Abstract
The tumor suppressor bridging integrator 1 (BIN1) is a corepressor of the transcription factor E2F1 and inhibits cell-cycle progression. BIN1 also curbs cellular poly(ADP-ribosyl)ation (PARylation) and increases sensitivity of cancer cells to DNA-damaging therapeutic agents such as cisplatin. However, how BIN1 deficiency, a hallmark of advanced cancer cells, increases cisplatin resistance remains elusive. Here, we report that BIN1 inactivates ataxia telangiectasia–mutated (ATM) serine/threonine kinase, particularly when BIN1 binds E2F1. BIN1 + 12A (a cancer-associated BIN1 splicing variant) also inhibited cellular PARylation, but only BIN1 increased cisplatin sensitivity. BIN1 prevented E2F1 from transcriptionally activating the human ATM promoter, whereas BIN1 + 12A did not physically interact with E2F1. Conversely, BIN1 loss significantly increased E2F1-dependent formation of MRE11A/RAD50/NBS1 DNA end-binding protein complex and efficiently promoted ATM autophosphorylation. Even in the absence of dsDNA breaks (DSBs), BIN1 loss promoted ATM-dependent phosphorylation of histone H2A family member X (forming γH2AX, a DSB biomarker) and mediator of DNA damage checkpoint 1 (MDC1, a γH2AX-binding adaptor protein for DSB repair). Of note, even in the presence of transcriptionally active (i.e. proapoptotic) TP53 tumor suppressor, BIN1 loss generally increased cisplatin resistance, which was conversely alleviated by ATM inactivation or E2F1 reduction. However, E2F2 or E2F3 depletion did not recapitulate the cisplatin sensitivity elicited by E2F1 elimination. Our study unveils an E2F1-specific signaling circuit that constitutively activates ATM and provokes cisplatin resistance in BIN1-deficient cancer cells and further reveals that γH2AX emergence may not always reflect DSBs if BIN1 is absent.
Keywords: ataxia telangiectasia, E2F transcription factor, DNA damage response, tumor suppressor gene, chemoresistance, genomic instability, cancer, p53, BIN1, double-stranded DNA breaks, MRN complex
Introduction
More than half a century ago, Rosenberg et al. (1) serendipitously discovered a potent cell growth-inhibiting property of cis-diamminedichloroplatinum (II) (cisplatin). During electrophoresis, cisplatin was released from a platinum electrode and dramatically changed the morphology of Escherichia coli and inhibited bacterial growth (1, 2). Because unlimited cell division is a typical feature commonly observed in bacterial and cancerous cells, they immediately applied this fascinating finding of bacteriology to cancer research (3). Inspired by the compelling anticancer activity of cisplatin originally documented in vitro and in vivo by Rosenberg et al. in the late 1960s (1–3), Einhorn and Donohue (4) conducted pioneering clinical trials using cisplatin and reported a tremendously improved survival rate of patients with deadly testicular cancer in the late 1970s. Platinum-based chemotherapy has since been recognized to be the first-line anticancer therapy (5).
Cisplatin is a chemically-unstable and highly-reactive compound in aqueous solution, so it easily cross-links two neighboring purine bases of one strand of a dsDNA molecule (6, 7). As a result, cisplatin forms platinum–DNA adducts, which then interfere with DNA replication, DNA transcription, and DNA repair in actively proliferating cells, such as cancer cells, hair follicle cells, and hematopoietic progenitor cells, and provoke cytostatic and cytotoxic effects (6–8). Severe side effects, such as nephrotoxicity, persistent hearing loss, and compromised immune systems, are observed in cisplatin-treated cancer patients (9, 10). Besides these adverse effects, acquired resistance to cisplatin of cancer cells is a major cause of treatment failure (6, 7). Some advanced (or late-stage) cancer cells tolerate cisplatin even before the cells are exposed to the drug, implying that cancer cells naturally develop cisplatin resistance by intrinsic mechanisms (6, 7). To maximize the anticancer efficacy, while minimizing the cytotoxic effects of cisplatin on healthy tissues, it is crucial to better understand how cancer cells elicit cisplatin resistance (8).
Platinum–DNA adducts are primarily removed by the nucleotide excision repair (NER)8 machinery. Impaired NER causes genomic instability mainly producing ssDNA breaks (SSBs) (11, 12). SSBs by themselves are not immediately detrimental, but unrepaired SSBs are easily converted to dsDNA breaks (DSBs), the most harmful form of DNA lesions, typically after the collapse of stalled replication forks (13). Therefore, in addition to the NER pathways, cellular DSB-repair mechanisms, such as homologous recombination and nonhomologous end-joining, are also believed to enable cancer cells to survive and grow in the presence of cisplatin.
When DSBs are produced by an environmental factor, such as γ-irradiation, the MRE11A/RAD50/NBS1 (MRN) protein complex immediately binds DNA ends, and then ataxia telangiectasia–mutated serine/threonine (Ser/Thr) protein kinase (ATM, EC 2.7.11.1), a member of the phosphatidylinositol 3-kinase superfamily, is recruited. Consequently, ATM protein is activated via autophosphorylation and triggers phosphorylation of a variety of the ATM effectors essential for DNA damage response (DDR) (14, 15), such as checkpoint kinase 2 (CHK2) (16), breast cancer type 1 susceptibility protein (BRCA1) (17), tumor protein p53 (TP53) (18–20), transcription factor E2F1 (21), histone H2AX (the member X of the core histone H2A family) (22, 23), and mediator of DNA damage checkpoint protein 1 (MDC1) (24, 25). Because ATM is essential for DSB repair (14–17), ATM could be a potential target of cancer chemotherapy (8, 26). In contrast, via TP53 phosphorylation, ATM could promote DNA damage-induced apoptosis (18–21). Thus, it is crucial to identify a genetic or epigenetic trait that determines what type of cancer is more likely eliminated by an ATM inhibitor in the presence of cisplatin.
Cisplatin resistance is promoted by activation of the cellular DNA repair machinery, but it can also be enhanced by inactivation of proapoptotic tumor suppressors (6, 7), such as TP53 (18–20) and the bridging integrator 1 protein (BIN1) (27–37). The proapoptotic activity of TP53 primarily depends on its own transcriptional activity (18–20). In contrast, little is known about how BIN1 increases DNA damage-induced apoptosis. BIN1 is the member of the BIN/amphiphysin/Rvs (BAR) family of proteins (27, 28) and is ubiquitously expressed in untransformed tissues (27, 33). In the nucleus, BIN1 interacts with and attenuates the two major cell-cycle–promoting transcription factors, MYC (27, 33, 34, 36) and E2F1 (27, 32, 35), and slows cell-cycle progression (27–36).
Intriguingly, unlike other orthodox tumor suppressors, such as TP53 and BRCA1, which normally preserve the integrity of the genome (38), BIN1 is the first documented tumor suppressor that continuously increases genomic instability (34). BIN1 physically interacts with poly(ADP-ribose) polymerase 1 (PARP1, EC 2.4.2.30) and inhibits the catalytic activity of the enzyme (34, 39, 40). In response to SSBs, PARP1 facilitates the base excision repair (BER) pathway to promote SSB repair (39–41). Because combination therapy using a PARP inhibitor, such as olaparib, with cisplatin has been reported to be clinically useful (8), it is reasonable to speculate that BIN1-mediated PARP1 inhibition is one of the potential mechanisms by which BIN1 increases genomic instability and cisplatin sensitivity (34).
In general, cancer cells, which inactivate tumor suppressors by gene mutation, gene deletion, or epigenetic gene silencing, obtain clonal advantage for growth and survival (38). Consistent with this conjecture, BIN1 expression or its structure is aberrantly regulated in cancer cells by, at least, the following two mechanisms. First, the human BIN1 gene promoter is transcriptionally repressed by the MYC oncoprotein (34). BIN1 was identified originally as a transcriptional corepressor of MYC (27, 28, 33), so the reduction in BIN1 levels by oncogenic MYC is believed to form a positive feedback loop to infinitely accelerate the MYC activity (34). The lack of BIN1 is highly related to the increase in cisplatin resistance (34), so we assume that MYC-dependent BIN1 repression is a mechanism by which MYC elicits cisplatin resistance (36).
Second, the BIN1 exon 12A mRNA, which encodes a unique 43-amino acid (aa) peptide, is aberrantly incorporated, and BIN1 + 12A is thus expressed (28, 31, 33). In the normal brain tissues, the BIN1 exons 12A–D are regularly spliced into the BIN1 mRNA to generate a brain-specific BIN1 isoform (also known as amphiphysin-II) (28). In some advanced cancer tissues, such as metastatic melanoma (31), the exon 12A mRNA is often incorporated next to a highly coiled-coil (CC) structural motif in the BIN1 BAR domain, named BIN1/BAR-CC. We previously identified almost the same region as a new BIN1 effector domain purely biochemically (33). It contains the BIN1 effector domain for cancer suppression (33). Hence, once inserted adjacent to the BIN1/BAR-CC domain, the exon 12A-encoding peptide would disrupt the proper three-dimensional structure of BIN1 protein and compromise its anticancer property (Fig. 1A) (31, 33). However, it remained unknown whether the exon-12A peptide insertion alters BIN1-dependent cisplatin sensitivity.
Figure 1.
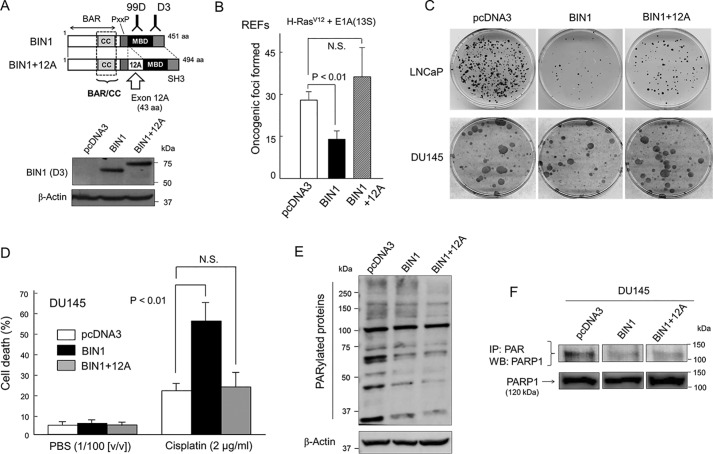
BIN1 and BIN1 + 12A decrease cellular PARylation, but only BIN1 inhibits cancer cell proliferation and increases cisplatin sensitivity. A, schematic diagrams of BIN1 and BIN1 + 12A proteins. The BIN1 exon 12A mRNA, which encodes 43 amino acids (aa) derived from the brain-specific BIN1 mRNA (which is also known as amphiphysin II) (27, 28, 31), is aberrantly incorporated by alternative splicing in several advanced cancers (31). Western blot analysis probed with an anti-BIN1 (clone D3) mAb detected both BIN1 and BIN1 + 12A proteins. BAR, BIN1/amphiphysin/Rvs-homologous domain (27, 28); BAR/CC, a BAR coiled-coil domain, which encodes the BIN1 effector domain (33–35). PxxP, a putative SH3-interacting domain (P is proline and x is any aa); MBD, MYC-binding domain; SH3, Src homology domain 3 (27, 28). B, primary REFs were transformed by cotransfection of two oncogenes, c-H-RasG12V and adenoviral E1A(13S) genes (27). Oncogenic foci were stained with Giemsa's solution and scored (27, 35). N.S. means (statistically) not significant. C, colony formation assays demonstrated that ectopically expressed BIN1 inhibited formation of G418-resistant colonies in LNCaP (a castration-sensitive prostate cancer cell line) but not in DU145 (a castration-resistant prostate cancer cell line). However, BIN1 + 12A transfection failed to suppress the colony-forming activity regardless of the cell lines. G418-resistant colonies were stained with Giemsa's solution and scored (33). D, at 24 h post-transfection with the indicated vectors, growing DU145 cells were treated with cisplatin (2.0 μg/ml) or phosphate-buffered saline (PBS) (1:100 (v/v)) for 72 h and were subjected to the trypan blue exclusion assays. E, Western blot analysis probed with an anti-poly(ADP-ribosyl)ated (PARylated) carbohydrate antibody (anti-PAR antibody) revealed reduced cellular PARylation after overexpression of BIN1 or BIN1 + 12A proteins in plain DU145 cells, which naturally express abundant PARP1 (34). The pcDNA3 empty vector was used as the negative control. F, immunoprecipitation probed with an anti-PAR antibody followed by Western blot analysis with an anti-PARP1 antibody demonstrated the inhibitory effect of overexpressed BIN1 and BIN1 + 12A proteins on the self-PARylation (i.e. automodification) of PARP1 in DU145 cell lysates.
Primary cancer cells are usually sensitive to cisplatin (3–5), but some of them eventually acquire cisplatin resistance (6, 7). To specifically block this malignant conversion, it is extremely important to better understand when and how cancer cells increase chemoresistance. In this study, we investigated the functional interplay between BIN1, E2F1, and ATM in the presence and absence of DNA lesions. In response to DNA damage, ATM activates the DDR pathways and controls chemoresistance in cancer cells (8, 14–16). However, little is known about a signaling path through which ATM is activated before chromosomal DNA is damaged (14, 15). Here, we report E2F1-specific signal circuits, by which ATM is constitutively activated and provokes cisplatin resistance when BIN1 is lacking.
Results
BIN1 and BIN1 + 12A inhibit cellular PARylation, but only BIN1 displays anticancer activities
As reported previously (27, 31), overexpression of BIN1 inhibited formation of oncogenic foci (i.e. piled masses of cells caused by the loss of contact inhibition and anchorage dependence) in rat embryonic primary fibroblasts (REFs). Oncogenic foci formation was routinely achieved by cotransfection of activated H-RasG12V and adenovirus E1A(13S) genes in rodent embryonic primary fibroblasts (27). However, BIN1 + 12A did not inhibit Ras/E1A cotransformation (Fig. 1B).
BIN1 inhibits cancer cell growth, at least in part, by inhibiting c-MYC (27) and E2F1 (35), two major cell-cycle–promoting transcription factors. In human LNCaP (castration-sensitive prostate cancer) cell line, ectopically expressed BIN1 markedly reduced colony-forming activity, and this anticancer property was evidently mitigated by the presence of exon 12A (Fig. 1C). In contrast, in human DU145 (castration-resistant prostate cancer) cell line, BIN1 did not inhibit the colony formation as efficiently as in LNCaP cells (Fig. 1C). This was not because BIN1 was entirely dormant in this advanced cancer cell line. Overexpressed BIN1 greatly increased cisplatin sensitivity of DU145 cells, but BIN1 + 12A did not change it (Fig. 1D). Thus, the mechanism by which BIN1 increases cisplatin sensitivity is compromised by the exon 12A–encoding peptide.
Inhibition of PARP1 was thought to be a likely way to increase cisplatin sensitivity of cancer cells (8). Therefore, we hypothesized that PARP1 inhibition could be a mechanism by which BIN1 (but not BIN1 + 12A) increases cisplatin sensitivity. The PARP1 activity is generally gauged by poly(ADP)-ribosylation (PARylation) of various nuclear proteins, including histone H1 and PARP1 itself (39, 40). To visualize the catalytic activity of PARP1, we performed a Western blot analysis probed with an anti-poly(ADP-ribose) (PAR) antibody. This is a semi-quantified method to roughly estimate global cellular PARylation (34, 35, 39, 40). As predicted, the intensity of the numerous PAR-positive bands vastly increased after brief treatment with hydrogen peroxide, H2O2 (a PARP activator), whereas those signals were greatly diminished by a PARP-specific inhibitor, such as PJ-34 and olaparib (Fig. S1) (34, 35). Importantly, the intensity of PAR-positive bands was visibly reduced by ectopically expressed BIN1, whereas the PAR-positive signals clearly increased when BIN1 was depleted (Fig. S2). Furthermore, in situ immunofluorescence signals probed with the anti-PAR antibody showed that the PAR-positive images detected exclusively in the nucleus were evidently enhanced by H2O2, particularly when BIN1 was deficient (Fig. S3). We concluded that the intensity of PAR-positive protein bands reflect global PARylation activity, which was counteracted by the presence of BIN1.
Interestingly, overexpressed BIN1 and BIN1 + 12A almost equally inhibited the cellular PARylation under optimal culture conditions (Fig. 1E). This suggests that BIN1 and BIN1 + 12A inhibit the catalytic activity of PARP1 to a similar extent, possibly because BIN1 and BIN1 + 12A commonly contain the BIN1/BAR-CC effector domain (see Fig. 1A), through which BIN1 binds PARP1 and inhibits its catalytic activity (34). We noticed that there were a few PARylated bands, including a 100-kDa band, the intensity of which was not lessened by ectopically expressed BIN1 and BIN1 + 12A (Fig. 1E). BIN1 is known to directly bind PARP1 by recognizing the PARP1 automodification domain (34). We therefore assumed that those PARylated proteins resistant to BIN1/BIN1 + 12A might be PARylated by a PARP1-related enzyme, such as PARP2, which does not have the automodification domain (39, 40). Because those BIN1/BIN1 + 12A–resistant PARylated proteins were not major, we concluded that the overall impact of those “BIN1-resistantly PARylated” proteins was limited.
Besides histone H1, PARP1 is one of the major substrates of PARP1-dependent PARylation in the nucleus (34, 35, 39, 40). As the self-PARylation (i.e. automodification) of PARP1 is a crucial step for initiation of the BER-mediated SSB-repair pathway (39, 40), we performed a co-immunoprecipitation (co-IP) experiment using an anti-PAR antibody, followed by Western blot analysis probed with an anti-PARP1 antibody in the presence of BIN1 or BIN1 + 12A. We found that BIN1 and BIN1 + 12A equally inhibited the PARP1 automodification (Fig. 1F). Because BIN1 + 12A did not increase cisplatin sensitivity (see Fig. 1D), it was apparent that inhibition of PARP1 activity by BIN1 + 12A was insufficient to elicit cisplatin sensitivity. We conclude that, in parallel to (or independent of) the PARP1 inhibition by BIN1 (34), BIN1 curbs a PARP1-independent function to elicit cisplatin sensitivity.
BIN1 inhibits the formation of γH2AX foci irrespective of PARP1, whereas BIN1 + 12A does not alter γH2AX
During DNA replication, SSBs are readily converted to DSBs after the collapse of replication forks (13). Because BIN1 and BIN1 + 12A similarly inhibit the catalytic activity of PARP1 (see Fig. 1, E and F), BIN1 and BIN1 + 12A are supposed to equally produce SSBs by limiting PARP1-dependent SSB repair (39–41). Thus, we hypothesized that BIN1 and BIN1 + 12A would similarly generate DSBs during DNA replication. To prove this theory, we monitored the generation of phosphorylated Ser-139 of histone H2AX (forming γH2AX) when BIN1 and BIN1 + 12A are ectopically expressed. The emergence of γH2AX foci is known to correspond to an increase in DSBs (22, 23). Paradoxically, under optimal conditions, ectopically expressed BIN1 significantly reduced the formation of γH2AX foci, whereas BIN1 + 12A did not alter it (Fig. 2A). Even in the presence of cisplatin, which robustly increased γH2AX foci, transfected BIN1 efficiently mitigated γH2AX formation in mouse fibroblasts (Fig. 2B) and DU145 cells (Fig. S4A).
Figure 2.
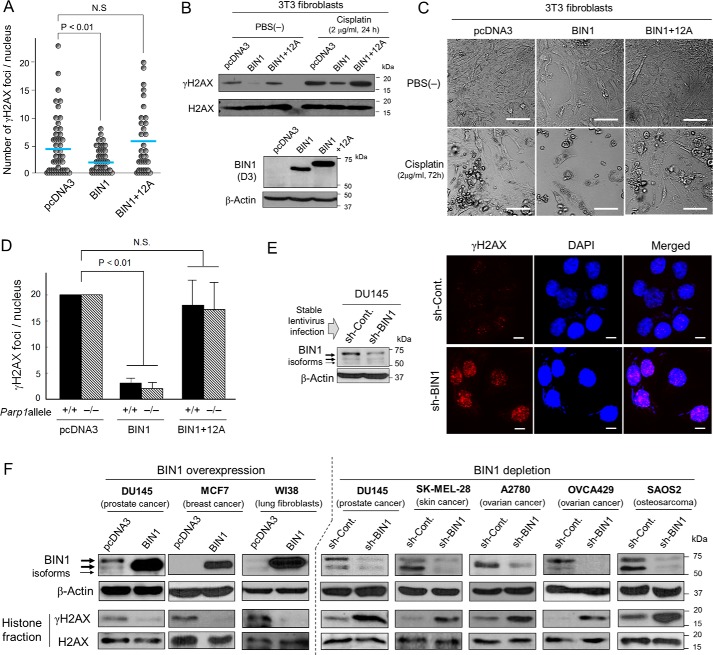
BIN1 suppresses the formation of γH2AX foci, but BIN1 + 12A does not. A, scatter plot analysis of the number of γH2AX foci per nucleus in mouse 3T3 fibroblasts, transfected with pcDNA3, BIN1, and BIN1 + 12A expression vectors. Horizontal bars indicate mean values. One of the three independent results is shown. N.S. means not significant. B, Western blot analysis of histone fractions of mouse 3T3 fibroblasts transiently transfected with the indicated vectors. Approximately 24 h post-transfection, cells were treated with/without cisplatin (2.0 μg/ml, 24 h). The amounts of heterologously expressed BIN1 and BIN1 + 12A proteins were verified by Western blot analysis probed with an anti-BIN1 (D3) antibody. C, morphology of mouse 3T3 fibroblasts transiently transfected with the indicated vectors in the presence and absence of cisplatin (2.0 μg/ml, 72 h). Scale bar, 100 μm. D, mouse 3T3 fibroblasts with (+/+) and without (−/−) Parp1 allele (35) were transfected with the indicated vectors. Approximately 48 h post-transfection, in situ fluorescence microscopy analyses were conducted using an anti-BIN1 (D3) antibody, which does not detect (endogenous) mouse Bin1 protein (W. P. Folk, A. Kumari, T. Iwasaki, and D. Sakamuro, unpublished observations.), and an anti-γH2AX antibody. The number of γH2AX foci only in BIN1 (or BIN1 + 12A)-positive nuclei was counted. E, Western blot analysis of DU145±sh-BIN1 cell lysates (left). The depletion of endogenous BIN1 was accomplished by stable infection of the recombinant lentivirus expressing BIN1 sh-RNA (short-hairpin RNA) (sh-BIN1). The scrambled (control) sh-RNA (sh-Cont.)-expressing lentivirus was used as the negative control. Under optimal culture conditions (right), in situ immunofluorescence analysis displayed γH2AX foci (red) in the DU145±sh-BIN1 cell nuclei, which were counterstained with DAPI (blue). Scale bar, 10 μm. F, Western blot analysis verified that overexpression of BIN1 by the transient transfection of pcDNA3-BIN1 for 48 h broadly reduced γH2AX formation, whereas depletion of endogenous BIN1 via stable expression of sh-BIN1 evidently increased it.
In general, heterologous expression of BIN1 does not cause apoptosis in untransformed cells unless the cells are under stress conditions, such as DNA damage or oncogenic stress (27, 32, 33). Consistent with this fact, the 3T3 fibroblasts did not display any damaged (i.e. apoptotic) morphology after transient transfection of BIN1 under optimal culture conditions (Fig. 2C). As shown above (see Fig. 1C), the transfection of BIN1 generally activates apoptosis in early-stage cancer cells, such as LNCaP (33), whereas this was not the case in some advanced cancer cells, such as DU145 (Fig. S4B). These results indicated that BIN1-induced γH2AX reduction was not a consequence of the impairment of cell viability by BIN1.
To determine whether BIN1 inhibits γH2AX formation in the absence of PARP1, BIN1 and BIN1 + 12A expression vectors were transiently transfected in the 3T3(−/−) fibroblasts, in which the Parp1 allele was genetically eliminated (39). The 3T3(+/+) fibroblasts express the intact Parp1 gene, thus being used as the control cell line. Irrespective of the status of the Parp1 allele, heterologous expression of BIN1 markedly attenuated the γH2AX formation, whereas BIN1 + 12A did not change it (Fig. 2D). We conclude that PARP1 is not involved in BIN1-dependent γH2AX suppression.
We next tested whether the formation of γH2AX foci increases when endogenous BIN1 decreases. For long-term BIN1 silencing, we used the replication-incompetent sh-BIN1–expressing lentivirus that constitutively expresses short-hairpin RNAs (sh-RNAs) directed against human BIN1 mRNA (28). For short-term BIN1 depletion, we transiently transfected si-BIN1 RNA duplexes, which were small-interfering RNAs (si-RNAs) designed to specifically cleave the corresponding three separate nucleotide (nt) sequences that the sh-BIN1–plasmid DNAs also cleave (Fig. S5A). The efficiency of BIN1 depletion by si-BIN1s was confirmed by separate transfection of si-BIN1 followed by Western blot analysis (Fig. S5B). The relevant phenotype caused by the si-BIN1 transfection (i.e. endogenous E2F1 activation) was verified in DU145 (BIN1-positive) cells. As the negative control, the MCF7 (BIN1-null) cell line was used (Fig. S5C). The E2F1 activity elicited by the BIN1 depletion was offset by the overexpression of BIN1 cDNA (Fig. S5D).
Endogenous BIN1 protein (including the minor alternative splicing isoforms) around 70 kDa was evidently reduced by the sh-BIN1–expressing lentivirus infection in DU145 cells, whereas formation of γH2AX foci was massively enhanced even under optimal culture conditions (Fig. 2E and Fig. S6). This was not a cell line-specific event or a by-chance result because, in various human cell lines, heterologously expressed BIN1 consistently decreased γH2AX formation. In contrast, the reduction in BIN1 always steadily increased γH2AX (Fig. 2F). We concluded that BIN1 generally inhibited γH2AX formation in a manner independent of PARP1 but vulnerable to the presence of the exon 12A-encoding peptide.
BIN1-dependent γH2AX inhibition is not attributable to apoptotic chromatin condensation or activation of DSB repair by BIN1
Because BIN1 is a proapoptotic protein (27, 32, 33, 35) and increases cisplatin sensitivity (see Fig. 1D) (34), we thought that the nuclear chromatin machinery essential for γH2AX formation might be instinctively injured by BIN1-dependent aberrant chromatin condensation, a typical morphological nuclear index of apoptosis (32, 34). However, in situ immunostaining assays revealed that, while ectopically expressed BIN1 effectively suppressed γH2AX formation even in the presence of cisplatin, the morphology of cellular nuclei stained by DAPI was almost intact (Fig. 3A). This suggests that BIN1-dependent γH2AX reduction occurs before apoptotic chromatin condensation emerges.
Figure 3.
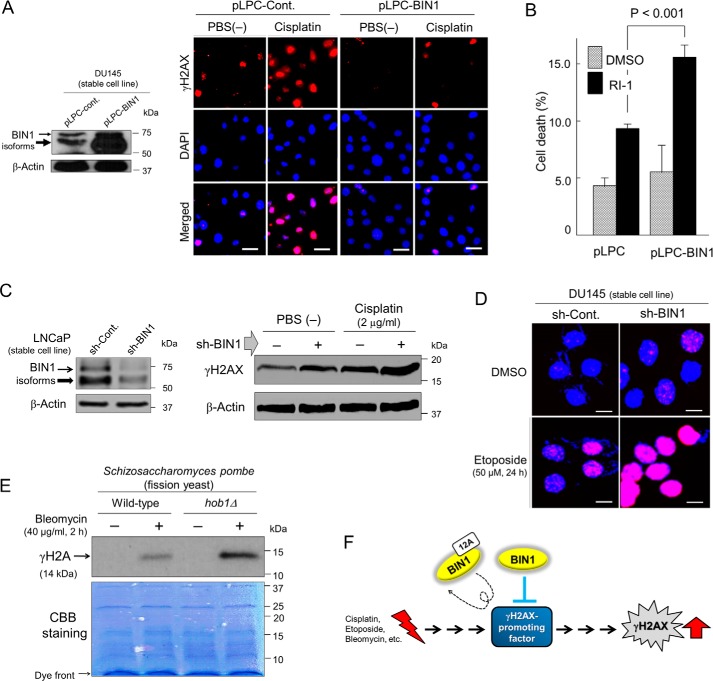
DNA damage-induced γH2AX formation is counterbalanced by BIN1. A, DU145/pLPC-BIN1 cell line stably expressing BIN1 (isoform 8) (27, 28) was established previously with recombinant pLPC-BIN1 retroviral infection (34). The cells were treated with cisplatin (2.0 μg/ml) or PBS (1:100 (v/v)) for 24 h and were subjected to in situ fluorescence microscopy analysis probed with an anti-γH2AX antibody (red). Nuclei were counter-stained with DAPI (blue). Scale bar, 25 μm. B, proliferating DU145±pLPC-BIN1 cell lines were incubated with RI-1 (30 μm), a chemical inhibitor of RAD51 (42), for 72 h. The loss of cellular viability was detected with trypan blue exclusion assay. C, LNCaP±sh-BIN1 cell lines were briefly exposed to cisplatin (2.0 μg/ml) for 24 h and were subjected to Western blot analysis probed with an anti-γH2AX antibody. D, DU145±sh-BIN1 cell lines were briefly exposed to etoposide (50 μm) for 24 h and were subjected to in situ fluorescence immunostaining probed with an anti-γH2AX antibody (pink). Nuclei were counter-stained with DAPI (blue). Scale bar, 10 μm. E, S. pombe (fission yeast) cells with intact hob1 allele (WT) or with hob1 deletion (hob1Δ) (43) were treated with bleomycin (40 μg/ml) (+) or PBS(−) (1:100 (v/v)) (−) for 2 h and were subjected to Western blotting analyses probed with an anti-(yeast) γH2A antibody (44). CBB, Coomassie Brilliant Blue. F, we hypothesize that BIN1 (not BIN1 + 12A) constitutively inhibits a “γH2AX-promoting (cellular) factor” that is responsible for the formation of γH2AX foci in response to DNA damage. Therefore, in the absence of BIN1, the “γH2AX-promoting factor” may be automatically hyper-activated even in the absence of DSBs.
It was unlikely that BIN1 (an inducer of genomic instability) would promote cellular DSB-repair activity. To prove this unlikelihood, we used RI-1, a small molecule inhibitor of RAD51, a DNA recombinase essential for DSB repair via homologous recombination (42). If BIN1 assists DSB repair, thereby reducing γH2AX, we assumed that DSB-induced cell death increased by RI-1 could be somehow mitigated by overexpressed BIN1. However, under optimal conditions, ectopically expressed BIN1 did not decrease the RI-1–induced cell death but rather promoted it (Fig. 3B). This result implies that BIN1-induced γH2AX silencing is not attributable to an increase in cellular DSB-repair activity. Consistently, the γH2AX formation caused by cisplatin (Fig. 3C) or etoposide (a chemical inhibitor of topoisomerase II) (Fig. 3D) (32) was enhanced further by the BIN1 loss. Our results indicate that BIN1 is directly involved in a mechanism silencing γH2AX. Given the central role of γH2AX formation in cellular DDR signals (8, 23), we investigated whether the effect of BIN1 on γH2AX reduction is evolutionarily conserved.
To do this, we used WT Schizosaccharomyces pombe (fission yeast), which carries the BIN1 gene ortholog, hob1 (homolog of BIN1), and the hob1-knockout fission yeast strain (hob1Δ) (43). We monitored γH2A (phosphorylated serine 129 in yeast H2A), which is equivalent to γH2AX in human (44). Western blot analysis (Fig. 3E) and in situ immunostaining assay (Fig. S7) showed that brief treatment with bleomycin, a radiomimetic chemical, visibly increased the formation of γH2A in hob1Δ strain more efficiently than in the WT yeast strain. We conclude that inhibition of γH2AX formation by BIN1/hob1 is evolutionarily conserved between yeast and human. We hypothesized a “γH2AX-promoting factor,” which is regularly activated by DSBs, is constitutively suppressed by BIN1/hob1 (Fig. 3F).
BIN1 physically interacts with E2F1 and prevents E2F1 from activating the human ATM promoter, whereas BIN1 + 12A does not bind E2F1
In response to DSBs, serine 139 in H2AX is phosphorylated, thus generating γH2AX (22). This process is catalyzed by the phosphatidylinositide 3-kinase family of enzymes, including ATM, ataxia-telangiectasia and Rad3-related kinase (ATR), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (22, 23). We found that BIN1 depletion increased the amount of ATM protein with minimal effects on the levels of ATR and DNA-PKcs (Fig. 4A). Quantitative RT-PCR (qRT-PCR) revealed that BIN1 depletion increased ATM transcription (Fig. 4B).
Figure 4.
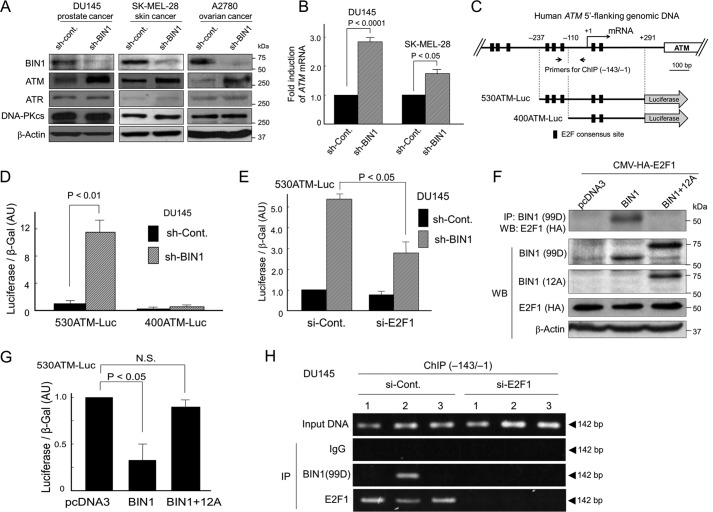
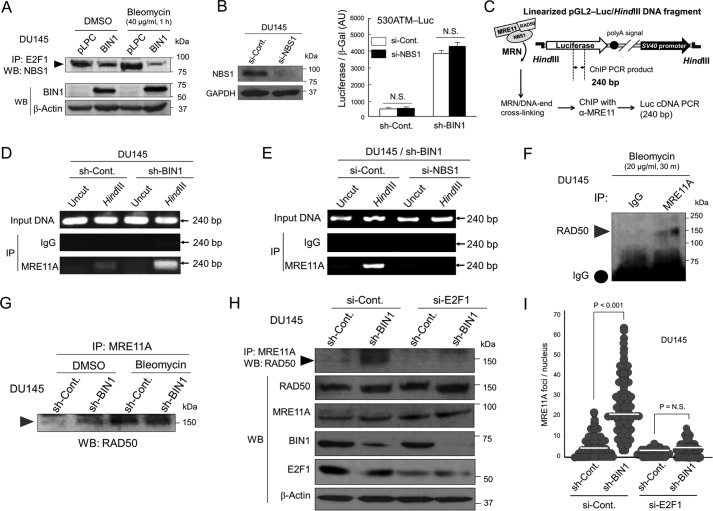
BIN1 physically interacts with E2F1 and prevents E2F1 from transcriptionally activating the human ATM promoter, whereas BIN1 + 12A does not bind E2F1. A, Western blot analysis determined the amounts of BIN1 (∼70 kDa), ATM (∼350 kDa), ATR (∼317 kDa), and DNA-PKcs (∼470 kDa) in the indicated cell lines stably expressing sh-BIN1 or scrambled (control) sh-RNA (sh-Cont). B, quantitative real-time RT-PCR (qRT-PCR) analysis of ATM mRNA expressed in the indicated cell lines±sh-BIN1 under optimal culture conditions. GAPDH mRNA was used as the internal control. C, schematic diagram of two human ATM promoter-driven luciferase reporters. The 530ATM promoter (−237/+291) contains three E2F-consensus sites upstream from the transcription initiation site (+1), whereas the 430ATM promoter (−110/+291) does not. For ChIP analysis, the (−143/−1) region of the human ATM promoter was amplified by genomic PCR. D, indicated Luc reporter vectors were transiently transfected in DU145±sh-BIN1 cells. The raw luciferase activities were normalized with the cotransfected β-gal (pcDNA3–β-gal: 1:10 (w/w)) activity. AU, arbitrary unit. E, E2F1 siRNA (si-E2F1) or scrambled control siRNA (si-Cont) was cotransfected with 530ATM-Luc in DU145±sh-BIN1 cells. F, co-IP/Western blotting (WB) analysis of transfected BIN1 (or BIN1 + 12A) and HA-tagged E2F1 proteins. For IP of BIN1 and BIN1 + 12A, an anti-BIN1 mAb (clone 99D), which recognizes the BIN1 exon 13, was used. To detect the exon 12A, we used an anti-BIN1 exon 12A-specific antibody. HA, hemagglutinin. G, 530ATM-Luc vector was cotransfected in growing DU145 cells with BIN1 and BIN1 + 12A expression vectors. AU, arbitrary unit. N.S., not significant. H, 530ATM-Luc vector was transiently cotransfected with the indicated vectors: 1) pcDNA3; 2) pcDNA3-BIN1; 3) pcDNA3-BIN1 + 12A, for 48 h. To deplete endogenous E2F1, si-E2F1 was cotransfected for 48 h. The scrambled siRNAs (si-Cont) were used as the negative control. The cell lysates treated with formaldehyde were subjected to IP probed with the indicated antibodies. The (−143/−1) region of the human 530ATM promoter region was amplified by genomic PCR.
The nt sequence of the human ATM promoter contains several E2F-consensus sites and can be directly transactivated by E2F1 (45). We previously reported that, in a manner dependent on the BIN1/BAR-CC effector region (see Fig. 1A), BIN1 physically interacts with the central region of E2F1, which contains the marked box domain of E2F1, thereby acting as an E2F1-binding corepressor (35). Even in the absence of the retinoblastoma 1 (RB1) transcriptional corepressor, the authentic E2F1-interacting corepressor BIN1 inhibits E2F1-dependent transactivation (35). Thus, we hypothesized that BIN1-dependent E2F1 repression is a mechanism by which BIN1 reduces ATM levels.
To test whether BIN1 attenuates the E2F1-sensitive ATM promoter, two luciferase reporter vectors, 530ATM-Luc and 400ATM-Luc, were generated with genomic PCR cloning. The 530ATM (−237/+291) DNA fragment contained three E2F-consensus sites upstream from the ATM transcription initiation site (+1), whereas the 400ATM (−110/+291) DNA fragment did not (Fig. 4C). In the DU145/sh-BIN1 cell line that constitutively expresses sh-BIN1 by recombinant lentivirus infection, the 530ATM-Luc activity was evidently increased, whereas the induction of 400ATM-Luc activity was minimal (Fig. 4D). Moreover, using the same stable BIN1-deficient cell system, the 530ATM-Luc activity was robustly activated by the BIN1 depletion but was significantly diminished by the cotransfection of the si-RNAs directed against E2F1 mRNA (si-E2F1) (Fig. 4E).
The BIN1/BAR-CC effector domain, which is required for BIN1/E2F1 binding (35), is commonly present in BIN1 and BIN1 + 12A (see Fig. 1A), implying that BIN1 + 12A might also inhibit E2F1 activity via physical binding. Interestingly, co-IP/Western blot analysis showed that BIN1 directly bound E2F1 in vivo, but BIN1 + 12A did not (Fig. 4F). Heterologous expression of BIN1 reduced the 530ATM-Luc activity, whereas BIN1 + 12A did not (Fig. 4G). Western blot analysis confirmed that ectopically expressed BIN1 + 12A was detectable as efficiently as BIN1 (see Figs. 1A and 2B). Furthermore, transfected BIN1 and BIN1 + 12A proteins were detected only in the nucleus (Fig. S8), suggesting that the exon 12A peptide would not alter the BIN1 protein stability and its nuclear localization. These results suggest that insertion of the exon 12A peptide at a position adjacent to the BIN1/BAR-CC effector domain perturbs a proper spatial interplay between BIN1 and E2F1 proteins in the nucleus.
Chromatin immunoprecipitation (ChIP) assays demonstrated that BIN1 protein was present on the 530ATM-Luc promoter, whereas BIN1 + 12A was not detectable on the same promoter. Furthermore, BIN1 protein was detected on the 530ATM-Luc promoter only when endogenous E2F1 was present there (Fig. 4H). We conclude that E2F1-dependent activation of the ATM promoter is repressed by BIN1 in a manner dependent on the E2F1/BIN1 interaction.
BIN1 loss promotes E2F1-dependent MRN formation in an E2F1 transcription-independent manner
It has been reported that E2F1 directly interacts with the N terminus of Nijmegen breakage syndrome protein 1 (NBS1), one of the key elements of the MRE11A/RAD50/NBS1 (MRN) DNA end-binding protein complex (46, 47). The central region of E2F1, which contains the E2F1 MB domain, is required not only for the BIN1/E2F1 interaction in vivo (35) but also for the NBS1/E2F1 binding (47–49). Therefore, we wondered whether BIN1 might interfere with the E2F1/NBS1 complex in vivo. As predicted, co-IP/Western blot analysis revealed that the endogenous E2F1/NBS1 protein complex, which was prominently stabilized by bleomycin, was lessened by ectopically expressed BIN1 (Fig. 5A).
Figure 5.
E2F1 is vital for MRN formation in BIN1-deficient nuclei under optimal culture conditions. A, co-IP/Western blot analysis of endogenous E2F1/NBS1 protein complex in DU145±pLPC-BIN1 cell lines in the presence and absence of bleomycin (40 μg/ml, 1 h). B, NBS1 siRNA (si-NBS1) or scrambled control siRNA (si-Cont) was cotransfected with 530ATM-Luc in DU145±sh-BIN1 cells. The raw luciferase activities were normalized with the cotransfected β-gal (pcDNA3–β-gal: 1:10 (w/w)) activity. AU, arbitrary unit. N.S., not significant. C, schematic diagram of ChIP-based in vivo DNA end-binding assay. Cells were transfected with the pGL2-control luciferase (Luc) plasmid DNA linearized by HindIII restriction (pGL2/HindIII-Luc DNA fragment). The super-coiled (uncut) pGL2-Luc plasmid DNA was used as the negative control. D, to determine whether a BIN1 loss enhances the MRE11A/DNA end-binding activity, the indicated 240-bp region of the Luc cDNA was amplified by genomic PCR after an immunoprecipitation with an anti-MRE11A antibody in the DU145±sh-BIN1 cell lysates treated with formaldehyde. E, ChIP-based DNA end-binding assays verified that endogenous NBS1 is vital for the MRE11/DNA-end interaction. To deplete endogenous NBS1 protein, si-NBS1 was cotransfected. F, co-IP/Western blot analysis demonstrated the physical binding of endogenous MRE11A with RAD50 in the presence of bleomycin (20 μg/ml, 30 min). G, co-IP/Western blot analysis verified that similarly to bleomycin treatment, the loss of BIN1 stabilizes the MRE11A/RAD50 protein complex in vivo. H, co-IP/Western blot analysis revealed that the BIN1 loss-mediated stabilization of the MRE11A/RAD50 protein complex was canceled by depleting E2F1. I, scatter plot analysis of the MRE11A foci per nucleus in the DU145±sh-BIN1 (stable) cell lines after transient transfection of si-E2F1 or si-Control. The cells were counterstained with an anti-E2F1 antibody, and the number of MRE11A foci in si-E2F1-transfected nuclei was counted. Horizontal bars indicate mean values.
BIN1 is a potent E2F1 corepressor, so the lack of BIN1 is sufficient to increase E2F1-dependent transcription (35). This means that, in the absence of BIN1, the E2F1 MB domain is not occupied by BIN1. If so, the NBS1/E2F1 binding via the E2F1 MB domain would be more stabilized. Therefore, we tested whether NBS1 controls E2F1-dependent ATM gene transcription in the absence of BIN1. To test this possibility, the si-RNA directed against the human NBS1 mRNA (si-NBS1) was transiently co-transfected with the 530ATM-Luc plasmid DNA in the DU145/sh-BIN1 cells. We observed that the reduction in NBS1 levels did not alter the ATM promoter activity in the absence of BIN1 (Fig. 5B). We conclude that the NBS1/E2F1 interaction does not participate in the E2F1-dependent ATM gene transcription.
Meiotic recombination-11 homolog-A (MRE11A) protein is a hub element of the MRN complex. It physically recognizes dsDNA termini, while binding both RAD50 and NBS1 proteins. Thus, MRE11A is believed to physically tether the entire MRN complex to DNA ends (46, 47). To study the effect of the E2F1/NBS1 direct interaction on the MRN formation in the absence of BIN1, we monitored the retention of endogenous MRE11A protein at the ends of broken DNA fragments in vivo in the presence and absence of NBS1 in BIN1-deficient cancer cells.
To visualize the presence of MRE11A protein at the DNA termini in transfected cells, we developed a ChIP-based in vivo DNA end-binding assay (Fig. 5C). The binding affinity of endogenous MRE11A protein to the linearized pGL2-Luc DNA fragments was prominently increased when endogenous BIN1 protein was depleted. The super-coiled (uncut) pGL2-Luc plasmid DNA, which did not have any broken DNA termini, was used as the negative control (Fig. 5D). Interestingly, the DNA end-binding efficiency of endogenous MRE11A protein in BIN1-deficient cells was almost completely abolished when si-NBS1 was cotransfected (Fig. 5E). These results suggest that the loss of BIN1 enhances the physical retention of MRE11A protein at DNA termini in an NBS1-dependent manner.
To study a possible inhibitory effect of BIN1 on the MRN formation through other MRN components, we tested whether the lack of BIN1 facilitates in vivo protein/protein interaction between MRE11A and RAD50, a coiled-coil adaptor protein containing a zinc hook domain through which the homodimerization of MRN is mediated (46). As expected, co-IP/Western blotting analyses revealed that the physical binding between endogenous MRE11A and RAD50 proteins in vivo was evidently enhanced by bleomycin (Fig. 5F). Notably, even in the absence of DSBs, the MRE11A/RAD50 binding was clearly promoted by BIN1 loss (Fig. 5G). Furthermore, the MRE11A/RAD50 complex in the absence of BIN1 was almost entirely eliminated by E2F1 depletion (Fig. 5H). These results suggest that, by directly interacting with NBS1, E2F1 stabilizes MRE11/RAD50 complex in vivo in BIN1-deficient cellular nuclei.
The E2F1-dependent stabilization of the protein/protein interaction between key elements of the MRN complex in BIN1-deficient cellular nuclei was also verified by in situ MRN/MRE11A foci in BIN1-deficient cells. In situ fluorescence immunostaining experiments demonstrated that, under optimal culture conditions, the lack of BIN1 robustly increased formation of MRE11A foci, which was evidently abolished by the cotransfection of si-E2F1 (Fig. 5I). BIN1 loss did not greatly increase the MRE11A mRNA, but slightly stabilized MRE11A protein (Fig. S9). Therefore, even in the absence of DSBs, E2F1 directly promotes the formation of MRN foci in a manner dependent on the E2F1/NBS1 binding but independent of E2F1-mediated transcription, particularly when BIN1 is deficient.
BIN1 loss constitutively promotes ATM autophosphorylation in an E2F1/MRE11A-dependent manner
In response to DSBs, the MRN complex physically recruits the ATM homodimers (the latent form), which is then activated via autophosphorylation at Ser-1981 to become an active ATM monomer (14, 15). In contrast, even in the absence of DSBs (this study), the ATM transcription is enhanced by E2F1 in BIN1-deficient cells (see Fig. 4). The formation of MRE11A foci is also directly promoted by E2F1, when BIN1 is deficient (see Fig. 5). Therefore, we examined whether ATM is autophosphorylated in BIN1-deficient cells under optimal culture conditions.
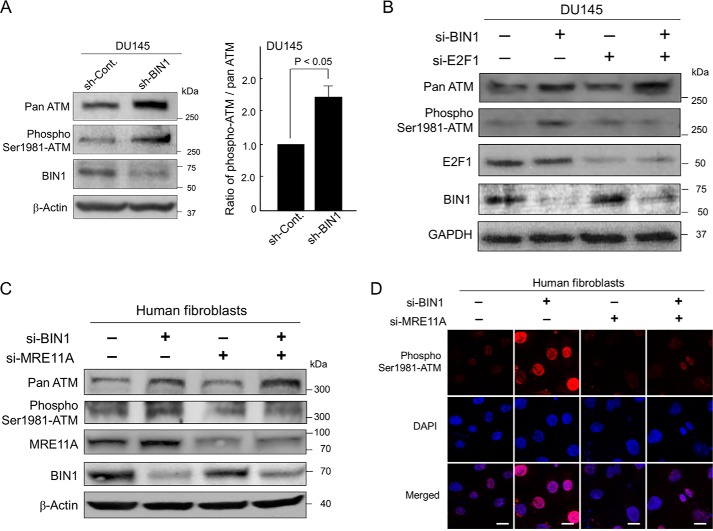
Western blotting experiments and subsequent densitometry analysis revealed that, in the absence of BIN1, ATM autophosphorylation augmented more efficiently than the total amount of ATM protein (Fig. 6A). Furthermore, the transient transfection of si-E2F1 evidently decreased ATM autophosphorylation in BIN1-depleted cells, whereas the high levels of pan-ATM protein were sustained after the si-E2F1 transfection for a while under the experimental condition we employed (Fig. 6B). This result was seemingly attributable to slow turnover of pooled ATM protein in response to transfected si-E2F1. These results suggest that, besides E2F1-dependent ATM transcription, a separate but still E2F1-mediated mechanism promotes ATM autophosphorylation.
Figure 6.
ATM autophosphorylation requires both E2F1 and MRE11A in BIN1-deficient cells. A, Western blotting (left) and densitometry analysis (right) of the amount of pan-ATM and pSer-1981 ATM proteins in DU145±sh-BIN1 cell lines cultured under optimal conditions. β-Actin was used as the loading control. B, Western blot analysis of pan-ATM, pSer-1981 ATM, E2F1, and BIN1 in DU145±sh-BIN1 cell lines transiently transfected with si-E2F1 or si-Cont. GAPDH was used as the loading control. C, Western blot analysis of pan-ATM, pSer-1981 ATM, MRE11A, and BIN1 in the human fibroblast cell line (GM00637) transiently transfected with si-BIN1, si-MRE11A, or si-Cont. β-Actin was used as the loading control. D, in human normal (GM00637) fibroblasts cultured under optimal conditions, the effects of transient transfection with si-BIN1 alone, si-MRE11 alone, and the combination of these two siRNAs on pSer-1981 ATM were analyzed with in situ immunofluorescence microscopy, probed with an anti-phospho-ATM (Ser-1981)-specific antibody (red). Nuclei were counter-stained with DAPI (blue). Scale bar, 10 μm.
According to the critical role of E2F1 in the formation of MRN foci in the absence of BIN1 (see above, Fig. 5), we investigated whether the impairment of E2F1-dependent MRN formation would reduce the ATM autophosphorylation in BIN1-deficient cells. Western blot analysis (Fig. 6C) and in situ immunostaining experiment (Fig. 6D) verified that the transient transfection of MRE11A si-RNA (si-MRE11A) clearly reduced the ATM autophosphorylation mediated by the BIN1 deficiency. We conclude that ATM autophosphorylation is directly promoted by E2F1 and subsequent MRN formation when BIN1 is absent, even under optimal culture conditions.
ATM is essential for BIN1 loss-dependent γH2AX formation and subsequent MDC1 phosphorylation, irrespective of phosphorylated Tyr-142 of H2AX
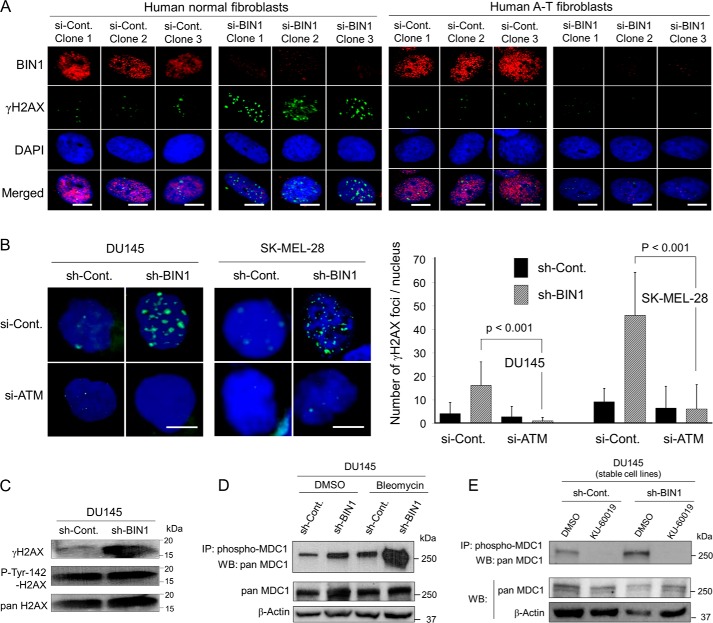
We determined whether the DSB-independent ATM activation mediated by a BIN1 loss is functionally meaningful. To do this, we investigated whether BIN1 deficiency promotes γH2AX formation in the absence of ATM. Under optimal culture conditions, the γH2AX formation by the BIN1 loss was clearly detectable in normal human fibroblasts, whereas it was undetectable in human ataxia telangiectasia (A-T) fibroblasts, in which endogenous ATM was hereditary-deficient (Fig. 7A). Similarly, in two independent human cancer cell lines, DU145 (prostate cancer) and SK-MEL-28 (skin cancer), BIN1 loss-induced γH2AX foci were markedly reduced by transiently transfected ATM siRNA (si-ATM) (Fig. 7B). We concluded that ATM was vital for DSB-independent γH2AX formation in BIN1-depleted cells.
Figure 7.
ATM is essential for both γH2AX formation and MDC1 phosphorylation in BIN1-depleted cells. A, human normal (GM00637) and ataxia-telangiectasia family-derived, i.e. ATM-deficient, GM16666 fibroblasts were transiently transfected with the BIN1 siRNA (si-BIN1) or the control siRNA (si-Cont). Approximately 48 h post-transfection, cells were analyzed with in situ fluorescence immunostaining. Endogenous BIN1 in the nuclei (red) and γH2AX foci (green) were detected with an anti-BIN1 antibody (99D) and an anti-γH2AX antibody, respectively. Nuclei were counterstained with DAPI (blue). Scale bar, 10 μm. Three representative images of each transfection are shown. B, γH2AX foci (green) were detected with in situ immunofluorescent staining after transient transfection of the ATM siRNA (si-ATM) or the control siRNA (si-Cont) in two cancer cell lines in the presence and absence of BIN1 (DU145±sh-BIN1 and SK-MEL-28±sh-BIN1). ∼100 nuclei in each transfection experiment were analyzed, and three independent transfection experiments were performed. The nuclei were counterstained with DAPI (dark blue). Scale bar, 10 μm. Western blotting analyses in the DU145±sh-BIN1 stable cell lines are demonstrated. C, induction of γH2AX by BIN1 depletion and phosphorylation of tyrosine 142 (pTyr-142) of H2AX. D, phosphorylation of MDC1 in the absence of BIN1 with/without brief treatment with bleomycin (20 μg/ml, 30 min); E, inhibitory effect of KU-60019, an ATM-selective chemical inhibitor (3 μm, 72 h), on the impaired BIN1-induced MDC1 phosphorylation.
If the level of DSBs is tolerable, cells generally promote DSB repair/cell survival but not apoptosis (23). To promote DSB repair and cell survival in response to DSBs, Mediator of DNA damage checkpoint protein 1 (MDC1) needs to physically bind γH2AX, thus being ATM-dependently phosphorylated and activated for DSB repair and subsequent cell survival (24, 25). However, if phosphorylated tyrosine 142 of histone H2AX remains phosphorylated, MDC1 would not physically bind γH2AX, probably because of steric interference between the pTyr-142 residue and the pSer-139 residue (i.e. γH2AX) within an H2AX molecule (50, 51). If so, ATM-dependent MDC1 phosphorylation (and subsequent activation of DSB repair) would not occur (50). Besides γH2AX, the phosphorylation status of Tyr-142 of H2AX seemed to be another determinant for cells to select either cell survival or apoptosis (50, 51).
In general, the loss of BIN1 is expected to protect cancer cells from DNA damage-induced apoptosis (32, 34, 35). Concurrently, BIN1 deficiency promotes the formation of γH2AX foci in the absence of DSBs (this study). Therefore, we assumed that Tyr-142 of H2AX would be already dephosphorylated in BIN1-deficient cells so that the cancer cells could ATM-dependently phosphorylate MDC1 to survive longer even under DNA-damaging conditions. However, Tyr-142 of H2AX remained phosphorylated even after BIN1 was diminished (Fig. 7C and Fig. S10A). Interestingly, when BIN1 was reduced, nearly all γH2AX foci coexisted with the pTyr-142 H2AX foci, whereas many pTyr-142 H2AX foci were observed even in the nuclear region where little or no γH2AX foci were present (Fig. S10B). Our results suggest that dephosphorylation of Tyr-142 of H2AX is not required for generating γH2AX foci in BIN1-deficient cells.
More importantly, even in the presence of pTyr-142 in H2AX, the loss of BIN1 markedly enhanced MDC1 phosphorylation, which was boosted further by brief treatment with bleomycin (Fig. 7D). The phosphorylation of MDC1 in BIN1-deficient cell nuclei was clearly abolished by KU-60019, a small molecule ATM-selective inhibitor (Fig. 7E and Fig. S11) (52). Our results were apparently inconsistent with the previous reports (50, 51). To reconcile this discrepancy, we wondered whether there might be two different chromatin structures in the vicinity of γH2AX. We presumed that the steric accessibility of MDC1 protein to γH2AX would be higher when γH2AX is formed by BIN1 deficiency than when γH2AX formation is triggered by DSBs (see “Discussion”). We conclude that, in BIN1-deficient cells, ATM is required for γH2AX formation and subsequent MDC1 phosphorylation, regardless of the phosphorylation status of Tyr-142 in H2AX.
E2F1-induced ATM autophosphorylation is required for acquisition of cisplatin resistance in BIN1-deficient cancer cells even in the presence of transcriptionally active TP53
The tumor suppressor TP53 is well-known to play a key role in promoting DNA damage-induced apoptosis by transcriptionally activating a number of apoptotic genes (20). Because BIN1 is also a proapoptotic protein in response to DNA damage (32, 34, 35), we wondered whether the cisplatin resistance provoked by a BIN1 loss is lessened by TP53. To do this, we used two TP53-positive cancer cell lines, LNCaP (prostate cancer) and U2OS (osteosarcoma), and a TP53-null cancer cell line, SAOS2 (osteosarcoma). These cell lines naturally express WT BIN1, thus being sensitive to cisplatin (32–35).
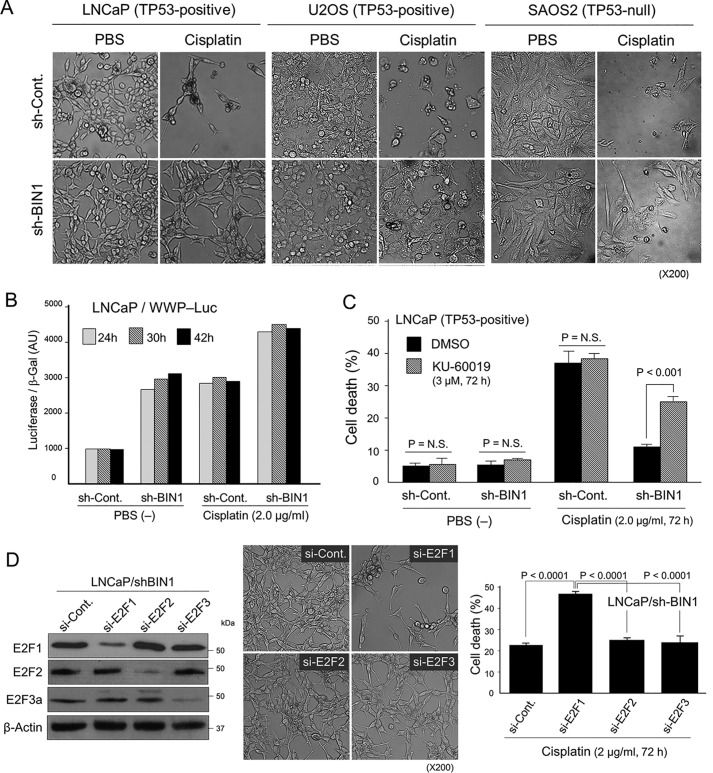
As expected, the majority of the sh-control cancer cells was eliminated by cisplatin within 72 h. However, the sh-BIN1–expressing cancer cells were resistant to cisplatin, regardless of the expression status of the TP53 gene (Fig. 8A). We noticed that the majority of cisplatin-treated LNCaP/sh-BIN1 and U2OS/sh-BIN1 cells were apparently bigger in size with more flattened morphology than untreated cells. In contrast, the cisplatin-treated SAOS2/sh-BIN1 cell line did not display such a morphological change. This implied that endogenous TP53 was certainly activated by cisplatin and elicited senescence-like growth arrest but not apoptosis in BIN1-deficient cancer cells. As predicted, the TP53-sensitive gene promoters were robustly activated when BIN1 was absent, particularly in the presence of cisplatin (Fig. 8B and Fig. S12). The TP53-dependent transactivation was likely due to stabilization of TP53 protein by ATM-dependent phosphorylation or E2F1-dependent induction of p14ARF (a TP53 stabilizer) or both (19, 20, 32). These results suggest that BIN1 loss promotes cisplatin resistance, even in the presence of transcriptionally active TP53.
Figure 8.
Deficiency of BIN1 increases cisplatin resistance in a manner dependent on ATM and E2F1 irrespective of the status of TP53. A, phase-contrast microscopy demonstrated the morphology of the indicated cancer cell lines±sh-BIN1 in the presence and absence of cisplatin (2.0 μg/ml, 72 h). B, WWP-Luc–transfected LNCaP±pLPC-BIN1 cells were subjected to luciferase assays in the presence or absence of cisplatin (2.0 μg/ml) at 24, 32, and 42 h. The WWP-Luc reporter vector is driven by the human p21WAF1 gene promoter, a direct transcriptional target of TP53 (20). AU, arbitrary unit. C, trypan blue exclusion assay of the LNCaP±sh-BIN1 cell lines treated with cisplatin (2.0 μg/ml, 72 h) in the presence of KU-60019 (3.0 μm, 72 h). Dimethyl sulfoxide (DMSO) was used as the vehicle control. N.S., not significant. D, LNCaP/sh-BIN1 cells were transiently transfected with si-Cont, si-E2F1, si-E2F2, or si-E2F3 for 72 h. The cells were then treated with cisplatin (2.0 μm, 72 h) and were subjected to phase-contrast microscopy and trypan blue exclusion assay. The si-E2Fs–transfected cell lysates in the absence of cisplatin were subjected to Western blot analysis.
We next examined whether activation of E2F1 and subsequent ATM autophosphorylation are essential for the cisplatin tolerance mediated by BIN1 deficiency. As shown in Fig. 8C, inhibition of endogenous ATM activity by KU-60019 significantly compromised cisplatin resistance in BIN1-depleted cancer cells. Similarly, the transfection of si-E2F1 prominently curbed the cisplatin resistance (Fig. 8D). Among the E2F family of transcription factors, E2F1, E2F2, and E2F3a are all transcriptional activators (32, 35). Interestingly, E2F2 or E2F3a depletion did not recapitulate the cisplatin sensitivity mediated by E2F1 diminution (Fig. 8D). We conclude that the E2F1-dependent ATM activation is extremely important for BIN1-deficient cancer cells to provoke cisplatin resistance, irrespective of the status of TP53.
Discussion
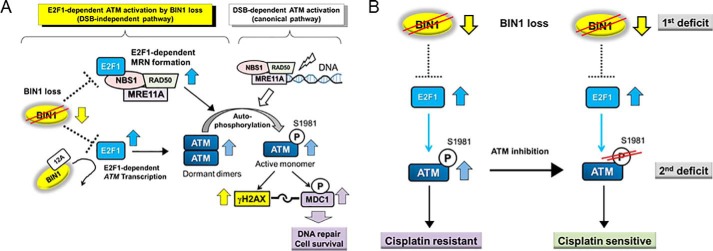
In this study, we have identified the new E2F1-dependent signaling circuit through which a deficiency in the BIN1 tumor suppressor promotes cisplatin resistance in an ATM-dependent manner. Because BIN1 is an E2F1 corepressor (35), BIN1 reduction constitutively reactivates E2F1-sensitive gene promoters (35), including the ATM promoter (45). In parallel, in a manner dependent on the E2F1/NBS1 association (47–49), E2F1 directly increases the formation of foci of MRE11/MRN DNA end-binding protein complex in BIN1-deficient cellular nuclei. Subsequently, ATM is autophosphorylated, thereby phosphorylating a variety of ATM effectors, such as H2AX, which forms γH2AX (a prevalent DSB biomarker, when phosphorylated) (22, 23) and the γH2AX-binding adaptor protein MDC1 (24, 25), regardless of the phosphorylation status of Tyr-142 of H2AX (50, 51). We also discovered that BIN1 loss–induced cisplatin resistance occurs even in the presence of transcriptionally active TP53, whereas the resistance to cisplatin is evidently abolished by E2F1 or ATM inactivation. We conclude that, in BIN1-deficient cancer cells, E2F1 enables ATM activation, thereby promoting subsequent DSB-independent γH2AX formation, MDC1 phosphorylation, and cisplatin resistance (Fig. 9A).
Figure 9.
BIN1 loss enables ATM activation by the nuclear protein E2F1. A, BIN1 is an E2F1 corepressor (35), so BIN1 loss, one of the hallmarks of advanced cancers, constitutively activates E2F1. In contrast, BIN1+12A, an aberrant BIN1 splicing variant, does not physically bind E2F1, so it does not participate in BIN1-dependent E2F1/ATM regulation. In BIN1-deficient cancer cells, E2F1 stimulates ATM transcription and increases the amount of ATM homodimers (the latent form). In parallel, by physically interacting with NBS1 (which is not involved in E2F1-dependent transcriptional machinery), E2F1 retains MRE11A at DNA termini and facilitates the MRE11A/RAD50 binding in vivo even in the absence of DNA damage. Consequently, E2F1 facilitates the formation of the MRN DNA end-binding complex, thereby promoting ATM autophosphorylation in BIN1-deficient cancer cells. Accordingly, BIN1 loss constitutively promotes ATM-dependent γH2AX formation (thus, producing a pseudo-DSB signal) and then ATM-dependent MDC1 phosphorylation, which is essential for activating the downstream DSB repair/cell survival signals (25, 50). We propose that late-stage cancer cells, which are frequently deficient in BIN1 expression, may develop cisplatin resistance through the E2F1/ATM-dependent pathway even before they are exposed to the DNA-damaging agent. B, loss of BIN1 enables E2F1 to activate ATM, thereby provoking cisplatin resistance. In BIN1-deficient cancer cells (which creates the 1st deficit), it may be recommended to specifically inhibit ATM autophosphorylation (thus, causing the 2nd deficit) to establish new synthetic lethality that increases cisplatin sensitivity.
In BIN1-deficient cancer cells, how does E2F1 not activate apoptosis by cisplatin but rather promote cisplatin resistance?
In response to DSBs, checkpoint kinase 2 (CHK2) is phosphorylated by ATM (16, 53). Subsequently, E2F1 is phosphorylated by ATM and CHK2. As a result, E2F1 protein is stabilized and becomes transcriptionally active to mediate DNA damage–induced apoptosis (53–55). Hence, apoptosis is an established biological outcome of activated E2F1 in response to DNA damage (32, 35, 55). Contradictorily, in this study, we found that endogenous E2F1, which is constitutively activated in BIN1-deficient cancer cells, never elicits cisplatin-induced apoptosis but rather actively protects cancer cells from cisplatin. Therefore, the status of BIN1 expression seems to be a key determinant that enables E2F1 to select cell survival or apoptosis following DNA damage.
BIN1 is a nucleocytoplasmic adaptor protein that is involved in the E2F1-dependent DDR pathways through, at least, two separate (but closely interconnected) pathways. First, the human BIN1 gene promoter is directly activated by E2F1 in response to DSBs, at least, in most human cancer cells and mouse fibroblasts (32). Second, among a number of E2F1-inducible proapoptotic proteins, such as p14ARF (a TP53-stabilizing factor), TP73, APAF1, caspases, and BIN1 (32, 55), only BIN1 protein also acts as an E2F1-binding transcriptional corepressor (35). This means that, in the presence of BIN1, there is a negative feedback loop that gradually curbs E2F1-dependent gene transcription (32, 35, 55). If so, in the absence of BIN1, we thought that E2F1-mediated apoptosis following DNA damage would be accelerated infinitely because of the disappearance of the E2F1-dependent negative feedback loop.
Nonetheless, as demonstrated above, in BIN1-deficient cancer cells, little or no apoptosis is elicited by cisplatin. This is not because endogenous E2F1 in the cancer model systems we used is latent or mutated, because forced elimination of E2F1 prominently mitigates the cisplatin tolerance mediated by BIN1 deficiency (this study). To better understand the antiapoptotic (i.e. cell survival) mechanism caused by E2F1 following DNA damage in the absence of BIN1, it would be reasonable to speculate that, in BIN1-deficient cancer cells, a cell survival factor (or mechanism) is activated and promotes the ATM-dependent cisplatin resistance. Consistent with this conjecture, the BIN1 loss offsets cisplatin-induced apoptosis even in the presence of transcriptionally competent (i.e. proapoptotic) TP53 (this study). It is possible that, when BIN1 is absent, activated E2F1 promotes various cellular activities essential for the development of cisplatin resistance, such as greater DSB repair activities, silenced apoptosis, efficient drug detoxification, and/or increased drug efflux (6, 7).
It would be ideal to specifically inactivate the aforesaid cell survival factor (or mechanism) for reversal of E2F1-dependent cisplatin resistance in BIN1-deficient cancer cells. However, in general, before we reach this goal, there remain many tasks to be accomplished, such as identification of a critical ATM effector responsible for DNA repair/cell survival and subsequent high-throughput screening for drug discovery. Meanwhile, it is noticed that several ATM-specific small-molecule inhibitors have already been approved and are commercially available for clinical trials in patients with a set of advanced cancers (8, 26, 52). Hence, as a feasible, practical, and quick approach to rebuilding cisplatin sensitivity in cisplatin-resistant cancer cells, in which BIN1 is frequently missing (1st deficit), we propose to create new synthetic lethality by generating the 2nd deficit using an ATM inhibitor (Fig. 9B).
How does BIN1 deficiency allow MDC1 to be phosphorylated by ATM regardless of pTyr-142 in H2AX?
The physical interaction between γH2AX and MDC1 and the subsequent ATM-dependent MDC1 phosphorylation are rate-limiting steps to initiate the downstream paths to DNA repair and cell survival (24, 25). Cook et al. (51) previously reported that the γH2AX/MDC1 protein/protein interaction occurs only when pTyr-142 in H2AX is dephosphorylated. In this study, we observed that BIN1 loss, which greatly enhances the formation of γH2AX foci in a manner independent of DSBs, stimulates ATM-dependent phosphorylation of MDC1 even in the presence of pTyr-142 in H2AX (see Fig. 7, C–E, and Figs. S9 and S10). This suggests that the steric accessibility of MDC1 to γH2AX in chromatin is blocked by pTyr-142 of H2AX when H2AX is provoked by DSBs, whereas the steric barrier would be drastically alleviated when γH2AX is formed by BIN1 deficiency.
Histone H1 is one of the major PARP1 substrates of PARylation (34, 39, 40). In general, PARP1 activity is associated with the deconcentration of higher-order chromatin structures (39, 40). Because BIN1 is the only natural PARP1-inhibitory nuclear protein documented so far (34), it is proposed that a BIN1 loss is likely to stimulate chromatin remodeling due to hyper-PARylation of histone H1 (see Fig. S3) (34). We presume that the chromatin structure close to the γH2AX foci caused by DSBs would be sterically different, at least, to some extent from the chromatin microenvironments adjacent to γH2AX foci triggered by a BIN1 loss. Although it remains unknown how PARylated histone H1 alters the spatial interplay between pTyr-142 of H2AX and pSer-139 of H2AX (i.e. γH2AX), we hypothesize that MDC1 is practically allowed to directly interact with γH2AX in hyper-PARylated chromatin, regardless of phosphorylated Tyr-142 of H2AX.
What biological messages are transmitted by the γH2AX foci formed by a BIN1 loss in the absence of actual DSBs?
Soutoglou and Misteli (56) reported that, when DSB repair-initiating nuclear proteins, such as MRE11A, NBS1, and MDC1, are artificially (i.e. experimentally) immobilized to the chromatin in the absence of DNA lesions, the cellular DDR effector pathways (including the formation of γH2AX foci) are promptly activated in a manner dependent on ATM and DNA-PKcs, as if there are real DNA lesions. This work was epoch-making because it evidently demonstrated that prolonged association of major DDR-initiation factors with chromatin is sufficient to stimulate the downstream DDR effector pathways even in the absence of DNA damage and that there is an ordered binding of DSB-repair proteins to amplify the DDR signal (56). However, the method used for prolonged immobilization of several key DDR-initiating factors fused with the E. coli lac-repressor was extremely artificial. Therefore, the physiological relevance of the DSB-independent DDR signal amplification (including γH2AX foci) and the biological message thereof have yet to be fully understood.
In this study, we reported that, even under optimal culture conditions, the loss of BIN1 is sufficient to stimulate the immobilization of the endogenous MRN complex and the subsequent ATM autophosphorylation in an E2F1-dependent manner. Consequently, the BIN1 loss constitutively promotes ATM-dependent cellular DDR-signaling events, including γH2AX formation and MDC1 phosphorylation regardless of the phosphorylation status of Tyr-142 of H2AX. Our results suggest that, even in the absence of DSBs, intrinsic cellular DDR-signaling machinery can be naturally activated if BIN1 is absent. It is well-known that the amount of BIN1 protein in the nucleus is altered according to cell-cycle progression (57) and cell differentiation (58). Thus, even in the absence of DNA lesions, γH2AX emergence due to a decrease in BIN1 levels in the nucleus is a physiological and dynamic phenomenon.
In response to DNA damage, the formation of γH2AX foci has been widely recognized not only as one of the most reliable DSB biomarkers but also as scaffold sites in chromatin on which the DDR downstream proteins are assembled to initiate either DNA repair/cell survival or apoptosis (23). Thus, we propose that the γH2AX appearance caused by a BIN1 loss creates a pseudo-DSB signal. Conversely, a reduction in γH2AX foci may not directly prove the repair of DSBs if BIN1 is abundant. The γH2AX foci that are increased by BIN1 deficiency in the absence of actual DSBs could be like a fire alarm that keeps ringing even though nothing is burning yet. Hence, the pseudo-DSB signals, such as γH2AX foci generated by the BIN1 loss, may automatically hypersensitize downstream DSB-repair machinery even through cancer cells have yet to be physically exposed to DNA-damaging therapeutic agents. If so, the increase in the formation of γH2AX foci by BIN1 deficiency under optimal conditions may create a self-defense message by which cancer cells constantly activate cellular DSB-repair machinery, thereby sustaining a DNA-damage–resistant phenotype.
How does BIN1 loss select the positions of γH2AX foci on undamaged chromatin?
Clearly separated numerous foci of γH2AX caused by DSBs in cellular nuclei are expected to correspond to broken sites of chromosomal DNA (22, 23). As shown in this study, even in the absence of DNA lesions, the γH2AX foci formed in BIN1-deficient nuclei also constantly display distinct multiple foci. This suggests that certain chromatin-associated mechanisms participate in the process of the formation of γH2AX foci on unbroken chromatin in BIN1-deficient nuclei. We hypothesize that an unknown chromatin-associated nuclear factor is stimulated by BIN1 loss and promptly assembles cellular DDR-initiating machinery, such as the MRN complex. Subsequently, downstream DDR effector pathways, including ATM activation and γH2AX formation, are naturally activated. If so, the positions of γH2AX foci produced by BIN1 loss on unbroken chromatin would be close to (or in the vicinity of) the hypothetical chromatin-associated nuclear factor.
One most likely candidate protein as the chromatin-associated nuclear factor is E2F1. We reported previously that the physical interaction between E2F1 and BIN1 is stabilized by endogenous PARP activity and that E2F1-dependent transactivation is attenuated by BIN1, particularly when E2F1 is PARylated (35). It is believed that an E2F1-dependent nuclear function, no matter whether it is transcription-dependent or not, could be controlled, to some extent, by E2F1's post-translational modification, such as phosphorylation, acetylation, and PARylation (21, 32, 35, 53, 55). It is also apparent that the time/space-specific E2F1 function would greatly depend on what nuclear proteins the E2F1 protein interacts with (35, 55). As discussed above, E2F1 physically binds NBS1 (a key component of MRN) (47–49) in a manner dependent on the E2F1 MB box (47) but independent of E2F1-mediated transcription (this study). Moreover, the E2F1 MB box is also necessary for the BIN1/E2F1 interaction (35). Because PARylation is one of the most likely post-translational modifications of E2F1 in the absence of BIN1 (34), it would be interesting to determine whether hyper-PARylated E2F1 modifies in situ E2F1/NBS1 binding and subsequent formation of γH2AX foci close to the NBS1 protein in BIN1-depleted cellular nuclei.
How does BIN1, an inducer of genomic instability, act as a tumor suppressor?
Cancer is a disease associated with genomic instability and aging (59). In general, oncoproteins (which act as drivers of tumorigenesis) increase genomic instability, whereas tumor suppressors (i.e. anti-oncoproteins) preserve the integrity of the genome (38). The BRCA1/2 tumor suppressors are required for DSB repair via homologous recombination (17, 38, 50). In response to DNA damage, the tumor suppressor TP53 activates cell-cycle checkpoints to halt replication of damaged genomic DNA. Therefore, TP53 is recognized to be the “guardian of the genome” (20).
Accumulating evidence indicates that BIN1 acts as a bona fide tumor suppressor in vitro and in vivo (27–37), insofar as the protein induces apoptosis or senescence in response to oncogenic stress (34, 35) and genotoxic stress (32, 34). However, unlike conventional tumor suppressors, BIN1 naturally increases genomic instability, at least partly, by constitutively inhibiting two major DNA repair-facilitating enzymes, PARP1 (34, 39–41) and ATM (this study and Refs. 14, 15). Intriguingly, similarly to BIN1, hob1 (homolog of BIN1) of fission yeast inhibits the formation of γH2A (yeast's γH2AX) (this study) and restricts the elongation of telomere length (by which the termini of the chromosome are protected) (60). These results indicate that BIN1/hob1 commonly decreases genomic and chromosomal stability (34).
In general, most likely biological reactions against accumulated genomic instability would be senescence-like growth arrest or apoptosis (38, 59). Because of the BIN1-dependent increase in genomic instability, we assume that senescence and/or apoptosis would be naturally triggered at a much earlier stage of genomic instability at which cancer would occur. In this respect, BIN1 certainly reduces the risk of spontaneous tumorigenesis by stimulating a DNA damage-sensitive cellular response (i.e. senescence and apoptosis). Hence, distinct from other traditional tumor suppressors, which are known to preserve the integrity of the genome (20, 38), the BIN1 tumor suppressor is categorized to be a “cancer-preventing” tumor suppressor that naturally destabilizes the genome. If a cancer-preventing factor like BIN1 is genetically or epigenetically inactivated, aged cells (even expressing WT TP53) would become less-sensitive to accumulated DNA damage. If so, it is anticipated that cells lacking BIN1 would gain biological privilege for further growing and surviving even after the extent of DNA damage exceeds the threshold for senescence and apoptosis.
In summary, we have identified the DSB-independent E2F1 signaling circuit through which the BIN1 loss constitutively promotes ATM autophosphorylation, γH2AX formation (i.e. pseudo-DSB signal), and cisplatin resistance (Fig. 9A). The survival of BIN1-deficient cancer cells in the presence of cisplatin primarily requires E2F1-mediated ATM activation. Therefore, it is reasonable to presume that ATM inhibition creates new synthetic lethality to reestablish cisplatin sensitivity in cisplatin-resistant cancer cells often lacking BIN1 (Fig. 9B). It is also advised to confirm the status of BIN1 expression to precisely interpret the biological message of γH2AX foci.
Experimental procedures
Mammalian cell lines
All primary, immortalized, and cancer cell lines used in this study were obtained from the American Type Culture Collection (ATCC, Manassas, VA) or have been described previously (27–35). The normal human (ATM-proficient, GM00637) and A-T (ATM-deficient, GM16666) fibroblasts were obtained from the Coriell Institute (Camden, NJ). All mammalian cell lines used in this study were maintained in 5% CO2 at 37 °C.
Yeast strains, media, and growth conditions
The S. pombe WT strain used in this study was FY71 (h-, ade6-M216, leu1-32, and ura4D18), and the hob1Δ mutant strain was derived from FY71 (h-, ade6-M216, leu1-32, ura4D18, hob1::kan) (43). These yeast cells were cultured nonselectively at 25–30 °C in YES medium, in which YE medium (0.5% yeast extract) was supplemented with 2.5% dextrose. DSBs were induced in WT cells and hob1Δ mutant cells with YES medium supplemented with 40 μg/ml bleomycin for 2 h. Yeast whole-cell extracts were prepared using 20% trichloroacetic acid and glass beads.
Oligonucleotides, antibodies, si-/sh-RNAs, and chemicals
The oligonucleotide primers, antibodies, si-RNAs (or sh-RNA-expression lentiviral plasmid DNAs), and chemicals used in this study are listed in the Tables S1–S4, respectively. According to the vendor's information (Santa Cruz Biotechnology., Santa Cruz, CA), the si-BIN1 RNA product (sc-29804) contains three different 19-nt RNA duplexes, which are designed to specifically cleave three different positions of the human BIN1 mRNA (28). Similarly, the sh-BIN1–expression lentivirus plasmid DNA (sc-29804-SH) is a mixture of three different sh-BIN1–expressing plasmid DNAs that cleave the three corresponding 19-nt sites on the human BIN1 mRNA (Fig. S5A). The Blast homology search of every single 19-nt sequence targeted by the si-/sh-BIN1 RNA molecules constantly identified the corresponding nt sequence of the human BIN1 mRNA (28). BIN2, BIN3, and amphiphysin-I mRNAs belong to the human BAR family and are structurally similar to BIN1 mRNA (28). However, the si-/sh-BIN1–targeting nt sequences did not detect any similar sequence with any of these BIN1-related mRNAs (data not shown). Transient transfection of every single si-BIN1 molecule effectively eliminated BIN1 protein (Fig. S5B). Transient transfection of si-BIN1s (sc-29804) robustly released endogenous E2F1 activity in DU145 (BIN1-positive) cells but not in MCF7 (BIN1-null) cells (Fig. S5C). Increased activities of endogenous E2F1 in sh-BIN1–expressing DU145 cells were reversed by the cotransfection of the pcDNA3-BIN1 expression vector (Fig. S5D). Thus, the si-/sh-BIN1 RNA molecules we used in this study effectively and specifically decreased endogenous BIN1 levels.
Plasmid DNAs
The expression vectors for the human BIN1, BIN1 + 12A, HA-tagged E2F1 (pRcCMV-HA-E2F1), and two TP53-sensitive luciferase reporter vectors (WWP-Luc and PIG3-Luc) have been described previously (27–35). To construct the human ATM promoter-driven luciferase reporter vectors, 530ATM-Luc and 400ATM-Luc, two human genomic DNA fragments, 530ATM (−237/+291), which contains the three E2F-consensus sites upstream from the transcription initiation site (+1), and 400ATM (−110/+291), which does not contain those sites (45), were amplified by PCR using genomic DNA extracted from the human DU145 cell line, respectively. The purified genomic PCR DNA fragments were restriction-digested with XhoI and HindIII and then subcloned into the pGL2-basic luciferase vector (Promega, Madison, WI).
BIN1 gene-silencing lentivirus production
The human BIN1 sh-RNA–expressing lentivirus plasmid DNA (sc-29804-SH, Santa Cruz Biotechnology) was cotransfected in the HEK293T cells (a packaging cell line) with the psPAX2 plasmid DNA (a lentivirus packaging vector) (Addgene, Cambridge, MA) and the pMD2.G plasmid DNA (a lentivirus VSV-G envelope–expressing vector) (Addgene). The sh-RNA plasmid vector that expressed scrambled sh-RNAs (sc-108060, Santa Cruz Biotechnology) was used as the negative control (sh-control) (32, 34, 35). Approximately 72 h post-transfection, the packaging culture supernatant (containing replication-incompetent, infectious sh-BIN1-expressing lentivirus particles) was harvested, precleared, and overlaid on the indicated host cell line in the presence of puromycin (a selection marker) for 72 h. All drug-resistant colonies were pooled and expanded for further biochemical and functional analysis.
BIN1-expressing retrovirus production
As described previously (32, 34, 35), the human BIN1 (isoform 8) cDNA-expressing retrovirus plasmid DNA, pLPC-BIN1, or its control vector, pLPC, was transfected in the Bing cell line (an amphotropic retroviral packaging cell line, ATCC) for 72 h. The supernatant of the culture medium, which contained the amphotropic, replication-incompetent retrovirus pLPC or pLPC-BIN1, was overlaid on an indicated actively proliferating host cell line. The infected cells were selected by puromycin (a selection marker) for 72 h. Drug-resistant colonies were pooled and expanded.
Immunoprecipitation and Western blot analysis
Approximately 1.5 mg of precleared cell lysates were incubated with 1.5 μg of an IP antibody at 4 °C for 4 h with gentle rocking. Purified preimmune IgG (Pierce) was used as the negative control. The immunoprecipitated complexes or precleared protein lysates were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for 1 h at 100 V in Towbin buffer (192 mm glycine, 20% methanol, and 25 mm Tris-HCl, pH 8.3). Protein-blotted membranes were blocked using 5% nonfat skim milk in PBST (PBS containing 0.1% Tween 20) overnight at 4 °C with gentle rocking, followed by hybridization with the appropriate primary and secondary antibodies. As the loading control, β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used.
ChIP
ChIP assays were performed as described previously (32, 34, 35). To detect endogenous BIN1 and E2F1 proteins on the human ATM promoter, a 142-bp DNA fragment (−143/−1) encompassing the promoter region was amplified using 35 cycles of PCR at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s.
ChIP-based in vivo DNA end–binding assay
We have established a ChIP-based in vivo DNA end–binding assay. The super-coiled pGL2-luciferase (Luc) plasmid DNA, which contained a unique HindIII restriction site at −29 bp upstream from the translational initiation site (+1) corresponding to the initial methionine of Luc, was restriction-digested with HindIII. A linearized pGL2-Luc DNA fragment (pGL2/HindIII-Luc) was transiently transfected. Super-coiled pGL2-Luc plasmid DNA (uncut) was used as the negative control. The sonicated DNA/protein complexes were subjected to a ChIP assay probed with a ChIP-grade anti-MRE11A antibody. The Luc open-reading frame (ORF) cDNA sequence does not naturally exist in untransfected cells. A 240-bp DNA fragment (+1053/+1293) encompassing the Luc ORF cDNA was amplified using 35 cycles of PCR at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s. A single 240-bp PCR product was confirmed on a 2% agarose gel.
qRT-PCR
qRT-PCR assays were carried out as described previously (32, 34, 35). Briefly, 1.0 μg of purified total RNA was used for first-strand cDNA synthesis using the iScript cDNA synthesis kit, according to the vendor's protocol (Bio-Rad). For qRT-PCR, we used iQ SYBR Green Supermix (Bio-Rad) according to the vendor's protocol.
Luciferase reporter assay
Luciferase reporter assays were performed as described previously (28, 32–35). To normalize transfection efficiency, a β-gal expression vector (pcDNA3–β-gal) was cotransfected with a one-tenth quantity of a luciferase reporter vector.
In situ immunofluorescence analysis
The formaldehyde-fixed cells were blocked in 3% bovine serum albumin (BSA) at room temperature for 1 h. The cells were hybridized with primary and secondary antibodies coupled to Alexa Fluor 488 (Molecular Probes, Eugene, OR) at room temperature in the dark. DAPI (Sigma) was used for nuclear counter-staining. The cell images were recorded with a Leica DM 5500 fluorescence microscope (Leica Microsystems, Buffalo Grove, IL) and a digital camera CoolSNAP HQ2 (Photometrics, Tucson, AZ).
Oncogenic transformation assay
REFs (ScienCell Research Laboratories, Carlsbad, CA) were cultured at a density of 0.5 × 105 cells in a 6-cm tissue-culture dish. Using the X-tremeGENE DNA transfection reagents (Roche Applied Science), the cells were transfected with 0.5 μg of the pT22 plasmid DNA (which expresses human H-RasG12V oncogene), 0.5 μg of the p1A/neo plasmid vector (which expresses the adenovirus E1A(13S) oncogene), and one of the following vectors: 1.0 μg of the pcDNA3 control vector, 1.0 μg of the pcDNA3-BIN1 expression vector, or 1.0 μg of the pcDNA3-BIN1 + 12A expression vector (27, 31, 33, 34). About 72 h post-transfection, the cells were trypsinized, transferred to a 10-cm dish, and were fed twice a week with fresh optimal growth medium for 2–3 weeks. All emerged foci (evidence of oncogenic transformation) were fixed with 100% methanol for 2 min and stained with Giemsa (Invitrogen) for 30 min.
Colony formation assay
The indicated cancer cells, which were freshly plated at a density of 5.0 × 103 cells/cm2 in a 6-cm tissue-culture dish, were transfected with 2.0 μg of the indicated plasmid DNA (G418-resistant) for 24 h at 37 °C. The cells were trypsinized and transferred to a 10-cm dish and were fed twice a week with fresh optimal growth medium containing G418 (500 μg/ml) (Invitrogen) for 2–3 weeks. The drug-resistant colonies were then fixed with 100% methanol and stained with Giemsa.
Trypan blue exclusion assay
Cell viability was determined by trypan blue exclusion assay as described previously (32–35). Floating (i.e. dead) cells and adherent (i.e. alive) cells were briefly suspended in PBS at room temperature and then gently mixed at a 1:1 ratio with the trypan blue solution (HyClone, Logan, UT) for 5 min at room temperature. 10 μl of this mixture was immediately used for cell counting to determine the percentage of blue-stained (dead) cells.
Statistical analysis
All biochemical, molecular biological, and cell-based results presented in this study were obtained after repeating essentially the same experiments at least three times to confirm rigor, reproducibility, and statistical significance. The data were presented as means ± S.E. Statistical significance was determined with Student's t test. Any p values less than 0.05 were considered statistically significant.
Author contributions
W. P. F., A. K., T. I., S. P., J. C. J., E. K. C., A. L. A.-C., and D. S. data curation; W. P. F., A. K., T. I., S. P., A. L. A.-C., and D. S. investigation; W. P. F., A. K., T. I., and D. S. validation; W. P. F., A. K., T. I., S. P., E. K. C., A. L. A.-C., and D. S. methodology; W. P. F., T. I., and D. S. conceptualization; A. L. A.-C. and D. S. resources; D. S. formal analysis; D. S. supervision; W. P. F., T. I., and D. S. funding acquisition; D. S. visualization; D. S. writing-original draft; D. S. project administration; D. S. writing-review and editing.
Supplementary Material
Acknowledgments
We thank S. Higashiyama, H. Takeda, and T. Tsuboi (Ehime University, Proteo-Science Center, Matsuyama, Japan), H. Harada (Virginia Commonwealth University, Philips Institute for Oral Health Research, Richmond, VA), and B. Xu (Southern Research Institute, Birmingham, AL) for helpful discussions and G. Prendergast (Lankenau Institute for Medical Research, Wynnewood, PA) for the hob1Δ yeast strain.
This work was supported in part by National Institutes of Health Grant R01CA140379, United States Army Department of Defense Prostate Cancer Program DAMD 17-02-1-0131, and the first Georgia Cancer Center Polatty Award for Innovative Cancer Research (2018–2019) (to D.S.). D. S. holds a 1999 United States Patent with G. Prendergast, Patent No. 6,410,238, “Box-dependent Myc-interacting protein (Bin1) compositions and uses thereof” (The Wistar Institute of Anatomy and Biology, Philadelphia, PA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S12, Tables S1–S4 and supporting Refs. S1–S6.
- NER
- nucleotide excision repair
- A-T
- ataxia-telangiectasia
- ATM
- ataxia-telangiectasia–mutated serine/threonine protein kinase
- ATR
- ataxia-telangiectasia and Rad3-related protein kinase
- BAR
- BIN/amphiphysin/Rvs-related
- BER
- base excision repair
- BIN1
- bridging integrator 1
- CC
- coiled-coil (domain)
- DAPI
- 4′-6-diamidino-2-phenylindole
- DDR
- DNA damage response
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- DSB
- double-stranded DNA break
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- γH2AX
- phosphorylated serine 139 in histone H2AX (human and mouse)
- γH2A
- phosphorylated serine 129 in histone H2A (yeast)
- MDC1
- mediator of DNA damage checkpoint protein 1
- MRE11A
- meiotic recombination 11 homolog-A
- PARylation
- poly(ADP-ribosyl)ation
- PAR
- anti-poly(ADP-ribose)
- PARP1
- poly(ADP-ribose) polymerase 1
- qRT-PCR
- quantitative reverse–transcription polymerase chain reaction
- sh-RNA
- short hairpin RNA
- SSB
- single-stranded DNA break
- nt
- nucleotide
- IP
- immunoprecipitation
- aa
- amino acid.
References
- 1. Rosenberg B., VanCamp L., and Krigas T. (1965) Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 205, 698–699 10.1038/205698a0 [DOI] [PubMed] [Google Scholar]
- 2. Rosenberg B., Van Camp L., Grimley E. B., and Thomson A. J. (1967) The inhibition of growth or cell division in Escherichia coli by different ionic species of platinum(IV) complexes. J. Biol. Chem. 242, 1347–1352 [PubMed] [Google Scholar]
- 3. Rosenberg B., VanCamp L., Trosko J. E., and Mansour V. H. (1969) Platinum compounds: a new class of potent antitumour agents. Nature 222, 385–386 10.1038/222385a0 [DOI] [PubMed] [Google Scholar]
- 4. Einhorn L. H., and Donohue J. (1977) Cis-diamminedichloroplatinum, vinblastine, and bleomycin combination chemotherapy in disseminated testicular cancer. Ann. Intern. Med. 87, 293–298 10.7326/0003-4819-87-3-293 [DOI] [PubMed] [Google Scholar]
- 5. Einhorn L. H., and Williams S. D. (1979) The role of cis-platinum in solid-tumor therapy. N. Engl. J. Med. 300, 289–291 10.1056/NEJM197902083000605 [DOI] [PubMed] [Google Scholar]
- 6. Galluzzi L., Senovilla L., Vitale I., Michels J., Martins I., Kepp O., Castedo M., and Kroemer G. (2012) Molecular mechanisms of cisplatin resistance. Oncogene 31, 1869–1883 10.1038/onc.2011.384 [DOI] [PubMed] [Google Scholar]
- 7. Shen D.-W., Pouliot L. M., Hall M. D., and Gottesman M. M. (2012) Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 64, 706–721 10.1124/pr.111.005637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basourakos S. P., Li L., Aparicio A. M., Corn P. G., Kim J., and Thompson T. C. (2017) Combination platinum-based and DNA damage response-targeting cancer therapy: evolution and future directions. Curr. Med. Chem. 24, 1586–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manohar S., and Leung N. (2018) Cisplatin nephrotoxicity: a review of the literature. J Nephrol. 31, 15–25 10.1007/s40620-017-0392-z [DOI] [PubMed] [Google Scholar]
- 10. Cunningham L. L., and Tucci D. L. (2017) Hearing loss in adults. N. Engl. J. Med. 377, 2465–2473 10.1056/NEJMra1616601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Earley J. N., and Turchi J. J. (2011) Interrogation of nucleotide excision repair capacity: impact on platinum-based cancer therapy. Antioxid. Redox Signal. 14, 2465–2477 10.1089/ars.2010.3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowden N. A. (2014) Nucleotide excision repair: why is it not used to predict response to platinum-based chemotherapy? Cancer Lett. 346, 163–171 10.1016/j.canlet.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 13. Kuzminov A. (2001) Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 98, 8241–8246 10.1073/pnas.131009198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shiloh Y., and Ziv Y. (2013) The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 10.1038/nrm3546 [DOI] [PubMed] [Google Scholar]
- 15. Paull T. T. (2015) Mechanisms of ATM activation. Annu. Rev. Biochem. 84, 711–738 10.1146/annurev-biochem-060614-034335 [DOI] [PubMed] [Google Scholar]
- 16. Smith J., Tho L. M., Xu N., and Gillespie D. A. (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112 10.1016/B978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- 17. Cortez D., Wang Y., Qin J., and Elledge S. J. (1999) Requirement of ATM-dependent phosphorylation of BRCA1 in the DNA damage response to double-strand breaks. Science 286, 1162–1166 10.1126/science.286.5442.1162 [DOI] [PubMed] [Google Scholar]
- 18. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., and Ziv Y. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 10.1126/science.281.5383.1674 [DOI] [PubMed] [Google Scholar]
- 19. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., and Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- 20. Liu Y., and Kulesz-Martin M. (2001) p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis 22, 851–860 10.1093/carcin/22.6.851 [DOI] [PubMed] [Google Scholar]
- 21. Lin W. C., Lin F. T., and Nevins J. R. (2001) Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 15, 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- 22. Burma S., Chen B. P., Murphy M., Kurimasa A., and Chen D. J. (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467 10.1074/jbc.C100466200 [DOI] [PubMed] [Google Scholar]
- 23. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., and Pommier Y. (2008) γH2AX and cancer. Nat. Rev. Cancer 8, 957–967 10.1038/nrc2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., and Jackson S. P. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- 25. Stucki M., and Jackson S. P. (2006) γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst). 5, 534–543 10.1016/j.dnarep.2006.01.012 [DOI] [PubMed] [Google Scholar]
- 26. Weber A. M., and Ryan A. J. (2015) ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 10.1016/j.pharmthera.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 27. Sakamuro D., Elliott K. J., Wechsler-Reya R., and Prendergast G. C. (1996) BIN1 is a novel MYC-interacting protein with features of a tumor suppressor. Nat. Genet. 14, 69–77 10.1038/ng0996-69 [DOI] [PubMed] [Google Scholar]
- 28. Wechsler-Reya R., Sakamuro D., Zhang J., Duhadaway J., and Prendergast G. C. (1997) Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem. 272, 31453–31458 10.1074/jbc.272.50.31453 [DOI] [PubMed] [Google Scholar]
- 29. Ge K., Duhadaway J., Sakamuro D., Wechsler-Reya R., Reynolds C., and Prendergast G. C. (2000) Losses of the tumor suppressor BIN1 in breast carcinoma are frequent and reflect deficits in programmed cell death capacity. Int. J. Cancer 85, 376–383 [DOI] [PubMed] [Google Scholar]
- 30. Ge K., Minhas F., Duhadaway J., Mao N. C., Wilson D., Buccafusca R., Sakamuro D., Nelson P., Malkowicz S. B., Tomaszewski J., and Prendergast G. C. (2000) Loss of heterozygosity and tumor suppressor activity of Bin1 in prostate carcinoma. Int. J. Cancer 86, 155–161 [DOI] [PubMed] [Google Scholar]
- 31. Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U., and Prendergast G. C. (1999) Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl. Acad. Sci. U.S.A. 96, 9689–9694 10.1073/pnas.96.17.9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cassimere E. K., Pyndiah S., and Sakamuro D. (2009) The c-MYC–interacting proapoptotic tumor suppressor BIN1 is a transcriptional target for E2F1 in response to DNA damage. Cell Death Differ. 16, 1641–1653 10.1038/cdd.2009.98 [DOI] [PubMed] [Google Scholar]
- 33. Lundgaard G. L., Daniels N. E., Pyndiah S., Cassimere E. K., Ahmed K. M., Rodrigue A., Kihara D., Post C. B., and Sakamuro D. (2011) Identification of a novel effector domain of BIN1 for cancer suppression. J. Cell. Biochem. 112, 2992–3001 10.1002/jcb.23222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pyndiah S., Tanida S., Ahmed K. M., Cassimere E. K., Choe C., and Sakamuro D. (2011) c-MYC suppresses BIN1 to release poly(ADP-ribose) polymerase 1: a mechanism by which cancer cells acquire cisplatin resistance. Sci. Signal. 4, ra19 10.1126/scisignal.2001556 [DOI] [PubMed] [Google Scholar]
- 35. Kumari A., Iwasaki T., Pyndiah S., Cassimere E. K., Palani C. D., and Sakamuro D. (2015) Regulation of E2F1-induced apoptosis by poly(ADP-ribosyl)ation. Cell Death Differ. 22, 311–322 10.1038/cdd.2014.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kumari A., Folk W. P., and Sakamuro D. (2017) The dual roles of MYC in genomic instability and cancer chemoresistance. Genes 8, 158 10.3390/genes8060158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang M. Y., Boulden J., Katz J. B., Wang L., Meyer T. J., Soler A. P., Muller A. J., Prendergast G. C. (2007) Bin1 ablation increases susceptibility to cancer during aging, particularly lung cancer. Cancer Res. 67, 7605–7612 10.1158/0008-5472.CAN-07-1100 [DOI] [PubMed] [Google Scholar]
- 38. Negrini S., Gorgoulis V. G., and Halazonetis T. D. (2010) Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- 39. de Murcia G., and Ménissier de Murcia J. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 19, 172–176 10.1016/0968-0004(94)90280-1 [DOI] [PubMed] [Google Scholar]
- 40. Bürkle A. (2001) Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 163, 1–5 10.1016/S0304-3835(00)00694-7 [DOI] [PubMed] [Google Scholar]
- 41. De Vos M., Schreiber V., and Dantzer F. (2012) The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 84, 137–146 10.1016/j.bcp.2012.03.018 [DOI] [PubMed] [Google Scholar]
- 42. Budke B., Logan H. L., Kalin J. H., Zelivianskaia A. S., Cameron McGuire W., Miller L. L., Stark J. M., Kozikowski A. P., Bishop D. K., and Connell P. P. (2012) RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 40, 7347–7357 10.1093/nar/gks353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Routhier E. L., Donover P. S., and Prendergast G. C. (2003) hob1+, the fission yeast homolog of Bin1, is dispensable for endocytosis or actin organization, but required for the response to starvation or genotoxic stress. Oncogene 22, 637–648 10.1038/sj.onc.1206162 [DOI] [PubMed] [Google Scholar]
- 44. Redon C., Pilch D. R., Rogakou E. P., Orr A. H., Lowndes N. F., and Bonner W. M. (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4, 678–684 10.1038/sj.embor.embor871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berkovich E., and Ginsberg D. (2003) ATM is a target for positive regulation by E2F-1. Oncogene 22, 161–167 10.1038/sj.onc.1206144 [DOI] [PubMed] [Google Scholar]
- 46. Stracker T. H., and Petrini J. H. (2011) The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maser R. S., Mirzoeva O. K., Wells J., Olivares H., Williams B. R., Zinkel R. A., Farnham P. J., and Petrini J. H. (2001) Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21, 6006–6016 10.1128/MCB.21.17.6006-6016.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frame F. M., Rogoff H. A., Pickering M. T., Cress W. D., and Kowalik T. F. (2006) E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene 25, 3258–3266 10.1038/sj.onc.1209352 [DOI] [PubMed] [Google Scholar]
- 49. Chen J., Zhu F., Weaks R. L., Biswas A. K., Guo R., Li Y., and Johnson D. G. (2011) E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle 10, 1287–1294 10.4161/cc.10.8.15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panier S., and Durocher D. (2013) Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 14, 661–672 10.1038/nrm3659 [DOI] [PubMed] [Google Scholar]
- 51. Cook P. J., Ju B. G., Telese F., Wang X., Glass C. K., and Rosenfeld M. G. (2009) Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458, 591–596 10.1038/nature07849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Golding S. E., Rosenberg E., Valerie N., Hussaini I., Frigerio M., Cockcroft X. F., Chong W. Y., Hummersone M., Rigoreau L., Menear K. A., O'Connor M. J., Povirk L. F., van Meter T., and Valerie K. (2009) Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 8, 2894–2902 10.1158/1535-7163.MCT-09-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stevens C., Smith L., and La Thangue N. B. (2003) Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5, 401–409 10.1038/ncb974 [DOI] [PubMed] [Google Scholar]
- 54. Carnevale J., Palander O., Seifried L. A., and Dick F. A. (2012) DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol. Cell. Biol. 32, 900–912 10.1128/MCB.06286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sakamuro D., Folk W. P., and Kumari A. (2015) To die, or not to die: E2F1 never decides by itself during serum starvation. Mol. Cell. Oncol. 2, e981447 10.4161/23723556.2014.981447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soutoglou E., and Misteli T. (2008) Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320, 1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kinney E. L., Tanida S., Rodrigue A. A., Johnson J. K., Tompkins V. S., and Sakamuro D. (2008) Adenovirus E1A oncoprotein liberates c-Myc activity to promote cell proliferation through abating Bin1 expression via an Rb/E2F1-dependent mechanism. J. Cell. Physiol. 216, 621–631 10.1002/jcp.21437 [DOI] [PubMed] [Google Scholar]
- 58. Wechsler-Reya R. J., Elliott K. J., and Prendergast G. C. (1998) A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell. Biol. 18, 566–575 10.1128/MCB.18.1.566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Campisi J. (2013) Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ramalingam A., Farmer G. E., Stamato T. D., and Prendergast G. C. (2007) Bin1 interacts with and restrains the DNA end-binding protein complex Ku. Cell Cycle 6, 1914–1918 10.4161/cc.6.15.4514 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.