Abstract
Modification of anticodon nucleotides allows tRNAs to decode multiple codons, expanding the genetic code. Additionally, modifications located in the anticodon loop, but outside the anticodon itself, stabilize tRNA–codon interactions, increasing decoding fidelity. Anticodon loop nucleotide 37 is 3′ to the anticodon and, in , is methylated at the N1 position in its nucleobase (m1G37). The m1G37 modification in stabilizes its interaction with the codon and maintains the mRNA frame. However, it is unclear how m1G37 affects binding at the decoding center to both cognate and +1 slippery codons. Here, we show that the m1G37 modification is important for the association step during binding to a cognate CCG codon. In contrast, m1G37 prevented association with a slippery CCC-U or +1 codon. Similar analyses of frameshift suppressor tRNASufA6, a derivative containing an extra nucleotide in its anticodon loop that undergoes +1 frameshifting, reveal that m1G37 destabilizes interactions with both the cognate CCG and slippery codons. One reason for this destabilization is the disruption of a conserved U32·A38 nucleotide pairing in the anticodon stem through insertion of G37.5. Restoring the tRNASufA6 U32·A37.5 pairing results in a high-affinity association on the slippery CCC-U codon. Further, an X-ray crystal structure of the 70S ribosome bound to tRNASufA6 U32·A37.5 at 3.6 Å resolution shows a reordering of the anticodon loop consistent with the findings from the high-affinity measurements. Our results reveal how the tRNA modification at nucleotide 37 stabilizes interactions with the mRNA codon to preserve the mRNA frame.
Keywords: translation, ribosome, transfer RNA (tRNA), RNA modification, mRNA, structural biology, decoding, mRNA frame maintenance
Introduction
Protein synthesis is performed by the ribosome, a conserved protein–RNA macromolecular machine where mRNA, tRNAs, and translation factors read the genetic information as presented on mRNA into proteins. There are four defined stages of protein synthesis: initiation, elongation, termination, and recycling (reviewed in Ref. 1). During elongation, three nucleotides of the mRNA codon are read (or decoded) by three anticodon nucleotides of a tRNA in the ribosomal aminoacyl site (A site)2 on the small 30S subunit. The three-nucleotide code on the mRNA defines a single amino acid delivered by the corresponding tRNA. The regulation of the mRNA frame is critically important to maintain the correct sequential addition of amino acids to the nascent chain (2). Despite the importance of accurate protein expression for cell viability, the molecular basis for how the ribosome maintains this three-nucleotide mRNA frame is not well-understood.
Because tRNAs decode mRNAs, these RNA molecules probably play a role in mRNA frame maintenance. tRNAs are ∼76–90 nucleotides in length and adopt an L-shaped tertiary structure allowing them to fit into ribosome-binding sites that span both subunits (Fig. 1). tRNAs undergo extensive post-transcriptional modifications important for the correct tertiary fold of the tRNA, including the conformation of the anticodon stem-loop (ASL) (3). RNA modifications that are located in the anticodon and neighboring nucleotides in the ASL contribute to the accuracy and speed of translation (4, 5) by stabilizing the interactions between the anticodon and codon (3, 6–8). After decoding, these tRNA modifications are also important during translocation of the mRNA-tRNA pairs (9) and have also been implicated in mRNA frame maintenance (6).
Figure 1.
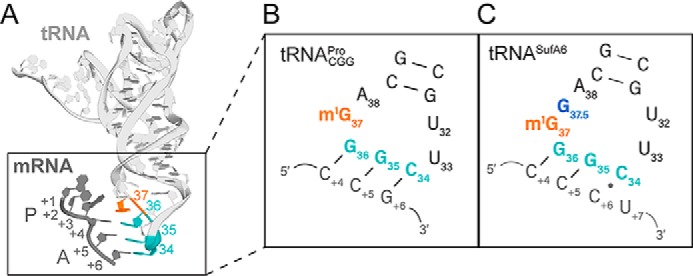
Frameshift suppressor tRNASufA6 is a derivative of . A, tertiary structure of tRNAPro with its anticodon depicted in cyan, anticodon loop nucleotide 37 in orange, and the mRNA in gray with the first P-site nucleotide denoted as +1; P-site codon nucleotides listed as +1, +2, and +3; and A-site codon nucleotides listed as +4, +5, and +6. B, secondary structure of the anticodon stem loop of tRNAPro bound to a cognate CCG codon containing three Watson–Crick base pairs. C, secondary structure of the anticodon stem loop of tRNASufA6 bound to a four-nucleotide slippery CCC-U codon and containing a C6-C34 wobble interaction. tRNASufA6 contains an extra guanosine between positions 37 and 38 (G37.5, blue).
The selection of the correct tRNA for each mRNA codon relies on the formation of Watson–Crick base pairs between the first two nucleotides of the codon and nucleotides 36 and 35 of the anticodon (Fig. 1A). The interaction between the third nucleotide of the codon and anticodon nucleotide 34 is not required to be Watson–Crick. Instead, a G·U wobble pair or a modified anticodon nucleotide 34–codon nucleotide pair can form. The modification of nucleotide 34 enables non-Watson–Crick interactions with the third nucleotide position of the mRNA that is accepted as cognate by the ribosome. The increased flexibility in codons that each tRNA can decode allows for the degeneracy of the genetic code where the 61 codons are decoded by fewer tRNAs (10). Therefore, tRNA modifications at nucleotide 34 have an important and essential role in the process of decoding.
Nucleotide 34 contains many diverse modifications that are typically required for accurate translation (11). Two examples include the uridine-5-oxyacetic acid (cmo5U34) in and the 5-methylaminomethyl-2-thiouridine (mnm5s2) U34 modification in . The cmo5U34 modification in stabilizes its interaction with C6 at the wobble position (12). The mnm5s2 U34 modification in allows for pairing with AAA or AAG codons (13, 14). However, a 6-threonylcarbamoyladenosine (t6A) or a 2-methylthio derivative (ms2t6A) at nucleotide 37 is required for recognition of the AAG codon (4, 7). Although all of these codon–anticodon pairings should be recognized by the ribosome, the instability of the anticodon loop of and, thus, its interactions with the AAA codon require both modifications at nucleotides 34 and 37. Nucleotide 37 of the tRNA is located 3′ to the anticodon, adjacent to the first position of the Watson–Crick base pair between the codon and anticodon nucleotide 36 (Fig. 1A). The codon–anticodon pairing between and its codon is weak in the absence of the modifications because of the three A-U base pairs and poor stacking of the UUU anticodon (15). Both the t6A and ms2t6A modifications contain planar heterocycle moieties that promote cross-strand stacking interactions between A38 of the tRNA and the first base in the mRNA codon to stabilize the codon–anticodon pairing (16). Because decoding relies on both the high-affinity binding of cognate tRNAs to the decoding center and conformational changes of the 30S known as domain closure (17–19), these modifications influence both aspects of decoding.
Nucleotide 37 is modified in >70% of all tRNAs and is typically a purine (20). Among the modified nucleosides, the t6A and methylated guanosine (m1G) are the most common (11). In the absence of the modification at nucleotide 37, the anticodon loops of human tRNALys and yeast tRNAAsp lack structural rigidity compared with their modified forms (21, 22). Likewise, the modification at nucleotide 37 in Escherichia coli tRNALys and tRNAPhe stabilizes the canonical uridine turn (U turn) in the ASL, which is required for high-affinity binding to the A site (23–25).
Approximately 75% of bacterial tRNAs containing the m1G37 modification decode CNN codons (where N indicates any nucleotide), including Leu, Pro, His, Gln, and Arg codons (11). The m1G37 modification is present in ∼95% of all known sequences of proline tRNAs (26). In bacteria, the essential methyltransferase TrmD (Trm5 in eukaryotes and archaea) catalyzes the N1-methylation of G37 in tRNAs. Furthermore, the m1G37 modification stabilizes the anticodon of the tRNA to prevent +1 ribosomal frameshift errors, whereas mutations in trmD also cause growth defects (6, 26–28). , the major isoacceptor for proline, decodes the CCG codon and, in the absence of the m1G37 modification, causes +1 frameshifting on CCC-N codons. It was previously thought that lacking the m1G37 modification would allow for a four-nucleotide interaction between the anticodon and the mRNA codon with G37 interacting with the mRNA codon (29). However, biochemical and structural studies of lacking the modification revealed that this four-nucleotide interaction does not occur in the A site during decoding (30, 31). Additionally, the mRNA is positioned in the unshifted or zero frame, indicating that the frameshift event occurs post-decoding, consistent with recent structures (32). Interestingly, the absence of the methylation at G37 causes a distortion of the tRNA on the opposite side of the anticodon loop at nucleotide U32 (31), leading to the disruption of interactions with A38. Collectively, these results suggest a previously unappreciated role of the stabilization of the 32·38 pairing in predicted to maintain the correct mRNA frame (31).
Frameshift suppressor tRNAs derived from tRNAPro contain an insertion between anticodon loop nucleotides 37 and 38 (referred to as 37.5) and decode CCC-N codons as proline (26, 33–36) (Fig. 1). These mutant tRNAs are genetic suppressors that perform noncanonical reading of the genetic code to restore the reading frame (37, 38). In this case, frameshift suppressor tRNASufA6, isolated from Salmonella enterica serovar Typhimurium, contains an eight-nucleotide anticodon loop by the addition of G37.5 that causes +1 frameshifting. The structure of 70S-tRNASufA6 bound to CCC-A/U/C codons at the decoding center that undergo +1 frameshifting revealed similarities to the structure of 70S– lacking the m1G37 bound to a near-cognate codon that also promotes +1 frameshifting (31). Both tRNAs decode the mRNA in the unshifted or zero frame, indicating that the shift into the new frame occurred post-decoding. Moreover, the inserted 37.5 nucleotide and the lack of m1G37 both cause destabilization of nucleotides on the opposite side of the anticodon loop that ablates a conserved, non-Watson–Crick U32·A38 pairing. The 32·38 pairing was restored in both the tRNAPro and tRNASufA6 in the context of recognizing a cognate, three-nucleotide codon. The disruption of the 32·38 pairing is particularly notable due to its universal significance in tuning the ribosomal binding across tRNAs (39). These results provide insight into how tRNA modifications and the 32·38 pairing in the anticodon loop together lead to mRNA frame maintenance.
Here, we tested how the m1G37 modification in and tRNASufA6 impacts binding at the decoding center to cognate and slippery +1 codons. Further, we engineer tRNASufA6 to contain a conserved U32·A38 pairing to attempt to restore high-affinity binding to the decoding center. Last, a 3.6 Å X-ray crystal structure of tRNASufA6 containing this engineered 32·38 pairing bound to the 70S ribosomal A site reveals a reordering of the 32·38 pair required for decoding.
Results
The m1G37 modification in stabilizes binding to the A site
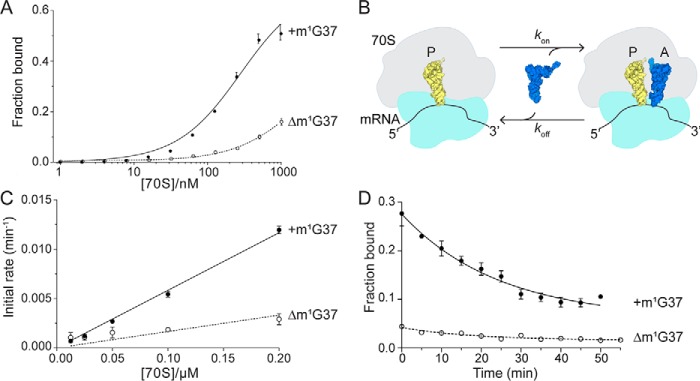
To assess the importance of the m1G37 modification in in decoding, we used established filter binding assays to determine binding kinetics to the A site (39). E. coli 70S ribosomes were programmed with mRNA containing a peptidyl-site (P-site) AUG start codon, an A-site proline CCG codon, and P-site E. coli tRNAfMet. We used a chemically synthesized ASL containing 18 nucleotides of and a m1G37 modification to ensure that the RNA was completely modified (Table S1). with the m1G37 modification binds to a cognate CCG codon in the A site with an equilibrium dissociation constant (Kd) of 284 nm (Fig. 2A and Table S2). This affinity is within the range of reported values for ASLs binding to the A site (33–500 nm) (9, 40). Removal of the m1G37 modification (Δm1G37) significantly reduced binding, by ∼6.5-fold (1.8 μm; Fig. 2A and Table S2).
Figure 2.
The m1G37 modification in is important for binding to a cognate CCG codon. A, equilibrium binding of with (+m1G37) or without (Δm1G37) the modification to a programmed 70S containing an A-site CCG codon. B, schematic of the association and dissociation of tRNA from the ribosomal A site. C and D, kon (C) and koff (D) rates of with (+m1G37) or without (Δm1G37) the modification to the 70S containing a cognate CCG codon in the A site. Data are the mean ± S.E. (error bars) of at least five independent experiments.
Although the data could be fit with reasonable confidence(Fig. S1 and Table S2), the low maximum binding was concerning, if not unprecedented (39–41). Furthermore, impractical 70S concentrations required to reach maximum binding for weaker interactions prevented us from attempting to continue with this approach. Therefore, we instead performed competition binding assays, which allow for the calculation of the equilibrium dissociation constant (Kd) based on measured association (kon) and dissociation (koff) rates (Fig. 2B) (42, 43). Unlike measuring the experimental Kd to observe equilibrium binding, the kon and koff measurements provide information regarding the influence of the m1G37 modification at each step of the binding event separately (i.e. the association with and dissociation from the ribosomal A site). Using this approach, we found a modest difference in koff between and Δm1G37, implying that the modification does not stabilize tRNA binding to ribosomes. Instead, the m1G37 modification is important for initial binding, as shown by the 2.6-fold slower association rate of Δm1G37 (kon = 0.012 μm−1 min−1) as compared with (kon = 0.031 μm−1 min−1) to the CCG A-site codon (Fig. 2 (C and D) and Table 1). These data are consistent with 70S structures demonstrating conformational distortion of the anticodon loop in the absence of the m1G37 modification (31). Furthermore, we found that the calculated dissociation constant (Kd) of m1G37 with the cognate CCG codon is 420 nm (Table 1), whereas Δm1G37 binds to the A-site CCG codon with a calculated Kd of 1.4 μm, consistent with directly measured Kd values (Fig. 2A and Table S2).
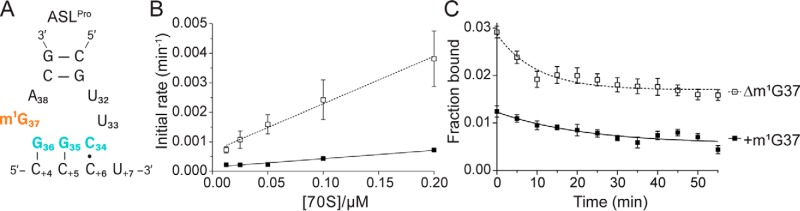
Table 1.
kon, koff, and calculated Kd values (best fit ± SE) for , ASLSufA6, and ASLSufA6 A37.5
Data from at least five replicates were fit in GraphPad Prism. Calculated Kd = koff/kon
Recognition of a +1 slippery CCC-U codon is enhanced by the lack of the m1G37 modification in
lacking the m1G37 modification undergoes high levels of +1 frameshifting on CCC-N codons (26, 28). Previous 70S structures of Δm1G37 bound to A-site CCC-N codons revealed that the tRNA decodes in the unshifted or zero frame (31). The three tRNA anticodon nucleotides C34-G35-G36 form three interactions with the C4-C5-C6 mRNA codon, respectively (where the first nucleotide of the P-site mRNA codon is denoted as +1 and the A-site nucleotides are +4, +5, and +6) (Fig. 1A). The interaction between the anticodon and the codon is near cognate as defined by a single mismatch between C34 and C6 (Fig. 3A). To test the impact of the m1G37 modification on the ability of to form a stable complex with the slippery CCC-U codon, we again measured binding kinetics. In the context of binding to a slippery codon, the m1G37 modification influences binding but in the opposite manner to binding to the cognate CCG codon (Fig. 2). lacking the m1G37 modification associates ∼4-fold faster to the slippery codon (kon = 0.017 versus 0.0039 μm−1 min−1); however, lack of the modification only has a moderate impact on koff (0.009 versus 0.014 min−1) (Fig. 3 (B and C) and Table 1). Calculated Kd measurements of 3.62 and 0.41 μm for and Δm1G37, respectively, indicate a 9-fold difference in binding affinity. In summary, the stabilizing effect observed in the cognate CCG context is reversed on a slippery CCC-U codon; the presence of the m1G37 modification actually impairs the association of to the A site programmed with a slippery CCC-U codon.
Figure 3.
The m1G37 modification prevents binding to the slippery CCC-U codon. A, secondary structure of (same color scheme as in Fig. 1) shown bound to its slippery CCC-U codon. B and C, kon (B) and koff (C) rates of with (+m1G37) or without (Δm1G37) the modification to a slippery CCC-U codon in the ribosomal A site. Data are the mean ± S.E. (error bars) of at least five independent experiments.
Nucleotide insertion in the ASL of tRNASufA6 counteracts the stabilization exerted by m1G37
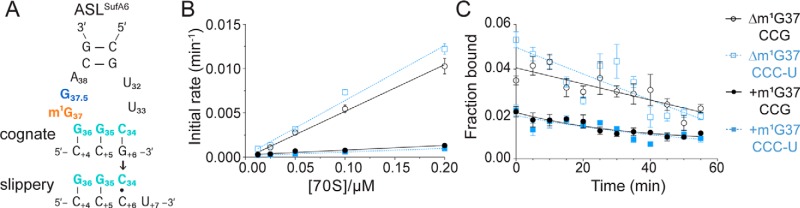
Frameshift suppressor tRNASufA6 undergoes +1 frameshifting similar to lacking the m1G37 modification (26, 31). Both tRNASufA6 and contain the m1G37 modification, and, in the case of tRNASufA6, this modification is located adjacent to the inserted nucleotide (30) (Fig. 4A). We next tested the importance of the m1G37 modification in the context of an eight-nucleotide anticodon loop in ASLSufA6, using the same kinetic binding assays as previously described. In contrast to the stabilizing effect observed with the m1G37 modification in on a cognate CCG codon, ASLSufA6 associates with the CCG codon 10-fold faster in the absence of the modification (0.053 versus 0.0053 μm−1 min−1) (Fig. 4B and Table 1). In contrast, the dissociation of ASLSufA6 was essentially unaffected by the absence or presence of the modification (koff = 0.013 and 0.014 min−1; Fig. 4C). The 10-fold difference in the calculated Kd between ASLSufA6 containing the m1G37 modification (2.5 μm) and lacking the m1G37 modification (0.26 μm) is thus reflective of the large changes in the tRNA association with the A site. One interpretation of these observations could be that, although the m1G37 modification imparts a stabilizing effect in anticodon loops of the canonical seven nucleotides (10, 44), increasing the anticodon loop to eight nucleotides, as seen in ASLSufA6, ablates any stabilization from the modification. Additionally, the overall trends of the ASLSufA6 association rates are similar to the rates seen with on the slippery CCC-U codon.
Figure 4.
The m1G37 modification in ASLSufA6 impairs binding to a cognate CCG or slippery CCC-U codon. A, secondary structure of ASLSufA6 (same color scheme as in Fig. 1) with either a cognate or slippery codon–anticodon interaction. B and C, kon (B) and koff (C) rates of ASLSufA6 with (+m1G37) or without (Δm1G37) the modification bound to either the cognate CCG (black) or slippery CCC-U (blue) codon in the ribosomal A site. Data are mean ± S.E. (error bars) of at least five independent experiments.
Next, we tested the binding of ASLSufA6 to the slippery CCC-U codon. We found that the influence of the modification status of ASLSufA6 follows similar trends regardless of whether ASLSufA6 is recognizing a cognate CCG or a slippery CCC-U codon (Table 1). The association rate of ASLSufA6 for a slippery CCC-U codon is 0.0041 μm−1 min−1 as opposed to 0.0053 μm−1 min−1 for binding to the CCG codon in the presence of m1G37 in the ASL. In the absence of the m1G37 modification, ASLSufA6 has a 10-fold greater association rate for both the CCG and slippery CCC-U codons (0.053 and 0.062 μm−1 min−1, respectively). The dissociation of ASLSufA6 from a cognate CCG or a slippery CCC-U codon are all very similar regardless of the G37 modification status (0.013–0.019 min−1). Together, these data indicate that the inserted G37.5 nucleotide in ASLSufA6 removes the dependence on the m1G37 modification required for tight association to the ribosome for the parent . Further, the G37.5 nucleotide also prevents the ribosome from distinguishing between cognate and near-cognate slippery codons, as evidenced by the similar calculated Kd values in the absence of the m1G37 modification (0.26 and 0.31 μm, respectively).
Engineering of the U32·A37.5 pairing in ASLSufA6 allows for tight association with the A site
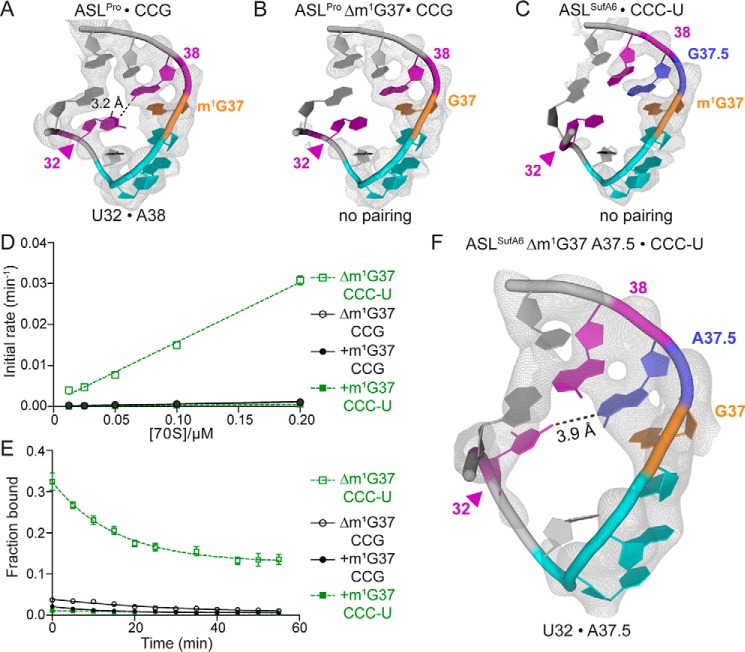
Our affinity assays show that frameshift suppressor ASLSufA6 is unable to bind with high affinity to a cognate CCG codon despite containing the same GGC anticodon as (Fig. 1). Therefore, the m1G37 modification in ASLSufA6 has a very different role in stabilizing the interactions between the anticodon and codon in contrast to . Although tRNASufA6 undergoes +1 frameshifting, it does so with low efficiencies because of its poor association with the slippery CCC-U codon (Fig. 3B) (30). These data lead us to question whether it is the inserted anticodon loop nucleotide alone that causes reduced binding affinity as seen with other frameshift suppressor ASL-binding studies (40). In the case of both and ASLSufA6 that bind poorly to the A site, 70S structures of these same tRNA-mRNA pairs bound have been solved (Fig. 5, A–C) (31). In the case of Δm1G37 bound to a cognate CCG codon, electron density is missing for nucleotide U32, which is located on the opposite side of the anticodon loop from A38 (Fig. 5B). The destabilization of the ASL is likely due to the apparent flexibility of the 5′ stem, which, in turn, disrupts the conserved U32·A38 interaction located at the base of the RNA stem. The 32·38 disruption is noteworthy because the identity of these nucleotides is universally important in fine-tuning tRNA affinity and therefore translation fidelity (39, 45). The same structural phenomenon is also observed in 70S structures containing ASLSufA6; ASLSufA6 binding to a slippery CCC-U codon results in local distortion of the 5′ stem disrupting the U32·A38 pairing (Fig. 5C). In both cases, the tRNA-mRNA pair undergoes +1 frameshifting. Therefore, we postulated that the frameshift event was directly influenced by the destabilization of the 32·38 pairing after tRNA selection but before movement to the P site.
Figure 5.
Reordering of the 32–38 pairing allows for high affinity binding of ASLSufA6 to a slippery CCC-U codon. A, 2Fo − Fc electron density maps from a structure containing the 70S ribosome with decoding a cognate CCG codon in the A site (PDB code 4LSK; color scheme is the same as in Fig. 1); B, 70S ribosome with Δm1G37 decoding a cognate CCG codon (PDB code 4P70); C, 70S ribosome with ASLSufA6 decoding a slippery CCC-U codon (PDB code 4L47). These structures demonstrate that the m1G37 modification stabilizes the U32·A38 interaction in on a cognate CCG codon (A) whereas the lack of the m1G37 modification results in disorder of the 3′ region of (B). A similar disordering is seen when ASLSufA6 containing an inserted nucleotide in its anticodon loop (G37.5) decodes a slippery CCC-U codon (C). D and E, kon (D) and koff (E) rates of ASLSufA6 with a mutated A37.5 with (+m1G37) or without (Δm1G37) the modification bound to either the cognate CCG (black) or slippery CCC-U (green) codon. Data are the mean ± S.E. (error bars) of at least five independent experiments. F, 2Fo − Fc electron density maps from a structure containing the 70S ribosome with ASLSufA6 A37.5 bound to an A-site slippery CCC-U codon. Mutation of G37.5 to A37.5 reorders the 3′ stem of the ASL, specifically nucleotides 31 and 32. 2Fo − Fc electron density maps are contoured at 1.5σ.
The G37.5 insertion in tRNASufA6 changes the potential base pairing interaction of U32·A38 to U32·G37.5 (Fig. 5C). In this context, the U32·G37.5 pairing should render the ribosome unable to distinguish a cognate from noncognate interaction as the 32–38 nucleotide identity is directly correlated to the anticodon sequence (39, 45). Indeed, ASLSufA6 binds to cognate CCG and near-cognate (i.e. slippery) CCC-U codon with similar affinities (calculated Kd of 0.31 and 0.26 μm, respectively, in the absence of the m1G37 modification; Table 1). We next tested whether changing G37.5 to A37.5 could restore high-affinity A-site binding due to the possible formation of a new U32·A37.5 pair. We found that potentially restoring the U32·A37.5 base pair does not result in high-affinity binding to a cognate CCG codon in the absence or presence of the m1G37 modification (calculated Kd of 7.4 and 15 μm, respectively; Fig. 5 (D and E) and Table 1). Notably, in contrast to , ASLSufA6A37.5 displays similar association rates both in the presence (0.0057 μm−1 min−1) and absence (0.0046 μm−1 min−1) of m1G37, but koff is reduced ∼3-fold (0.09 and 0.034 min−1, respectively) (Fig. 5, D and E).
In binding to the slippery CCC-U codon, ASLSufA6 A37.5 has a ∼2-fold higher affinity (calculated Kd = 1.8 μm) than ASLSufA6 containing G37.5 (calculated Kd = 3.7 μm) in the presence of the m1G37 modification (Table 1). Removal of m1G37 results in ASLSufA6 A37.5 binding with high affinity, similar to ASLSufA6 G37.5 (calculated Kd = 0.31 μm for ASLSufA6 G37.5 and 0.45 μm for ASLSufA6 A37.5). The Kd for ASLSufA6 A37.5 Δm1G37 binding to a slippery CCC-U codon is comparable with modified binding to a cognate CCG codon (calculated Kd = 0.42 μm). For the ASLSufA6 A37.5 Δm1G37 binding to a slippery CCC-U codon, both the kon and koff rates are higher than those of other ASLs (kon = 0.146 μm−1 min−1, koff = 0.06 min−1), implying that the recognition mechanism is altered. The increase in affinity implies that in the case of ASLSufA6 with the restored U32–A37.5, the lack of m1G37 enables high-affinity binding and recognition of the ASL when the slippery CCC-U codon is presented in the A site. Most importantly, we demonstrate that by changing the identity of the base insertion and controlling the modification at position 37 in the anticodon stem loop, we can tune the affinity of the ASL to the ribosomal A site. This has significant implications for understanding how the ribosome interacts with rationally engineered tRNAs.
Engineering the 32·38 pairing in ASLSufA6 to U32·A37.5 reorders the 5′ stem of the anticodon loop
To determine whether the engineered ASLSufA6 A37.5 Δm1G37 does indeed reorder the ASL as suggested from the binding kinetics, we solved a 3.6 Å resolution X-ray crystal structure of ASLSufA6 A37.5 Δm1G37 bound to the Thermus thermophilus 70S ribosome (Fig. 5F and Table 2). The ASL and mRNA density are well-ordered and unambiguously demonstrate a change in the anticodon loop (Fig. 5F) as compared with other ASLSufA6 structures bound to the ribosome (Fig. 5, A–C) (31). The ASLSufA6 A37.5 Δm1G37 has good density for U32 in contrast to the previous structures that showed distortion of the 5′ stem of the ASL (Fig. 5, B and C). The phosphate backbone of nucleotide A37.5 shifts by 2.8 Å as compared with WT bound to its cognate CCG codon and by 6.2 Å when compared with ASLSufA6 with the G37.5 (Fig. 5F, S3A). This movement places A37.5 across from U32, allowing the possible formation of a single hydrogen bond similar to the 32·38 orientation observed in other tRNAs (Fig. 5A, S2) (31, 46, 47). Overall, the A37.5 insertion in ASLSufA6 seems to orient the ASL to a conformation more similar to that of than that of WT ASLSufA6 (Fig. S3, B and C). 16S rRNA nucleotides A1492 and A1493 flip from their internal position in helix 44, and G530 is positioned close to A1492, demonstrating recognition by the ribosome (Fig. S4).
Table 2.
Data collection and refinement statistics
Values for the highest-resolution shell are shown in parentheses.
| ASLSufA6 A37.5Δm1G37 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Wavelength (Å) | 1.00000 |
| Cell dimensions | |
| a, b, c (Å) | 208.91, 445.91, 617.31 |
| α, β, γ (degrees) | 90, 90, 90 |
| Resolution (Å) | 49.17–3.64 (3.77–3.64) |
| Rpim (%) | 11.5 (62.8) |
| I/σI | 4.77 (1.14) |
| Completeness (%) | 93.31 (87.06) |
| Redundancy | 5.0 (4.2) |
| CC1/2 | 0.986 (0.395) |
| Refinement | |
| Reflections | 596,260 (55,238) |
| Rwork/Rfree (%) | 21.5/26.2 |
| No. of atoms | 291,830 |
| B-factors (Å2) | |
| Overall | 150.84 |
| Macromolecule | 151.28 |
| Ligand/ion | 94.30 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (degrees) | 0.89 |
Discussion
Modification of tRNAs adds an important layer of regulation during translation. These modifications are so functionally important that more genes are devoted to tRNA modification pathways than to the expression of tRNAs themselves (48). Modifications in the ASLs of tRNAs are critical, given that only 7 of the 61 sense codons are decoded by tRNAs that lack modifications at nucleotide 34 or 37 in E. coli (49, 50). In this work, we determine that the m1G37 modification in is required for high-affinity binding to a cognate CCG codon in the decoding center (Fig. 2). The absence of the modification results in low-affinity binding, and specifically, the association (kon) is reduced almost 3-fold, whereas koff is unaffected (Fig. 2). These results indicate that the m1G37 modification in provides stability in association with the decoding center rather than causing A-site drop-off. Consistent with these data are our previous structural studies that showed destabilization of the anticodon loop when lacks the m1G37 modification and interacts with a cognate CCG codon (31) (Fig. 5B).
Both and isoacceptors lacking the m1G37 modification undergo +1 frameshifting on CCC-N codons (26, 29, 51). Although containing the m1G37 modification significantly impairs binding to a slippery CCC-U codon (3.62 μm; Fig. 3A), its removal causes a ∼4-fold enhancement in association with the slippery codon (Fig. 3B). This association results in tighter binding (calculated Kd of 0.41 μm) that is comparable with binding of WT to a cognate CCG codon (0.42 μm). These data suggest that the additional stability that the m1G37 modification imparts in binding to a cognate CCG codon is lost in the context of a non-Watson–Crick C34-C4 pair at the third or wobble position. Collectively, these results show that the m1G37 modification in stabilizes high-affinity interactions in the cognate case but prevents recognition of slippery codons that would result in +1 frameshifting.
tRNAs containing expanded anticodon stem loops can cause “slipping” on mRNA codons, resulting in frameshifts (37). An extra nucleotide insertion in the anticodon loop of was identified in a frameshift suppressor tRNA (named tRNASufA6) that reverts a +1 frameshift. Primer extension analyses revealed that tRNASufA6 was also modified at nucleotide 37, similar to all three tRNAPro isoacceptors (30, 52), but the extent of modification was not determined. It is unclear what role, if any, the m1G37 modification has in tRNASufA6-mediated +1 frameshifting. We find that the presence of the m1G37 modification renders ASLSufA6 unable to bind to both cognate CCG and slippery CCC-U codons (Fig. 4B). In contrast, ASLSufA6 lacking m1G37 binds with high affinity to either a cognate CCG or a slippery CCC-U codon. These data support the notion that the m1G37 modification and the inserted G37.5 nucleotide likely stabilize the anticodon loop in similar ways. In support of the functional similarities of m1G37 and G37.5, structures of Δm1G37 or tRNASufA6 decoding codons that allow for +1 frameshifting reveal structural similarities. The 3′ stem of the ASL, in particular nucleotides 30–32 on the opposite side of the G37.5/G37, is conformationally dynamic in both structures, strongly suggesting that +1 frameshifts induced by these two tRNAs occur by a similar mechanism (Fig. 5, B and C) (31).
In , nucleotide U32 normally forms a single hydrogen bond with A38 and thus is not a Watson–Crick base pair (Fig. 5A and Fig. S2). The nucleotide identity of the 32·38 pairing in all tRNAs is inversely correlated to the strength of the codon–anticodon interaction (39, 41, 45). For example, the anticodon of E. coli is considered strong because of the three GC pairs between the codon and the anticodon. Therefore, in this strong case, the 32·38 pairing needs to be correspondingly weak to counterbalance the strength of the codon–anticodon. Changing the 32·38 pairing in from a weak, conserved U32·A38 pair to a strong pair, such as C32·A38, prevents the ribosome from being able to distinguishing correct from incorrect tRNA-mRNA pairs (41, 53). In the context of tRNASufA6, the inserted G37.5 displaces A38, preventing a U32·A38 pairing (31). Binding of ASLSufA6 to a cognate CCG or a +1 slippery CCC-U codon is extremely weak as indicated by both the kon and koff rates (Fig. 4). We attempted to restore the WT U32·A38 found in by changing G37.5 in tRNASufA6 to an adenosine. ASLSufA6 A37.5 binds poorly to a CCG codon regardless of the m1G37 modification status (Fig. 5D). Interestingly, the A37.5 mutant bound tightly to the slippery CCC-U codon, but only in the absence of the m1G37 modification, similar to ASLSufA6. An X-ray crystal structure of the ribosome with an A-site ASLSufA6 A37.5 (lacking the m1G37 modification) bound to a slippery CCC-U codon reveals a reordering of the 3′ stem such that U32 regains rigidity as assessed by its electron density (Fig. 5F). We predict that this engineered tRNASufA6 does not undergo +1 frameshifting because of ordering of the ASL, but further studies are required to test this.
The studies here demonstrate that the m1G37 modification of tRNAPro influences recognition of both cognate and near-cognate, slippery codons. lacking the m1G37 modification undergoes +1 frameshifting, but our previous structures, along with other structures of extended ASLs that frameshift, show that the shift into the new frame does not occur in the decoding center (31, 54, 55). At what stage of elongation does lacking the m1G37 modification cause a +1 frameshift? Kinetic analyses of movement through the ribosome reveal that the shift can occur at two distinct stages: a fast mechanism during translocation of the tRNA-mRNA pairs to the P site and a slower mechanism when is stalled in the P site while waiting for A-site tRNA delivery (28). A recent structure of ASLSufA6 bound to a +1 codon in the P site demonstrates the ASL alone is sufficient for the +1 mRNA frameshift consistent with the slow mechanism presented above (32). Another possibility is what is also observed in the structure of lacking the m1G37 modification in the A site (31): the 3′ anticodon stem is destabilized, which, in turn, may influence how elongation factor G (EF-G) recognizes the tRNA in the A site to initiate translocation. This second possibility is consistent with the fast mechanism as observed in the kinetic analyses.
Although the data presented focus on the impact of the m1G37 modification on decoding of single proline codons, polyproline sequences in protein-coding genes are prone to +1 frameshifts (56–60). The unique nature of proline where it is both a poor donor and acceptor during the peptidyltransferase reaction results in a slow rate of peptide bond formation. Therefore, consecutive prolines cause ribosome stalling (60, 61). Additionally, the nucleotide repeats in the proline codons present the same codon–anticodon interactions regardless of whether the tRNA binds in the zero or +1 frame (62). These events can collectively lead to the shifty nature of but are counterbalanced by the action of elongation factor P (EF-P) that helps to stabilize peptidyl-tRNAPro located in the P site of the ribosome (28, 60, 61, 63). EF-P binds in the exit site of the ribosome on both the small and large ribosomal subunits and abuts against P-site peptidyl-tRNAPro (63). A modified Lys residue of EF-P protrudes into the 50S P site and stabilizes peptidyl-tRNAPro to help resume protein synthesis stalled at a stretch of polyprolines (60, 64, 65). Cryo-EM structures of ribosomes bound to peptidyl-tRNAPro reveal the flexibility of the peptidyl-tRNAPro in the absence of EF-P. EF-P binding orders peptidyl-tRNAPro to facilitate efficient peptide bond formation of the cyclic proline moiety (63). In addition to this function, EF-P also can suppress +1 frameshifts, suggesting a previously unappreciated role in helping to maintain the mRNA frame (28). Conserved EF-P residues that interact with anticodon stem nucleotides 41 and 42 are essential for function (63). In the absence of the m1G37 modification, the interaction between EF-P and tRNAPro may be destabilized due to the flexibility of the 3′ stem (31). The interplay between the m1G37 modification in tRNAPro and EF-P suggests that this elongation factor has synergistic roles in translational fidelity dependent on tRNA metabolism.
Experimental procedures
Ribosome purification
E. coli 70S ribosomes were purified as described previously (66). E. coli MRE600 cells were grown to an A600 of ∼0.7 in Luria broth (LB) medium at 37 °C and then cooled on ice for 20 min. All centrifugation steps were performed at 4 °C. Cells were pelleted by centrifugation and washed with buffer 1 (10 mm HEPES-KOH, pH 7.6, 10 mm MgCl2, 1 m NH4Cl, 6 mm β-mercaptoethanol (β-Me)) twice and then resuspended in buffer 2 (10 mm HEPES-KOH, pH 7.6, 10 mm MgCl2, 100 mm NH4Cl, 6 mm β-Me). Cells were lysed using an EmulsiFlex-C5 high-pressure homogenizer (Avestin), and cell debris was removed by centrifuging at 13,000 × g for 15 min. The lysate was further centrifuged at 27,000 × g for 30 min to obtain the S30 fraction. Ribosomes were pelleted by centrifuging at 42,000 × g for 17 h. The pellets were resuspended in buffer 2, and ribosomes were further purified over a 10–40% sucrose gradient in buffer 2 at 70,000 × g for 12 h. 70S ribosomes were separated from polysomes and subunits using a Brandel gradient fractionator. The 70S fractions were pooled, pelleted, resuspended in buffer 2, and stored at −80 °C.
70S complex formation
ASLs and mRNAs were chemically synthesized (Integrated DNA Technologies), and purified E. coli tRNAfMet was purchased from Chemical Block (Table S1). mRNAs contained either an CCG or CCC-U in the A site. The E. coli 70S ribosome complex was formed by incubation with a 2-fold molar excess of mRNA for 5 min followed by a 2-fold molar excess of tRNAfMet for 30 min at 37 °C. A-site ASLs were 5′-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) using T4 PNK enzyme (New England Biolabs).
Kinetic binding assays
A modified 96-well Bio-Rad dot-blot apparatus with two membranes was used to study binding kinetics of tRNAs to ribosomes (67). An upper nitrocellulose membrane and a lower nylon membrane (Amersham Biosciences Hybond-N+, GE Healthcare) were pre-equilibrated in cold buffer 3 (5 mm HEPES-KOH, pH 7.5, 50 mm KCl, 10 mm NH4Cl, 10 mm Mg(CH3COO)2, 6 mm β-Me). All binding experiments were performed in buffer 3. Nonspecific binding was controlled for by having a [32P]ASL-only sample with each experiment. Reactions were filtered by vacuum and immediately washed using 100 μl of cold buffer 3. After incubations, membranes were dried and exposed to a PhosphorImager screen (GE Healthcare) and imaged on a Typhoon FLA 7000. Quantification was performed using ImageQuantTL software and analyzed using GraphPad Prism. The fraction of A-site ASL bound was calculated as the ratio between nitrocellulose counts and the total counts on both membranes after correcting for nonspecific binding.
Measuring Kd values
Serial dilutions of the ribosome complex (70S, mRNA, P-site tRNAfMet) were performed to generate a range of 70S concentrations from 0.98 nm to 1 μm. [32P]ASL was added and incubated for 3 h at 25 °C. Reaction volumes of 10 μl were applied to the filters, washed, and then quantified as described above. The ASL fraction bound was fit using a one-site–specific binding equation in GraphPad Prism as described previously (fraction bound = (Bmax × [70S])/(Kd + [70S]) (39, 68).
Measuring kon values
Association with the 70S A site was measured as described previously (42, 43). Briefly, 15 μl of 4.5 nm 32P-labeled ASL was added to 15 μl of increasing concentrations of 70S complex programmed with mRNA and tRNAfMet. Three-μl aliquots were removed at different times (0.5, 1, 2, 3, 4, 5, 6, and 7 min), immediately filtered, and washed with 100 μl of cold buffer 3. Initial association rates were obtained using different concentrations of the 70S complex (final concentrations of 12.5, 25, 50, 100, and 200 nm). kon was derived as the slope of the linear regression performed on the initial rates versus [70S] plot.
Measuring koff values
Dissociation of the ASL from the 70S A site was initiated by a 1:100 dilution of the equilibrium binding reaction (1 μm 70S, 3 μm mRNA, 5 μm tRNAfMet, 0.05 μm [32P]ASL) in buffer 3 containing 0.3 μm unlabeled ASL. At 5-min intervals, 10-μl aliquots of the reaction were removed, filtered, and washed. The ASL fraction bound was normalized to t = 0. The natural log of the normalized fraction bound was fitted with a linear regression versus time, and koff was derived as the negative of the slope.
Crystallization, X-ray data collection, and structural determination
Purification of T. thermophilus 70S ribosomes, formation of complexes with mRNA and tRNAs, and initial screening conditions followed previously established protocols (46, 68). Two μl of the ribosome complex (4.4 μm 70S, 8.8 μm mRNA, 11 μm tRNAfMet, 22 μm ASLSufA6, 11 μm CC-puromycin (Dharmacon), and 2.8 μm deoxy-BigCHAP (Hampton Research)) were mixed with 2.4 μl of reservoir condition (0.1 m Tris-HOAc, pH 7.0, 0.2 m KSCN, 4.5–5.5% (w/v) PEG 20K, 4.5–5.5% (w/v) PEG 550 MME, 10 mm Mg(OAc)2). Crystals were grown by sitting drop at 20 °C in 2 weeks. Crystals were cryoprotected using increasing amounts of PEG 550 MME to a final concentration of 35%, with the final solution containing 22 μm ASLSufA6 and 11 μm CC-puromycin. The crystals were screened at the SER-CAT 22-ID and NE-CAT 24ID-C/E beamlines, and data sets were collected at the SER-CAT 22-ID beamline, all at the Advanced Photon Source, Argonne National Laboratory (Table 2). Data sets were integrated and scaled using XDS (69), and the structure was solved by molecular replacement using 70S coordinates from PDB entry 4Y4O. Crystallographic refinements were performed with PHENIX (70) followed by manual model building in Coot (71). Figures were generated in PyMOL (72).
Author contributions
H. A. N., E. D. H., and C. M. D. conceptualization, data curation, formal analysis, validation, investigation, methodology, writing-original draft, project administration, writing-review and editing, and supervision; C. M. D. resources and funding acquisition.
Supplementary Material
Acknowledgments
We thank Dunham laboratory members for advice and encouragement, Dr. Kurt Fredrick for advice on kinetic binding assays, Stacey Miles for technical assistance, Dr. Tatsuya Maehigashi for crystallization advice, and Dr. Graeme Conn for comments on the manuscript. Crystals were screened and X-ray crystallography data sets were collected at the NE-CAT beamlines (funded by NIGMS, National Institutes of Health, Grant P30 GM124165), using a Pilatus detector (RR029205) and an Eiger detector (OD021527), and at the SER-CAT beamlines (funded by its member institutions and National Institutes of Health Equipment Grants RR25528 and RR028976). This research used resources of the APS, a United States Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contracts DE-AC02-06CH11357 (NE-CAT) and W-31-109-Eng-38 (SER-CAT).
This work was supported by NIGMS, National Institutes of Health, Grant R01 GM093278 and National Science Foundation Grant CHE 1808711 (to C. M. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Tables S1 and S2 and Figs. S1–S4.
The atomic coordinates and structure factors (code 6NDK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- A site
- aminoacyl site
- P site
- peptidyl site
- ASL
- anticodon stem-loop
- cmo5U34
- uridine-5-oxyacetic acid
- mnm5s2
- 5-methylaminomethyl-2-thiouridine
- t6A
- 6-threonylcarbamoyladenosine
- ms2t6A
- 2-methylthio derivative
- EF
- elongation factor
- β-Me
- β-mercaptoethanol
- PDB
- Protein Data Bank.
References
- 1. Schmeing T. M., and Ramakrishnan V. (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- 2. Dunkle J. A., and Dunham C. M. (2015) Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code. Biochimie 114, 90–96 10.1016/j.biochi.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Agris P. F., Eruysal E. R., Narendran A., Väre V. Y. P., Vangaveti S., and Ranganathan S. V. (2018) Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15, 537–553 10.1080/15476286.2017.1356562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yarian C., Townsend H., Czestkowski W., Sochacka E., Malkiewicz A. J., Guenther R., Miskiewicz A., and Agris P. F. (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277, 16391–16395 10.1074/jbc.M200253200 [DOI] [PubMed] [Google Scholar]
- 5. Krüger M. K., Pedersen S., Hagervall T. G., and Sørensen M. A. (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284, 621–631 10.1006/jmbi.1998.2196 [DOI] [PubMed] [Google Scholar]
- 6. Urbonavicius J., Qian Q., Durand J. M., Hagervall T. G., and Björk G. R. (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 10.1093/emboj/20.17.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yarian C., Marszalek M., Sochacka E., Malkiewicz A., Guenther R., Miskiewicz A., and Agris P. F. (2000) Modified nucleoside dependent Watson–Crick and wobble codon binding by tRNALysUUU species. Biochemistry 39, 13390–13395 10.1021/bi001302g [DOI] [PubMed] [Google Scholar]
- 8. Weixlbaumer A., Murphy F. V. 4th, Dziergowska A., Malkiewicz A., Vendeix F. A., Agris P. F., and Ramakrishnan V. (2007) Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 14, 498–502 10.1038/nsmb1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phelps S. S., Jerinic O., and Joseph S. (2002) Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell 10, 799–807 10.1016/S1097-2765(02)00686-X [DOI] [PubMed] [Google Scholar]
- 10. Hou Y. M., Gamper H., and Yang W. (2015) Post-transcriptional modifications to tRNA–a response to the genetic code degeneracy. RNA 21, 642–644 10.1261/rna.049825.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boccaletto P., Machnicka M. A., Purta E., Piatkowski P., Baginski B., Wirecki T. K., de Crécy-Lagard V., Ross R., Limbach P. A., Kotter A., Helm M., and Bujnicki J. M. (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kothe U., and Rodnina M. V. (2007) Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell 25, 167–174 10.1016/j.molcel.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 13. Murphy F. V. 4th, and Ramakrishnan V. (2004) Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 11, 1251–1252 10.1038/nsmb866 [DOI] [PubMed] [Google Scholar]
- 14. Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., and Yusupova G. (2016) Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 7, 10457 10.1038/ncomms10457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agris P. F. (1991) Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 73, 1345–1349 10.1016/0300-9084(91)90163-U [DOI] [PubMed] [Google Scholar]
- 16. Murphy F. V. 4th, Ramakrishnan V., Malkiewicz A., and Agris P. F. (2004) The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 11, 1186–1191 10.1038/nsmb861 [DOI] [PubMed] [Google Scholar]
- 17. Ogle J. M., Murphy F. V., Tarry M. J., and Ramakrishnan V. (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 10.1016/S0092-8674(02)01086-3 [DOI] [PubMed] [Google Scholar]
- 18. Demeshkina N., Jenner L., Westhof E., Yusupov M., and Yusupova G. (2012) A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 10.1038/nature10913 [DOI] [PubMed] [Google Scholar]
- 19. Hoffer E. D., Maehigashi T., Fredrick K., and Dunham C. M. (2019) Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding. Nucleic Acids Res. 47, 1557–1563 10.1093/nar/gky1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Machnicka M. A., Olchowik A., Grosjean H., and Bujnicki J. M. (2014) Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 11, 1619–1629 10.4161/15476286.2014.992273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Durant P. C., and Davis D. R. (1999) Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 285, 115–131 10.1006/jmbi.1998.2297 [DOI] [PubMed] [Google Scholar]
- 22. Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., and Giegé R. (1990) Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie 72, 735–743 10.1016/0300-9084(90)90158-D [DOI] [PubMed] [Google Scholar]
- 23. Ashraf S. S., Ansari G., Guenther R., Sochacka E., Malkiewicz A., and Agris P. F. (1999) The uridine in “U-turn”: contributions to tRNA-ribosomal binding. RNA 5, 503–511 10.1017/S1355838299981931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sundaram M., Durant P. C., and Davis D. R. (2000) Hypermodified nucleosides in the anticodon of tRNA(Lys) stabilize a canonical U-turn structure. Biochemistry 39, 12575–12584 10.1021/bi005120y [DOI] [PubMed] [Google Scholar]
- 25. Cabello-Villegas J., Winkler M. E., and Nikonowicz E. P. (2002) Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe). J. Mol. Biol. 319, 1015–1034 10.1016/S0022-2836(02)00382-0 [DOI] [PubMed] [Google Scholar]
- 26. Björk G. R., Wikström P. M., and Byström A. S. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244, 986–989 10.1126/science.2471265 [DOI] [PubMed] [Google Scholar]
- 27. Li J., Esberg B., Curran J. F., and Björk G. R. (1997) Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 271, 209–221 10.1006/jmbi.1997.1176 [DOI] [PubMed] [Google Scholar]
- 28. Gamper H. B., Masuda I., Frenkel-Morgenstern M., and Hou Y. M. (2015) Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 6, 7226 10.1038/ncomms8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hagervall T. G., Tuohy T. M., Atkins J. F., and Björk G. R. (1993) Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 232, 756–765 10.1006/jmbi.1993.1429 [DOI] [PubMed] [Google Scholar]
- 30. Qian Q., Li J. N., Zhao H., Hagervall T. G., Farabaugh P. J., and Björk G. R. (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1, 471–482 10.1016/S1097-2765(00)80048-9 [DOI] [PubMed] [Google Scholar]
- 31. Maehigashi T., Dunkle J. A., Miles S. J., and Dunham C. M. (2014) Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc. Natl. Acad. Sci. U.S.A. 111, 12740–12745 10.1073/pnas.1409436111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hong S., Sunita S., Maehigashi T., Hoffer E. D., Dunkle J. A., and Dunham C. M. (2018) Mechanism of tRNA-mediated +1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 115, 11226–11231 10.1073/pnas.1809319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Riddle D. L., and Roth J. R. (1970) Suppressors of frameshift mutations in Salmonella typhimurium. J. Mol. Biol. 54, 131–144 10.1016/0022-2836(70)90451-1 [DOI] [PubMed] [Google Scholar]
- 34. Yourno J. (1972) Externally suppressible +1 “glycine” frameshift: possible quadruplet isomers for glycine and proline. Nat. New Biol 239, 219–221 10.1038/newbio239219a0 [DOI] [PubMed] [Google Scholar]
- 35. Sroga G. E., Nemoto F., Kuchino Y., and Björk G. R. (1992) Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 20, 3463–3469 10.1093/nar/20.13.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Connor M. (2002) Imbalance of tRNA(Pro) isoacceptors induces +1 frameshifting at near-cognate codons. Nucleic Acids Res. 30, 759–765 10.1093/nar/30.3.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Atkins J. F., and Björk G. R. (2009) A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. 73, 178–210 10.1128/MMBR.00010-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., and Baranov P. V. (2016) Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 10.1093/nar/gkw530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olejniczak M., Dale T., Fahlman R. P., and Uhlenbeck O. C. (2005) Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 12, 788–793 10.1038/nsmb978 [DOI] [PubMed] [Google Scholar]
- 40. Walker S. E., and Fredrick K. (2006) Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J. Mol. Biol. 360, 599–609 10.1016/j.jmb.2006.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ledoux S., Olejniczak M., and Uhlenbeck O. C. (2009) A sequence element that tunes Escherichia coli to ensure accurate decoding. Nat. Struct. Mol. Biol. 16, 359–364 10.1038/nsmb.1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fahlman R. P., and Uhlenbeck O. C. (2004) Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43, 7575–7583 10.1021/bi0495836 [DOI] [PubMed] [Google Scholar]
- 43. Shoji S., Abdi N. M., Bundschuh R., and Fredrick K. (2009) Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 37, 4033–4042 10.1093/nar/gkp296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konevega A. L., Soboleva N. G., Makhno V. I., Semenkov Y. P., Wintermeyer W., Rodnina M. V., and Katunin V. I. (2004) Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10, 90–101 10.1261/rna.5142404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Olejniczak M., and Uhlenbeck O. C. (2006) tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88, 943–950 10.1016/j.biochi.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 46. Selmer M., Dunham C. M., Murphy F. V. 4th, Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., and Ramakrishnan V. (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- 47. Jenner L. B., Demeshkina N., Yusupova G., and Yusupov M. (2010) Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 10.1038/nsmb.1790 [DOI] [PubMed] [Google Scholar]
- 48. Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., and Wikström P. M. (1987) Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287 10.1146/annurev.bi.56.070187.001403 [DOI] [PubMed] [Google Scholar]
- 49. Sprinzl M., Horn C., Brown M., Ioudovitch A., and Steinberg S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 10.1093/nar/26.1.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Agris P. F. (2004) Decoding the genome: a modified view. Nucleic Acids Res. 32, 223–238 10.1093/nar/gkh185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Björk G. R., Jacobsson K., Nilsson K., Johansson M. J., Byström A. S., and Persson O. P. (2001) A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239 10.1093/emboj/20.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kuchino Y., Yabusaki Y., Mori F., and Nishimura S. (1984) Nucleotide sequences of three proline tRNAs from Salmonella typhimurium. Nucleic Acids Res. 12, 1559–1562 10.1093/nar/12.3.1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murakami H., Ohta A., and Suga H. (2009) Bases in the anticodon loop of prevent misreading. Nat. Struct. Mol. Biol. 16, 353–358 10.1038/nsmb.1580 [DOI] [PubMed] [Google Scholar]
- 54. Dunham C. M., Selmer M., Phelps S. S., Kelley A. C., Suzuki T., Joseph S., and Ramakrishnan V. (2007) Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA 13, 817–823 10.1261/rna.367307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fagan C. E., Dunkle J. A., Maehigashi T., Dang M. N., Devaraj A., Miles S. J., Qin D., Fredrick K., and Dunham C. M. (2013) Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state. Proc. Natl. Acad. Sci. U.S.A. 110, 9716–9721 10.1073/pnas.1301585110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wohlgemuth I., Brenner S., Beringer M., and Rodnina M. V. (2008) Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 283, 32229–32235 10.1074/jbc.M805316200 [DOI] [PubMed] [Google Scholar]
- 57. Muto H., and Ito K. (2008) Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem. Biophys. Res. Commun. 366, 1043–1047 10.1016/j.bbrc.2007.12.072 [DOI] [PubMed] [Google Scholar]
- 58. Pavlov M. Y., Watts R. E., Tan Z., Cornish V. W., Ehrenberg M., and Forster A. C. (2009) Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 106, 50–54 10.1073/pnas.0809211106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johansson M., Ieong K. W., Trobro S., Strazewski P., Åqvist J., Pavlov M. Y., and Ehrenberg M. (2011) pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 79–84 10.1073/pnas.1012612107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., and Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- 61. Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., and Jung K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 10.1126/science.1228985 [DOI] [PubMed] [Google Scholar]
- 62. Yourno J., and Tanemura S. (1970) Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature 225, 422–426 10.1038/225422a0 [DOI] [PubMed] [Google Scholar]
- 63. Huter P., Arenz S., Bock L. V., Graf M., Frister J. O., Heuer A., Peil L., Starosta A. L., Wohlgemuth I., Peske F., Nováček J., Berninghausen O., Grubmüller H., Tenson T., Beckmann R., Rodnina M. V., Vaiana A. C., and Wilson D. N. (2017) Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol. Cell 68, 515–527.e6 10.1016/j.molcel.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 64. Navarre W. W., Zou S. B., Roy H., Xie J. L., Savchenko A., Singer A., Edvokimova E., Prost L. R., Kumar R., Ibba M., and Fang F. C. (2010) PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 39, 209–221 10.1016/j.molcel.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bullwinkle T. J., Zou S. B., Rajkovic A., Hersch S. J., Elgamal S., Robinson N., Smil D., Bolshan Y., Navarre W. W., and Ibba M. (2013) (R)-β-lysine-modified elongation factor P functions in translation elongation. J. Biol. Chem. 288, 4416–4423 10.1074/jbc.M112.438879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y., Hong S., Ruangprasert A., Skiniotis G., and Dunham C. M. (2018) Alternative mode of E-site tRNA binding in the presence of a downstream mRNA stem loop at the entrance channel. Structure 26, 437–445.e3 10.1016/j.str.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wong I., and Lohman T. M. (1993) A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 10.1073/pnas.90.12.5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fagan C. E., Maehigashi T., Dunkle J. A., Miles S. J., and Dunham C. M. (2014) Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA 20, 1944–1954 10.1261/rna.046953.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 10.1107/S0108768110007202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 72. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.