The transformation of normal tissue to malignant tumours is driven by the widespread reprogramming of gene expression. Historically, the majority of research efforts have focused on the alternations of protein-coding genes as they were thought to be the only biologically functional feature in the human genome. Consequently, transcripts from the noncoding regions were viewed as transcriptional noise and overlooked despite reports of their aberrant expression in various cancers (1,2). In the past decade through emerging technologies, studies began to reveal that noncoding RNAs (ncRNAs) can also have important biological functions and are implicated in diverse cellular processes and disease progressions (3-5). Ever since, ncRNAs have gained significant research interest and a large number of studies have been conducted to elucidate their functions and roles.
ncRNAs can be grouped into two classes based on transcript size: small ncRNAs (smRNAs) and long ncRNAs (lncRNAs). smRNAs are generally 18 to 200 nucleotides in length while lncRNAs are greater than 200 nucleotides (6). smRNAs have diverse cellular functions and consist of several subclasses such as transfer RNA (tRNA), microRNA (miRNA) and piwi interacting RNA (piRNA). They have been extensively studied, especially miRNAs, which are one of the first post-transcriptional regulators implicated in many cancer progressions, such as breast and prostate cancer (7,8). miRNAs are suppressors of gene expression and indispensable components of the gene regulatory network. The functional importance of miRNAs led to the investigation of other classes of regulatory ncRNAs. However, despite the number of studies, the function of many ncRNAs remains unknown and unclear.
A recent article in Nature Medicine by Fish et al. (9) characterized a previous unknown class of smRNAs, which they termed ‘orphan’ noncoding RNAs (oncRNAs), and explored how they may play a role in cancer progression. These oncRNAs are novel as they are specifically expressed in cancer cells and do not fall into any existing classes of smRNAs. Cancer cells are capable of utilizing various strategies to alter gene expression patterns, including somatic mutations, gene amplifications and deletions, and epigenetic changes (10,11). However, the majority of current studies has been focused on how regulatory molecules in normal cells are altered as cells go through oncogenic transformation. Fish et al. (9) hypothesized that cancer cells may engineer novel regulators absent in normal cells as an additional strategy to achieve malignancy and these oncRNAs may be such regulators. They proposed that two steps are necessary for the emergence of such neo-regulators. First is the existence of a large pool of diverse macromolecules with regulatory potential. Second is the exploitation of these molecules for novel function by cancer cells. smRNAs are good candidates for neo-regulators since they are sufficiently abundant in cancer cells and many have been shown to possess regulatory abilities (12). The goal of the study (9) was to screen for cancer-specific oncRNAs and explore their potential functions.
Fish et al. (9) performed a systematic screen for smRNAs that exist in breast cancer but are absent from healthy normal. Through the screening of breast cancer cell lines, The Cancer Genome Atlas (TCGA) dataset of breast cancer biopsies and patient-derived xenograft (PDX) mouse models, the authors identified a pool of 201 smRNAs that are strongly associated with breast cancer but mostly undetectable in normal cells (Figure 1A). These smRNAs fit the definition of oncRNAs proposed by the authors as they are cancer-associated but functionally unknown. Their existence supported the hypothesis that tumours may have a novel repertoire of molecules with regulatory potential. Out of the 201 oncRNAs, one was found to have significantly higher expression in the metastatic breast cancer cells compared to the metastatic ones. The authors named it T3p as it maps to the 3’ end of the TERC gene which encodes for the RNA component of telomerase. Clinically, T3p level correlated with poor patient survival in breast cancer. The downregulation or overexpression of T3p resulted in significant changes in gene expression landscapes which were comparable to those caused by miRNA modulations, suggesting that T3p may be a broad gene expression regulator as well. Interestingly, the knockdown of full-length TERC transcript did not alter gene expression to the same extent. This finding is significant as it revealed the functional divergence of T3p from its parental gene. When tumour cells with T3p knockdown were injected into mice, reduced metastatic capacity was observed, confirming that T3p functions as a metastatic promoter.
Figure 1.
Schematic illustration of the identification and functional characterization of oncRNAs. (A) Cancer specific oncRNAs in breast cancer. oncRNAs are only expressed in cancer cells but not normal cells. Breast cancer oncRNAs were discovered by screening for smRNAs expressed in breast cancer but not mammary epithelial cell lines. The oncRNAs were further annotated in TCGA smRNA expression profiles of breast cancer biopsies and adjacent normal tissues and validated in breast cancer PDX models. (B) Characterization of oncRNA T3p. T3p is formed from TERC RNA nucleolytic activity in interaction with TARBP2 and DROSHA. It inhibits the activity of the RISC-complex by competing with miR-10b and miR-378c targets for binding. As a result, T3p expression elicits broad gene expression changes, and specifically the elevation of NUPR1 and PANX2 levels, leading to the promotion of metastasis.
Fish et al. (9) next investigated the mechanisms of T3p, specifically how it is produced and how it regulates gene expression. They showed that T3p is a by-product of TERC RNA digestion by the RNA binding protein TARBP2 and RNA-specific endoribonuclease DROSHA, which both had elevated levels in breast cancer. The finding suggests that cancer cells can hijack existing machineries to generate a repertoire of RNAs that may be consequently adopted for function. T3p was found to be an inhibitor of the miRNA-RISC interference pathway by competing with miRNAs for target gene binding. T3p could directly interact with miRNA-10b and miRNA-378c through partial complementary base pairing. Interestingly, full length TERC RNA had no such interactions, again demonstrating the novel function of T3p. It is known that ncRNAs especially lncRNAs can act as molecular sponge to sequester miRNAs from binding to target mRNAs (13). However, very limited smRNAs have been reported with miRNA decoy functions (13,14). The article by Fish et al. (9) is one of the first to show that smRNAs such as T3p can inhibit miRNAs by forming stable duplexes. This also reveals an additional strategy engineered by cancer cells to reprogram gene expression. In highly metastatic breast cancer cells, T3p was upregulated and led to suppressed activity of miRNA-10b and miRNA-378c. As a consequence, downstream target genes NUPR1 and PANX2 were upregulated and high expression levels of these two genes were associated with metastasis and worse patient outcome (Figure 1B).
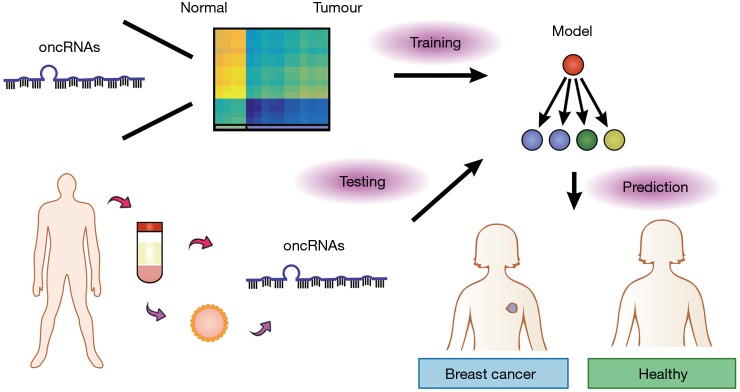
Since T3p and other oncRNAs are cancer-specific and mostly absent in normal tissues, they have the potential to be good biomarkers for cancer detection. Importantly, Fish et al. (9) found that nearly half of the oncRNAs screened in the study, including T3p, can be detected in the extracellular vesicles isolated from the conditioned medium of breast cancer cell lines as well as in patient serum. This reveals the potential of oncRNAs for liquid biopsy (Figure 2). Traditionally, cancer diagnosis is done through biopsy sampling of tumor tissue. However, tissue biopsy is invasive as it requires a needle incision or surgery, which can lead to procedural complications and sampling bias (15). Since cancer-specific molecules such as DNA and RNA are present in blood and other body fluids, the concept of liquid biopsy emerges (16). In comparison to traditional biopsy, the sampling of blood or other body fluids to analyze tumour materials is minimally invasive and may allow for earlier diagnosis. Moreover, it can allow for real-time monitoring of disease progression, treatment response and relapse. The challenge in the field is to identify suitable biomarkers for robust cancer diagnosis. Current strategies include detecting tumour DNA with specific genetic or epigenetic mutations, tumour-specific mRNAs, miRNA and other ncRNA expression patterns (16-18). The study by Fish et al. (9) is the first to show that cancer-specific oncRNAs have clinical diagnostic potential and they may be superior biomarkers. The authors utilized a machine-learning approach, specifically a gradient-boosted classifier (GBC), to assess how well oncRNAs can be used to classify breast cancer patients from healthy individuals. The GBC is a popular and powerful ensemble technique where a series of weak predictors, regression trees as used in the paper, is trained on the error residuals of previous predictors and is combined to make a final prediction. Thus, GBCs offer the advantage of being able to learn complex non-linear boundaries for classification and is very applicable to unbalanced biological datasets almost right off the shelf (19). Although the model built by Fish et al. (9) was trained on expression levels of oncRNAs in tumour biopsies, it was directly transferable to patient classification using serum oncRNA expression levels and achieved a high performance. More importantly, the GBC model trained on oncRNA levels was notably superior to a GBC model trained on the expression levels of miRNAs previously proposed as biomarkers, highlighting how serum oncRNA levels reflect that of the underlying tumour tissue, but not miRNAs. The application of GBCs to assess biomarker validity in liquid biopsy by Fish et al. (9) represents a promising direction in the inter-disciplinary union of machine learning and biomedical sciences, though there is still a long journey ahead in interpretability and clinical utilization.
Figure 2.
oncRNAs in liquid biopsy for diagnosis. oncRNAs are released specifically by tumour cells into the bloodstream directly or in extracellular vesicles. Models trained using oncRNA expression levels can be used to classify individuals as healthy or with cancer, enabling diagnosis through liquid biopsy.
Fish et al. (9) focused on breast cancer, however, it is unknown whether oncRNAs are universal regulators utilized by other cancers. The question can be answered by applying similar screening strategies to other tumour types. Heterogeneity is a common characteristic of cancer. It is unclear how much oncRNA expression varies within a heterogenous tumour. This raises the question of which oncRNAs and how many different oncRNA species are needed for accurate and robust cancer diagnosis through liquid biopsy. A strategy for oncRNA detection through liquid biopsy may help overcome the inherent biases in sequencing studies of tumours with high heterogeneity, which is typically underestimated due to sampling. Fish et al. (9) screened for oncRNAs that only exist in cancer. However, some were shown to be detectable in low levels in healthy normal, including T3p. This weakens the claim that oncRNAs are ‘binary’—they are found in cancer cells exclusively. As a result, the use of oncRNAs for cancer diagnosis may necessarily extend beyond a simple measure of absence or presence. It may be necessary to determine at what expression levels oncRNAs can be considered indicative of cancer. However, it may be difficult to establish such cut-offs as the levels may vary in individuals due to heterogeneity of the disease, though luckily this process will be aided with further adoption of machine learning and ever growing publicly available genomic datasets. The concept of truly cancer-specific oncRNAs is certainly not without possibility, and with further discoveries and validations they may one day move into the clinic.
The study by Fish et al. (9) explored a class of previously unknown smRNAs and proposed novel views of tumourigenesis. Instead of analyzing alternations of existing gene expression regulators, the authors explored the possibility of cancer cells engineering new players to promote malignancy. They established the novel concept of ‘orphan’ RNAs and hypothesized that these cancer-specific smRNAs with unknown function may be neo-regulators of gene expression. As exemplified in T3p, this smRNA adopted functions drastically different from its parental gene. It was shown to be a broad gene expression regulator by inhibiting the activity of the RISC complex. The findings revealed novel functions of smRNAs, specifically how they can act as miRNA decoys and this knowledge may help in understanding other ncRNAs. Clinically, oncRNAs present an advantage for liquid biopsy over other biomarkers such as miRNA or DNA since they predominantly exist in cancer cells. As studies continue, these ‘orphan’ RNAs may no longer be orphans with unknown functions as their biological roles are further revealed and identified.
Acknowledgements
Funding: This study was supported by the Princess Margaret Cancer Foundation (886012001223 to HH He). JT Hua was supported by CIHR Graduate Student Doctoral Fellowship. S Chen was supported by a Faculty of Medicine Award, a University of Toronto Fellowship and a STARS21 Training program fellowship. LY Liu was supported by a Faculty of Medicine Award.
Provenance: This is an Invited article commissioned by Section Editor Dr. Clive R Da Costa (Principal Laboratory Research Scientist, Adult Stem Cell Laboratory, The Francis Crick Institute, London, UK).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 2007;14:103-5. 10.1038/nsmb0207-103 [DOI] [PubMed] [Google Scholar]
- 2.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 2008;17:642-55. 10.1093/hmg/ddm336 [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Kertesz M, Wang JK, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007;129:1311-23. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015;34:5003-11. 10.1038/onc.2014.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 6.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol 2009;21:416-25. 10.1016/j.ceb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70. 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- 8.Ozen M, Creighton CJ, Ozdemir M, et al. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008;27:1788-93. 10.1038/sj.onc.1210809 [DOI] [PubMed] [Google Scholar]
- 9.Fish L, Zhang S, Yu JX, et al. Cancer cells exploit an orphan RNA to drive metastatic progression. Nat Med 2018;24:1743-51. 10.1038/s41591-018-0230-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998;396:643-9. 10.1038/25292 [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011;17:330-9. 10.1038/nm.2305 [DOI] [PubMed] [Google Scholar]
- 12.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012;6:590-610. 10.1016/j.molonc.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci 2018;75:467-84. 10.1007/s00018-017-2626-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazalla D, Yario T, Steitz J. Down-regulation of a host MicroRNA by a herpesvirus saimiri noncoding RNA. Science 2010;328:1563-6. 10.1126/science.1187197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J Clin Oncol 2013;31:17-22. 10.1200/JCO.2012.43.1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold B, Cankovic M, Furtado LV, et al. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? J Mol Diagn 2015;17:209-24. 10.1016/j.jmoldx.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono S, Lam S, Nagahara M, et al. Circulating microRNA Biomarkers as Liquid Biopsy for Cancer Patients: Pros and Cons of Current Assays. J Clin Med 2015;4:1890-907. 10.3390/jcm4101890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol Cancer 2016;15:39. 10.1186/s12943-016-0524-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frery J, Habrard A, Sebban M, et al. Efficient Top Rank Optimization with Gradient Boosting for Supervised Anomaly Detection. In: Ceci M, Hollmén J, Todorovski L, et al. editors. Machine Learning and Knowledge Discovery in Databases. New York City: Springer International Publishing, 2917. [Google Scholar]