Abstract
Inherited disorders of oxidative phosphorylation cause the clinically and genetically heterogeneous diseases known as mitochondrial energy generation disorders, or mitochondrial diseases. Over the last three decades, mutations causing these disorders have been identified in almost 290 genes, but many patients still remain without a molecular diagnosis. Moreover, while our knowledge of the genetic causes is continually expanding, our understanding into how these defects lead to cellular dysfunction and organ pathology is still incomplete. Here, we review recent developments in disease gene discovery, functional characterization, and shared pathogenic parameters influencing disease pathology that offer promising avenues toward the development of effective therapies.
Keywords: genetic disease, genomics, mitochondria, mitochondrial disease, respiratory chain, OXPHOS
Introduction
Mitochondrial energy generation disorders (hereafter termed mitochondrial diseases) are a heterogeneous group of rare disorders involving defective oxidative phosphorylation (OXPHOS).2 The OXPHOS system comprises five enzyme complexes, situated in the mitochondrial inner membrane. The first four complexes and two electron carriers form the respiratory chain, which generates a proton gradient used by complex V (F1F0-ATP synthase) to generate the majority of cellular ATP. For the purpose of this review, we have focused on genes and mechanisms that have a demonstrated defect in primary energy generation (e.g. components of the OXPHOS system) or a clear expected role in mitochondrial homeostasis and the ability to generate energy (e.g. components of the TCA cycle and pyruvate dehydrogenase complex (PDC) that feed into OXPHOS, or those affecting mitochondrial membranes and structure).
Mitochondrial diseases are clinically diverse and can present at any age (1). They can manifest in a tissue-specific or a multisystemic manner, but most often they affect organs with the highest energy demands such as brain, skeletal muscle, eyes, and heart (2). Mitochondrial diseases can show any mode of inheritance: maternal; X-linked; autosomal recessive; autosomal dominant; and de novo. This is due to the dual involvement of both the mitochondrial and nuclear genomes in maintaining the mitochondrial proteome. Mitochondrial DNA (mtDNA) is present in thousands of copies in most cells, which means the heteroplasmic load of a mutation can vary from 0 to 100% and may also differ between tissues in an individual. The threshold level above which the proportion of mutant mtDNA causes cellular dysfunction varies between mutations and tissues, further complicating the pathogenesis of mtDNA disease, as described in detail elsewhere (1, 2).
Disorders of energy generation have a minimum birth prevalence of 1 in 5000 live births (3, 4). Mutations in mtDNA appear to account for ∼75% of adult-onset disease (4) but only ∼25% of childhood-onset disease (5, 6).
Gene discoveries
In 1988, the first genetic causes of human mitochondrial disease were published when large deletions in mtDNA and the m.11778G→A point mutation in MT-ND4 were identified as causing mitochondrial myopathy (MIM 251900) and Leber's hereditary optic neuropathy (LHON, MIM 535000), respectively (7, 8). The following year, the first nuclear cause of mitochondrial disease was identified by the finding of pathogenic mutations in the X-chromosome–encoded PDC subunit gene, PDHA1 (9).
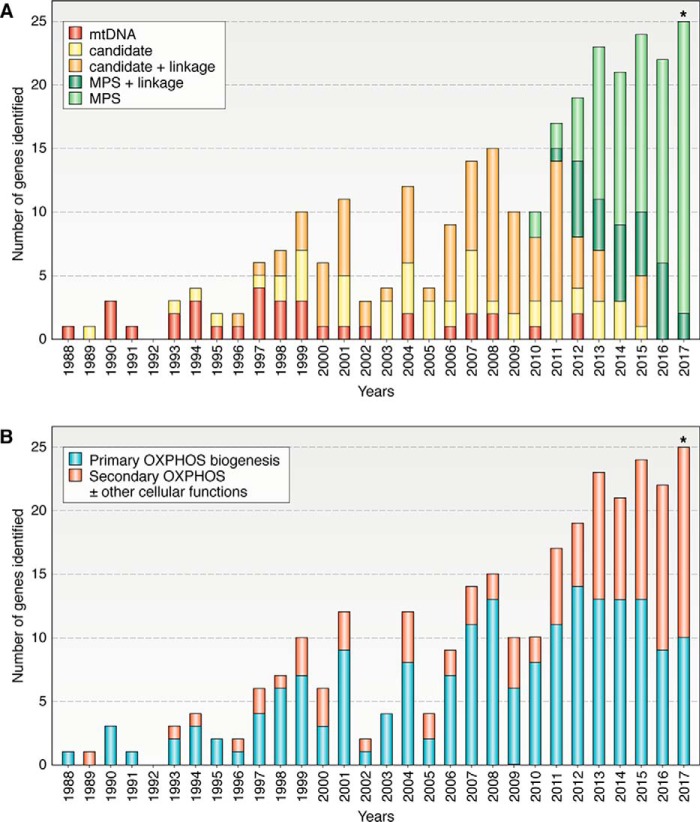
In the following 4 years, mutations in another six mtDNA genes and a second nuclear encoded PDC subunit gene (DLD) were shown to cause mitochondrial disorders (Fig. 1 and Table S1) (10–16). This signaled the beginning of a 16-year period where 94 additional nuclear encoded gene defects were identified by candidate gene sequencing (often in conjunction with linkage studies or homozygosity mapping from 1996) (Fig. 1A). The advent of massively parallel (or next generation) sequencing (MPS) led to the identification of two genes encoding the mitochondrial complex I assembly factors, NUBPL and FOXRED1, as disease genes in 2010 (17). This technology has now enabled the discovery of another 116 novel mitochondrial disease genes. Candidate gene sequencing and linkage studies continue to be used, leading to the discovery of an additional 42 nuclear disease genes. The utility of linkage studies and homozygosity mapping is now successfully being combined with MPS for mutation identification, currently accounting for about 20–30% of yearly gene discoveries.
Figure 1.
Discovery of genes linked to mitochondrial energy generation disorders: mode of identification and link to OXPHOS biogenesis by year (1988–2017*). A, because mtDNA was first linked to human disease in 1988, all 37 genes encoded by the mitochondrial genome have had mutations reported; however, only 35 of these have had adequate levels of evidence reported to confirm pathogenicity (32). The first nuclear gene to be linked to mitochondrial energy generation disorders was identified in 1989 by candidate gene sequencing; an additional 138 genes have been identified by this technique to date (with or without the assistance of linkage or homozygosity studies). Since 2010, MPS technology (including targeted sequencing panels and whole-exome or whole-genome sequencing and RNAseq) has identified 116 additional nuclear genes. Linkage or homozygosity studies have aided in the identification of 119 (or 47%) of the 254 nuclear genes associated with mitochondrial disease. B, in recent years, the number of genes linked to secondary defects in OXPHOS as well as to additional cellular functions has been steadily increasing, most likely due to the relatively unbiased nature of MPS in identifying gene defects. *, numbers are complete up to Nov. 23, 2017.
It was been suggested that the rate of novel gene discoveries in Mendelian disorders has been declining in recent years (post-2013) (18). However, this does not yet seem to hold true for mitochondrial diseases, which have averaged 22 novel gene discoveries per year since 2012 (Fig. 1A), including 25 for 2017 thus far (listed in Table S1). As of November 23 2017, 289 mitochondrial disease genes have been identified (35 mtDNA encoded genes and 254 nuclear encoded disease genes); numbers refined and updated from Refs. 19, 20. Recent studies of mitochondrial disease cohorts undergoing mtDNA and whole-exome sequencing typically report finding a molecular diagnosis in 30–60% of patients (21–24). Although some patients will have had mutations in known disease genes missed for technical reasons, the general consensus in the field is that known disease genes account for only ∼60% of patients with suspected mitochondrial diseases.
Categorizing OXPHOS disease genes: direct versus wider cellular impact
The 289 genes we have categorized as causing mitochondrial disease can be divided into those that have a primary role specific to OXPHOS biogenesis and those whose impacts on OXPHOS are indirect or involve other cellular functions. Interestingly, as disease gene discovery has become more reliant on MPS, the number of genes with secondary or more indirect roles in OXPHOS has increased to around half of all new findings (Fig. 1B). Based on their function, the encoded proteins can be roughly grouped into the following categories: 1) OXPHOS subunits, assembly factors, and electron carriers; 2) mtDNA maintenance; 3) mtDNA expression; 4) enzyme cofactors; 5) mitochondrial homeostasis and quality control; and 6) general metabolism (Fig. 2). Individual references for genes are listed in Table S1 or detailed in Refs. 19, 20, 25–29.
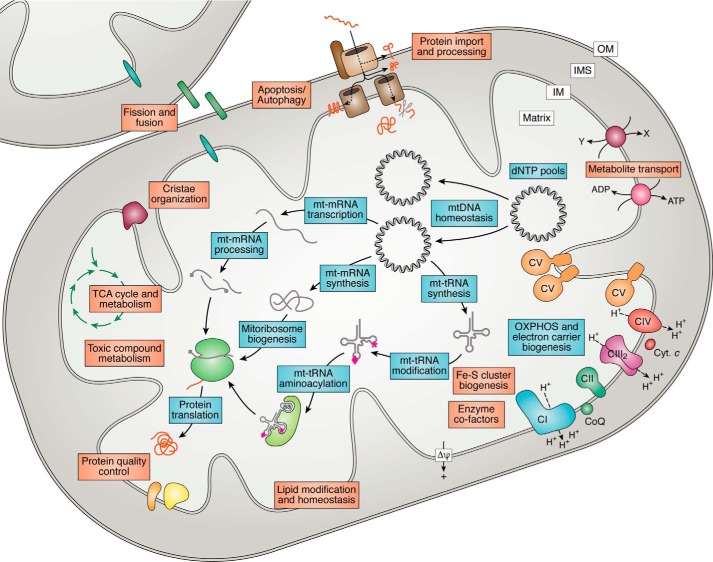
Figure 2.
Functional categories of genes impacting mitochondrial energy generation. Mutations in genes functioning in a wide variety of pathways have been linked to mitochondrial diseases and can be separated into those with a primary role specific to OXPHOS biogenesis (blue boxes) or those with a secondary impact on OXPHOS that may also involve other cellular functions (orange boxes). These pathways include the following: 1) OXPHOS subunits, assembly factors, and electron carriers; 2) mtDNA maintenance (including dNTP homeostasis); 3) mtDNA expression (including synthesis, processing, and modification of mt-rRNAs, mt-tRNAs, mt-mRNAs, mitoribosome biogenesis, and translation); 4) enzyme cofactors (including Fe-S cluster biogenesis); 5) mitochondrial homeostasis and quality control (including mitochondrial protein import, lipid modification and homeostasis, mitochondrial morphology comprising fission/fusion and cristae organization factors, protein quality control, and apoptosis/autophagy); and 6) general metabolism (e.g. TCA cycle and metabolite transport). CI–CV, OXPHOS complexes I–V; Cyt. c, cytochrome c; CoQ, ubiquinone/coenzyme Q; Δψ, mitochondrial membrane potential; aa, amino acid; OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
Genes with a primary role specific to OXPHOS biogenesis
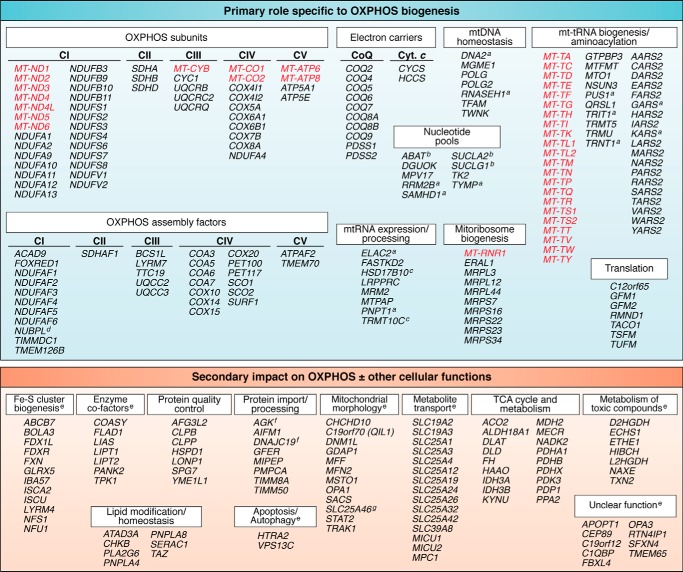
The larger portion (185 of 289) of the mitochondrial disease genes falls into one of the first three categories, with a primary role specifically related to OXPHOS biogenesis (Fig. 3). Mitochondrial DNA itself encodes 13 subunits of OXPHOS complexes I and III–V, along with 22 tRNAs and 2 rRNAs that are essential for expression of the 13 subunits. The dual genetic nature of mitochondria means that many additional nuclear encoded proteins are required, not just for the biogenesis of the OXPHOS complexes themselves but for the maintenance and expression of mtDNA (30, 31). Pathogenic mutations have been reported in all 37 mtDNA genes (26, 32); however, for two genes (MT-CO3 and MT-RNR2), the evidence for pathogenicity has yet to be sufficiently validated (32).
Figure 3.
Genes linked to mitochondrial energy generation disorders. This list includes all of the genes reported to date in which mutations have been shown to cause mitochondrial diseases. They are categorized according to whether their role is primarily linked to OXPHOS biogenesis (185 genes of 289) or whether their impact on OXPHOS is secondary, likely involving additional cellular functions (104 genes). Genes listed in red are encoded by mtDNA and in black by nuclear DNA. a, genes encoding proteins that either function in the cytosol/nucleus or localize to both the mitochondria and cytosol/nucleus (19, 27, 34 and references therein). b, genes encoding dual-function enzymes that affect mitochondrial nucleoside salvage pathways along with their primary function (42, 98, 99). c, proteins encoded by TRMT10C and HSD17B10 (MRPP1 and MRPP2) are subunits of the mitochondrial RNase P enzyme, along with MRPP3 encoded by KIAA0391, and are responsible for processing and maturation of mt-tRNAs from polycistronic transcripts. Together, they also form a subcomplex involved in tRNA modification (m1R9 methylation). HSD17B10 encodes 17β-hydroxysteroid dehydrogenase type 10, a multifunction enzyme that catalyzes the oxidation of a wide variety of fatty acids and steroids, in addition to RNA processing (100). d, NUBPL plays a role in the incorporation of Fe-S clusters into OXPHOS complex I (CI) subunits, so it could also be classified as having a role in Fe-S biogenesis (17). e, some of the genes within these groups encode proteins that are either cytosolic or recruited from the cytosol to mitochondrial membranes or are reported to have alternative isoforms that are targeted to the mitochondria or cytosol. f, although classified as having a role in mitochondrial protein import, AGK and DNAJC19 are also implicated in mitochondrial lipid homeostasis (56, 57, 59). g, although classified as a member of the carrier family, SLC25A46 localizes to the mitochondrial outer membrane where it functions in mitochondrial cristae architecture (61).
Disorders of mtDNA maintenance can cause mtDNA depletion or multiple deletions of mtDNA (27). Many of these genes are involved in the regulation of mitochondrial or cytosolic nucleotide pools, as mitochondria rely on cytosolic de novo synthesis or salvage pathways to maintain a balanced supply of dNTPs (27). Other disease genes impacting on mtDNA homeostasis encode proteins involved in replication and repair pathways, as well as mtDNA nucleoid packaging (27, 31). This includes the mitochondrial polymerase-γ, encoded by POLG, in which almost 200 mutations have been reported, making it the most common nuclear cause of mitochondrial disease (33).
Numerous nuclear encoded and mtDNA encoded factors are required for transcription and translation of the 13 mtDNA-encoded OXPHOS polypeptides. Hence, genes linked to OXPHOS disorders include those vital for expression and stability of the long polycistronic mtDNA transcripts, which require post-transcriptional processing and modification to produce stable functional mt-mRNAs, mt-rRNAs, and mt-tRNAs (29, 31). Likewise, mitochondria require 19 aminoacyl tRNA synthetases to attach cognate amino acids to the appropriate tRNA, with mutations in all 19 now identified (19, 34–36). Mitochondrial ribosomes consist of 80 nuclear encoded subunits, with mutations identified in only eight so far (31, 37). In addition to the two mtDNA-encoded rRNAs, it had long been assumed that mitochondrial ribosomes required a 5S rRNA imported from the cytosol. However, recent studies showed that mtDNA-encoded mt-tRNAVal plays this role in humans (38). Other mtDNA expression defects are caused by mutations in factors required for mitochondrial ribosome biogenesis and translational regulation (19, 31).
The largest group of primary defect genes encode structural subunits of one of the five OXPHOS complexes, factors required for their assembly, or are involved in the biogenesis of the electron carriers cytochrome c and ubiquinone (coenzyme Q) (19, 26, 28). Probably due to its large size (1 MDa) and complexity, the majority of genes in this category are subunits or assembly factors of OXPHOS complex I (NADH:ubiquinone oxidoreductase), with mutations now identified in all 14 subunits of its catalytic core, along with 13 of the 30 accessory subunits, and 11 of at least 15 assembly factors (19, 39–41). In fact, isolated complex I deficiency is the most common mitochondrial enzyme deficiency, accounting for ∼30% of pediatric presentations (26).
Some of the genes we have categorized as having a primary effect on OXPHOS have dual roles or subcellular localizations that may contribute to their impact on OXPHOS and disease pathology. For example, mutations in SUCLA2 and SUCLG1 leading to mtDNA depletion syndromes are thought to result from a nucleotide imbalance due to their role in generating ATP and GTP, rather than their function as subunits of the TCA cycle enzyme succinate-CoA ligase (42). Additionally, some of the genes implicated in mitochondrial RNA processing and modification (ELAC2, encoding RNase Z; PUS1, encoding pseudouridylate synthase 1; TRIT1, encoding tRNA isopentenyltransferase; TRNT1, encoding CCA-adding tRNA nucleotidyltransferase) have been found to also function in the nucleus (43–46).
Genes with a secondary impact on OXPHOS biogenesis as well as other cellular functions
Mutations in genes in this category often impact other cellular functions, including enzyme cofactors, metabolism, or mitochondrial homeostasis and quality control (Fig. 3). A significant proportion of these genes are those involved in Fe-S cluster biogenesis. Fe-S clusters are found in OXPHOS complexes I–III, thereby directly affecting OXPHOS biogenesis, but also in other mitochondrial and cellular enzymes, such as aconitase (TCA cycle), electron transfer flavoprotein-ubiquinone oxidoreductase (electron transfer to ubiquinone, e.g. fatty acid oxidation), and isopropylmalate isomerase (amino acid synthesis) (47). These defects can thus have a wider impact on diverse cellular pathways, along with OXPHOS function.
Other genes in this category may influence general mitochondrial homeostasis or biogenesis, thereby impacting OXPHOS function via a number of mechanisms. In general, these genes can be categorized into mitochondrial protein import, mitochondrial lipid or membrane homeostasis, mitochondrial fission/fusion and cristae organization, and/or mitochondrial protein quality control (19, 48, 49). For some, a mechanism for their effect on OXPHOS function has been elucidated. An example is TAZ, encoding a phospholipid transacylase involved in remodeling the mitochondrial inner membrane phospholipid cardiolipin, in which mutations cause Barth syndrome (MIM 302060) (48). In patient cell lines, supercomplexes consisting of OXPHOS complexes I, III, and IV in various stoichiometries are destabilized, resulting in impaired OXPHOS efficiency (48, 50). In contrast, the ATAD3 locus, recently identified to underlie diverse clinical presentations often involving cerebellar defects (51–53), is linked to mitochondrial functions that include regulation of mtDNA maintenance and translation. ATAD3 patient cell lines show defects in cellular cholesterol and mtDNA homeostasis, providing a basis for the disease pathology, yet a clear link between ATAD3 and OXPHOS impairment is still lacking (52).
Additional genes responsible for mitochondrial disorders affect mitochondrial and cellular metabolism, including components of the PDC and TCA cycle, enzymes involved in toxic compound metabolism, and carrier proteins involved in metabolite and cofactor transport across cellular and mitochondrial membranes (19, 54). We have elected not to include genes involved in other metabolic pathways, such as mitochondrial fatty acid oxidation, in which secondary inhibition of OXPHOS function may result from altered metabolite concentrations or cellular signaling pathways (55).
Assigning many of these secondary genes to a distinct category is difficult as they can have dual or unresolved roles. An example is AGK, encoding a mitochondrial inner membrane lipid-modifying acylglycerol kinase, in which defects lead to Sengers syndrome (MIM 212350) (48). Recent studies show that AGK also functions in the carrier protein import pathway of mitochondria, independent of its kinase activity (56, 57). Likewise, mutations in the mitochondrial ATP/ADP transporter (ANT1), encoded by SLC25A4, can impact not only on the ADP/ATP ratio but also on mtDNA homeostasis by altering mitochondrial dNTP equilibrium (58).
Other genes that could be classified into several categories include DNAJC19 (protein import and cardiolipin homeostasis) (59) and MECR (mitochondrial fatty acid synthesis and synthesis of the cofactor lipoic acid) (54, 60). Similarly, although considered a member of the carrier family, SLC25A46 localizes to the mitochondrial outer membrane where it functions in mitochondrial cristae architecture and endoplasmic reticulum/mitochondrial contacts implicated in lipid transfer (61). It is interesting to note that complex V and the associated mitochondrial contact site and cristae-organizing system (MICOS) play roles in inner membrane cristae structure and curvature (62, 63), and some patients with mutations affecting complex V show altered mitochondrial morphology (64).
Generally, an isolated OXPHOS enzyme deficiency (e.g. complex I deficiency) is suggestive of a mutation in either a structural subunit or assembly factor of the affected OXPHOS complex. However, there are many exceptions to this, with mutations in genes encoding OXPHOS subunits and assembly factors sometimes resulting in a combined OXPHOS deficiency. These pleiotropic effects may arise due to secondary alterations in mitochondrial membranes or mtDNA, changes in cofactor or metabolic regulation, or secondary impact due to changes in OXPHOS supercomplexes. In contrast, mutations in the other mitochondrial disease genes would be assumed to affect multiple OXPHOS complexes, and this is observed in 30–60% of patients (3, 19). Yet, isolated OXPHOS deficiency has been found for genes that would be expected to have pleiotropic effects, such as mutations in ETHE1, encoding a mitochondrial sulfur dioxygenase, which results in complex IV deficiency and ethylmalonic encephalopathy (MIM 602473) due to a toxic build up of hydrogen sulfide (65).
From genotypes to phenotypes: underlying mechanisms
In recent years, our knowledge of the genetic architecture of mitochondrial disorders has increased rapidly and will continue to do so as additional genes are identified by genomic analyses in patients, in parallel with studies in model systems that identify or validate novel candidate genes based on proteomic analyses, genetic screens, or other approaches (41, 66–68). However, our understanding of how these basic defects lead to specific types of cellular dysfunction, organ involvement, and disease severity remains fragmentary.
The pathogenesis of mitochondrial disorders shows both genetic heterogeneity and pleiotropy. The former is demonstrated by Leigh syndrome (MIM 256000), an infantile neurodegenerative disorder with characteristic lesions in the basal ganglia and brainstem caused by mutations in >75 genes to date (69). These genes are split across most of the categories listed in Fig. 3, suggesting that defects in many different processes can trigger a common tissue-specific pathology. In contrast, a number of the most common genes underlying mitochondrial disease show pleiotropy. For example, autosomal recessive mutations in POLG can cause a range of syndromes such as Leigh syndrome, mitochondrial DNA depletion syndrome 4A (Alpers type, MIM 203700), mitochondrial recessive ataxia syndrome (including MIRAS and SANDO, MIM 607459) and others that differ in age of onset, severity, and in the combination of diverse neurological symptoms plus muscle, gut, liver, pancreas, or renal involvement (33). Autosomal dominant POLG mutations can cause many of the same features plus Parkinsonism, hypogonadism, and cataracts, but there is only weak correlation between the specific POLG mutations and phenotype (33). Similarly, the m.3243A→G mutation in the mtDNA MT-TL1 gene is best known for causing MELAS. However, many patients lack stroke-like episodes, instead suffering from one or more symptoms such as seizures, short stature, myopathy, cardiomyopathy, ophthalmoplegia, deafness, diabetes, gastrointestinal or renal symptoms (70).
Many factors contribute to the tissue specificity of mitochondrial disorders. Mitochondrial quantity varies by up to 30-fold in different tissues, based on cytochrome c levels, whereas large-scale proteomic analyses suggest that ∼25% of mitochondrial proteins are not shared between any given pair of tissues (71). This variation means that the control of mitochondrial metabolic flux can be determined by different components of the OXPHOS system in different tissues (72).
Inadequate ATP synthesis is an obvious issue for cells with constant or intermittently high energy demands such as neurons, cardiomyocytes, and renal tubular cells, which need to propagate action potentials or reabsorb metabolites (73). Similarly, pancreatic beta cells must increase the ATP/ADP ratio above a threshold level to trigger insulin release (74), and astrocytes must recycle neurotransmitters released by neurons, a process that likely accounts for a large proportion of the brain's energy budget (75). Apart from an ATP deficit, inadequate ATP synthesis can also increase the AMP/ATP ratio, leading to activation of AMP-activated protein kinase and multiple signaling cascades (76). Some organs show marked increases in energy demand during growth and development, e.g. brain glucose consumption increases 3–4-fold per cm3 during the first few years of life before declining to adult levels (77). Thresholds for ATP requirement, and how they respond to changes in demand, undoubtedly contribute to different susceptibility to OXPHOS dysfunction between and within tissues, and to why conditions like Leigh syndrome can show substantial presymptomatic periods of good health in early life.
The mitochondrial membrane potential is the second obvious parameter impacted in bioenergy disorders and might be expected to decrease (i.e. become more electroneutral) if electron transfer and proton pumping are restricted. Apart from driving ATP production, the mitochondrial membrane potential is also required for biogenesis of Fe-S clusters, to drive mitochondrial import of many nuclear encoded proteins (78), and for import of calcium from the cytosol and endoplasmic reticulum to sequester transient increases and prevent aberrant calcium signaling or induction of cell death by opening of the mitochondrial permeability pore (73, 79). Mitochondria with decreased membrane potential are prone to cleavage of OPA1, triggering mitochondrial fission, and to accumulation of PINK1 on the outer membrane, which induces degradation by mitophagy following recruitment of Parkin and LC3 (49, 80). It is important to note that mitochondrial membrane potential can also be increased by mitochondrial dysfunction. This has been reported for the MT-ATP6 m.8993T→G mutation, which leads to a leucine to arginine change that appears to block the proton channel of complex V, resulting in increases in membrane potential, reactive oxygen species (ROS) production, lipid peroxidation, and susceptibility to apoptosis (81, 82).
Although ATP and mitochondrial membrane potential could be considered primary targets of OXPHOS defects, mitochondria are also key components of metabolic signaling pathways, and a range of metabolites play key roles in influencing cellular function. A detailed review of mitochondrial signaling pathways is beyond the scope of this review, but excellent reviews of pathways involved in nutrient sensing and stress responses to energy deficit, oxidative stress, and mitochondrial unfolded proteins are available (49, 76, 80). Mitochondrial metabolism is also now recognized to play key roles in regulating inflammation and immune responses (49, 83). We will therefore focus on some of the key metabolites that are dysregulated by mitochondrial dysfunction and may jointly contribute to triggering a range of signaling pathways mediating cellular homeostasis and pathology, as summarized in Table 1 and discussed below.
Table 1.
Mechanisms linking genotype to phenotype
| Abnormalities triggering cellular dysfunction | Cellular consequences |
|---|---|
| ATP synthesis and adenine nucleotide pools | ↓ ATP levels |
| Altered AMP/ATP and ADP/ATP ratios → changes in cellular signaling/function | |
| Mitochondrial membrane potential | Changes to ATP levels, Fe-S cluster biogenesis, and mitochondrial protein import |
| Altered Ca2+ homeostasis → aberrant cell signaling and/or cell death | |
| Changes in mitochondrial morphology and turnover | |
| ROS | Changes to ROS signaling pathways |
| Damage to protein, lipids, nucleic acids → cellular stress responses, and/or cell death | |
| NAD+/NADH and redox balance | Altered cellular signaling pathways and effects on mitochondrial biogenesis |
| TCA cycle metabolites | Altered cellular signaling pathways and epigenetic changes |
| Amino acids and one-carbon metabolism | Changes to nutrient sensing signaling networks and transcriptional responses |
OXPHOS complexes I and III are regarded as the major sites of mitochondrial ROS generation under normal conditions, and ROS are key mediators of homeostatic mechanisms regulating cell proliferation, maintenance of stemness, differentiation, and stress responses (49, 78). Modest increases in ROS levels can trigger a protective reaction via activation of antioxidant response elements (76), but if this is exceeded then oxidation of proteins, lipids, and nucleic acids can trigger other stress responses or cell death (73). Damaging increases in ROS production can occur in defects affecting not just complexes I and III (49, 84) but complex V, as described above, and other processes leading to increased mitochondrial membrane potential. Mutations in proteins that are key components of ROS homeostasis, such as thioredoxin 2, can also prevent the cell from adequately buffering increased ROS production (85).
Primary or secondary defects of complex I inhibit NADH oxidation, and cell lines and tissues from complex I–deficient mouse models show a decreased ratio of NAD+/NADH, which can sometimes be reflected in plasma by an increase in the ratio of hydroxyacylcarnitines to acylcarnitines (84, 86, 87). Decreased NAD+ levels inhibit SIRT1-PGC1α signaling, potentially down-regulating mitochondrial biogenesis.
A number of TCA cycle intermediates influence cellular signaling, and primary defects in TCA cycle enzymes or secondary alteration caused by OXPHOS disorders can impact a range of processes. Citrate exported from the mitochondria to the cytosol is converted by ATP citrate lyase to acetyl-CoA, which can be used for protein lysine acetylation, and a decrease of citrate levels can decrease acetylation of specific histone H3 marks (78). Increased 2-ketoglutarate levels result in increased generation of the “oncometabolite” 2-hydroxyglutarate, which can inhibit enzymes such as hypoxia-inducible factor prolyl hydroxylases and Jumonji-domain family histone lysine demethylases, leading to activation of hypoxic signaling and marked epigenetic changes (79). Increased levels of succinate or fumarate can also result in protein succinylation and transcriptional changes, contributing to a number of TCA cycle defects that result in predisposition to tumors such as paragangliomas, pheochromocytomas, myomas, and gliomas (49).
Amino acid metabolism can also be dysregulated in mitochondrial disease. Although the specific responses vary, muscle of “deletor” mice shows an amino acid starvation-like response, with activation of Akt signaling and release of FGF21, a hormone-like cytokine into the blood, leading to mobilization of lipids from storage fat (49). Serine levels are particularly elevated in “deletor” mouse muscle and in a HEK-293 cell model of mtDNA depletion (88, 89). In the cell model, this involved an ATF4-mediated increase in serine biosynthesis and a decrease in serine consumption reflecting decreased formate production (89). This defect in cellular one-carbon metabolism was predicted to impact purine and thymidylate synthesis as well as cellular methylation reactions. More broadly, amino acid abnormalities influence the nutrient-sensing signaling network and can lead to global transcriptional responses in a highly tissue-specific manner (90).
Conclusions and future directions
Investigations over the last 30 years have led to the identification of almost 290 genetic causes for disorders of mitochondrial energy generation, making these among the most genetically diverse group of inherited diseases. Since the widespread uptake of MPS technologies from 2012, there has been an average of 22 new disease genes identified each year. Challenges remain in detecting and interpreting pathogenic DNA variants, particularly in non-coding regions. Analytical approaches continue to evolve and are complemented by improvements in data sharing and comprehensive databases cataloguing population-based genetic variation (91), together with transcriptomic and proteomic analyses (37, 92, 93). It therefore seems likely that the current rate of gene discovery will continue for some years.
Whether they affect OXPHOS directly due to their primary function, or in a secondary manner that may also affect other cellular functions, categorizing these genes according to their functional roles emphasizes the wide array of cellular pathways that converge at OXPHOS biogenesis. In many cases, the roles can be ambiguous, as the proteins may have dual functions, alternative cellular localizations, or less clear roles in mitochondrial pathways. However, for some, their identification as a disease gene has aided in their functional characterization, resulting in new insights into mitochondrial and cellular pathways and disease pathology (e.g. ATAD3A and SLC25A46). In others, it has led to a treatment, as in the case of biotin and thiamine supplementation in SLC19A3 mutations (94).
Although our understanding of the heterogeneity and pleiotropy of mitochondrial disorders is still incomplete, a number of parameters influencing disease pathology have been identified that span many of the gene classification categories and affect different facets of cell signaling. The last Cochrane review of mitochondrial therapy in 2012 concluded: “there is currently no clear evidence supporting the use of any intervention in mitochondrial disorders” (95). However, a series of preclinical studies have provided encouraging evidence supporting the use of small molecule therapies targeting cellular signaling pathways related to NAD bioavailability and mitochondrial biogenesis, quality control, and stress responses (96, 97). Although for the vast majority of mitochondrial diseases there are currently no effective treatments or cures, the continued identification of disease genes, their mitochondrial and cellular functions, and their shared pathogenic parameters are leading to a greater understanding and an increasing number of clinical trials (2).
Supplementary Material
This work was supported by the Australian National Health and Medical Research Council (GNT1022896, GNT1068409, GNT1107094) and the Victorian Government's Operational Infrastructure Support Program. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
- OXPHOS
- oxidative phosphorylation
- PDC
- pyruvate dehydrogenase complex
- TCA
- tricarboxylic acid
- ROS
- reactive oxygen species
- MPS
- massively parallel sequencing
- MIM
- Online Mendelian Inheritance in Man (https://omim.org/).
References
- 1. DiMauro S. (2013) Mitochondrial encephalomyopathies–fifty years on: the Robert Wartenberg Lecture. Neurology 81, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorman G. S., Chinnery P. F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D. R., Zeviani M., and Turnbull D. M. (2016) Mitochondrial diseases. Nat. Rev. Dis. Primers 2, 16080 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- 3. Skladal D., Halliday J., and Thorburn D. R. (2003) Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain 126, 1905–1912 10.1093/brain/awg170 [DOI] [PubMed] [Google Scholar]
- 4. Gorman G. S., Schaefer A. M., Ng Y., Gomez N., Blakely E. L., Alston C. L., Feeney C., Horvath R., Yu-Wai-Man P., Chinnery P. F., Taylor R. W., Turnbull D. M., and McFarland R. (2015) Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 10.1002/ana.24362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lebon S., Chol M., Benit P., Mugnier C., Chretien D., Giurgea I., Kern I., Girardin E., Hertz-Pannier L., de Lonlay P., Rötig A., Rustin P., and Munnich A. (2003) Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J. Med. Genet. 40, 896–899 10.1136/jmg.40.12.896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thorburn D. R. (2004) Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 27, 349–362 10.1023/B:BOLI.0000031098.41409.55 [DOI] [PubMed] [Google Scholar]
- 7. Holt I. J., Harding A. E., and Morgan-Hughes J. A. (1988) Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 10.1038/331717a0 [DOI] [PubMed] [Google Scholar]
- 8. Wallace D. C., Singh G., Lott M. T., Hodge J. A., Schurr T. G., Lezza A. M., Elsas L. J. 2nd., and Nikoskelainen E. K. (1988) Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242, 1427–1430 10.1126/science.3201231 [DOI] [PubMed] [Google Scholar]
- 9. Endo H., Hasegawa K., Narisawa K., Tada K., Kagawa Y., and Ohta S. (1989) Defective gene in lactic acidosis: abnormal pyruvate dehydrogenase E1 α-subunit caused by a frameshift. Am. J. Hum. Genet. 44, 358–364 [PMC free article] [PubMed] [Google Scholar]
- 10. Liu T. C., Kim H., Arizmendi C., Kitano A., and Patel M. S. (1993) Identification of two missense mutations in a dihydrolipoamide dehydrogenase-deficient patient. Proc. Natl. Acad. Sci. U.S.A. 90, 5186–5190 10.1073/pnas.90.11.5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prezant T. R., Agapian J. V., Bohlman M. C., Bu X., Oztas S., Qiu W. Q., Arnos K. S., Cortopassi G. A., Jaber L., and Rotter J. I. (1993) Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4, 289–294 10.1038/ng0793-289 [DOI] [PubMed] [Google Scholar]
- 12. Moraes C. T., Ciacci F., Bonilla E., Ionasescu V., Schon E. A., and DiMauro S. (1993) A mitochondrial tRNA anticodon swap associated with a muscle disease. Nat. Genet. 4, 284–288 10.1038/ng0793-284 [DOI] [PubMed] [Google Scholar]
- 13. Huoponen K., Vilkki J., Aula P., Nikoskelainen E. K., and Savontaus M. L. (1991) A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am. J. Hum. Genet. 48, 1147–1153 [PMC free article] [PubMed] [Google Scholar]
- 14. Shoffner J. M., Lott M. T., Lezza A. M., Seibel P., Ballinger S. W., and Wallace D. C. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61, 931–937 10.1016/0092-8674(90)90059-N [DOI] [PubMed] [Google Scholar]
- 15. Holt I. J., Harding A. E., Petty R. K., and Morgan-Hughes J. A. (1990) A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 46, 428–433 [PMC free article] [PubMed] [Google Scholar]
- 16. Goto Y., Nonaka I., and Horai S. (1990) A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 10.1038/348651a0 [DOI] [PubMed] [Google Scholar]
- 17. Calvo S. E., Tucker E. J., Compton A. G., Kirby D. M., Crawford G., Burtt N. P., Rivas M., Guiducci C., Bruno D. L., Goldberger O. A., Redman M. C., Wiltshire E., Wilson C. J., Altshuler D., Gabriel S. B., et al. (2010) High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 42, 851–858 10.1038/ng.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boycott K. M., Rath A., Chong J. X., Hartley T., Alkuraya F. S., Baynam G., Brookes A. J., Brudno M., Carracedo A., den Dunnen J. T., Dyke S. O. M., Estivill X., Goldblatt J., Gonthier C., Groft S. C., et al. (2017) International cooperation to enable the diagnosis of all rare genetic diseases. Am. J. Hum. Genet. 100, 695–705 10.1016/j.ajhg.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mayr J. A., Haack T. B., Freisinger P., Karall D., Makowski C., Koch J., Feichtinger R. G., Zimmermann F. A., Rolinski B., Ahting U., Meitinger T., Prokisch H., and Sperl W. (2015) Spectrum of combined respiratory chain defects. J. Inherit. Metab. Dis. 38, 629–640 10.1007/s10545-015-9831-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Craven L., Alston C. L., Taylor R. W., and Turnbull D. M. (2017) Recent advances in mitochondrial disease. Annu. Rev. Genomics Hum. Genet. 18, 257–275 10.1146/annurev-genom-091416-035426 [DOI] [PubMed] [Google Scholar]
- 21. Taylor R. W., Pyle A., Griffin H., Blakely E. L., Duff J., He L., Smertenko T., Alston C. L., Neeve V. C., Best A., Yarham J. W., Kirschner J., Schara U., Talim B., Topaloglu H., Baric I., et al. (2014) Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 312, 68–77 10.1001/jama.2014.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wortmann S. B., Koolen D. A., Smeitink J. A., van den Heuvel L., and Rodenburg R. J. (2015) Whole exome sequencing of suspected mitochondrial patients in clinical practice. J. Inherit. Metab. Dis. 38, 437–443 10.1007/s10545-015-9823-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y., Hirata T., Yatsuka Y., Yamashita-Sugahara Y., Nakachi Y., Kato H., Okuda A., Tamaru S., Borna N. N., Banshoya K., et al. (2016) A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 12, e1005679 10.1371/journal.pgen.1005679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pronicka E., Piekutowska-Abramczuk D., Ciara E., Trubicka J., Rokicki D., Karkucińska-Więckowska A., Pajdowska M., Jurkiewicz E., Halat P., Kosińska J., Pollak A., Rydzanicz M., Stawinski P., Pronicki M., Krajewska-Walasek M., and Płoski R. (2016) New perspective in diagnostics of mitochondrial disorders: two years' experience with whole-exome sequencing at a national paediatric centre. J. Transl. Med. 14, 174 10.1186/s12967-016-0930-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koopman W. J., Willems P. H., and Smeitink J. A. (2012) Monogenic mitochondrial disorders. N. Engl. J. Med. 366, 1132–1141 10.1056/NEJMra1012478 [DOI] [PubMed] [Google Scholar]
- 26. Alston C. L., Rocha M. C., Lax N. Z., Turnbull D. M., and Taylor R. W. (2017) The genetics and pathology of mitochondrial disease. J. Pathol. 241, 236–250 10.1002/path.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Hattab A. W., Craigen W. J., and Scaglia F. (2017) Mitochondrial DNA maintenance defects. Biochim. Biophys. Acta 1863, 1539–1555 10.1016/j.bbadis.2017.02.017 [DOI] [PubMed] [Google Scholar]
- 28. Acosta M. J., Vazquez Fonseca L., Desbats M. A., Cerqua C., Zordan R., Trevisson E., and Salviati L. (2016) Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta 1857, 1079–1085 10.1016/j.bbabio.2016.03.036 [DOI] [PubMed] [Google Scholar]
- 29. Pearce S. F., Rebelo-Guiomar P., D'Souza A. R., Powell C. A., Van Haute L., and Minczuk M. (2017) Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem. Sci. 42, 625–639 10.1016/j.tibs.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stewart J. B., and Chinnery P. F. (2015) The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542 10.1038/nrg3966 [DOI] [PubMed] [Google Scholar]
- 31. Hällberg B. M., and Larsson N. G. (2014) Making proteins in the powerhouse. Cell Metab. 20, 226–240 10.1016/j.cmet.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 32. Lott M. T., Leipzig J. N., Derbeneva O., Xie H. M., Chalkia D., Sarmady M., Procaccio V., and Wallace D. C. (2013) MtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics 44, 1.23.1–26 10.1002/0471250953.bi0123s44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohen B. H., Chinnery P. F., and Copeland W. C. (2014) in Gene Reviews (Pagon R. A., Adam M. P., Ardinger H. H., Wallace S. E., Amemiya A., Bean L. J. H., Bird T. D., Ledbetter N., Mefford H. C., Smith R. J. H., and Stephens K., eds), University of Washington at Seattle, Seattle [Google Scholar]
- 34. Diodato D., Ghezzi D., and Tiranti V. (2014) The mitochondrial aminoacyl tRNA synthetases: genes and syndromes. Int. J. Cell Biol. 2014, 787956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Musante L., Püttmann L., Kahrizi K., Garshasbi M., Hu H., Stehr H., Lipkowitz B., Otto S., Jensen L. R., Tzschach A., Jamali P., Wienker T., Najmabadi H., Ropers H. H., and Kuss A. W. (2017) Mutations of the aminoacyl-tRNA synthetases SARS and WARS2 are implicated in the etiology of autosomal recessive intellectual disability. Hum. Mutat. 38, 621–636 10.1002/humu.23205 [DOI] [PubMed] [Google Scholar]
- 36. Theisen B. E., Rumyantseva A., Cohen J. S., Alcaraz W. A., Shinde D. N., Tang S., Srivastava S., Pevsner J., Trifunovic A., and Fatemi A. (2017) Deficiency of WARS2, encoding mitochondrial tryptophanyl tRNA synthetase, causes severe infantile onset leukoencephalopathy. Am. J. Med. Genet. A 173, 2505–2510 10.1002/ajmg.a.38339 [DOI] [PubMed] [Google Scholar]
- 37. Lake N. J., Webb B. D., Stroud D. A., Richman T. R., Ruzzenente B., Compton A. G., Mountford H. S., Pulman J., Zangarelli C., Rio M., Boddaert N., Assouline Z., Sherpa M. D., Schadt E. E., Houten S. M., et al. (2017) Biallelic mutations in MRPS34 lead to instability of the small mitoribosomal subunit and Leigh syndrome. Am. J. Hum. Genet. 101, 239–254 10.1016/j.ajhg.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rorbach J., Gao F., Powell C. A., D'Souza A., Lightowlers R. N., Minczuk M., and Chrzanowska-Lightowlers Z. M. (2016) Human mitochondrial ribosomes can switch their structural RNA composition. Proc. Natl. Acad. Sci. U.S.A. 113, 12198–12201 10.1073/pnas.1609338113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Friederich M. W., Erdogan A. J., Coughlin C. R. 2nd, Elos M. T., Jiang H., O'Rourke C. P., Lovell M. A., Wartchow E., Gowan K., Chatfield K. C., Chick W. S., Spector E. B., Van Hove J. L. K., and Riemer J. (2017) Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Hum. Mol. Genet. 26, 702–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sánchez-Caballero L., Guerrero-Castillo S., and Nijtmans L. (2016) Unraveling the complexity of mitochondrial complex I assembly: a dynamic process. Biochim. Biophys. Acta 1857, 980–990 10.1016/j.bbabio.2016.03.031 [DOI] [PubMed] [Google Scholar]
- 41. Stroud D. A., Surgenor E. E., Formosa L. E., Reljic B., Frazier A. E., Dibley M. G., Osellame L. D., Stait T., Beilharz T. H., Thorburn D. R., Salim A., and Ryan M. T. (2016) Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126 10.1038/nature19754 [DOI] [PubMed] [Google Scholar]
- 42. Miller C., Wang L., Ostergaard E., Dan P., and Saada A. (2011) The interplay between SUCLA2, SUCLG2, and mitochondrial DNA depletion. Biochim. Biophys. Acta 1812, 625–629 10.1016/j.bbadis.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 43. Brzezniak L. K., Bijata M., Szczesny R. J., and Stepien P. P. (2011) Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 8, 616–626 10.4161/rna.8.4.15393 [DOI] [PubMed] [Google Scholar]
- 44. Patton J. R., Bykhovskaya Y., Mengesha E., Bertolotto C., and Fischel-Ghodsian N. (2005) Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 280, 19823–19828 10.1074/jbc.M500216200 [DOI] [PubMed] [Google Scholar]
- 45. Yarham J. W., Lamichhane T. N., Pyle A., Mattijssen S., Baruffini E., Bruni F., Donnini C., Vassilev A., He L., Blakely E. L., Griffin H., Santibanez-Koref M., Bindoff L. A., Ferrero I., Chinnery P. F., et al. (2014) Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 10, e1004424 10.1371/journal.pgen.1004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sasarman F., Thiffault I., Weraarpachai W., Salomon S., Maftei C., Gauthier J., Ellazam B., Webb N., Antonicka H., Janer A., Brunel-Guitton C., Elpeleg O., Mitchell G., and Shoubridge E. A. (2015) The 3′ addition of CCA to mitochondrial tRNASer(AGY) is specifically impaired in patients with mutations in the tRNA nucleotidyl transferase TRNT1. Hum. Mol. Genet. 24, 2841–2847 10.1093/hmg/ddv044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stehling O., Wilbrecht C., and Lill R. (2014) Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 100, 61–77 10.1016/j.biochi.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 48. Lu Y. W., and Claypool S. M. (2015) Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front. Genet. 6, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nunnari J., and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McKenzie M., Lazarou M., Thorburn D. R., and Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361, 462–469 10.1016/j.jmb.2006.06.057 [DOI] [PubMed] [Google Scholar]
- 51. Cooper H. M., Yang Y., Ylikallio E., Khairullin R., Woldegebriel R., Lin K. L., Euro L., Palin E., Wolf A., Trokovic R., Isohanni P., Kaakkola S., Auranen M., Lönnqvist T., Wanrooij S., and Tyynismaa H. (2017) ATPase-deficient mitochondrial inner membrane protein ATAD3A disturbs mitochondrial dynamics in dominant hereditary spastic paraplegia. Hum. Mol. Genet. 26, 1432–1443 10.1093/hmg/ddx042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Desai R., Frazier A. E., Durigon R., Patel H., Jones A. W., Dalla Rosa I., Lake N. J., Compton A. G., Mountford H. S., Tucker E. J., Mitchell A. L. R., Jackson D., Sesay A., Di Re M., van den Heuvel L. P., et al. (2017) ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain 140, 1595–1610 10.1093/brain/awx094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harel T., Yoon W. H., Garone C., Gu S., Coban-Akdemir Z., Eldomery M. K., Posey J. E., Jhangiani S. N., Rosenfeld J. A., Cho M. T., Fox S., Withers M., Brooks S. M., Chiang T., Duraine L., et al. (2016) Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am. J. Hum. Genet. 99, 831–845 10.1016/j.ajhg.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sperl W., Fleuren L., Freisinger P., Haack T. B., Ribes A., Feichtinger R. G., Rodenburg R. J., Zimmermann F. A., Koch J., Rivera I., Prokisch H., Smeitink J. A., and Mayr J. A. (2015) The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis. 38, 391–403 10.1007/s10545-014-9787-3 [DOI] [PubMed] [Google Scholar]
- 55. Touw C. M., Derks T. G., Bakker B. M., Groen A. K., Smit G. P., and Reijngoud D. J. (2014) From genome to phenome–simple inborn errors of metabolism as complex traits. Biochim. Biophys. Acta 1842, 2021–2029 10.1016/j.bbadis.2014.05.032 [DOI] [PubMed] [Google Scholar]
- 56. Kang Y., Stroud D. A., Baker M. J., De Souza D. P., Frazier A. E., Liem M., Tull D., Mathivanan S., McConville M. J., Thorburn D. R., Ryan M. T., and Stojanovski D. (2017) Sengers syndrome-associated mitochondrial acylglycerol kinase is a subunit of the human TIM22 protein import complex. Mol. Cell 67, 457–470 10.1016/j.molcel.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 57. Vukotic M., Nolte H., König T., Saita S., Ananjew M., Krüger M., Tatsuta T., and Langer T. (2017) Acylglycerol kinase mutated in Sengers syndrome is a subunit of the TIM22 protein translocase in mitochondria. Mol. Cell 67, 471–483.e7 10.1016/j.molcel.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 58. Kaukonen J., Juselius J. K., Tiranti V., Kyttälä A., Zeviani M., Comi G. P., Keränen S., Peltonen L., and Suomalainen A. (2000) Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289, 782–785 10.1126/science.289.5480.782 [DOI] [PubMed] [Google Scholar]
- 59. Richter-Dennerlein R., Korwitz A., Haag M., Tatsuta T., Dargazanli S., Baker M., Decker T., Lamkemeyer T., Rugarli E. I., and Langer T. (2014) DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab. 20, 158–171 10.1016/j.cmet.2014.04.016 [DOI] [PubMed] [Google Scholar]
- 60. Heimer G., Kerätär J. M., Riley L. G., Balasubramaniam S., Eyal E., Pietikäinen L. P., Hiltunen J. K., Marek-Yagel D., Hamada J., Gregory A., Rogers C., Hogarth P., Nance M. A., Shalva N., Veber A., et al. (2016) MECR mutations cause childhood-onset dystonia and optic atrophy, a mitochondrial fatty acid synthesis disorder. Am. J. Hum. Genet. 99, 1229–1244 10.1016/j.ajhg.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Janer A., Prudent J., Paupe V., Fahiminiya S., Majewski J., Sgarioto N., Des Rosiers C., Forest A., Lin Z. Y., Gingras A. C., Mitchell G., McBride H. M., and Shoubridge E. A. (2016) SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med. 8, 1019–1038 10.15252/emmm.201506159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wollweber F., von der Malsburg K., and van der Laan M. (2017) Mitochondrial contact site and cristae organizing system: a central player in membrane shaping and crosstalk. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1481–1489 10.1016/j.bbamcr.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 63. Strauss M., Hofhaus G., Schröder R. R., and Kühlbrandt W. (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 10.1038/emboj.2008.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jonckheere A. I., Smeitink J. A., and Rodenburg R. J. (2012) Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit. Metab. Dis. 35, 211–225 10.1007/s10545-011-9382-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., Tiveron C., Levitt M. D., Prelle A., Fagiolari G., Rimoldi M., and Zeviani M. (2009) Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 15, 200–205 10.1038/nm.1907 [DOI] [PubMed] [Google Scholar]
- 66. Guarani V., Paulo J., Zhai B., Huttlin E. L., Gygi S. P., and Harper J. W. (2014) TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex. Mol. Cell Biol. 34, 847–861 10.1128/MCB.01551-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Floyd B. J., Wilkerson E. M., Veling M. T., Minogue C. E., Xia C., Beebe E. T., Wrobel R. L., Cho H., Kremer L. S., Alston C. L., Gromek K. A., Dolan B. K., Ulbrich A., Stefely J. A., Bohl S. L., et al. (2016) Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell 63, 621–632 10.1016/j.molcel.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Arroyo J. D., Jourdain A. A., Calvo S. E., Ballarano C. A., Doench J. G., Root D. E., and Mootha V. K. (2016) A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 24, 875–885 10.1016/j.cmet.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lake N. J., Compton A. G., Rahman S., and Thorburn D. R. (2016) Leigh syndrome: one disorder, more than 75 monogenic causes. Ann. Neurol. 79, 190–203 10.1002/ana.24551 [DOI] [PubMed] [Google Scholar]
- 70. Nesbitt V., Pitceathly R. D., Turnbull D. M., Taylor R. W., Sweeney M. G., Mudanohwo E. E., Rahman S., Hanna M. G., and McFarland R. (2013) The UK MRC mitochondrial disease patient cohort study: clinical phenotypes associated with the m.3243A>G mutation–implications for diagnosis and management. J. Neurol. Neurosurg. Psychiatry 84, 936–938 10.1136/jnnp-2012-303528 [DOI] [PubMed] [Google Scholar]
- 71. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., and Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rossignol R., Letellier T., Malgat M., Rocher C., and Mazat J. P. (2000) Tissue variation in the control of oxidative phosphorylation: implication for mitochondrial diseases. Biochem. J. 347, 45–53 10.1042/0264-6021:3470045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smeitink J. A., Zeviani M., Turnbull D. M., and Jacobs H. T. (2006) Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 3, 9–13 10.1016/j.cmet.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 74. Wallace D. C. (2001) Mouse models for mitochondrial disease. Am. J. Med. Genet. 106, 71–93 10.1002/ajmg.1393 [DOI] [PubMed] [Google Scholar]
- 75. Mergenthaler P., Lindauer U., Dienel G. A., and Meisel A. (2013) Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci. 36, 587–597 10.1016/j.tins.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Quirós P. M., Mottis A., and Auwerx J. (2016) Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- 77. Chugani H. T., Phelps M. E., and Mazziotta J. C. (1987) Positron emission tomography study of human brain functional development. Ann. Neurol. 22, 487–497 10.1002/ana.410220408 [DOI] [PubMed] [Google Scholar]
- 78. Martínez-Reyes I., Diebold L. P., Kong H., Schieber M., Huang H., Hensley C. T., Mehta M. M., Wang T., Santos J. H., Woychik R., Dufour E., Spelbrink J. N., Weinberg S. E., Zhao Y., DeBerardinis R. J., and Chandel N. S. (2016) TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell 61, 199–209 10.1016/j.molcel.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bohovych I., and Khalimonchuk O. (2016) Sending out an SOS: mitochondria as a signaling hub. Front. Cell Dev. Biol. 4, 109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sun N., Youle R. J., and Finkel T. (2016) The mitochondrial basis of aging. Mol. Cell 61, 654–666 10.1016/j.molcel.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mattiazzi M., Vijayvergiya C., Gajewski C. D., DeVivo D. C., Lenaz G., Wiedmann M., and Manfredi G. (2004) The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum. Mol. Genet. 13, 869–879 10.1093/hmg/ddh103 [DOI] [PubMed] [Google Scholar]
- 82. Geromel V., Kadhom N., Cebalos-Picot I., Ouari O., Polidori A., Munnich A., Rötig A., and Rustin P. (2001) Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum. Mol. Genet. 10, 1221–1228 10.1093/hmg/10.11.1221 [DOI] [PubMed] [Google Scholar]
- 83. Mehta M. M., Weinberg S. E., and Chandel N. S. (2017) Mitochondrial control of immunity: beyond ATP. Nat. Rev. Immunol. 17, 608–620 10.1038/nri.2017.66 [DOI] [PubMed] [Google Scholar]
- 84. Valsecchi F., Monge C., Forkink M., de Groof A. J., Benard G., Rossignol R., Swarts H. G., van Emst-de Vries S. E., Rodenburg R. J., Calvaruso M. A., Nijtmans L. G., Heeman B., Roestenberg P., Wieringa B., Smeitink J. A., et al. (2012) Metabolic consequences of NDUFS4 gene deletion in immortalized mouse embryonic fibroblasts. Biochim. Biophys. Acta 1817, 1925–1936 10.1016/j.bbabio.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 85. Holzerova E., Danhauser K., Haack T. B., Kremer L. S., Melcher M., Ingold I., Kobayashi S., Terrile C., Wolf P., Schaper J., Mayatepek E., Baertling F., Friedmann Angeli J. P., Conrad M., Strom T. M., et al. (2016) Human thioredoxin 2 deficiency impairs mitochondrial redox homeostasis and causes early-onset neurodegeneration. Brain 139, 346–354 10.1093/brain/awv350 [DOI] [PubMed] [Google Scholar]
- 86. Nagana Gowda G. A., Abell L., Lee C. F., Tian R., and Raftery D. (2016) Simultaneous analysis of major coenzymes of cellular redox reactions and energy using ex vivo 1H NMR spectroscopy. Anal. Chem. 88, 4817–4824 10.1021/acs.analchem.6b00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ke B. X., Pepe S., Grubb D. R., Komen J. C., Laskowski A., Rodda F. A., Hardman B. M., Pitt J. J., Ryan M. T., Lazarou M., Koleff J., Cheung M. M., Smolich J. J., and Thorburn D. R. (2012) Tissue-specific splicing of an Ndufs6 gene-trap insertion generates a mitochondrial complex I deficiency-specific cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 109, 6165–6170 10.1073/pnas.1113987109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tyynismaa H., Carroll C. J., Raimundo N., Ahola-Erkkilä S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjøsund K. E., Tulkki V., Oresic M., Moraes C. T., Pietiläinen K., Hovatta I., and Suomalainen A. (2010) Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 19, 3948–3958 10.1093/hmg/ddq310 [DOI] [PubMed] [Google Scholar]
- 89. Bao X. R., Ong S. E., Goldberger O., Peng J., Sharma R., Thompson D. A., Vafai S. B., Cox A. G., Marutani E., Ichinose F., Goessling W., Regev A., Carr S. A., Clish C. B., and Mootha V. K. (2016) Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 5, e10575 10.7554/eLife.10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang Z., Tsukikawa M., Peng M., Polyak E., Nakamaru-Ogiso E., Ostrovsky J., McCormack S., Place E., Clarke C., Reiner G., McCormick E., Rappaport E., Haas R., Baur J. A., and Falk M. J. (2013) Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PLoS ONE 8, e69282 10.1371/journal.pone.0069282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ramoni R. B., Mulvihill J. J., Adams D. R., Allard P., Ashley E. A., Bernstein J. A., Gahl W. A., Hamid R., Loscalzo J., McCray A. T., Shashi V., Tifft C. J., Undiagnosed Diseases Network, and Wise A. L. (2017) The undiagnosed diseases network: accelerating discovery about health and disease. Am. J. Hum. Genet. 100, 185–192 10.1016/j.ajhg.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cummings B. B., Marshall J. L., Tukiainen T., Lek M., Donkervoort S., Foley A. R., Bolduc V., Waddell L. B., Sandaradura S. A., O'Grady G. L., Estrella E., Reddy H. M., Zhao F., Weisburd B., Karczewski K. J., et al. (2017) Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 9, eaal5209 10.1126/scitranslmed.aal5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kremer L. S., Bader D. M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T. B., Graf E., Schwarzmayr T., Terrile C., Koňaříková E., Repp B., Kastenmüller G., Adamski J., Lichtner P., et al. (2017) Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 8, 15824 10.1038/ncomms15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Haack T. B., Klee D., Strom T. M., Mayatepek E., Meitinger T., Prokisch H., and Distelmaier F. (2014) Infantile Leigh-like syndrome caused by SLC19A3 mutations is a treatable disease. Brain 137, e295 10.1093/brain/awu128 [DOI] [PubMed] [Google Scholar]
- 95. Pfeffer G., Majamaa K., Turnbull D. M., Thorburn D., and Chinnery P. F. (2012) Treatment for mitochondrial disorders. Cochrane Database Syst. Rev. CD004426 10.1002/14651858.CD004426.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nightingale H., Pfeffer G., Bargiela D., Horvath R., and Chinnery P. F. (2016) Emerging therapies for mitochondrial disorders. Brain 139, 1633–1648 10.1093/brain/aww081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Koopman W. J., Beyrath J., Fung C. W., Koene S., Rodenburg R. J., Willems P. H., and Smeitink J. A. (2016) Mitochondrial disorders in children: toward development of small-molecule treatment strategies. EMBO Mol. Med. 8, 311–327 10.15252/emmm.201506131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Donti T. R., Stromberger C., Ge M., Eldin K. W., Craigen W. J., and Graham B. H. (2014) Screen for abnormal mitochondrial phenotypes in mouse embryonic stem cells identifies a model for succinyl-CoA ligase deficiency and mtDNA depletion. Dis. Model. Mech. 7, 271–280 10.1242/dmm.013466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Besse A., Wu P., Bruni F., Donti T., Graham B. H., Craigen W. J., McFarland R., Moretti P., Lalani S., Scott K. L., Taylor R. W., and Bonnen P. E. (2015) The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab. 21, 417–427 10.1016/j.cmet.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Deutschmann A. J., Amberger A., Zavadil C., Steinbeisser H., Mayr J. A., Feichtinger R. G., Oerum S., Yue W. W., and Zschocke J. (2014) Mutation or knock-down of 17β-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum. Mol. Genet. 23, 3618–3628 10.1093/hmg/ddu072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.