Abstract
α-Ketoglutarate is an important metabolic intermediate that acts as a cofactor for several chromatin-modifying enzymes, including histone demethylases and the Tet family of enzymes that are involved in DNA demethylation. In this review, we focus on the function and genomic localization of these α-ketoglutarate–dependent enzymes in the maintenance of pluripotency during cellular reprogramming to induced pluripotent stem cells and in disruption of pluripotency during in vitro differentiation. The enzymatic function of many of these α-ketoglutarate–dependent proteins is required for pluripotency acquisition and maintenance. A better understanding of their specific function will be essential in furthering our knowledge of pluripotency.
Keywords: induced pluripotent stem cell (iPS cell) (iPSC), pluripotency, histone demethylase, DNA demethylation, stem cells, epigenetics, chromatin, reprogramming, cell differentiation, α-KG enzymes, cell fate
Introduction
Pluripotent stem cells have the ability to self-renew indefinitely and give rise to all the cell types of a multicellular organism. Pluripotency appears transiently during early embryonic development and can be captured with the correct culture conditions to obtain embryonic stem cells (ESCs).3 Pluripotent stem cells can also be derived from somatic cells either by the transfer of a somatic cell nucleus to an oocyte (SCNT) (1, 2) or through reprogramming, which is the overexpression of a small set of factors, usually Oct4, Sox2, Klf4, and c-Myc (OSKM) to generate induced pluripotent stem cells (iPSCs) (3–6) (Fig. 1). In vitro pluripotency is thought to exist in a continuum that is profoundly affected by growth conditions (7) For example, when ESCs are grown in the presence of signaling inhibitors to mitogen-activated protein kinase and glycogen synthase kinase (2i), their transcriptional profile better resembles the in vivo equivalent from the blastocyst stage of early embryonic development than that of ESCs grown in serum. Both the maintenance of pluripotency and its acquisition from somatic cells are affected by culture conditions. α-Ketoglutarate (α-KG)–dependent enzymes are important regulators of chromatin structure and are particularly sensitive to levels of intracellular metabolites as well as external components of the growth medium. Pluripotent cells can be grown in serum replacement medium, which contain vitamin C (Vc), that can affect the rate of catalysis of α-KG enzymes (Fig. 2) (8, 9). In the 2i conditions mentioned above, mouse ESCs (mESCs) utilize both glucose and glutamine in the medium to maintain high levels of α-KG to alter chromatin modifications (10, 11).
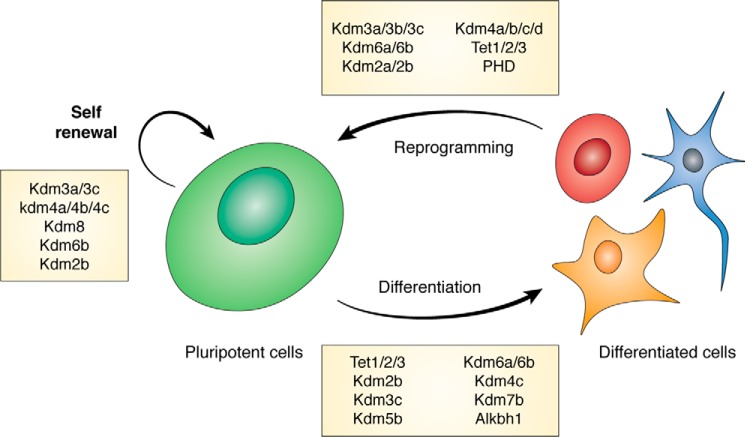
Figure 1.
Pluripotent stem cells can self-renew indefinitely and differentiate into a multitude of cells. Pluripotent cells can be isolated both from the inner cell mass of the blastocyst and the reprogramming of differentiated cells. In the boxes are the α-ketoglutarate–dependent proteins that are known to modulate each process.
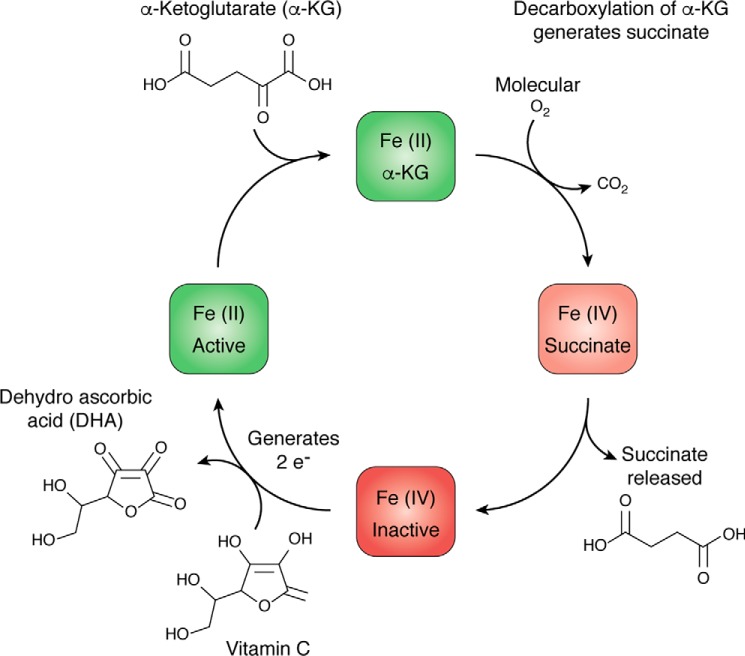
Figure 2.
Fe(II) recycling of α-KG–dependent dioxygenases. The catalytic activity of α-KG dioxygenases utilizes oxygen for the decarboxylation of α-KG and oxidation of Fe(II) to Fe(IV), rendering the enzyme inactive. Vitamin C can be used to regenerate iron back to the Fe(II) state, thus restoring catalytic activity.
The plasticity of pluripotent stem cells is associated with a hyperdynamic chromatin structure where histones and heterochromatin-associated proteins have a higher mobility than in somatic cells (12). Pluripotent stem cells also have a reduced amount of heterochromatin, usually associated with gene repression, which is enriched for histone modifications such as histone H3 lysine 9 methylation (H3K9me2 and H3K9me3) (13–15). Another chromatin feature that is more prevalent in pluripotent cells are the “bivalent” domains. These are regulatory regions that contain both an activating histone modification, histone H3 lysine 4 methylation (H3K4me3), and a repressive modification, histone H3 lysine 27 methylation (H3K27me3), that is mediated by the polycomb repressive complex 2 (PRC2) (1, 16). Several lineage specification genes are held in such a bivalent state and are resolved into either expression by retaining the H3K4me3 mark or repression by retaining the H3K27me3 mark (1, 2). Thus, the bivalent state can be considered poised for gene transcription. The H3K27me3 mark is usually found in opposition to H3K36me, a mark for active transcription elongation in gene bodies (3–5). In addition to histone modifications, DNA can also be methylated. In general, DNA methylation correlates with gene repression when present at regulatory regions. A pathway to active DNA demethylation can be provided by the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (4–6). The levels of 5hmC are higher in ESCs than most differentiated cells. α-KG–dependent enzymes include the jumonji domain–containing histone demethylases, Tet proteins that modify DNA methylation, and RNA-modifying enzymes of the Alkbh family, thus modulating the delicate state of pluripotency itself.
In this review, we focus on the changes to genomic localization of the cognate histone or DNA modifications resulting from perturbation of levels of α-KG–dependent enzymes as well as the subsequent changes in transcriptional output. We organize the review based on the phenotypes obtained during 1) the maintenance of the pluripotent state, 2) the acquisition of pluripotency from somatic cells, and 3) its disruption during in vitro differentiation.
Jumonji domain (Jmjd)-containing histone demethylases
Kdm5 family: Demethylases of histone H3K4
Kdm5a, Kdm5b, and Kdm5c share specificity for demethylating H3K4me2/3. Because H3K4me3 is enriched at the transcription start sites (TSSs) of poised or active genes, it is expected that a loss of these enzymes would result in increased gene expression.
Kdm5a was found to be an interacting partner of the PRC2 complex that contains the H3K27 methyltransferase Ezh2 (17). Thus, there is an association between the methyltransferase and demethylase of histone marks that are correlated with opposing effects on gene expression. When bivalent domains are resolved during differentiation of pluripotent cells, the regions that become silenced and retain H3K27me3 have the potential to simultaneously be demethylated at H3K4me3. In mESCs, Kdm5a preferentially binds the TSS of genes that are activated during differentiation (8, 9). Kdm5a-depleted mESCs are pluripotent but display increased expression of specific PRC2 target genes (17). Kdm5a depletion does not impact PRC2 binding or the levels of H3K27me3; however, knockdown (KD) of PRC2 component Suz12 leads to a reduction of Kdm5a binding at shared promoters and a subsequent increase in H3K4me3 (17) (Fig. 3). Thus, although Kdm5a is recruited to shared locations by PRC2, it does not appear to affect the function of PRC2 (10, 11).
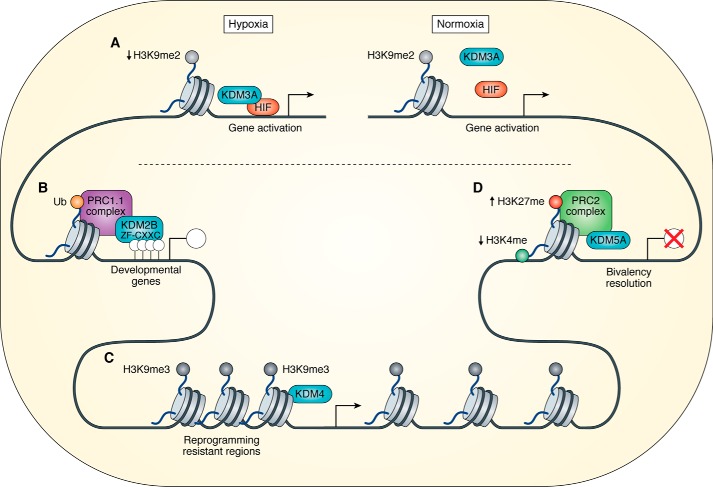
Figure 3.
A, HIF1 recruits Kdm3a under hypoxic conditions, regulating gene expression by the removal of H3K9me2. B, Kdm2b binds unmethylated CpG islands via a ZF-CXXC domain. Kdm2b is part of the PRC1.1 complex that mediates gene repression through the ubiquitination (Ub) of histone H2A. C, the Kdm4 family of proteins reduce the levels of H3K9me3 at reprogramming-resistant regions, opening the condensed heterochromatin and allowing transcription. D, Kdm5a interacts with the PRC2 complex, removing H3K4me at bivalent genes, leading to gene repression.
There have been conflicting reports on the role of Kdm5b in pluripotency. Although overexpression of Kdm5b in mESCs led to increased proliferation (1, 16), there are contradictory reports on the effects of Kdm5b depletion as well as its genomic localization. The KD of Kdm5b compromised self-renewal of ESCs and led to spontaneous differentiation (18). In this study, Kdm5b was found localized intragenically and seemed to be recruited by MRG15, a protein that binds to transcription elongation–associated H3K36me. Therefore, a role for Kdm5b activity in suppressing cryptic transcription initiation from within the gene body was proposed (18).
In contrast to these results, genomic deletion of Kdm5b in ESCs was compatible with self-renewal and pluripotency, but differentiation toward ectoderm was compromised (19). Kdm5b was found enriched at both the TSS and intragenic regions that did not coincide with H3K36me. The depletion of Kdm5b increased H3K4me3 at the TSS but did not cause many gene expression changes. Kdm5b-localized promoters had lower levels of H3K4me3, suggesting that it functioned in maintaining levels rather than eliminating this mark (19). Corroborating this idea, Kdm5b KD ESCs were refractory to embryoid body differentiation due to continued pluripotency gene expression as well as maintenance of H3K4me3 at bivalent genes (20). During the acquisition of pluripotency by SCNT, the KD of Kdm5b by siRNA led to an increase of cloned embryos arresting in the four-cell stage of the blastocyst, suggesting an essential role for appropriate localization of H3K4me3 (21).
Another study found that Kdm5b was localized to promoters, intragenic regions, and enhancers. Kdm5b KD led to an increase of H3K4me3 at the gene body, a decrease at promoters, and spreading at enhancers (20). Thus, Kdm5b may function in restricting distribution of H3K4me3. Kdm5b KD also increased the expression of alternatively spliced exons in ESCs, suggesting a role for intragenic localization (22).
Kdm5c binds to both proximal promoter regions and distal intergenic enhancer regions in mESCs. Upon KD of Kdm5c, pluripotency is maintained, and global H3K4me3 levels remained unchanged. However, at loci that are greatly enriched for Kdm5c binding, H3K4me1 levels were gained at the expense of H3K4me3 levels. This effect was magnified at regions bound by the transcription factor c-Myc, which is a direct interacting partner of Kdm5c (23).
Thus, the Kdm5 family is involved in fine-tuning both the degree and localization of H3K4me3. The loss of each individual enzyme seems compatible with pluripotency, suggesting redundancy in function. Site specificity may be conferred by interacting factors other than c-Myc and PRC2. In this regard, it is interesting that in nonpluripotent cells, Kdm5a interacts with the Sin3 histone deacetylase complex specifically at sites that are bound by the transcription factor E2F4 (24). The role of such an interaction in pluripotent cells has not been determined.
Kdm6 family: Demethylases of histone H3K27 methylation
Kdm6a (UTX) and Kdm6b (JMJD3) can demethylate H3K27me2/me3, whereas the mouse Kdm6a paralog, Kdm6c (UTY), is located on the Y chromosome and lacks this activity (25). Similar to the reciprocal recruitment observed in the case of the Kdm5b–PRC2 complex, Kdm6a, the H3K27 demethylase, is part of the mixed-lineage leukemia H3K4 methyltransferase complex (26).
Both mouse and human Kdm6a, as well as mKdm6b, are dispensable for maintenance of self-renewal as knockout (KO) ESCs display normal morphology, cell proliferation rates, and high levels of pluripotency gene expression (27–34). Male Kdm6b KO mESCs up-regulate lineage commitment genes upon differentiation and contribute toward adult chimeras (27). Unlike the Kdm5 family, there are nonredundant functions of Kdm6a and Kdm6b in pluripotency. Overexpression of the Kdm6b catalytic domain (but not that of Kdm6a) leads to spontaneous differentiation in human ESCs (hESCs) and is accompanied by depleted global levels of H3K27me3 (18, 35). This is surprising given that the catalytic Jumonji domains are the most conserved regions between members of the same subfamily (36). Remarkably, ectopic Kdm6b expression in conjunction with lineage-defining transcription factors promotes hESC differentiation into multiple lineages (18, 35). Because directed differentiation of ESCs into specific lineages remains a goal of regenerative therapy, the transient overexpression of Kdm6b could be a useful tool for accelerating differentiation into desirable cell types.
Kdm6a and Kdm6b also have opposing roles in generating iPSCs with well-defined mechanisms. Kdm6a promotes the reprogramming of both epiblast stem cells (EpiSCs) to naïve pluripotency (Box 1) and somatic cells to iPSCs in a catalysis-dependent manner (19, 27). In the absence of Kdm6a, several important pluripotency genes (e.g. Sall4) are not activated due to retention of the repressive H3K27me3 that silences their expression in somatic cells such as mouse embryonic fibroblasts (MEFs). Kdm6a is likely to be targeted to these loci by direct interaction with the Oct4, Sox2, and Klf4 reprogramming factors (19, 27). In contrast to Kdm6a, Kdm6b depletion improves reprogramming of MEFs to iPSCs by two independent activities. First, its depletion decreases Ink4/ARF transcription, removing a senescence block and leading to increased reprogramming (20, 37). Second, it promotes the TRIM26 ubiquitin ligase–mediated degradation of a particular scaffold protein, Phf20. Phf20 is required to assemble a transcription activation complex at the Oct4 locus during reprogramming (20, 37).
In contrast to their opposing roles in gaining pluripotency, Kdm6a and Kdm6b enhance in vitro differentiation from ESCs (32, 38, 39). Hox clusters are large bivalent domains that are important for patterning the body plan during development. Surprisingly, the deletion of either Kdm6a alone (22, 40) or combined with Kdm6b (23, 31) in mESCs is accompanied by a loss of H3K27me3 rather than a gain as would be expected when deleting a demethylase. This suggests that the down-regulation of PRC2 expression may contribute to passive dilution of the H3K27me3 mark during differentiation.
Both the Kdm6a and Kdm6b KOs decrease the levels of the mesoderm-specifying gene Brachyury during differentiation. Interestingly, the introduction of a catalytically inactive mutant of Kdm6a restores Brachyury expression, suggesting functions beyond H3K27 demethylation (25, 28, 29, 34). Thus, during differentiation, the function of Kdm6a and Kdm6b may be linked to both H3K27 demethylation and the structural role they provide as part of protein complexes.
Kdm2 family: Demethylases of histone H3K36 methylation
H3K36me1/2, a histone modification associated with active transcription elongation, can be demethylated by Kdm2a (Jhdm1a) and Kdm2b (Jhdm1b). In mESCs, both Kdm2a and Kdm2b are preferentially recruited to non-DNA–methylated CpG islands (27–34, 41, 42). The depletion of Kdm2b, but not Kdm2a, leads to spontaneous differentiation of mESCs, indicating a nonredundant role for Kdm2b in maintaining pluripotency (42).
Both Kdm2a and Kdm2b share a ZF-CXXC domain that targets the enzyme to unmethylated CpG islands throughout the genome; however, they interact with different partners to mediate gene regulation (41, 43). Kdm2b is part of the noncanonical PRC1 complex, PRC1.1, which has E3 ubiquitin ligase activity that mediates monoubiquitination of histone H2A, leading to gene repression (Fig. 3). In mESCs, Kdm2b recruits PRC1.1 to targets that are not specifically depleted for H3K36me enrichment (44). Furthermore, spontaneous differentiation of mESC upon Kdm2b depletion can be rescued by the catalytic mutant but not the CXXC-deleted Kdm2b mutant.
Interestingly, the reverse phenomenon where a PRC1 component, Phf19, recruits H3K36me demethylase NO66 to polycomb-related regions also occurs in mESCs. However, the deletion of NO66 does not have effects on pluripotency or differentiation (27, 45).
The role of Kdm2a and Kdm2b has also been assessed in several reprogramming systems. The overexpression of Kdm2b enhances OSKM reprogramming by promoting the mesenchymal-to-epithelial transition (MET) (46), an early event in the reprogramming of MEFs in serum (FBS)-containing medium (Box 1). This enhancement occurs in a catalysis-dependent manner by demethylating H3K36me at epithelial genes such as E-cadherin (46). The addition of Vc in general can improve reprogramming efficiency, which is further enhanced by the overexpression of either Kdm2a or Kdm2b. However, unlike the FBS condition, under Vc conditions, Kdm2b seems to function in the suppression of the Ink4/Arf senescence block (47). In fact, in the presence of Kdm2b and Vc, Oct4 alone (as compared with the entire set of OSKM) is sufficient to generate iPSCs through the activation of the ESC cell cycle–specific microRNA miR-302 cluster (47). Signaling through BMP4 counteracts this Oct4/Vc/Kdm2b-mediated reprogramming in a PRC1-dependent manner because under these conditions the levels of H2A ubiquitination genome-wide are significantly reduced (48). Taken together, depending on the conditions of reprogramming, the exact targets that are causal for increased reprogramming efficiency may differ, but the activity of Kdm2b is important for this process.
The Kdm2 family has been less characterized during in vitro differentiation, although Kdm2b-depleted ESCs cannot fully silence pluripotency genes. This may lead to the impaired activation of lineage-specific genes (42).
Kdm3: Demethylases of H3K9me1/me2
Kdm3a, Kdm3b, and Kdm3c share specificity for demethylating the repressive histone modifications H3K9me1/2 but play different roles during reprogramming and differentiation. The depletion of any of the three Kdm3 family members seems to compromise self-renewal of ESCs (49). Kdm3a depletion down-regulates pluripotency gene Tcl1 expression, which may be causal for the self-renewal phenotype (50). When ESCs are exposed to Vc, germline-associated genes of the blastocyst are up-regulated in a Kdm3a- and Kdm3b-dependent manner (51).
Given the responsiveness of the Jmjd enzymes to the antioxidant Vc, it is not surprising that they may also act as oxygen sensors. Under conditions of low oxygen tension, i.e. hypoxia, proteins called hypoxia-inducible factors (HIFs) are stabilized. Kdm3a has been shown to impact gene expression in an interaction with HIF1 in both cell culture and in vivo systems (52) (Fig. 3). The environmental milieu of early development is hypoxic and instructive for placental development. Under such hypoxic conditions, HIF1 is able to recruit Kdm3a to demethylate the promoter of MMP12, a gene important for placental development, spiral artery remodeling, and trophoblast migration. This leads to activation of gene expression specifically under hypoxic conditions (53).
Depletion of Kdm3c in ESCs leads to disruption of self-renewal and a decrease in the expression of microRNAs of the miR-200 family and the miR-290/295 cluster (49). This miR dysregulation leads to an increase in ERK/MAPK signaling and drives cells toward epithelial-to-mesenchymal transition that can be partially rescued by ectopic miR expression (49). Although the catalytic activity of Kdm3c has been disputed (54), there is a global increase of H3K9me1/2/3 upon Kdm3c KD in mESCs.
Overexpression of Kdm3a improved the efficiency of Oct4 reactivation in ESC fusion-induced reprogramming of neural stem cells (55). This improvement depended on the catalytic activity of Kdm3a as the overexpressing cells exhibited a widespread loss of H3K9me2 (55). Interestingly, only Kdm3b is essential in the Vc-mediated conversion of partially reprogrammed intermediates (pre-iPSCs) to iPSCs (56, 57). Kdm3b was also recently shown to have activity in demethylating H4R3 arginine methylation in hematopoietic stem cells (58). It needs to be determined whether these modifications on H3 and H4 are coordinately regulated. Kdm3c KD in OSKM-mediated reprogramming of MEFs leads to a decrease in obtaining bona fide iPSC colonies (49, 59). These effects could be mediated through the regulation of the Oct4 locus because Kdm3c is enriched at its distal enhancer, reducing H3K9me2 and promoting FACT (facilitates chromatin transcription) chaperone recruitment (59). Thus, although all three Kdm3 enzymes are required for reprogramming, they seem to contribute to the process in distinct mechanisms.
Contrary to the phenotype in mESCs, when Kdm3c is knocked down in hESCs, Oct4 and Nanog levels as well as pluripotency are maintained. However, neuronal differentiation is much more rapid due to the reduced expression of ESC-specific miR-302 in these cells (60).
Kdm4 family: Demethylases of histone H3K9me2/me3 and H3K36me3 methylation
The Kdm4 family consists of four members, Kdm4a–d, which have demethylase activity toward H3K9me2/3 with Kdm4a–c also having specificity to H3K36me3. This substrate specificity is interesting as H3K9me2/3 are repressive modifications, whereas H3K36me3 is associated with actively transcribed genes.
Conflicting studies complicate the roles of the Kdm4 family in pluripotency. KD of both Kdm4b and Kdm4c in mESCs led to morphological changes that could be partially rescued by the catalytic mutant proteins, suggesting additional roles beyond H3K9 demethylation (61). Kdm4c was thought to function through demethylating the promoter of the pluripotency gene Nanog (50). However, this was not replicated in another study that instead found a connection between Kdm4b and Nanog target genes (61). Although Kdm4b and Kdm4c share some locations in ESCs, the unique targets of each protein show differential association: Kdm4c is localized with bivalent and PRC2-occupied promoters in ESCs, whereas Kdm4b is associated with H3K4me and activating marks (61).
The phenotypic results of Kdm4 depletion are disputed because individual-KO ESCs of Kdm4a, Kdm4b, and Kdm4c remain pluripotent (62–64). Only the combined deletion of Kdm4a and Kdm4c (DKO) impaired proliferation of mESCs in a catalytic activity–dependent manner (62). Nonetheless, all three proteins are localized to H3K4me3-enriched regions. At highly enriched Kdm4a/4c sites, there is an increase in H3K9me3 levels upon Kdm4a/4c deletion. Furthermore, the DKO cells display a propensity to express endoderm markers when grown in serum but not 2i conditions (62). This sensitivity to culture conditions was further borne out when the localization of Kdm4c switched from largely TSS-enriched in 2i to more distally located in serum-grown ESCs (64). This switch may be related to the post-transcriptional regulation of Kdm4c because the mitogen-activated protein kinase inhibitor in 2i medium prevents the phosphorylation and degradation of Kdm4c (65).
Although the roles of the Kdm4 proteins in the maintenance of pluripotency remain conflicting, the overexpression of several Kdm4 family members promotes acquisition of pluripotency in several systems. In mice, injection of catalytically competent Kdm4d mRNA during SCNT increases efficiency and improves the developmental potential of SCNT embryos (66). Excitingly, the inclusion of the primate Kdm4d resulted in the recent cloning by SCNT of monkeys that survived to term (67). The function of Kdm4d seems to be to decrease H3K9me3 levels at so-called “reprogramming-resistant regions” that are repressed in SCNT at the two-cell stage, leading to their expression (21, 66) (Fig. 3). Kdm4a performs a similar function when ectopically expressed during human SCNT (68). Kdm4b can also improve the developmental potential of SCNT-derived embryos (69, 70). Furthermore, depletion of Kdm4b or Kdm4c reduces Vc-mediated conversion of pre-iPSCs to iPSCs, whereas overexpression of Kdm4b promotes MEF reprogramming to iPSCs (57).
Deletion of Kdm4c resulted in the misregulation of lineage-specific genes upon differentiation (64). Interestingly, at distal regions, Kdm4c colocalizes with the H3K9me2 methyltransferase G9a. However, these sites lack H3K9me2, suggesting constant turnover of the modification at such loci (64).
Kdm7 family
The Kdm7 family consists of three family members, Kdm7a (KIAA1718), Kdm7b (PHF8), and Kdm7c (PHF2), that can all demethylate H3K9me2; however, Kdm7b has additional specificity to H4K20me1, and Kdm7c has additional specificity to H3K27me1/2. Kdm7a promotes neural differentiation from mESCs by demethylating H3K27me2 at the FGF4 locus (71). Deletion of Kdm7b in mESCs does not compromise maintenance of pluripotency, but upon differentiation, Kdm7b KO mESCs showed an increase in expression of mesodermal and cardiac lineage genes by controlling apoptosis (72).
Kdm8
The demethylase activity of Kdm8 toward H3K36me2 is disputed. Depletion of Kdm8 in hESCs compromises pluripotency and up-regulates lineage specification markers. KD of Kdm8 in hESCs leads to a higher percentage of cells in G1 phase due to elevated levels of the cell cycle gene CDKN1A, but it is unclear whether this is due to direct regulation of H3K36me2 levels. The changes in cell cycle precede down-regulation of pluripotency gene expression upon Kdm8 depletion, suggesting that the disruption of the cell cycle may lead to pluripotency defects. This corroborates the idea that certain phases of the cell cycle are more conducive to exit from pluripotency (73). Additional regulation of pluripotency by Kdm8 comes from direct binding and regulation of the pluripotency-related miR-302 cluster (74).
Taken together, several common themes emerge from the studies on the α-KG–dependent histone demethylases in pluripotency thus far. 1) They can have overlapping functions that may rely on interacting proteins. 2) They can have functions that do not rely on catalytic activity. 3) They can localize to the same genomic locations as the mark that they modify. Technically confounding results are obtained between KD and KO studies. With the advent of CRISPR-Cas9 technology it may be preferable to perform KOs to obtain clearer phenotypic results.
Tet: DNA 5-methylcytosine modifiers
As mentioned above, DNA can be methylated on the 5th position of cytosine, known as 5mC, which is usually associated with gene repression when found at regulatory regions. Its removal can occur by the iterative oxidation by the ten-eleven-translocation family of enzymes (Tet1, Tet2, and Tet3) from 5mC to 5hmC to 5-formylcytosine (5fC) to 5-carboxylcytosine (5caC). 5fC and 5caC can be excised by thymine-DNA glycosylase to generate cytosine. 5mC is also passively diluted during DNA replication (75). Tet1 and Tet2 can also oxidize thymine to 5-hydroxymethyluracil (5hmU), although the functional role of this modification is unknown (76).
Tet1 and Tet2 are highly expressed in ESCs (75). Global levels of 5hmC are maintained at higher levels in pluripotent cells as compared with most somatic cells. This suggests that the 5hmC modification may function as an epigenetic mark in addition to being an intermediate for demethylation (75). Genome-wide 5hmC profiling in mESCs revealed localization at both active and repressed genes. Increased gene expression was associated with low 5hmC at the promoter and high enrichment at gene bodies (77–81). At distal enhancers, 5hmC, 5fc, and 5caC are enriched at regions bound by pluripotency factors (77, 78, 80, 82).
Initial KD of Tet1 alone or both Tet1 and Tet2 in mESCs decreased the expression of pluripotency genes (77, 83). However, both the Tet1 KO or Tet1 and Tet2 DKO mESCs remain pluripotent despite a global loss of 5hmC (84, 85). Both mouse and human triple-Tet KO ESCs are able to self-renew but have impaired differentiation potential (86, 87). Further functional diversity is also provided by a full-length pluripotency Tet1 isoform and a shorter isoform that lacks the CXXC DNA-binding domain that is up-regulated during differentiation (88).
Tet proteins and 5hmC are enriched at bivalent domains (77, 78, 80, 81). Upon deletion of Tet proteins, an increase of 5mC at bivalent domains is observed (84, 85, 87). Interestingly, depletion of Tet1 in mESCs decreased PRC2 binding at bivalent regions (80). Furthermore, the Tet DKO resulted in the loss of H3K27me3 in a large subset of bivalent CpG islands, whereas H3K4me3 remained unchanged (89). These observations may explain the differentiation defects observed in the Tet KO ESCs. Deletion of Tet2 in mESCs led to hypermethylation at a few enhancers, a subset of which also lost H3K27 acetylation, leading to reduced expression of associated genes (90). Tet1 depletion led to a reduction of 5hmC at both the TSS and gene body, whereas Tet2 KD decreased 5hmC within the gene body but increased levels at the TSS. Furthermore, 5hmC enrichment is reduced at exon boundaries in high- and low-expressed genes upon Tet2 and Tet1 depletion, respectively. However, there is no specific pattern of 5hmC that correlates with gene expression changes upon Tet1 or Tet2 depletion, suggesting a complex relationship with transcription (91).
In addition to protein-coding gene expression, the Tet enzymes also regulate telomere stability, although the telomere length in Tet TKO is variable (92). Tet1 is specifically enriched at young L1 transposable elements and regulates their silencing in a 5hmC-independent manner through the recruitment of Sin3a that is independent of catalytic function (93).
The Tet proteins also seem to regulate transition to cell types that resemble other developmental stages in vitro. Tet TKO mESCs show increased expression of Zscan4, which is a marker for the totipotent two-cell stage of development. ESCs in culture go through a two cell–like state at low frequency (94) that is enhanced in the Tet TKO (92). The addition of Vc increases global levels of 5hmC in mESCs and up-regulates a subset of germline-related genes through Tet-mediated 5hmC conversion at these genes (95). Supplementing retinol in 2i-containing medium can also modulate 5hmC through up-regulation of Tet2 and promote the reprogramming of EpiSCs to naïve iPSCs. Moreover, low retinol concentrations and Vc can synergistically promote EpiSC reprogramming (96) (Box 1).
Tet function has a supportive role in reprogramming of somatic cells to iPSCs. Ectopic expression of Tet1 promotes reprogramming that depends on catalytic function. Tet1 is localized to both the promoter and enhancer region of Oct4, leading to elevated 5hmC and decreased 5mC levels, resulting in endogenous Oct4 activation. Interestingly, Tet1 can replace Oct4 in the OSKM mixture to generate TSKM-derived pluripotent iPSCs (97). Global analysis of TSKM reprogramming showed a gradual increase of 5hmC during reprogramming, whereas 5mC levels transiently increase followed by a reduction in the late stage of reprogramming (97). Furthermore, depletion of Tet1 also reduces human OSKM reprogramming efficiency (98). Tet2 can promote reprogramming by binding to regulatory regions of pluripotency genes Nanog and Esrrb to promote 5hmC accumulation (99). Tet TKO MEFs could not be reprogrammed as they fail to undergo MET but can be rescued by ectopic expression of the catalytic domain of any Tet protein. Failed activation of MET in TKO is due to lack of DNA demethylation of the miR-200 family that down-regulate mesenchymal genes (100). Surprisingly, overexpression of Tet1 (but not Tet2) in conjunction with Vc inhibited OSK reprogramming by prohibiting MET (101). Tet proteins also promote reprogramming of pre-iPSCs, which have undergone MET. Tet1 and Tet2 can interact with Nanog, and the overexpression of both Nanog and Tet1 synergistically promotes pre-iPSC to iPSC conversion. This is mediated at least in part by elevating 5hmC levels at the Esrrb locus (102). Pre-iPSCs can be robustly converted into iPSCs in the presence of Vc and 2i (56), and this is diminished by combined depletion of Tet1 and Tet2 (56).
Tet enzymes can interact with a multitude of different proteins that alter their localization and function. Tet proteins in conjunction with pluripotency-related protein Prdm14 can promote active demethylation at germline genes (103). Idax can modulate Tet2 stability during differentiation by interacting with Tet2 and triggers its degradation through caspase activation (104). Interestingly, certain proteins can bind to 5hmC and regulate Tet function. Mbd3 is reported to bind 5hmC, and deletion of Mbd3 disrupts localization of Tet proteins (105, 106). In addition, pluripotency-related Sall4 binds 5hmC and promotes 5hmC oxidation and active demethylation (107). Tet regulation can also occur at the expression level as several enzymes and microRNA have been shown to regulate Tet expression and influence pluripotency and differentiation (108–110). Non-5hmC–related functions of the Tets can be mediated by their association with O-linked GlcNAc-transferase (Ogt) and the repressor Sin3a complex in mESCs (79, 93, 111–113). Ogt alters the post-translational modification of the Tet proteins, which could impact the function of these enzymes. Although inhibition of Ogt does not alter global 5hmC levels, the interaction of Ogt and Tet proteins has a significant role in targeting these proteins to the genome. In mESCs, Ogt can recruit Tet1 to regulate developmental genes. The depletion of Ogt diminishes Tet1 recruitment and decreases 5hmC levels at targeted genes. By contrast, Tet2, recruits Ogt to the genome and mediates gene regulation through O-GlcNAc on histone H2B, which is independent of the catalytic activity of Tet2. Depletion of Tet2 abolishes Ogt recruitment to these specific sites and leads to gene repression. Furthermore, in nonpluripotent cells, the recruitment of Ogt by Tet2 can promote binding of SET1/COMPASS (Complex of Proteins Associated with Set1) to promote H3K4me3 and gene activation. Whether similar mechanisms are utilized in mESCs requires further investigation.
Alkbh1: DNA N6-methyladenine modifier
Beyond cytosine methylation, adenine methylation on DNA N6-methyladenine (N6-mA) was recently identified in mESCs (114). Alkbh1 has been shown to demethylate N6-mA in mESCs (114). Alkbh1 KO ESCs maintain pluripotent traits, exhibiting unchanged (114) or higher (115) Nanog and Oct4 expression. However, Alkbh1 KO ESCs show defects in their ability to differentiate, especially to the neuroectoderm lineage (115, 116). This could be due to the inability of the Alkbh1 KO ESCs to remove N6-mA from pluripotent genes during differentiation because higher levels of N6-mA correlate with gene silencing (114).
Prolyl-hydroxylase domain enzyme (PHD) and factor inhibiting HIF (FIH): Regulators of hypoxia-inducible factor
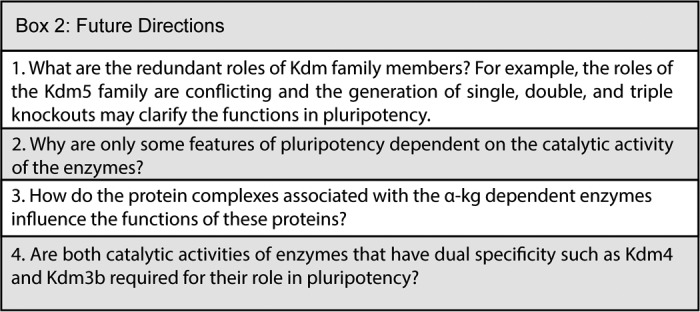
The HIF proteins are regulated by the α-KG–dependent dioxygenases PHD1–3 and FIH-1. HIF protein stability has both positive and negative effects on pluripotency (117–119). PHDs hydroxylate proline residues within the oxygen-dependent domains, resulting in degradation of the HIF proteins. In contrast, FIH-1 hydroxylates asparagine residues within the C-terminal transactivation domain of HIF, which prohibits the interaction with its coactivator, p300. Under hypoxic conditions, PHD and FIH-1 function is blocked, allowing HIF nuclear translocation to regulate hypoxia-related genes. Chemical inhibition of PHD or the α-KG analog dimethyloxalylglycine can regulate Oct4 expression (120), and PHD inhibitors can increase OSKM-mediated human reprogramming (121, 122). Taken together, the myriad functions of α-KG–dependent enzymes in pluripotency are only beginning to be uncovered and will be interesting to investigate in the future (Box 2).
Acknowledgments
We thank Coral Willie, Nur Zafirah Zaidan, Stefan Pietrzak, Edwin Neumann, Simone Shen, and Nikita Patel for critical reading of the manuscript.
This work was supported by the Shaw Scientist Award and National Institutes of Health Grant RO1GM113033 (to R. S.). This research was also supported by the National Institute of General Medical Sciences, National Institutes of Health Award T32GM081061. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ESC
- embryonic stem cell
- SCNT
- somatic cell nuclear transfer
- OSKM
- Oct4, Sox2, Klf4, and c-Myc
- iPSC
- induced pluripotent stem cell
- 2i
- inhibitors to mitogen-activated protein kinase and glycogen synthase kinase
- α-KG
- α-ketoglutarate
- Vc
- vitamin C
- mESC
- mouse ESC
- PRC
- polycomb repressive complex
- 5mC
- 5-methylcytosine
- 5hmC
- 5-hydroxymethylcytosine
- Jmjd
- Jumonji domain
- TSS
- transcription start site
- H3K9me
- histone H3 lysine 9 methylation
- KD
- knockdown
- KO
- knockout
- DKO
- double KO
- TKO
- triple KO
- hESC
- human ESC
- EpiSC
- epiblast stem cell
- MEF
- mouse embryonic fibroblast
- ARF
- ADP-ribosylation factor
- ZF
- zinc finger
- MET
- mesenchymal-to-epithelial transition
- FBS
- fetal bovine serum
- HIF
- hypoxia-inducible factor
- miR
- microRNA
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- 5fC
- 5-formylcytosine
- 5caC
- 5-carboxylcytosine
- Tet
- ten-eleven-translocation
- 5hmU
- 5-hydroxymethyluracil
- TSKM
- Tet1, Sox2, Klf, and c-Myc
- Ogt
- O-linked GlcNAc-transferase
- N6-mA
- N6-methyladenine
- PHD
- prolyl-hydroxylase domain enzyme
- FIH
- factor inhibiting HIF.
References
- 1. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 2. Yu J., and Thomson J. A. (2008) Pluripotent stem cell lines. Genes Dev. 22, 1987–1997 10.1101/gad.1689808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan W., Xu M., Huang C., Liu N., Chen S., and Zhu B. (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 10.1074/jbc.M110.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 5. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- 6. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., and Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinberger L., Ayyash M., Novershtern N., and Hanna J. H. (2016) Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- 8. Beshiri M. L., Holmes K. B., Richter W. F., Hess S., Islam A. B., Yan Q., Plante L., Litovchick L., Gévry N., Lopez-Bigas N., Kaelin W. G. Jr., and Benevolenskaya E. V. (2012) Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 18499–18504 10.1073/pnas.1216724109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monfort A., and Wutz A. (2013) Breathing-in epigenetic change with vitamin C. EMBO Rep. 14, 337–346 10.1038/embor.2013.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pasini D., Cloos P. A., Walfridsson J., Olsson L., Bukowski J.-P., Johansen J. V., Bak M., Tommerup N., Rappsilber J., and Helin K. (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 10.1038/nature08788 [DOI] [PubMed] [Google Scholar]
- 11. Carey B. W., Finley L. W., Cross J. R., Allis C. D., and Thompson C. B. (2015) Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., and Misteli T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cel. 10, 105–116 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soufi A., Donahue G., and Zaret K. S. (2012) Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994–1004 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M., Carey M., Garcia B. A., and Plath K. (2013) Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 15, 872–882 10.1038/ncb2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaidan N. Z., Walker K. J., Brown J. E., Schaffer L. V., Scalf M., Shortreed M. R., Iyer G., Smith L. M., and Sridharan R. (2018) Compartmentalization of HP1 proteins in pluripotency acquisition and maintenance. Stem Cell Rep. 10, 627–641 10.1016/j.stemcr.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dey B. K., Stalker L., Schnerch A., Bhatia M., Taylor-Papidimitriou J., and Wynder C. (2008) The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol. Cell. Biol. 28, 5312–5327 10.1128/MCB.00128-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pasini D., Hansen K. H., Christensen J., Agger K., Cloos P. A., and Helin K. (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 22, 1345–1355 10.1101/gad.470008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., and Impey S. (2011) KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 30, 1473–1484 10.1038/emboj.2011.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitz S. U., Albert M., Malatesta M., Morey L., Johansen J. V., Bak M., Tommerup N., Abarrategui I., and Helin K. (2011) Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 30, 4586–4600 10.1038/emboj.2011.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kidder B. L., Hu G., and Zhao K. (2014) KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol. 15, R32 10.1186/gb-2014-15-2-r32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu W., Liu X., Wang C., Gao Y., Gao R., Kou X., Zhao Y., Li J., Wu Y., Xiu W., Wang S., Yin J., Liu W., Cai T., Wang H., Zhang Y., and Gao S. (2016) Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2, 16010 10.1038/celldisc.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He R., and Kidder B. L. (2017) H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells. Nucleic Acids Res. 45, 6427–6441 10.1093/nar/gkx251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Outchkourov N. S., Muiño J. M., Kaufmann K., van IJcken W. F., Groot Koerkamp M. J., van Leenen D., de Graaf P., Holstege F. C., Grosveld F. G., and Timmers H. T. (2013) Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep. 3, 1071–1079 10.1016/j.celrep.2013.02.030 [DOI] [PubMed] [Google Scholar]
- 24. van Oevelen C., Wang J., Asp P., Yan Q., Kaelin W. G. Jr., Kluger Y., and Dynlacht B. D. (2008) A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32, 359–370 10.1016/j.molcel.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shpargel K. B., Sengoku T., Yokoyama S., and Magnuson T. (2012) UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 10.1371/journal.pgen.1002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee M. G., Villa R., Trojer P., Norman J., Yan K.-P., Reinberg D., Di Croce L., and Shiekhattar R. (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450 10.1126/science.1149042 [DOI] [PubMed] [Google Scholar]
- 27. Mansour A. A., Gafni O., Weinberger L., Zviran A., Ayyash M., Rais Y., Krupalnik V., Zerbib M., Amann-Zalcenstein D., Maza I., Geula S., Viukov S., Holtzman L., Pribluda A., Canaani E., et al. (2012) The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488, 409–413 10.1038/nature11272 [DOI] [PubMed] [Google Scholar]
- 28. Morales Torres C., Laugesen A., and Helin K. (2013) Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 8, e60020 10.1371/journal.pone.0060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C., Lee J.-E., Cho Y.-W., Xiao Y., Jin Q., Liu C., and Ge K. (2012) UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 109, 15324–15329 10.1073/pnas.1204166109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dhar S. S., Lee S.-H., Chen K., Zhu G., Oh W., Allton K., Gafni O., Kim Y. Z., Tomoiga A. S., Barton M. C., Hanna J. H., Wang Z., Li W., and Lee M. G. (2016) An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 44, 3659–3674 10.1093/nar/gkv1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shpargel K. B., Starmer J., Yee D., Pohlers M., and Magnuson T. (2014) KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 10, e1004507 10.1371/journal.pgen.1004507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang W., Wang J., and Zhang Y. (2013) Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 23, 122–130 10.1038/cr.2012.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burgold T., Voituron N., Caganova M., Tripathi P. P., Menuet C., Tusi B. K., Spreafico F., Bévengut M., Gestreau C., Buontempo S., Simeone A., Kruidenier L., Natoli G., Casola S., Hilaire G., et al. (2012) The H3K27 Demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2, 1244–1258 10.1016/j.celrep.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 34. Ohtani K., Zhao C., Dobreva G., Manavski Y., Kluge B., Braun T., Rieger M. A., Zeiher A. M., and Dimmeler S. (2013) Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ. Res. 113, 856–862 10.1161/CIRCRESAHA.113.302035 [DOI] [PubMed] [Google Scholar]
- 35. Akiyama T., Wakabayashi S., Soma A., Sato S., Nakatake Y., Oda M., Murakami M., Sakota M., Chikazawa-Nohtomi N., Ko S. B. and Ko M. S. (2016) Transient ectopic expression of the histone demethylase JMJD3 accelerates the differentiation of human pluripotent stem cells. Development 143, 3674–3685 10.1242/dev.139360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cloos P. A., Christensen J., Agger K., and Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao W., Li Q., Ayers S., Gu Y., Shi Z., Zhu Q., Chen Y., Wang H. Y., and Wang R.-F. (2013) Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 152, 1037–1050 10.1016/j.cell.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kartikasari A. E., Zhou J. X., Kanji M. S., Chan D. N., Sinha A., Grapin-Botton A., Magnuson M. A., Lowry W. E., and Bhushan A. (2013) The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 32, 1393–1408 10.1038/emboj.2013.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Li Y., Guo C., Lu Q., Wang W., Jia Z., Chen P., Ma K., Reinberg D., and Zhou C. (2016) ISL1 and JMJD3 synergistically control cardiac differentiation of embryonic stem cells. Nucleic Acids Res. 44, 6741–6755 10.1093/nar/gkw301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welstead G. G., Creyghton M. P., Bilodeau S., Cheng A. W., Markoulaki S., Young R. A., and Jaenisch R. (2012) X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. U.S.A. 109, 13004–13009 10.1073/pnas.1210787109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blackledge N. P., Zhou J. C., Tolstorukov M. Y., Farcas A. M., Park P. J., and Klose R. J. (2010) CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 38, 179–190 10.1016/j.molcel.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu X., Johansen J. V., and Helin K. (2013) Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 49, 1134–1146 10.1016/j.molcel.2013.01.016 [DOI] [PubMed] [Google Scholar]
- 43. Blackledge N. P., Farcas A. M., Kondo T., King H. W., McGouran J. F., Hanssen L. L., Ito S., Cooper S., Kondo K., Koseki Y., Ishikura T., Long H. K., Sheahan T. W., Brockdorff N., Kessler B. M., et al. (2014) Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 157, 1445–1459 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Z., Yang X., He J., Liu J., Wu F., Yu S., Liu Y., Lin R., Liu H., Cui Y., Zhou C., Wang X., Wu J., Cao S., Guo L., et al. (2017) Kdm2b regulates somatic reprogramming through variant PRC1 complex-dependent function. Cell Rep. 21, 2160–2170 10.1016/j.celrep.2017.10.091 [DOI] [PubMed] [Google Scholar]
- 45. Brien G. L., Gambero G., O'Connell D. J., Jerman E., Turner S. A., Egan C. M., Dunne E. J., Jurgens M. C., Wynne K., Piao L., Lohan A. J., Ferguson N., Shi X., Sinha K. M., Loftus B. J., et al. (2012) Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 19, 1273–1281 10.1038/nsmb.2449 [DOI] [PubMed] [Google Scholar]
- 46. Liang G., He J., and Zhang Y. (2012) Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat. Cell Biol. 14, 457–466 10.1038/ncb2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M. A., Pan G., and Pei D. (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9, 575–587 10.1016/j.stem.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 48. Zhou Z., Liu Y.-T., Ma L., Gong T., Hu Y.-N., Li H.-T., Cai C., Zhang L.-L., Wei G., and Zhou J.-Q. (2017) Independent manipulation of histone H3 modifications in individual nucleosomes reveals the contributions of sister histones to transcription. Elife 6, e30178 10.7554/eLife.30178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao F., Liao B., Hu J., Li S., Zhao H., Sun M., Gu J., and Jin Y. (2017) JMJD1C ensures mouse embryonic stem cell self-renewal and somatic cell reprogramming through controlling microRNA expression. Stem Cell Rep. 9, 927–942 10.1016/j.stemcr.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loh Y.-H., Zhang W., Chen X., George J., and Ng H.-H. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 10.1101/gad.1588207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ebata K. T., Mesh K., Liu S., Bilenky M., Fekete A., Acker M. G., Hirst M., Garcia B. A., and Ramalho-Santos M. (2017) Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenetics Chromatin 10, 36 10.1186/s13072-017-0143-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mimura I., Nangaku M., Kanki Y., Tsutsumi S., Inoue T., Kohro T., Yamamoto S., Fujita T., Shimamura T., Suehiro J., Taguchi A., Kobayashi M., Tanimura K., Inagaki T., Tanaka T., et al. (2012) Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell. Biol. 32, 3018–3032 10.1128/MCB.06643-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chakraborty D., Cui W., Rosario G. X., Scott R. L., Dhakal P., Renaud S. J., Tachibana M., Rumi M. A., Mason C. W., Krieg A. J., and Soares M. J. (2016) HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 113, E7212–E7221 10.1073/pnas.1612626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brauchle M., Yao Z., Arora R., Thigale S., Clay I., Inverardi B., Fletcher J., Taslimi P., Acker M. G., Gerrits B., Voshol J., Bauer A., Schübeler D., Bouwmeester T., and Ruffner H. (2013) Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One 8, e60549 10.1371/journal.pone.0060549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma D. K., Chiang C.-H., Ponnusamy K., Ming G.-L., and Song H. (2008) G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells 26, 2131–2141 10.1634/stemcells.2008-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tran K. A., Jackson S. A., Olufs Z. P., Zaidan N. Z., Leng N., Kendziorski C., Roy S., and Sridharan R. (2015) Collaborative rewiring of the pluripotency network by chromatin and signalling modulating pathways. Nat. Commun. 6, 6188 10.1038/ncomms7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., et al. (2013) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 10.1038/ng.2491 [DOI] [PubMed] [Google Scholar]
- 58. Li S., Ali S., Duan X., Liu S., Du J., Liu C., Dai H., Zhou M., Zhou L., Yang L., Chu P., Li L., Bhatia R., Schones D. E., Wu X., et al. (2018) JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403 10.1016/j.celrep.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shakya A., Callister C., Goren A., Yosef N., Garg N., Khoddami V., Nix D., Regev A., and Tantin D. (2015) Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Mol. Cell. Biol. 35, 1014–1025 10.1128/MCB.01105-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang J., Park J. W., Drissi H., Wang X., and Xu R.-H. (2014) Epigenetic regulation of miR-302 by JMJD1C inhibits neural differentiation of human embryonic stem cells. J. Biol. Chem. 289, 2384–2395 10.1074/jbc.M113.535799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Das P. P., Shao Z., Beyaz S., Apostolou E., Pinello L., De Los Angeles A., O'Brien K., Atsma J. M., Fujiwara Y., Nguyen M., Ljuboja D., Guo G., Woo A., Yuan G.-C., Onder T., et al. (2014) Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol. Cell 53, 32–48 10.1016/j.molcel.2013.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pedersen M. T., Kooistra S. M., Radzisheuskaya A., Laugesen A., Johansen J. V., Hayward D. G., Nilsson J., Agger K., and Helin K. (2016) Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35, 1550–1564 10.15252/embj.201593317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pedersen M. T., Agger K., Laugesen A., Johansen J. V., Cloos P. A., Christensen J., and Helin K. (2014) The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 34, 1031–1045 10.1128/MCB.00864-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tomaz R. A., Harman J. L., Karimlou D., Weavers L., Fritsch L., Bou-Kheir T., Bell E., Del Valle Torres I., Niakan K. K., Fisher C., Joshi O., Stunnenberg H. G., Curry E., Ait-Si-Ali S., Jørgensen H. F., et al. (2017) Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation. Development. 144, 567–579 10.1242/dev.142489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sim Y.-J., Kim M.-S., Nayfeh A., Yun Y.-J., Kim S.-J., Park K.-T., Kim C.-H., and Kim K.-S. (2017) 2i maintains a naive ground state in ESCs through two distinct epigenetic mechanisms. Stem Cell Rep. 8, 1312–1328 10.1016/j.stemcr.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matoba S., Liu Y., Lu F., Iwabuchi K. A., Shen L., Inoue A., and Zhang Y. (2014) Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu Z., Cai Y., Wang Y., Nie Y., Zhang C., Xu Y., Zhang X., Lu Y., Wang Z., Poo M., and Sun Q. (2018) Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 172, 881–887.e7 10.1016/j.cell.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 68. Chung Y. G., Matoba S., Liu Y., Eum J. H., Lu F., Jiang W., Lee J. E., Sepilian V., Cha K. Y., Lee D. R., and Zhang Y. (2015) Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17, 758–766 10.1016/j.stem.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 69. Wei J., Antony J., Meng F., MacLean P., Rhind R., Laible G., and Oback B. (2017) KDM4B-mediated reduction of H3K9me3 and H3K36me3 levels improves somatic cell reprogramming into pluripotency. Sci. Rep. 7, 7514 10.1038/s41598-017-06569-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Antony J., Oback F., Chamley L. W., Oback B., and Laible G. (2013) Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos. Mol. Cell. Biol. 33, 974–983 10.1128/MCB.01014-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huang C., Xiang Y., Wang Y., Li X., Xu L., Zhu Z., Zhang T., Zhu Q., Zhang K., Jing N., and Chen C. D. (2010) Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165 10.1038/cr.2010.5 [DOI] [PubMed] [Google Scholar]
- 72. Tang Y., Hong Y.-Z., Bai H.-J., Wu Q., Chen C. D., Lang J.-Y., Boheler K. R., and Yang H.-T. (2016) Plant homeo domain finger protein 8 regulates mesodermal and cardiac differentiation of embryonic stem cells through mediating the histone demethylation of pmaip1. Stem Cells 34, 1527–1540 10.1002/stem.2333 [DOI] [PubMed] [Google Scholar]
- 73. Pauklin S., and Vallier L. (2013) The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu H., Hu S., and Baker J. (2014) JMJD5 regulates cell cycle and pluripotency in human embryonic stem cells. Stem Cells 32, 2098–2110 10.1002/stem.1724 [DOI] [PubMed] [Google Scholar]
- 75. Wu H., and Zhang Y. (2014) Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 10.1016/j.cell.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S. K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O., Schuermann D., Michalakis S., Kosmatchev O., Schiesser S., Steigenberger B., et al. (2014) Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 10, 574–581 10.1038/nchembio.1532 [DOI] [PubMed] [Google Scholar]
- 77. Ficz G., Branco M. R., Seisenberger S., Santos F., Krueger F., Hore T. A., Marques C. J., Andrews S., and Reik W. (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 10.1038/nature10008 [DOI] [PubMed] [Google Scholar]
- 78. Pastor W. A., Pape U. J., Huang Y., Henderson H. R., Lister R., Ko M., McLoughlin E. M., Brudno Y., Mahapatra S., Kapranov P., Tahiliani M., Daley G. Q., Liu X. S., Ecker J. R., Milos P. M., et al. (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 10.1038/nature10102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Williams K., Christensen J., Pedersen M. T., Johansen J. V., Cloos P. A., Rappsilber J., and Helin K. (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 10.1038/nature10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu H., D'Alessio A. C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y. E., and Zhang Y. (2011) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 10.1038/nature09934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A. J., Zheng L., Zhang H., Huang S., Min J., Nicholson T., Chen T., Xu G., Shi Y., et al. (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464 10.1016/j.molcel.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A. C., Fung H.-L., Zhang K., and Zhang Y. (2013) Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 10.1016/j.cell.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., and Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dawlaty M. M., Ganz K., Powell B. E., Hu Y.-C., Markoulaki S., Cheng A. W., Gao Q., Kim J., Choi S.-W., Page D. C., and Jaenisch R. (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 10.1016/j.stem.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dawlaty M. M., Breiling A., Le T., Raddatz G., Barrasa M. I., Cheng A. W., Gao Q., Powell B. E., Li Z., Xu M., Faull K. F., Lyko F., and Jaenisch R. (2013) Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323 10.1016/j.devcel.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dawlaty M. M., Breiling A., Le T., Barrasa M. I., Raddatz G., Gao Q., Powell B. E., Cheng A. W., Faull K. F., Lyko F., and Jaenisch R. (2014) Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111 10.1016/j.devcel.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Verma N., Pan H., Doré L. C., Shukla A., Li Q. V., Pelham-Webb B., Teijeiro V., González F., Krivtsov A., Chang C. J., Papapetrou E. P., He C., Elemento O., and Huangfu D. (2018) TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 50, 83–95 10.1038/s41588-017-0002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang W., Xia W., Wang Q., Towers A. J., Chen J., Gao R., Zhang Y., Yen C.-A., Lee A. Y., Li Y., Zhou C., Liu K., Zhang J., Gu T.-P., Chen X., et al. (2016) Isoform switch of TET1 regulates DNA demethylation and mouse development. Mol. Cell 64, 1062–1073 10.1016/j.molcel.2016.10.030 [DOI] [PubMed] [Google Scholar]
- 89. Kong L., Tan L., Lv R., Shi Z., Xiong L., Wu F., Rabidou K., Smith M., He C., Zhang L., Qian Y., Ma D., Lan F., Shi Y., and Shi Y. G. (2016) A primary role of TET proteins in establishment and maintenance of de novo bivalency at CpG islands. Nucleic Acids Res. 44, 8682–8692 10.1093/nar/gkw529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hon G. C., Song C.-X., Du T., Jin F., Selvaraj S., Lee A. Y., Yen C.-A., Ye Z., Mao S.-Q., Wang B.-A., Kuan S., Edsall L. E., Zhao B. S., Xu G.-L., He C., et al. (2014) 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297 10.1016/j.molcel.2014.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang Y., Chavez L., Chang X., Wang X., Pastor W. A., Kang J., Zepeda-Martínez J. A., Pape U. J., Jacobsen S. E., Peters B., and Rao A. (2014) Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 1361–1366 10.1073/pnas.1322921111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lu F., Liu Y., Jiang L., Yamaguchi S., and Zhang Y. (2014) Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 28, 2103–2119 10.1101/gad.248005.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. de la Rica L., Deniz Ö., Cheng K. C., Todd C. D., Cruz C., Houseley J., and Branco M. R. (2016) TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 17, 234 10.1186/s13059-016-1096-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D., Firth A., Singer O., Trono D., and Pfaff S. L. (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Blaschke K., Ebata K. T., Karimi M. M., Zepeda-Martínez J. A., Goyal P., Mahapatra S., Tam A., Laird D. J., Hirst M., Rao A., Lorincz M. C., and Ramalho-Santos M. (2013) Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 10.1038/nature12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hore T. A., von Meyenn F., Ravichandran M., Bachman M., Ficz G., Oxley D., Santos F., Balasubramanian S., Jurkowski T. P., and Reik W. (2016) Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. U.S.A. 113, 12202–12207 10.1073/pnas.1608679113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y., Yao C., Liu X., Lu Z., Xu Z., Kang L., et al. (2013) Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 10.1016/j.stem.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 98. Wang T., Wu H., Li Y., Szulwach K. E., Lin L., Li X., Chen I.-P., Goldlust I. S., Chamberlain S. J., Dodd A., Gong H., Ananiev G., Han J. W., Yoon Y.-S., Rudd M. K., et al. (2013) Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol. 15, 700–711 10.1038/ncb2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Doege C. A., Inoue K., Yamashita T., Rhee D. B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W. B., Shih A., Levine R. L., Nik S., Chen E. I., and Abeliovich A. (2012) Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 10.1038/nature11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hu X., Zhang L., Mao S.-Q., Li Z., Chen J., Zhang R.-R., Wu H.-P., Gao J., Guo F., Liu W., Xu G.-F., Dai H.-Q., Shi Y. G., Li X., Hu B., et al. (2014) Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14, 512–522 10.1016/j.stem.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 101. Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., Liu J., Liu J., Wu H., Mao S.-Q., Mo K., et al. (2013) Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45, 1504–1509 10.1038/ng.2807 [DOI] [PubMed] [Google Scholar]
- 102. Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., Das S., Levasseur D. N., Li Z., Xu M., Reik W., et al. (2013) NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 10.1038/nature11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Okashita N., Sakashita N., Ito K., Mitsuya A., Suwa Y., and Seki Y. (2015) PRDM14 maintains pluripotency of embryonic stem cells through TET-mediated active DNA demethylation. Biochem. Biophys. Res. Commun. 466, 138–145 10.1016/j.bbrc.2015.08.122 [DOI] [PubMed] [Google Scholar]
- 104. Ko M., An J., Bandukwala H. S., Chavez L., Aijö T., Pastor W. A., Segal M. F., Li H., Koh K. P., Lähdesmäki H., Hogan P. G., Aravind L., and Rao A. (2013) Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497, 122–126 10.1038/nature12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yildirim O., Li R., Hung J.-H., Chen P. B., Dong X., Ee L.-S., Weng Z., Rando O. J., and Fazzio T. G. (2011) Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510 10.1016/j.cell.2011.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hainer S. J., McCannell K. N., Yu J., Ee L.-S., Zhu L. J., Rando O. J., and Fazzio T. G. (2016) DNA methylation directs genomic localization of Mbd2 and Mbd3 in embryonic stem cells. Elife 5, e21964 10.7554/eLife.21964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xiong J., Zhang Z., Chen J., Huang H., Xu Y., Ding X., Zheng Y., Nishinakamura R., Xu G.-L., Wang H., Chen S., Gao S., and Zhu B. (2016) Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol. Cell 64, 913–925 10.1016/j.molcel.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 108. Fidalgo M., Huang X., Guallar D., Sanchez-Priego C., Valdes V. J., Saunders A., Ding J., Wu W.-S., Clavel C., and Wang J. (2016) Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19, 355–369 10.1016/j.stem.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Etchegaray J.-P., Chavez L., Huang Y., Ross K. N., Choi J., Martinez-Pastor B., Walsh R. M., Sommer C. A., Lienhard M., Gladden A., Kugel S., Silberman D. M., Ramaswamy S., Mostoslavsky G., Hochedlinger K., et al. (2015) The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 17, 545–557 10.1038/ncb3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tu J., Ng S. H., Shui Luk A. C., Liao J., Jiang X., Feng B., Lun Mak K. K., Rennert O. M., Chan W.-Y., and Lee T.-L. (2015) MicroRNA-29b/Tet1 regulatory axis epigenetically modulates mesendoderm differentiation in mouse embryonic stem cells. Nucleic Acids Res. 43, 7805–7822 10.1093/nar/gkv653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., and Pasini D. (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 10.1016/j.molcel.2012.12.019 [DOI] [PubMed] [Google Scholar]
- 112. Yamaguchi S., Hong K., Liu R., Inoue A., Shen L., Zhang K., and Zhang Y. (2013) Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339 10.1038/cr.2013.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chen Q., Chen Y., Bian C., Fujiki R., and Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 10.1038/nature11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wu T. P., Wang T., Seetin M. G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S. D., Mackintosh S. G., Zhong M., Tackett A., Wang G., Hon L. S., Fang G., Swenberg J. A., et al. (2016) DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532, 329–333 10.1038/nature17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ougland R., Lando D., Jonson I., Dahl J. A., Moen M. N., Nordstrand L. M., Rognes T., Lee J. T., Klungland A., Kouzarides T., and Larsen E. (2012) ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells 30, 2672–2682 10.1002/stem.1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wu J., Ocampo A., and Belmonte J. C. I. (2016) Cellular metabolism and induced pluripotency. Cell 166, 1371–1385 10.1016/j.cell.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 117. Mathieu J., Zhou W., Xing Y., Sperber H., Ferreccio A., Agoston Z., Kuppusamy K. T., Moon R. T., and Ruohola-Baker H. (2014) Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell 14, 592–605 10.1016/j.stem.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jeong C.-H., Lee H.-J., Cha J.-H., Kim J. H., Kim K. R., Kim J.-H., Yoon D.-K., and Kim K.-W. (2007) Hypoxia-inducible factor-1α inhibits self-renewal of mouse embryonic stem cells in vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. J. Biol. Chem. 282, 13672–13679 10.1074/jbc.M700534200 [DOI] [PubMed] [Google Scholar]
- 119. Covello K. L., Kehler J., Yu H., Gordan J. D., Arsham A. M., Hu C.-J., Labosky P. A., Simon M. C., and Keith B. (2006) HIF-2α regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 20, 557–570 10.1101/gad.1399906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Binó L., Kučera J., Štefková K., Šindlerová L. Š., Lánová M., Kudová J., Kubala L., and Pacherník J. (2016) The stabilization of hypoxia inducible factor modulates differentiation status and inhibits the proliferation of mouse embryonic stem cells. Chem. Biol. Interact. 244, 204–214 10.1016/j.cbi.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 121. Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., and Ding S. (2010) Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655 10.1016/j.stem.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Prigione A., Rohwer N., Hoffmann S., Mlody B., Drews K., Bukowiecki R., Blümlein K., Wanker E. E., Ralser M., Cramer T., and Adjaye J. (2014) HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1–3 and PKM2. Stem Cells 32, 364–376 10.1002/stem.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]