli-responsive bonds to conjugate drugs to carrier materials provides another strategy.[8] For example, various polyprodrugs that conjugate drugs to the polymer chains through responsive linkers have been developed for self-assembly, resulting in nanomedicines with high drug loading content and loading stability.[9]
Despite application potential of the nanomedicines based on stimuli-responsive polyprodrugs, the targeting effect of most nanomedicines is currently suboptimal. To improve the targeting effect of nanomedicines, the surface properties of the nanomedicines should be rationally designed. However, in different delivery stages, the nanomedicines are expected to have different properties. For instance, during blood circulation, neutral surface and ‘stealth’ coating are preferred for prolonged circulation time and effective tumor accumulation;[10] while in tumor tissue, positive charge and surface ligand are necessary to achieve enhanced tumor retention and intracellular drug delivery.[11] To address this problem, various programmed targeting strategies have been developed.[12] In general, the design of programmed targeting strategies is mainly based on stimuli-responsive nanomaterials which can undergo property changes (e.g. surface charge conversion, ligand activation) under special stimuli in the tumor microenvironment.[13] Thus the programmed targeting strategies combining the advantages of passive targeting and active targeting show great potential to achieve improved tumor targeting efficiency.[14] In order to endow nanomedicines with the ability to change their surface properties, the most common method is the use of responsive chemical bonds; however, most of these chemical bonds suffer from low stability under physiological condition or low sensitivity to tumor microenvironment.[15] Previous studies have proven that poly(2-(hexamethyleneimino) ethyl methacrylate) (PC7A) is an ultra pH sensitive polymer with a transition pH of about 6.9.[16] At physiological pH, PC7A is a stable hydrophobic polymer; while it effectively converts to hydrophilic polymer with positive charge at tumor extracellular pH.[17] Therefore, the PC7A shows great potential for the development of tumor-responsive programmed targeting nanomedicines.
Herein, we developed a hierarchical responsive nanomedicine (HRNM) for programmed CARIR (circulation, accumulation, retention, internalization and drug release) chemotherapeutic delivery (Figure 1). The nanomedicine is formed by a cyclic RGD peptide conjugated triblock copolymer, RGD-PC7A-POEG-PssCPT, consisting of hydrophobic PC7A chain, hydrophilic poly(oligo-(ethylene glycol) monomethyl ether methacrylate) (POEG) segment and hydrophobic poly reduction-responsive camptothecin (CPT) prodrug (PssCPT) chain. At physiological pH (approximately 7.4), both the RGD-PC7A and PssCPT segments were trapped inside the hydrophobic core of HRNMs and thus the targeting function of RGD peptides were shielded by the POEG coating, resulting in high stability and effective tumor accumulation. Once the HRNMs accumulated in the tumor tissue (pH < 6.8), the protonated PC7A changed to hydrophilic segments with positive charge, permitting charge conversion of the HRNMs and exposure of RGD peptides. Thus, the exposed positive charge and RGD peptide could bind to tumor cells, resulting in enhanced tumor retention and cellular internalization of HRNMs. Afterwards, in the presence of intracellular glutathione (GSH), CPT moieties in their active form were released through a cascade elimination reaction. Compared to conventional nanomedicines, the HRNMs demonstrate three distinct features: i) the polyprodrug segment allows high drug loading content and loading stability; ii) the PC7A segment permits high stability under physiological condition and high sensitivity to tumor microenvironment; iii) the programmed targeting effect and intracellular GSH-triggered drug release lead to potent antitumor effect and reduced systemic toxicity. Therefore, this hierarchical responsive strategy provides a promising approach for enhanced chemotherapeutic delivery.
Figure 1.
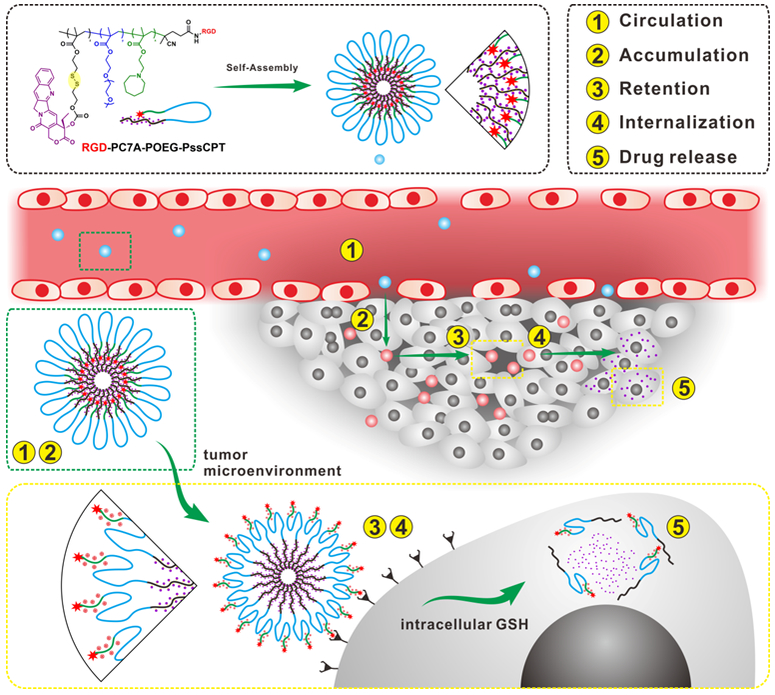
Schematic illustration of HRNMs for programmed CARIR chemotherapeutic delivery. The HRNMs show high stability in blood circulation (①) due to their nanoscale particle size and neutral POEG surface; therefore, they can effectively accumulate in the tumor via the EPR effect (②). In tumor microenvironment, acidic pH will cause charge conversion of the HRNMs and exposure of RGD peptides, permitting enhanced tumor retention (③) and cellular internalization (④). Thereafter, intracellular GSH will trigger CPT release for cancer chemotherapy (⑤).
The pH-responsive C7A monomer and reduction-responsive CPT prodrug monomer (ssCPT) were firstly synthesized and their chemical structures were confirmed by NMR and liquid chromatography–mass spectrometry (LC-MS) analysis (Scheme S1 and Figures S1-S5, Supporting Information). Then PC7A-POEG-PssCPT was obtained through reversible addition fragmentation chain transfer (RAFT) polymerization (Scheme S2 and Figures S6-S8, Supporting Information). To serve as control groups, the non-responsive polymer (PCHA-POEG-PccCPT), pH-responsive polymer (PC7A-POEG-PccCPT) and reduction-responsive polymer (PCHA-POEG-PssCPT) were also synthesized, respectively (Schemes S3-S4 and Figures S9-S12, Supporting Information). The CPT content in PC7A-POEG-PssCPT was determined to be 30.1% by NMR analysis. The absorption spectra showed that the PC7A-POEG-PssCPT and all three control polymers had the typical peak of CPT (~370 nm), indicating successful polymerization (Figure S13, Supporting Information). The CPT content of PC7A-POEG-PssCPT was determined to be 29.3% according to a standard curve (Figures S14-S15, Supporting Information), which was consistent with the 1H NMR result. The critical micelle concentration (CMC) values of the PC7A-POEG-PssCPT and PCHA-POEG-PssCPT were measured to be 2.05 mg L−1 and 3.09 mg L−1 by using pyrene as fluorescence probe (Figure S16, Supporting Information), indicating good stability in water. Afterwards, RGD peptide was conjugated to the polymers by the NHS/NH2 coupling reaction.
The RGD-PC7A-POEG-PssCPT self-assembled into nanoparticles in aqueous solution to afford HRNMs. Similarly, NRNMs, pRNMs and rRNMs, were also prepared as control groups by assembly of RGD-PCHA-POEG-PccCPT, RGD-PC7A-POEG-PccCPT and RGD-PCHA-POEG-PssCPT, respectively. As shown in the transmission electron microscopy (TEM) image of HRNMs (Figure 2a), spherical nanoparticles with a diameter of 85.6 ± 11.9 nm can be observed. The hydrodynamic diameter of HRNMs (in hydrated state) measured by dynamic light scattering (DLS) was 99.8 ± 17.9 nm (Figure 2b), which was slightly larger than the diameter in dry state measured by TEM. Moreover, the HRNMs exhibited excellent colloidal stability in phosphate buffered solution (PBS) or culture medium with fetal bovine serum (FBS) (Figure 2c). This nanosized diameter and good colloidal stability make the HRNMs suitable for passive tumor targeting. The morphologies and hydrodynamic diameters of the control groups were similar to those of HRNMs (Figure S17, Supporting Information). The zeta potentials of nanoparticles prepared by PC7A-POEG-PssCPT, RGD-PC7A-POEG-PssCPT, PCHA-POEG-PssCPT and RGD-PCHA-POEG-PssCPT were measured by DLS. As shown in Figure S18 (Supporting Information), all the nanoparticles showed slight negative surface charge due to POEG surface coating.
Figure 2.

a) TEM image and b) particle diameter of HRNMs. c) Colloidal stability of HRNMs in different conditions. Zeta potential changes of HRNMs (d) and rRNMs (e) at pH 7.4, 7.0 or 6.6. f) Mechanism of GSH-triggered CPT release from the nanomedicines. In vitro CPT release profiles of HRNMs (g) and pRNMs (h) in different conditions. i) Representative TEM images recorded for HRNMs and pRNMs after 24 h incubation with 10 mM GSH.
To demonstrate the pH-responsiveness of HRNMs in acidic environment, the HRNMs were incubated with PBS at different pH values (7.4, 7.0 or 6.6), and their zeta potentials were monitored by DLS. As shown in Figure 2d, the HRNMs had slight negative surface charge at pH 7.4 and 7.0 due to the POEG coating. However, the zeta potential of the HRNMs increased rapidly and reached about 10 mV after 2 h of incubation at pH 6.6, attributing to the hydrophobic-to-hydrophilic conversion and the following exposure of RGD-PC7A segment. In contrast, the rRNMs showed stable surface charge at all pH values, demonstrating no pH responsiveness of the rRNMs (Figure 2e). To investigate the GSH-triggered CPT release behaviors of HRNMs and pRNMs in vitro, the samples were dialyzed against PBS with or without 10.0 mM GSH. As shown in Figure 2g, less than 5% of CPT release was observed for HRNMs in the absence of GSH after 48 h incubation. However, the CPT was released rapidly in the presence of GSH, in which about 80% of CPT release was achieved upon 48 h incubation. The released products from HRNMs were analyzed by LC-MS. As shown in Figure S19 (Supporting Information), after treating the nanomedicines with GSH, free CPT was generated and released due to the GSH-induced cascade elimination reaction (Figure 2f). After incubating with GSH for 24 h, the morphologies of HRNMs were observed by TEM, which confirmed disassembly of nanostructures (Figure 2i). Considering the low GSH concentration (2−20 μM) under physiological condition and high GSH concentration (2−10 mM) in cancer cells, the HRNMs is promising to achieve reduced side effects caused by leaked drugs. In contrast, for the pRNMs, the drug release rates in all four conditions showed no significant difference (Figure 2h).
The RGD peptide can bind to αvβ3 integrin, which is overexpressed on some tumor cells (e.g. U87MG).[18] Therefore, under acidic condition, the exposure of RGD peptides is expected to achieve enhanced cellular internalization of HRNMs. To demonstrate this, U87MG cells treated with HRNMs or rRNMs at different pH were analyzed using confocal fluorescence microscopy and flow cytometry (FCM). As shown in Figure 3, weak FITC fluorescence (green) was observed at pH 7.4 for the cells treated with HRNMs and rRNMs, confirming low cellular internalization of the nanomedicines under this condition. However, much higher fluorescence intensity in cells incubated with HRNMs at pH 6.6 was observed, demonstrating the cellular internalization of HRNMs was remarkably enhanced in acidic environment. In contrast, the fluorescence intensity in cells incubated with rRNMs at pH 7.4 or 6.6 showed no obvious difference. To observe the CPT fluorescence inside cells, the U87MG cells were incubated with HRNMs or rRNMs and then stained with SYTO 13 (green). At pH 6.6, higher CPT fluorescence intensity demonstrated more amount of CPT molecules were delivered into cells by HRNMs (Figure S20, Supporting Information). Then A549 cells (αvβ3-negative) were also used to evaluate cellular uptake of the HRNMs. As shown in Figure S21 (Supporting Information), when treated with FITC-labeled HRNMs at pH 6.6, the cellular uptake of HRNMs was slightly higher than that at pH 7.4. These results demonstrated that the pH-responsive cellular internalization of HRNMs was due to the pH-induced exposure of RGD peptides and positive charges.
Figure 3.
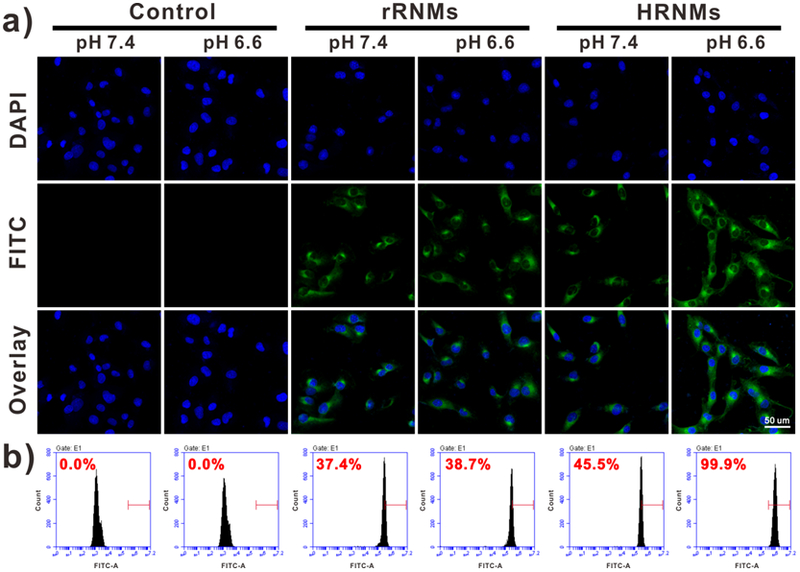
a) Confocal fluorescence images of U87MG cells upon incubation with FITC-labeled HRNMs and rRNMs at different pH for 6 h. b) Cellular internalization analysis of U87MG cells by FCM.
We further investigated the in vitro anticancer activities of CPT and nanomedicines by using MTT assay. Neither PC7A-POEG nor PCHA-POEG showed any obvious cytotoxicity to U87MG cells even at a high concentration (Figure S22, Supporting Information). However, as shown in Figure 4a, both free CPT and nanomedicines showed dose-dependent anticancer activities. The pRNMs and NRNMs inhibited cell proliferation only at high CPT concentrations due to the inefficient CPT release of these nanomedicines. However, HRNMs showed efficient anticancer activities at pH 6.6, with an IC50 value of 1.61 μM, which was 4.2-fold lower than that of HRNMs at pH 7.4 (Figure 4b). This enhanced anticancer effect is attributed to the higher cellular internalization of the HRNMs in acidic condition. In contrast, the rRNMs without pH-responsive PC7A chain didn’t show significant difference in the IC50 values when incubated at different pH values. After treatments, the live/dead cells were stained by calcein AM (green)/propidium iodide (PI, red). As shown in Figure 4c, when treated with HRNMs at pH 6.6, fewer live cells were observed when compared to that at pH 7.4, in good agreement with the result obtained from MTT assay.
Figure 4.
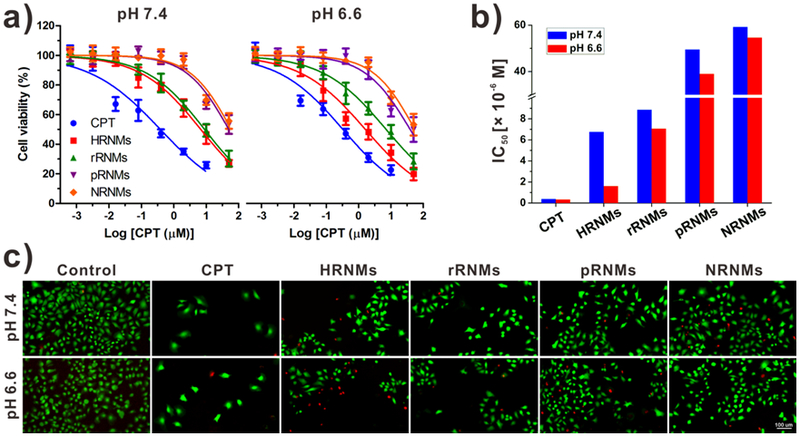
a) Cell viability of U87MG cells measured by MTT assay. b) The IC50 values of different samples. Cells were treated with free CPT, HRNMs, pRNMs, rRNMs or NRNMs at pH 7.4 or 6.6. c) Fluorescence images of U87MG cells after treatments (CPT concentration: 2 μM) and Calcein AM (green)/PI (red) costaining.
To investigate the in vivo drug delivery efficiency of the nanomedicines, the tumor accumulation and biodistribution of HRNMs and rRNMs were studied by positron emission tomography (PET) imaging. For 64Cu radionuclide labeling, 1,4,7-triazacyclononane-1,4,7-triacetate (NOTA) conjugated poly CPT (NOTA-PCPT) was synthesized (Scheme S5, Supporting Information) and encapsulated in nanomedicines. Then the 64Cu-HRNMs and 64Cu-rRNMs were intravenously injected into U87MG tumor-bearing mice. As shown in the PET images (Figure 5a), at 24 h after intravenous injection of 64Cu-HRNMs or 64Cu-rRNMs, obvious tumor signals were observed. The distribution of nanomedicines in the heart area and tumor was determined by using quantitative region-of-interest (ROI) analysis. As shown in Figure S23 (Supporting Information), the blood concentration of nanomedicines in heart slowly decreased over time. As shown in Figure 5b, both 64Cu-HRNMs and 64Cu-rRNMs had effective tumor accumulations, which was mainly due to the EPR effect. More importantly, compared to 64Cu-rRNMs, 64Cu-HRNMs showed higher tumor accumulation and retention. The tumor uptakes of 64Cu-HRNMs were 5.07 and 3.94 %ID g−1 at 24 and 48 h post-injection, respectively. This enhanced tumor accumulation and retention demonstrated that the exposure of RGD peptides and positive charges in tumor microenvironment could enhance the cellular internalization of the HRNMs. At 48 h post-injection, the mice were sacrificed to collect tumors and major organs for quantification of radioactivity by using a γ-counter. The tumor uptake of HRNMs at 48 h post-injection was 3.83 %ID g−1, which was 1.32-fold higher than that of rRNMs (Figure 5c). The biodistribution result also confirmed high tumor accumulation and prolonged tumor retention of the HRNMs.
Figure 5.
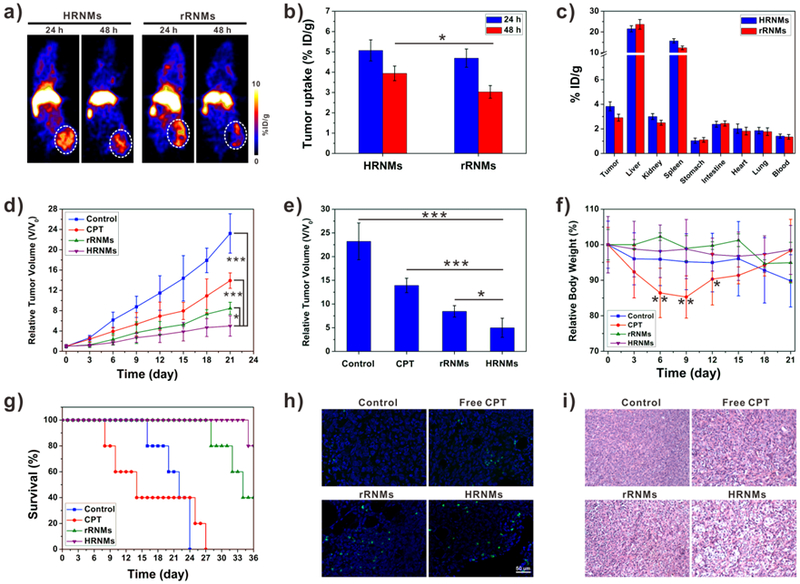
a) PET images of U87MG tumor-bearing mice after intravenous injection of 64Cu-HRNMs or 64Cu-rRNMs. b) Tumor uptake of 64Cu-HRNMs or 64Cu-rRNMs at 24 and 48 h post-injection. (*P < 0.05). c) Biodistribution of tumor and primary organs at 48 h post-injection. d) Tumor growth curves of the mice upon different treatments. e) Tumor sizes on the 21th day (*P < 0.05, ***P < 0.001). f) Mice body weight changes during the treatments (*P < 0.05, **P < 0.01). g) Survival curves of the mice. h) TUNEL and i) H&E analyses of tumor tissues after different treatments.
Encouraged by the relatively high tumor accumulation and triggered drug release behavior, the HRNMs were further employed for in vivo antitumor therapy on U87MG tumor-bearing mice. Mice were treated with saline, free CPT, HRNMs or rRNMs every 3 days (CPT-equivalent dose: 3 mg kg−1). As shown in Figure 5d, tumors grew rapidly for the mice administered with saline, the relative tumor volume was more than 20 after 21 days. Compared to the saline control group, the free CPT and nanomedicine treatments delayed tumor growth. Particularly, the HRNMs group showed the most potent antitumor effect. On day 21, the average tumor volume in HRNMs group was 4.65-fold lower than that of the control group (Figure 5e). Furthermore, no obvious body weight loss was caused by the treatment of nanomedicines (Figure 5f). The hematoxylin and eosin (H&E) staining results further confirmed that nanomedicines did not cause noticeable tissue damages in major organs (heart, liver, spleen, lung, and kidneys) of mice (Figure S24, Supporting Information). These results demonstrated that the nanomedicines exhibited low systemic toxicity and were safe for in vivo applications. However, the free CPT treated mice suffered from obvious weight loss and organ damage, indicating serious systemic toxicity of free CPT. The reduced side effect of nanomedicines was attributed to the controlled drug release behavior. The CPT in the nanomedicines was in its inactive state with low toxicity. The CPT release rate of nanomedicines was relatively low, especially in the environment with low GSH concentration. Therefore, a large amount of CPT will be metabolized by liver and spleen before releasing. In accordance with the antitumor effect, when treated with HRNMs, the survival time of mice was greatly prolonged (Figure 5g). To further investigate the CPT-induced tumor cell apoptosis, one mouse in each group was euthanized after treatment, and the tumors were collected for terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and H&E staining.[19] The tumors treated with HRNMs showed much more TUNEL-apoptotic cells and tumor necrosis (Figure 5h and 5i), indicating enhanced therapeutic efficacy.
In summary, a hierarchical responsive nanomedicine (HRNM) was developed for programmed targeting and triggered drug release. At physiological pH, RGD peptides were trapped inside the hydrophobic core of HRNMs, resulting in high stability in blood circulation and effective tumor accumulation through EPR effect. Once the HRNMs reached acidic tumor microenvironment, the rapid hydrophobic-to-hydrophilic conversion of PC7A chains led to the exposure of RGD peptides. The exposed RGD peptides could achieve enhanced tumor retention and cellular internalization of HRNMs. Then active CPT moieties would be released in response to the intracellular GSH. The in vivo results demonstrated effective tumor targeting, potent antitumor effect and reduced systemic toxicity of the HRNMs. Therefore, this HRNM holds great promise for enhanced chemotherapeutic delivery.
Supplementary Material
Acknowledgements
This work was supported by the Shaanxi Science & Technology Coordination & Innovation Project (2016KTCQ03–09), The International Cooperation Program of Xijing Hospital (XJZT15G01), Shaanxi Innovation Capability Support Plan (2018PT-08) and the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH).
Contributor Information
Sheng Wang, Department of Nuclear Medicine, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, China; Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Guocan Yu, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Zhantong Wang, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Orit Jacobson, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Rui Tian, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Li-Sen Lin, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Fuwu Zhang, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
Jing Wang, Department of Nuclear Medicine, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, China.
Xiaoyuan Chen, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland 20892, United States.
References
- [1] a).Hanahan D, Weinberg RA, Cell 2000, 100, 57; [DOI] [PubMed] [Google Scholar]; b) Cheng L, Wang C, Feng L, Yang K, Liu Z, Chem. Rev 2014, 114, 10869; [DOI] [PubMed] [Google Scholar]; c) Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, The Lancet 2013, 380, 2095. [Google Scholar]
- [2] a).Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R, Nat. Nanotechnol 2007, 2, 751; [DOI] [PubMed] [Google Scholar]; b) Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A, Science 2012, 338, 903; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Nat. Mater 2011, 10, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3] a).Lou JW, Philp L, Hou W, Walsh CD, Liu J, Charron DM, Zheng G, Nanomedicine 2018, 13, 977; [DOI] [PubMed] [Google Scholar]; Nie S, Xing Y, Kim GJ, Simons JW, Annu. Rev. Biomed. Eng 2007, 9, 257; [DOI] [PubMed] [Google Scholar]; b) Ferrari M, Nat. Rev. Cancer 2005, 5, 161; [DOI] [PubMed] [Google Scholar]; c) Overchuk M, Zheng G, Biomaterials 2018, 156, 217. [DOI] [PubMed] [Google Scholar]
- [4] a).Shen Z, Chen T, Ma X, Ren W, Zhou Z, Zhu G, Zhang A, Liu Y, Song J, Li Z, Ruan H, Fan W, Lin L, Munasinghe J, Chen X, Wu A, ACS Nano 2017, 11, 10992; [DOI] [PubMed] [Google Scholar]; b) Wang H, Zhao P, Liang X, Gong X, Song T, Niu R, Chang J, Biomaterials 2010, 31, 4129; [DOI] [PubMed] [Google Scholar]; c) Wang S, Yang W, Cui J, Li X, Dou Y, Su L, Chang J, Wang H, Zhang B, Biomater. Sci 2016, 4, 338. [DOI] [PubMed] [Google Scholar]
- [5] a).Li X, Schumann C, Albarqi HA, Lee CJ, Alani AWG, Bracha S, Milovancev M, Taratula O, Theranostics 2018, 8, 767; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mura S, Nicolas J, Couvreur P, Nat. Mater 2013, 12, 991; [DOI] [PubMed] [Google Scholar]; c) Karimi M, Ghasemi A, Sahandi Zangabad P, Rahighi R, Moosavi Basri SM, Mirshekari H, Amiri M, Shafaei Pishabad Z, Aslani A, Bozorgomid M, Ghosh D, Beyzavi A, Vaseghi A, Aref AR, Haghani L, Bahrami S, Hamblin MR, Chem. Soc. Rev 2016, 45, 1457; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ge Z, Liu S, Chem. Soc. Rev 2013, 42, 7289; [DOI] [PubMed] [Google Scholar]; e) Zhou L, Wang H, Li Y, Theranostics 2018, 8, 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6] a).Rim HP, Min KH, Lee HJ, Jeong SY, Lee SC, Angew. Chem. Int. Ed 2011, 50, 8853; [DOI] [PubMed] [Google Scholar]; b) Xiao Z, Ji C, Shi J, Pridgen EM, Frieder J, Wu J, Farokhzad OC, Angew. Chem. Int. Ed 2012, 51, 11853; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen G, Jaskula‐Sztul R, Esquibel CR, Lou I, Zheng Q, Dammalapati A, Harrison A, Eliceiri KW, Tang W, Chen H, Adv. Funct. Mater 2017, 27, 1604671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7] a).Cheng R, Meng F, Deng C, Klok H-A, Zhong Z, Biomaterials 2013, 34, 3647; [DOI] [PubMed] [Google Scholar]; b) Zhang CY, Yang YQ, Huang TX, Zhao B, Guo XD, Wang JF, Zhang LJ, Biomaterials 2012, 33, 6273; [DOI] [PubMed] [Google Scholar]; c) Wang S, Wang H, Liu Z, Wang L, Wang X, Su L, Chang J, Nanoscale 2014, 6, 7635. [DOI] [PubMed] [Google Scholar]
- [8] a).Xu X, Saw PE, Tao W, Li Y, Ji X, Bhasin S, Liu Y, Ayyash D, Rasmussen J, Huo M, Shi J, Farokhzad OC, Adv. Mater 2017, 29, 1700141; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheetham AG, Chakroun RW, Ma W, Cui H, Chem. Soc. Rev 2017, 46, 6638; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Duan X, Xiao J, Yin Q, Zhang Z, Yu H, Mao S, Li Y, ACS Nano 2013, 7, 5858; [DOI] [PubMed] [Google Scholar]; d) Yang D, Dai Y, Liu J, Zhou Y, Chen Y, Li C, Ma P. a., Lin J, Biomaterials 2014, 35, 2011; [DOI] [PubMed] [Google Scholar]; e) Cheetham AG, Lin YA, Lin R, Cui H, Acta Pharmacol. Sin 2017, 38, 874; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Zhu L, Wang D, Wei X, Zhu X, Li J, Tu C, Su Y, Wu J, Zhu B, Yan D, Control J. Release 2013, 169, 228; [DOI] [PubMed] [Google Scholar]; g) Rao N V, Mane S, Kishore A, Das Sarma J, Shunmugam R, Biomacromolecules 2011, 13, 221; [DOI] [PubMed] [Google Scholar]; h) Ma W, Su H, Cheetham AG, Zhang W, Wang Y, Kan Q, Cui H, Control J. Release 2017, 263, 102; [DOI] [PubMed] [Google Scholar]; i) Su H, Zhang P, Cheetham AG, Koo JM, Lin R, Masood A, Schiapparelli P, Quinones-Hinojosa A, Cui H, Theranostics 2016, 6, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9] a).Zhang F, Zhu G, Jacobson O, Liu Y, Chen K, Yu G, Ni Q, Fan J, Yang Z, Xu F, Fu X, Wang Z, Ma Y, Niu G, Zhao X, Chen X, ACS nano 2017, 11, 8838; [DOI] [PubMed] [Google Scholar]; b) Hu X, Liu G, Li Y, Wang X, Liu S, J. Am. Chem. Soc 2015, 137, 362; [DOI] [PubMed] [Google Scholar]; c) Guo D, Xu S, Huang Y, Jiang H, Yasen W, Wang N, Su Y, Qian J, Li J, Zhang C, Zhu X, Biomaterials 2018, 177, 67; [DOI] [PubMed] [Google Scholar]; d) Hu X, Hu J, Tian J, Ge Z, Zhang G, Luo K, Liu S, J. Am. Chem. Soc 2013, 135, 17617; [DOI] [PubMed] [Google Scholar]; e) Li J, Ke W, Wang L, Huang M, Yin W, Zhang P, Chen Q, Ge Z, J. Control. Release 2016, 225, 64; [DOI] [PubMed] [Google Scholar]; f) Jin H, Sun M, Shi L, Zhu X, Huang W, Yan D, Biomater. Sci 2018, 6, 1403; [DOI] [PubMed] [Google Scholar]; g) Li J, Li Y, Wang Y, Ke W, Chen W, Wang W, Ge Z, Nano Letters 2017, 17, 6983; [DOI] [PubMed] [Google Scholar]; h) Sun P, Wang N, Jin X, Zhu X, ACS Appl. Mater. Interfaces 2017, 9, 36675. [DOI] [PubMed] [Google Scholar]
- [10] a).Martinez JO, Molinaro R, Hartman KA, Boada C, Sukhovershin R, De Rosa E, Kirui D, Zhang S, Evangelopoulos M, Carter AM, Theranostics 2018, 8, 1131; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Parodi A, Molinaro R, Sushnitha M, Evangelopoulos M, Martinez JO, Arrighetti N, Corbo C, Tasciotti E, Biomaterials 2017, 147, 155; [DOI] [PubMed] [Google Scholar]; c) Gullotti E, Yeo Y, Mol. Pharmaceutics 2009, 6, 1041; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lankveld DP, Rayavarapu RG, Krystek P, Oomen AG, Verharen HW, Van Leeuwen TG, De Jong WH, Manohar S, Nanomedicine 2011, 6, 339; [DOI] [PubMed] [Google Scholar]; e) Fang RH, Kroll AV, Gao W, Zhang L, Adv. Mater 2018, 30, 1706759; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Hu CM, Fang RH, Wang KC, Luk BT, Thamphiwatana S, Dehaini D, Nguyen P, Angsantikul P, Wen CH, Kroll AV, Carpenter C, Ramesh M, Qu V, Patel SH, Zhu J, Shi W, Hofman FM, Chen TC, Gao W, Zhang K, Chien S, Zhang L, Nature 2015, 526, 118; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Dehaini D, Wei X, Fang RH, Masson S, Angsantikul P, Luk BT, Zhang Y, Ying M, Jiang Y, Kroll AV, Gao W, Zhang L, Adv. Mater 2017, 29, 1606209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM, Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12] a).Sun Q, Sun X, Ma X, Zhou Z, Jin E, Zhang B, Shen Y, Van Kirk EA, Murdoch WJ, Lott JR, Lodge TP, Radosz M, Zhao Y, Adv. Mater 2014, 26, 7615; [DOI] [PubMed] [Google Scholar]; b) Wang S, Huang P, Chen X, Adv. Mater 2016, 28, 7340; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Du JZ, Du XJ, Mao CQ, Wang J, J. Am. Chem. Soc 2011, 133, 17560; [DOI] [PubMed] [Google Scholar]; d) Wang S, Zhang S, Liu J, Liu Z, Su L, Wang H, Chang J, ACS Appl. Mater. Interfaces 2014, 6, 10706; [DOI] [PubMed] [Google Scholar]; e) Sun Q, Zhou Z, Qiu N, Shen Y, Adv. Mater 2017, 29, 1606628. [DOI] [PubMed] [Google Scholar]
- [13] a).Wang S, Huang P, Chen X, ACS Nano 2016, 10, 2991; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mizuhara T, Saha K, Moyano DF, Kim CS, Yan B, Kim Y-K, Rotello VM, Angew. Chem. Int. Ed 2015, 54, 6567; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang X-Z, Du J-Z, Dou S, Mao C-Q, Long H-Y, Wang J, ACS Nano 2011, 6, 771; [DOI] [PubMed] [Google Scholar]; d) Chien YH, Chou YL, Wang SW, Hung ST, Liau MC, Chao YJ, Su CH, Yeh CS, ACS Nano 2013, 7, 8516. [DOI] [PubMed] [Google Scholar]
- [14] a).Sun CY, Shen S, Xu CF, Li HJ, Liu Y, Cao ZT, Yang XZ, Xia JX, Wang J, J. Am. Chem. Soc 2015, 137, 15217; [DOI] [PubMed] [Google Scholar]; b) Sun CY, Liu Y, Du JZ, Cao ZT, Xu CF, Wang J, Angew. Chem. Int. Ed 2016, 55, 1010; [DOI] [PubMed] [Google Scholar]; c) Yuan Z, Zhao D, Yi X, Zhuo R, Li F, Adv. Funct. Mater 2014, 24, 1799. [Google Scholar]
- [15] a).Zhou Z, Shen Y, Tang J, Fan M, Van Kirk EA, Murdoch WJ, Radosz M, Adv. Funct. Mater 2009, 19, 3580; [Google Scholar]; b) Yu L, Xie M, Li Z, Lin C, Zheng Z, Zhou L, Su Y, Wang X, J. Mater. Chem. B 2016, 4, 6081. [DOI] [PubMed] [Google Scholar]
- [16] a).Wang Y, Zhou K, Huang G, Hensley C, Huang X, Ma X, Zhao T, Sumer BD, DeBerardinis RJ, Gao J, Nat. Mater 2014, 13, 204; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xu X, Saw PE, Tao W, Li Y, Ji X, Yu M, Mahmoudi M, Rasmussen J, Ayyash D, Zhou Y, Farokhzad OC, Shi J, Nano Letters 2017, 17, 4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Li HJ, Du JZ, Liu J, Du XJ, Shen S, Zhu YH, Wang X, Ye X, Nie S, Wang J, ACS nano 2016, 10, 6753; [DOI] [PubMed] [Google Scholar]; b) Zhou K, Liu H, Zhang S, Huang X, Wang Y, Huang G, Sumer BD, Gao J, J. Am. Chem. Soc 2012, 134, 7803; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhou K, Wang Y, Huang X, Luby-Phelps K, Sumer BD, Gao J, Angew. Chem. Int. Ed 2011, 50, 6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18] a).Yu G, Yang Z, Fu X, Yung BC, Yang J, Mao Z, Shao L, Hua B, Liu Y, Zhang F, Fan Q, Wang S, Jacobson O, Jin A, Gao C, Tang X, Huang F, Chen X, Nat. Commun 2018, 9, 766; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang K, Liu Y, Zhang Q, Kong C, Yi C, Zhou Z, Wang Z, Zhang G, Zhang Y, Khashab NM, Chen X, Nie Z, J. Am. Chem. Soc 2018, 140, 4666. [DOI] [PubMed] [Google Scholar]
- [19] a).Tang W, Yang Z, Wang S, Wang Z, Song J, Yu G, Fan W, Dai Y, Wang J, Shan L, Niu G, Fan Q, Chen X, ACS Nano 2018, 12, 2610; [DOI] [PubMed] [Google Scholar]; b) Guo X, Wang L, Duval K, Fan J, Zhou S, Chen Z, Adv. Mater 2018, 30, 1705436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


