Abstract
Mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs) are essential components of the mitochondrial translation machinery. The correlation of mitochondrial disorders with mutations in these enzymes has raised the interest of the scientific community over the past several years. Most surprising has been the wide-ranging presentation of clinical manifestations in patients with mt-aaRS mutations, despite the enzymes' common biochemical role. Even among cases where a common physiological system is affected, phenotypes, severity, and age of onset varies depending on which mt-aaRS is mutated. Here, we review work done thus far and propose a categorization of diseases based on tissue specificity that highlights emerging patterns. We further discuss multiple in vitro and in cellulo efforts to characterize the behavior of WT and mutant mt-aaRSs that have shaped hypotheses about the molecular causes of these pathologies. Much remains to do in order to complete our understanding of these proteins. We expect that futher work is likely to result in the discovery of new roles for the mt-aaRSs in addition to their fundamental function in mitochondrial translation, informing the development of treatment strategies and diagnoses.
Keywords: aminoacyl tRNA synthetase, mitochondria, genetic disease, central nervous system (CNS), mutant
Introduction
Translation in mammalian mitochondria is unusual in many ways compared with the process in the cytosol and even compared with mitochondrial translation in simpler eukaryotes. The organelles, thought to be descendants of alphaproteobacteria, retain a DNA genome distinct from the eukaryotic cells (1), but extant mitochondrial genomes, especially in animals, are considerably smaller than those of bacteria. Mammalian mitochondrial genomes contain just 13 protein-encoding genes, all of which are components of the oxidative phosphorylation pathway (2). Production of even this small number of proteins requires a distinct mitochondrial translation apparatus (Fig. 1). Mammalian mitochondrial genomes encode all of the RNA components of this machinery: 22 mitochondrial tRNAs and two mitochondrial ribosomal RNAs. In contrast, all of the protein components, including tRNA maturation and modification enzymes, initiation and elongation factors, ribosomal proteins, and aminoacyl-tRNA synthetases, are encoded by the nuclear genome, translated in the cytosol, and then imported into the mitochondria (3, 4).
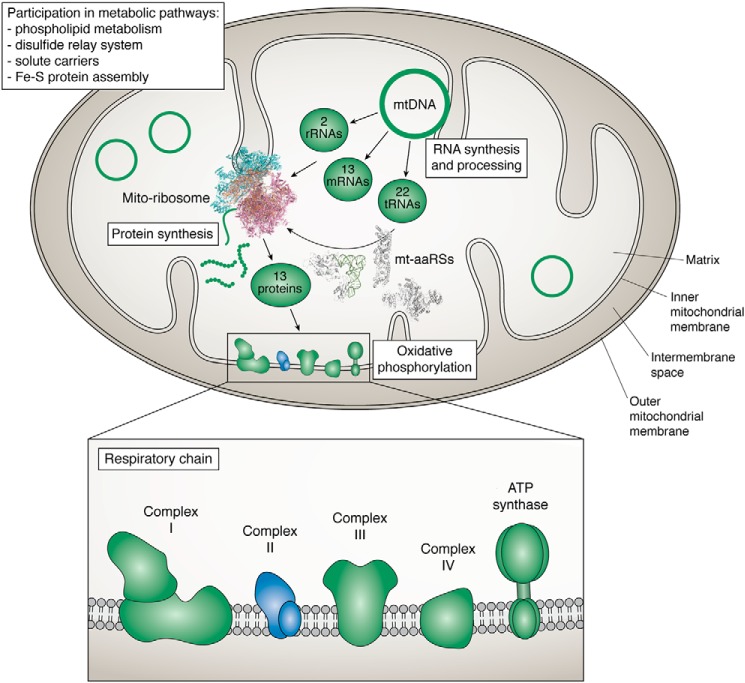
Figure 1.
Schematic representation of a mitochondrion. Mitochondria host numerous metabolic pathways. They are double-membrane organelles, hosting a distinct genome (mtDNA) and translation machinery, dedicated to the synthesis of 13 proteins, all subunits of the respiratory chain complexes (with representatives in all complexes except complex II). All additional proteins required for mtDNA maintenance and expression are encoded in the nucleus, synthesized in the cytosol, and subsequently imported into the mitochondria. This is, for instance, the case for proteins for the mito-ribosome and for mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs). Represented structures are the large (PBD code 4V19) and small (PBD code 5AJ3) subunits of the Sus scrofa mito-ribosome and the human mt-PheRS in complex with tRNA (PBD code 3TUP), mt-TyRS (PDB code 2PID), and mt-AspRS (PDB code 4AH6).
Some of the peculiarities of mitochondrial translation derive from the high mutation rate in the oxidizing mitochondrial environment and the correspondingly high mutation rate of mitochondrial DNA. For example, mammalian mitochondrial ribosomal RNAs are considerably truncated relative to their cytosolic homologs. Apparently to compensate for this change, the mitochondrial ribosomes contain increased numbers of proteins, resulting in a 2:1 protein/RNA ratio, inverted from the ratio typically found in bacteria (5–7). Mammalian mitochondrial tRNAs are also truncated and lack many conserved features typical of tRNAs in the rest of evolution (Fig. 2). In some cases, one of the arms of the classic cloverleaf secondary structure is lost, most frequently in mitochondrial tRNASer (8, 9). All mammalian mitochondrial RNAs are A:U-rich, probably as a consequence of the relative ease of oxidation of guanine nucleotides.
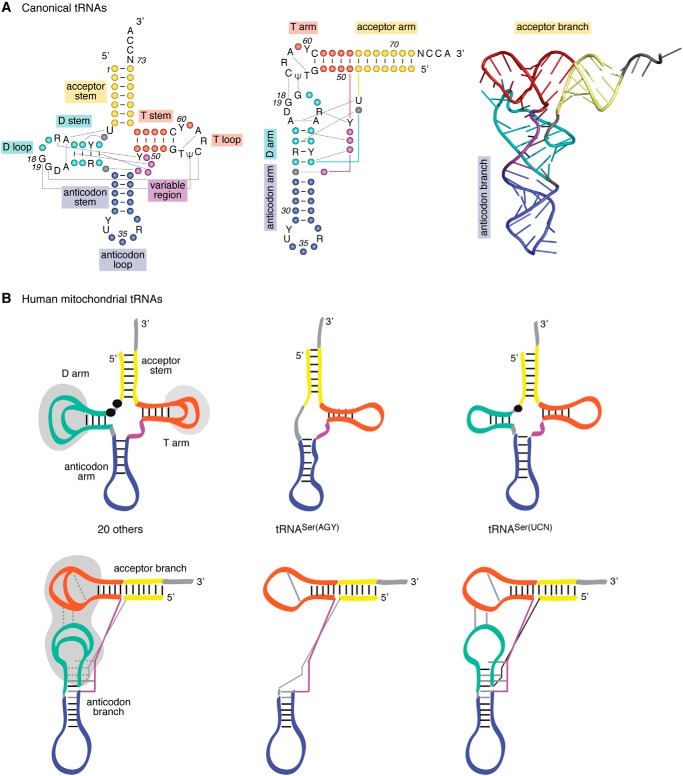
Figure 2.
Canonical tRNAs versus human mitochondrial tRNAs. A, secondary and tertiary structures of canonical tRNAs. The different structural domains are named and colored. The network of tertiary interactions at the origin of the three-dimensional folding is represented by black dashed lines. The nucleotides indicated in black are those conserved in all tRNAs. Y, pyrimidine; R, purine; A, adenosine; C, cytosine; G, guanosine; T, thymine; U, uridine; and Ψ, pseudouridine. Left, cloverleaf consensus secondary structure of canonical tRNAs; middle, two-dimensional representation of tertiary refolding of tRNA; right, crystallographic structure of S. cerevisiae tRNAPhe (PBD code 1EHZ). B, schematic representations of cloverleaf secondary structures (upper part) and 3D structures (lower part) of human mt-tRNAs. Schematic representations of the general structure of 20 tRNAs (left), of tRNASer(AGY) missing the D-arm (middle), and of tRNASer(UCN) displaying a shorter connector between the acceptor stem and the D-arm (right). Dashed lines correspond to nonstrictly conserved triple interactions. Gray zones highlight domains where variations in the number and type of interactions differ from tRNA to tRNA. This figure is adapted from Ref. 9.
Given the mitochondria's central role in ATP synthesis via oxidative phosphorylation, it is not surprising that errors in mitochondrial translation have been linked with human disease (10–12). Mutations within mitochondrially encoded molecules of the translational machinery have been identified in patients since the late 1980s, leading to the presently recognized concept of “mitochondrial translation disorders,” which include a large spectrum of clinical presentations, particularly muscular and neurological disorders (13–16). Initial work focused on mutations within the mtDNA, in either the rRNAs, the 22 mitochondrial tRNAs, or the 13 mRNAs. Although some correlations between particular mutations and distinct disease states were made (17, 18), tissue specificity and differences in symptoms and time of onset are most readily explained by heteroplasmy, nonhomogeneous mitochondrial populations in cells and tissues. The penetrance of a particular mutation within the multiple copies of mtDNA in any cell can vary from tissue to tissue in a random manner, resulting in idiosyncratic phenotypes (14).
More recently, it has been recognized that mutations in nuclearly-encoded mitochondrial proteins involved in translation are also correlated with mitochondrial diseases. In this review, we focus on the aminoacyl-tRNA synthetases (aaRS),3 which play the crucial role of specifically aminoacylating mitochondrial tRNAs with their cognate amino acid. In humans, mitochondria-specific aaRSs exist for 17 of the 20 proteogenic amino acids (19). Genes for these proteins are generally designated as ARS2; for example, the mitochondrial alanyl-tRNA synthetase is designated AARS2. Exceptions are the glycyl-tRNA synthetase gene (GARS), which uses an alternate start sequence to encode both the cytosolic and mitochondrial enzymes (20, 21), and the lysyl-tRNA synthetase gene (KARS), which uses alternate splicing to generate distinct mRNAs (22). In both cases, the cytosolic and mitochondrial enzymes differ mainly in the presence or absence of an N-terminal mitochondrial-targeting sequence. Mitochondrial Gln-tRNAGln is formed by transamidation of Glu-tRNAGln by a tRNA-dependent amidotransferase (23).
Pathogenic mutations in each of the 19 nuclear genes coding for a mitochondrial aaRS have been reported (24–29). Defects in the exclusively mitochondrial enzymes all have either homozygous or compound heterozygous presentations, giving rise to autosomal recessive disorders. Mutations in the dual-localized GARS and KARS genes have been reported with both recessive and dominant inheritance, giving rise to different clinical presentations. Autosomal dominant mutations in GARS and KARS affect the peripheral nervous system and are correlated with Charcot-Marie-Tooth disease type 2 (CMT2) (30). Recessive mutations in these genes, however, have been reported to produce phenotypes similar to those reported by mutations in exclusively mt-aaRSs (31, 32). Information about all reported pathogenic mutations in human mt-aaRSs has been compiled in a knowledge base we recently developed (33). The entry page of the website illustrates the apparently random distribution of the disease-related mutations within the different human mt-aaRSs.
Despite the fact that genes for mitochondrial aaRSs are nuclearly encoded and ubiquitously expressed, mutations give rise to a variety of distinct phenotypes (24–29). With a few exceptions detailed below, all mutations in a particular synthetase result in similar disease states. These effects are manifested mostly in the central nervous system but also in a variety of other tissues. The available data present a number of surprising contrasts that complicate simple hypotheses based on the linkage between defects in mitochondrial translation and a reduction in cellular ATP production. Tissue-specific developmental differences in energy requirements, connections with pathways for mitochondrial homeostasis associated with differences in intraorganellar localization, and alternative functions of the mitochondrial aaRS proteins are among the hypotheses currently under investigation.
Diversity of clinical manifestations
Disorders correlated with mutations in mitochondrial aminoacyl-tRNA synthetases span a broad range, including diseases characterized by defined symptoms and/or neuroradiological features (e.g. LBSL), isolated clinical signs (e.g. nonsyndromic hearing loss) to described syndromes (e.g. Perrault syndrome). Since the first description of a correlation between mutations in mt-aaRS–encoding gene and a human disease (34), the number of reported cases has increased steadily (33). In this section, we categorize mt-aaRSs according to the affected tissues and organ systems, including some details described in the medical reports that introduced these mutations into the literature. This physiological classification is intended to highlight similarities and differences in the pathological phenotypes that are not easily explained at a molecular level.
Four main groups emerge (Table 1 and Fig. 3): mt-aaRSs with mutations leading to clinical manifestations (i) exclusively in the central nervous system (CNS); (ii) in the CNS and another system; (iii) in the CNS or another system, and (iv) a system other than the CNS.
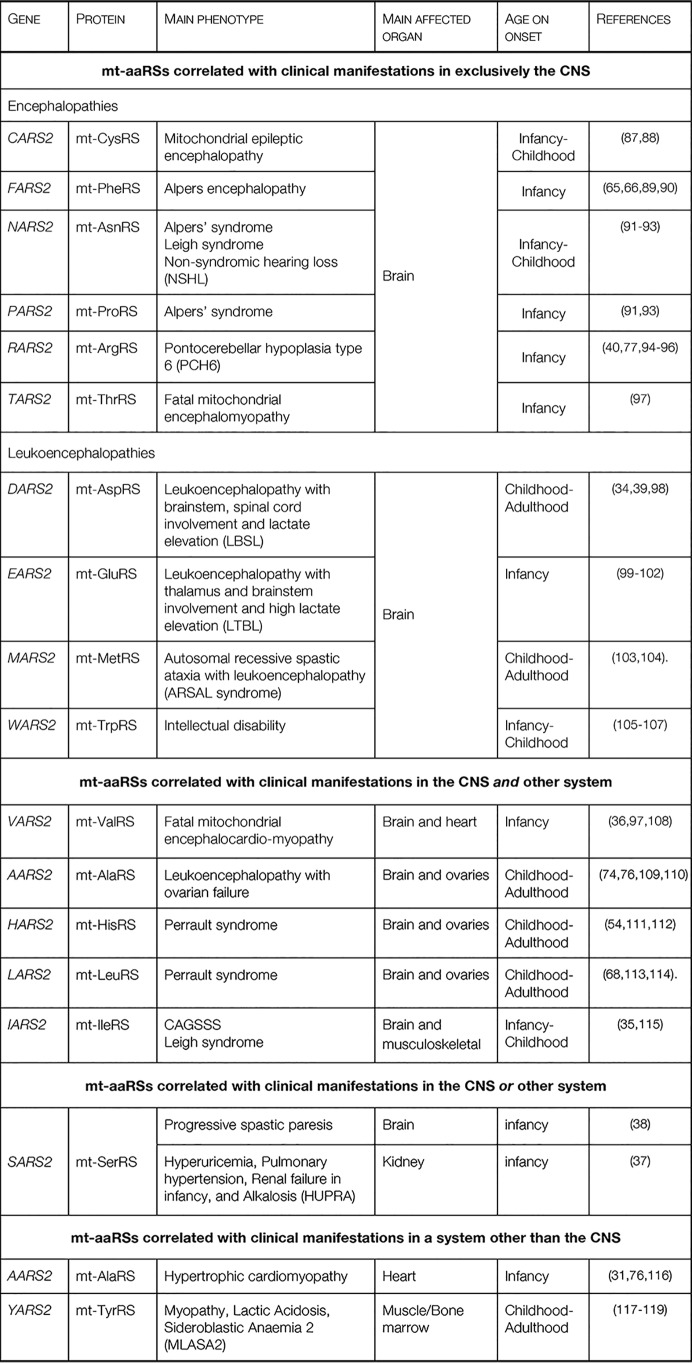
Table 1.
Classification of the pathologies produced by mutations on human mitochondrial aminoacyl-tRNA synthetases
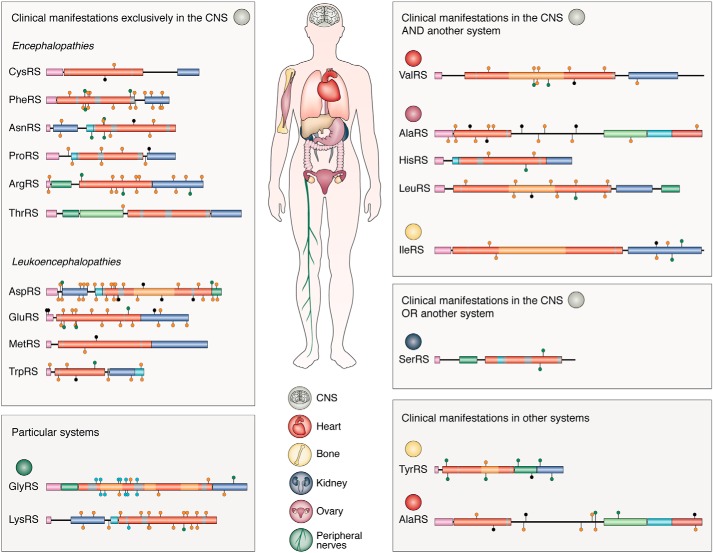
Figure 3.
Classification of the mt-aaRSs according to the clinical manifestations identified in mt-aaRS–related patients. The mt-aaRSs are shown as graphical representations of their modular organizations. Putative mitochondrial targeting sequences, catalytic domains, and anticodon-binding domains are represented by purple, red, and dark blue bars, respectively. Other system-specific domains are also colored. Data are taken from misynpat.org (33). Disease-associated mutations are represented by colored lollipops, corresponding to homozygous (orange) and compound heterozygous (green) recessive disease-associated missense and nonsense (black) mutations.
The first group, containing mt-aaRSs with mutations leading to clinical manifestations in exclusively the CNS, is further subdivided into those causing mainly epileptic encephalopathies and those causing leukoencephalopathies. Epileptic encephalopathies are observed in patients with mutations in CARS2, FARS2, NARS2, PARS2, RARS2, and TARS2. Epilepsy, which can present either as myoclonus, spastic, or focal seizures, is the common clinical manifestation. Leukoencephalopathies are observed in patients with mutations in DARS2, EARS2, MARS2, and WARS2. Changes in the white matter are the main hallmark in the diagnosis of the disease. These mutations manifest in patients mostly as ataxia, predominantly in the lower limbs. The appearance of these neurological clinical symptoms may be due to demyelination.
The second group, where defects are observed in both the CNS and another organ system, includes some patients with AARS2 mutations (those leading to leukodystrophy) and all reported patients with mutations in HARS2, LARS2, IARS2, and VARS2. Although this group is clinically distinct, it has been suggested that secondary symptoms are the result of a primary defect in the CNS (28). For example, the ovarian failures in Perrault syndrome (correlated with mutations in HARS2 and LARS2) and the ovarian failure in female patients with AARS2 mutations correlated with leukodystrophy are likely induced by a primary dysfunction in the pituitary gland, the hormonal center responsible for the correct ovarian function. In the case of patients with mutations in IARS2, defects in growth hormone production (by the pituitary gland at the level of the CNS) may cause injuries in the musculoskeletal system, explaining the skeletal dysplasia syndrome (35). In VARS2-related patients, cardiomyopathy is proposed to result from an encephalopathy that primarily produces hypotony (36). We suggest that this hypotonia causes stronger heart contractions, which is the underlying cause of the hypertrophic cardiomyopathy.
The third group, where mutations lead to effects in either the CNS or another system, includes only SARS2 mutations. Some of the SARS2-related patients have the HUPRA syndrome (hyperuricemia, pulmonary hypertension, renal failure, and alkalosis), with injuries in the kidneys that in most cases lead to renal failure (37). Other SARS2-related patients manifest with neurological clinical symptoms (not shown in the Fig. 3 because the reported mutations are splicing defects for all those patients), which lead to progressive spastic paresis (increased muscle tone) (38). Interestingly, the two sub-groups of patients do not present overlapping clinical symptoms and have distinct sets of mutations.
The last group includes YARS2-related patients and some of the AARS2-related patients, with presentations of myopathy and cardiomyopathy, respectively. None of these patients have clinical manifestations in the CNS. Again, cardiomyopathy-related mutations of mt-AlaRS are distinct from the leukodystrophy-related mutations.
As mentioned above, heterogeneity exists within these four groups. For example, among the mutations that affect the CNS, there is a strong correlation between early onset of disease and the severity of the clinical symptoms, illustrated by the contrast between DARS2-associated leukoencephalopathies, which present as LBSL disease, and RARS2-associated epileptic encephalopathy, which presents as Pontocerebellar hypoplasia type 6 (PCH6). LBSL patients usually develop movement problems during childhood or adolescence, but in some cases, the clinical manifestations do not appear until adulthood. Symptoms presented by individuals with LBSL are mainly spasticity (muscular stiffness) and ataxia (difficulty with coordinating movements). These conditions tend to affect the legs more than the arms. In the most severely affected patients, the use of wheelchair assistance is required (39). In contrast, PCH6 patients manifest the symptoms soon after birth with, in most cases, intractable seizures and recurrent apnea (40). Other neurological signs include generalized hypotonia, microcephaly (unusually small head size, caused by impaired growth of some parts of the brain), lethargy, poor suckling, and poor feeding. The most heavily affected patients live only into infancy or childhood, and they never achieve developmental milestones (41). Patients with RARS2 mutations usually manifest symptoms soon after birth, with severe seizures that tend to evolve into epileptic status. In contrast, the later the symptoms become present in LBSL patients, the milder the symptoms (e.g. weakness in the lower limbs).
This relationship between early onset and severity of symptoms is observed in other cases as well. In patients with YARS2 mutations that present mitochondrial myopathy, lactic acidosis, and sideroblastic anemia (MLASA) mortality was usually a consequence in patients with early onset. However, some exceptions have been noted; for instance, one YARS2-related patient with early onset showed spontaneous improved muscle strength and stamina at the age of 17 years and no longer required blood transfusions (which had previously been given every 6 weeks) (42).
Although our categorization is meant to point out distinct classes of mt-aaRS–related disease, it remains unclear whether the enzymes belonging to each of the groups described above have similar cellular properties that explain similarities in clinical phenotypes. Indeed, our categorization may need to modified as further ARS2-related patients are identified.
In vitro characterization of mitochondrial aaRSs
The unique features of the mammalian mitochondrial aminoacyl-tRNA synthetases, not least their ability to recognize their unusual tRNA substrates, have made them the subject of structural and functional studies since the late 1990s (43). Technical challenges in handling these proteins, including identification and removal of the N-terminal mitochondrial targeting tags (44, 45), needed to be overcome (46), but crystal structures have now been solved of human mt-AspRS (47), mt-PheRS (48, 49), mt-TrpRS (PDB code 5EKD), and mt-TyrRS (50), as well as the closely related Bos taurus mt-SerRS (51, 52). Other enzymes have also been heterologously overexpressed, purified, and characterized (including mt-AlaRS (53); mt-HisRS (54); mt-LeuRS (55); and mt-ThrRS (56)). Several common trends have been noted. These include a relatively high level of promiscuity with respect to tRNA recognition, correlated with a general reduction in the number and effect of tRNA identity elements (19, 56–60). A similar promiscuity with respect to amino acid substrates is also observed (61). In some human mitochondrial synthetases, the editing domains, which correct misacylated or misactivated amino acids, are either impaired (in the case of e.g. mt-LeuRS (62)) or missing (in the case of e.g. PheRS (63)). In others, however, robust editing activity is retained (e.g. the mt-AlaRS (53) and the mt-ThrRS (56)). In some cases, significant structural differences between the cytoplasmic and mitochondrial homologs are observed. The most dramatic example is PheRS, which is an α2β2 tetramer in the cytosol and across most of evolution, but a monomer in mitochondria (43). Another peculiar example is mt-SerRS. Mammals possess two mt-tRNASer isoacceptors, one of which lacks the D stem-loop. Mammalian mt-SerRS has acquired additional N- and C-terminal extensions (relative to its bacterial homolog) so as to be able to recognize and aminoacylate both mt-tRNASer isoacceptors (51). Other, more subtle but still significant, insertions and structural rearrangements associated with tRNA binding have been observed in crystal structures of human mt-AspRS (47) and mt-TyrRS (50).
Systematic in vitro characterization of pathogenic mutants is a more recent development, which has tested the hypothesis that these mutations would result in a reduction in tRNA aminoacylation. Available aminoacylation data reveal diverse or only weak effects of the disease-associated mutations on the level of aminoacylation. One of our laboratories has carried several investigations to decipher the relationship between mutations on the enzymatic activity of the mt-AspRS. A series of mutations showed various impacts on the aminoacylation rates, ranging from no effect at all to a decrease of ∼80-fold (34, 64). Similarly, variable results have been obtained for mutants of mt-PheRS, with some pathogenic mutations resulting in drastic (∼4000-fold) reductions in aminoacylation rates, whereas others had virtually no effect (65–67). In other enzymes, targeted characterization of single mutants has shown weak impacts. For example, the G191D mutant of mt-TyrRS displayed a 38-fold loss in catalytic efficiency compared with the WT enzyme (42), and two mutations of the mt-LeuRS, T522N and A430V, resulted in a 9- and 18-fold loss of catalytic efficiency, respectively (68).
Recombinant expression of the mutant enzymes has also allowed for the comparison of additional characteristics extending beyond aminoacylation rates. Biophysical characterization of six mt-AspRS mutants showed diverse effects on dimer formation, protein stability, and aggregation, with only mild correlations between these effects and reductions in in vitro aminoacylation rates (64, 69). Similarly, extensive work has been done with a series of pathogenic mt-PheRS mutants (67). Crystal structures of four of these were solved and were very similar to the WT protein, with main-chain root mean square deviation values less than 0.5 Å. The authors used molecular dynamics simulations to explain the failure of four other mutants to crystallize under similar conditions, because these are predicted to adopt an alternative tertiary structure that would both prevent crystal lattice contacts and disrupt protein–substrate interactions. These latter mutants are also particularly inactive in aminoacylation reactions.
Both of the mt-AspRS and mt-PheRS studies made preliminary comparisons between the extent of effects on in vitro enzymatic and biophysical properties and the type, severity, and age of onset of patients' disease symptoms. Although some correlations were noted, each study documents cases in which relatively mild in vitro effects are observed for a mutation found in a patient with severe symptoms (40, 67). These comparisons are, of course, complicated by potential differences in heterologously expressed protein behavior in vitro and in human cells. Post-translational modifications and interactions with other proteins may lead to cellular effects that would be invisible in vitro.
A puzzling aspect of pathogenic mutations identified thus far is the under-representation of mutations at highly conserved positions. Because aminoacyl-tRNA synthetases are present in every living organism, the identification of positions with key functional or structural roles is straightforward, because those amino acids are strictly conserved across the entire phylogeny. Surprisingly, mitochondrial disease-associated mutations rarely occur at these positions. Only 10 of the missense and nonsense mutations reported to date occur at positions that are 100% conserved across bacteria, archaea, and eukarya, including organelles (28, 33). More than half (129 mutations) affect nonconserved residues, whereas the remainder (69 mutations) affect positions with lower degrees of conservation. Interestingly, only two enzymes, mt-AlaRS and mt-PheRS, are impacted by mutations in highly conserved positions. In the case of mt-AlaRS, mutations at these positions are associated with the less severe leukodystrophy phenotype rather than with the more severe cardiomyopathy phenotype (although, again, it should be noted that these mutations are always part of a compound heterozygous presentation). Although initial work (65) suggested a correlation between conservation of positions mutated in mt-PheRS and physiological impact, more recent work (67) has ruled out this view. Mutants with drastic impacts on aminoacylation (observed losses of ∼3000- to ∼4000-fold) affect positions that are less conserved or not conserved, and those with the least significant impact (from 1.2- to 2.3-fold) are in highly or strictly conserved positions. More narrow phylogenetic comparisons may prove more informative. For example, LBSL-related mutations in mt-AspRS are found in positions that are not strictly conserved across all organisms, but are strictly conserved within the subphylum of mammals (69). This suggests a selective pressure at these positions, possibly restricted to mammals, and may be indicative of roles unrelated to tRNA aminoacylation.
In vivo characterization of mitochondrial synthetases
Given the incomplete, and in some cases completely absent, correlation between the mutations' effects on in vitro activity and pathogenic presentation, efforts to gain a deeper understanding of the in cellulo and in vivo biology of the mitochondrial aaRSs are of particular importance.
One area of current exploration is the sub-mitochondrial localization of the enzymes. As components of the oxidative phosphorylation pathway, all of the 13 proteins encoded by the human mitochondrial genome are membrane-bound, so the membrane-bound localization of many elements of the mitochondrial translational apparatus is not surprising (70). The mitochondrial ribosome is tethered to the matrix side of the mitochondrial inner membrane (71) through MRPL45, the mitochondrial homolog of ribosomal protein L45 (5, 6). Mitochondrial EF-Tu is also independently associated with the inner mitochondrial membrane (72). Recent work has shown that at least two mitochondrial synthetases are also associated with mitochondrial membranes (73). mt-ArgRS is exclusively associated with the membrane and is only dissociated from the membrane upon treatment with urea, indicating a hydrophobic mode of interaction. mt-AspRS is found both in membrane-bound and soluble forms. The membrane-bound fraction is dissociated with high salt, indicating an electrostatic interaction. Meanwhile, no mitochondrially localized LysRS was found to be membrane-associated. The differences in sub-mitochondrial localization of these three enzymes raise questions about potential secondary functions for the proteins. The dual localization of mt-AspRS is especially interesting, as it may result from differential processing of the cytoplasmically translated protein by the mitochondrial import machinery (45). Thus far, no pathogenic mutations have been shown to completely switch sub-mitochondrial localization (73), but effects on interactions with potential molecular partners remain to be elucidated, as does the sub-mitochondrial localization of the remaining 16 enzymes.
Another observation resulting from in cellulo work is the relationship between mitochondrial aaRS mutants and steady-state tRNA levels. In several instances, clinicians have been able to characterize mitochondrial function in patient-derived cells, usually fibroblasts. In many of these cases, cognate mitochondrial tRNA levels are specifically reduced relative to other tRNA species. Examples include experiments performed on fibroblasts from mt-SerRS–related patients, presenting with either progressive spastic paresis (38) or with HUPRA syndrome (37) or mt-ArgRS–related patients presenting with PCH6 syndrome (40). These results suggest either that the mitochondrial aaRSs serve as tRNA chaperones or that aminoacylation protects tRNA species from degradation. Because tRNA modification enzymes should affect stability, correlating stability effects with the action of those enzymes may unearth new understanding.
Cultures of patient fibroblast cells have also facilitated analysis of the mutants' effects on mitochondrial protein synthesis, total levels of aaRSs present in the mitochondria, and mitochondrial concentrations of free amino acids. Unfortunately, these data cannot fully address the tissue specificity with which mitochondrial ARS mutations manifest disease. Experiments with differentiated patient-derived stem cells, transfection of appropriate cell culture lines, or the use of animal models may be necessary to fully address these questions.
Toward the origins of tissue specificity
Although the in vitro and in vivo work described above offers some hints, at this point, aside from the fact that all mt-aaRSs are correlated with pathological disorders, there is no common combination of molecular mechanisms that explains the various phenotypic expressions observed in patients. The categorization of the disorders proposed in this review suggests that different molecular mechanisms may be at play in different tissues. To date, hypotheses explaining tissue specificity fall into two broad categories. In one set of explanations, the connection between mitochondrial protein synthesis and oxidative phosphorylation is paramount. Tissue specificity is thought to be due to differences in energy requirements in different organs, which may change depending on developmental stage. These hypotheses are most reasonable in cases where there is a clear correlation between a mutant's effect on aaRS function and the resulting disease state. A second set of hypotheses suggests that mt-aaRSs have different functions or roles, in addition to tRNA aminoacylation, that have yet to be uncovered. These hypotheses are most attractive in cases where there is little or no correlation between the pathogenicity of a mutation and its effect on aminoacylation.
As mentioned above, a complication in correlating pathogenicity and enzymatic function arises because of the heterozygous presentation of many patients, making it difficult to correlate disease severity with one or the other of two potentially deleterious mutations. Euro et al. (74) have provided an approach to this problem in their analysis of AARS2 mutations. Ten pathogenic point mutations are predicted, based on structural modeling, to have various effects on protein stability, tRNA binding, aminoacylation, and editing. The predicted effects are classified with respect to severity, from “loss of function” to “moderate.” Heterozygous patients with two “severe” or “loss of function” mutations suffered from more severe and earlier onset disease (infantile cardiomyopathy), whereas those with at least one allele predicted to have “moderate” effects suffered from symptoms with later onset (leukoencephalopathy with ovarian failure). Although the structural predictions remain to be experimentally validated, this approach to correlating molecular and organismal phenotypes is promising.
Several other hypotheses have been invoked to explain these tissue-specific differences in mt-aaRS function (24). One specific case involves a mutation in intron 2 of DARS2 found in many LBSL patients, which affects the splicing of the third exon (34). Using a slicing reporter construct, van Berge et al. (75) found cell-type–specific differences in the sensitivity to these mutations: the mutations have a larger effect on the exclusion of exon 3 in the neuronal cell lines than in non-neuronal cell lines, suggesting that the particular disease state results from tissue-specific differences in the splicing apparatus. A similarly narrow but functionally very different hypothesis was initially proposed by Götz et al. (76) when studying clinical manifestations of the cardiomyopathy produced by mutations in AARS2. They suggested that variable amino acid concentrations in different tissues, especially those of glycine and serine, might influence the misincorporation rate of serine and glycine in RC complexes in the case of proofreading-deficient mtAlaRS (76). Neuronal cells are an especially important case. Because neuronal cells present a great diversity of morphologies and an extremely high-energy demand, they face exceptional challenges in maintaining energy homeostasis. Neurons require specialized mechanisms to efficiently distribute mitochondria to distal areas where energy is in high demand, such as synaptic terminals, active growth cones, and axonal branches. Variations in mitochondrial translation in these exceptional cells have yet to be fully understood.
Validating these hypotheses, especially work on understanding variations in developmental stages, poses challenges. For example, some mutations in mt-aaRSs may participate at some points in the development of embryonic tissues. Fully understanding these effects might require the development of animal models in which mutant genes are conditionally expressed in a tissue-specific manner. Cessation of embryonic processes at different stages in these models, followed by examination of affected organs, would elucidate the involvement of mutations in embryonic development. PCH6 is one condition that we can imagine being investigated in this manner. PCH6 is produced by mutations in RARS2 (40) and has a very early onset, in most cases within the few first hours after birth (77). The hallmark of this pathology is a hypoplasia of the pons (78), which is relatively easy to identity using neuroimaging analysis. Experiments of this type would help establish whether there is a specific moment during embryogenesis when participation of the mt-ArgRS is most essential.
Explanations of tissue specificity based on energy requirements, and by extension mitochondrial protein synthesis, are intuitive. However, the generally poor correlation between the mutants' effects on in vitro behavior and severity or type of disease and the relative lack of pathogenic mutations at highly conserved positions strongly suggest that some other functions of the enzymes, unrelated to protein synthesis, are being impacted. Recent discoveries that human cytosolic aaRSs and fragments thereof are involved in multiple signaling pathways and linked to numerous diseases (79–83), ranging from cancer to Charcot-Marie-Tooth disease, strengthen this view. These enzymes have been found to act as procytokines (84, 85) and partners in pathways, including tumorigenesis, immune response, and inflammation (81, 82, 86). Given this functional expansion among their cytosolic homologs, it is likely that similar noncanonical functions have also evolved in the mt-aaRSs. The distinct sub-mitochondrial localizations of some enzymes, described above, may provide a route to identifying new functions and protein partners. Assuming that mitochondrial translation is integrated with cell metabolism and actively participates as an environmental sensor, it is likely that connections between mt-aaRS and cellular homeostasis exist. More detailed hypotheses have been discussed elsewhere (28–29).
Further exploration of these hypotheses will not only answer questions about the tissue specificity of mitochondrial diseases, but also broaden our understanding of the complex biological processes mediated by the mitochondrial aaRSs. We expect that additional roles and new connections to these “housekeeping” enzymes will continue to be uncovered, improving our ability to predict, diagnose, and treat the diseases caused by their mutation.
This work was supported in part by CNRS, Université de Strasbourg (UNISTRA), and the French National Program “Investissement d'Avenir” (Labex MitoCross), administered by the “Agence National de la Recherche,” and referenced as ANR-11-LABX-0057_MITOCROSS, by Carleton College, and by the Towsley Foundation (to J. W. C.). This is the second article in the “tRNAs and aminoacyl-tRNA synthetases in human disease” JBC Reviews series. The authors declare that they have no conflicts of interest with the contents of this article.
- aaRS
- aminoacyl-tRNA synthetase
- mt-aaRS
- mitochondrial aminoacyl-tRNA synthetase
- CNS
- central nervous system
- PDB
- Protein Data Bank
- LBSL
- leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation.
References
- 1. Gray M. W., Burger G., and Lang B. F. (2001) The origin and early evolution of mitochondria. Genome Biol. 2, REVIEWS1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- 3. Christian B. E., and Spremulli L. L. (2012) Mechanism of protein biosynthesis in mammalian mitochondria. Biochim. Biophys. Acta 1819, 1035–1054 10.1016/j.bbagrm.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hällberg B. M., and Larsson N. G. (2014) Making proteins in the powerhouse. Cell Metab. 20, 226–240 10.1016/j.cmet.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 5. Greber B. J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., and Ban N. (2015) Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 10.1126/science.aaa3872 [DOI] [PubMed] [Google Scholar]
- 6. Amunts A., Brown A., Toots J., Scheres S. H. W., and Ramakrishnan V. (2015) Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98 10.1126/science.aaa1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hosseini M., Roy P., Sissler M., Zirbel C. L., Westhof E., and Leontis N. (2018) How to fold and protect mitochondrial ribosomal RNA with fewer guanines. Nucleic Acids Res. 46, 10946–10968 10.1093/nar/gky762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Helm M., Brulé H., Friede D., Giegé R., Pütz D., and Florentz C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6, 1356–1379 10.1017/S1355838200001047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Florentz C., Sohm B., Tryoen-Tóth P., Pütz J., and Sissler M. (2003) Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 60, 1356–1375 10.1007/s00018-003-2343-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chinnery P. F. (2015) Mitochondrial disease in adults: what's old and what's new? EMBO Mol. Med. 7, 1503–1512 10.15252/emmm.201505079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng Y. S., and Turnbull D. M. (2016) Mitochondrial disease: genetics and management. J. Neurol. 263, 179–191 10.1007/s00415-015-7884-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alston C. L., Rocha M. C., Lax N. Z., Turnbull D. M., and Taylor R. W. (2017) The genetics and pathology of mitochondrial disease. J. Pathol. 241, 236–250 10.1002/path.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DiMauro S., and Moraes C. T. (1993) Mitochondrial encephalomyopathies. Arch. Neurol. 50, 1197–1208 10.1001/archneur.1993.00540110075008 [DOI] [PubMed] [Google Scholar]
- 14. Larsson N.-G., and Clayton D. A. (1995) Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 29, 151–178 10.1146/annurev.ge.29.120195.001055 [DOI] [PubMed] [Google Scholar]
- 15. Wallace D. C. (1999) Mitochondrial diseases in man and mouse. Science 283, 1482–1488 10.1126/science.283.5407.1482 [DOI] [PubMed] [Google Scholar]
- 16. Rötig A. (2011) Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta 1807, 1198–1205 10.1016/j.bbabio.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 17. Kelley S., Steinberg S., and Schimmel P. (2000) Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat. Struc. Biol. 7, 862–865 10.1038/79612 [DOI] [PubMed] [Google Scholar]
- 18. Levinger L., Mörl M., and Florentz C. (2004) Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 32, 5430–5441 10.1093/nar/gkh884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonnefond L., Fender A., Rudinger-Thirion J., Giegé R., Florentz C., and Sissler M. (2005) Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry 44, 4805–4816 10.1021/bi047527z [DOI] [PubMed] [Google Scholar]
- 20. Shiba K., Schimmel P., Motegi H., and Noda T. (1994) Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J. Biol. Chem. 269, 30049–30055 [PubMed] [Google Scholar]
- 21. Mudge S. J., Williams J. H., Eyre H. J., Sutherland G. R., Cowan P. J., and Power D. A. (1998) Complex organisation of the 5′-end of the human glycine tRNA synthetase gene. Gene 209, 45–50 10.1016/S0378-1119(98)00007-9 [DOI] [PubMed] [Google Scholar]
- 22. Tolkunova E., Park H., Xia J., King M. P., and Davidson E. (2000) The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual splicing of the primary transcript. J. Biol. Chem. 275, 35063–35069 10.1074/jbc.M006265200 [DOI] [PubMed] [Google Scholar]
- 23. Echevarría L., Clemente P., Hernández-Sierra R., Gallardo M. E., Fernández-Moreno M. A., and Garesse R. (2014) Glutamyl-tRNAGln amidotransferase is essential for mammalian mitochondrial translation in vivo. Biochem. J. 460, 91–101 10.1042/BJ20131107 [DOI] [PubMed] [Google Scholar]
- 24. Konovalova S., and Tyynismaa H. (2013) Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol. Genet. Metab. 108, 206–211 10.1016/j.ymgme.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 25. Boczonadi V., and Horvath R. (2014) Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell Biol. 48, 77–84 10.1016/j.biocel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diodato D., Ghezzi D., and Tiranti V. (2014) The mitochondrial aminoacyl tRNA synthetases: genes and syndromes. Int. J. Cell Biol. 2014, 787956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwenzer H., Zoll J., Florentz C., and Sissler M. (2014) in Topics in Current Chemistry-Aminoacyl-tRNA Synthetases: Applications in Chemistry, Biology and Medicine (Kim S., ed) pp. 247–292, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 28. Sissler M., González-Serrano L. E., and Westhof E. (2017) Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 23, 693–708 10.1016/j.molmed.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 29. Ognjenović J., and Simonović M. (2018) Human aminoacyl-tRNA synthetases in diseases of the nervous system. RNA Biol. 15, 623–634 10.1080/15476286.2017.1330245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seburn K. L., Nangle L. A., Cox G. A., Schimmel P., and Burgess R. W. (2006) An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 51, 715–726 10.1016/j.neuron.2006.08.027 [DOI] [PubMed] [Google Scholar]
- 31. Taylor R. W., Pyle A., Griffin H., Blakely E. L., Duff J., He L., Smertenko T., Alston C. L., Neeve V. C., Best A., Yarham J. W., Kirschner J., Schara U., Talim B., Topaloglu H., et al. (2014) Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 312, 68–77 10.1001/jama.2014.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verrigni D., Diodato D., Di Nottia M., Torraco A., Bellacchio E., Rizza T., Tozzi G., Verardo M., Piemonte F., Tasca G., D'Amico A., Bertini E., and Carrozzo R. (2017) Novel mutations in KARS cause hypertrophic cardiomyopathy and combined mitochondrial respiratory chain defect. Clin. Genet. 91, 918–923 [DOI] [PubMed] [Google Scholar]
- 33. Moulinier L., Ripp R., Castillo G., Poch O., and Sissler M. (2017) MiSynPat: an integrated knowledge base linking clinical, genetic, and structural data for the disease-causing mutations on human mitochondrial aminoacyl-tRNA synthetase. Hum. Mutat. 38, 1316–1324 10.1002/humu.23277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scheper G. C., van der Klok T., van Andel R. J., van Berkel C. G., Sissler M., Smet J., Muravina T. I., Serkov S. V., Uziel G., Bugiani M., Schiffmann R., Krägeloh-Mann I., Smeitink J. A., Florentz C., Coster R. V., et al. (2007) Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 39, 534–539 10.1038/ng2013 [DOI] [PubMed] [Google Scholar]
- 35. Moosa S., Haagerup A., Gregersen P. A., Petersen K. K., Altmüller J., Thiele H., Nürnberg P., Cho T. J., Kim O. H., Nishimura G., Wollnik B., and Vogel I. (2017) Confirmation of CAGSSS syndrome as a distinct entity in a Danish patient with a novel homozygous mutation in IARS2. Am. J. Med. Genet. A 173, 1102–1108 10.1002/ajmg.a.38116 [DOI] [PubMed] [Google Scholar]
- 36. Bruni F., Di Meo I., Bellacchio E., Webb B. D., McFarland R., Chrzanowska-Lightowlers Z. M. A., He L., Skorupa E., Moroni I., Ardissone A., Walczak A., Tyynismaa H., Isohanni P., Mandel H., Prokisch H., et al. (2018) Clinical, biochemical, and genetic features associated with VARS2-related mitochondrial disease. Hum. Mutat. 39, 563–578 10.1002/humu.23398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Belostotsky R., Ben-Shalom E., Rinat C., Becker-Cohen R., Feinstein S., Zeligson S., Segel R., Elpeleg O., Nassar S., and Frishberg Y. (2011) Mutations in the mitochondrial seryl-tRNA synthetase cause hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis, HUPRA syndrome. Am. J. Hum. Genet. 88, 193–200 10.1016/j.ajhg.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linnankivi T., Neupane N., Richter U., Isohanni P., and Tyynismaa H. (2016) Splicing defect in mitochondrial Seryl-tRNA synthetase gene causes progressive spastic paresis instead of HUPRA syndrome. Hum. Mutat. 37, 884–888 10.1002/humu.23021 [DOI] [PubMed] [Google Scholar]
- 39. van Berge L., Hamilton E. M., Linnankivi T., Uziel G., Steenweg M. E., Isohanni P., Wolf N. I., Krägeloh-Mann I., Brautaset N. J., Andrews P. I., de Jong B. A., al Ghamdi M., van Wieringen W. N., Tannous B. A., Hulleman E., et al. (2014) Leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation: clinical and genetic characterization and target for therapy. Brain 137, 1019–1029 10.1093/brain/awu026 [DOI] [PubMed] [Google Scholar]
- 40. Edvardson S., Shaag A., Kolesnikova O., Gomori J. M., Tarassov I., Einbinder T., Saada A., and Elpeleg O. (2007) Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 81, 857–862 10.1086/521227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joseph J. T., Innes A. M., Smith A. C., Vanstone M. R., Schwartzentruber J. A., Bulman D. E., Majewski J., Daza R. A., Hevner R. F., Michaud J., Boycott K. M., and FORGE Canada Consortium. (2014) Neuropathologic features of pontocerebellar hypoplasia type 6. J. Neuropathol. Exp. Neurol. 73, 1009–1025 10.1097/NEN.0000000000000123 [DOI] [PubMed] [Google Scholar]
- 42. Riley L. G., Menezes M. J., Rudinger-Thirion J., Duff R., de Lonlay P., Rotig A., Tchan M. C., Davis M., Cooper S. T., and Christodoulou J. (2013) Phenotypic variability and identification of novel YARS2 mutations in YARS2 mitochondrial myopathy, lactic acidosis and sideroblastic anaemia. Orphanet. J. Rare Dis. 8, 193 10.1186/1750-1172-8-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bullard J. M., Cai Y.-C., Demeler B., and Spremulli L. L. (1999) Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J. Mol. Biol. 288, 567–577 10.1006/jmbi.1999.2708 [DOI] [PubMed] [Google Scholar]
- 44. Gaudry A., Lorber B., Messmer M., Neuenfeldt A., Sauter C., Florentz C., and Sissler M. (2012) Redesigned N terminus enhances expression, solubility, and crystallisability of mitochondrial enzyme. Protein Eng. 25, 473–481 10.1093/protein/gzs046 [DOI] [PubMed] [Google Scholar]
- 45. Carapito C., Kuhn L., Karim L., Rompais M., Rabilloud T., Schwenzer H., and Sissler M. (2017) Two proteomic methodologies for defining N-termini of mature human mitochondrial aminoacyl-tRNA synthetases. Methods 113, 111–119 10.1016/j.ymeth.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 46. Sissler M., Lorber B., Messmer M., Schaller A., Pütz J., and Florentz C. (2008) Handling mammalian mitochondrial tRNAs and aminoacyl-tRNA synthetases for functional and structural characterization. Methods 44, 176–189 10.1016/j.ymeth.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 47. Neuenfeldt A., Lorber B., Ennifar E., Gaudry A., Sauter C., Sissler M., and Florentz C. (2013) Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res. 41, 2698–2708 10.1093/nar/gks1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klipcan L., Levin I., Kessler N., Moor N., Finarov I., and Safro M. (2008) The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase. Structure 16, 1095–1104 10.1016/j.str.2008.03.020 [DOI] [PubMed] [Google Scholar]
- 49. Klipcan L., Moor N., Finarov I., Kessler N., Sukhanova M., and Safro M. G. (2012) Crystal structure of human mitochondrial PheRS complexed with tRNA(Phe) in the active “open” state. J. Mol. Biol. 415, 527–537 10.1016/j.jmb.2011.11.029 [DOI] [PubMed] [Google Scholar]
- 50. Bonnefond L., Frugier M., Touzé E., Lorber B., Florentz C., Giegé R., Sauter C., and Rudinger-Thirion J. (2007) Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features. Structure 15, 1505–1516 10.1016/j.str.2007.09.018 [DOI] [PubMed] [Google Scholar]
- 51. Chimnaronk S., Gravers Jeppesen M., Suzuki T., Nyborg J., and Watanabe K. (2005) Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 24, 3369–3379 10.1038/sj.emboj.7600811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chimnaronk S., Jeppesen M. G., Shimada N., Suzuki T., Nyborg J., and Watanabe K. (2004) Crystallization and preliminary X-ray diffraction study of mammalian mitochondrial seryl-tRNA synthetase. Acta Crystallgr. D Biol. Crystallgr. 60, 1319–1322 10.1107/S0907444904011217 [DOI] [PubMed] [Google Scholar]
- 53. Hilander T., Zhou X. L., Konovalova S., Zhang F. P., Euro L., Chilov D., Poutanen M., Chihade J., Wang E. D., and Tyynismaa H. (2018) Editing activity for eliminating mischarged tRNAs is essential in mammalian mitochondria. Nucleic Acids Res. 46, 849–860 10.1093/nar/gkx1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pierce S. B., Chisholm K. M., Lynch E. D., Lee M. K., Walsh T., Opitz J. M., Li W., Klevit R. E., and King M. C. (2011) Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc. Natl. Acad. Sci. U.S.A. 108, 6543–6548 10.1073/pnas.1103471108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bullard J. M., Cai Y.-C., and Spremulli L. L. (2000) Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim. Biophys. Acta 1490, 245–258 10.1016/S0167-4781(99)00240-7 [DOI] [PubMed] [Google Scholar]
- 56. Wang Y., Zhou X. L., Ruan Z. R., Liu R. J., Eriani G., and Wang E. D. (2016) A human disease-causing point mutation in mitochondrial threonyl-tRNA synthetase induces both structural and functional defects. J. Biol. Chem. 291, 6507–6520 10.1074/jbc.M115.700849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sohm B., Sissler M., Park H., King M. P., and Florentz C. (2004) Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 339, 17–29 10.1016/j.jmb.2004.03.066 [DOI] [PubMed] [Google Scholar]
- 58. Sissler M., Helm M., Frugier M., Giege R., and Florentz C. (2004) Aminoacylation properties of pathology-related variants of human mitochondrial tRNALys variants. RNA 10, 841–853 10.1261/rna.5267604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bonnefond L., Frugier M., Giegé R., and Rudinger-Thirion J. (2005) Human mitochondrial TyrRS disobeys the tyrosine idenity rules. RNA 11, 558–562 10.1261/rna.7246805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fender A., Sauter C., Messmer M., Pütz J., Giegé R., Florentz C., and Sissler M. (2006) Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 281, 15980–15986 10.1074/jbc.M511633200 [DOI] [PubMed] [Google Scholar]
- 61. Messmer M., Blais S. P., Balg C., Chênevert R., Grenier L., Lagüe P., Sauter C., Sissler M., Giegé R., Lapointe J., and Florentz C. (2009) Peculiar inhibition of human mitochondrial aspartyl-tRNA synthetase by adenylate analogs. Biochimie 91, 596–603 10.1016/j.biochi.2009.02.005 [DOI] [PubMed] [Google Scholar]
- 62. Lue S. W., and Kelley S. O. (2005) An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry 44, 3010–3016 10.1021/bi047901v [DOI] [PubMed] [Google Scholar]
- 63. Klipcan L., Moor N., Kessler N., and Safro M. G. (2009) Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc. Natl. Acad. Sci. U.S.A. 106, 11045–11048 10.1073/pnas.0905212106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van Berge L., Kevenaar J., Polder E., Gaudry A., Florentz C., Sissler M., van der Knaap M. S., and Scheper G. C. (2013) Pathogenic mutations causing LBSL affect mitochondrial aspartyl-tRNA synthetase in diverse ways. Biochem. J. 450, 345–350 10.1042/BJ20121564 [DOI] [PubMed] [Google Scholar]
- 65. Elo J. M., Yadavalli S. S., Euro L., Isohanni P., Götz A., Carroll C. J., Valanne L., Alkuraya F. S., Uusimaa J., Paetau A., Caruso E. M., Pihko H., Ibba M., Tyynismaa H., and Suomalainen A. (2012) Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum. Mol. Genet. 21, 4521–4529 10.1093/hmg/dds294 [DOI] [PubMed] [Google Scholar]
- 66. Walker M. A., Mohler K. P., Hopkins K. W., Oakley D. H., Sweetser D. A., Ibba M., Frosch M. P., and Thibert R. L. (2016) Novel compound heterozygous mutations expand the recognized phenotypes of FARS2-linked disease. J. Child Neurol. 31, 1127–1137 10.1177/0883073816643402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kartvelishvili E., Tworowski D., Vernon H., Moor N., Wang J., Wong L. J., Chrzanowska-Lightowlers Z., and Safro M. (2017) Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2. Protein Sci. 26, 1505–1516 10.1002/pro.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Riley L. G., Rudinger-Thirion J., Schmitz-Abe K., Thorburn D. R., Davis R. L., Teo J., Arbuckle S., Cooper S. T., Campagna D. R., Frugier M., Markianos K., Sue C. M., Fleming M. D., and Christodoulou J. (2016) LARS2 variants associated with hydrops, lactic acidosis, sideroblastic anemia, and multisystem failure. JIMD Rep. 28, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sauter C., Lorber B., Gaudry A., Karim L., Schwenzer H., Wien F., Roblin P., Florentz C., and Sissler M. (2015) Neurodegenerative disease-associated mutants of a human mitochondrial aminoacyl-tRNA synthetase present individual molecular signatures. Sci. Rep. 5, 17332 10.1038/srep17332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ott M., and Herrmann J. M. (2010) Co-translational membrane insertion of mitochondrially encoded proteins. Biochim. Biophys. Acta 1803, 767–775 10.1016/j.bbamcr.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 71. Liu M., and Spremulli L. (2000) Interaction of mammalian mitochondrial ribosomes with the inner membrane. J. Biol. Chem. 275, 29400–29406 10.1074/jbc.M002173200 [DOI] [PubMed] [Google Scholar]
- 72. Suzuki H., Ueda T., Taguchi H., and Takeuchi N. (2007) Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J. Biol. Chem. 282, 4076–4084 [DOI] [PubMed] [Google Scholar]
- 73. González-Serrano L. E., Karim L., Pierre F., Schwenzer H., Rötig A., Munnich A., and Sissler M. (2018) Three human aminoacyl-tRNA synthetases have distinct sub-mitochondrial localizations that are unaffected by disease-associated mutations. J. Biol. Chem. 293, 13604–13615 10.1074/jbc.RA118.003400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Euro L., Konovalova S., Asin-Cayuela J., Tulinius M., Griffin H., Horvath R., Taylor R. W., Chinnery P. F., Schara U., Thorburn D. R., Suomalainen A., Chihade J., and Tyynismaa H. (2015) Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects predicts differential effects on aminoacylation. Front. Genet. 6, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. van Berge L., Dooves S., van Berkel C. G., Polder E., van der Knaap M. S., and Scheper G. C. (2012) Leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation is associated with cell-type-dependent splicing of mtAspRS mRNA. Biochem. J. 441, 955–962 10.1042/BJ20110795 [DOI] [PubMed] [Google Scholar]
- 76. Götz A., Tyynismaa H., Euro L., Ellonen P., Hyötyläinen T., Ojala T., Hämäläinen R. H., Tommiska J., Raivio T., Oresic M., Karikoski R., Tammela O., Simola K. O., Paetau A., Tyni T., and Suomalainen A. (2011) Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am. J. Hum. Genet. 88, 635–642 10.1016/j.ajhg.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang J., Zhang Z., Zhang Y., and Wu Y. (2018) Distinct magnetic resonance imaging features in a patient with novel RARS2 mutations: a case report and review of the literature. Exp. Ther. Med. 15, 1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kastrissianakis K., Anand G., Quaghebeur G., Price S., Prabhakar P., Marinova J., Brown G., and McShane T. (2013) Subdural effusions and lack of early pontocerebellar hypoplasia in siblings with RARS2 mutations. Arch. Dis. Child. 98, 1004–1007 10.1136/archdischild-2013-304308 [DOI] [PubMed] [Google Scholar]
- 79. Park S. G., Schimmel P., and Kim S. (2008) Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 10.1073/pnas.0802862105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo M., Yang X. L., and Schimmel P. (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 10.1038/nrm2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim S., You S., and Hwang D. (2011) Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer 11, 708–718 10.1038/nrc3124 [DOI] [PubMed] [Google Scholar]
- 82. Guo M., and Schimmel P. (2013) Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153 10.1038/nchembio.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Blocquel D., Li S., Wei N., Daub H., Sajish M., Erfurth M. L., Kooi G., Zhou J., Bai G., Schimmel P., Jordanova A., and Yang X. L. (2017) Alternative stable conformation capable of protein misinteraction links tRNA synthetase to peripheral neuropathy. Nucleic Acids Res. 45, 8091–8104 10.1093/nar/gkx455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wakasugi K., and Schimmel P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 10.1126/science.284.5411.147 [DOI] [PubMed] [Google Scholar]
- 85. Son S. H., Park M. C., and Kim S. (2013) in Aminoacyl-tRNA Synthetases in Biology and Medicine (Kim S., ed) pp. 145–166, Springer, New York [Google Scholar]
- 86. Lee E. Y., Kim S., and Kim M. H. (2018) Aminoacyl-tRNA synthetases, therapeutic targets for infectious diseases. Biochem. Pharmacol. 154, 424–434 10.1016/j.bcp.2018.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hallmann K., Zsurka G., Moskau-Hartmann S., Kirschner J., Korinthenberg R., Ruppert A. K., Ozdemir O., Weber Y., Becker F., Lerche H., Elger C. E., Thiele H., Nürnberg P., Sander T., and Kunz W. S. (2014) A homozygous splice-site mutation in CARS2 is associated with progressive myoclonic epilepsy. Neurology 83, 2183–2187 10.1212/WNL.0000000000001055 [DOI] [PubMed] [Google Scholar]
- 88. Coughlin C. R. 2nd., Scharer G. H., Friederich M. W., Yu H. C., Geiger E. A., Creadon-Swindell G., Collins A. E., Vanlander A. V., Coster R. V., Powell C. A., Swanson M. A., Minczuk M., Van Hove J. L., and Shaikh T. H. (2015) Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J. Med. Genet. 52, 532–540 10.1136/jmedgenet-2015-103049 [DOI] [PubMed] [Google Scholar]
- 89. Yang Y., Liu W., Fang Z., Shi J., Che F., He C., Yao L., Wang E., and Wu Y. (2016) A newly identified missense mutation in FARS2 causes autosomal-recessive spastic paraplegia. Hum. Mutat. 37, 165–169 10.1002/humu.22930 [DOI] [PubMed] [Google Scholar]
- 90. Cho J. S., Kim S. H., Kim H. Y., Chung T., Kim D., Jang S., Lee S. B., Yoo S. K., Shin J., Kim J. I., Kim H., Hwang H., Chae J. H., Choi J., Kim K. J., and Lim B. C. (2017) FARS2 mutation and epilepsy: possible link with early-onset epileptic encephalopathy. Epilepsy Res. 129, 118–124 10.1016/j.eplepsyres.2016.11.022 [DOI] [PubMed] [Google Scholar]
- 91. Sofou K., Kollberg G., Holmström M., Dávila M., Darin N., Gustafsson C. M., Holme E., Oldfors A., Tulinius M., and Asin-Cayuela J. (2015) Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol. Genet. Genomic Med. 3, 59–68 10.1002/mgg3.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Simon M., Richard E. M., Wang X., Shahzad M., Huang V. H., Qaiser T. A., Potluri P., Mahl S. E., Davila A., Nazli S., Hancock S., Yu M., Gargus J., Chang R., Al-Sheqaih N. Newman W. G., et al. (2015) Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh Syndrome. PLoS Genet. 11, e1005097 10.1371/journal.pgen.1005097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mizuguchi T., Nakashima M., Kato M., Yamada K., Okanishi T., Ekhilevitch N., Mandel H., Eran A., Toyono M., Sawaishi Y., Motoi H., Shiina M., Ogata K., Miyatake S., Miyake N., et al. (2017) PARS2 and NARS2 mutations in infantile-onset neurodegenerative disorder. J. Hum. Genet. 62, 525–529 10.1038/jhg.2016.163 [DOI] [PubMed] [Google Scholar]
- 94. Namavar Y., Barth P. G., Kasher P. R., van Ruissen F., Brockmann K., Bernert G., Writzl K., Ventura K., Cheng E. Y., Ferriero D. M., Basel-Vanagaite L., Eggens V. R., Krägeloh-Mann I., De Meirleir L., King M., et al. (2011) Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain 134, 143–156 10.1093/brain/awq287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cassandrini D., Cilio M. R., Bianchi M., Doimo M., Balestri M., Tessa A., Rizza T., Sartori G., Meschini M. C., Nesti C., Tozzi G., Petruzzella V., Piemonte F., Bisceglia L., Bruno C., et al. (2013) Pontocerebellar hypoplasia type 6 caused by mutations in RARS2: definition of the clinical spectrum and molecular findings in five patients. J. Inherit. Metab. Dis. 36, 43–53 10.1007/s10545-012-9487-9 [DOI] [PubMed] [Google Scholar]
- 96. Lühl S., Bode H., Schlötzer W., Bartsakoulia M., Horvath R., Abicht A., Stenzel M., Kirschner J., and Grünert S. C. (2016) Novel homozygous RARS2 mutation in two siblings without pontocerebellar hypoplasia–further expansion of the phenotypic spectrum. Orphanet. J. Rare Dis. 11, 140 10.1186/s13023-016-0525-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Diodato D., Melchionda L., Haack T. B., Dallabona C., Baruffini E., Donnini C., Granata T., Ragona F., Balestri P., Margollicci M., Lamantea E., Nasca A., Powell C. A., Minczuk M., Strom T. M., et al. (2014) VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum. Mutat. 35, 983–989 10.1002/humu.22590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Uluc K., Baskan O., Yildirim K. A., Ozsahin S., Koseoglu M., Isak B., Scheper G. C., Gunal D. I., and van der Knaap M. S. (2008) Leukoencephalopathy with brain stem and spinal cord involvement and high lactate: a genetically proven case with distinct MRI findings. J. Neurol. Sci. 273, 118–122 10.1016/j.jns.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 99. Steenweg M. E., Ghezzi D., Haack T., Abbink T. E., Martinelli D., van Berkel C. G., Bley A., Diogo L., Grillo E., Te Water Naudé J., Strom T. M., Bertini E., Prokisch H., van der Knaap M. S., and Zeviani M. (2012) Leukoencephalopathy with thalamus and brainstem involvement and high lactate 'LTBL' caused by EARS2 mutations. Brain 135, 1387–1394 10.1093/brain/aws070 [DOI] [PubMed] [Google Scholar]
- 100. Talim B., Pyle A., Griffin H., Topaloglu H., Tokatli A., Keogh M. J., Santibanez-Koref M., Chinnery P. F., and Horvath R. (2013) Multisystem fatal infantile disease caused by a novel homozygous EARS2 mutation. Brain 136, e228 10.1093/brain/aws197 [DOI] [PubMed] [Google Scholar]
- 101. Şahin S., Cansu A., Kalay E., Dinçer T., Kul S., Çakır İ. M, Kamaşak T., and Budak G. Y. (2016) Leukoencephalopathy with thalamus and brainstem involvement and high lactate caused by novel mutations in the EARS2 gene in two siblings. J. Neurol. Sci. 365, 54–58 10.1016/j.jns.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 102. Oliveira R., Sommerville E. W., Thompson K., Nunes J., Pyle A., Grazina M., Chinnery P. F., Diogo L., Garcia P., and Taylor R. W. (2017) Lethal neonatal LTBL associated with Biallelic EARS2 variants: case report and review of the reported neuroradiological features. JIMD Rep. 33, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bayat V., Thiffault I., Jaiswal M., Tétreault M., Donti T., Sasarman F., Bernard G., Demers-Lamarche J., Dicaire M. J., Mathieu J., Vanasse M., Bouchard J. P., Rioux M. F., Lourenco C. M., Li Z., et al. (2012) Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 10, e1001288 10.1371/journal.pbio.1001288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Webb B. D., Wheeler P. G., Hagen J. J., Cohen N., Linderman M. D., Diaz G. A., Naidich T. P., Rodenburg R. J., Houten S. M., and Schadt E. E. (2015) Novel, compound heterozygous, single-nucleotide variants in MARS2 associated with developmental delay, poor growth, and sensorineural hearing loss. Hum. Mutat. 36, 587–592 10.1002/humu.22781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Musante L., Püttmann L., Kahrizi K., Garshasbi M., Hu H., Stehr H., Lipkowitz B., Otto S., Jensen L. R., Tzschach A., Jamali P., Wienker T., Najmabadi H., Ropers H. H., and Kuss A. W. (2017) Mutations of the aminoacyl-tRNA-synthetases SARS and WARS2 are implicated in the etiology of autosomal recessive intellectual disability. Hum. Mutat. 38, 621–636 10.1002/humu.23205 [DOI] [PubMed] [Google Scholar]
- 106. Theisen B. E., Rumyantseva A., Cohen J. S., Alcaraz W. A., Shinde D. N., Tang S., Srivastava S., Pevsner J., Trifunovic A., and Fatemi A. (2017) Deficiency of WARS2, encoding mitochondrial tryptophanyl tRNA synthetase, causes severe infantile onset leukoencephalopathy. Am. J. Med. Genet. A 173, 2505–2510 10.1002/ajmg.a.38339 [DOI] [PubMed] [Google Scholar]
- 107. Burke E. A., Frucht S. J., Thompson K., Wolfe L. A., Yokoyama T., Bertoni M., Huang Y., Sincan M., Adams D. R., Taylor R. W., Gahl W. A., Toro C., and Malicdan M. C. V. (2018) Biallelic mutations in mitochondrial tryptophanyl-tRNA synthetase cause levodopa-responsive infantile-onset Parkinsonism. Clin. Genet. 93, 712–718 10.1111/cge.13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Baertling F., Alhaddad B., Seibt A., Budaeus S., Meitinger T., Strom T. M., Mayatepek E., Schaper J., Prokisch H., Haack T. B., and Distelmaier F. (2017) Neonatal encephalocardiomyopathy caused by mutations in VARS2. Metab. Brain Dis. 32, 267–270 10.1007/s11011-016-9890-2 [DOI] [PubMed] [Google Scholar]
- 109. Dallabona C., Diodato D., Kevelam S. H., Haack T. B., Wong L. J., Salomons G. S., Baruffini E., Melchionda L., Mariotti C., Strom T. M., Meitinger T., Prokisch H., Chapman K., Colley A., Rocha H., et al. (2014) Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology 82, 2063–2071 10.1212/WNL.0000000000000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lakshmanan R., Adams M. E., Lynch D. S., Kinsella J. A., Phadke R., Schott J. M., Murphy E., Rohrer J. D., Chataway J., Houlden H., Fox N. C., and Davagnanam I. (2017) Redefining the phenotype of ALSP and AARS2 mutation-related leukodystrophy. Neurol. Genet. 3, e135 10.1212/NXG.0000000000000135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lieber D. S., Hershman S. G., Slate N. G., Calvo S. E., Sims K. B., Schmahmann J. D., and Mootha V. K. (2014) Next generation sequencing with copy number variant detection expands the phenotypic spectrum of HSD17B4-deficiency. BMC Med. Genet. 15, 30 10.1186/1471-2350-15-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lerat J., Jonard L., Loundon N., Christin-Maitre S., Lacombe D., Goizet C., Rouzier C., Van Maldergem L., Gherbi S., Garabedian E. N., Bonnefont J. P., Touraine P., Mosnier I., Munnich A., Denoyelle F., and Marlin S. (2016) An application of NGS for molecular investigations in Perrault syndrome: study of 14 families and review of the literature. Hum. Mutat. 37, 1354–1362 10.1002/humu.23120 [DOI] [PubMed] [Google Scholar]
- 113. Pierce S. B., Gersak K., Michaelson-Cohen R., Walsh T., Lee M. K., Malach D., Klevit R. E., King M. C., and Levy-Lahad E. (2013) Mutations in LARS2, encoding mitochondrial leucyl-tRNA synthetase, lead to premature ovarian failure and hearing loss in Perrault syndrome. Am. J. Hum. Genet. 92, 614–620 10.1016/j.ajhg.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Soldà G., Caccia S., Robusto M., Chiereghin C., Castorina P., Ambrosetti U., Duga S., and Asselta R. (2016) First independent replication of the involvement of LARS2 in Perrault syndrome by whole-exome sequencing of an Italian family. J. Hum. Genet. 61, 295–300 10.1038/jhg.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Schwartzentruber J., Buhas D., Majewski J., Sasarman F., Papillon-Cavanagh S., Thiffault I., Sheldon K. M., Massicotte C., Patry L., Simon M., Zare A. S., McKernan K. J., Consortium F. C., Michaud J., Boles R. G., et al. (2014) Mutation in the nuclear encoded mitochondrial isoleucyl tRNA-synthetase IARS2 in patients with cataracts, growth hormone deficiency with short stature, partial sensorineural deafness and peripheral neuropathy or with Leigh syndrome. Hum. Mutat. 35, 1285–1289 [DOI] [PubMed] [Google Scholar]
- 116. Mazurova S., Magner M., Kucerova-Vidrova V., Vondrackova A., Stranecky V., Pristoupilova A., Zamecnik J., Hansikova H., Zeman J., Tesarova M., and Honzik T. (2017) Thymidine kinase 2 and alanyl-tRNA synthetase 2 deficiencies cause lethal mitochondrial cardiomyopathy: case reports and review of the literature. Cardiol. Young 27, 936–944 10.1017/S1047951116001876 [DOI] [PubMed] [Google Scholar]
- 117. Riley L. G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S. C., Thorburn D., Ryan M. T., Giegé R., Bahlo M., and Christodoulou J. (2010) Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia-MLASA syndrome. Am. J. Hum. Genet. 87, 52–59 10.1016/j.ajhg.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ardissone A., Lamantea E., Quartararo J., Dallabona C., Carrara F., Moroni I., Donnini C., Garavaglia B., Zeviani M., and Uziel G. (2015) A novel homozygous YARS2 mutation in two Italian siblings and a review of literature. JIMD Rep. 20, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sommerville E. W., Ng Y. S., Alston C. L., Dallabona C., Gilberti M., He L., Knowles C., Chin S. L., Schaefer A. M., Falkous G., Murdoch D., Longman C., de Visser M., Bindoff L. A., Rawles J. M., et al. (2017) Clinical features, molecular heterogeneity, and prognostic implications in YARS2-related mitochondrial myopathy. JAMA Neurol. 74, 686–694 10.1001/jamaneurol.2016.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]