Abstract
Besides intrinsic changes, malignant cells also release soluble signals that reshape their microenvironment. Among these signals is WNT1-inducible signaling pathway protein 1 (WISP1), a secreted matricellular protein whose expression is elevated in several cancers, including melanoma, and is associated with reduced survival of patients diagnosed with primary melanoma. Here, we found that WISP1 knockout increases cell proliferation and represses wound healing, migration, and invasion of mouse and human melanoma cells in multiple in vitro assays. Metastasis assays revealed that WISP1 knockout represses tumor metastasis of B16F10 and YUMM1.7 melanoma cells in both C57BL/6Ncrl and NOD-scid IL2Rγnull (NSG) mice. WT B16F10 cells having an invasion phenotype in a transwell assay possessed a gene expression signature similar to that observed in the epithelial–mesenchymal transition (EMT), including E-cadherin repression and fibronectin and N-cadherin induction. Upon WISP1 knockout, expression of these EMT signature genes went in the opposite direction in both mouse and human cell lines, and EMT-associated gene expression was restored upon exposure to media containing WISP1 or to recombinant WISP1 protein. In vivo, Wisp1 knockout–associated metastasis repression was reversed by the reintroduction of either WISP1 or snail family transcriptional repressor 1 (SNAI1). Experiments testing EMT gene activation and inhibition with recombinant WISP1 or kinase inhibitors in B16F10 and YUMM1.7 cells suggested that WISP1 activates AKT Ser/Thr kinase and that MEK/ERK signaling pathways shift melanoma cells from proliferation to invasion. Our results indicate that WISP1 present within the tumor microenvironment stimulates melanoma invasion and metastasis by promoting an EMT-like process.
Keywords: melanoma, metastasis, invasion, epithelial-mesenchymal transition (EMT), tumor microenvironment, cancer, Wnt signaling, AKT signaling, secreted protein, WNT1-inducible signaling pathway protein 1 (WISP1/CCN4)
Introduction
Tumor metastasis is a multistep cascade that starts with local invasion into the surrounding tissue and culminates in colonizing distant tissues (1, 2). Classically, melanoma is thought to progress linearly, whereby the growth of the primary tumor progressively increases the propensity for metastasis (3). However, 4–12% of patients with metastatic melanoma do not have a clinically identifiable primary tumor, and the excised primary melanoma can still recur at different sites in the body as metastatic lesions (4). Observed early dissemination and metastasis of melanoma suggest a more complex, parallel progression model of metastasis in melanoma (4). The basis for this parallel progression model is attributed to reversible phenotype switching of melanoma between proliferative and invasive phenotypes and the resulting intratumoral heterogeneity, driven by oncogenic signaling and environmental cues (5, 6).
The switch in malignant melanocytes between proliferative and invasive phenotypes resembles the epithelial–mesenchymal transition (EMT),2 a key process of phenotypic change that is associated with the metastatic progression of epithelial cancers through the control of EMT-inducing transcription factors (EMT-TFs) such as SNAI1/2, ZEB1/2, and TWIST (5, 6). Whereas the specific EMT-TFs that control the phenotypic state depend on cellular context (7–9), this core network is regulated by various signaling pathways that integrate information from environmental cues, including TGF-β, fibroblast growth factor, epidermal growth factor, hepatocyte growth factor, NF-κB, Wnt/β-catenin, and Notch pathways (4–6, 10). Of these signaling pathways, genetically engineered mouse models and samples from melanoma patients provide strong evidence for the essential role of the Wnt/β-catenin pathway for melanoma development, phenotype switching/EMT, metastasis, and drug resistance (4, 11, 12). Whereas it is generally accepted that altered β-catenin signaling is critical for melanoma initiation and proliferation, conflicting roles of β-catenin have been reported for melanoma metastasis (4, 11). Using BrafV600E/Pten−/− and BrafV600E/Pten−/−/CAT-STA mice as melanoma models (13, 14), Damsky et al. (14) found that β-catenin activation substantially increased melanoma lung metastasis, and Spranger et al. (15) revealed that melanoma-intrinsic active Wnt/β-catenin signaling prevented anti-tumor immunity via T-cell exclusion, thus facilitating tumor progression and metastasis. On the other hand, using the BrafV600E/Cdk2a−/−/Pten−/− mouse-derived YUMM1.7 melanoma cell line, Kaur et al. discovered that a fibroblast-secreted Wnt antagonist, sFRP2, increased tumor metastasis by repressing β-catenin activity and the expression of MITF, the melanoma differentiation marker microphthalmia-associated transcription factor (16).
Propagation of environmental cues initiated by aberrant signaling within malignant cells, like β-catenin, to reshape the tissue microenvironment is important yet poorly understood (17). Interestingly, activated nuclear β-catenin directly promotes the transcription of a variety of Wnt/β-catenin signaling effectors, including WNT1-inducible signaling pathway protein 1 (WISP1/CCN4) (18–20). WISP1/CCN4 is a secreted matricellular protein that belongs to the CCN family (originally abbreviated from the first three members CYR61/CCN1, CTGF/CCN2, and NOV/CCN3 and recently officially renamed as cellular communication network factors) (21). Except for WISP2, all CCN proteins contain a short N-terminal signal peptide, followed by four conserved structural domains (IGFBP, VWC, TSP, and CT) to mediate their interactions with extracellular proteins and cell surface receptors (22). As matricellular proteins, CCNs do not interact with specific membrane receptors; rather, they bind multiligand receptors, primarily integrins, to regulate the intracellular signaling (22, 23). The canonical and noncanonical integrin signaling from CCNs mediate a variety of downstream events, depending on the specific cellular context (23, 24).
Depending on context, WISP1 activates a variety of downstream signaling, including focal adhesion kinase, RAS/RAF/MEK/ERK, NF-κB, TGF-β, and PI3K/AKT pathways (25–37). Functionally, WISP1-initiated signals regulate various biological processes, including cell adhesion, proliferation, differentiation, survival, motility, and wound healing/tissue repair (38, 39). Compared with CCN1–3, the components and steps of WISP1 signaling are less characterized, but putative integrin recognition sites exist within VWC, TSP, and CT domains (22). In vitro binding assays and functional assays with integrin-blocking antibodies implicated that α5β1, αvβ3, and αvβ5 were involved in WISP1 signaling, and these integrins were essential for WISP1-induced activation of focal adhesion kinase, Rac, RAS/RAF/MEK/ERK, JNK, or NF-κB pathways in epithelial cells, fibroblasts, bone marrow stromal cells, or cancer cells (26, 30, 31, 33–35, 37).
In humans, elevated WISP1 expression correlates with poor prognosis in the majority of cancers studied, and WISP1 promotes tumor cell proliferation, survival, migration/invasion, and tumor metastasis in a variety of malignant tumors, such as brain, breast, colorectal, lung, pancreatic, and prostate cancers (38, 39). For its role in tumor cell dissemination, WISP1 was shown to induce EMT to promote cell migration and invasion in lung epithelial, gastric cancer, and breast cancer cells (40–43). In human glioblastoma, the WISP1-activated MEK/ERK pathway might be responsible for the EMT of the tumor cells (44). The activation of various signaling, including PI3K/AKT, MEK/ERK, NF-κB, or JNK/p38 pathways, has been shown to be essential for WISP1-induced cell migration and/or invasion in vascular smooth muscle cells, cholangiocarcinoma, chondrosarcoma, oral squamous cell carcinoma, osteosarcoma, and colorectal cancer cells (30, 33, 34, 45–48).
Despite the reports in other cancers, the role of WISP1 in melanoma appears to be contradicted, and an intracellular signaling basis for these observations remains unclear (18, 49–51). Recently, we showed that WISP1 from melanoma cells contributed to tumor immunosuppression (52) and that WISP1 expression correlated with tumor cell invasion in both melanoma and breast cancer (52, 53). Furthermore, disrupting adherens junctions induced the synthesis and release of WISP1 via noncanonical activation of β-catenin (54). Taken together, these findings led us to investigate whether WISP1 is a paracrine effector of Wnt/β-catenin signaling that coordinates EMT/phenotype switching and metastasis, as described below.
Results
WISP1 expression is increased in primary melanoma and is associated with reduced overall survival of patients diagnosed with primary melanoma
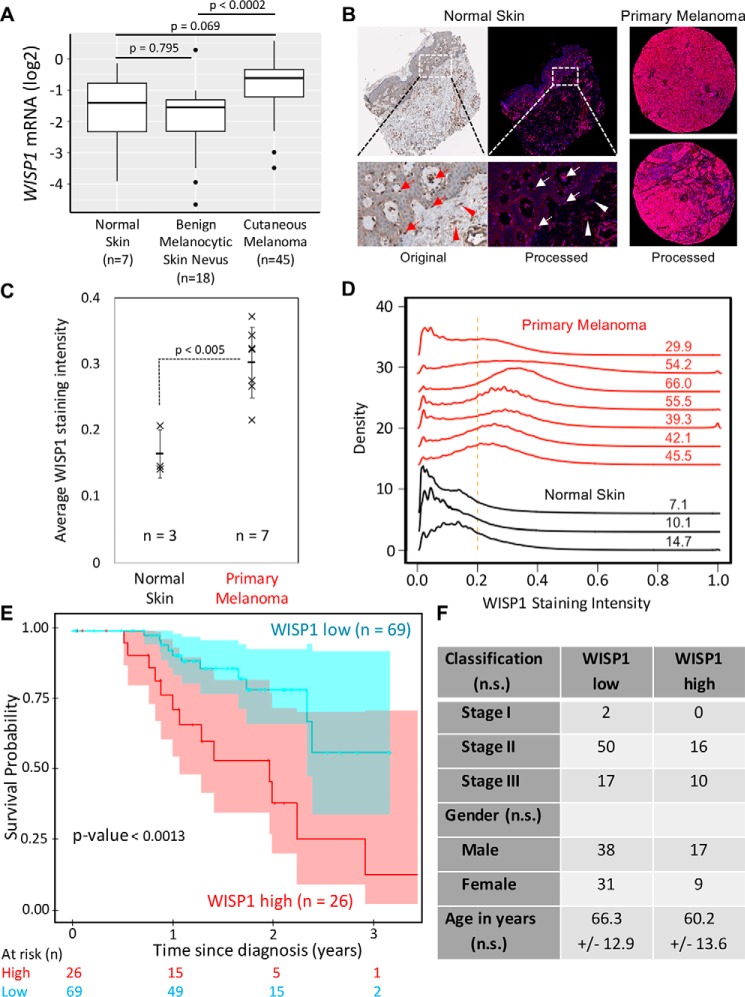
To ground our study clinically, we first reviewed public databases for gene expression profiles in human primary melanoma samples. Analysis of a study comparing primary melanoma and skin nevi (55) showed that the WISP1 mRNA level was almost doubled in primary melanoma samples as compared with benign melanocytic skin nevi (p < 0.0002; Fig. 1A). Expression of WISP1 mRNA was not significantly different in benign melanocytic skin relative to normal skin (p = 0.795). At the protein level, an independent tissue microarray containing samples from normal epithelial tissue (n = 3) and primary melanoma (n = 7) tissue were used to characterize WISP1 expression. Signal deconvolution and image segmentation were used to quantify differences in WISP1 staining in melanocytes and other cells present among architectural features of the skin. In both primary melanoma and normal skin, the protein is located in the cytoplasm (Fig. 1B). In melanoma samples, almost all tumor cells (>75%) exhibited medium to high WISP1 intensity (Fig. 1B, right), whereas in normal skin, there was little or no WISP1 in epidermal keratinocytes but medium WISP1 staining in melanocytes within both basal layer of epidermis and hair follicles (Fig. 1B, left). Medium WISP1 expression was observed in the fibroblasts in skin dermis (stroma) as well (Fig. 1B, left). The average intensity of WISP1 staining within a tissue sample suggested that an increase in WISP1 also correlates with oncogenic transformation (Fig. 1C; p < 0.005). Whereas an increase in average intensity could be explained by a change in cellular composition of the tissue sample, a quantitative analysis of the intensity of WISP1 staining suggests that more of the tissue area stains positive for WISP1, which suggests that more WISP1-producing cells are present, and the staining intensity is greater in primary melanoma than in normal skin, which suggests that WISP1-positive cells are producing more WISP1 (Fig. 1D). The results from this quantitative IHC analysis are consistent with the mRNA data presented in Fig. 1A such that WISP1 expression was increased in primary melanoma compared with benign skin conditions.
Figure 1.
WISP1 expression is increased in melanoma and is associated with reduced overall survival of patients diagnosed with primary melanoma. A, WISP1 mRNA expression in benign skin conditions (normal skin and benign melanocytic skin nevus) compared with primary melanoma. The original expression data set (GSE3189) was deposited by Talantov et al. (55). p values were calculated using analysis of variance with the post hoc Tukey honest significant difference test. B, representative original and deconvoluted color images derived from human normal skin and melanoma tissue microarray probed using a WISP1 antibody (HPA007121) and imaged using 3,3′-diaminobenzidine and stained using hematoxylin for normal skin (left) and two melanoma (right) tissue samples. Original tissue microarray images were obtained from www.proteinatlas.org3 (77). Deconvoluted intensity of WISP1 staining is shown in red, whereas the cellular structures stained using hematoxylin are shown in blue. Arrows, melanocytes in epidermis; arrowheads, fibroblasts in dermis (stroma). C, average WISP1 staining within normal skin and primary melanoma tissue samples. D, distributions in nonzero pixel intensity values of WISP1 staining for normal skin (black curves) and primary melanoma (red curves) tissue samples. Numbers, percentage of the distribution that has pixel intensity values greater than a normalized pixel intensity of 0.2. E, Kaplan–Meier estimate of overall survival of melanoma patients stratified by WISP1 transcript abundance. The original data set was from the Cancer Genome Atlas. Sample numbers and p values calculated using the Peto and Peto modification of the Gehan–Wilcoxon test are indicated. F, patient population characteristics of WISP1 high and WISP1 low groups. Statistical differences among categorical data and age were assessed using Fisher's exact test and Student's t test, respectively (n.s., p > 0.05).
As the IHC and gene expression analyses suggest that malignant transformation of melanocytes is associated with an increase in WISP1 production, we explored genetic mutations, including both coding sequence changes and changes in copy number via structural alterations, that are enriched in melanoma. As WISP1 expression can be induced by noncanonical β-catenin signaling resulting from dynamic turnover of adherens junctions (54), we found that mutations associated with melanoma suggest enhanced malignant cell production of WISP1 (Table S1). Whereas mutations in BRAF, NRAS, and CDKN2A are highly prevalent in melanoma, mutations also frequently occur in PTEN, TP53, MITF, KIT, CTNNB1, and WISP1. Notably, the mutation rate (8%) for WISP1 in melanoma is equal to or higher than those for CTNNB1, MITF, and KIT, which are considered as promising therapeutic targets (4, 56, 57). Mutations impacting either WISP1 or CTNNB1 comprised 13% of the samples. We also noted that mutations in WISP1 were mainly copy number amplifications (q = 2.9E−5) and in CTNNB1 were mainly single-nucleotide variants associated with exon 3. Mutations in exon 3 of CTNNB1, which inhibits proteasomal degradation, and copy number amplifications of WISP1 both favor increased transcriptional response to dynamic turnover of adherens junctions.
To assess the clinical implications of WISP1 overexpression, overall survival of patients diagnosed with primary melanoma stratified by WISP1 expression was estimated using RNA-Seq data obtained from 95 patient samples from the TCGA with corresponding survival data. Kaplan–Meier analysis was performed by separating the population into two groups based upon a WISP1 expression cutoff of 1.0 FPKM (WISP1 low n = 69, WISP1 high n = 26) (Fig. 1, E and F). Whereas the staging, age, and gender profiles of these two groups are not statistically significantly different, the WISP1 high group patients have a lower 3-year survival rate of only 14% compared with a rate of 58% in the WISP1 low group (p < 0.0013). The median survival time is about 44 months for WISP1 low group patients but only 24 months for WISP1 high group patients (Fig. 1E). In addition, a multivariate Cox proportional hazards regression analysis using WISP1 classification (high versus low), tumor stage, and gender as potential covariates of overall survival as the outcome variable indicated that WISP1 classification was the only covariate with a significant association with overall survival (likelihood ratio test p = 0.0061), such that a low value of WISP1 (FPKM < 1) reduces the risk of death by a factor of 0.287 (95% confidence interval: 0.1279–0.6442). Collectively, these results suggested that WISP1 expression was elevated in human melanomas and that WISP1 may potentially serve as a biomarker for worse patient prognosis and survival. However, as other genes are co-amplified in conjunction with WISP1, we decided to explore the functional impact of WISP1 on melanoma cells to identify a mechanistic underpinning for this difference in patient survival.
WISP1 knockout in mouse/human melanoma inhibited tumor cell migration and invasion
Using B16 mouse melanoma cell models, we previously reported a role for WISP1 in immunosuppression and its synthesis and secretion following β-catenin release from adherens junctions (52, 54). Following from these studies, ELISA revealed that WISP1 was secreted into media conditioned in 2D culture by nonmetastatic B16F0 cells (605 ± 15 pg/ml) and metastatic B16F10 cells (1300 ± 35 pg/ml) as well as immortalized melanocyte Melan-A cells (1018 ± 32 pg/ml) (Table S2). In comparison, the mouse fibroblast cell line NIH3T3 expressed lower WISP1 (83 ± 2.2 pg/ml), whereas WISP1 was almost undetectable (<20 pg/ml) in media conditioned by another two mouse tumor cells: Lewis lung carcinoma LLC1 cells and breast cancer E0771 cells (Table S2).
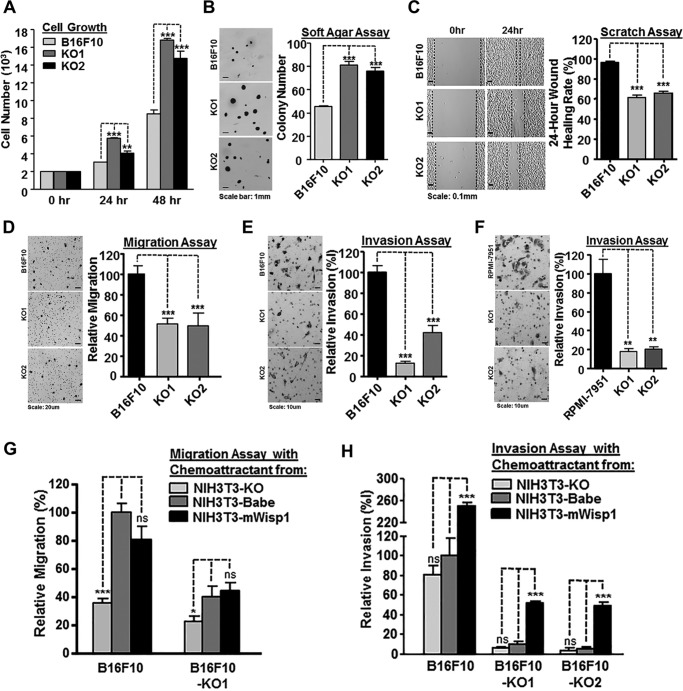
To investigate the roles of WISP1 in melanoma progression and metastasis, we knocked out the Wisp1 gene in metastatic B16F10 cells and evaluated the phenotype of the resulting cell lines using an ensemble of in vitro assays that capture aspects of metastasis including cell–matrix and cell–cell interactions, migration, and invasion. Using two CRISPR/Cas9 (double nickase) systems to target the Wisp1 gene at two different locations, we cloned two Wisp1-knockout cells from B16F10 (Table S2). In 2D culture, the knockout cells outgrew the parental cells by 96.7 ± 2.5% (F10-KO1) and 73.1 ± 9.5% (F10-KO2) in a 2-day period (Fig. 2A). In 3D culture, WT B16F10 and Wisp1-knockout cells were used to evaluate the effect of WISP1 on anchorage-independent growth (Fig. 2B). A soft agar assay showed that Wisp1 knockout in B16F10 cells increased its colony formation by 78.7 ± 6.7% (F10-KO1) and 66.9 ± 7.0% (F10-KO2), respectively. In vivo, Wisp1 knockout did not affect subcutaneous growth of tumors in NSG mice (Fig. S6A). In addition, a wound healing assay was used to test the effect of Wisp1 knockout on tissue repair in vitro. For B16F10 cells, the wound healing rate was reduced from 96.3 ± 0.7% (F10) to 61.4 ± 1.4% (F10-KO1) and 65.9 ± 1.0% (F10-KO2) (Fig. 2C). These in vitro results suggested that reducing WISP1 expression increased melanoma proliferation but repressed tumor cell migration, which was consistent with the previous reports that WISP1 repressed melanoma growth both in cell culture and in a mouse model (18, 50).
Figure 2.
WISP1 knockout in mouse and human melanoma cells inhibited tumor cell migration and invasion. A, 48-h 2D growth of mouse metastatic melanoma cell line B16F10 and two B16F10 Wisp1-knockout cells (-KO1 and -KO2). B, anchorage-independent growth assay of B16F10 and the two knockout cells in soft agar. Colonies were fixed and counted after 14 days. A representative staining image for each sample is shown on the left, and colony counts are plotted on the right. C, wound healing assay of B16F10 and the two knockout cells. Scratches were created on 6-well plates in biological triplicate, and the healing rate was calculated after 24 h. D, Boyden transwell migration assay of B16F10 and the two knockout cells. A representative staining image for each sample is shown on the left, and relative migration efficiency is graphed on the right. E, Boyden transwell invasion assay of B16F10 and the two knockout cells. F, Boyden transwell invasion assay of human metastatic melanoma cell line RPMI-7951 and its two WISP1-knockout cells (-KO1 and -KO2). G, transwell migration assay of B16F10 and its knockout cell (-KO1) using conditioned media with different concentrations of WISP1 as chemoattractant. B16F10 migrated cells with conditioned medium from NIH3T3-Babe were set up as 100% of relative migration efficiency and compared with other cells. H, transwell invasion assay of B16F10 and the two knockout cells using conditioned media with different concentrations of WISP1 as chemoattractant. B16F10 invaded cells with conditioned medium from NIH3T3-Babe were set up as 100% of relative invasion efficiency and compared with other cells. Statistical significance was determined by Student's t test, where p < 0.05 was considered significant, and asterisks were used to indicate calculated range in p values. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. Error bars, S.D.
We used Boyden chamber transwell assays to characterize the effects of WISP1 on both mouse and human melanoma cells. For B16F10, the migration rate of knockout cells was only 51.4 ± 3.2% (F10-KO1) and 49.3 ± 7.4% (F10-KO2) as compared with the parental cells (Fig. 2D), and the invasion rate was reduced even lower to 12.5 ± 1.4% (F10-KO1) and 41.7 ± 7.1% (F10-KO2), relative to the parental cells (Fig. 2E). Among several human melanoma lines we obtained, including RPMI-7951, SH-4, SK-MEL-3, and SK-MEL-24, WISP1 was detected only in medium from RPMI-7951 cells (1331 ± 34 pg/ml) (Table S2). After we knocked out WISP1 in RPMI-7951 using similar CRISPR/Cas9 (double nickase) systems as described above (Table S2), we found that the invasion rate was repressed significantly to 18.1 ± 2.6% (RPMI-7951-KO1) and 20.6 ± 2.2% (RPMI-7951-KO2), as compared with the parental cells (Fig. 2F).
As illustrated by the tissue microarray IHC results, secretion of WISP1 may also come from tumor stromal cells, such as cancer-associated fibroblasts (38, 39, 58, 59). Interestingly, different from most other tumor cells that can use culture medium as a chemoattractant, melanoma cells need conditioned medium from mouse fibroblast NIH3T3 cells as a chemoattractant for migration and invasion in the in vitro transwell assays (Fig. 2, D–F). As WISP1 was also present in NIH3T3-conditioned media, we next asked whether paracrine WISP1 would affect melanoma cell behavior in our assay systems by creating three variants of NIH3T3 cells that had Wisp1 knocked out using a CRISPR/Cas9 construct, Wisp1 overexpressed using a retrovirus, and been transduced using a control retrovirus. The knockout cell NIH3T3-KO secreted no detectable WISP1, and the control cell NIH3T3-pBabe provided a similar concentration of WISP1 compared with parental cells (90.3 ± 2.8 pg/ml) (Table S2). The overexpressing cell line NIH3T3-mWisp1 produced about 10 times the concentration of mouse WISP1 protein (920 ± 15.6 pg/ml) relative to NIH3T3-pBabe, but its concentration was still lower than what we measured in B16F10 cells.
Using the three different conditioned media as chemoattractants, we evaluated the effect of varying levels of WISP1 below the transwell insert on the migration and invasion of B16F10, B16F10-KO1, and B16F10-KO2 cells (Fig. 2, G and H). Generally, existing in chemoattractants, WISP1 dose-dependently increased the migration and invasion of all three cell lines (Fig. 2, F and G). The presence of WISP1, rather than its concentration, appeared to be more important in tumor cell migration (Fig. 2G), whereas the high concentration of WISP1, from NIH3T3-mWisp1 cells, seemed to be more decisive in promoting tumor cell invasions no matter whether or not the melanoma cells expressed WISP1 by themselves (Fig. 2G). Collectively, WISP1 exhibited both autocrine and paracrine effects to stimulate the in vitro migration and invasion of melanoma cells. In addition, melanoma invasion, compared with its migration property, responded more drastically to the increased concentration of WISP1 in its microenvironment.
Wisp1 knockout repressed mouse melanoma metastasis in vivo
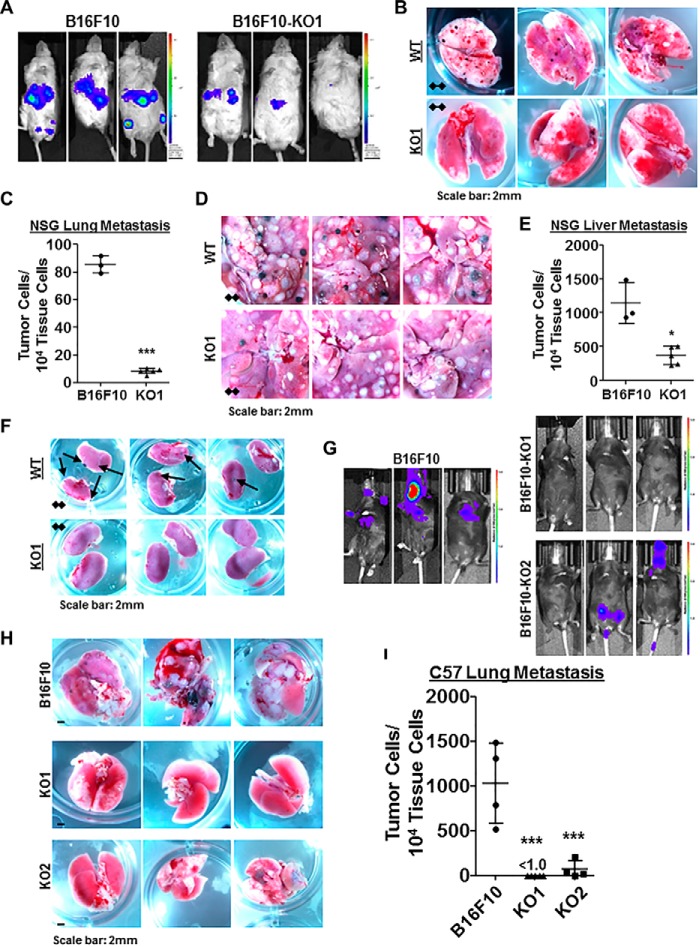
Given the effect of WISP1 on melanoma cell migration and invasion in vitro, we next tested the in vivo effect of WISP1 on mouse melanoma metastasis using an experimental metastasis assay that directly delivers B16F10 cells into the circulation through mouse tail vein injection. Before injection, all B16F10 and Wisp1-knockout cells were transduced with lentivirus expressing a codon-optimized luciferase reporter gene Luc2 to quantify tumor burden in vivo. A real-time quantitative PCR (qPCR) method was also developed to quantify the number of metastatic tumor cells (with inserted Luc2 gene) within defined mouse organs such as the lung and liver (60). This method enabled us to detect mouse organ tumor load for as low as one tumor cell within a total of 104 tissue cells (60).
To avoid a confounding influence of host immunity on the response to Wisp1 knockout (52), we used immunodeficient NSG mice for the first sets of experiments (Fig. 3 (A–F) and Figs. S1–S3). After tail vein injection, WT B16F10 cells disseminated widely and grew rapidly. Bioluminescence imaging showed that the main tumor burden was located in the abdomen (Fig. 3A, left), whereas the metastatic tumor signals at a similar location from B16F10-KO1 cells were much weaker (Fig. 3A, right). Upon dissection, each pair of lungs from B16F10-injected mice was covered with dozens of metastatic tumor colonies, compared with clear lungs from mice receiving B16F10-KO1 cells (Fig. 3B and Fig. S1). Real-time qPCR revealed that there was an average tumor burden of 85.3 ± 4.2 metastatic tumor cells among 104 lung cells for B16F10-injected mice, compared with an average tumor burden of 7.9 ± 1.1, a reduction of more than 90%, for lungs from B16F10-KO1–injected mice (Fig. 3C). However, lung metastases were a small portion of the overall tumor burden. The majority of B16F10 metastases were observed in the abdomen region, including livers (Fig. 3D and Fig. S2), intestines (not shown), and kidney (Fig. 3F and Fig. S3). In mice injected with B16F10-KO1 cells, metastatic lesions were either reduced in size and number (livers and intestines; Fig. 3D) or not observed (kidneys; Fig. 3F). Real-time qPCR revealed that the average tumor burden in livers from B16F10-injected mice was 1141 ± 136 metastatic tumor cells among 104 liver cells, whereas the average tumor burden was reduced to 370 ± 26, about a 70% repression, for livers from B16F10-KO1–injected mice (Fig. 3E).
Figure 3.
Wisp1 knockout repressed the experimental metastasis of melanoma cell line B16F10 in immunodeficient NSG mice and immunocompetent C57BL/6Ncrl mice. Experimental metastasis assays were performed in NSG mice (A–F) and C57BL/6Ncrl mice (G–I) using B16F10 and the indicated knockout cells with injection through mouse tail veins. Each group contained five duplicates (n = 5), and only mice surviving the whole experiments were analyzed at the same time for imaging, photography, and qPCR (final n ≥ 3). These experiments were repeated, and similar results were achieved. A, bioluminescence imaging performed 1 day before NSG mice were euthanized. All animals were compared with the same bioluminescence scale. B and C, tumor lung metastases (black colonies) of NSG mice as captured by photography (B) and real-time genomic qPCR (C). Quantitative tumor lung metastatic burden was assayed and presented as tumor cell number within 10,000 mouse tissue cells. D–E, tumor liver metastases (black and white nodules) of NSG mice as captured by photography (D) and real-time genomic qPCR (E). Quantitative tumor liver metastatic burden was assayed and presented as tumor cell number within 10,000 mouse tissue cells. F, tumor kidney metastases (black colonies) of NSG mice as captured by photography. G, bioluminescence imaging performed 1 day before C57BL/6Ncrl mice were euthanized. All animals were compared with the same bioluminescence scale. H–I, tumor lung metastases of C57BL/6Ncrl mice as captured by photography (H) and real-time genomic qPCR (I). Four high-resolution images for B, D, F, and H are provided as Figs. S1–S4. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.D.
We next used immunocompetent C57BL/6Ncrl mice for similar experimental metastasis assays (Fig. 3 (G–I) and Figs. S4–S6). After tail vein injection of WT B16F10 cells, bioluminescence imaging showed that tumor metastases developed in the chest region of all mice (Fig. 3G). In individual mice, signals derived from WT B16F10 cells were also observed in lymph nodes and brains (Fig. 3G, left). In mice injected with one of two Wisp1-knockout cells (B16F10-KO1 and -KO2), the metastatic tumor signals were consistently absent in the chest region, although individual mice did show signals originating from either the lower abdomen or head (Fig. 3G, right). Dissection confirmed that the majority of tumor metastases associated with WT B16F10 cells were in the lungs (Fig. 3H and Fig. S4). In mice injected with Wisp1-knockout cells, metastatic nodules were either completely absent (B16F10-KO1) or significantly reduced in size and number (B16F10-KO2) (Fig. 3H). Real-time qPCR calculated that the average tumor burden in lungs dropped from 1159 ± 349 metastatic tumor cells among 104 lung cells for B16F10-injected mice to less than 1.0 tumor cell (>99.9% reduction) and 57.8 ± 38.7 tumor cells for KO1- and KO2-injected mice, respectively (Fig. 3I). No metastatic nodules were observed on livers from any group of mice (Fig. S5A). Although qPCR detected a small number of tumor cells in livers, no difference was found between livers from B16F10-injected mice and those from B16F10-KO1 mice (Fig. S5B).
As experimental metastasis assays suggested that Wisp1-knockout B16F10 cells had a reduced potential to extravasate and colonize vital organs, we also assessed invasion potential using spontaneous metastasis assays in C57BL/6Ncrl mice with subcutaneous injection of mouse melanoma B16F10 and its Wisp1-knockout counterpart (B16F10-KO2). Interestingly, the tumors with Wisp1 knockout grew slower than WT tumors (Fig. S6B), which was opposite to the 2D and 3D growth results observed in vitro (Fig. 2, A and B). This difference might be explained by the loss of WISP1-mediated repression of host anti-tumor immune response, which follows from in vitro studies and retrospective analysis of clinical data (52, 53). This hypothesis was supported by the fact that tumors derived from WT B16F10 and Wisp1-knockout variants grew at a similar speed in NSG mice (Fig. S6A). More focused studies are on-going to clarify this observation. Upon dissection at the humane end point of the C57BL/6Ncrl and NSG mice, no metastatic colonies were observed visually in the lungs and liver of mice injected subcutaneously with either WT or knockout cells (Fig. S6C). However, qPCR surveys of the lungs and livers from C57BL/6Ncrl mice did reveal small micrometastases from B16F10 cells but not from B16F10-KO2 cells (Fig. S6D).
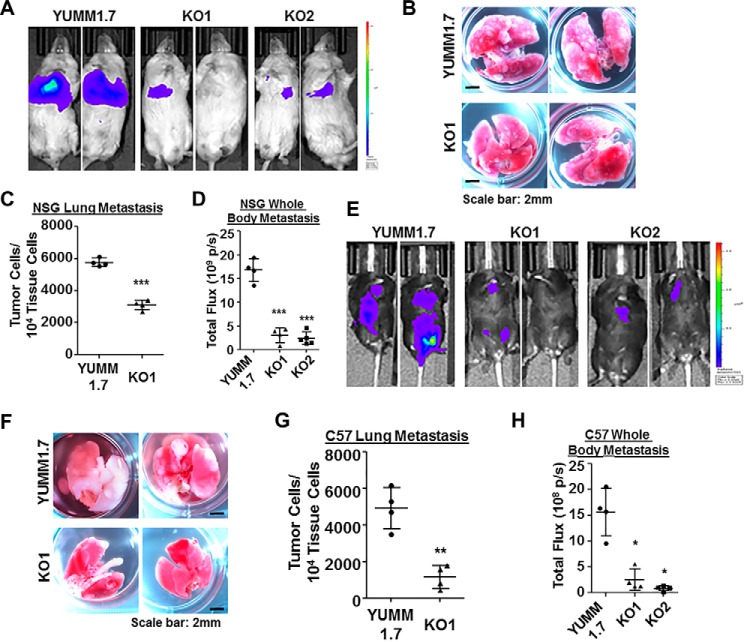
B16 variants are relatively unique, chemically induced melanoma models without a defined genetic background and lack BrafV600E mutations that are prevalent in human melanomas (61, 62). We then tried to reproduce our in vivo metastasis assay with more clinically relevant melanoma models. As WT and two WISP1-knockout variants of RPMI-7951 cells failed to survive tail vein injection, we focused next on a series of mouse melanoma cell lines (Yale University Mouse Melanoma, YUMM) recently developed with defined and stable human-relevant driver mutations from genetically engineered C57BL/6 mouse models (62). We tested two of these lines, YUMM1.1 and YUMM1.7 (genotypes: BrafV600E/WT Pten−/− Cdkn2−/−) and found that YUMM1.7 secreted a relatively high amount of WISP1 (451 ± 25 pg/ml), whereas YUMM1.1 secreted barely detectable WISP1 in conditioned medium (Table S2). We then created two Wisp1-knockout cells in YUMM1.7 (-KO1 and -KO2) with two sets of CRISPR/Cas9 (double nickase) plasmids and used them in our experimental metastasis assays (Table S2 and Fig. 4).
Figure 4.
Wisp1 knockout repressed the experimental metastasis of melanoma cell line YUMM1.7 in NSG and C57BL/6Ncrl mice. Experimental metastasis assays were performed in NSG (A–D) and C57BL/6Ncrl (E–H) mice using YUMM1.7 and the indicated knockout cells with injection through mouse tail veins. Each group contained five duplicates (n = 5), and two representative images are shown. A, bioluminescence imaging performed 1 day before NSG mice were euthanized. All animals were compared with the same bioluminescence scale. B, tumor lung metastases (white nodules) of NSG mice as captured by photography. C, real-time genomic qPCR quantitatively comparing tumor lung metastatic burdens (tumor cell number within 10,000 mouse tissue cells). D, the whole-body metastasis of tumor cells in NSG mice was plotted and compared using bioluminescence intensity detected in A. Total flux is presented as photons/s (p/s). E, bioluminescence imaging performed 1 day before C57BL/6Ncrl mice were euthanized. All animals were compared with the same bioluminescence scale. F, tumor lung metastases (white nodules) of C57BL/6Ncrl mice as captured by photography. G, real-time genomic qPCR quantitatively comparing tumor lung metastatic burdens. H, whole-body metastasis of tumor cells in C57BL/6Ncrl mice was plotted and compared using bioluminescence intensity detected in E. Two high-resolution images for B and F are provided as Figs. S7 and S8. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.D.
In NSG mice, the main metastatic tumor burden from WT YUMM1.7 was still located in the abdomen, with much weaker signals at a similar location from knockout cells (Fig. 4A). Upon dissection, we found that the lungs from YUMM1.7-injected mice were covered with numerous white metastatic tumor nodules, but nodules on the lungs from knockout cell–injected mice were much less in number and smaller in size (Fig. 4B and Fig. S7). Few visible macrometastatic nodules or colonies were discovered on the liver surfaces from YUMM1.7-injected mice, whereas nothing was visible on the livers from knockout cell–injected mice. This observation is different from B16F10-injected NSG mice, in which liver metastasis from either WT or knockout B16F10 cells took the majority of overall tumor metastatic burden (Fig. 3D). The quantitative comparison of lung metastasis and whole-body metastasis based on real-time genomic qPCR and bioluminescence intensity was calculated and plotted in Fig. 4 (C and D).
In C57BL/6Ncrl mice, YUMM1.7 metastasized to a variety of internal organs, including lungs, intestines, pancreas, ovary, and lymph nodes (Fig. 4E). In mice injected with Wisp1-knockout cells (-KO1 and -KO2), the metastatic tumor signals were much weaker and were detected in much fewer locations in individual mice (Fig. 4E). The surface of the lungs from knockout cell–injected mice was covered with far fewer and smaller metastatic white tumor nodules, as compared with YUMM1.7-injected mice (Fig. 4F and Fig. S8). Again, the quantitative comparison of lung metastasis and whole-body metastasis supported the significance difference observed visually between the WT cell–injected mice and knockout cell–injected mice (Fig. 4, G and H). In general, these in vivo results suggest that WISP1 stimulates melanoma metastasis.
WISP1 stimulated melanoma cell invasion and metastasis through promoting EMT
Following from these in vitro and in vivo observations, we next focused on identifying a mechanistic basis for how WISP1 promotes a metastatic phenotype. The collective effect of WISP1, one of the Wnt/β-catenin downstream effectors, to inhibit proliferation of melanoma cells while simultaneously promoting migration and invasion is reminiscent of EMT-like phenotype switching. In melanoma, the EMT switch starts with the up-regulation of EMT-associated transcriptional factors and repression of E-cadherin, as well as the loss of MITF, among other changes in EMT marker genes at the development of melanoma metastasis (5, 6, 57). Therefore, we asked whether an EMT gene signature is influenced by WISP1 and whether EMT-related transcription factors induced by WISP1 regulate tumor cell invasion and metastasis.
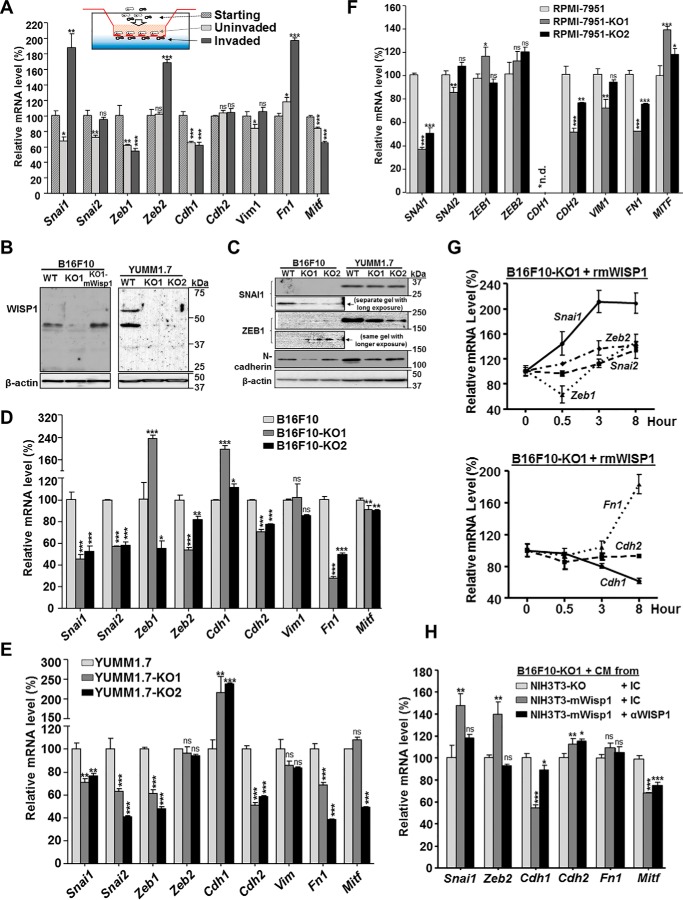
Given that the specific genes controlling an EMT switch may depend on cellular context, we first established a gene signature mapped to phenotype by comparing the EMT gene expression profiles between invaded and uninvaded mouse B16F10 melanoma cells in standard Boyden chamber transwell assays (Fig. 5A). Compared with starting cells and uninvaded cells, the invaded B16F10 cells exhibited a typical EMT gene signature, including the activation of EMT transcription factor Snai1 and Zeb2, up-regulation of extracellular matrix mesenchymal marker fibronectin (Fn1), and down-regulation of epithelial marker E-cadherin (Cdh1) as well as melanoma differentiation marker Mitf (Fig. 5A). The expression of another main EMT transcription factor, Zeb1, was very low and not observed by Western blotting (Fig. 5C) in B16F10 cells, which suggested that the observed change in mRNA may not be physiologically relevant. It is unclear whether Zeb1 plays certain context-specific roles other than promoting EMT or if its function is simply compensated by the redundancy of other EMT-TFs in B16F10 cells. More detailed work is needed to clarify its role associated with B16F10 cell invasion.
Figure 5.
WISP1 induced an EMT gene signature in mouse/human melanoma cells. Unless otherwise specified, all cells were plated on 6-well plates in complete growth medium for 48 h before they were harvested for RNA analysis or treated with the indicated conditioned medium or recombinant protein. A, mRNA expression, revealed by real-time quantitative RT-PCR, of select EMT marker genes and Mitf in uninvaded and invaded B16F10 cells from a Boyden transwell invasion assay. B, immunoblot analysis of WISP1 protein to confirm the disruption of Wisp1 gene in B16F10 and YUMM1.7 knockout cells. 20 μg of whole-cell lysate was loaded in each lane, and β-actin was used as an internal loading control. B16F10-KO1-mWisp1 cells, in which mouse WISP1 expression was resumed with retroviral transduction, were used as a positive control. C, immunoblot analysis of certain EMT marker proteins in B16F10 and YUMM1.7 knockout cells. 20 μg of whole-cell lysate was loaded in each lane, and all cells were compared on the same gel to reveal the relative intensity of each protein. D, comparison of EMT marker gene expression in mouse melanoma B16F10 and its two Wisp1-knockout cells (-KO1 and -KO2). E, comparison of EMT marker gene expression in mouse melanoma YUMM1.7 and its two Wisp1-knockout cells (-KO1 and -KO2). F, comparison of EMT marker gene expression in human melanoma RPMI-7951 and its two WISP1-knockout cells (-KO1 and -KO2). G, stimulation of EMT marker gene expression with recombinant mouse WISP1 protein (rmWISP1). B16F10-KO1 cells were treated with rmWISP1 (final concentration 5 μg/ml) and harvested at the indicated time point for real-time quantitative RT-PCR analysis. H, stimulation of EMT marker gene expression with WISP1-overexpressed or WISP1-immunodepleted conditioned medium (CM). The conditioned media were pretreated with the indicated antibodies for 30 min before they were used on Wisp1-knockout B16F10 cells (-KO1). The cells were collected for real-time qRT-PCR after 3 h of treatment. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. Error bars, S.D.
We then set out to compare the expression of genes associated with this EMT signature in parental cells and WISP1-knockout cells from mouse B16F10, YUMM1.7, and human RPMI-7951 melanoma lines. First, in addition to ELISA (Table S2), we used immunoblotting to confirm the knockout of WISP1 protein in mouse B16F10 and YUMM1.7 cells after the disruption of the Wisp1 gene (Fig. 5B). Interestingly, multiple WISP1 bands were detected in YUMM1.7 cells, suggesting the existence of covalent modification (glycosylation) or WISP1 oligomers. Another set of immunoblotting with available antibodies showed the reduction of EMT transcription factor SNAI1 and mesenchymal marker N-cadherin upon Wisp1 knockout in B16F10 and YUMM1.7 cells (Fig. 5C). Whereas ZEB1 went up in B16F10, it did decrease in YUMM1.7 after Wisp1 knockout (Fig. 5C).
Using real-time qPCR, we found that, in B16F10, those invasion-associated EMT signature genes as determined in Fig. 5A changed in the opposite direction after Wisp1 knockout (Fig. 5D). The pattern observed upon WISP1 knockout is consistent with a mesenchymal–epithelial transition (MET) type switch, which included up-regulation of epithelial marker E-cadherin (Cdh1); down-regulation of EMT-TFs such as Snai1, Snai2, and Zeb2; and down-regulation of mesenchymal markers such as N-cadherin (Cdh2) and fibronectin (Fn1) (Fig. 5D). The only exception was the Mitf expression from B16F10 knockout cells, which was slightly reduced in both of the B16F10 knockout cells. This may suggest that Mitf repression is involved in melanoma cell invasion but is not directly regulated by WISP1 in the context of B16F10 cells. Similar gene expression profiles were discovered in mouse YUMM1.7 and human RPMI-7951 melanoma lines upon WISP1 knockout (Fig. 5, E and F). Although we observed subtle differences in the expression of specific genes among the three WT and six knockout melanoma cells, WISP1 knockout consistently up-regulated the epithelial marker E-cadherin (CDH1) and down-regulated the EMT transcription factor SNAI1, the mesenchymal marker N-cadherin (CDH2), and fibronectin (FN1) (Fig. 5, D–F). The result strongly suggested that WISP1 stimulates melanoma invasion and metastasis through promoting EMT of tumor cells, whereas WISP1 knockout decreases melanoma invasion by causing MET of tumor cells.
A rescue experiment with B16F10-KO1 cells was performed using recombinant mouse WISP1 protein (rmWISP1) to track the change of these EMT signature genes in real time (Fig. 5G). Within 30 min of rmWISP1 treatment, an immediate increase of Snai1 and decrease of Zeb1 were observed. Over an 8-h period, Snai1 continued to increase and then maintained at a high level, which was followed by the increase of other EMT-TFs, the increase of mesenchymal marker Fn1, and the decrease of epithelial marker E-cadherin (Cdh1) (Fig. 5G). A similar EMT rescue response in B16F10 knockout cells was observed using conditioned medium from mouse fibroblast NIH3T3-mWisp1 that overexpressed mouse WISP1 (Fig. 5H; only the result from -KO1 is shown). Immunodepleting WISP1 in conditioned media prior to treatment of knockout cells abolished such rescue effects (except for Mitf), confirming the functional role of WISP1 from the media. Collectively, these results support a notion that WISP1 stimulates tumor invasion and metastasis through promoting an EMT-like process within melanoma cells, and SNAI1 plays a major role as a transcription factor in this transition process.
SNAI1 overexpression in Wisp1-knockout melanoma cells reversed the repression of tumor invasion in vitro and metastasis in vivo
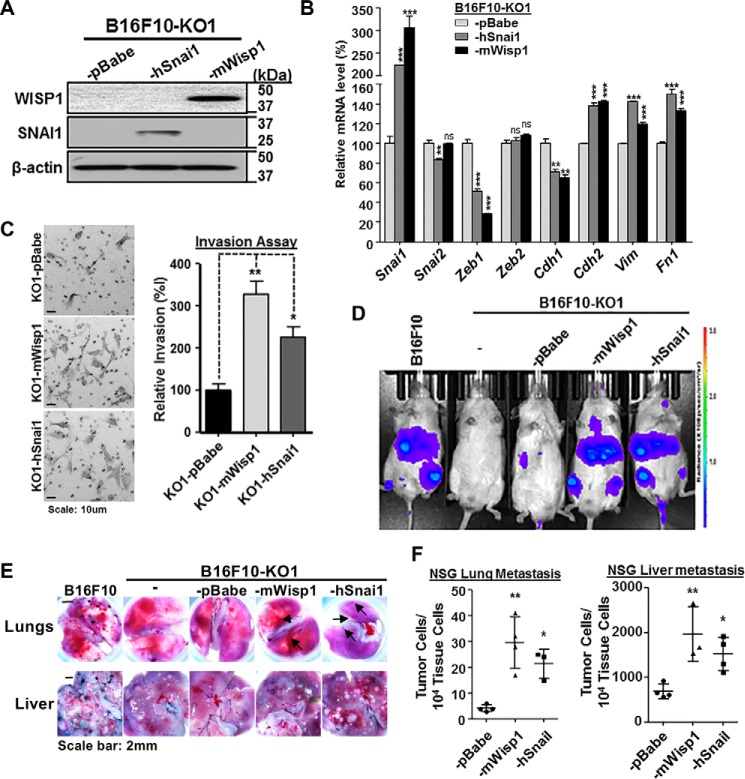
The dynamic results described above strongly suggest that SNAI1 is one of main primary effectors downstream of WISP1 signaling to stimulate EMT in melanoma cells and hence to promote tumor invasion and metastasis. The idea followed that reintroduction of SNAI1 into Wisp1-knockout melanoma cells would reverse, at least in part, back to the WT genotype and phenotype. For such a purpose, B16F10 knockout cell was transduced with retroviral vector to overexpress human SNAI1 protein (-KO1-hSnai1) (Fig. 6A). Another two cells from -KO1 were also created either with retroviral vector control (-KO1-pBabe) or with vector overexpressing mouse WISP1 protein (-KO1-mWisp1) (Fig. 6A).
Figure 6.
SNAI1 overexpression in B16F10 Wisp1-knockout cell rescued the repression of tumor invasion in vitro and metastasis in vivo. A, immunoblot analysis of WISP1 and SNAI1 using B16F10-KO1 cell that were transduced with retroviral vector control (-pBabe) or retrovirus expressing either mouse WISP1 (-mWisp1) or human SNAI1 (-hSnai1). B, comparison of EMT marker gene expression after overexpression of SNAI1 or reintroduction of WISP1 in B16F10-KO1 cells. Cells were plated on 6-well plates in complete growth medium for 48 h before they were harvested for RNA analysis. C, Boyden transwell invasion assay after overexpression of SNAI1 or reintroduction of WISP1 in B16F10-KO1 cells. A representative staining image for each sample is shown on the left, and relative invasion efficiency is graphed on the right. D, experimental metastasis assay in NSG mice using the indicated cells. Each group contained 3–4 mice. All mice were imaged 1 day before the end of the assay, and representative bioluminescence images are shown. E, representative lung and liver images from NSG mice in the experimental metastasis assay described in D. Metastatic tumor colonies on the lung surface from mice with -mWisp1 or -hSnai1 cells are indicated by arrows. F, real-time genomic qPCR for lungs and livers from the experimental metastasis assay in D. The quantitative tumor metastatic burdens were presented as tumor cell number within 10,000 mouse tissue cells. A high-resolution image for E is provided as Fig. S9. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. Error bars, S.D.
Real-time qPCR showed that overexpression of either human SNAI1 or mouse WISP1 in knockout cells recovered the gene expression pattern toward EMT (Fig. 6B). In addition to promoting the expression of endogenous Snai1, they both enhanced the expression of mesenchymal markers N-cadherin (Cdh2), vimentin (Vim), and fibronectin (Fn1) and repressed the epithelial marker E-cadherin (Cdh1) (Fig. 6B). The highly similar rescue effects also suggested that the two protein factors may work on the same signaling cascades. In the transwell assay, -KO1-hSnai1 cells showed an increase in invasion by more than 126.2 ± 25.2% compared with the control cell -KO1-pBabe, and -KO1-mWisp1 cells exhibited an increase of invasion efficiency by over 228.6 ± 29.7% (Fig. 6C).
In vivo experimental metastasis assays were performed via intravenous injection using NSG mice. As shown in Fig. 6D, bioluminescence imaging detected very weak metastatic signals in the abdominal region of mice receiving either B16F10-KO1 cells or its retroviral vector control, B16F10-KO1-pBabe cells. Expression of either WISP1 or SNAI1 in knockout cells restored the metastatic phenotype, with a similar intensity as observed with WT B16F10 cells. Mouse dissection revealed that metastatic tumor colonies on the lung and tumor nodules on the liver, which were absent or significantly reduced after Wisp1 knockout (Fig. 3, B and D), were restored upon re-expression of either WISP1 or SNAI1 in knockout cells (Fig. 6E and Fig. S9). Real-time qPCR confirmed the significant increase in metastasis after WISP1 or SNAI1 were re-expressed in knockout cells (Fig. 6F). In these experiments, the average lung tumor burden was 4.3 ± 0.6 metastatic tumor cells among 104 lung cells for mice receiving -KO1-pBabe cells, and the numbers were increased to 30.0 ± 7.0 and 21.3 ± 3.2 for mice with -KO1-mWisp1 and -KO1-hSnai1 cells, respectively (Fig. 6F, left). Similarly, the average liver tumor burden was 715 ± 110 metastatic tumor cells for mice with -KO1-pBabe cells and increased to 1944 ± 249 and 1391 ± 186 for mice with -KO1-mWisp1 and -KO1-hSnai1 cells, respectively (Fig. 6F, right). Collectively, these in vitro and in vivo results supported our proposed role of SNAI1 as a downstream effector of WISP1 signaling and illustrate the role of this signaling pathway in promoting melanoma cell metastasis.
WISP1 activated AKT/MAPK signaling to promote EMT in mouse melanoma cells
WISP1 activates AKT signaling pathway to promote a variety of cellular functions, such as proliferation, survival, migration, and invasion in normal tissue and cancer cells (25, 27, 29, 32, 36, 47, 48). It also stimulates the MEK/ERK pathway to enhance tumor migration and invasion (30, 34, 45, 46), possibly through the induction of MEK/ERK signaling–induced EMT (44). Because AKT signaling and MEK/ERK signaling are the intracellular signaling cascades known to induce EMT and tumor metastasis (4–6), we hypothesized that these two signaling pathways are essential for EMT in melanoma cells and that WISP1 activates these signaling pathways to promote EMT.
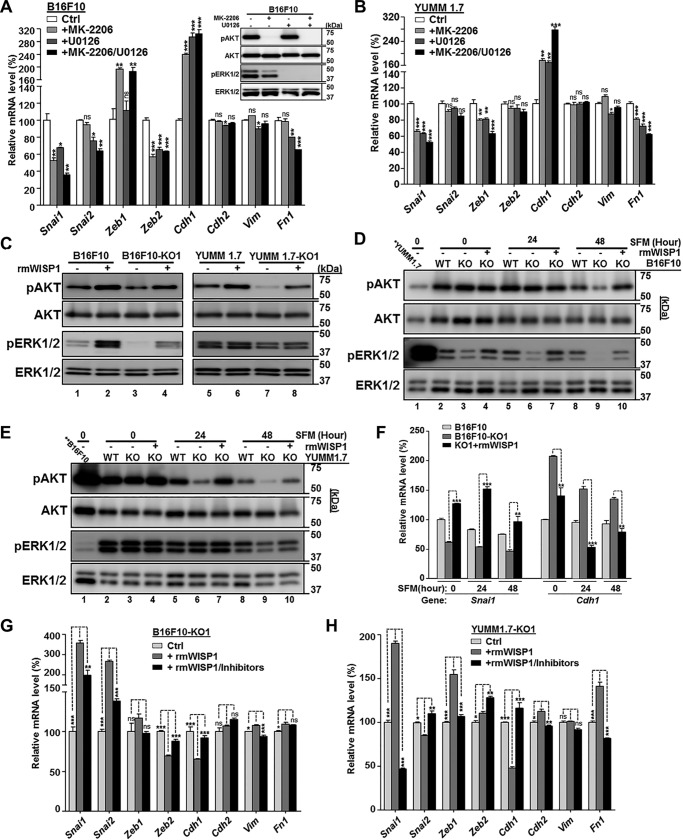
To test this, we blocked either AKT, MEK, or both pathways in B16F10 cells using kinase inhibitors and compared the change in EMT marker gene expression after 3 h (Fig. 7A). As expected, we observed a shift in gene expression toward MET whenever AKT or MEK signaling was inhibited, with at least an additive effect when both were blocked. Changes included the reduction of EMT-TFs, such as Snai1, Snai2, and Zeb2, and the increase of epithelial marker E-cadherin (Cdh1) (Fig. 7A). Among the three mesenchymal markers tested, only fibronectin (Fn1) showed significant decreases, probably because the 3-h treatment was not long enough to exhibit any difference on other genes (Fig. 7A). A similar MET pattern was observed in YUMM1.7 cells upon treatment with kinase inhibitors (Fig. 7B). We also saw the reduction of Zeb1, which was consistent with the EMT gene expression pattern we observed for YUMM1.7 in Fig. 5E.
Figure 7.
WISP1 activated AKT and MEK/ERK signaling and promoted EMT marker gene expression in mouse melanoma cells. Unless otherwise specified, cell treatment for kinase immunoblot analysis was maintained for 30 min before cells were lysed for protein extraction, whereas cell treatment for comparison of EMT marker gene expression was maintained for 3 h before cells were harvested for RNA extraction. A, comparison of EMT marker gene expression after inhibition of AKT and/or MEK/ERK signaling in B16F10 cells. DMSO was used for control cells. Immunoblotting for phospho-AKT (pAKT) and phospho-ERK1/2 (pERK1/2) is shown in the top right corner. Pan-AKT and total ERK1/2 were probed as loading control. B, comparison of EMT marker gene expression after inhibition of AKT and/or MEK/ERK signaling in YUMM1.7 cells. C, immunoblot analysis of AKT and ERK1/2 activation in the indicated mouse melanoma cells with treatment of recombinant mouse WISP1 protein (rmWISP1; final concentration 5 μg/ml). All cells were grown on 6-well plates in complete DMEM for 48 h and SFM for another 48 h before rmWISP1 was added. D, immunoblot analysis of AKT and ERK1/2 activation in B16F10 knockout cell (-KO1) by rmWISP1 under different basal phosphokinase levels. All cells were grown on 6-well plates in complete DMEM for 48 h (0-h point for SFM) and switched to SFM for 24 or 48 h. The indicated cells were treated with rmWISP1 at the 0-, 24-, and 48-h time points (of SFM) for 30 min before they were lysed for kinase analysis. The first lane on the gels was loaded with YUMM1.7 at the 0-h point to compare the relative kinase level between B16F10 and YUMM1.7 cells. E, immunoblot analysis of AKT and ERK1/2 activation in YUMM1.7 knockout cells (-KO1) by rmWISP1 under different basal phosphokinase levels. All cells were treated similarly as described in D. The first lane on the gels was loaded with B16F10 at the 0-h point to compare the relative kinase level between B16F10 and YUMM1.7 cells. F, comparison of SNAI11 activation and E-cadherin repression in B16F10 knockout cells (-KO1) by rmWISP1 under different basal phosphokinase levels. All cells were treated similarly as described in D except that rmWISP1 treatment at each point was maintained for 3 h. G and H, comparison of EMT marker gene expression after AKT/ERK1/2 activation in B16F10-KO1 (G) or YUMM1.7-KO1 (H) by rmWISP1 was blocked. rmWISP1 with DMSO or inhibitors was added after the indicated cells were grown on 6-well plates in complete DMEM for 48 h and in SFM for 24 h. The relative protein levels of pAKT and AKT and of pERK1/2 and ERK1/2 in C–E were measured, and they are listed in Table S3. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. Error bars, S.D.
To assay gene expression, melanoma cells, including B16F10 and YUMM1.7, were normally plated in complete DMEM (10% fetal bovine serum (FBS)) for 48 h before they were harvested for RNA extraction. However, to detect maximal AKT and MEK/ERK signaling activation with minimal background noise from FBS, we grew these melanoma cells (WT and knockouts) in serum-free medium (SFM; 0.1% FBS) for another 48 h before we treated cells with rmWISP1 for 30 min and lysed cells for immunoblotting analysis (Fig. 7C). Both B16F10 and YUMM1.7, with autocrine WISP1 secretion, maintained a higher basic level of phospho-AKT and phospho-ERK1/2 than their knockouts (Fig. 7C, compare lane 1 with lane 3 and lane 5 with lane 7), and paracrine rmWISP1 treatment similarly elevated phospho-AKT and phospho-ERK1/2 of both WT and knockout cells (Fig. 7C, compare lane 2 with lane 4 and lane 6 with lane 8). Collectively, WISP1 rapidly and efficiently activated AKT and ERK signaling pathways that were critical signal transducers for inducing an EMT-like gene expression signature in both B16F10 and YUMM1.7 cells.
Whereas these results are consistent with our hypothesis, we noticed some subtle difference in how these two melanoma models responded to different growth conditions. We designed experiments to explore how preconditioning in serum-free medium and the presence of the BrafV600E mutation in YUMM1.7 cells impacted the signaling response to WISP1. In two sets of time-course experiments, we grew the indicated cells in complete DMEM for 48 h (SFM, 0-h time point) and switched to SFM for 24 or 48 h. At the SFM time point (post-incubation) 0, 24, or 48 h, cells were treated with rmWISP1 for 30 min before assaying for kinase activation. Immunoblotting revealed that B16F10 and its knockout cells exhibited relative high phospho-AKT but low phospho-ERK1/2 background level (Fig. 7D, compare lane 1 with lanes 2–4), whereas YUMM1.7 and its knockout cells exhibited relatively low phospho-AKT but high phospho-ERK1/2 background level (Fig. 7E, compare lane 1 with lanes 2–4). Hence for B16F10-KO1 cells, rmWISP1 readily stimulated ERK signaling at time point 0 but showed stimulation of AKT signaling at a later time (time point 48 h) (Fig. 7D). Certainly, for YUMM1.7-KO1 cells, rmWISP1 mainly stimulated AKT signaling because of their BrafV600E mutation and activated BRAF/MEK/ERK pathway (Fig. 7E). The relative activation of kinases in Fig. 7 (C–E) was calculated after protein densitometry measurement and presented in Table S3.
Using similar experimental conditions, a time-course gene expression analysis was also performed for Snai1 and Cdh1 (Fig. 7F). The result confirmed the effect of rmWISP1 on EMT switch at all three time points and also suggested the best time for maximal stimulation, which was 24 h in SFM (Fig. 7F). Under this optimal condition, we stimulated B16F10-KO1 and YUMM1.7-KO1 cells with rmWISP1 in the presence or absence of both AKT and MEK inhibitors to test whether activation of AKT and MEK/ERK signaling was essential in melanoma cells for a WISP1-mediated EMT switch (Fig. 7, G and H). Inhibiting these two signaling pathways dramatically repressed the elevation of EMT transcription factors, including Snai1 in B16F10-KO1 and Snai1/Zeb1 in YUMM1.7-KO1, and reversed the repression of Cdh1 expression from those EMT-TFs (Fig. 7, G and H). Similar to the results shown in Fig. 7 (A and B) we did not see much change (except for Fn1 in YUMM1.7-KO1) with the three mesenchymal markers (Cdh2, Vim, and Fn1), due to the short period of treatment (Fig. 7, G and H). In short, we think that WISP1 promotes melanoma EMT by stimulating AKT and MEK/ERK signaling pathways.
Discussion
For melanoma invasion and metastasis, revealing factors present within the tumor microenvironment that regulate these processes has important implications for the diagnosis, prognosis, and treatment of melanoma. In this work, analysis of molecular and survival data derived from patients diagnosed with primary melanoma showed that the expression of Wnt-inducible signaling protein 1 (WISP1), a downstream effector of the Wnt/β-catenin pathway, is increased in melanoma and is associated with reduced overall survival. Functionally, we found that WISP1 enhanced tumor invasion and metastasis by promoting melanoma EMT using metastatic mouse and human melanoma cell lines. Results from experimental metastasis assays in both NSG and C57BL/6Ncrl mice with either B16F10 or YUMM1.7 melanoma cells supported the functional role of WISP1 in vivo. Collectively, these observations for WISP1 revealed a connection back to aberrant Wnt/β-catenin signaling and provide insight into the context-dependent role of the Wnt/β-catenin pathway in melanoma metastasis.
Clarifying relations in vivo between aberrant Wnt/β-catenin signaling and functional implications of WISP1 signaling help to explain the prevalence of early metastatic dissemination in melanoma. As a secreted signal, WISP1 connects intrinsic cell signaling pathways with biological cues released into the tissue microenvironment to restore homeostasis following tissue damage (63–65) and to sustain a mesenchymal stem cell niche (66). Whereas these studies focus on bone and cartilage homeostasis, the expression of WISP1 by normal melanocytes and dermal fibroblasts (Fig. 1, A–D) and the induction of WISP1 expression upon disruption of adherens junctions (54) suggest that WISP1 plays a similar role in the skin. Given similarities between stromal–epithelial cross-talk in wounds and tumors (67), it follows then that melanocytes are poised for metastasis, which is realized by acquiring mutations that amplify the production of this environmental cue. This model is supported by the genetic evidence that WISP1 gene amplification was enriched in melanomas and that β-catenin gene amplification and stabilizing exon 3 mutations were also enriched (Table S1). Interestingly, the majority of these changes associated with aberrant β-catenin signaling were independent of WISP1 amplification in patient samples (Table S1).
Mechanistically, melanoma invasion and metastasis is connected with an EMT-like process via up-regulating EMT-related transcription factors and repressing E-cadherin (4–6). Our results showed that WISP1 up-regulated EMT transcription factors and mesenchymal markers and repressed the epithelial marker E-cadherin as well as the melanocyte differentiation marker MITF (Figs. 5–7). The observed changes in gene expression were largely conserved between mouse and human cell lines and consistent with conceptual models of EMT, especially with the coincidental reduction in the epithelial marker gene E-cadherin and induction of the mesenchymal marker genes fibronectin and N-cadherin (4–6). One intriguing observation was the different levels of basal expression of ZEB1 and the change in expression during EMT induction in three melanoma cell lines we tested. When we knocked out WISP1, we found that SNAI1 expression decreased in all cases but that ZEB1 was reduced only in YUMM1.7 from a high basal level of expression, barely changed in RPMI-7951, and even increased in B16F10 cells from a low basal level of expression. Such counterintuitive response has been reported previously during the switch of primary melanoma to a mesenchymal-like invasive phenotype for another EMT-inducing transcription factor, ZEB2 (7–9). Whereas the observed dynamics of EMT transcription factor response to WISP1 suggest that SNAI1 induction is a primary response and that ZEB1 appears to be a secondary response, a more systematic analysis of the dynamics of the underlying network of EMT transcription factors under these different conditions, as well as the individual activation of the downstream signaling pathways, may help reveal how environmental and contextual differences collectively influence EMT.
The regulation of EMT marker genes by WISP1 was achieved, at least partially, through activating AKT and MEK/ERK signaling pathways in B16F10 and YUMM1.7 melanoma cells (Fig. 7). As mentioned in the Introduction, the signal is likely transduced via integrin signaling that, upon binding to CCN family members, activates various downstream signaling pathways, including PI3K/AKT and RAS/RAF/MEK/ERK (22–24). Whereas activating additional pathways such as Rac/JNK and NF-κB downstream of integrin signaling cannot be excluded, the changes in EMT marker gene expression upon AKT and MEK/ERK kinase inhibition (Fig. 7, panels A and B and panels G and H, respectively) suggested that these two signaling pathways were strongly involved in mouse melanoma cells. However, the quantitative contribution from each signaling pathway is different, depending on context. As we observed in Fig. 7 (C–E), rmWISP1 enhanced both AKT and MEK/ERK signaling in B16F10, YUMM1.7, and their knockout cells, but the relative activation of each signaling was different in different cell lines and growth conditions. With a background level of relatively high phospho-AKT but low phospho-ERK, B16F10 and its KO cells responded more drastically to ERK activation. With high background MEK/ERK signaling from the BrafV600E mutation in YUMM1.7 and its knockout cells, the relative phospho-AKT level increased much more than its phospho-ERK level with rmWISP1 stimulation (Table S3). Our result in YUMM1.7 cells also demonstrated that, even with basal activation of MEK/ERK signaling, the relative MEK/ERK level could still be enhanced or reduced, depending on the conditions (Fig. 7, C–E), and such regulation would still produce biological consequences (Fig. 7, B and H). Interestingly, Herlyn's group showed that, when Notch1 signaling was activated in human WM278 melanoma cells (with the BrafV600E allele), PI3K/AKT and ERK signaling could be elevated to enhance melanoma proliferation, survival, and metastasis (68).
WISP1's role in melanoma has also been studied in the context of fibroblasts and Notch signaling. The Notch signaling pathway is an intercellular signaling cascade that is activated in human melanoma cells and is essential for melanoma growth and metastasis (69). In this context, adjacent differentiated keratinocytes or endothelial cells are responsible for the cell surface Notch ligands, and neither Notch ligands nor active Notch signaling are detected in the stromal fibroblasts (70, 71). Interestingly, when constitutively active Notch signaling was ectopically engineered into fibroblast cells by overexpressing a NOTCH1 intracellular domain (NIC), WISP1 expression was elevated through increased transcription, suggesting that WISP1 is a downstream target of Notch signaling (50, 51). Using engineered primary human dermal fibroblasts, Shao et al. (50) showed in vivo, complemented by in vitro studies using conditioned media, that these cells repressed melanoma growth and angiogenesis but showed no effect on tumor migration. Similarly, using engineered mesenchymal stem cell–derived fibroblasts, Shao et al. (51) also showed that these cells had no effect on melanoma growth but repressed tumor migration, invasion, and metastasis. The authors attributed the repressive effects from these engineered fibroblasts on melanoma progression to the elevated WISP1 secretion after activated Notch signaling. This is different from what we observed using immortalized mouse fibroblast NIH3T3 cells and derivatives (Fig. 2). In those two reports, the authors showed the increase of WISP1 in fibroblasts with NIC by RT-PCR and Western blotting, but the increase of the secreted, functional form of WISP1 was never confirmed by ELISA. The gene expression profiles and the functional changes in the engineered fibroblasts were also not reported. Without understanding how these engineered fibroblasts reconfigured the tumor microenvironment, generalizing these results to infer a role for WISP1 in repressing melanoma progression is difficult.
Targeting the Wnt/β-catenin pathway has been proposed to treat human cancers, including melanoma, yet a few conceptual and safety concerns challenge development of this therapeutic approach (72–74). Some of these challenges related to β-catenin stem from it being an intracellular target that plays various roles depending on context, such as in cancer progression, organismal development, and adult tissue homeostasis (72). As a secreted downstream effector of aberrant Wnt/β-catenin in melanoma, targeting WISP1 has several advantages. First, targeting WISP1 may provide more specificity in reshaping the melanoma microenvironment to favor anti-tumor immunity and to inhibit metastasis than pleiotropic effects of inhibiting β-catenin. Second, a secreted target opens more options for developing therapeutic reagents, including humanized monoclonal antibodies against WISP1 to block its activities, siRNA and other oligonucleotides to repress WISP1 expression, or small molecules and peptides to inhibit WISP1 signaling. Whereas there is some evidence suggesting receptors for WISP1 (22, 23), clarifying membrane-proximal signaling events that lead to activation of EMT-related transcription factors, including SNAI1, may provide additional extracellular targets. Future preclinical studies with animal models exhibiting the full spectrum of melanoma progression will be of great interest for translating WISP1 as a target into the clinic to limit metastatic dissemination for patients diagnosed with melanoma.
Experimental procedures
Cell culture, WISP1 ELISA, and conditioned medium preparation
Mouse melanoma line B16F0 (purchased in 2008, RRID: CVCL_0604), B16F10 (purchased in 2008, RRID: CVCL_0159), mouse fibroblast line NIH3T3 (purchased in 2007, RRID: CVCL_0594), mouse Lewis lung carcinoma line LLC1 (purchased in May 2017, RRID: CVCL_4358), HEK293T (purchased in 2005, RRID: CVCL_0063), human metastatic melanoma cell lines RPMI-7951 (purchased in July 2015, RRID: CVCL_1666), SK-MEL-3 (purchased in July 2015, RRID: CVCL_0550), and SH-4 (purchased in July 2015, RRID: CVCL_1692) were purchased from American Type Culture Collection (ATCC) on the indicated dates. Mouse breast cancer line E0771 was kindly provided by Dr. Linda Vona-Davis (received in August 2015, RRID: CVCL_GR23; West Virginia University). Mouse melanoma lines YUMM1.1 (received in September 2017, RRID: CVCL_JK10) and YUMM1.7 (received in September 2017, RRID: CVCL_JK16) were gifts from Drs. William E. Damsky and Marcus W. Bosenberg (Yale University) (62). B16F0, B16F10, NIH3T3, 293T, YUMM1.1, and YUMM1.7 cells were cultured in high-glucose DMEM supplemented with l-glutamine, penicillin–streptomycin, and 10% FBS. They may also be cultured in the same medium with only 0.1% FBS (SFM). Other cells were grown as recommended by ATCC. All cell lines were revived from frozen stock, used within 10–15 passages that did not exceed a period of 6 months, and routinely tested for mycoplasma contamination by PCR.
To measure WISP1 secretion from each line, cells were grown for 48 h to reach about 90% confluence, and the medium was filtered for ELISA analysis using the human WISP-1/CCN4 DuoSet ELISA Development Kit (R&D Systems, Minneapolis, MN). To prepare conditioned media with different concentrations of WISP1, WT and derivative NIH3T3 cells (NIH3T3-KO, -pBabe, and -mWisp1) were counted and seated on 100-mm plates with the same density. Cells were grown in DMEM with 0.1% FBS or 10% FBS for 48 h to reach about 70% confluence. The medium was then filtered, aliquoted, and frozen at −80 °C for future use. Generally, conditional media with 0.1% FBS were used for transwell migration and invasion assays, whereas conditional media with 10% FBS were used for gene expression stimulation (Fig. 5G).
Retroviral/lentiviral plasmids and virus transduction
Mouse WISP1 DNA sequence encoding the total 367 amino acids was amplified by PCR from B16F0 cDNA using primers with a BamHI site on each side. A retroviral expression vector for mouse WISP1 (pBabe-mWisp1) was created by inserting the above coding sequence into the BamHI site of pBabe-puro retroviral vector, and was verified by sequencing. Another retroviral vector for human SNAIL (pBabe puro Snail, or pBabe-hSnai1) was from Addgene (plasmid 23347, gift of Bob Weinberg). Lentiviral vector pLU-Luc2, expressing a codon-optimized luciferase reporter gene Luc2, was kindly provided by Dr. Alexey V. Ivanov (West Virginia University) and was described previously (60).
Retroviruses were packaged and transduced into the indicated cells. The stable cells were achieved with puromycin selection. Lentiviruses were produced by transfecting pLU-Luc2 and two packaging plasmids, psPAX2 (Addgene plasmid 12260) and pCMV-VSG-G (Addgene plasmid 8454), into HEK293T cells. Virus soup was aliquoted and used to transduce the indicated cells at constant conditions.
Creation of WISP1-knockout cells using the CRISPR/Cas9 system
To achieve high specificity and reduce variability in genetic backgrounds, CRISPR/double nickase systems were selected to knock out the WISP1 gene. Two pairs of mouse Wisp1 double nickase plasmids (sc-423705-NIC and sc-423705-NIC-2), targeting the mouse Wisp1 gene at different locations, were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX) and used in B16F10, YUMM1.7, and NIH3T3 cells. Another two sets of WISP1 double nickase plasmids against the human WISP1 gene (sc-402559-NIC and sc-402559-NIC-2) were also from Santa Cruz Biotechnology and used in RPMI-7951 cells.
Following the manufacturer's instructions, cells were transfected with an individual set of plasmids, which also express a puromycin resistance gene. Cells were selected by puromycin for 5 days to achieve 100% transfection efficiency. Surviving cells were counted and plated onto a 96-well plate with a density of 0.5 cells/well. After 1 week, single clones were isolated and expanded on 6-well plates. The cell culture media from those wells were used for WISP1 ELISA to characterize knockout clones. The identified WISP1-knockout cells were further expanded and used for the next steps.
2D cell growth assay, soft agar assay, and wound healing assay
Two-dimensional cell growth was tested on 96-well plates in biological triplicate using the ATPlite Luminescence Assay System (PerkinElmer Life Sciences) following the manufacturer's instructions. Anchorage-independent cell growth (soft agar assay) was performed on 6-well plates in biological triplicates as described (75).
For wound healing assays, all cells were prepared on 6-well plates in biological triplicates and allowed to reach 95% confluence. A wound in each well was created by scratching straight though the middle of the well with a 200-μl pipette tip. Plates were washed to remove dislodged cells and debris, refed with fresh media, and incubated at 37 °C for 24 h. The center of each scratch was photographed at the 0- and 24-h time points, the relative wound width was measured with ImageJ (National Institutes of Health), and the healing rate was calculated.
Transwell migration and invasion assays and collection of invaded cells
BioCoat control inserts for the migration assay and BioCoat Matrigel invasion chambers for the invasion assay were from Corning, Inc. The assays were performed on 24-well plates in biological triplicates following the manufacturer's instructions. Briefly, cells were serum-starved for 24 h before they were trypsinized and resuspended in DMEM with 0.5% BSA. Each well was filled with 0.75 ml of serum-free conditioned medium from either NIH3T3 or another indicated cell as chemoattractant. The chamber inserts were then placed onto wells, and 5.0 × 104 cells in a 0.5-ml suspension were loaded into the inserts. The plates were incubated at 37 °C for 24 h, the cells on the upper surface of the insert PET membrane were carefully removed with a cotton swab, and the cells that migrated or invaded through the membrane were stained with the Hema 3 Staining System (Thermo Fisher Scientific). The membrane was peeled off with a razor blade and mounted on a glass slide. The cells were then quantified by a microscope.
To collect uninvaded and invaded B16F10 cells in the transwell assay for RNA isolation and gene expression analysis (Fig. 5A), a practice similar to that described above was performed on 24-well plates. After 24 h of incubation, the Matrigel chamber inserts were washed with PBS on both sides and put back into wells that were filled with 0.75 ml of trypsin/EDTA solution (0.05%), followed by the addition of another 0.5 ml of trypsin/EDTA inside each insert. The trypsinized uninvaded cells (from above the Matrigel) were removed from the interior of each Matrigel chamber insert into a new 15-ml tube, and the trypsinized invaded cells (from below the membrane) were removed from each well into a new 15-ml tube. Both cells were then washed with complete growth medium and PBS before RNA was extracted.
In vivo metastasis assays and bioluminescence imaging
Animal experiments described in this study were approved by the West Virginia University Institutional Animal Care and Use Committee and were performed at the West Virginia University Animal Facility. 6–8-week-old female C57BL/6Ncrl mice were purchased from Charles River Laboratories, and 6–8-week-old male NOD-scid IL2Rγnull (NSG, stock no. 005557) mice were from the Jackson Laboratory. Generally, metastasis assays were performed with five mice in each group and replicated at least twice with independent cohorts, whereby results similar to those described were achieved each time.
For experimental metastasis assays, mice were injected intravenously with the indicated cells with Luc2 expression. For B16F10 cells, 6 × 104 cells/mouse were used for NSG and 2 × 105 cells/mouse for C57BL/6Ncrl. Mice were euthanized on day 15 post-injection for NSG and on day 21 post-injection for C57BL/6Ncrl. For YUMM1.7 cells, 1.5 × 105 cells/mouse were used for both NSG and C57BL/6Ncrl. Mice were euthanized on day 24 post-injection for NSG and day 32 post-injection for C57BL/6Ncrl. Lungs, livers, and other organs (kidneys, brains, etc.) were dissected, and images were taken using an Olympus MVX10 microscope. All organs were collected and frozen at −80 °C for real-time qPCR analysis. For tumor growth and spontaneous metastasis of B16F10 cells, C57BL/6Ncrl or NSG mice were injected subcutaneously with the indicated cells with Luc2 expression (1.2 × 105 cells/mouse). Tumor volumes were recorded every other day from day 7 or day 8 post-injection to day 21. All mice were then euthanized, and organs were dissected for imaging and qPCR analysis.
Bioluminescence imaging was performed to quantify tumor burden in vivo 1 day before experimental animals were euthanized. Briefly, mice were injected intraperitoneally with d-luciferin (Caliper Life Sciences, 150 mg/kg), and all images were taken between 10 and 20 min post-injection using the IVIS Lumina-II imaging system (PerkinElmer Life Sciences) with 1.0-min capture and medium binning. Living Image version 4.0 software was used to process the captured images. Signal intensity was quantified as the sum of all detected photon counts within the region of interest after subtraction of background luminescence.
Genomic DNA extraction and determination of metastatic tumor burden
The method was described previously (60). It utilized two pairs of primers targeting the firefly luciferase Luc2 gene (only from injected tumor cells) and the mouse Ptger2 gene (from injected tumor cells and also mouse tissues) to calculate the relative ratio of metastatic melanoma cells within 104 tissue cells. The number was used as a quantitative measurement of tumor burden in this work. Briefly, genomic DNA from mouse organs was extracted using Proteinase K digestion followed by ethanol precipitation. Each biological sample was then amplified in technical triplicate on a StepOnePlus real-time PCR system (Thermo Fisher Scientific) for Luc2 and Ptger2 fragments. On each plate, serial dilutions of B16F0-Luc2 or YUMM1.7-Luc2 genomic DNA were used for Luc2 and Ptger2 fragment amplification to create standard curves for the calculation of relative Luc2 DNA and total mouse DNA. Microsoft Excel 2013 was used to establish gene amplification standard curves (Ct versus log DNA) for Luc2 and Ptger2. The relative Luc2 DNA amount (QLuc2) and total mouse (Ptger2) DNA amount (Qmm) for each genomic DNA sample were then calculated. The Luc2 cell ratio is calculated as r = QLuc2/Qmm. r is presented as the Luc2 cell number in 104 tissue (lung, liver, etc.) cells.
RNA isolation and gene expression analysis by real-time qRT-PCR
All samples for RNA analysis were prepared in biological triplicates. Unless otherwise specified, all cells were plated on 6-well plates in complete growth medium for 48 h before they were harvested for gene expression analysis. Some samples were switched to SFM for additional indicated time before RNA extraction. Total RNA was isolated using the GeneJET RNA purification kit (Thermo Fisher Scientific) except for the invaded and uninvaded B16F10 cells in the transwell assay (Fig. 5A), from which RNA was extracted using the PicoPure RNA isolation kit (Thermo Fisher Scientific). 50–500 ng of each RNA was reverse-transcribed using the High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). Real-time quantitative RT-PCR was performed on a StepOnePlus real-time PCR system with Brilliant II SYBR Green qPCR master mix (Agilent Technologies, Santa Clara, CA). Glyceraldehyde-3-phosphate dehydrogenase served as the internal control for the reactions, and the normalized results were analyzed by GraphPad Prism version 5. The primer pairs for the indicated genes in this work were adopted from PrimerBank (76) and verified before assay use. The PrimerBank IDs will be provided upon request.
Western blot analysis
Whole-cell lysates for immunoblotting were obtained by extraction in ice-cold radioimmune precipitation buffer (no SDS) with protease and phosphatase inhibitor mixtures (Thermo Fisher Scientific). Proteins were normalized by a BCA assay (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific), separated by SDS-PAGE, transferred to Bio Trace polyvinylidene difluoride membrane (PALL Life Sciences, Pensacola, FL), probed by the indicated antibodies, revealed using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific), and detected with an Amersham Biosciences Imager 680 (GE Healthcare). The relative protein levels were analyzed using ImageJ (National Institutes of Health). Rabbit anti-WISP-1 (H-55, sc-25441) was from Santa Cruz Biotechnology, Inc. The other rabbit polyclonal antibodies were purchased from Cell Signaling Technology (Danvers, MA): anti-β-actin (13E5), anti-Snail (C15D3), anti-ZEB1 (D80D3), anti-N-cadherin (D4R1H), anti-phospho-Akt (Ser-473) (D9E), anti-Akt (pan) (C67E7), anti-phospho-p44/42 MAPK (Erk1/2) (Thr-202/Tyr-204) (D13.14.4E), and anti-p44/42 MAPK (Erk1/2) (137F5).
Cell treatment with conditioned medium, recombinant WISP1, and kinase inhibitors
To treat cells with conditioned medium containing overexpressed WISP1 for EMT gene stimulation (Fig. 5H), B16F10-KO1 cells were seated on 6-well plates for 24 h and grown in conditioned medium from Wisp1-knockout NIH3T3-KO cells for another 24 h. Thirty minutes before stimulation treatment, three groups of media were prepared. The first group was conditioned media from Wisp1-knockout NIH3T3-KO cells, with antibody isotype control (normal rat IgG, from Sigma-Aldrich) at a final concentration of 20 μg/ml. The second group was conditioned media from WISP1-overexpressed NIH3T3-mWisp1 cells, with the same antibody isotype control. The third group was conditioned media from WISP1-overexpressed NIH3T3-mWisp1 cells, with rat anti-Wisp1 (MAB1680, R&D Systems) at a final concentration of 20 μg/ml. All three groups of media were incubated at 37 °C for 30 min and then used to replace the media for B16F10-KO1 cells. The stimulation treatments were performed in biological triplicates for 3 h, and cells were harvested for RNA extraction and real-time qRT-PCR analysis.
Recombinant mouse WISP1 (rmWISP1, 1680-WS-050), produced in mouse myeloma NS0 cells, was from R&D Systems and was used at a final concentration of 5 μg/ml following the manufacturer's instructions. AKT inhibitor MK-2206 (final concentration 2.0 μg/ml) was from Sigma-Aldrich, and MEK inhibitor U0126 (final concentration 10 μm) was from Cell Signaling Technology (Danvers, MA). DMSO with the same volume was used for control cells. Unless otherwise specified, cell treatment for kinase immunoblot analysis was maintained for 30 min, whereas cell treatment for comparison of EMT marker gene expression was maintained for 3 h.
Data sets and statistical analysis
To compare melanoma with benign skin samples (55), the gene expression profiles of Affymetrix arrays (GSE3189) were downloaded from Gene Expression Omnibus, and WISP1 mRNA expression levels were compared. The protein abundance of WISP1 in primary melanoma and normal skin was quantified by immunohistological analysis using a tissue microarray derived from de-identified human skin tissue samples, as provided by the Human Protein Atlas (www.proteinatlas.org3 (77)) and in accordance with approval from the Uppsala University Hospital Ethics Committee. The tissue microarray analysis included samples from seven primary melanomas and three normal epithelial tissues that represented both male and female patients ranging in age from 46 to 87 years. The tissue microarrays were processed and analyzed as similarly described previously (53). In brief, processed tissue microarrays were probed using a rabbit polyclonal antibody against WISP1 (Sigma-Aldrich, catalog no. HPA007121, RRID: AB_1858844) that was validated by providing partly consistent staining patterns with previously reported gene/protein data, a weak band of the predicted size in Western blotting validation, and passing protein array validation tests. WISP1 staining was visualized using diaminobenzidine, and microscopic tissue features were visualized by counterstaining with Harris hematoxylin. Immunohistochemically stained tissue microarrays were scanned at ×20 resolution (1-mm diameter) and provided as an 8-bit RGB JPEG image. The average intensity of WISP1 staining per tissue sample was quantified by deconvoluting the intensity of WISP1 staining from nonspecific hematoxylin tissue staining in R using the EBImage package. Following color deconvolution, the image was segmented into tissue and nontissue regions. A tissue mask used for segmenting IHC images was determined based on nonzero staining in any of the RGB channels, following background image correction. To address whether more of the tissue microarray image stains positive for WISP1 (i.e. there are more cells that produce WISP1 within the tissue sample) but that the intensity of WISP1 staining is the same (i.e. WISP1 production per cell is not increased) in melanoma samples, we calculated the distribution in WISP1 staining intensity and the fraction of the total tissue area that stains strongly for WISP1. To compare the gene expression profiles of WISP1 in primary melanoma with overall survival, level 3 skin cutaneous melanoma RNAseqV2 mRNA expression results (FPKM-normalized) and clinical profiles for patients diagnosed with primary melanoma that had not metastasized (i.e. stage I–III) were obtained from the Cancer Genome Atlas. Individual statistical methods are indicated in the figure legend.
Unless specified, all analyses were performed with GraphPad Prism version 5. Individual quantitative results were shown as mean ± S.D. Box plots indicate median and interquartile range (box) and 5th and 95th percentiles (whiskers). Data sets were compared using the unpaired Student's t test (two-tailed) or one-way analysis of variance followed by Tukey's multiple-comparison ad hoc post-test. To estimate cumulative survival probability, Kaplan–Meier survival curves were estimated from the cohort overall survival data. Statistical significance associated with a difference in survival between two groups was estimated using the Peto and Peto modification of the Gehan–Wilcoxon test and the Cox proportional hazards regression model, as implemented in the R survival package. A p value of <0.05 was considered statistically significant. Asterisks are used to indicate the numerical value as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Author contributions
W. D. and D. J. K. conceptualized the study; W. D., A. F., and S. L. M. performed experiments; W. D., A. F., and D. J. K. analyzed data; and W. D. and D. J. K. drafted the original manuscript. All authors edited and approved of the final version of the manuscript.
Supplementary Material
Acknowledgments
Small animal imaging and image analysis were performed in the West Virginia University Animal Models and Imaging Facility, which has been supported by the West Virginia University Cancer Institute and National Institutes of Health Grants P20 RR016440 and P30RR032138/P30GM103488.
This work was supported by National Science Foundation Grant CBET-1644932 (to D. J. K.) and NCI, National Institutes of Health, Grant R01CA193473 (to D. J. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1–S9.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- EMT
- epithelial–mesenchymal transition
- EMT-TF
- EMT-inducing transcription factor
- qPCR and qRT-PCR
- quantitative PCR and quantitative RT-PCR, respectively
- TGF
- transforming growth factor
- CCN
- cellular communication network
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- PI3K
- phosphatidylinositol 3-kinase
- JNK
- c-Jun N-terminal kinase
- IHC
- immunohistochemistry
- FPKM
- fragments per kilobase of transcript per million mapped reads
- 2D and 3D
- two- and three-dimensional, respectively
- YUMM
- Yale University Mouse Melanoma
- MET
- mesenchymal–epithelial transition
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- SFM
- serum-free medium.
References
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Lambert A. W., Pattabiraman D. R., and Weinberg R. A. (2017) Emerging biological principles of metastasis. Cell 168, 670–691 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clark W. H. Jr., Elder D. E., Guerry D. 4th, Epstein M. N., Greene M. H., and Van Horn M. (1984) A study of tumor progression: the precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 15, 1147–1165 10.1016/S0046-8177(84)80310-X [DOI] [PubMed] [Google Scholar]
- 4. Damsky W. E., Theodosakis N., and Bosenberg M. (2014) Melanoma metastasis: new concepts and evolving paradigms. Oncogene 33, 2413–2422 10.1038/onc.2013.194 [DOI] [PubMed] [Google Scholar]
- 5. Vandamme N., and Berx G. (2014) Melanoma cells revive an embryonic transcriptional network to dictate phenotypic heterogeneity. Front. Oncol. 4, 352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li F. Z., Dhillon A. S., Anderson R. L., McArthur G., and Ferrao P. T. (2015) Phenotype switching in melanoma: implications for progression and therapy. Front. Oncol. 5, 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wels C., Joshi S., Koefinger P., Bergler H., and Schaider H. (2011) Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial–mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 131, 1877–1885 10.1038/jid.2011.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caramel J., Papadogeorgakis E., Hill L., Browne G. J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J. H., Ansieau S., and Tulchinsky E. (2013) A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 10.1016/j.ccr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 9. Denecker G., Vandamme N., Akay O., Koludrovic D., Taminau J., Lemeire K., Gheldof A., De Craene B., Van Gele M., Brochez L., Udupi G. M., Rafferty M., Balint B., Gallagher W. M., Ghanem G., et al. (2014) Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 21, 1250–1261 10.1038/cdd.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., and Weinberg R. A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 11. Webster M. R., Kugel C. H. 3rd, and Weeraratna A. T. (2015) The Wnts of change: how Wnts regulate phenotype switching in melanoma. Biochim. Biophys. Acta 1856, 244–251 10.1016/j.bbcan.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spranger S., and Gajewski T. F. (2016) Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology 5, e1086862 10.1080/2162402X.2015.1086862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dankort D., Curley D. P., Cartlidge R. A., Nelson B., Karnezis A. N., Damsky W. E. Jr., You M. J., DePinho R. A., McMahon M., and Bosenberg M. (2009) BrafV600E cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 41, 544–552 10.1038/ng.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Damsky W. E., Curley D. P., Santhanakrishnan M., Rosenbaum L. E., Platt J. T., Gould Rothberg B. E., Taketo M. M., Dankort D., Rimm D. L., McMahon M., and Bosenberg M. (2011) β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 20, 741–754 10.1016/j.ccr.2011.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spranger S., Bao R., and Gajewski T. F. (2015) Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 16. Kaur A., Webster M. R., Marchbank K., Behera R., Ndoye A., Kugel C. H. 3rd, Dang V. M., Appleton J., O'Connell M. P., Cheng P., Valiga A. A., Morissette R., McDonnell N. B., Ferrucci L., Kossenkov A. V., et al. (2016) sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 532, 250–254 10.1038/nature17392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Tape C. J., Ling S., Dimitriadi M., McMahon K. M., Worboys J. D., Leong H. S., Norrie I. C., Miller C. J., Poulogiannis G., Lauffenburger D. A., and Jørgensen C. (2016) Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165, 910–920 10.1016/j.cell.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto Y., Shindo-Okada N., Tani M., Nagamachi Y., Takeuchi K., Shiroishi T., Toma H., and Yokota J. (1998) Expression of the Elm1 gene, a novel gene of the CCN (connective tissue growth factor, Cyr61/Cef10, and neuroblastoma overexpressed gene) family, suppresses in vivo tumor growth and metastasis of K-1735 murine melanoma cells. J. Exp. Med. 187, 289–296 10.1084/jem.187.3.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pennica D., Swanson T. A., Welsh J. W., Roy M. A., Lawrence D. A., Lee J., Brush J., Taneyhill L. A., Deuel B., Lew M., Watanabe C., Cohen R. L., Melhem M. F., Finley G. G., Quirke P., et al. (1998) WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc. Natl. Acad. Sci. U.S.A. 95, 14717–14722 10.1073/pnas.95.25.14717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu L., Corcoran R. B., Welsh J. W., Pennica D., and Levine A. J. (2000) WISP-1 is a Wnt-1- and β-catenin-responsive oncogene. Genes Dev. 14, 585–595 [PMC free article] [PubMed] [Google Scholar]
- 21. Perbal B., Tweedie S., and Bruford E. (2018) The official unified nomenclature adopted by the HGNC calls for the use of the acronyms, CCN1–6, and discontinuation in the use of CYR61, CTGF, NOV and WISP 1–3 respectively. J. Cell Commun. Signal. 12, 625–629 10.1007/s12079-018-0491-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holbourn K. P., Acharya K. R., and Perbal B. (2008) The CCN family of proteins: structure–function relationships. Trends Biochem. Sci. 33, 461–473 10.1016/j.tibs.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau L. F. (2016) Cell surface receptors for CCN proteins. J. Cell Commun. Signal. 10, 121–127 10.1007/s12079-016-0324-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosanò L., Spinella F., and Bagnato A. (2013) Endothelin 1 in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 13, 637–651 10.1038/nrc3546 [DOI] [PubMed] [Google Scholar]
- 25. Su F., Overholtzer M., Besser D., and Levine A. J. (2002) WISP-1 attenuates p53-mediated apoptosis in response to DNA damage through activation of the Akt kinase. Genes Dev. 16, 46–57 10.1101/gad.942902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soon L. L., Yie T. A., Shvarts A., Levine A. J., Su F., and Tchou-Wong K. M. (2003) Overexpression of WISP-1 down-regulated motility and invasion of lung cancer cells through inhibition of Rac activation. J. Biol. Chem. 278, 11465–11470 10.1074/jbc.M210945200 [DOI] [PubMed] [Google Scholar]
- 27. Colston J. T., de la Rosa S. D., Koehler M., Gonzales K., Mestril R., Freeman G. L., Bailey S. R., and Chandrasekar B. (2007) Wnt-induced secreted protein-1 is a prohypertrophic and profibrotic growth factor. Am. J. Physiol. Heart Circ. Physiol. 293, H1839–H1846 10.1152/ajpheart.00428.2007 [DOI] [PubMed] [Google Scholar]
- 28. Inkson C. A., Ono M., Kuznetsov S. A., Fisher L. W., Robey P. G., and Young M. F. (2008) TGF-β1 and WISP-1/CCN-4 can regulate each other's activity to cooperatively control osteoblast function. J. Cell. Biochem. 104, 1865–1878 10.1002/jcb.21754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Venkatesan B., Prabhu S. D., Venkatachalam K., Mummidi S., Valente A. J., Clark R. A., Delafontaine P., and Chandrasekar B. (2010) WNT1-inducible signaling pathway protein-1 activates diverse cell survival pathways and blocks doxorubicin-induced cardiomyocyte death. Cell. Signal. 22, 809–820 10.1016/j.cellsig.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hou C.-H., Chiang Y.-C., Fong Y.-C., and Tang C.-H. (2011) WISP-1 increases MMP-2 expression and cell motility in human chondrosarcoma cells. Biochem. Pharmacol. 81, 1286–1295 10.1016/j.bcp.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 31. Ono M., Inkson C. A., Kilts T. M., and Young M. F. (2011) WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J. Bone Miner. Res. 26, 193–208 10.1002/jbmr.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang S., Chong Z. Z., Shang Y. C., and Maiese K. (2012) Wnt1 inducible signaling pathway protein 1 (WISP1) blocks neurodegeneration through phosphoinositide 3 kinase/Akt1 and apoptotic mitochondrial signaling involving Bad, Bax, Bim, and Bcl-xL. Curr. Neurovasc. Res. 9, 20–31 10.2174/156720212799297137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chuang J. Y., Chang A. C., Chiang I. P., Tsai M. H., and Tang C. H. (2013) Apoptosis signal-regulating kinase 1 is involved in WISP-1-promoted cell motility in human oral squamous cell carcinoma cells. PLoS One 8, e78022 10.1371/journal.pone.0078022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu C.-L., Tsai H.-C., Chen Z.-W., Wu C.-M., Li T.-M., Fong Y.-C., and Tang C.-H. (2013) Ras activation mediates WISP-1-induced increases in cell motility and matrix metalloproteinase expression in human osteosarcoma. Cell. Signal. 25, 2812–2822 10.1016/j.cellsig.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 35. Stephens S., Palmer J., Konstantinova I., Pearce A., Jarai G., and Day E. (2015) A functional analysis of Wnt inducible signalling pathway protein-1 (WISP-1/CCN4). J. Cell Commun. Signal. 9, 63–72 10.1007/s12079-015-0267-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang H., Luo H., Hu Z., Peng J., Jiang Z., Song T., Wu B., Yue J., Zhou R., Xie R., Chen T., and Wu S. (2015) Targeting WISP1 to sensitize esophageal squamous cell carcinoma to irradiation. Oncotarget 6, 6218–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ono M., Masaki A., Maeda A., Kilts T. M., Hara E. S., Komori T., Pham H., Kuboki T., and Young M. F. (2018) CCN4/WISP1 controls cutaneous wound healing by modulating proliferation, migration and ECM expression in dermal fibroblasts via α5β1 and TNFα. Matrix Biol. 68, 533–546 10.1016/j.matbio.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berschneider B., and Königshoff M. (2011) WNT1 inducible signaling pathway protein 1 (WISP1): a novel mediator linking development and disease. Int. J. Biochem. Cell Biol. 43, 306–309 10.1016/j.biocel.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 39. Gurbuz I., and Chiquet-Ehrismann R. (2015) CCN4/WISP1 (WNT1 inducible signaling pathway protein 1): a focus on its role in cancer. Int. J. Biochem. Cell Biol. 62, 142–146 10.1016/j.biocel.2015.03.007 [DOI] [PubMed] [Google Scholar]
- 40. Königshoff M., Kramer M., Balsara N., Wilhelm J., Amarie O. V., Jahn A., Rose F., Fink L., Seeger W., Schaefer L., Günther A., and Eickelberg O. (2009) WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J. Clin. Invest. 119, 772–787 10.1172/JCI33950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heise R. L., Stober V., Cheluvaraju C., Hollingsworth J. W., and Garantziotis S. (2011) Mechanical stretch induces epithelial-mesenchymal transition in alveolar epithelia via hyaluronan activation of innate immunity. J. Biol. Chem. 286, 17435–17444 10.1074/jbc.M110.137273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia S., Qu T., Feng M., Ji K., Li Z., Jiang W., and Ji J. (2017) Association of Wnt1-inducible signaling pathway protein-1 with the proliferation, migration and invasion in gastric cancer cells. Tumor Biol. 39, 1010428317699755 10.1177/1010428317699755 [DOI] [PubMed] [Google Scholar]
- 43. Chiang K.-C., Yeh C.-N., Chung L.-C., Feng T.-H., Sun C.-C., Chen M.-F., Jan Y.-Y., Yeh T.-S., Chen S.-C., and Juang H.-H. (2015) WNT-1 inducible signaling pathway protein-1 enhances growth and tumorigenesis in human breast cancer. Sci. Rep. 5, 8686 10.1038/srep08686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jing D., Zhang Q., Yu H., Zhao Y., and Shen L. (2017) Identification of WISP1 as a novel oncogene in glioblastoma. Int. J. Oncol. 51, 1261–1270 10.3892/ijo.2017.4119 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka S., Sugimachi K., Kameyama T., Maehara S., Shirabe K., Shimada M., Wands J. R., and Maehara Y. (2003) Human WISP1v, a member of the CCN family, is associated with invasive cholangiocarcinoma. Hepatology 37, 1122–1129 10.1053/jhep.2003.50187 [DOI] [PubMed] [Google Scholar]
- 46. Chuang J.-Y., Chen P.-C., Tsao C.-W., Chang A.-C., Lein M.-Y., Lin C.-C., Wang S.-W., Lin C.-W., and Tang C.-H. (2015) WISP-1, a novel angiogenic regulator of the CCN family, promotes oral squamous cell carcinoma angiogenesis through VEGF-A expression. Oncotarget 6, 4239–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu S., Liu H., Lu L., Wan H., Lin Z., Qian K., Yao X., Chen Q., Liu W., Yan J., and Liu Z. (2016) WISP1 overexpression promotes proliferation and migration of human vascular smooth muscle cells via AKT signaling pathway. Eur. J. Pharmacol. 788, 90–97 10.1016/j.ejphar.2016.06.027 [DOI] [PubMed] [Google Scholar]
- 48. Liu Y. D., Ji C. B., Li S. B., Yan F., Gu Q. S., Balic J. J., Yu L., and Li J. K. (2018) Toll-like receptor 2 stimulation promotes colorectal cancer cell growth via PI3K/Akt and NF-κB signaling pathways. Int. Immunopharmacol. 59, 375–383 10.1016/j.intimp.2018.04.033 [DOI] [PubMed] [Google Scholar]
- 49. Nagai Y., Watanabe M., Ishikawa S., Karashima R., Kurashige J., Iwagami S., Iwatsuki M., Baba Y., Imamura Y., Hayashi N., and Baba H. (2011) Clinical significance of Wnt-induced secreted protein-1 (WISP-1/CCN4) in esophageal squamous cell carcinoma. Anticancer Res. 31, 991–997 [PubMed] [Google Scholar]
- 50. Shao H., Cai L., Grichnik J. M., Livingstone A. S., Velazquez O. C., and Liu Z.-J. (2011) Activation of Notch1 signaling in stromal fibroblasts inhibits melanoma growth by upregulating WISP-1. Oncogene 30, 4316–4326 10.1038/onc.2011.142 [DOI] [PubMed] [Google Scholar]
- 51. Shao H., Cai L., Moller M., Issac B., Zhang L., Owyong M., Moscowitz A. E., Vazquez-Padron R., Radtke F., and Liu Z.-J. (2016) Notch1-WISP-1 axis determines the regulatory role of mesenchymal stem cell-derived stromal fibroblasts in melanoma metastasis. Oncotarget 7, 79262–79273 10.18632/oncotarget.13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kulkarni Y. M., Chambers E., McGray A. J. R., Ware J. S., Bramson J. L., and Klinke D. J. 2nd (2012) A quantitative systems approach to identify paracrine mechanisms that locally suppress immune response to interleukin-12 in the B16 melanoma model. Integr. Biol. 4, 925–936 10.1039/c2ib20053h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klinke D. J., 2nd (2014) Induction of Wnt-inducible signaling protein-1 correlates with invasive breast cancer oncogenesis and reduced type 1 cell-mediated cytotoxic immunity: a retrospective study. PLoS Comput. Biol. 10, e1003409 10.1371/journal.pcbi.1003409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klinke D. J. 2nd, Horvath N., Cuppett V., Wu Y., Deng W., and Kanj R. (2015) Interlocked positive and negative feedback network motifs regulate β-catenin activity in the adherens junction pathway. Mol. Biol. Cell. 26, 4135–4148 10.1091/mbc.E15-02-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Talantov D., Mazumder A., Yu J. X., Briggs T., Jiang Y., Backus J., Atkins D., and Wang Y. (2005) Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin. Cancer Res. 11, 7234–7242 10.1158/1078-0432.CCR-05-0683 [DOI] [PubMed] [Google Scholar]
- 56. Carvajal R. D., Antonescu C. R., Wolchok J. D., Chapman P. B., Roman R. A., Teitcher J., Panageas K. S., Busam K. J., Chmielowski B., Lutzky J., Pavlick A. C., Fusco A., Cane L., Takebe N., Vemula S., et al. (2011) KIT as a therapeutic target in metastatic melanoma. JAMA 305, 2327–2334 10.1001/jama.2011.746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aladowicz E., Ferro L., Vitali G. C., Venditti E., Fornasari L., and Lanfrancone L. (2013) Molecular networks in melanoma invasion and metastasis. Future Oncol. 9, 713–726 10.2217/fon.13.9 [DOI] [PubMed] [Google Scholar]
- 58. Lee J. T., and Herlyn M. (2007) Microenvironmental influences in melanoma progression. J. Cell. Biochem. 101, 862–872 10.1002/jcb.21204 [DOI] [PubMed] [Google Scholar]
- 59. Zhou L., Yang K., Andl T., Wickett R. R., and Zhang Y. (2015) Perspective of targeting cancer-associated fibroblasts in melanoma. J. Cancer 6, 717–726 10.7150/jca.10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deng W., McLaughlin S. L., and Klinke D. J. (2017) Quantifying spontaneous metastasis in a syngeneic mouse melanoma model using real time PCR. Analyst 142, 2945–2953 10.1039/C7AN00623C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herlyn M., and Fukunaga-Kalabis M. (2010) What is a good model for melanoma? J. Invest. Dermatol. 130, 911–912 10.1038/jid.2009.441 [DOI] [PubMed] [Google Scholar]
- 62. Meeth K., Wang J. X., Micevic G., Damsky W., and Bosenberg M. W. (2016) The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 29, 590–597 10.1111/pcmr.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. French D. M., Kaul R. J., D'Souza A. L., Crowley C. W., Bao M., Frantz G. D., Filvaroff E. H., and Desnoyers L. (2004) WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am. J. Pathol. 165, 855–867 10.1016/S0002-9440(10)63348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maeda A., Ono M., Holmbeck K., Li L., Kilts T. M., Kram V., Noonan M. L., Yoshioka Y., McNerny E. M. B., Tantillo M. A., Kohn D. H., Lyons K. M., Robey P. G., and Young M. F. (2015) WNT1-induced secreted protein-1 (WISP1), a novel regulator of bone turnover and Wnt signaling. J. Biol. Chem. 290, 14004–14018 10.1074/jbc.M114.628818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoshioka Y., Ono M., Maeda A., Kilts T. M., Hara E. S., Khattab H., Ueda J., Aoyama E., Oohashi T., Takigawa M., Young M. F., and Kuboki T. (2016) CCN4/WISP-1 positively regulates chondrogenesis by controlling TGF-β3 function. Bone 83, 162–170 10.1016/j.bone.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schlegelmilch K., Keller A., Zehe V., Hondke S., Schilling T., Jakob F., Klein-Hitpass L., and Schütze N. (2014) WISP 1 is an important survival factor in human mesenchymal stromal cells. Gene 551, 243–254 10.1016/j.gene.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 67. Schäfer M., and Werner S. (2008) Cancer as an overhealing wound: an old hypothesis revisited. Nat. Rev. Mol. Cell Biol. 9, 628–638 10.1038/nrm2455 [DOI] [PubMed] [Google Scholar]
- 68. Liu Z. J., Xiao M., Balint K., Smalley K. S. M., Brafford P., Qiu R., Pinnix C. C., Li X., and Herlyn M. (2006) Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 66, 4182–4190 10.1158/0008-5472.CAN-05-3589 [DOI] [PubMed] [Google Scholar]
- 69. Bedogni B. (2014) Notch signaling in melanoma: interacting pathways and stromal influences that enhance Notch targeting. Pigment Cell Melanoma Res. 27, 162–168 10.1111/pcmr.12194 [DOI] [PubMed] [Google Scholar]
- 70. Howard J. D., Moriarty W. F., Park J., Riedy K., Panova I. P., Chung C. H., Suh K. Y., Levchenko A., and Alani R. M. (2013) Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment Cell Melanoma Res. 26, 697–707 10.1111/pcmr.12131 [DOI] [PubMed] [Google Scholar]
- 71. Golan T., Messer A. R., Amitai-Lange A., Melamed Z., Ohana R., Bell R. E., Kapitansky O., Lerman G., Greenberger S., Khaled M., Amar N., Albrengues J., Gaggioli C., Gonen P., Tabach Y., Sprinzak D., Shalom-Feuerstein R., and Levy C. (2015) Interactions of melanoma cells with distal keratinocytes trigger metastasis via Notch signaling inhibition of MITF. Mol. Cell. 59, 664–676 10.1016/j.molcel.2015.06.028 [DOI] [PubMed] [Google Scholar]
- 72. Kahn M. (2014) Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 10.1038/nrd4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang X., and Hao J. (2015) Development of anticancer agents targeting the Wnt/β-catenin signaling. Am. J. Cancer Res. 5, 2344–2360 [PMC free article] [PubMed] [Google Scholar]
- 74. Xue G., Romano E., Massi D., and Mandalà M. (2016) Wnt/β-catenin signaling in melanoma: preclinical rationale and novel therapeutic insights. Cancer Treat. Rev. 49, 1–12 10.1016/j.ctrv.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 75. Deng W., Vanderbilt D. B., Lin C.-C., Martin K. H., Brundage K. M., and Ruppert J. M. (2015) SOX9 inhibits β-TrCP-mediated protein degradation to promote nuclear GLI1 expression and cancer stem cell properties. J. Cell Sci. 128, 1123–1138 10.1242/jcs.162164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Spandidos A., Wang X., Wang H., and Seed B. (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 10.1093/nar/gkp1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., et al. (2015) Proteomics: Tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.