Abstract
Cumulative evidence suggests that the heat shock protein 90 (Hsp90) co-chaperone UNC-45 myosin chaperone A (UNC45A) contributes to tumorigenesis and that its expression in cancer cells correlates with proliferation and metastasis of solid tumors. However, the molecular mechanism by which UNC45A regulates cancer cell proliferation remains largely unknown. Here, using siRNA-mediated gene silencing and various human cells, we report that UNC45A is essential for breast cancer cell growth, but is dispensable for normal cell proliferation. Immunofluorescence microscopy, along with gene microarray and RT-quantitative PCR analyses, revealed that UNC45A localizes to the cancer cell nucleus, where it up-regulates the transcriptional activity of the glucocorticoid receptor and thereby promotes expression of the mitotic kinase NIMA-related kinase 7 (NEK7). We observed that UNC45A-deficient cancer cells exhibit extensive pericentrosomal material disorganization, as well as defects in centrosomal separation and mitotic chromosome alignment. Consequently, these cells stalled in metaphase and cytokinesis and ultimately underwent mitotic catastrophe, phenotypes that were rescued by heterologous NEK7 expression. Our results identify a key role for the co-chaperone UNC45A in cell proliferation and provide insight into the regulatory mechanism. We propose that UNC45A represents a promising new therapeutic target to inhibit cancer cell growth in solid tumor types.
Keywords: cancer biology, mitosis, molecular chaperone, centrosome, glucocorticoid receptor, Heat shock protein 90, Hsp90, KEK7, UNC-45A, UNC45A
Introduction
UNC453 (UNCoordinated (1)) is a member of the UCS (UNC-45/Cro1/She4p) family of myosin-interacting proteins. In vertebrates, two genes encode two UNC45 isoforms that share ∼55% sequence identity (2). Isoform A is ubiquitous, whereas isoform B is restricted to skeletal and cardiac muscle expression. The UCS domain of UNC45B is organized in helical armadillo repeats (3, 4), which cooperate with the N-terminal tetratricopeptide domain to recruit the heat shock protein 90 (Hsp90) for myosin folding (5–10). Mutations in UNC45B result in cardiomyopathies and defects in skeletal muscle (11, 12). Although in vitro studies suggest that UNC45 isoforms share overlapping functions (e.g. myosin folding), zebrafish studies show that UNC45A is not required for myogenesis (13). A recent study using the U2OS osteosarcoma cell line has shown that UNC45A promotes myosin folding and stress fibers assembly (14). Biochemically, both isoforms interact with Hsp90 (5, 15), but UNC45A showed higher specificity toward Hsp90β (16).
Cumulative evidence suggests that UNC45A contributes to tumorigenesis (15, 17–19); its expression in cancer cells is correlated with the stage and the grade of the disease (17, 18). UNC45A co-localizes with nonmuscle myosin II (NMII) in the cleavage furrow during cytokinesis (17) and to centrosomes, to which it helps recruit checkpoint kinase 1 (ChK1) (20). Recent work from Bazzaro's group (21) showed that UNC45A is a microtubule-associated protein that modulates the sensitivity of ovarian cancer cells to paclitaxel. UNC45A expression in reporter gene systems (chloramphenicol acetyltransferase and luciferase) has shown that UNC45A can regulate transcription of the progesterone (PR) (15), retinoic acid α, and peroxisome proliferator-activated receptor-α and -γ (19). The molecular mechanisms underlying the function and localization of UNC45A in cancer cells, however, has not been well studied.
Here, we report that UNC45A is largely dispensable for the proliferation of immortalized, nontransformed mammary cell lines, but is essential for breast cancer cell proliferation in vitro and in vivo. In cancer cells, UNC45A localizes to the cell nucleus, where it promotes glucocorticoid receptor (GR) transcription of the mitotic kinase gene NEK7. Previous research established that NEK7 is required for cell cycle progression because of its ability to regulate aspects of mitotic spindle formation and cytokinesis (22–28). Cancer cells lacking UNC45A expression exhibit a strikingly similar phenotype to NEK7 knockout cells, suggesting that UNC45A control of the cell cycle in cancer cells is mediated by NEK7 signaling.
Results
UNC45A is essential for breast cancer cell proliferation in vitro and in vivo
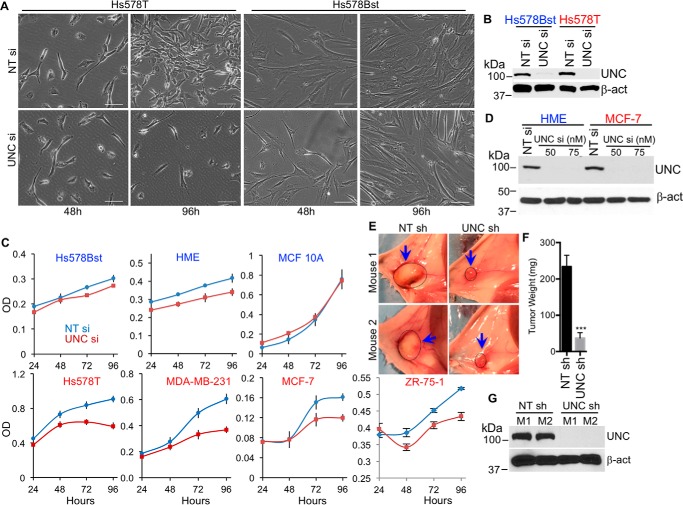
We investigated the role of UNC45A in the proliferation of breast cancer cell lines in vitro. Microscopic analysis indicates that silencing UNC45A using siRNA causes cell proliferation arrest and ultimately cell death in Hs578T cells (Fig. 1, A and B). In contrast, loss of UNC45A expression had no noticeable effect on the shape or proliferation of Hs578Bst cells; these results were confirmed using an MTT cell proliferation assay. Fig. 1C shows that silencing UNC45A did not affect the proliferation of any nontransformed cell line tested, including the Hs578Bst, HME, and MCF-10A mammary epithelial lines. In contrast, loss of UNC45A significantly reduced the proliferation of all transformed cell lines tested, including Hs578T and MDA-MB-231 (both triple negative), (MCF-7 (ER/PR-positive), and the metastatic ZR-75-1, further supporting the concept that UNC45A expression may be required for the growth of various breast cancer subtypes.
Figure 1.
UNC45A is essential for proliferation of cancer, but is dispensable for normal cell proliferation. A, bright light microscopic images of Hs587T and Hs587Bst cells transfected with 75 nm nontargeting siRNA (NT si) or UNC45A siRNA (UNC si). Scale bar represents 100 μm. B, Western blot analysis of lysates from cells in A at 96 h post-transfection. C, MTT assay monitoring cell proliferation of the indicated breast cell lines transfected with nontargeting siRNA (blue) or UNC45A siRNA (red). The experiment was repeated twice. All data points are done sextuplicates. Error bars represent mean ± S.D. D, Western blot analysis of lysates from HME and MCF-7 cells at 96 h after transfection with 50 or 75 nm UNC45A siRNA. 75 nm NT siRNA was used as control. Images in A–D are representative of 3 independent experiments. E–G, groups of female NOD/SCID mice (n = 6) were implanted with 105 MDA-MB-231 cells harboring UNC45A shRNA (UNC sh) or nontargeting shRNA (NT sh) into the mammary fat pad. Forty-eight hours after implantation, mice were fed doxycycline-containing food for 6 weeks. Mice were sacrificed, tumor images were taken (E), and tumors were extracted, weighed (F), and used for Western blot analysis (G). Error bars represent mean ± S.D. Unpaired two-tailed t test was used for significance. ***, p < 0.001. β-Actin (β-act) and Hsp90β were used as loading controls.
To investigate the specific role of UNC45A in cancer cell proliferation in vivo, we used the non-obese diabetic/severe combined immunodeficiency (NOD-SCID) mouse model (Fig. 1, E–G), surgically implanted with 105 MDA-MB-231 cells stably harboring lentivirus-based doxycycline-inducible UNC45A shRNA (or nontargeting shRNA control) into the mammary fat pads. Two days later, mice were fed doxycycline-containing food for 6 weeks. UNC45A was efficiently silenced in tumors harboring UNC45A versus control shRNA (Fig. 1G). Loss of UNC45A expression inhibited tumor growth, as illustrated by ex vivo images of tumors in control versus knockdown animals (Fig. 1E, left and right panels, respectively), as well as by the average tumor weights (Fig. 1F). Together, these findings demonstrate that UNC45A is essential for cancer cell proliferation in vitro and in vivo.
UNC45A is required for NEK7 gene expression in cancer cells
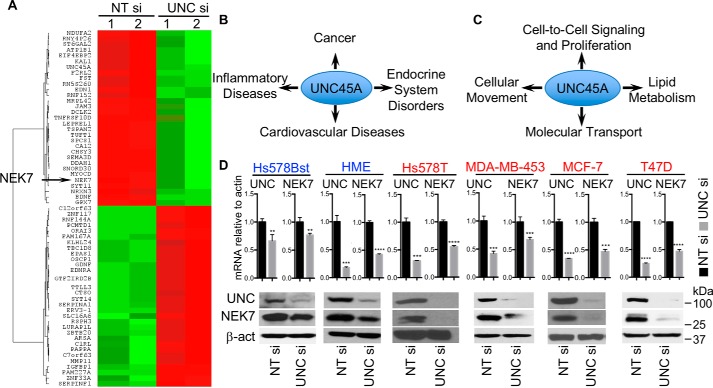
To better understand how UNC45A might regulate cancer cell proliferation, we performed microarray analysis using mRNAs from Hs578T cells treated with either UNC45A siRNA or control nontargeting siRNA. As predicted, UNC45A mRNA was significantly reduced in UNC45A siRNA-treated Hs578T cells, which correlated with changes in the expression of 121 other genes, of which 62 exhibit at least a 1.64-fold or greater change (Fig. 2A, Table S1). Ingenuity Pathway Analysis indicates that these differentially expressed genes are involved in different pathologies, including: cancer, endocrine disorders, and inflammatory and cardiovascular diseases (Fig. 2B). Further analysis indicated that these genes are also connected to molecular networks involved in: cell-to-cell signaling and proliferation, cellular movement, molecular transport, and lipid metabolism (Fig. 2C). Some of the genes identified are also associated with control of cell cycle, cell morphology, multicellular organization, and development of the nervous system and its function (Fig. S1A). Based on its relevance to cell division, we noted that loss of UNC45A was associated with a 2-fold decrease in the mRNA encoding the mitotic kinase NEK7 (Fig. 2A, Table S1).
Figure 2.
NEK7 expression requires UNC45A. A, microarray analysis of mRNA isolated from Hs578T 96 h post-transfection with 75 nm UNc45A siRNA (UNC si) or nontargeting siRNA (NT si). Experiments were done in two biological duplicates. The heat map shows the top 62 genes with 1.64-fold change or higher in their expression. B and C, functional annotation using the database for annotation, visualization, and integrated discovery (DAVID) version 6.8 using the genes listed in A. B shows the potential pathologies in which UNC45A could be involved, and C shows cellular functions and molecular networks that could be affected by loss of UNC45A. D, RT-qPCR (upper) and Western blotting (lower) analyses of UNC45A and NEK7 in the indicated breast cell lines. Images are representative of 3 independent experiments. Error bars represent mean ± S.D. Unpaired two-tailed t test was used for significance. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01. β-Actin was used as a control. Images are representative of 2 independent experiments.
Loss of NEK7 was validated by RT-qPCR and immunoblot analyses of a panel of cancer-derived cell lines. Silencing UNC45A drastically reduced mRNA and protein expression of NEK7 in Hsp578T, MDA-MB-453, MCF-7, and T47D breast cancer cells (Fig. 2D). Loss of NEK7 was also observed in HeLa cells and MDA-MB-231 (Fig. S1, C and E). Remarkably, whereas loss of UNC45A in nontransformed, immortalized breast epithelial cells (Hs578Bst and HME) significantly reduced NEK7 mRNA expression, this resulted in only a 50–70% decrease in NEK7 protein expression (Fig. 2D), suggesting that in normal cells, NEK7 expression is not totally dependent on UNC45A, whereas in cancer cells, UNC45A appears to be required for NEK7 expression. Indeed, the remaining NEK7 protein (30–50%) observed in normal cells might contribute to the relative insensitivity of these cells to the requirement of UNC45A for cell proliferation that we observed in cancer cells (Fig. 1C).
UNC45A selectively regulates NEK7 expression
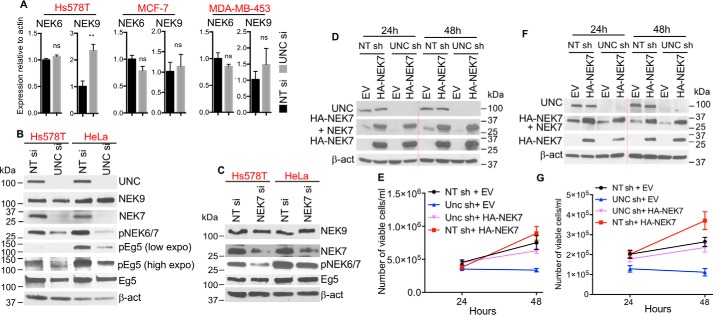
Although NEK7 is one of 11 NEK family members (29), our microarray data showed that only NEK7 expression is altered when UNC45A is silenced (Fig. 2A, Table S1). Loss of UNC45A did not significantly affect the transcription of other key components of the NEK signaling axis. This was validated by RT-qPCR showing that silencing UNC45A did not alter NEK6 mRNA levels in Hs578T, MCF-7, or MDA-MB-453 cells (Fig. 3A). Similarly, NEK9 patterns of expression were unchanged after UNC45A silencing in both MCF-7 and MDA-MB-453 cells. In Hs578T cells, however, loss of UNC45A significantly increased NEK9 mRNA expression, but it did not alter NEK9 protein expression (Fig. 3B). Similar results were obtained when HeLa cells were used (Fig. 3B).
Figure 3.
UNC45A selectively regulates NEK7 expression. A, RT-qPCR analysis of NEK6 and NEK9 in cells treated with 100 nm nontargeting siRNA (NT si) or UNC45A siRNA (UNC si) for 96 h (n = 3). B, Western blot analysis of lysates from Hs578T and HeLa cells treated for 96 h with nontargeting siRNA and UNC45A siRNA. NEK9, total NEK9; Eg5, total Eg5; pEg5, phosphor-EG5-S1033; low expo, low exposure of the film; high expo, high exposure of the film. C, Western blot analysis of lysates from Hs578T and HeLa cells treated 72 h with nontargeting siRNA and NEK siRNA (NEK7 si). β-Actin was used as a loading control. Images of B and C are representative of 2 independent experiments. D–G, expression of HA-mNEK7 rescues HeLa (D and E) and MDA-MB-231 (F and G) cell growth. Cells stably expressing doxycycline-inducible UNC45A shRNA (UNC sh) or nontargeting shRNA (NT sh) were treated for 48 h with doxycycline and then transfected with HA-mNEK7-expressing vector (HA-NEK7) or empty vector (EV). E and G, cells were counted at 24 and 48 h after transfection and used for Western blot analysis (D and F). Red dotted lines indicate a removal of a protein standard lane between 24- and 48-h samples on the same gel.
We next examined the impact of UNC45A loss on NEK7 signaling. We tested whether loss of UNC45A alters the phosphorylation/activation of the mitotic kinesin Eg5, an established downstream target of NEK6 and NEK7. Eg5 is a member of molecular motor proteins that play essential roles in mitosis through regulation of spindle assembly and function. Eg5 contributes to the establishment and balance of forces in the mitotic spindles and drives the sliding of microtubules (30, 31). NEK6/7 phosphorylation of Eg5 at Ser-1033 is essential for Eg5 accumulation at the centrosomes, which is necessary for centrosomal separation and normal mitotic spindle formation (27, 32).
As expected, UNC45A deletion drastically reduced NEK7 protein expression in Hs578T and HeLa cells (Fig. 3B). Accordingly, we also observed a drastic loss of phosphorylation as determined by the only available phosphospecific antibody recognizing active forms of both NEK7 and -6 (Fig. 3B). The loss of NEK7 translates into a significant decrease in phosphorylation of Eg5 at Ser-1033, previously shown to be the target site of NEK6/7 kinases (27). These findings suggest that UNC45A's function lies upstream of NEK7 and the signaling pathway controlling Eg5. Supporting this conclusion, knocking down NEK7 in Hs578T and HeLa cells did not affect UNC45A protein levels (Fig. S2A), and had no noticeable decrease in total Eg5 or NEK9 protein levels (Fig. 3C).
To test the hypothesis that NEK7 might play a key role in UNC45A function during cell proliferation, we assessed whether expression of heterologous NEK7 could rescue the growth of cancer cells lacking UNC45A. pcDNA3 vector expressing mouse HA-tagged NEK7 (HA-mNEK7) was transfected into HeLa and MDA-MB-23 cells harboring doxycycline-inducible UNC45A shRNA. We initially treated cells with doxycycline for 5 days to completely silence endogenous UNC45A expression before transfecting them with the heterologous HA-mNEK7 construct. As shown in Fig. S1, B–E), transient expression of HA-mNEK7 in HeLa and MDA-MB-231 cells treated with doxycycline resulted in a significant rescue of cell proliferative capacity. In fact, when heterologous HA-mNEK7 is introduced earlier (48 h), before complete loss of UNC45A and NEK7 has occurred, cell proliferation capacity is even further enhanced (Fig. 3, D–G). These results strongly suggest that NEK7 is a major mediator of the cancer cell proliferation defect we observed in UNC45A-silenced cells.
Loss of UNC45A generates similar phenotype as NEK7 deletion
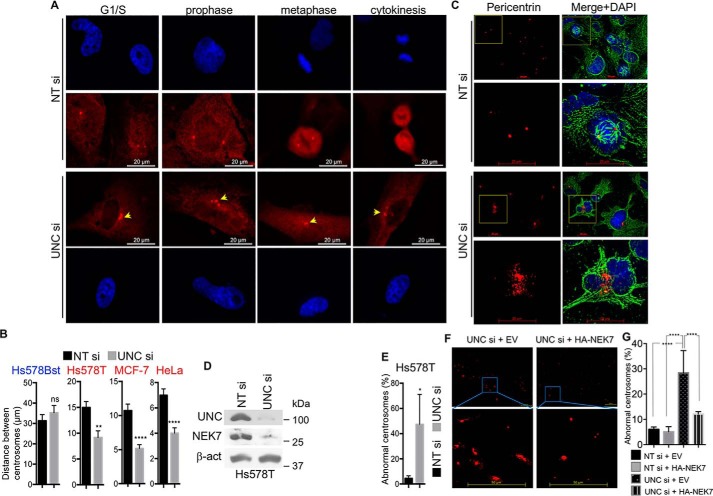
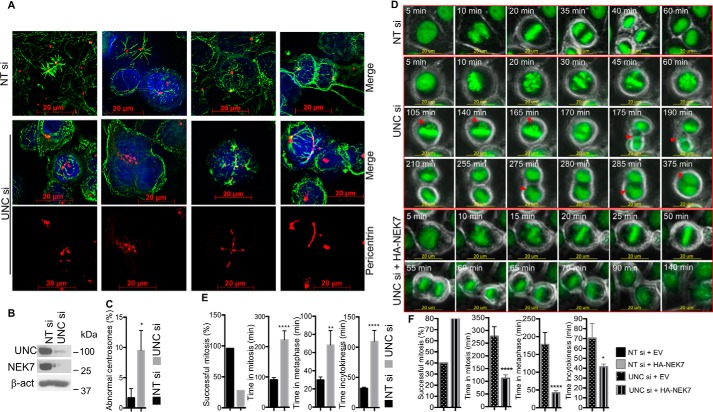
Because silencing UNC45A results in a significant decrease in NEK7 mRNA and protein levels selectively in cancer cells, as well as reduced phosphorylation of NEK7's downstream target Eg5 in cancer cells, we reasoned that lack of UNC45A might lead to centrosomal separation defects and mitotic abnormalities, two phenotypes previously associated with NEK7 deletion in various cell lines (22–27). To test this hypothesis, we used a pericentrin antibody and immunofluorescence microscopy to examine the number and pattern of centrosomes in cells lacking UNC45A expression. As illustrated in Fig. 4A (lower panels), loss of UNC45A expression results in centrosome amplification (cells with more than 2 centrosomes), as well as defects in centrosome separation in Hs578T cells compared with control cells (Fig. 4A, upper panels). The overall average distance between centrosomes is significantly smaller in Hs578T, MCF-7, and HeLa cells lacking UNC45A (Fig. 4B). In contrast, centrosome number and separation in the nontransformed Hs578Bst cells is not significantly affected (Fig. 4B), consistent with the hypothesis that “residual” NEK7 expression in UNC45A-silenced normal cells might be protective (Fig. 1C).
Figure 4.
Silencing UNC45A causes centrosomal abnormalities and generates polynucleated Hs578T cells. A, immunocytochemistry analysis of centrosomes using pericentrin antibody in Hs578T cells treated with UNC45A siRNA or nontargeting siRNA for 96 h. Images are representative of 3 independent experiments. B, the overall distance between centrosomes in the indicated unsynchronized cell lines. In each case, 300 cells were counted. The experiment was repeated three times. Error bars represent mean ± S.D. Unpaired two-tailed t test was used for significance. ****, p < 0.0001; **, p < 0.01. C, immunocytochemistry analysis of Hs578T cells treated with nontargeting siRNA (NT si) or UNC45A siRNA (UNC si) for 96 h using pericentrin (red) and α-tubulin (green) antibodies. DAPI (blue) was used to visualize nuclei. The bottom panels correspond to enlargement of the yellow squared areas. Scale bar represent 20 μm. D, Western blot analysis extracts from cells in C. Images are representative of 3 independent experiments. E, statistical analysis of abnormal centrosomes in NT si- and UNC si-treated Hs587T cells in C. From each sample, 40 cells were counted in triplicate. Experiments were repeated more than three times. F, immunocytochemistry analysis of Hs578T cells lacking UNC45A transfected with empty pcDNA3 or expressing HA-mNEK7. G, statistical analysis of cells (n = 20) lacking UNC45A and transfected with empty vector or pcDNA3 expressing HA-mNEK7 from F, in addition to Hs578T cell transfected with NT siRNA and Empty vector or HA-NEK7 vector (Fig. S3).
Further immunofluorescence analysis showed a strikingly disorganized pattern of pericentrosomal material in Hs578T cells lacking UNC45A compared with control cells (Fig. 4, C–E). The observed centrosomal abnormalities in UNC45A-deficient cancer cells correlated with loss of mitotic activity and an accumulation of polynucleated cells (Fig. 4C). As expected, silencing NEK7 in Hs578T (Fig. S2B) also induced accumulation of polynucleated cells (Fig. S2C, white arrowheads, and D) with micronuclei (Fig. S2C, red arrowheads). Further analysis showed that loss of NEK7 caused a drastic disorganization of centrosomal material in Hs578T cells (Fig. S2, E and F).
Importantly, the abnormal centrosome phenotype induced by loss of UNC45A was significantly rescued when exogenous HA-mNEK7 was overexpressed in Hs578T cells lacking UNC45A (Fig. 4, F and G, and Fig. S3). Together, these findings suggest that UNC45A is essential for normal centrosome homeostasis, and provide further support for the notion that these functions are mediated by NEK7, a known centrosome regulator, in cancer cells.
Similar results were obtained when HeLa cells were used. Indeed, loss of UNC45A causes drastic centrosomal abnormalities in HeLa cells leading to accumulation of multinucleated cells (Fig. 5, A–C). Additional insight into the specific role that UNC45A might play during cell division was obtained by time-lapse microscopy using HeLa cells expressing the fusion protein histone H2B-GFP (33). HeLa cells were transiently transfected with UNC45A siRNA (or nontargeting siRNA control), followed by time-lapse fluorescence microscopy after 72 h post-transfection (Movies S1 and S2). Cells treated with control siRNA progress to anaphase within 20–30 min of initiating metaphase, completing mitosis within 60–90 min (Movie S1, Fig. 5D, upper panel, and E, Fig. S4). In contrast, cells lacking UNC45A are delayed in mitosis and form multinucleated cells, which frequently undergo apoptosis (Movie S2, Fig. 5D, middle panels, and E, Fig. S4). During this aberrant, prolonged metaphase (∼120 min), chromosomes failed to align (Fig. 5D, middle panels, 30–165 min). In some cases, cells were unable to enter anaphase and instead retrogress to form an early metaphase-like state trying unsuccessfully to align their chromosomes. Ultimately, a few chromosomes remained lagging (Fig. 5D, middle panel, 105–165 min) and formed extra micronuclei (Fig. 5D, middle panel, 175–375 min). After cycles of failed attempts to partition chromosomes, these cells ultimately entered cytokinesis, where they stalled for ∼105 min. Most of them did not complete cytokinesis, forming multinucleated cells (see Movie S2, and images from 105 to 375 min in Fig. 5D, middle panel) before ultimately undergoing apoptosis. These observations are nearly identical to time-lapse results demonstrating significant mitotic abnormalities in cells lacking NEK7 (26), further supporting the hypothesis that the role of UNC45A during cancer cell division is mainly mediated by NEK7. This conclusion is strengthened by the fact that overexpression of HA-mNEK7 in H2B-GFP HeLa cells lacking UNC45A rescued cell proliferation and reduced cell death (Movies S3 and S4). Exogenous HA-mNEK7 significantly improved the rate of successful mitosis, reduced the time spent in mitosis as a whole, and reduced the time of metaphase and cytokinesis (Fig. 5D, lower panel, and F, and Fig. S5). Together, these results established that UNC45A is required for normal mitotic progression through regulation of NEK7 expression in cancer cells; specifically, silencing of UNC45A in cancer cells results in mitotic catastrophe.
Figure 5.
Time course analysis of HeLa cells lacking UNC45A. A, immunocytochemistry analysis of HeLa cells transfected with nontargeting siRNA (NT si) or UNC45A siRNA (UNC si) for 96 h using pericentrin (red) and α-tubulin (green) antibodies. B, Western blot analysis of cells from A. C, statistical analysis of cells (n = 20) from A. D, HeLa cells expressing the histone H2B-GFP fusion protein were transfected with UNC45A si or NT si for 72 h and analyzed by time-lapse microscopy for 16 h using overlay of green fluorescence and phase-contrast channels. Images were extracted from Movie S1 (upper panels) and Movie S2 (middle panels). Images represent prometaphase onwards. Images are representative of 2 independent experiments. Lower panels show images of H2B-GFP HeLa cells expressing HA-mNEK7 extracted from Movie S4. E, statistical analysis of 25 control and 18 UNC45A si-treated cells from Movies S1 and S2, respectively. F, statistical analysis of cells (n = 20) lacking UNC45A and transfected with empty vector or pcDNA3 expressing HA-mNEK7. Images are from Movies S3 and S4, respectively. Error bars represent mean ± S.E. One-way ANOVA or unpaired two-tailed t test were used for significance. ****, p < 0.0001; **, p < 0.01; *, p < 0.05.
UNC45A controls NEK7 transcription by activating GR transcriptional activity
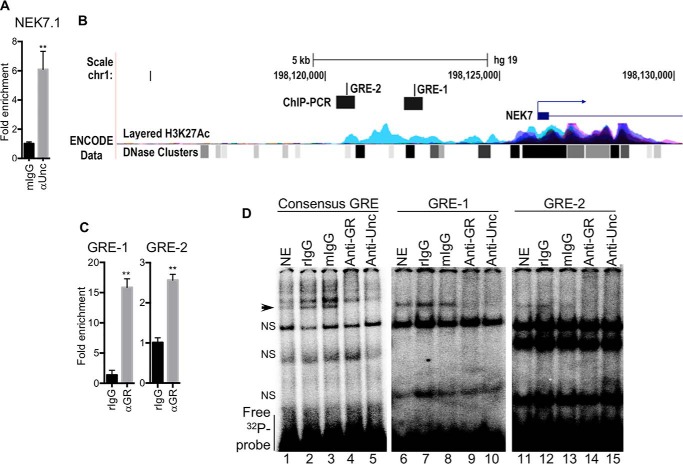
Next, we sought to determine how UNC45A regulates NEK7 expression. The observed loss of NEK7 mRNA upon UNC45A silencing (Fig. 2, A and D) suggested that UNC45A might mediate NEK7 gene transcription. This hypothesis was supported by ChIP data showing that UNC45A binds to the NEK7 promoter in Hs578T cells (Fig. 6A). UNC45A is not a transcription factor, but it is known to regulate the transcriptional activity of nuclear receptors such as PR, retinoic acid α, and peroxisome proliferator-activated receptor-α and -γ (15, 19). Thus, we investigated whether NEK7 transcription could be regulated by any of these. Computational analysis using Qiagen and the University of California Santa Cruz genome browser, Gateway, identified two novel glucocorticoid response elements (GREs), i.e. GR-binding consensus sequences, upstream of the NEK7 transcription start site (TSS) (Fig. 6B). We named these two putative GR-binding sites GRE-1 and GRE-2, located at positions 3094 and 5495 bp, respectively, upstream of the NEK7 TSS. The potential significance of these response elements is suggested by a positive correlation between NEK7 and GR (NR3C1) expression in the TCGA breast invasive carcinoma RNA-seq data set (Fig. S6A). The possibility that these two GREs might regulate NEK7 expression downstream of UNC45A is also supported by a clear nuclear co-localization of UNC45A and GR (Fig. S6B). This hypothesis was also tested by performing ChIP-qPCR experiments using a GR antibody and NEK7 promoter primers that included either GRE-1 or GRE-2 (Fig. 6C). Our results demonstrated that GR binds to both GREs in the NEK7 regulatory regions upstream of the TSS in Hs578T cells, but GR may have greater affinity for GRE-1 compared with GRE-2 (Fig. 6C, notice the scale). We further tested the functionality of these binding sites using an electrophoretic mobility shift assay (EMSA). As shown in Fig. 6D, 32P radiolabeled GRE-1 and GRE-2 both bind to GR from nuclear extracts (NE) of Hs587T cells and yield DNA:protein complex bands with the expected electrophoretic mobility (Fig. 6D, lanes 1, 6, and 11, arrowhead). This binding is sequence-specific because: 1) it appears when the GRE consensus sequence is used as a positive control (Fig. 6D, left panel); 2) it is absent when the mutated consensus sequence is used as a negative control (Fig. S6C, left panel); and 3) this band is competed off with cold GRE sequence probes (Fig. S6C, right panel). Furthermore, addition of a rabbit polyclonal antibody against GR (anti-GR) induced a complete supershift of GR:Consensus GRE (lane 4), GR:GRE-1 (lane 9), and GR:GRE-2 (lane 14) bands (Fig. 6D). However, a rabbit nonimmune antibody control (rIgG) did not affect the electrophoretic mobility of GR:GRE complexes (Fig. 6D, lanes 2, 7, and 12). Importantly, addition of anti-UNC45A antibody to the nuclear extract also caused a supershift of GR:GRE complexes (lanes 5, 10, and 15), but a mouse nonimmune antibody control (mIgG) had no effect on the electrophoretic mobility of GR:GRE complexes (lanes 3, 8, and 13). The band's total shift with individual specific antibodies suggests that UNC45A and GR simultaneously exist in complex with both GRE-1 and GRE-2, as well as with the consensus GRE sequence. Our results do suggest that the GR:UNC45A complex may have greater affinity for GRE-1 than for GRE-2 (Fig. 6C, notice the scale). In addition, silencing GR using siRNA caused a significant reduction in NEK7 mRNA (Fig. 7A) and protein levels (Fig. 7B), which is translated into a significant reduction in tumor cell proliferation (Fig. S6, D and E).
Figure 6.
UNC45A and glucocorticoid receptor bind to NEK7 gene promoter. A, ChIP RT-qPCR showing that UNC45A occupies the NEK7 gene promoter in Hs578T cells. The experiment was repeated twice. B, NEK7 promoter map generated based on information in UCSC Genome Browser, showing the localizations of GRE1 and GRE2. ChIP-PCR product 2 (Chr1: 198,120,583–198,120,765); GRE-2 (Chr: 198,120,604–198,120,619); ChIP-PCR product 1 (Chr1: 198,122,941–198,123,063); GRE-1 (Chr: 198,123,999–198,123,014); TSS (Chr1: 198,126,108). C, ChIP RT-qPCR showing that GR occupies NEK7 gene promoter in Hs578T cells. Two pairs of primers covering GRE-1 and GRE-2 upstream of the NEK7 TSS were used (n = 3). D, EMSA experiment showing GRE1 and GRE2 interact with GR in the nuclear extract of Hs578T cells. The GRE consensus sequence was used as a positive control. Arrowhead indicates the specific band of GR and UNC45A in complex with 32P-labeled GREs. αGR or αUNC caused this specific band to be supershifted, which indicates that UNC45A and GR simultaneously exist in complex with both of these GREs. NS indicates nonspecific band. Images are representative of 3 independent experiments.
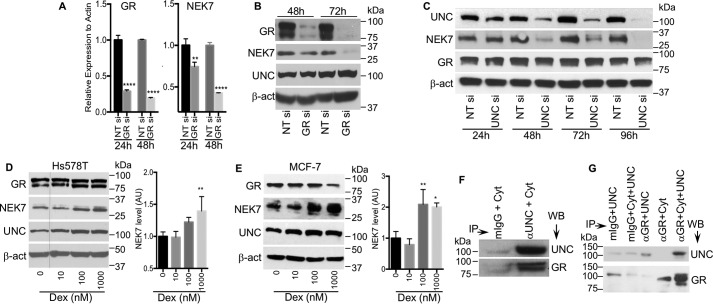
Figure 7.
The glucocorticoid receptor regulates NEK7 gene transcription. Silencing GR significantly reduces NEK7 mRNA (A) and protein (B) levels in a time-dependent manner. Images are representative of 2 independent experiments. C, time course analysis of UNC45A silencing with siRNA and its impact on NEK7 and GR protein levels. Images are representative of 2 independent experiments. D and E, Western blot analysis of cell extracts from Hs578T and MCF-7 cells treated with dexamethasone for 24 h. Red dotted line in D indicates that lane 0 nm Dex was moved from the right to the left of the same gel. Graph bars are densitometry analysis of Western blots images from 3 independent experiments. Error bars represent mean ± S.D. Data were tested for significance by one-way ANOVA or unpaired two-tailed t test. ****, p < 0.0001; **, p < 0.01. F, immunoprecipitation analysis showing that UNC45A and GR form endogenous complexes in Hs578T cells. G, UNC45A interacts physically with GR. GR was immunoprecipitated from T47D cell lysates using anti-GR bound to protein A-Sepharose. GR was then stripped from its associated proteins and incubated with 5 μg of purified UNC45A. Mouse Igs (mIgG) were used as a negative control.
We then verified whether loss of UNC45 affects GR protein levels (Fig. 7C). A time course experiment shows that, as expected, NEK7 protein levels are reduced over time with UNC45A knockdown, but GR protein levels remain unchanged (Fig. 7C). This was confirmed in a double knockdown experiment where GR and UNC45A are silenced individually or in combination (Fig. S6F). Loss of both UNC45A and GR causes a more efficient reduction of NEK7 protein level (Fig. S6F). Together these findings strongly suggest that, in cancer cells, UNC45A might function as a GR-positive regulator to promote NEK7 gene transcription, which correlates with changes in the expression of several known GR target genes identified in our microarray analysis (Fig. 2A and Fig. S6G). Additional experimental support for this hypothesis is provided by testing the impact of activating the transcriptional activity of GR using its agonist dexamethasone. As shown in Fig. 7, D and E, dexamethasone increased the NEK7 protein level in a concentration-dependent manner in Hs578T and MCF-7 cell lines. Finally, immunoprecipitation experiments using an UNC45A antibody showed that UNC45A is a component of the GR endogenous complex in Hs578T cells (Fig. 7F). Furthermore, purified UNC45A can physically interact with GR isolated from T47D cells (Fig. 7G).
NEK7 transcriptional regulation by UNC45A is consistent with UNC45A nuclear localization in breast tumor tissues and cellular models
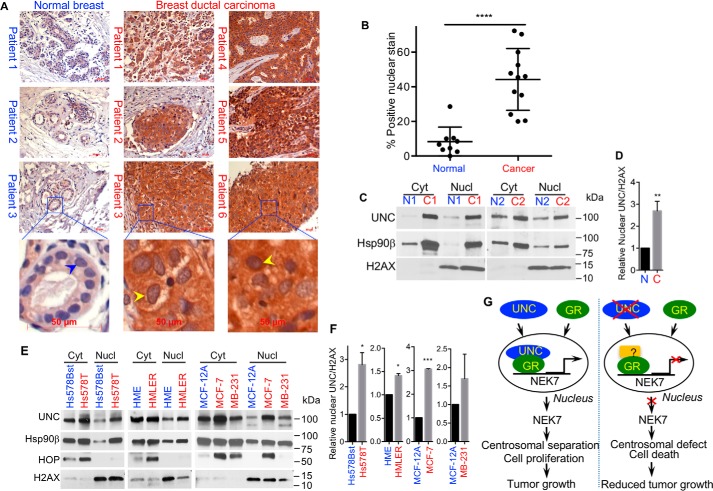
The question then becomes whether the subcellular localization of UNC45A is consistent with the nuclear GR transcriptional regulation in cancer cells. UNC45A overexpression has been previously shown to positively correlate with grade and stage in ovarian and breast cancers (17, 18). Immunohistochemistry analysis using a mAb against UNC45A confirmed that UNC45A is highly overexpressed in human breast-infiltrating ductal carcinomas compared with normal breast tissues (Fig. 8A). Furthermore, in breast ductal carcinoma cells, UNC45A also exhibits highly increased nuclear localization compared with normal breast cells (Fig. 8A). Statistical analysis showed that this increase in nuclear localization is highly significant (Fig. 8B). This observation was confirmed by Western blot analysis of cytoplasmic and nuclear fractions of normal and cancerous human breast tissues (Fig. 8, C and D). Similar distribution was also observed in vitro using breast cancer cell lines compared with nontransformed breast cells. Indeed, immunoblot analyses demonstrate that UNC45A is significantly more nuclear in cancer Hs578T, HMLER, MCF-7 and MDA-MB-231 cell lines compared with nontransformed Hs578Bst, HME, and MCF-10A, MCF-12A mammary epithelial cells (Fig. 8, E and F). Interestingly, Hsp90β nuclear localization correlates with that of UNC45A (Fig. 8E), which was also observed in breast cancer patients (Fig. 8C). In contrast, another co-chaperone protein, the Hsp70/Hsp90 organizing protein (HOP), is overexpressed in cancer cells, but remain characteristically cytoplasmic in all cell lines tested, except in MCF-7 (Fig. 8E). Together, these findings support the premise that UNC45A controls cancer cell proliferation and centrosomal separation by acting as a positive regulator of GR in the nucleus to promote NEK7 gene transcription.
Figure 8.
UNC45A becomes more nuclear in human breast cancer. A, immunohistochemical analysis of human breast tissue sections counterstained with hematoxylin. UNC45A is overexpressed in human breast-infiltrating ductal carcinomas (right panel) as compared with normal breast tissues (left panel). Yellow arrowheads indicate a significant increase in UNC45A nuclear localization in human breast-infiltrating ductal carcinomas as compared with normal breast tissues (blue arrowheads). B, quantification of UNC45A in the nucleus of normal versus cancer tissues used in A. C, UNC45A nuclear localization in normal versus cancer breast tissues (n = 10). Representative Western blot analysis of breast tissue cytoplasmic (Cyt) and nuclear (Nucl) fractions of two normal patients (N1 and N2) and cancer patients (C1 and C2) tissues. D, quantification of UNC45A in the nucleus of normal versus cancer cells in patient breast tissues relative to H2AX. E, Western blot analysis of cytoplasmic and nuclear fractions from normal (Hs578Bst, HME, and MCF-12A) and cancer (Hs578T, HMLER, MCF-7, and MDA-MB-231) breast cell lines stained for UNC45A (UNC), Hsp90β, and HOP. The histone H2AX was used as a nuclear marker. F, quantification of nuclear UNC45A in normal versus cancer in E relative to H2AX in the indicated breast cancer cell lines. Results are expressed as average of three experiments. G, model for UNC45A:GR regulation of NEK7 gene transcription, centrosomal separation, and cell proliferation.
Discussion
In this report, we describe a molecular mechanism underlying UNC45A regulation of cancer growth. UNC45A has been shown to be overexpressed in breast (18) and ovarian (17) cancers. Consistent with these reports, we show that UNC45A is selectively required for cancer cell proliferation in various breast cancer-derived cell lines in vitro, as well as for tumor growth in vivo using a NOD-SCID xenograft mouse model and the triple-negative breast cancer MDA-MB231 cell line. Furthermore, these data corroborate the results of recent studies in HeLa cells, showing that loss of UNC45A expression results in reduced cell proliferation in vitro and tumor growth in vivo (20). Together these findings consolidate UNC45A as an important driver of tumorigenesis in various malignant tumors including breast (18), ovarian (17), cervical (20), and neuroblastoma (19).
It was, therefore, somewhat surprising to observe that UNC45A expression is dispensable for normal epithelial cell proliferation (Fig. 1, A–D); yet this observation is consistent with studies showing that UNC45A silencing does not affect natural killer (NK) cell proliferation, but does regulate exocytosis of their lytic granules (34). UNC45A is also dispensable for neuron survival but required for neurite elongation by controlling NMII activation (35). Together, these studies suggest that during oncogenic transformation, UNC45A assumes novel function(s) that are essential for cancer cell proliferation and survival. Supporting this hypothesis, here we show that, like some other oncogenic proteins (e.g. Rel) (36), the cellular localization of UNC45A become more nuclear in malignant versus normal cells. In transformed epithelial cells, the UNC45A protein assumes a nuclear localization, whereas in normal epithelial cells and tissues, this protein mostly resides in the cytoplasm (Fig. 8). Interestingly, Hsp90β also shows a higher nuclear localization in breast cancer cells lines, which is consistent with a previous observation that UNC45A has higher affinity for Hsp90β over Hsp90α (16).
Although additional studies are needed to fully understand the putative nuclear function(s) of UNC45A in cancer cells, and to elucidate the role of the Hsp90β/Un45A interaction in these processes, our results further suggest that at least one selective function of nuclear UNC45A in cancer cells is regulating transcription of endogenous genes via nuclear receptors, such as GR. This is consistent with the known function of UNC45A as a positive factor for PR activity (15). Here we show that UNC45A is essential for GR-mediated transcription of the mitotic kinase gene, NEK7. Rescue experiments suggest that loss of NEK7 expression, as a consequence of UNC45A deletion, causes the observed reduction in cancer cell proliferation and survival. Specifically, we show that HeLa cells lacking UNC45A are stalled in metaphase and cytokinesis, form multinucleated cells, and ultimately undergo mitotic catastrophe, phenotypes identical to those displayed by HeLa cells either lacking NEK7 or expressing kinase-inactive NEK7 mutants (26, 28). Importantly, these phenotypes are significantly reversed by overexpression of exogenous HA-mNEK7 in Hs578T and HeLa cells lacking UNC45A (Figs. 4, F and G, and Fig. 5, D and F).
Although the contributions of the centrosome to the transformed phenotype are complex (37), both structure and function of this organelle are regulated by the NEK family of protein kinases, including control of pericentriolar material accumulation and centriole assembly, duplication, and separation (29, 38, 39). Here we show that UNC45A regulates centrosome structure/function via NEK7 in multiple ways, which is consistent with a previous report showing that UNC45A is a component of the centrosome complex and regulates ChK1 recruitment into the centrosome (20). In this study, we demonstrate that UNC45A also functions as a key regulator of NEK7 expression, which itself is a key regulator of centrosome function. Interestingly, NEK7 has been shown to accelerate the microtubule dynamic instability (40), which is consistent with the newly described role for UNC45A as mitotic spindle-associated protein that destabilizes microtubules (21). UNC45A also plays a direct role in the later phases of mitosis; specifically, it co-localizes with NMII in the cleavage furrow during cytokinesis (17).
Interestingly, the effect of UNC45A on NEK7 transcription is apparently selective for NEK7, even though NEK7 and NEK6 share a high degree of sequence identity (29). The notion that these two proteins share overlapping (41) but distinct regulatory functions is supported by the observations that NEK7 and NEK6 exhibit distinct tissue-specific patterns of localization during embryogenesis (42), their activities are differentially regulated by serum deprivation (43), and they do not substitute for each other functionally in cell (26, 27) or animal models (25). Our results provide further support for the hypothesis that these two kinases have distinct regulatory functions; indeed, NEK6 transcription is independent of UNC45A (Fig. 3A), whereas NEK7 requires UNC45A for transcriptional activation of GR (Fig. 7).
In conclusion, Fig. 8G depicts a simplified model integrating our observations from UNC45A-deficient cells, demonstrating UNC45A's ability to selectively regulate NEK7 transcription in cancer cells, resulting in selective promotion of cancer cell survival and proliferation. This selective function of NEK7 in cancer cells involves differential subcellular localization of NEK7 in normal versus malignant epithelial cells. This difference in localization results in profound functional differences for UN45A in normal versus malignant cells. In malignant cells, UNC45A has the capacity to regulate NEK7 expression indirectly, via transcriptional regulation of GR transcription. This novel mechanism of NEK7 regulation by UNC45A in cancer cells is just one example of an important gene that becomes differentially expressed when UNC45A is silenced, as determined by microarray analysis. Future studies will determine the contribution of other UNC45A-regulated pathways (Fig. 2A) and to determine the mechanism by which UNC45A selectively localizes to the nucleus in cancer cells. Our findings reveal a new dimension of complexity for the role of co-chaperone proteins in the regulation of cancer cell proliferation, and suggest that molecular targets that transcend cancer types without being required for normal cell proliferation not only exist, but may be attractive targets for therapeutic intervention in diseases such as cancer. This concept is supported by recent findings showing a selective cooperation between Hsp72 and NEK6 to regulate centrosome clustering in cancer cells (44). Yet our study suggests a potential therapeutic advantage in targeting UNC45A to block cancer cell proliferation while preserving NEK7 expression and function (cell proliferation) in normal cells.
Experimental procedures
The primers used were: NEK7.1.F, GCCTTACGACCGGATATGGG; NEK7.1.R, CACTAAATTGTCCGCGACCAA; NEK6.F, CATCCCAACACGCTGTCTTTT; NEK6.R, TACACCTCGCTGAACTGTCCT; NEK9.F, TACGAGCGACACTGCGATTC; NEK9.R, ACGCGGATGGGGATGTAGT; GR.F, ACAGCATCCCTTTCTCAACAG; GR.R, AGATCCTTGGCACCTATTCCAAT; Actin.F, TCCCTGGAGAAGAGCTACGA; Actin.R, AGCACTGTGTTGGCGTACAG; Eg5.F, TCCCTTGGCTGGTATAATTCCA; Eg5.R, GTTACGGGGATCATCAAACATCT; UNC45A.F, GCAAAAGGCTTCTCAGAACCTG; and UNC45A.R, AACGTTGCAAGAGCTGAACC.
Antibodies
Rabbit polyclonal antibody against GR was obtained from Dr. Michael Garabedian (New York University School of Medicine, New York). Monoclonal anti-rat GR (BuGR2, ab2768, Abcam); Eg5 (BD Biosciences, 611187); phosphor-EG5-S1033 (45); phopho-NEK6/7 (41); NEK7 (Cell Signaling, 3057S); NEK9 (46); HA tag (C29F4, rabbit mAb number 3724S, Cell signaling); β-actin (Santa Cruz, sc-47778); Hop (F5); Hsp90β (H90.10); anti-pericentrin (ab4448) and anti-α-tubulin for immunofluorescence were obtained as indicated. Monoclonal UNC45A antibody (ADI-SRA-1800-F, Enzo) was used for Western blotting and ChIP experiments. Rabbit polyclonal anti-UNC45A provided by Dr. Kathryn B. Horwitz (University of Colorado, Denver, CO) was used for immunofluorescence. Histone H2AX antibody was from GenScript (Ab-139).
Cell lines
HeLa, MDA-MB-231, MDA-MB-453, Hs578T, Hs578Bst, HME, T47D MCF7, ZR-75–1, MCF12A, MCF10A, RWPE1, LNCaP, DU145, and PC3 cell lines were obtained from ATCC within the last 5 years. Large stocks were made and stored in liquid nitrogen. For the cells that are less frequently used, we are still using cells from the original stocks. H2B-GFP HeLa cells were provided by Dr. Geoffrey M. Wahl (The Salk Institute for Biological Studies, La Jolla, CA). DU145 and PC3 cell lines were provided by Dr. Balakrishna Lokeshwar (Augusta University). MCF10A and MCF12A were provided by Dr. Muthusamy Thangaraju (Augusta University). All cell lines were authenticated twice by morphologic and isoenzyme analysis during the study period. All cell lines are regularly tested at least quarterly for mycoplasma using the Genetica Inc. mycoplasma test and MycoAlert PLUS Mycoplasma Detection Kits. The latest mycoplasma testing was performed in May 2018. Cancer cells were discarded after 10 to 12 passages. Normal cells were discarded after 4–6 passages.
Silencing UNC45A, NEK7, and GR
Normal and cancer cell lines were transfected with 75–100 nm concentration of siRNA against UNC45A (15). Nontargeting siRNA#1 (Dharmacon/GE) was used as a negative control. Cells were harvested 96 h after transient transfection. Stable transfection of cancer cells with UNC45A shRNA or nontargeting shRNA control were performed using lentivirus system (pHR-H1-EF-tetO-Puro). pHR-H1-EF-tetO-Puro with nontargeting shRNA was used as control. UNC45A depletion was achieved by exposing cells stably harboring UNC45A shRNA to 2 μg/ml of doxycycline for 96–144 h. The following siRNA sequence was used for GR: sense sequence, GCAUGUACGACCAAUGUAATT; antisense sequence, UUACAUUGGUCGUACAUGCAG. The following siRNA sequence was used to silence NEK7: sense sequence, CCAGAAUGAUCAAGCAUUUTT; antisense sequence, AAAUGCUUGAUCAUUCUGGAT.
Nuclear fractionation
Hs578Bst, Hs578T, HME, HMLER, MCF-12A, MCF-7, MDA-MB-231, RWPE-1, and LNCaP cells were grown on 100 × 20-mm tissue culture dishes (Corning number 353003) to 70–80% confluence. Cells were harvested and washed in cold PBS, and spun down at 2000 rpm for 10 min at 4 °C. Nuclear extraction was carried out following EpiQuickTM Nuclear Extraction Kit standard protocol (EPIGENTEK number OP-00021). Both cytoplasmic and nuclear extract fractions were collected, assayed for protein concentration following the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, number 23225) standard protocol, and stored in −80 °C for further Western blotting analyses.
Immunoprecipitation
Cell extracts (300 μg of protein) from Hs578T or T47D cells were incubated with homemade UNC45A antibody or the anti-GR antibody (BuGR2, Abcam ab2768) overnight 4 °C with gentle rotation. The next day, a 30-μl slurry of protein A/protein G-agarose beads (Pierce® Recombinant Protein A-agarose, catalogue number 20366 and Pierce® Protein G-agarose, catalogue number 20399) was added to each sample, and the samples were incubated at 4 °C with gentle rotation for 2 h. The samples were then washed three times with 20 mm Tris, pH 7.4, 150 mm KCl, 10 mm 1-thioglycerol, 0.03% Nonidet P-40, 10% glycerol, protease inhibitors (Roche Applied Science), 10 mm NaF, 2 mm sodium pyrophosphate, 2 mm β-glycerophosphate, and 1 mm sodium orthovanadate. To strip GR from its endogenous protein complexes, immunoprecipitated GR was incubated with the above buffer supplemented with 300 mm KCl for 30 min at 4 °C. The resin was then washed 3 times before incubation with purified UNC45A for 30 min at 30 °C. Protein complexes were then washed and prepared for Western blotting.
Immunofluorescence microscopy
Cells were grown in 24-well plates (Corning number 3337) on microcover glasses (Electron Microscopy Sciences) to about 50% confluence in MEM, 1× (Cellgro number 10-010-CV) medium supplemented with 10% fetal bovine serum. Hs578T or HeLa cells were treated with 100 nm control siRNA or siRNA against UNC45A for 96 h. For rescue experiments, Hs578T cells were treated with 100 nm control siRNA or UNC45A siRNA for 48 h, transfected with empty pcDNA3 or pcDNA3 expressing HA-mNEK7, and then collected 48 h post-transfection. Cells were washed with microtubule stabilizing buffer (3 mm EGTA, 50 mm Pipes buffer, 1 mm MgSO4, 25 mm KCl) and fixed in methanol for 10 min. Cells were then allowed to dry, were blocked with 10% goat serum with 5% glycerol for 1 h, and stored at 4 °C. Primary and secondary antibodies were prepared in the blocking buffer. Cells were visualized using Zeiss Imager 1.1 and Keyence BZ-X710 microscopes.
Immunohistochemistry and tissue analysis
Commercial tissue microarrays (TMAs) and patient tissues obtained from Georgia Cancer Center Biorepository (Augusta University) were stained with anti-UNC45A (1/100) antibodies with chromogenic signal amplification (DAB) and hematoxylin counterstain following standard immunohistochemistry protocols. Slides were incubated for 30 min at 37 °C for UNC45A. Representative regions of interest (ROIs) were selected from TMA for semi-quantitative analysis of DAB percent staining using inform 2.4 (PerkinElmer Life Sciences) software as follows. 1) Optical density conversion from RGB images depicting a white area as a background; 2) reference library built from images expressing only DAB or hematoxylin; 3) algorithm (built in inForm) based unmixing of colors with pseudocolor formation and normalization; 4) segmentation of nuclei based on hematoxylin pseudocolor using a training algorithm; 5) scoring of expression based on intensity (unit: normalized counts) correlating with visually identified threshold (supervised by research pathologist); and 6) few cases were removed from final analysis due to inadequate image quality or absence of representative tissue. Expression of UNC45A was quantified after color un-mixing and nuclear/cytoplasmic segmentation was performed using inForm 2.4 based. Unpaired t test was performed to compare the means of different groups. For breast tissues, we used TMA T8235714D-5 (BioChain). UNC45A expression was evaluated in 12 breast cancer cases (12 ROIs; 85 cells/ROI) and compared with 9 normal breast cases (9 ROIs; 64 cells/ROI). Percentage of cells expressing nuclear UNC45A in both normal and cancer cases is represented. Percentage of cells expressing UNC45A in both normal and cancer cases is represented.
Cell proliferation assay
To monitor proliferation, cells were grown to 10–20% confluence on 96-well tissue culture plates (Corning number 3599) followed by transient transfection with 75 nm UNC45 siRNA or nontargeting siRNA control. Cell proliferation was measured using The CellTiter 96® AQueous One Solution Cell Proliferation Assay reagent (Promega number G3580).
Human tumor cell implantation
Six-week-old female NOD-SCID immunocompromised mice were purchased from The Jackson Laboratory. MDA-MB-231 triple negative human cells (105) harboring nontargeting shRNA or UNC45A shRNA were implanted into the fat pads of animals. Two days later, mice were fed doxycycline-containing food throughout the time of the experiment. Six animals per condition were used. All mouse procedures were conducted in accordance with the University Committee on the Use and Care of Animals at Augusta University.
RNA extraction and real-time RT-qPCR
Total RNA was extracted using QIAzol lysis reagent (Qiagen). The RNA extracts were analyzed by a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA quality was determined by the ratios of A260/A280 (close to 2) and A260/A230 (close to 2). RNA (1–5 μg) was used for making cDNA using RNA to cDNA EcoDryTM Premix (Double Primed) (Clontech Laboratories, Inc., a Takara Bio Company). cDNA was analyzed in triplicate using iQTM SYBR Green Supermix (Bio-Rad) in a Bio-Rad CFX96 system. Eurofins genomics primers were used for the following genes: UNC45A, NEK7, NEK6, NEK9, Eg5, and GR. mRNA expression was normalized against that of β-actin (internal control) (ΔCt = Ct (target gene) − Ct (internal control gene)). The relative fold-change was measured by the 2−ΔΔCt formula compared with the control cells. Means and differences of the means with 95% confidence intervals were obtained using GraphPad Prism (GraphPad Software Inc.). Two-tailed Student's t test was used for unpaired analysis comparing average expression between conditions. p values <0.05 were considered statistically significant.
Gene expression analysis
RNA extracts were first analyzed by a Nanodrop 2000 spectrophotometer. RNA quality was determined as described above. Qualified RNAs were further tested using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and samples with RIN >7 were selected for microarray analysis using the Affymetrix MTA 1.0 (Affymetrix). The labeling, hybridization, scanning, and data extraction of the microarray analysis were performed according to the recommended Affymetrix protocols. Briefly, the fluorescence signals of the microarray were scanned and saved as DAT image files. The AGCC software (Affymetrix GeneChip Command Console) transformed DAT files into CEL files to change image signals into digital signals, which recorded the fluorescence density of probes. Next, we used Affymetrix Expression Console software to pretreat CEL files through the Robust Multichip Analysis algorithm, including background correction, probeset signal integration, and quantile normalization. After pretreatment, the obtained chp files were analyzed by Affymetrix Transcriptome Analysis Console software to detect differentially expressed genes. The microarray data have been submitted to NCBI's Gene Expression Omnibus (GEO) (accession number GSE108258).
ChIP RT-qPCR assay
Chromatin immunoprecipitation (ChIP) assay was carried out according to protocols from the EMD Millipore ChIP Assay Kit (catalogue number 17-295). Sheared chromatin was immunoprecipitated using homemade GR and UNC45A (ADI-SRA-1800-F, Enzo) antibodies and a slurry of Protein A/Protein G-agarose beads (Pierce® Recombinant Protein A-agarose, number 20366, and Pierce® Protein G-agarose, number 20399). Mouse and rabbit nonimmune Igs (mIgG and rIgG) were used as controls. The protein:DNA complexes were eluted from beads, and the cross-linking was reversed. The DNA was purified from the eluted solution using Thermo Scientific GeneJet Genomic DNA purification kit (catalogue number K0721) and used for RT-qPCR with forward primer 5′-TTCATGAAGCCAAACAAGCA-3′ and reverse primer 5′-CAGAAAAACCTGGCCAAAGA-3′ for GRE-1 and forward primer 5′-TGTTTGCCTTCTACCCCTTG-3′ and reverse primer 5′-TTGATAACCTGTGGGGGAAA-3′ for GRE-2. For UNC45A, forward primer (Promo.NEK7.1) 5′-CATACTGCCTCCTAGCCGTCTC-3′ and reverse primer 5′-GTAAACTCCCTGGTGTGGTACG-3′ were used. β-Actin was used as control.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from Hs578T tumor cells as previously described (47). The following double-stranded oligonucleotides (Santa Cruz Biotechnology, sc-2545) were used as probes in the gel shift assay: GRE probe corresponds to the consensus binding site for glucocorticoid receptor 5′-GACCCTAGAGGATCTGTACAGGATGTTCTAGAT-3′ and reverse complement, 3′-CTGGGATCTCCTAGACATGTCCTACAAGATCTA-5′; GRE NEK7 probe 1 (GRE 1), which is 3094 upstream of the transcription starting site: sense, 5′-CTAGATTCTGTTCTAAAGAT-3′, and antisense, 5′-ATCTTTAGAACAGAATCTAG-3′; and GRE NEK7 probe 2 (GRE 2) is located 5495 upstream of the transcription starting site: sense, 5′-CTTGAAAGAACAGATTATCCCATAAG-3′, antisense, 5′-CTTATGGGATAATCTGTTCTTTCAAG-3′. The probes (50 ng) were end-labeled with [γ-32P]ATP using T4 DNA polynucleotide kinase (Promega) and purified with Micro G-25 spin columns (Santa Cruz). The end-labeled probes were incubated with nuclear extracts (25 μg) in protein-DNA binding buffer (10 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm DTT, 50 mm NaCl, 4% glycerol, and 0.05 mg/ml of poly(dI-dC)) for 20 min at room temperature. For specificity controls, the GR mutant oligonucleotide (catalogue number sc-2546, Santa Cruz) identical to (sc-2545) with the exception of two “GT” → “CA” substitutions in the GR binding motif was used: 5′-GACCCTAGAGGATCTCAACAGGATCATCTAGAT-3′; 3′-CTGGGATCTCCTAGAGTTGTCCTAGTAGATCTA-5′. We also used either ×100 cold GR-WT or cold GRE-1 to compete the radioactive probes. For the supershift assay, 1 μl of homemade anti-GR, anti-UNC45A, mIgG, and rIgG were preincubated with nuclear extracts for 14–16 h at 4 °C prior to addition of the labeled probe, and the same procedure as described above was followed. DNA:protein complexes were separated by electrophoresis in 6% polyacrylamide gels in 45 mm Tris borate, 1 mm EDTA, pH 8.3. The gels were dried and exposed to a phosphorimaging screen (Molecular Dynamics) for 2 days; the resulting images were acquired using a Personal Molecular Imager (Bio-Rad).
Dexamethasone treatment experiments
MCF-7 and Hs578T cells were grown to 50–70% confluence in minimal essential medium (MEM) supplemented with regular 10% fetal bovine serum. Medium was switched for MEM supplemented with 10% charcoal:dextran-stripped fetal bovine serum (catalog number 100–119, GEMINI) for 24 h. Cells were treated for 24 h with increasing concentrations of dexamethasone (10, 100, and 1000 nm). Cells were then harvested following the 24-h treatment and stored at −80 °C for further Western blot analysis. B-Hs578T cells were seeded at 30–50% confluence in MEM supplemented with regular 10% fetal bovine serum. The next day, cells were treated with 100 nm control siRNA or siRNA against UNC45A. 24 h later, the medium was switched for MEM supplemented with 10% charcoal:dextran-stripped fetal bovine serum (catalog number 100–119, GEMINI). 72 h post-transfection, cells were treated for an additional 24 h with dexamethasone (1000 nm) before collecting and storing them at −80 °C for further Western blot analysis.
NEK7 overexpression cell growth rescue experiment
MDA-MB-231 or HeLa cells harboring either doxycycline-inducible UNC45A shRNA or nontargeting shRNA control were gown in 12-well plates (Falcon, number 351143). Cells were treated with 2 μg/ml of doxycycline for 120 h, and then transfected with either pcDNA3 vector expressing mouse HA-tagged NEK7 (HA-mNEK7) or pcDNA3 empty vector as control following the LipofectamineTM 3000 Transfection Reagent (Thermo Fisher Scientific, number L3000008) standard protocol. Cells were collected 48 h post-transfection, counted using HyCloneTM Trypan Blue Solution, 0.4% phosphate-buffered saline exclusion method (GE Healthcare Life Sciences, number SV30084.01), and then pelleted for further Western blot analysis.
To test the impact of overexpressing NEK7 at early time on cell proliferation, cells were treated with 2 μg/ml of doxycycline for 48 h, and then the cells were transfected with either pcDNA3 vector expressing mouse HA-mNEK7 or pcDNA3 empty vector as indicated above. Cells were collected 24 and 48 h post-transfection, counted, and then pelleted for further Western blot analysis.
Statistics
All statistical analyses were performed using GraphPad Prism 6. Data were expressed as the mean ± S.D. as indicated. Data were analyzed by unpaired two-tailed Student's t test or one-way ANOVA, as indicated. p values of statistical significance are represented as: ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; and *, p < 0.05.
Data availability
Data presented in this manuscript have been deposited in NCBI's Gene expression Omnibus under the GEO Series accession code GSE108258. The TCGA data referenced in this paper are based upon the data generated by the TCGA Research Network and are available in a public repository from the cBioportal for Cancer Genomics website (http://www.cbioportal.org/) (48, 49).4 All the other data are available within the article and the supplementary files.
Author contributions
N. H. E., Y. J., K. K., P. R., S. L., O. B., H. A. S., C. A. P., A. S., H. K., and A. C. data curation; N. H. E., H. A. S., N. M. E., L. A. E., M. M. E.-S., J. R., and A. C. formal analysis; N. H. E., H. S., and A. C. validation; N. H. E. and A. C. investigation; N. H. E., N. M. E., L. A. E., M. M. E.-S., N. M., J. R., H. K., J. K. C., and A. C. writing-review and editing; Y. J., K. K., S. L., H. A. S., and A. C. methodology; H. S., K. L., A. H., R. B., N. M., J. R., H. K., J. K. C., and A. C. resources; A. C. conceptualization; A. C. software; A. C. supervision; A. C. funding acquisition; A. C. visualization; A. C. writing-original draft; A. C. project administration.
Supplementary Material
Acknowledgments
We thank the Georgia Cancer Center core facilities, especially the integrated genomics core, and Kimya Johns, from the Pathology Department, for excellent technical assistance in histology. We thank Dr. Lisa Middleton for the thorough editing of the manuscript. We thank Dr. Robert Weinberg for providing HMLER cells. We also thank Dr. Geoffrey M. Wahl (The Salk Institute for Biological Studies, La Jolla, CA) for providing H2B-GFP HeLa cells and Dr. Muthusamy Thangaraju (Augusta University) for sharing MCF10A and MCF12A cells.
This work was supported by National Institutes of Health Grant R01 GM102443-01 and Augusta University Startup Funds (to A. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Movies S1–S4, Table S1, and Figs. S1–S6.
Data presented in this manuscript have been deposited in NCBI's Gene expression Omnibus under GEO Series accession code GSE108258.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- UNC45
- uncoordinated 45/Cro1/She4p
- Hsp
- heat shock protein
- NMII
- nonmuscle myosin II
- ChK1
- checkpoint kinase 1
- PR
- progesterone
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element
- TSS
- transcription start site
- qPCR
- quantitative PCR
- NE
- nuclear extract
- NEK7
- NIMA-related kinase 7
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NOD-SCID
- non-obese diabetic/severe combined immunodeficiency
- shRNA
- short hairpin RNA
- HA
- hemagglutinin
- TMA
- tissue microarray
- DAB
- diaminobenzidine
- ROI
- regions of interest
- MEM
- minimal essential medium
- ANOVA
- analysis of variance
- HOP
- Hsp70/Hsp90 organizing protein
- GFP
- green fluorescent protein.
References
- 1. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price M. G., Landsverk M. L., Barral J. M., and Epstein H. F. (2002) Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 115, 4013–4023 10.1242/jcs.00108 [DOI] [PubMed] [Google Scholar]
- 3. Lee C. F., Hauenstein A. V., Fleming J. K., Gasper W. C., Engelke V., Sankaran B., Bernstein S. I., and Huxford T. (2011) X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster. Structure 19, 397–408 10.1016/j.str.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi H., and Blobel G. (2010) UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc. Natl. Acad. Sci. U.S.A. 107, 21382–21387 10.1073/pnas.1013038107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barral J. M., Hutagalung A. H., Brinker A., Hartl F. U., and Epstein H. F. (2002) Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295, 669–671 10.1126/science.1066648 [DOI] [PubMed] [Google Scholar]
- 6. Landsverk M. L., Li S., Hutagalung A. H., Najafov A., Hoppe T., Barral J. M., and Epstein H. F. (2007) The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J. Cell Biol. 177, 205–210 10.1083/jcb.200607084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Etard C., Behra M., Fischer N., Hutcheson D., Geisler R., and Strähle U. (2007) The UCS factor Steif/UNC-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev. Biol. 308, 133–143 10.1016/j.ydbio.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 8. Wohlgemuth S. L., Crawford B. D., and Pilgrim D. B. (2007) The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev. Biol. 303, 483–492 10.1016/j.ydbio.2006.11.027 [DOI] [PubMed] [Google Scholar]
- 9. Gazda L., Pokrzywa W., Hellerschmied D., Löwe T., Forné I., Mueller-Planitz F., Hoppe T., and Clausen T. (2013) The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans. Cell 152, 183–195 10.1016/j.cell.2012.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ni W., Hutagalung A. H., Li S., and Epstein H. F. (2011) The myosin-binding UCS domain but not the Hsp90-binding TPR domain of the UNC-45 chaperone is essential for function in Caenorhabditis elegans. J. Cell Sci. 124, 3164–3173 10.1242/jcs.087320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janiesch P. C., Kim J., Mouysset J., Barikbin R., Lochmüller H., Cassata G., Krause S., and Hoppe T. (2007) The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 9, 379–390 10.1038/ncb1554 [DOI] [PubMed] [Google Scholar]
- 12. Walker M. G. (2001) Pharmaceutical target identification by gene expression analysis. Mini. Rev. Med. Chem. 1, 197–205 10.2174/1389557013407034 [DOI] [PubMed] [Google Scholar]
- 13. Comyn S. A., and Pilgrim D. (2012) Lack of developmental redundancy between UNC45 proteins in zebrafish muscle development. PLoS ONE 7, e48861 10.1371/journal.pone.0048861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lehtimäki J. I., Fenix A. M., Kotila T. M., Balistreri G., Paavolainen L., Varjosalo M., Burnette D. T., and Lappalainen P. (2017) UNC-45a promotes myosin folding and stress fiber assembly. J. Cell Biol. 216, 4053–4072 10.1083/jcb.201703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chadli A., Graham J. D., Abel M. G., Jackson T. A., Gordon D. F., Wood W. M., Felts S. J., Horwitz K. B., and Toft D. (2006) GCUNC-45 is a novel regulator for the progesterone receptor/hsp90 chaperoning pathway. Mol. Cell. Biol. 26, 1722–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chadli A., Felts S. J., and Toft D. O. (2008) GCUNC45 is the first Hsp90 co-chaperone to show α/β isoform specificity. J. Biol. Chem. 283, 9509–9512 10.1074/jbc.C800017200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bazzaro M., Santillan A., Lin Z., Tang T., Lee M. K., Bristow R. E., Shih IeM., and Roden R. B. (2007) Myosin II co-chaperone general cell UNC-45 overexpression is associated with ovarian cancer, rapid proliferation, and motility. Am. J. Pathol. 171, 1640–1649 10.2353/ajpath.2007.070325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo W., Chen D., Fan Z., and Epstein H. F. (2011) Differential turnover of myosin chaperone UNC-45A isoforms increases in metastatic human breast cancer. J. Mol. Biol. 412, 365–378 10.1016/j.jmb.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 19. Epping M. T., Meijer L. A., Bos J. L., and Bernards R. (2009) UNC45A confers resistance to histone deacetylase inhibitors and retinoic acid. Mol. Cancer Res. 7, 1861–1870 10.1158/1541-7786.MCR-09-0187 [DOI] [PubMed] [Google Scholar]
- 20. Jilani Y., Lu S., Lei H., Karnitz L. M., and Chadli A. (2015) UNC45A localizes to centrosomes and regulates cancer cell proliferation through ChK1 activation. Cancer Lett. 357, 114–120 10.1016/j.canlet.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mooneyham A., Iizuka Y., Yang Q., Coombes C., McClellan M., Shridhar V., Emmings E., Shetty M., Chen L., Ai T., Meints J., Lee M. K., Gardner M., and Bazzaro M. (2019) UNC-45A is a novel microtubule-associated protein and regulator of paclitaxel sensitivity in ovarian cancer cells. Mol. Cancer Res. 17, 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yissachar N., Salem H., Tennenbaum T., and Motro B. (2006) Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 580, 6489–6495 10.1016/j.febslet.2006.10.069 [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Lee K., and Rhee K. (2007) NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 360, 56–62 10.1016/j.bbrc.2007.05.206 [DOI] [PubMed] [Google Scholar]
- 24. Kim S., Kim S., Kim S., and Rhee K. (2011) NEK7 is essential for centriole duplication and centrosomal accumulation of pericentriolar material proteins in interphase cells. J. Cell Sci. 124, 3760–3770 10.1242/jcs.078089 [DOI] [PubMed] [Google Scholar]
- 25. Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., and Motro B. (2010) Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene 29, 4046–4057 10.1038/onc.2010.162 [DOI] [PubMed] [Google Scholar]
- 26. O'Regan L., and Fry A. M. (2009) The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29, 3975–3990 10.1128/MCB.01867-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bertran M. T., Sdelci S., Regué L., Avruch J., Caelles C., and Roig J. (2011) Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30, 2634–2647 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cullati S. N., Kabeche L., Kettenbach A. N., and Gerber S. A. (2017) A bifurcated signaling cascade of NIMA-related kinases controls distinct kinesins in anaphase. J. Cell Biol. 216, 2339–2354 10.1083/jcb.201512055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fry A. M., O'Regan L., Sabir S. R., and Bayliss R. (2012) Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125, 4423–4433 10.1242/jcs.111195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., and Schmidt C. F. (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- 31. Ferenz N. P., Gable A., and Wadsworth P. (2010) Mitotic functions of kinesin-5. Semin. Cell Dev. Biol. 21, 255–259 10.1016/j.semcdb.2010.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sdelci S., Bertran M. T., and Roig J. (2011) Nek9, Nek6, Nek7 and the separation of centrosomes. Cell Cycle 10, 3816–3817 10.4161/cc.10.22.18226 [DOI] [PubMed] [Google Scholar]
- 33. Kanda T., Sullivan K. F., and Wahl G. M. (1998) Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- 34. Iizuka Y., Cichocki F., Sieben A., Sforza F., Karim R., Coughlin K., Isaksson Vogel R., Gavioli R., McCullar V., Lenvik T., Lee M., Miller J., and Bazzaro M. (2015) UNC-45A is a nonmuscle myosin IIA chaperone required for NK cell cytotoxicity via control of lytic granule secretion. J. Immunol. 195, 4760–4770 10.4049/jimmunol.1500979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iizuka Y., Mooneyham A., Sieben A., Chen K., Maile M., Hellweg R., Schütz F., Teckle K., Starr T., Thayanithy V., Vogel R. I., Lou E., Lee M. K., and Bazzaro M. (2017) UNC-45A is required for neurite extension via controlling NMII activation. Mol. Biol. Cell 28, 1337–1346 10.1091/mbc.e16-06-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilmore T. D., and Temin H. M. (1986) Different localization of the product of the v-rel oncogene in chicken fibroblasts and spleen cells correlates with transformation by REV-T. Cell 44, 791–800 10.1016/0092-8674(86)90845-7 [DOI] [PubMed] [Google Scholar]
- 37. Godinho S. A., and Pellman D. (2014) Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130467 10.1098/rstb.2013.0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fu J., Hagan I. M., and Glover D. M. (2015) The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7, a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gupta A., Tsuchiya Y., Ohta M., Shiratsuchi G., and Kitagawa D. (2017) NEK7 is required for G1 progression and procentriole formation. Mol. Biol. Cell 28, 2123–2134 10.1091/mbc.e16-09-0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cohen S., Aizer A., Shav-Tal Y., Yanai A., and Motro B. (2013) Nek7 kinase accelerates microtubule dynamic instability. Biochim. Biophys. Acta 1833, 1104–1113 10.1016/j.bbamcr.2012.12.021 [DOI] [PubMed] [Google Scholar]
- 41. Belham C., Roig J., Caldwell J. A., Aoyama Y., Kemp B. E., Comb M., and Avruch J. (2003) A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278, 34897–34909 10.1074/jbc.M303663200 [DOI] [PubMed] [Google Scholar]
- 42. Feige E., and Motro B. (2002) The related murine kinases, Nek6 and Nek7, display distinct patterns of expression. Mech. Dev. 110, 219–223 10.1016/S0925-4773(01)00573-1 [DOI] [PubMed] [Google Scholar]
- 43. Minoguchi S., Minoguchi M., and Yoshimura A. (2003) Differential control of the NIMA-related kinases, Nek6 and Nek7, by serum stimulation. Biochem. Biophys. Res. Commun. 301, 899–906 10.1016/S0006-291X(03)00049-4 [DOI] [PubMed] [Google Scholar]
- 44. Sampson J., O'Regan L., Dyer M. J. S., Bayliss R., and Fry A. M. (2017) Hsp72 and Nek6 cooperate to cluster amplified centrosomes in cancer cells. Cancer Res. 77, 4785–4796 10.1158/1538-7445.AM2017-4785 [DOI] [PubMed] [Google Scholar]
- 45. Rapley J., Nicolàs M., Groen A., Regué L., Bertran M. T., Caelles C., Avruch J., and Roig J. (2008) The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 121, 3912–3921 10.1242/jcs.035360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roig J., Mikhailov A., Belham C., and Avruch J. (2002) Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 16, 1640–1658 10.1101/gad.972202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andrews N. C., and Faller D. V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499 10.1093/nar/19.9.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this manuscript have been deposited in NCBI's Gene expression Omnibus under the GEO Series accession code GSE108258. The TCGA data referenced in this paper are based upon the data generated by the TCGA Research Network and are available in a public repository from the cBioportal for Cancer Genomics website (http://www.cbioportal.org/) (48, 49).4 All the other data are available within the article and the supplementary files.