Abstract
In all eukaryotes, a functional mitotic spindle is essential for distributing duplicated chromosomes into daughter cells. Mitotic spindle assembly involves highly ordered arrangement of microtubules (MTs). The Augmin protein complex recruits γ-tubulin ring complex (γ-TuRC) to MTs and thereby promotes MT-based MT nucleation and mitotic spindle assembly. However, several factors that may promote Augmin recruitment to MTs remain unknown. Here, we show that echinoderm microtubule-associated protein-like 3 (EML3), an MT-associated protein, facilitates binding between MTs and Augmin/γ-TuRC and recruiting the latter to MTs for proper mitotic spindle assembly and kinetochore–MT connections. Using immunofluorescence microscopy, live-cell imaging, and immunoprecipitation assays, we found that EML3 recruits Augmin/γ-TuRC to the MTs to enhance MT-based MT nucleation in both spindle and small acentrosomal asters. We also noted that the EML3-mediated recruitment is controlled by cyclin-dependent kinase 1 (CDK1), which phosphorylated EML3 at Thr-881 and promoted its binding to Augmin/γ-TuRC. RNAi-mediated EML3 knockdown in HeLa cells reduced spindle localization of Augmin/γ-TuRC, which resulted in abnormal spindle assembly and caused kinetochore–MT misconnection. The introduction of exogenous WT or a Thr-881 phosphorylation mimic EML3 variant into the EML3 knockdown cells restored normal Augmin/γ-TuRC localization and spindle assembly. The EML3 knockdown also affected the spindle assembly checkpoint, delaying chromosome congression and cell division. Taken together, our results indicate that EML3 regulates mitotic spindle assembly and the kinetochore–MT connection by regulating MT-based MT nucleation and recruiting Augmin/γ-TuRC to MTs.
Keywords: mitotic spindle, microtubule, microtubule-associated protein (MAP), checkpoint control, mitosis, cell division, cytoskeleton, Augmin, chromosome congression, EML3, microtubule nucleation, spindle assembly
Introduction
The distribution of duplicated chromosomes into daughter cells requires a functional mitotic spindle in all eukaryotes. The assembly of mitotic spindle involves a highly ordered arrangement of MT2 arrays in association with many MT-associated proteins (MAPs) such as TPX2 (1), NuMA (2), TACC3 (3, 4), and MCRS1 (5), which regulate MT nucleation, function, and dynamics (1, 6–9). Work in different model organisms has shown that MT nucleation takes place at multiple sites. In higher eukaryotes, these MT arrays emanate primarily from the main MT organization center, the centrosomes. During mitotic spindle assembly, the γ-tubulin ring complex (γ-TuRC) anchors to the centrosomes to initiate centrosome-based MT nucleation under the regulation of many cell cycle regulators, including kinases CDK1 and Plk1 and Auroras A/B (10–15). In addition to the centrosomes, many small acentrosomal MT asters formed adjacent to the chromosomes also contribute to bipolar spindle formation in mammalian somatic cells (4, 16). In plants and animal oocytes that have no equivalent centrosomes, many small MT organization centers with unclear structural features are responsible for organizing MT arrays by regulating the assembly of small acentrosomal MT asters. These small asters interact with each other, connect with the chromosomes, and gradually organize to form the bipolar spindle (17, 18).
MT nucleation also occurs on existing spindle body MTs to contribute to mitotic spindle assembly (19–21). In this MT-based MT nucleation process, the multi-subunit protein complex Augmin recruits γ-TuRC to the spindle MTs to initiate daughter MT nucleation from the existing mother MTs at a low branch angle and at the same polarity as the mother MTs (22–24). Human Augmin consists of eight subunits, and reconstitution of the complex with recombinant proteins has shown that Augmin has a Y-shaped configuration and can adopt multiple conformations (25). Hice1 and hDgt6 are core components of the complex and can bind MTs in vitro (25–27). Using Xenopus egg extracts, it has been shown that MT-based MT nucleation is stimulated by Ran-GTP and its co-effector, TPX2 (22). However, whether other factors regulate Augmin recruitment to the MTs for MT-based MT nucleation remains unknown.
EML3 (echinoderm MT-associated protein-like protein 3) is a MAP that is required for correct chromosome alignment in metaphase (28); however, the underlying mechanism is unknown. In this work, we found that EML3 regulates the MT-based MT nucleation for proper MT density in the mitotic spindle body in mammalian cells. We reveal that EML3 recruits Augmin and γ-TuRC to existing MTs in a CDK1 phosphorylation-dependent manner to initiate MT-based MT nucleation. EML3 RNAi knockdown in cells leads to the reduction of spindle-localized Augmin and γ-TuRC, a decrease in MT density in the spindle body, and chromosome congression failure. Taken together, our data reveal a novel mechanism of how EML3 regulates mitotic spindle assembly and the kinetochore–MT connection via recruitment of Augmin and γ-TuRC to MT for MT-based MT nucleation.
Results
EML3 recruits Augmin and γ-TuRC complex to spindle MTs
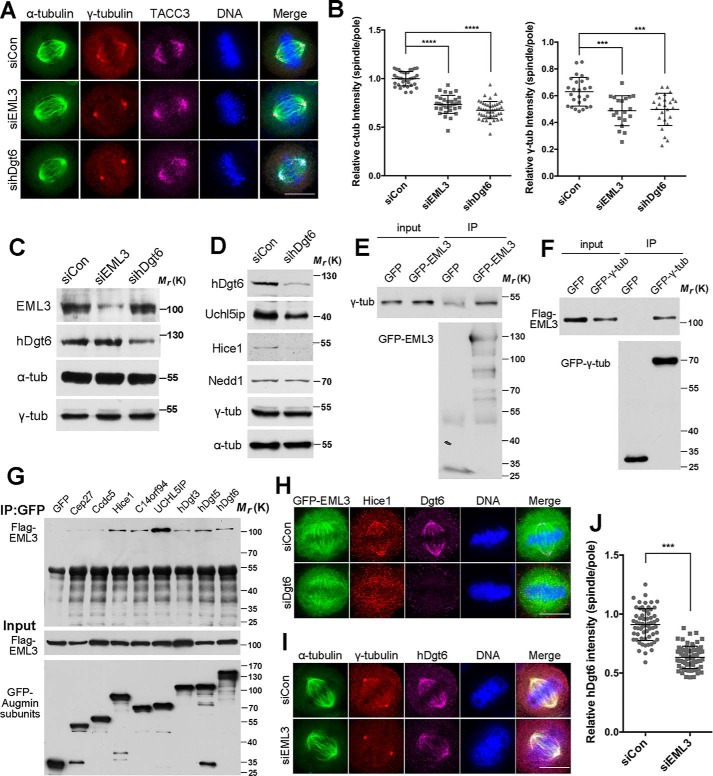
First, to reveal the functions of EML3 in mitosis, we performed siRNA knockdown experiments in HeLa cells (Fig. 1, A–C) and investigated the effects of EML3 knockdown on mitotic spindle dynamics. Through immunofluorescence microscopy (IFM), we observed that EML3 knockdown resulted in chromosome misalignment and MT density decrease (Fig. 1, A and B) as measured according to the reports (Fig. S1B) (19, 21). Because γ-TuRC nucleates MTs at both the centrosomes and the spindle MTs, and loss of γ-TuRC on the spindle MTs results in reduced MT density within the spindle body (10), we immunostained cells with a specific antibody against γ-tubulin, the main component of γ-TuRC. The results showed that the γ-tubulin signal on the spindle body MTs was significantly reduced in EML3 knockdown cells compared with the control (Fig. 1, A and B). Meanwhile, the microtubule-associated protein TACC3 was retained on the spindle (Fig. 1A). We also performed time-lapse microscopy in EML3 knockdown cells and found that, unlike the dramatic reduction of the spindle body MTs in mitosis, the situation of MTs in interphase was not significantly changed (Fig. S1A and Movie S1). As several reports have shown that Augmin recruits γ-TuRC to the MT lattice to take part in MT amplification within the spindle body in different cell types (19, 21, 29, 30), we performed siRNA knockdown of hDgt6, one of the core Augmin subunits, to investigate the correlations between EML3 and Augmin. Interestingly, we observed a MT density reduction in hDgt6 knockdown cells similar to that found in EML3 knockdown cells (Fig. 1, A–C). In addition, we found that hDgt6 knockdown led to reduced protein levels of Uchil5ip and Hice1 (two subunits of Augmin), whereas the protein expression of Nedd1 and γ-tubulin (γ-TuRC components) was not affected (Fig. 1D), which indicated that hDgt6 knockdown may substitute knockdown of the whole Augmin complex. Then, we constructed a GFP-tagged full-length EML3 expression vector (GFP-EML3) for the first time by joining an N-terminal truncate (EML3 aa1–116) cloned from a cDNA library and an EML3 truncate (EML3 aa116–896, a kind gift from Dr. Gruss) and expressed it in HeLa cells. The results show that GFP-EML3 associated with the spindle body MTs but not the poles in mitosis and additionally with the interphase MTs during the cell cycle (Fig. S1C). We also observed that Flag-EML3 co-localized with hDgt6 on the mitotic spindle (Fig. S1D). Through co-expression of Flag-EML3 with GFP-tagged Augmin subunits in HEK293 cells followed by co-IP, we found that EML3 interacted with multiple subunits of Augmin (Fig. 1G). Through co-IP assays, we also showed that EML3 and γ-tubulin were co-immunoprecipitated with each other (Fig. 1, E and F). IFM revealed that EML3 and Augmin subunits were co-localized on the spindle body MTs during mitosis (Fig. S1E). Because of the absence of centrosomal localization of EML3, we speculated that EML3 bound with Augmin and γ-TuRC only on the mitotic spindle body MTs. By knocking down hDgt6 in GFP-EML3–expressing cells, we found that although hDgt6 knockdown led to dissociation of Hice1 from the spindle body MTs, this knockdown had little effect on the spindle body MT localization of EML3 (Fig. 1H). This suggested that EML3 situates between Augmin and the spindle body MTs and that EML3 can localize alone to the spindle body MTs without Augmin. To confirm this proposition, we immunostained hDgt6 and γ-tubulin in EML3 knockdown cells, revealing that EML3 knockdown significantly reduced hDgt6 and γ-tubulin signals on spindle body MTs and the MT density of the spindle body (Fig. 1, I and J). This demonstrated that EML3 mediates the localization of Augmin and γ-TuRC to the spindle body MTs. Taking these findings together, we propose that EML3 functions upstream of Augmin and γ-TuRC and participates in MT-based MT nucleation of the spindle body through recruitment of Augmin and γ-TuRC to spindle MTs.
Figure 1.
EML3 recruits Augmin and γ-TuRC complex to spindle MTs. A, EML3 knockdown cells were analyzed by IFM. Scale bar, 10 μm. Note that the α-tubulin and γ-tubulin signals on spindle MTs decreased following EML3 knockdown. B, quantification of the relative γ-tubulin and α-tubulin as shown in Fig. S1B. Error bars, S.D. The number of control, EML3 RNAi, and hDgt6 RNAi cells from three independent experiments is 32, 30, and 35, respectively. C, EML3 knockdown efficiency in HeLa cells was analyzed by Western blotting. D, hDgt6 knockdown in HeLa cells was analyzed by Western blotting. E, co-immunoprecipitation assay of exogenous EML3 with endogenous γ-tubulin. HEK293 cells transiently expressing GFP or GFP-EML3 were subjected to an immunoprecipitation (IP) assay using an anti-GFP antibody. F, HEK293 cells co-expressing Flag-EML3 with GFP or GFP-γ-tubulin were subjected to an immunoprecipitation assay using an anti-GFP antibody. G, HEK293 cells co-transfected with Flag-EML3, and GFP-tagged Augmin subunits were arrested in mitosis with nocodazole. The cell lysates were immunoprecipitated using an anti-GFP antibody. H, HeLa cells were co-transfected with the indicated siRNA with GFP-EML3 for 48 h followed by immunofluorescence labeling using anti-Hice1 and anti-hDgt6 antibodies. Scale bar, 10 μm. I, HeLa cells were transfected with the indicated siRNA for 72 h followed by immunofluorescence labeling using anti-γ-tubulin and anti-hDgt6 antibodies. Scale bar, 10 μm. Note that the hDgt6 signal on spindle MTs decreased following knockdown of EML3 at the protein level. J, analysis of the relative hDgt6 intensity on spindles and poles shown in H. Error bars, S.D. The number of control and EML3 RNAi cells was 62 and 71, respectively. Three independent experiments were performed. On all graphs, significance was assessed by performing t tests. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Fig. S1.
EML3 promotes MT amplification within the spindle body
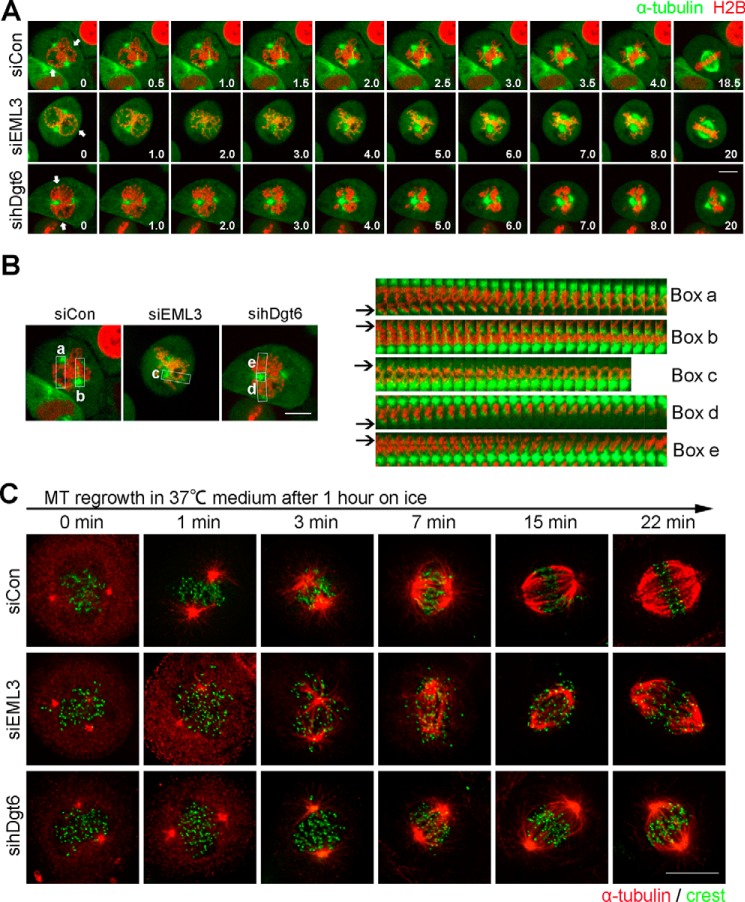
In mammalian cells, Augmin recruits γ-TuRC to spindle MTs to initiate daughter MTs at the same polarity as mother MTs (22–24). Because daughter MTs can also serve as mother MTs, Augmin-dependent MT nucleation can rapidly generate fan-shaped MT arrays that interact and fuse to form a “plump” mitotic spindle (22–24). To confirm the EML3 function in mitotic spindle assembly, we performed time-lapse microscopy using a cell line stably expressing GFP-α-tubulin (Fig. 2A). In control cells we observed that, in addition to the large centrosomal MT aster assembly around the centrosomes, many small acentrosomal MT asters also assembled around the disassembling nucleus and grew quickly during mitotic entry (Fig. 2A and Movie S2, marked by arrows). After nuclear envelope breakdown (NEBD), these small acentrosomal MT asters were connected with the condensed chromosomes and quickly sorted into large MT asters to form the bipolar spindle (Fig. 2, A and B, boxes a and b). Importantly, we observed that the MTs within the spindle increased dramatically and were sorted into the bipolar spindle until a normal high spindle body MT density was achieved (Fig. 2A and Movie S2). In contrast, in EML3 knockdown cells, we observed a significant reduction in MT density in the spindle body and a decrease in the growth rate of the small acentrosomal MT asters (Fig. 2, A and B, box c, and Movie S3). Most likely because the MT density of the spindle body was much lower, the chromosome alignment was obviously abnormal in EML3 knockdown cells (Fig. 2A). Consistently, in hDgt6 knockdown cells we also observed very similar results to that in EML3 knockdown cells (Fig. 2, A and B, boxes d and e, and Movie S4). We also performed a MT regrowth assay in HeLa cells to verify the role of EML3 in regulating mitotic spindle assembly. The cells were treated on ice for 1 h to depolymerize their spindle MTs followed by release into a warm medium to allow MT regrowth. We observed that whereas MTs in control cells rapidly grew to reform the bipolar spindle with proper chromosome alignment, the growth of the MTs in EML3 or hDgt6 knockdown cells showed a significant reduction, resulting in an obvious decrease in both spindle reformation efficiency and spindle body MT density (Fig. 2C). More importantly, we revealed that the chromosomes did not align well on the equatorial plate in EML3 knockdown cells (Fig. 2C). Together, these results show that EML3 is required for rapid MT amplification, efficient spindle assembly, and proper chromosome alignment during mitosis.
Figure 2.
EML3 promotes MT amplification within the spindle body. A, EML3 knockdown inhibited MT amplification. RFP-H2B–expressing HeLa cells were co-transfected with GFP-α-tubulin and the indicated siRNA. Then, the cells were subjected to live-cell imaging. Images were captured every 5 s with 4 slices of 4-μm Z-section thickness in total. Scale bar, 10 μm. B, acentrosomal MT nucleation and sorting analysis of live cells with EML3 or Dgt6 knockdown as described in A. High-magnification images of the MT nucleation and sorting regions in the upper panels (boxes a–e) were captured at 10 s/frame and assembled into the lower panels. Arrows indicate the MT nucleation and sorting regions. C, control, EML3, or hDgt6 knockdown cells were treated on ice for 1 h and then released into fresh medium at 37 °C. Cells were fixed at the indicated times and stained with α-tubulin (red) and Crest (green) antibodies. Scale bar, 10 μm.
EML3 also regulates small acentrosomal MT aster formation through Augmin and γ-TuRC
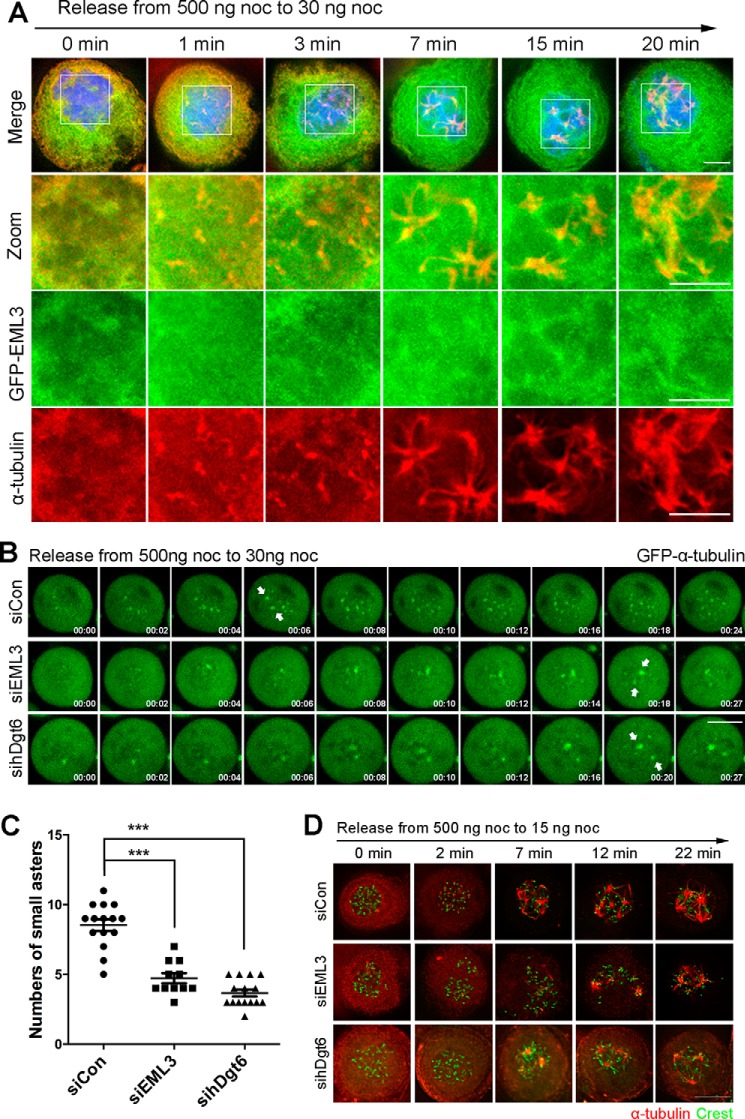
In addition to the big centrosomal MT asters, the small acentrosomal MT asters also contribute to spindle assembly through binding and sorting into the big centrosomal MT asters (4). To verify whether EML3 also regulates the small acentrosomal MT aster formation, we treated cells with 500 ng/ml nocodazole for 2 h to disassemble their MTs followed by releasing these cells into fresh medium containing a low concentration of nocodazole (30 ng/ml) to induce the acentrosomal MT nucleation (4). We revealed that many small acentrosomal MT asters were assembled in the control cells and that EML3 and Augmin subunits associated with these small acentrosomal MT asters (Figs. 3A and S2A). We also observed that, whereas the centrosome-based MT nucleation was partially inhibited in the presence of this low concentration of nocodazole, the acentrosomal MT nucleation was induced, and these nucleated MTs rapidly assembled into many small acentrosomal asters (Fig. 3, A and B, and Movie S5). In contrast, in EML3 or hDgt6 knockdown cells we found that the numbers of small acentrosomal MT asters were significant reduced (Fig. 3, B and C, and Movies S6 and S7). Through immunostaining of the fixed cells, we also observed the acentrosomal microtubule assembly, and the results were consistent with that in the live cells (Fig. 3D). Through immunostaining for γ-tubulin, Nedd1, and TACC3, we revealed that, although γ-tubulin and Nedd1 localized mainly to the aster centers, most TACC3 were situated on the aster MTs (Fig. S2B). Immunoprecipitation assay showed that the Augmin subunit Hice1 bound with EML3 and γ-tubulin but not with TACC3 (Fig. S2C). These results were consistent with our previous reports that γ-tubulin and Nedd1 take parts in MT nucleation, whereas TACC3 and short MTs bind each other to form small TACC3-MT seeds near the kinetochores along with the assembly of the big centrosomal asters (3, 4, 13). Taken together, these results indicate that both EML3 and Augmin also regulate MT nucleation of the small acentrosomal MT asters.
Figure 3.
EML3 also regulates small acentrosomal MT aster formation through Augmin and γ-TuRC. A, HeLa cells transiently expressing GFP-EML3 were arrested with 500 ng/ml nocodazole and then released into medium with 30 ng/ml nocodazole followed by fixation at the indicated times. The DNA was stained with DAPI (blue). Scale bar, 5 μm. Note that many small acentrosomal MT asters were organized and EML3 was associated with them. B, EML3 is required for acentrosomal MT aster formation. HeLa cells expressing GFP-α-tubulin were transfected with the indicated siRNA and treated with 500 ng/ml nocodazole for 2 h. Then, the treated cells were released into medium with 30 ng/ml nocodazole followed by live-cell imaging. The positions of centrosome-dependent MT asters are marked by arrows. Images were captured every 1 min with 4 slices of 4-μm Z-section thickness in total. Scale bar, 10 μm. C, quantification of the number of small acentrosomal MT asters when cells were released at the time point of 24 min (shown in B). ***, p < 0.001. D, HeLa cells were transfected with the indicated siRNA and treated with 500 ng/ml nocodazole for 2 h. Then, the treated cells were released into medium with 30 ng/ml nocodazole and fixed at the indicated time. See also Fig. S2.
EML3-regulated acentrosomal MT nucleation contributes to kinetochore–MT connection
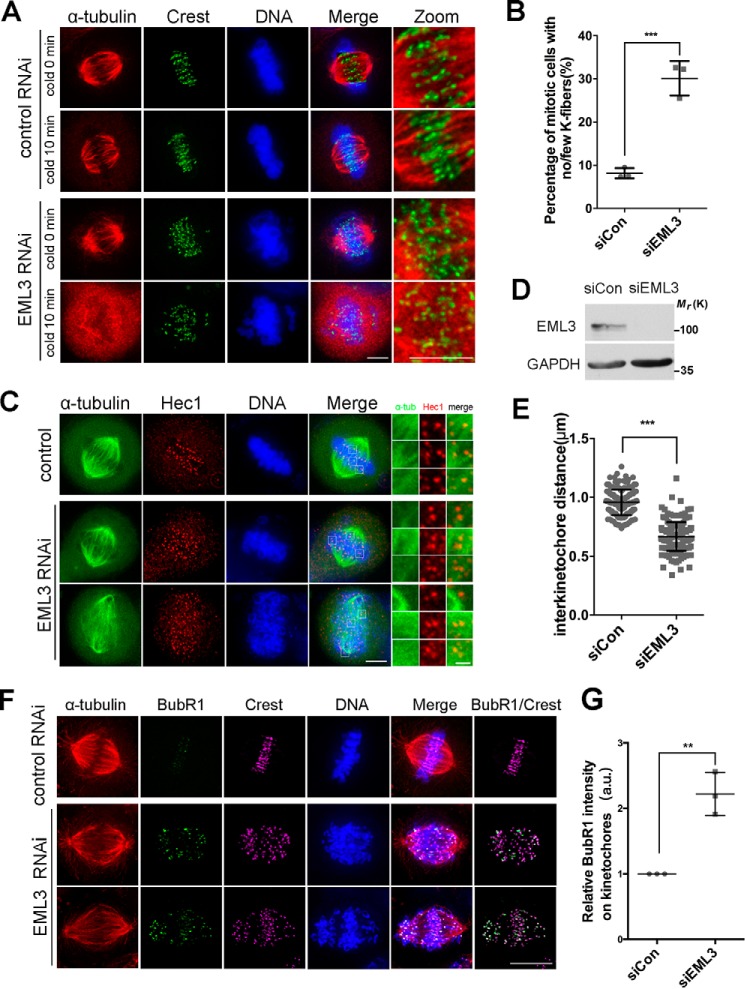
Proper kinetochore–MT connection is essential for accurate chromosome congression and segregation (31, 32). We have demonstrated previously that the small acentrosomal MT asters capture kinetochores and contribute to kinetochore–MT fiber formation (4). Here, we further investigated whether EML3 contributes to the kinetochore–MT connection during mitosis. First, HeLa cells were transfected with control or EML3 siRNA, treated with MG132 to achieve fully assembled bipolar spindles, and placed on ice to depolymerize non-K-fiber MTs (33). With fluorescence microscopy we observed that, under this condition, the intact end-on attached K-fibers were stably preserved in normal control cells; and in contrast, few intact MTs were stably preserved after the cold treatment in EML3 knockdown cells (Fig. 4, A and B), suggesting that EML3 knockdown abolished the end-on attachment between MTs and kinetochores during the mitotic spindle assembly. Consistently, we uncovered that EML3 knockdown also resulted in a remarkable decrease in mean inter-kinetochore distance with 0.957 ± 0.008 μm in the control and 0.668 ± 0.009 μm in EML3 knockdown cells (Fig. 4, C–E), indicating that EML3 was also required for maintenance of proper inter-kinetochore tension.
Figure 4.
EML3-regulated acentrosomal MT nucleation contributes to kinetochore–MT connection. A, HeLa cells transfected with control or EML3 siRNA for 72 h were treated with ice for 10 min to depolymerize non-K-fiber MTs and processed for IFM using anti-α-tubulin and anti-Crest antibodies. The DNA was stained by DAPI. Scale bar, 5 μm. The connection areas of kinetochores and MTs are magnified in the rightmost panels (Zoom). Note that EML3 knockdown reduced the K-fibers, resulting in significant chromosome misalignment. B, quantification of the percentage of mitotic cells with no/few K-fibers. Error bars, S.D. 100 cells were counted in each sample from three independent experiments. C, GFP-α-tubulin–expressing HeLa cells with EML3 knockdown were immunostained with Hec1 (red) antibodies. Scale bar, 5 μm. The box areas were zoomed into the right three columns; scale bar, 1 μm. D, detection of EML3 RNAi knockdown efficiency in GFP-α-tubulin HeLa cells by Western blotting. E, measurement of interkinetochore (labeled by Hec1) distances. Error bars, S.D.; at least 50 pairs of kinetochores from 5 cells were measured in each of the three independent experiments. F, knockdown of EML3 by siRNA in HeLa cells led to activation of the spindle assembly checkpoint. HeLa cells with siRNA knockdown of EML3 were stained with anti-α-tubulin (red), anti-BubR1 (green), and anti-Crest (magenta). Scale bar, 10 μm. Note that EML3 knockdown led to chromosome misalignment and BubR1-positive staining. G, analysis of BubR1 localization on kinetochores. Three independent experiments were performed, and at least 50 pairs of kinetochores in different cells were analyzed in each sample. Error bars, S.D. The average cytoplasmic immunofluorescence intensity was subtracted as background. **, p < 0.01; ***, p < 0.001. See also Fig. S3.
To understand the underlying mechanism, we stained the cells with a specific antibody against the spindle checkpoint protein BubR1. The results showed that BubR1 was maintained at the kinetochores in EML3 knockdown cells (Fig. 4, F and G), suggesting that a proper stable connection between kinetochores and spindle MTs was not established. Accordingly, the numbers of cells showing misaligned chromosomes near the spindle poles was significantly higher in EML3 knockdown (40.67%) than in the control knockdown samples (∼8%) (Fig. S3, A and B). This result indicated that chromosome congression problems existed in EML3 knockdown cells.
We also tested whether the localization of CENP-E is abnormal in EML3 knockdown cells, because the chromosome congression at the metaphase plate also depends on the plus end-directed motor protein CENP-E (34), which also functions in maintaining the spindle assembly checkpoint (35, 36). The results showed that CENP-E could still localize to the un-congressed chromosome kinetochores in EML3 knockdown cells (Fig. S3A), suggesting that the spindle assembly checkpoint was still engaged. We also carried out live-cell imaging, and revealed abnormal spindle assembly and chromosome misalignment in EML3 knockdown cells. Although the mean time required for NEBD to achieve proper chromosome alignment was 30.28 ± 1.90 min in control RNAi cells, this time was significantly prolonged to 169.3 ± 12.54 min in EML3 knockdown cells (Fig. S3, C and D and Movies S8 and S9).
Taking all above findings together, we conclude that EML3-regulated MT-based MT nucleation on both small acentrosomal and large centrosomal MT asters contributes to the spindle body MT density and the kinetochore–MT attachment during mitotic spindle assembly and chromosome congression.
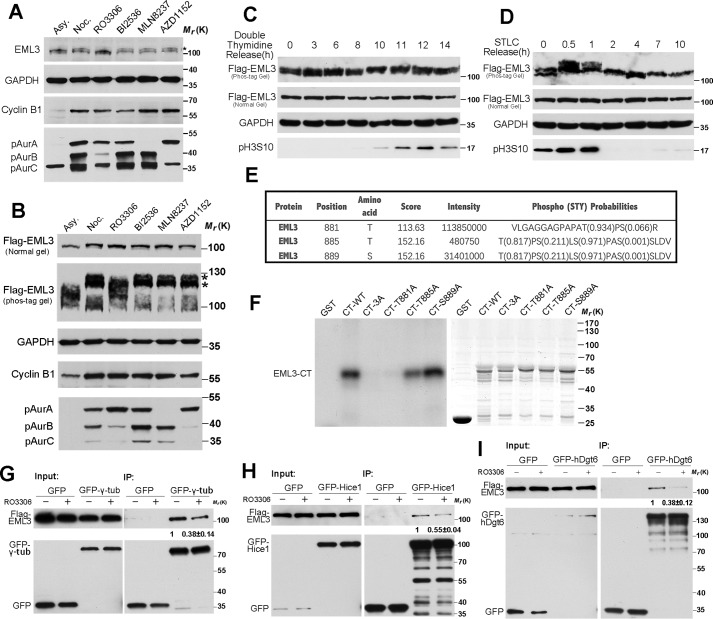
CDK1-mediated phosphorylation of EML3 is required for the binding with Augmin and γ-TuRC
To investigate how the function of EML3 is regulated, we screened its posttranslational modifications. First, through Western blot analysis using an antibody against an EML3, we showed that this antibody recognized a clear band at ∼100 kDa in interphase, and this band was up-shifted in mitosis (Fig. 5A), indicating that EML3 was post-translationally modified during mitotic entry. Unfortunately, this antibody is not suitable for IFM. Then, we constructed and expressed Flag-EML3 in HeLa cells, followed by SDS-PAGE and Western blot analysis. The result also showed that the exogenous EML3 was up-shifted in mitosis (Fig. 5B). Through treating mitotic cells with a number of mitotic kinase inhibitors in combination with MG132, which inhibits proteasome activity and arrests cells at metaphase, we revealed that treatment with the specific inhibitor RO3306 for kinase CDK1 significantly reduced the higher molecular weight band of endogenous EML3 or Flag-EML3 (Fig. 5, A and B); in contrast, Plk1 kinase inhibition by BI2536 or Aurora kinases A/B inhibition by MLN8237/AZD1152 did not down-shift the EML3 band (Fig. 5, A and B). These results indicated that EML3 might be phosphorylated by CDK1 in mitosis. To verify this, we expressed and separated Flag-EML3 on a Phos-tag gel, which specifically retards phosphoproteins. Through Western blot analysis, we revealed that the majority of EML3 proteins in nocodazole-arrested cell lysate were in up-shifted bands, and the up-shifted bands were remarkably reduced when CDK1 was inhibited (Fig. 5B). Through a kinase assay, we also revealed that EML3 proteins could be phosphorylated by CDK1-cyclin B1 in vitro (Fig. S4A). Taken together, these results demonstrate that EML3 is phosphorylated by CDK1 kinase in mitosis.
Figure 5.
CDK1-mediated phosphorylation of EML3 is required for binding with Augmin and γ-TuRC. A, HeLa cells were arrested with nocodazole (Noc.) for 17 h and then treated with selected kinase inhibitors (9 μm RO3306 for 15 min, 0.25 μm MLN8237 for 30 min, 0.1 μm AZD1152 for 30 min, and 0.1 μm BI2536 for 30 min, respectively). Asy, asynchronized cell samples. Samples were analyzed on normal SDS-PAGE and immunoblotted with the indicated antibodies. B, HeLa cells transfected with Flag-EML3 were treated by the same method as described in A. Samples were analyzed by normal SDS-PAGE and Phos-tag acrylamide gels. C, HeLa cells transfected with Flag-EML3 were blocked at G1/S by double thymidine treatment and then released and harvested at the indicated time point. D, HeLa cells transfected with Flag-EML3 were blocked at prometaphase by STLC treatment and then released and harvested at the indicated time point. E, mitotic cells arrested by nocodazole were processed for MS analysis. Three amino acids, Thr-881, Thr-885, and Ser-889, that are consistent with the CDK1 phosphorylation consensus sequence were found to be phosphorylated. F, GST-tagged EML3 C-terminal truncated proteins with point mutations were subjected to CDK1/cyclin B kinase assay in vitro followed by autoradiography (left). Coomassie Blue (right) staining shows the loading of the GST-tagged EML3 mutant proteins in the reactions. The experiment was performed three times independently. G—I, inhibition of the kinase activity of CDK1 disrupted binding of EML3 with γ-tubulin/Hice1/hDgt6. Mitotic HEK293 cells transfected with GFP or GFP-γ-tubulin/hDgt6/Hice1 plasmids were treated with 9 μm RO3306 or DMSO as a control for 20 min followed by a co-IP assay with GFP-Trap beads and Western blot analysis using anti-Flag or anti-GFP antibodies. The experiment was performed twice independently. The protein–protein interaction discrepancy was quantified by ImageJ. The value of the immunoprecipitated Flag-EML3 band was quantified by ImageJ and then divided by the value of immunoprecipitated GFP-γ-tubulin/hDgt6/Hice1 band. The ratio of the control group was set as 1, and then the ratio of RO3306-treated group was calculated. See also Fig. S4.
Next, we collected cells from different phases of the cell cycle to see the phosphorylation state of EML3. We expressed Flag-EML3 in HeLa cells, then blocked the cells at G1/S phase by double thymidine treatment, released the cells and collected samples at different time point. Through Phos-tag gel and Western blot analysis, we found that a portion of EML3 proteins were in up-shifted bands when cells were released to 10 and 11 h (Fig. 5C), which suggested that EML3 might be phosphorylated during mitosis (indicated by Histone H3pS10 antibody). To further observe the phosphorylation state of EML3 in mitosis, we harvested prometaphase cells by STLC (an inhibitor of Eg5) block and released them into fresh medium. The results showed that the majority of EML3 proteins in mitotic cell lysate were in up-shifted bands. And the up-shifted bands of EML3 were remarkably reduced when cells exit mitosis (Fig. 5D). To identify the phosphorylation site(s) of CDK1 on EML3, we expressed and immunoprecipitated GFP-EML3 from mitotic cell lysates using a GFP antibody, prepared the sample as described (37), and performed MS to identify the possible phosphorylation sites of EML3 by CDK1. The results revealed that Thr-881, Thr-885, and Ser-889, which are consistent with the consensus sequence (S/T*)P or (S/T*)PX(K/R) of CDK1 (38), were phosphorylated (Fig. 5E). The phosphorylation of Thr-881 in mitosis but not in G1/S phase was also found through a large scale MS screening (39). To verify this phosphorylation, we expressed and purified a long truncate of EML3 (aa 106-end) and performed an in vitro kinase assay. The results showed that this truncate was phosphorylated by CDK1 kinase in vitro (Fig. S4A). Then, we constructed the C-terminal EML3 truncate mutant proteins (aa 417-end) with single or triple point mutations and performed the in vitro kinase assay. The results showed that although EML3-CT-WT and mutants EML3-CT-T885A and EML3-CT-S889A were phosphorylated by CDK1 kinase, the mutants EML3-CT-3A and EML3-CT-T881A were not phosphorylated (Fig. 5F), indicating that EML3 was phosphorylated by CDK1 kinase at Thr-881. Then, we examined the function of EML3 phosphorylation by CDK1, which revealed that the binding of EML3 with γ-tubulin decreased to about 38% when cells co-expressing Flag-EML3 and GFP-γ-tubulin were treated with RO3306 compared with control cells (Fig. 5G). Similarly, in cells with GFP-hDgt6 or GFP-Hice1 expression, the binding of EML3 with hDgt6 or Hice1 was also hampered by the inhibition of CDK1 kinase activity (Fig. 5, H and I).
Collectively, these results demonstrate that EML3 is phosphorylated at Thr-881 in mitosis by CDK1, and this phosphorylation regulates its function via promoting its binding with Augmin and γ-TuRC.
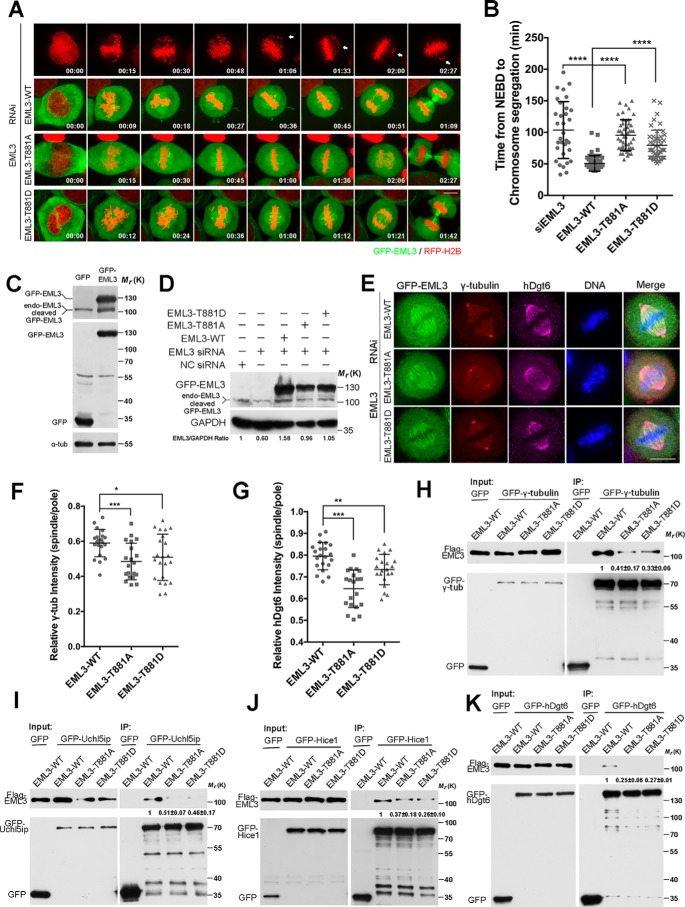
Thr-881 phosphorylation of EML3 is required for recruiting Augmin and γ-TuRC complex to spindle MTs
To study the regulation of EML3 function further, we performed an EML3 siRNA knockdown-and-rescue assay in HeLa cells using the siRNA-resistant mutants GFP-EML3-WT, GFP-EML3-T881A, and GFP-EML3-T881D (Fig. 6, A–D, and Movies S10–S13). An efficient expression of exogenous EML3 was confirmed by Western blot analysis, whereas the endogenous EML3 was knocked down by siRNA (Fig. 6, C and D). Then, through time-lapse microscopy, we found that the cells went through a severe chromosomal segregation upon EML3 knockdown (Fig. 6A and Movie S10), and GFP-tagged EML3-WT was able to successfully rescue the chromosomal segregation defects (Fig. 6, A and B, and Movie S11). In comparison, EML3-T881D was also able to rescue this defect, although it needed a bit longer time (Fig. 6, A and B, and Movie S13). In contrast, EML3-T881A required a much longer time to rescue this defect (Fig. 6, A and B, and Movie S12). Through immunostaining for Augmin and γ-tubulin in EML3 knockdown-and-rescue cells, we further tested how phosphorylation of EML3 at Thr-881 influences its function in stabilizing Augmin and γ-tubulin on spindle MTs. The results showed that whereas GFP-tagged EML-WT fully rescued the losses of hDgt6 and γ-tubulin on spindle MTs, and EML3-T881D largely rescued these losses, EML3-T881A was weak to rescue these losses (Fig. 1, A, B, H, and I, and Fig. 6, E–G). Co-IP assays also showed that, compared with EML3-WT, EML3-T881A showed defects in binding with γ-tubulin (Fig. 6H) and Augmin subunits (Fig. 6, I–K, and Fig. S4, B–F). Moreover, we tested the binding of EML3-WT and mutants with Augmin subunits and revealed that, except for the subunit ccdc5, the other seven Augmin subunits showed binding patterns consistent with EML3 (Fig. 6, I–K, and Fig. S4, B–F). Together, these results demonstrate that the phosphorylation of EML3 at Thr-881 by CDK1 not only regulates the binding of EML3 with Augmin and γ-TuRC but also regulates the recruitment of Augmin and γ-TuRC to MTs for MT-based MT nucleation, kinetochore–MT connection, and chromosome congression.
Figure 6.
Thr-881 phosphorylation of EML3 is required for the recruitment of Augmin and γ-TuRC complex to spindle MTs. A, RFP-H2B HeLa cells with EML3 siRNA knockdown were processed for rescue experiments with none, GFP-tagged EML3-WT, EML3-T881A, or EML3-T881D followed by live-cell imaging. Images were captured every 3 min with 4 slices of 6-μm Z-section thickness in total. Scale bar, 10 μm. B, quantification of the time from NEBD to chromosome segregation shown in A. Error bars, S.D. 32 cells without rescue, 55 cells rescued with EML3-WT, 47 cells rescued with EML3 T881A, and 47 cells rescued with EML3-T881D in three independent experiments were analyzed. C, GFP or GFP-EML3 expression. Note that GFP-EML3 was expressed efficiently, and a portion of it was cleaved. D, Western blot analysis of EML3 siRNA knockdown-and-rescue efficiency. Note that endogenous EML3 were efficiently knocked down and the siRNA-resistant EML3 and mutants were efficiently expressed, whereas a portion of the GFP-tagged EML3 was cleaved. E, HeLa cells were co-transfected with EML3 siRNA oligos and siRNA-resistant GFP-tagged EML3-WT, EML3-T881A, or EML3-T881D and then fixed and stained with anti-γ-tubulin and anti-hDgt6 antibodies. DNA was stained with DAPI. Scale bar, 10 μm. F and G, quantification of relative γ-tubulin (F) and hDgt6 intensity (G) on spindle bodies and spindle poles in E. Error bars, S.D. 21 cells co-transfected with EML3 siRNA oligos and siRNA-resistant GFP-tagged EML3-WT, 20 cells co-transfected with EML3 siRNA oligos and siRNA-resistant GFP-tagged EML3-T881A, and 21 cells co-transfected with EML3 siRNA oligos and siRNA-resistant GFP-tagged EML3-T881D in two independent experiments were analyzed. H—K, HEK293 cells co-transfected with the indicated GFP-tagged protein and Flag-tagged EML3 or mutants were arrested in mitosis with nocodazole and processed for IP assay with GFP-Trap beads and mouse anti-Flag antibodies. The experiment was performed twice independently. The protein–protein interaction discrepancy was quantified as described in the legend for Fig. 5. In all graphs, significance was assessed by performing t tests. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Fig. S4.
Discussion
In this work, we investigated underlying mechanism of the mitotic spindle assembly. We found that EML3 regulates mitotic spindle assembly, kinetochore–MT connection, and chromosome separation via recruitment of Augmin and γ-TuRC to the existing MTs and promoting MT-based MT nucleation. We reveal that EML3 is phosphorylated when the cell enters mitosis, and this phosphorylation regulates its binding with Augmin and γ-TuRC and their recruitment to the MTs. It has been reported that MT-based Augmin mediates MT-based MT nucleation through binding and recruitment of γ-TuRC (22–24). However, it is difficult to know whether Augmin and γ-TuRC bind MT tubulins or MAPs directly, although Augmin subunits Hice1 (26) and hDgt6 (27) associate with the MTs. Hice1 and hDgt6 proteins form a stable heterodimer, which then assembles with other subunits into a functional subcomplex (25). In the association of Augmin with the MTs, the phosphorylation of Hice1 by Plk1 (Polo-like kinase 1) is required (40). Here, we reveal that recruitment of Augmin to the MTs depends on MT-localized EML3 under the regulation of CDK1, which phosphorylates EML3 and enhances the binding of MT-localized EML3 with Augmin and γ-TuRC.
Although Augmin localizes to both the spindle body MT and the spindle pole, EML3 only localizes to and co-localizes with Augmin on the spindle body MTs of the mitotic spindle. EML3 knockdown remarkably reduces the intensity of Augmin on the spindle body but not on the centrosomal region, leading to abnormal assembly of the mitotic spindle that loses its plump MT density and stable MT-kinetochore attachment. Therefore, EML3 may be responsible for MT-based MT nucleation and recruiting Augmin to the MTs but not for the centrosome-based MT nucleation. Because knocking down EML3 or Augmin did not significantly influence the centrosome-based MT nucleation and the large centrosomal aster assembly, both EML3 and Augmin may not be essential for MT nucleation on the centrosomes.
In addition to spindle body MT-based MT nucleation contributing to the kinetochore–MT connection, the small acentrosomal aster MT nucleation also contributes to the kinetochore–MT connection and chromosome alignment (4, 41). It has been shown that the small acentrosomal aster forms spontaneously near the chromosomes, and TACC3 may be responsible for the initiation of the small acentrosomal aster formation (4). In this work, although we have revealed that the small acentrosomal aster could not form without EML3, we did not find EML3 binding with TACC3. As it is thought that TACC3 bundles MTs in combination with clathrin (3, 42), we suggest that EML3 initiates MT-based MT nucleation through recruitment of Augmin and γ-TuRC to the MTs, whereas TACC3, in cooperation with clathrin, bundles the MTs during spindle assembly and chromosome alignment.
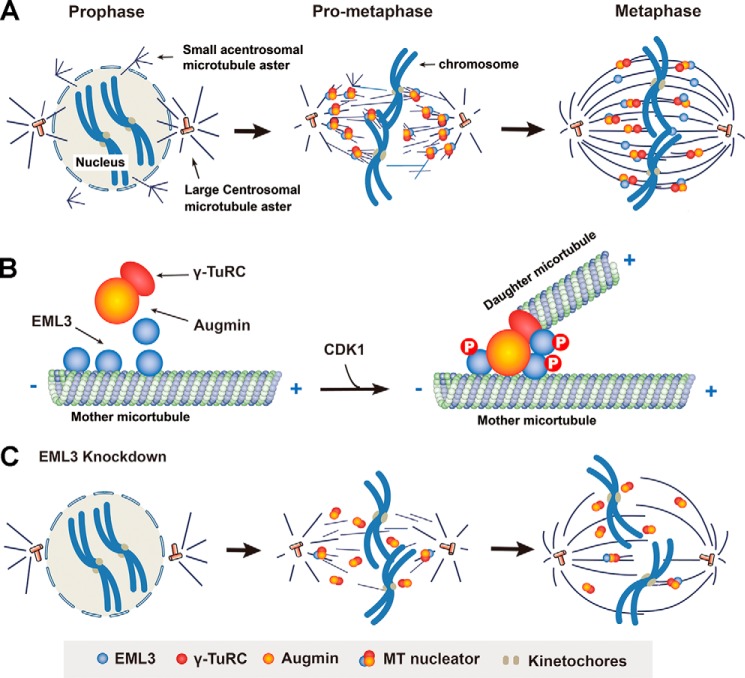
In summary, in this work we have revealed that EML3 regulates spindle assembly and kinetochore–MT connection. Based on our present data and previous reports, we propose a working model to illustrate EML3-regulated processes of the mitotic spindle assembly and kinetochore–MT connection under cell cycle control. During the G2–M phase transition, although the duplicated centrosomes separate from each other and induce MT nucleation around them to form two large centrosomal asters for bipolar mitotic spindle assembly, many small acentrosomal MT asters are organized around the disassembling nucleus. Along with the nuclear envelope breakdown in prophase, both the centrosomal and acentrosomal MTs penetrate into the nuclear area and catch the kinetochores on the chromosomes to establish the kinetochore–MT connection. Then, by generating more MTs on both the large centrosomal and the small acentrosomal asters, these MTs are joined, and the small asters are subsequently sorted into the large centrosomal asters to form the bipolar spindle with high MT density within the spindle body. Meanwhile, the MTs from both the small and large asters catch the kinetochores to establish proper kinetochore–MT attachment leading to accurate chromosome alignment and segregation (Fig. 7A). During this process, EML3, as a MAP, recruits Augmin and γ-TuRC to the existing MTs of both the large centrosomal and small acentrosomal asters in a CDK1 phosphorylation-dependent manner. Once recruited to the existing mother MTs, Augmin and γ-TuRC start to nucleate daughter MTs at an appropriate angle to their mother MTs (Fig. 7B). If EML3 is knocked down, the recruitment of γ-TuRC to the existing MTs is reduced accordingly, leading to a dramatic decrease in MT density of the spindle body and MT-kinetochore attachment defects and causing severe chromosome congression problems (Fig. 7C). In conclusion, in this work we show that EML3 regulates mitotic spindle assembly and kinetochore–MT connection for proper chromosome alignment through the recruitment of Augmin and γ-TuRC under the regulation of CDK1 to existing MTs of both small acentrosomal and large centrosomal asters and by regulating the MT-based MT nucleation.
Figure 7.
A working model for the role of EML3 in regulating mitotic spindle assembly and kinetochore–MT connection for chromosome congression. A, centrosomal and acentrosomal aster formation contribute to bipolar mitotic spindle assembly. Along with MT nucleation around the separating centrosomes to form the two large centrosomal asters, many small acentrosomal asters are also organized around the disassembling nucleus. Once the nuclear envelope breaks down, the MTs from both large and small asters penetrate into the nuclear area and catch the kinetochores of the condensing chromosomes to establish the kinetochore–MT connection. Through sorting of the small acentrosomal asters into the large centrosomal asters and acentrosomal MT-based MT nucleation, a mature metaphase bipolar spindle with high MT density assembles. B, EML3 recruits Augmin and γ-TuRC to existing mother MTs for MT-based MT nucleation during the mitotic spindle assembly. When the cell enters mitosis, EML3 is phosphorylated by CDK1, and this phosphorylation promotes EML3 binding with Augmin and γ-TuRC, recruiting them to the existing MTs for daughter MT nucleation. C, when EML3 is knocked down or dysfunctions, the acentrosomal MT aster formation is blocked and the MT-based MT nucleation on both large centrosomal and small acentrosomal asters is impaired, leading to a decrease in spindle MT density and chromosomal misalignment.
Experimental procedures
Cell culture, transfection, and synchronization
HeLa and HEK293 cells were cultured in DMEM (Gibco) supplemented with 10% (v/v) bovine calf serum (CellMax) at 37 °C in a 5% CO2 atmosphere. For plasmid or siRNA transfection in HeLa cells, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's instructions. The following siRNA duplexes were used: siEML3–1, 5′-CCAGCUUCAUCACUCAUCUTT-3′; siEML3–2, 5′-UCACGCUCAUGAAGGUUCUTT-3′; and 5′-sihDgt6, CAGUUAAGCAGGUACGAAATT-3′ (Uehara et al. (21)). To synchronize to metaphase for immunofluorescence analysis, HeLa cells were treated with thymidine for 16–20 h, released for 9 h, and then incubated with 10 μm MG132 for 1 h to fully establish the bipolar spindle. To synchronize to mitosis for immunoprecipitation, HeLa or HEK293 cells were treated with thymidine for 20 h, released for 3 h, and then supplemented with 100 ng/ml nocodazole for another 10 h. To establish stable GFP-α-tubulin– or RFP-H2B–expressing cell lines, HeLa cells were transfected with GFP-α-tubulin or RFP-H2B plasmids, and single colonies were screened using 500 μg/ml G418.
Plasmid construction, protein purification, and antibody preparation
The full-length EML3 gene was overlapped with a cDNA library (from the J. Han laboratory, Xiamen University, China) and an EML3 truncate (EML3–106-896 from the Oliver J. Gruss laboratory, Zentrum für Molekulare Biologie der Universität Heidelberg, Germany). Augmin subunits were cloned from the cDNA library. Point mutation and truncation mutants of EML3 were generated by PCR with specific primers. To obtain recombinant proteins, EML3 and hDgt6 truncates were inserted into the pGEX-4T-1 vector and expressed in Escherichia coli BL21 cells (DE3 pLys). GST-tagged proteins were affinity-purified using GSH–Sepharose 4B beads (GE Healthcare). EML3 and hDgt6 polyclonal antibodies were obtained by immunizing mice (Balb/c) with the recombinant proteins EML3-aa293–584 and hDgt6-aa448–955, respectively. The EML3 antibody was found to be applicable for Western blotting but not immunostaining. The hDgt6 antibody showed excellent specificity in both Western blotting and immunostaining. The anti-GFP antibody used for immunoprecipitation was generated by immunizing rabbits with bacterially expressed recombinant His-tagged GFP. All animal experiments were performed according to the approved guidelines. Other antibodies used in this study were as follows: mouse anti-α-tubulin (Sigma, T9026), mouse anti-γ-tubulin (Sigma, T6557), rabbit anti-γ-tubulin (Sigma, T3559), rabbit anti-Hice1 (Sigma, SAB2700752), mouse anti-Flag (MBL, M185-3L), mouse anti-Hec1 (Abcam, ab3613), human anti-Crest (Antibodies Inc., 15-234-0001), rabbit anti-cyclin B1 (Santa Cruz Biotechnology, sc-25764), mouse anti-GAPDH (Proteintech, 60004-1-Ig), mouse anti-Nedd1 (Zhang et al. (13)), and rabbit anti-TACC3 (Santa Cruz Biotechnology, sc-22773). Alexa Fluor–conjugated secondary antibodies were all from Invitrogen (Life Technology).
IFM, live-cell imaging, and statistical analysis
Cells were grown on coverslips and fixed in precooled methanol for 5 min on ice followed by incubation with primary antibodies (diluted in PBS containing 3% BSA) overnight at 4 °C. After three washes in PBS, the cells were incubated with secondary antibodies for 1 h at room temperature. Coverslips were mounted by Mowiol containing 1 μg/ml DAPI and analyzed on a DeltaVision imaging system (Applied Precision) equipped with an Olympus IX-71 inverted microscope and 100×/1.4 N.A. oil objective lens. The images were captured by a CoolSnap HQ2 CCD camera. All immunofluorescence images were captured with a 6-μm Z-section thickness by 6 slices and processed for maximum intensity projection. To determine the γ-tubulin distribution ratio of spindle MTs and the spindle pole, the fluorescence intensity was measured using Volocity software. All statistical analysis was performed in GraphPad Prism 6 software.
For live-cell imaging, GFP-α-tubulin or RFP-H2B stably expressing cells were plated on a glass-bottom dish. Before imaging, the dishes were locked in a heated chamber (37 °C) supplemented with 5% CO2. Images were acquired using a microscope (ECLIPSE Ti, Nikon) equipped with a 60×/1.4 N.A. oil objective lens an EM CCD (Hamamatsu Photonics, Inc.) followed by processing in Volocity software.
Immunoprecipitation
Mitotic HeLa cells transfected with the indicated constructs were shaken off the dish and lysed on ice in lysis buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EGTA, 0.5 mm EDTA, 0.5% NP-40, 5 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and protease mixture inhibitor) for 30 min. Lysates were centrifuged at 15,000 × g for 15 min, and supernatants were incubated with beads conjugated with GFP antibodies (4 μg/sample) or GFP-Trap beads (Chromo Tek, gtc-20) for 2 h at 4 °C. After five washes with lysis buffer, the beads were suspended in gel sample buffer, and the bound proteins were analyzed by Western blotting.
Phos-tag gel electrophoresis and in vitro kinase assays
For kinase inhibitor assays, mitotic cells arrested by nocodazole were treated with the indicated kinase inhibitors before harvest. Kinase inhibitor treatments used in this study include the following: 9 μm RO3306 for 15 min, 0.25 μm MLN8237 for 30 min, 0.2 μm AZD1152 for 30 min, and 0.2 μm BI2536 for 30 min. To analyze phosphorylation by Phos-tag gel, cells were collected and rinsed with TBS (20 mm Tris, pH 7.4, 150 mm NaCl). Protein samples were analyzed on 8% SDS-PAGE containing 25 mm Phos-tag acrylamide (Wako, Japan). After electrophoresis, the gels were soaked in transfer buffer with 1 mm EDTA for 10 min, followed by transfer buffer without EDTA for another 10 min before wet transfer.
For in vitro kinase assays, 2 μg of GST-tagged EML3 truncates or point mutants were incubated with 50 ng of human recombinant CDK1-cyclin B (New England Biolabs) for 30 min at 30 °C in kinase buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2 and 10 mm DTT) plus 100 μm ATP and 6000 Ci/mmol γ-[32P]ATP. Reactions were quenched with SDS sample buffer and analyzed by SDS-PAGE and autoradiography.
Author contributions
J. L., Q. J., and C. Z. conceptualization; J. L., B. Y., G. X., M. S., B. Z., X. G., and Q. J. data curation; J. L. and C. Z. writing-original draft; J. L., Q. J., and C. Z. writing-review and editing; Q. J. project administration; C. Z. resources; C. Z. supervision; C. Z. writing final manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Olive Gruss (University of Bonn) for EML3 truncate plasmids and Dr. Jiahuai Han (University of Xiamen) for the Augmin cDNA library, Dr. Si Li for help with drawing the model figure, and Drs. Hongxia Lv and Xiaochen Li and other colleagues at the Core Facilities at the College of Life Sciences, Peking University, for their help with live-cell imaging and data analysis.
This work was supported by the National Natural Science Foundation of China and the State Key Basic Research and Development Plan of the Ministry of Science and Technology of China (Grants 2016YFA0100501, 31430051, 2016YFA0500201, and 31520103906). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Movies S1–S13.
- MT
- microtubule
- MAP
- MT-associated protein
- γ-TuRC
- γ-tubulin ring complex
- IFM
- immunofluorescence microscopy
- co-IP
- co-immunoprecipitation
- NEBD
- nuclear envelope breakdown
- DAPI
- 4′,6-diamidino-2-phenylindole
- aa
- amino acid.
References
- 1. Walczak C. E., and Heald R. (2008) Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265, 111–158 10.1016/S0074-7696(07)65003-7 [DOI] [PubMed] [Google Scholar]
- 2. Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., and Weis K. (2001) Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- 3. Fu W., Tao W., Zheng P., Fu J., Bian M., Jiang Q., Clarke P. R., and Zhang C. (2010) Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123, 3645–3651 10.1242/jcs.075911 [DOI] [PubMed] [Google Scholar]
- 4. Fu W., Chen H., Wang G., Luo J., Deng Z., Xin G., Xu N., Guo X., Lei J., Jiang Q., and Zhang C. (2013) Self-assembly and sorting of acentrosomal microtubules by TACC3 facilitate kinetochore capture during the mitotic spindle assembly. Proc. Natl. Acad. Sci. U.S.A. 110, 15295–15300 10.1073/pnas.1312382110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meunier S., and Vernos I. (2011) K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 13, 1406–1414 10.1038/ncb2372 [DOI] [PubMed] [Google Scholar]
- 6. Helmke K. J., Heald R., and Wilbur J. D. (2013) Interplay between spindle architecture and function. Int. Rev. Cell Mol. Biol. 306, 83–125 10.1016/B978-0-12-407694-5.00003-1 [DOI] [PubMed] [Google Scholar]
- 7. Sauer G., Körner R., Hanisch A., Ries A., Nigg E. A., and Silljé H. H. (2005) Proteome analysis of the human mitotic spindle. Mol. Cell Proteomics 4, 35–43 10.1074/mcp.M400158-MCP200 [DOI] [PubMed] [Google Scholar]
- 8. Nousiainen M., Silljé H. H., Sauer G., Nigg E. A., and Körner R. (2006) Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U.S.A. 103, 5391–5396 10.1073/pnas.0507066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malik R., Lenobel R., Santamaria A., Ries A., Nigg E. A., and Körner R. (2009) Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8, 4553–4563 10.1021/pr9003773 [DOI] [PubMed] [Google Scholar]
- 10. Lüders J., Patel U. K., and Stearns T. (2006) GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- 11. Takahashi M., Yamagiwa A., Nishimura T., Mukai H., and Ono Y. (2002) Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol. Biol. Cell 13, 3235–3245 10.1091/mbc.e02-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zimmerman W. C., Sillibourne J., Rosa J., and Doxsey S. J. (2004) Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657 10.1091/mbc.e03-11-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang X., Chen Q., Feng J., Hou J., Yang F., Liu J., Jiang Q., and Zhang C. (2009) Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the γTuRC to the centrosome. J. Cell Sci. 122, 2240–2251 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- 14. Wang G., Jiang Q., and Zhang C. (2014) The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell Sci. 127 4111–4122 [DOI] [PubMed] [Google Scholar]
- 15. Fu J., Bian M., Jiang Q., and Zhang C. (2007) Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 5, 1–10 10.1158/1541-7786.MCR-06-0208 [DOI] [PubMed] [Google Scholar]
- 16. Tulu U. S., Rusan N. M., and Wadsworth P. (2003) Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr. Biol. 13, 1894–1899 10.1016/j.cub.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 17. Schuh M., and Ellenberg J. (2007) Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- 18. Masoud K., Herzog E., Chabouté M. E., and Schmit A. C. (2013) Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J. 75, 245–257 10.1111/tpj.12179 [DOI] [PubMed] [Google Scholar]
- 19. Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R. D. (2008) Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawo S., Bashkurov M., Mullin M., Ferreria M. G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J. R., Hegemann B., Pinchev D., Buchholz F., Peters J. M., Hyman A. A., Gingras A. C., and Pelletier L. (2009) HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- 21. Uehara R., Nozawa R. S., Tomioka A., Petry S., Vale R. D., Obuse C., and Goshima G. (2009) The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 6998–7003 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petry S., Groen A. C., Ishihara K., Mitchison T. J., and Vale R. D. (2013) Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777 10.1016/j.cell.2012.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamasaki T., O'Toole E., Kita S., Osumi M., Usukura J., McIntosh J. R., and Goshima G. (2013) Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 202, 25–33 10.1083/jcb.201304031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song J. G., King M. R., Zhang R., Kadzik R. S., Thawani A., and Petry S. (2018) Mechanism of how augmin directly targets the γ-tubulin ring complex to microtubules. J. Cell Biol., 217, 2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsia K. C., Wilson-Kubalek E. M., Dottore A., Hao Q., Tsai K. L., Forth S., Shimamoto Y., Milligan R. A., and Kapoor T. M. (2014) Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol. 16, 852–863 10.1038/ncb3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu G., Lin Y. T., Wei R., Chen Y., Shan Z., and Lee W. H. (2008) Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell Biol. 28, 3652–3662 10.1128/MCB.01923-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu H., Coppinger J. A., Jang C. Y., Yates J. R. 3rd, Fang G. (2008) FAM29A promotes microtubule amplification via recruitment of the NEDD1-γ-tubulin complex to the mitotic spindle. J. Cell Biol. 183, 835–848 10.1083/jcb.200807046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tegha-Dunghu J., Neumann B., Reber S., Krause R., Erfle H., Walter T., Held M., Rogers P., Hupfeld K., Ruppert T., Ellenberg J., and Gruss O. J. (2008) EML3 is a nuclear microtubule-binding protein required for the correct alignment of chromosomes in metaphase. J. Cell Sci. 121, 1718–1726 10.1242/jcs.019174 [DOI] [PubMed] [Google Scholar]
- 29. Petry S., Pugieux C., Nédélec F. J., and Vale R. D. (2011) Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. U.S.A. 108, 14473–14478 10.1073/pnas.1110412108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu T., Tian J., Wang G., Yu Y., Wang C., Ma Y., Zhang X., Xia G., Liu B., and Kong Z. (2014) Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 24, 2708–2713 10.1016/j.cub.2014.09.053 [DOI] [PubMed] [Google Scholar]
- 31. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., and Desai A. (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 32. Cheeseman I. M., and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 33. Meunier S., and Vernos I. (2012) Microtubule assembly during mitosis: From distinct origins to distinct functions? J. Cell Sci. 125, 2805–2814 10.1242/jcs.092429 [DOI] [PubMed] [Google Scholar]
- 34. Kapoor T. M., Lampson M. A., Hergert P., Cameron L., Cimini D., Salmon E. D., McEwen B. F., and Khodjakov A. (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weaver B. A., Bonday Z. Q., Putkey F. R., Kops G. J., Silk A. D., and Cleveland D. W. (2003) Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo Y., Kim C., Ahmad S., Zhang J., and Mao Y. (2012) CENP-E–dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J. Cell Biol. 198, 205–217 10.1083/jcb.201202152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu S., Cao Y., Fan S. B., Chen Z. L., Fang R. Q., He S. M., and Dong M. Q. (2018) Mapping disulfide bonds from sub-micrograms of purified proteins or micrograms of complex protein mixtures. Biophys. Rep. 4, 68–81 10.1007/s41048-018-0050-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johmura Y., Soung N. K., Park J. E., Yu L. R., Zhou M., Bang J. K., Kim B. Y., Veenstra T. D., Erikson R. L., and Lee K. S. (2011) Regulation of microtubule-based microtubule nucleation by mammalian Polo-like kinase 1. Proc. Natl. Acad. Sci. U.S.A. 108, 11446–11451 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu X. W., Luo J., Jiang Q., and Zhang C. M. (2016) Mitotic spindle assembly: The roles of microtubule nucleation and assembly, in eLS, Wiley Online Library, John Wiley & Sons, Ltd., Chichester, United Kingdom, 10.1002/9780470015902.a0022518 10.1002/9780470015902.a0022518 [DOI] [Google Scholar]
- 42. Fu W., Jiang Q., and Zhang C. (2011) Novel functions of endocytic player clathrin in mitosis. Cell Res. 21, 1655–1661 10.1038/cr.2011.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.