Abstract
Background
To determine the number of cores adequate for histopathologic diagnosis as well as evaluate the success rate of molecular analyses in CT-guided percutaneous core needle biopsy (PCNB) for malignant pulmonary lesions using a 20-guage coaxial needle.
Methods
Biopsy records of 196 malignant lung lesions were reviewed. Core obtained from each needle pass was put in a separate container for individual pathological analysis. Types of molecular analysis attempted and their success rates were recorded for each patient. We categorized each patient into one of six groups according to the number of cores (n=1, n=2, n=3, n=4, n=5, n≥6) acquired, and diagnostic sensitivity for histopathologic diagnosis was calculated for each core in each group. In order to assess the increase in cumulative sensitivity up to 4th core, the data from 1st to 4th needle passes in 4-, 5-, and ≥6-core groups were pooled and cumulative diagnostic sensitivities up to 4th core were calculated.
Results
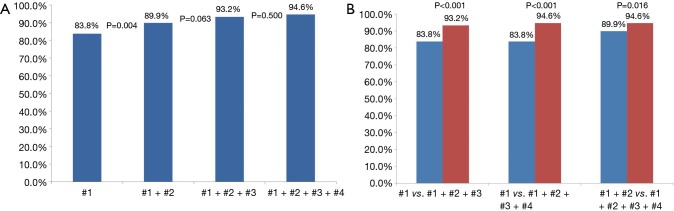
Of 196 cases of lung malignancies, five different types of molecular studies (EGFR mutation, ALK translocation, KRAS mutation, RET and ROS1 rearrangements) were attempted with PCNB specimens in 100 cases and successfully done in 96 cases (96.0%). In ≥4-core group (4-, 5-, and ≥6-core groups combined; n=148), cumulative sensitivity increased from 83.8% to 89.9% between 1st and 2nd cores, 89.9% to 93.2% between 2nd and 3rd cores, and 93.2% to 94.6% between 3rd and 4th cores.
Conclusions
The cumulative diagnostic sensitivity for the histopathologic diagnosis increases significantly between the second and fourth sampling. Multiple samples obtained with a 20-guage coaxial needle are adequate and have a high success rate for various molecular studies for lung malignancy.
Keywords: Lung cancer, CT-guided biopsy, molecular diagnosis, core needle biopsy
Introduction
CT-guided percutaneous core needle biopsy (PCNB) is a minimally invasive method to diagnose malignant lung lesions, providing tissue for evaluation of tumor architecture, immunohistochemical staining, and molecular analyses in patients with lung malignancy (1,2). Although molecular analyses can be performed on cytology samples, they are more successfully done on samples with a larger volume of tissue and number of tumor cells, such as those from a core biopsy; therefore, the use of a core biopsy technique is preferred over aspiration technique for the detection of genetic mutations in lung cancer patients (3-5). Mutation studies have become just as important as the diagnosis of cancer due to the wide availability of molecularly targeted therapies, which represent a significant advance in lung cancer treatment in the recent years (6). Mutation studies help decide whether or not to use a first-line epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) such as erlotinib and gefitinib. In addition, the mechanism of resistance can also be identified when those patients progress after receiving EGFR-TKI (7).
Large portions of these mutation studies (as well as histological diagnostic procedures) are done with small biopsy samples; however, no clear guideline regarding needle gauge and number of cores adequate for molecular studies or histological diagnosis exist despite the establishment of PCNB using a coaxial technique as a standard procedure for the small samples. Some institutions provide rapid on-site evaluation (ROSE) to help determine the number of samplings. However, there are also many hospitals without ROSE, where the number of core samples and needle size are largely decided by the established routine or preference of the practicing radiologist. For example, 20-guage needle, which is the finest core needle available, has gained popularity in the PCNB of lung for histopathological analysis over the last three decades in order to reduce bleeding complications (8-12).
One study showed that majority (66%) of the surveyed radiologists obtain at least three cores (13), probably because radiologists believe that greater amount of tissue maximizes success rates in gene mutation analysis. However, the survey results only show a trend and are not based on any scientific methods. There was one study claiming that the optimal number of cores in 20-guage coaxial lung biopsy is three; however we believe that the study was faulty because all core samples in any single patient were pooled together for one pathological analysis (14). Such circumstances make it impossible to measure the contribution of later samples to the improvement of pathological accuracy in a given patient. A study by Lim et al. (15) showed the increments in cumulative diagnostic accuracy by examining each core sample separately, with a conclusion that the optimal number of cores was three. However, the majority of the involved patients in the study had three or fewer cores acquired, and we believed that it did not fully explore the possible benefit of obtaining four or more cores.
Although specimens obtained with 20-guage core needles are widely used for molecular analyses due to its popularity, there have only been two studies that have actually explored the adequacy of 20-guages needles for molecular analysis specimens (16,17).
We ourselves use a 20-guage needle for PCNB; it has been our policy over the past few years, to acquire four or more core samples and add one (safety margin) to the optimal number of three generated from the study by Lim et al. (15). For quality control audit purposes, each core is put in a separately numbered formalin container for separate histopathological analysis in our hospital. We sought to analyze this retrospective data to confirm whether three is indeed the optimal number of cores for histopathological diagnosis for 20-guage coaxial PCNB and whether samples obtained using such technique were adequate for molecular analyses.
The purpose of this study is to determine the number of cores adequate for histopathologic diagnosis as well as evaluate the success rate of molecular analyses in PCNB for malignant pulmonary lesions using a 20-guage coaxial needle.
Methods
The institutional review board approved this retrospective study, with a waiver of informed consent.
Study population
We reviewed 307 consecutive patients who underwent CT-guided PCNB of thoracic lesions from March 2014 to February 2015. Biopsies with benign pathological results, obtained from extrapulmonary locations, done with aspiration technique only, or conducted for clinical trials (pathology not reviewed at our hospital) were excluded from the study.
Daily practice: biopsy procedure
Multidetector CT scanner (Siemens Definition AS Plus, Siemens Healthcare, Erlangen, Germany) with no CT-fluoroscopy function was used for the procedure. PCNB was performed by one chest radiologist with 13 years’ experience, using a 20-gauge coaxial needle system with a fixed 1.5-cm cutting trough (Stericut®, TSK Laboratory, Tochigi, Japan). Many of the cases with overtly clear indications for PCNB (e.g., stage 3 or 4 lung cancers either with mediastinal lymph node or distant metastases) usually underwent PCNB without discussion in the multidisciplinary team, with PCNBs requested by pulmonologists and oncologists directly to the radiologists. Other difficult-to-decide cases underwent discussion in the multidisciplinary team (lung cancer board) to decide whether PCNB or surgical excision should be performed. At our institution, pure ground-glass opacity (GGO) or part-solid nodules with a high suspicion of adenocarcinoma usually undergo surgical excision without biopsy; the radiologists recommend surgical excision for such lesions due to fear of false-negative results and inadequate specimens. Likewise, the radiologists also usually decline requests for biopsy of a very small lesion (longest diameter <0.8 cm), because such lesions are difficult to target using CT without CT-fluoroscopy function. In cases with severe COPD, only those patients decided suitable to undergo PCNB by the pulmonologists underwent PCNB. If the tumor was too close to the vessels, deemed too risky by the operating radiologist, then he declined the request for PCNB. From preprocedural CT images, the optimal needle path (that ensures avoiding major vessels and bronchi) and patient position (prone or supine) were determined. After skin sterilization and a local infiltration of 2% lidocaine, an 18-gauge coaxial introducer (outer needle) was inserted into the chest wall with no skin incision. The introducer was inserted into the target lesion under intermittent CT guidance (about 5 cm length of short volume scans). The stylet was removed after confirming the proper placement of about 3 mm of the needle tip within the lesion and checking the direction of the needle trajectory to be within the lesion. Multiple core samplings were done by passing a 20-gauge semi-automated cutting needle (inner needle) via the introducer. A curved needle technique was routinely used to direct needles toward various areas of the lesion without repositioning the outer sheath, to decrease sampling error and increase diagnostic yield (18). Between core samplings, the hub of the outer needle was blocked with a 1-cc syringe to prevent fatal pulmonary venous air embolism. Each core was put in a separately numbered formalin container (Figure 1), and the pathology department provided individual histopathological results for each container number. Four core samples were routinely acquired unless the procedure was interrupted by cough or hemoptysis, or if multiple samplings were deemed unadvisable due to the proximity of a critical structure (such as the heart) to the lesion. The routine number of core sampling was determined based on the study of PCNB by Lim et al., which showed that three samplings are adequate for histopathologic diagnosis of malignant lung lesions (15). We added one more sampling as a safety margin and for the sake of ensuring adequate specimen for molecular analyses, which was not studied in the study by Lim et al. (15). More than four core samples were acquired if core specimens from the first four samplings were deemed macroscopically inadequate (hemorrhage with no particulate material or only small fragments of solid tissue) (19) by the radiologist.
Figure 1.
Numbered formalin containers. Each core was placed in a separately numbered formalin container. The pathology department then provided with individual histopathological results for each container number.
CT scans were obtained immediately after the procedure to detect any occurrence of complications such as pneumothorax or perilesional hemorrhage. Patients with asymptomatic pneumothorax or intrapulmonary hemorrhage were treated conservatively. A drainage catheter or chest tube was inserted if the patient was symptomatic and showed signs of respiratory distress.
Daily practice: molecular analysis
Molecular analysis [EGFR mutation, anaplastic lymphoma receptor tyrosine kinase (ALK) translocation, Kirsten rat sarcoma viral oncogene homolog (KRAS) mutation, RET and ROS1 rearrangements], when requested, was done by the pathology department at our hospital, and the details are: EGFR and KRAS mutation tests were done using a peptide nucleic acid (PNA) clamping method. DNA was isolated from 10 formalin-fixed, paraffin-embedded (FFPE), 5-µm-thick tissue sections using a High Pure PCR Template Preparation Kit (Roche Applied Science, Mannheim, Germany). Mutations were detected by real-time PCR using PNAClamp™ EGFR Mutation Detection Kit and the PNAClamp™ KRAS Kit (PANAGENE, Inc., Daejeon, Korea) (20).
ALK, ROS1, and RET fluorescence in situ hybridization (FISH) were performed using the dual-color break-apart probe (Abbott Molecular, Abbott Park, IL, USA) according to the manufacturer’s instructions. The testing procedure and the interpretation of results have been previously described (21). The ALK FISH assay was independently captured and scored with the automated BioView Duet scanning system (BioView, Rehovot, Israel). A minimum of 50 tumor nuclei were counted. ALK, ROS1, and RET FISH-positive cases were defined as more than 25 (50%) break-apart (BA) signals or an isolated signal (IRS) in tumor cells. ALK, ROS1, and RET FISH-negative samples were defined as less than 5 (10%) BA or IRS cells. ALK, ROS1, and RET FISH cases were considered borderline if 5–25 (10–50%) cells were positive. In cases of borderline results, a second reader evaluated the slide, added cell count readings from the automatically captured images, and calculated a percentage out of 100 cells. The sample was considered negative if the percentage of positive cells was lower than 15%, and the sample was considered positive if the percentage of positive cells was higher or equal to 15%.
Data collection for the study
For each patient, the following data were collected from medical records, pre-procedural diagnostic CT images taken mean 12 days before (range, 0–112 days), and procedural and post-procedural CT images: age, sex, lesion size and location, distance from the pleura to the lesion (measured along the needle length from the pleura to the lesion on procedural CT images), number of cores acquired, histopathological results, and development of complications (pneumothorax, pulmonary hemorrhage, hemoptysis, or blood-tinged sputum). Lesion size was defined as the maximum diameter of the lesion on axial diagnostic CT images. Pneumothorax was diagnosed on post-procedural CT or on subsequent chest radiography taken three hours after the biopsy if asymptomatic. Pulmonary hemorrhage was defined as newly noted airspace opacity (GGO or consolidation) around the needle track or within 20 cm of the lesion.
Assessment of number of acquired cores as related to sample adequacy for histopathologic analysis
We assigned each patient into one of six groups according to the number of cores (n=1, n=2, n=3, n=4, n=5, n≥6) acquired. Diagnostic sensitivity and cumulative diagnostic sensitivity were calculated for each core in each group. Diagnostic sensitivity for nth core was defined as the percentage of patients within a group with a positive result for malignancy from nth core. Cumulative diagnostic sensitivity up to nth core was defined as the percentage of patients within the group with positive results for malignancy from one or more cores obtained up until the nth sampling. In order to assess the increase in cumulative sensitivity up to 4th core, the data from 1st to 4th needle passes in 4-, 5-, and ≥6-core groups were pooled, and the cumulative diagnostic sensitivities up to 4th core were calculated.
Assessment of success of molecular analysis
For each patient, we assessed if there was an attempt for molecular analysis using PCNB specimens. If the patient did not undergo molecular analyses with PCNB specimens, we recorded the reasons for not doing so. We also recorded the type and number of successful molecular testing performed on PCNB specimen in each patient. Successful molecular testing was defined as the ability to report the presence or absence of EGFR mutation, KRAS mutation, ALK translocation, or other miscellaneous (ROS1 or RET) mutations. We then compared the clinical and biopsy-related factors between the group with adequate specimen for molecular analysis and those with inadequate specimen for molecular analysis.
Statistical analysis
The data were recorded on worksheets (Excel; Microsoft, Redmond, Wash) and analyzed using Excel. McNemar’s test was done to analyze whether there is a significant increment in the cumulative sensitivity up to 4th core with addition of each core in the pooled dataset (4-, 5-, and ≥6-core groups). In order to analyze factors determining diagnostic sensitivity, univariate logistic regression analysis was performed. In order to analyze factors determining biopsy-related complication rates, univariate logistic regression analysis was firstly performed, and multivariate logistic regression analysis was performed for variables that showed a P value of less than 0.10 at univariate analysis. Statistical analyses were performed at a significance level of 0.05 using SPSS 24.0 (IBM SPSS Statistics, Armonk, NY, USA).
Results
Of 307 CT-guided PCNB cases, 111 cases were excluded according to exclusion criteria, leaving 196 cases of lung malignancies for the study. The excluded cases were: 58 benign lesions (tuberculosis, inflammation, fibrosis, organizing pneumonia, and pneumoconiotic nodule), 28 extrapulmonary targets (16 in the pleura or chest wall, 11 in the mediastinum, and 1 in the axilla), 12 cases conducted for clinical trials (pathology not reviewed at our hospital), and 13 procedures performed with aspiration technique only.
Patient demographics, biopsy-related factors, and diagnostic sensitivity
Of 196 cases of lung malignancies (female:male=72:124, mean age 67.7 years old, range, 31–92 years old), the final diagnosis of malignancy was established at initial CT-guided PCNB in 186 cases (overall sensitivity =94.9%), and at surgery or repeat CT-guided PCNB in 10 patients.
There were 177 (90.3%) primary lung cancers and 19 metastatic carcinoma or sarcoma (9.7%). The histologic types of primary lung cancer were 118 adenocarcinoma (66.7%), 37 squamous cell carcinoma (20.9%), 11 small cell carcinoma (6.2%), 5 poorly differentiated carcinoma (2.8%), 2 combined small cell carcinoma and adenocarcinoma, 1 large cell carcinoma, 1 undifferentiated carcinoma, 1 adenosquamous carcinoma, and 1 neuroendocrine carcinoma. Of 177 primary lung cancers, there were 40 (22.6%) stage I, 12 (6.8%) stage II, 42 (23.7%) stage III, and 83 (46.9%) stage IV diseases. Mean size of the biopsied tumor was 4.1 cm (range, 0.9–12.9 cm), with mean pleura-to-target distance of 1.4 cm. Mean number of cores obtained was 4.1. There were 157 (80.1%) solid and 39 (19.9%) subsolid tumors, of which 38 were part-solid and one was pure GGO. There were 17 (8.7%) lesions with presence of necrotic portion on CT images. On univariate analysis, female sex (P=0.038) and presence of necrotic portion (P=0.026) were significantly associated with diagnostic sensitivity. Tumor size (P=0.055) or the pleura-to-target distance (P=0.788) was not significantly associated with diagnostic sensitivity. Table 1 summarizes the clinical characteristics and biopsy-related factors of the study population as well as comparison of these factors between the true positive and the false negative group and the corresponding P values.
Table 1. Patient demographics and factors determining diagnostic sensitivity and univariate regression analysis.
| Parameter | Total study population (n=196) | True positive (n=186) | False negative (n=10) | P |
|---|---|---|---|---|
| Age (years), mean [range] | 67.7 [31–92] | 67.7 [31–92] | 68.1 [61–84] | 0.907 |
| Sex, n (%) | 0.038 | |||
| Male | 124 (63.3) | 121 (65.1) | 3 (30.0) | |
| Female | 72 (36.7) | 65 (34.9) | 7 (70.0) | |
| Smoking history, n (%) | 0.554 | |||
| Current smoker | 9 (4.6) | 8 (4.3) | 1 (10.0) | |
| Former smoker | 106 (54.1) | 102 (54.8) | 4 (40.0) | |
| Never smoker | 81 (41.3) | 76 (40.9) | 5 (50.0) | |
| Histology, n (%) | 0.402 | |||
| Primary lung cancer | 177 (90.3) | 169 (90.9) | 8 (80.0) | |
| Adenocarcinoma | 118 (66.7) | 113 (66.9) | 5 (62.5) | |
| Squamous cell carcinoma | 37 (20.9) | 35 (20.7) | 2 (25.0) | |
| Poorly differentiated carcinoma | 5 (2.8) | 4 (2.4) | 1 (12.5) | |
| Small cell carcinoma | 11 (6.2) | 11 (6.5) | 0 (0.0) | |
| Others | 6 (3.4) | 6 (3.6) | 0 (0.0) | |
| Metastatic carcinoma or sarcoma | 19 (9.7) | 17 (9.1) | 2 (20.0) | |
| Emphysema, n (%) | 60 (30.6) | 58 (31.2) | 2 (20.0) | 0.461 |
| Tumor size (cm), mean [range] | 4.1 [0.9–12.9] | 4.1 [0.9–12.9] | 5.5 [1.5–12.2] | 0.055 |
| CT findings of the tumor, n (%) | 0.993 | |||
| Solid | 157 (80.1) | 149 (80.1) | 8 (80.0) | |
| Subsolid | 39 (19.9) | 37 (19.9) | 2 (20.0) | |
| Presence of necrotic portion, n (%) | 17 (8.7) | 14 (7.5) | 3 (30.0) | 0.026 |
| Pleura-to-target distance (cm), mean [range] | 1.4 [0–7.9] | 1.4 [0–7.9] | 1.6 [0–4.1] | 0.788 |
| Number of cores, mean [range] | 4.0 [1–9] | 4.0 [1–9] | 3.6 [1–5] | 0.350 |
| Biopsy location, n (%) | 0.740 | |||
| Upper lobes | 108 (55.1) | 103 (55.4) | 5 (50.0) | |
| Middle and lower lobes | 88 (44.9) | 83 (44.6) | 5 (50.0) | |
| Biopsy position, n (%) | 0.905 | |||
| Supine | 90 (45.9) | 86 (46.2) | 4 (40.0) | |
| Prone | 102 (52.1) | 96 (51.6) | 6 (60.0) | |
| Others (oblique or decubitus) | 4 (2.0) | 4 (2.2) | 0 (0.0) | |
| Complications, n (%) | 61 (31.1) | 57 (30.6) | 4 (40.0) | 0.536 |
| Pneumothorax, n (%) | 40 (20.4) | 38 (20.4) | 2 (20.0) | 0.974 |
| Pulmonary hemorrhage, n (%) | 15 (7.7) | 14 (7.5) | 1 (10.0) | 0.720 |
| Blood-tinged sputum or hemoptysis, n (%) | 9 (4.6) | 8 (4.3) | 1 (10.0) | 0.417 |
Number of acquired cores and the cumulative sensitivity of histopathologic diagnosis
Of 196 cases of lung malignancies, there were 9 cases with 1 core sampling, 15 cases with 2 core samplings, 24 with 3 core samplings, 99 cases with 4 core samplings, 31 cases with 5 core samplings, and 18 cases with ≥6 core samplings. Table 2 shows the sensitivity for each core in each core group, and Table 3 shows the cumulative sensitivity for each core. There were 148 cases when data from 4-core, 5-core, and ≥6-core groups were pooled for analysis of cumulative sensitivities up to 4th core. Cumulative sensitivity significantly increased from 83.8% to 89.9% (increment 6.1%, P=0.004) between 1st and 2nd cores, 89.9% to 93.2% (increment 3.3%, P=0.063) between 2nd and 3rd cores, and 93.2% to 94.6% (increment 1.4%, P=0.500) between 3rd and 4th cores. Cumulative sensitivity significantly increased from 83.8% to 93.2% (increment 9.4%, P<0.001) between 1st and 3rd cores, 83.8% to 94.6% (increment 10.8%, P<0.001) between 1st and 4th cores, and 89.9% to 94.6% between 2nd and 4th cores (increment 4.7%, P=0.016) (Figure 2).
Table 2. Sensitivity for histopathologic diagnosis for each core.
| Core number | Sensitivity, % | |||||
|---|---|---|---|---|---|---|
| 1-core group (n=9) | 2-core group (n=15) | 3-core group (n=24) | 4-core group (n=99) | 5-core group (n=31) | ≥6-core group (n=18) | |
| #1 | 88.9 | 73.3 | 83.3 | 86.9 | 71.0 | 88.9 |
| #2 | – | 86.7 | 75.0 | 83.8 | 87.1 | 66.7 |
| #3 | – | – | 91.7 | 83.8 | 74.2 | 72.2 |
| #4 | – | – | – | 78.8 | 77.4 | 83.3 |
| #5 | – | – | – | – | 71.0 | 83.3 |
| #6 | – | – | – | – | – | 94.4 |
Table 3. Cumulative sensitivity for histopathologic diagnosis for each group.
| Core number | Cumulative sensitivity, % | ||||
|---|---|---|---|---|---|
| 2-core group (n=15) | 3-core group (n=24) | 4-core group (n=99) | 5-core group (n=31) | ≥6-core group (n=18) | |
| #1 | 73.3 | 83.3 | 86.9 | 71.0 | 88.9 |
| #1 + #2 | 93.3 | 87.5 | 90.9 | 87.1 | 88.9 |
| #1 + #2 + #3 | – | 95.8 | 94.9 | 90.3 | 88.9 |
| #1 + #2 + #3 + #4 | – | – | 94.9 | 93.5 | 94.4 |
| #1 + #2 + #3 + #4 + #5 | – | – | – | 93.5 | 94.4 |
| #1 + #2 + #3 + #4 + #5 + #6 | – | – | – | – | 100.0 |
Figure 2.
Comparison of cumulative sensitivities for histopathological diagnosis up to 4th core (n=148). (A) Cumulative sensitivity significantly increased from 83.8% to 89.9% (increment 6.1%, P=0.004) between 1st and 2nd cores, 89.9% to 93.2% (increment 3.3%, P=0.063) between 2nd and 3rd cores, and 93.2% to 94.6% (increment 1.4%, P=0.500) between 3rd and 4th cores; (B) cumulative sensitivity significantly increased from 83.8% to 93.2% (increment 9.4%, P<0.001) between 1st and 3rd cores, 83.8% to 94.6% (increment 10.8%, P<0.001) between 1st and 4th cores, and 89.9% to 94.6% between 2nd and 4th cores (increment 4.7%, P=0.016).
Molecular analysis
Of 196 cases of lung malignancies, molecular analyses were attempted using PCNB specimens in 100 patients with primary lung cancers. Molecular analyses were not attempted with PCNB specimens in 96 patients for following reasons: 32 cases underwent molecular analyses using either bronchial biopsy specimens (8 cases) or surgical specimens (24 cases); 10 were false negative cases whose PCNB specimens were negative for malignancy; 14 were primary lung cancers but not non-small cell lung carcinoma (11 small cell carcinoma, two combined small cell carcinoma and adenocarcinoma, and one neuroendocrine carcinoma); 13 were metastases without the clinical need for undergoing molecular analyses; 6 were prebiopsy cases for entering into clinical trials with only the histopathology of PCNB sample reviewed at our hospital; three were recurrence cases; two cases with molecular testing done at PCNB specimens from another site in the lung; three transferred to another hospital with only the basic histopathological results confirmed at our hospital; one terminal case who expired shortly after undergoing PCNB, one patient refusal; 11 cases (9 squamous cell carcinoma and 2 adenocarcinoma) due to confusion in clinical practice pattern because standards for performing molecular analyses were not firmly established in 2014.
Among the 100 patients whose PCNB specimens were submitted for molecular analysis, molecular analyses were successfully done in 96 patients (96.0%). The other four patients had PCNB specimens with insufficient tumor component for DNA analysis; three of them used alternative specimens (subsequent surgical specimens in two and bone biopsy specimen in one) for successful molecular analyses. Comparison of the characteristics of adequate versus inadequate biopsies for molecular analysis is shown in Table 4. Of the patients who had inadequate biopsy specimen for molecular analysis, three of the patients were men and the mean age was 73.8 years (range, 62–89 years). Mean tumor size of the group with adequate specimen was 4.1 cm (range, 1.0–10.0 cm), compared with a mean tumor size of 2.9 cm (range, 1.9–3.7 cm) in the group with inadequate specimen. The average number of core biopsy samples obtained in the group with adequate specimen was 4.1 (range, 1–9), compared with an average of 3.3 core biopsy samples (range, 1–5) in the group with inadequate specimen. Three out of 4 (75.0%) patients in the inadequate group and 27 out of 96 patients (28.1%) in the adequate group developed biopsy-related complications. Due to the small number of inadequate biopsies, statistical analysis for the factors that contributed to adequacy or inadequacy of biopsies could not be performed.
Table 4. Molecular analysis: adequacy and associated factors.
| Parameter | Adequate for molecular analysis (n=96) | Inadequate for molecular analysis (n=4) |
|---|---|---|
| Sex, n (%) | ||
| Female | 38 (39.6) | 1 (25.0) |
| Male | 58 (60.4) | 3 (75.0) |
| Age (years), mean [range] | 69.1 [44–92] | 73.8 [62–89] |
| Emphysema, n (%) | 28 (29.2) | 1 (25.0) |
| Tumor size (cm), mean [range] | 4.1 [1.0–10.0] | 2.9 [1.9–3.7] |
| CT findings of the tumor, n (%) | ||
| Subsolid nodule | 21 (21.9) | 0 (0.0) |
| Presence of necrotic portion | 8 (8.3) | 0 (0.0) |
| Pleura-to-target distance (cm), mean [range] | 1.4 [0–5.7] | 1.9 [0.6–4.5] |
| Number of cores, mean [range] | 4.1 [1–9] | 3.3 [1–5] |
| Biopsy location, n (%) | ||
| Upper lobes | 49 (51.0) | 2 (50.0) |
| Middle and lower lobes | 47 (49.0) | 2 (50.0) |
| Biopsy position, n (%) | ||
| Supine | 43 (44.8) | 1 (25.0) |
| Prone | 50 (52.1) | 3 (75.0) |
| Others | 3 (3.1) | 0 (0.0) |
| Complications, n (%) | 27 (28.1) | 3 (75.0) |
| Pneumothorax | 19 (19.8) | 1 (25.0) |
| Pulmonary hemorrhage | 6 (3.2) | 2 (50.0) |
| Hemoptysis or blood-tinged sputum | 4 (4.2) | 0 (0.0) |
Five different types of molecular studies were done: EGFR mutation, ALK translocation, KRAS mutation, ROS1 fusion, and RET fusion. EGFR mutation and ALK translocation were prioritized according to recommendation (22) and KRAS, ROS1, RET were tested in EGFR negative and ALK negative cases. Table 5 shows the number of each study conducted and their results. The number of different types of molecular studies done in a patient using PCNB specimens were: one in 17 cases, two in 46 cases, three in 25 cases, four in 4 cases, and five in 6 cases.
Table 5. Number of each type of molecular study and the results.
| Type of study | Values |
|---|---|
| EGFR, n (%) | 92 |
| Positive | 31 (33.7) |
| Exon 19 deletion | 20 (21.7) |
| Exon 21 p.L858R or p.L861Q mutation | 9 (9.8) |
| Exon 18 G719 mutation and exon 20 p.T790M mutation | 1 (1.1) |
| Exon 20 20 Ser768Ile (c.2303G>T) mutation | 1 (1.1) |
| Negative | 61 (66.3) |
| ALK FISH, n (%) | 86 |
| Positive | 4 (4.7) |
| Negative | 82 (95.3) |
| KRAS, n (%) | 34 |
| Positive | 2 (5.9) |
| Negative | 32 (94.1) |
| RET, n (%) | 11 |
| Positive | 1 (9.1) |
| Negative | 10 (90.9) |
| ROS1, n (%) | 11 |
| Positive | 0 (0.0) |
| Negative | 11 (100.0) |
EGFR, epidermal growth factor receptor; ALK FISH, anaplastic lymphoma receptor tyrosine kinase fluorescence in situ hybridization; KRAS, Kirsten rat sarcoma viral oncogene homolog.
Complications
Complications occurred in 61 (31.1%) of 196 biopsies. Pneumothorax was observed in 40 biopsies (20.4%), and 4 of them required a drainage procedure (10.0%). Asymptomatic pulmonary hemorrhage, defined as new GGO lesion seen on post-procedural CT images without evidence of blood-tinged sputum or hemoptysis was observed in 15 biopsies (7.7%); two of these 15 cases also showed pneumothorax. Blood-tinged sputum or hemoptysis was observed in 9 biopsies (4.6%), and one of them also showed pneumothorax. Except for four cases of pneumothorax that required a drainage procedure and thus were grade 2 complications according to the Common Terminology Criteria for Adverse Events (CTCAE) (23), all complications were CTCAE grade 1. There weren’t any cases of infection or air embolism. Statistical analysis to determine significant contributing factors was performed for biopsies without and with complications. Comparison of the characteristics of biopsies without and with complications and univariate P values are shown in Table 6. Mean tumor size was smaller (P=0.003) and the target lesion was located farther from the pleura (P=0.024) in biopsies with complications. Variables included in the multivariate analysis and corresponding p values are shown in Table 7. Tumor size (P=0.021) and number of cores acquired (P=0.007) remained significantly associated with increased complication rates in the multivariate analysis.
Table 6. Factors determining the biopsy-related complication rates and univariate regression analysis.
| Parameter | No complication (n=135) | Complication (n=61) | P |
|---|---|---|---|
| Sex | 0.407 | ||
| Female | 47 | 25 | |
| Male | 88 | 36 | |
| Age (years), mean [range] | 67.7 [31–92] | 67.9 [40–92] | 0.897 |
| Histology | 0.635 | ||
| Primary lung cancer | 121 | 56 | |
| Metastasis | 14 | 5 | |
| Emphysema | 38 | 22 | 0.267 |
| Tumor size (cm), mean [range] | 4.5 [1.1–12.9] | 3.4 [0.9–10.0] | 0.003 |
| CT findings of the tumor | |||
| Subsolid nodule | 26 | 13 | 0.739 |
| Presence of necrotic portion | 14 | 3 | 0.220 |
| Pleura-to-target distance (cm), mean [range] | 1.3 [0–7.9] | 1.8 [0–4.8] | 0.024 |
| Number of cores, mean [range] | 4.2 [1–9] | 3.5 [1–6] | 0.001 |
| Biopsy location | 0.904 | ||
| Upper lobes | 74 | 34 | |
| Middle and lower lobes | 61 | 27 | |
| Biopsy position | 0.214 | ||
| Supine | 61 | 29 | |
| Prone | 73 | 29 | |
| Others | 1 | 3 |
Table 7. Multivariate analysis of risk factors for biopsy-related complications.
| Parameter | Odds ratio (95% CI) | P |
|---|---|---|
| Tumor size | 0.808 (0.675, 0.969) | 0.021 |
| Pleura-to-target distance | 1.211 (0.987, 1.485) | 0.067 |
| Number of cores | 0.684 (0.518, 0.902) | 0.007 |
Discussion
The overall sensitivity of CT-guided PCNB in the histopathologic diagnosis of malignant lung lesions was 94.9% in the present study. This is comparable to the reported range of success for thoracic/pulmonary needle biopsy (77–96%), higher than the pooled mean success rate (89%) or the suggested quality improvement threshold (75%) from JVIR Quality Improvement Guidelines (24), and similar to the reported overall sensitivity of ROSE-assisted PCNB for lung lesions (94.1–96.4%) (25). This diagnostic sensitivity of PCNB using 20-gauge needles in our study (94.9%) is also comparable to the reported diagnostic accuracy of PCNB using 18-gauge needles (92.9–95.9%) (26,27). Our result is at the higher end of the reported range of diagnostic accuracy (64–97%) for aspiration lung biopsy (28). The sensitivities for each core in each group were in the 71–94% range, also comparable to the reported range of success for thoracic/pulmonary needle biopsy (77–96%) (24).
Before conducting this study, we suspected that the yield from the first core may be significantly higher than that from the later cores. The results show that the highest sensitivities for 3-core group and ≥6-core group came from the last cores (3rd and 6th cores, respectively), not from the first one. Our results also indicate that repetitive coaxial sampling using 20-guage needle for malignant lung lesions yields a sequential increase in cumulative sensitivity, from 83.8% at the first core sampling to 89.9% at the second, to 93.2% at the third, and 94.6% at the fourth core samplings, although the increase was statistically significant only from the first core to the second core (P=0.004). However, compared with the second core, the fourth core did show a statistically significant increase in cumulative sensitivity, from 89.9% to 94.6% (P=0.016). These results can serve as reference data for what radiologists should expect after the acquisition of a certain number of core samples via a 20-guage needle, especially in a situation where ROSE is unavailable. Our results indicate that the acquisition of four cores is recommended to achieve cumulative sensitivity of close to 95% for histopathologic diagnosis in a 20-gauge coaxial needle biopsy.
The vast majority of our study population was primary lung cancer, and 70.6% of them had either stage III or stage IV advanced disease. Despite the growing importance of molecular analysis in the management of advanced lung cancer, only a few studies have been conducted to analyze the adequacy of core needle samples for molecular analysis (16,17,29,30). Three studies with a relatively small number of patients have shown that molecular analyses (each examining EGFR; EGFR and KRAS; and EGFR, KRAS, and ALK) can be adequately done using 18-gauge or 20-gauge needle biopsy samples. However, the number of patients with samples from the 20-guage needle was fewer than 20 in all three studies combined (16,29,30). There was another study in 2013 with a larger number of cases (n=170) using a 20-guage needle for biomarker analysis with a success rate of 82.9% (17). Our success rate in molecular analysis using PCNB samples was 96.0%, which is higher than the results (82.9%) from the 2013 study by Tam et al. (17). In the study by Tam et al., “at least two cores” with a median of three were acquired; however, we acquired a mean of four cores in our study. We believe the difference in the number of cores obtained may be a factor contributing to the difference in the success rate between the two studies as both studies use the same type of imaging guidance and the same needle size. Also, although the number of inadequate samples for molecular analysis was too small (n=4) to perform a statistical analysis, those with inadequate samples tended to have smaller number of cores than those with adequate samples (3.3 vs. 4.1). Therefore, we believe that four core samplings are desirable in order to obtain adequate samples for molecular analysis.
The complication rates of our study—pneumothorax in 20.4% biopsies, pulmonary hemorrhage in 7.7%, and blood-tinged sputum or hemoptysis in 4.6%—were similar or lower to the pooled complication rates of CT-guided transthoracic lung biopsy in a meta-analysis study (31), which were: 25.3% pneumothorax, 18.0% pulmonary hemorrhage, and 4.1% hemoptysis. Although Lalji et al. claimed that biopsy with 10-guage needles is safe with comparable complication rates as in biopsies with 18-guage needles, the rates of pneumothorax and parenchymal bleeding were 40% and 45.7% (5), which were much higher than the complication rates in our study or the pooled complication rates in the meta-analysis study (31). There may be worries that acquiring more cores would result in an increased risk of complications, however our study has demonstrated an opposite picture. Complication rates were significantly higher in those with fewer core samplings, and number of cores was conversely related to the development of complications in a multivariate analysis. We believe such a result is due to the fact that the operating radiologist stopped acquiring more cores and finished the procedure when the patient showed development of complication. However, the result also means that acquiring more cores does not increase the risk of complications. Since cumulative sensitivity is significantly increased from 89.9% to 94.6% between the 2nd and 4th core and adequate specimens for different molecular tests tended to be acquired with more cores (4.1 vs. 3.3) with 20-gauge needles without increased complication rates, we believe we can confidently recommend acquiring four cores with 20-gauge needles. 20-gauge needles with four cores would be safer yet yield adequate and satisfactory results, and therefore using large-bore needles with such high complication rates, as in the study by Lalji et al. (5), doesn’t seem to be justifiable.
Our study has a few limitations. First, this is a retrospective study focused on assessing the increase in cumulative sensitivity as number of cores increases, so we could not compare the results to the control group. Second, this is a single-center study, and a single radiologist performed all procedures, so the results may be biased by the preference, skill, and expertise of the operating radiologist. Also, because we do not usually perform CT-guided biopsies for pure GGO and very small nodules (longest diameter <0.8 cm), this might have influenced the results. However, potential problems with inadequate sampling and false-negative results are established as limitations for biopsy of GGO lesions and very small nodules (32), and some recommend bypassing biopsy for highly suspicious GGO nodules (33). Therefore, our practice is not deviated from the recommendations; rather, we recommend that biopsy of GGO and very small nodules be declined in order to achieve high diagnostic accuracy as ours. Third, we did not have a chance to assess the correlation between the number of samplings and specimen adequacy for molecular testing. Separate histopathologic diagnosis was made for each core sample, but records were unavailable on how many samples were required or exactly which specimens were used for molecular analyses. Fourth, successful histopathologic or molecular analyses not only depends on the quantity and quality of the biopsy sample but also on the tissue handling process by the pathology technicians or the pathologists in the pathology department (i.e., large amount of tissue may be lost due to excessive and multiple trimming). Therefore, the current results may reflect the process in our pathology department and remain difficult to generalize. However, it could also suggest that three to four cores may be adequate for molecular analyses even when excessive tissue trimming practice is rampant. Fifth, almost half of the included patients did not undergo molecular analyses. However, this is not due to the inferiority or inadequacy of the PCNB specimens, but due to the clinical practice pattern at our hospital. At our institution, bronchoscopy and PCNB are usually performed on the same day or within a few days of interval, and pathologists choose the specimen for molecular analysis according to the availability, sample size, and sample adequacy. This may explain the eight cases of performing molecular analysis using bronchial biopsy specimens. Also, the guidelines for EGFR and ALK testing came out in 2013 (22), and standards for performing molecular analysis on small biopsy specimens were not firmly established at our institution and varied greatly among clinicians back in 2014 and up until 2015. This would explain 24 cases of molecular testing performed on surgical specimens and 11 cases of not performing molecular analyses altogether.
Conclusions
In conclusion, our study shows that the cumulative diagnostic sensitivity for the histopathologic diagnosis increases significantly between the second and fourth sampling. Multiple samples obtained with a 20-guage coaxial needle are adequate and have a high success rate for various molecular studies for lung malignancy. Accordingly, we recommend that radiologists routinely acquire four 20-guage core samples in the PCNB of potentially malignant lung lesions.
Acknowledgements
None.
Ethical Statement: The institutional review board approved this retrospective study, with a waiver of informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Lal H, Neyaz Z, Nath A, et al. CT-guided percutaneous biopsy of intrathoracic lesions. Korean J Radiol 2012;13:210-26. 10.3348/kjr.2012.13.2.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider F, Smith MA, Lane MC, et al. Adequacy of core needle biopsy specimens and fine-needle aspirates for molecular testing of lung adenocarcinomas. Am J Clin Pathol 2015;143:193-200; quiz 306. 10.1309/AJCPMY8UI7WSFSYY [DOI] [PubMed] [Google Scholar]
- 3.Kim-Son Nguyen JN. Lung Cancer. In: Ajay K. Singh JL, editor. The Brigham Intensive Review of Internal Medicine. 2nd edition ed. Oxford University Press, 2014:100. [Google Scholar]
- 4.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med 2013;137:828-60. 10.5858/arpa.2012-0720-OA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalji UC, Wildberger JE, Hausen AZ, et al. CT-Guided Percutaneous Transthoracic Needle Biopsies Using 10G Large-Core Needles: Initial Experience. Cardiovasc Intervent Radiol 2015;38:1603-10. 10.1007/s00270-015-1098-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen JE, Cascone T, Gerber DE, et al. Targeted therapies for lung cancer: clinical experience and novel agents. Cancer J 2011;17:512-27. 10.1097/PPO.0b013e31823e701a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuiper JL, Heideman DA, Thunnissen E, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer 2014;85:19-24. 10.1016/j.lungcan.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 8.Geraghty PR, Kee ST, McFarlane G, et al. CT-guided transthoracic needle aspiration biopsy of pulmonary nodules: needle size and pneumothorax rate. Radiology 2003;229:475-81. 10.1148/radiol.2291020499 [DOI] [PubMed] [Google Scholar]
- 9.Westcott JL. Percutaneous transthoracic needle biopsy. Radiology 1988;169:593-601. 10.1148/radiology.169.3.3055026 [DOI] [PubMed] [Google Scholar]
- 10.Perlmutt LM, Johnston WW, Dunnick NR. Percutaneous transthoracic needle aspiration: a review. AJR Am J Roentgenol 1989;152:451-5. 10.2214/ajr.152.3.451 [DOI] [PubMed] [Google Scholar]
- 11.Sinner WN. Complications of percutaneous transthoracic needle aspiration biopsy. Acta Radiol Diagn (Stockh) 1976;17:813-28. 10.1177/028418517601700609 [DOI] [PubMed] [Google Scholar]
- 12.Herman PG, Hessel SJ. The diagnostic accuracy and complications of closed lung biopsies. Radiology 1977;125:11-4. 10.1148/125.1.11 [DOI] [PubMed] [Google Scholar]
- 13.Lee C, Guichet PL, Abtin F. Percutaneous Lung Biopsy in the Molecular Profiling Era: A Survey of Current Practices. J Thorac Imaging 2017;32:63-7. 10.1097/RTI.0000000000000237 [DOI] [PubMed] [Google Scholar]
- 14.Wehrschuetz M, Wehrschuetz E, Portugaller HR. Number of biopsies in diagnosing pulmonary nodules. Clin Med Insights Circ Respir Pulm Med 2010;4:9-14. 10.4137/CCRPM.S4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim C, Lee KY, Kim YK, et al. CT-guided core biopsy of malignant lung lesions: how many needle passes are needed? J Med Imaging Radiat Oncol 2013;57:652-6. 10.1111/1754-9485.12054 [DOI] [PubMed] [Google Scholar]
- 16.Cheung YC, Chang JW, Hsieh JJ, et al. Adequacy and complications of computed tomography-guided core needle biopsy on non-small cell lung cancers for epidermal growth factor receptor mutations demonstration: 18-gauge or 20-gauge biopsy needle. Lung Cancer 2010;67:166-9. 10.1016/j.lungcan.2009.04.007 [DOI] [PubMed] [Google Scholar]
- 17.Tam AL, Kim ES, Lee JJ, et al. Feasibility of image-guided transthoracic core-needle biopsy in the BATTLE lung trial. J Thorac Oncol 2013;8:436-42. 10.1097/JTO.0b013e318287c91e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh AK, Leeman J, Shankar S, et al. Core biopsy with curved needle technique. AJR Am J Roentgenol 2008;191:1745-50. 10.2214/AJR.08.1165 [DOI] [PubMed] [Google Scholar]
- 19.Williams SM, Gray W, Gleeson FV. Macroscopic assessment of pulmonary fine needle aspiration biopsies: correlation with cytological diagnostic yield. Br J Radiol 2002;75:28-30. 10.1259/bjr.75.889.750028 [DOI] [PubMed] [Google Scholar]
- 20.Kim TJ, Park CK, Yeo CD, et al. Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non-small-cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage. J Surg Oncol 2014;110:245-51. 10.1002/jso.23646 [DOI] [PubMed] [Google Scholar]
- 21.Thunnissen E, Bubendorf L, Dietel M, et al. EML4-ALK testing in non-small cell carcinomas of the lung: a review with recommendations. Virchows Arch 2012;461:245-57. 10.1007/s00428-012-1281-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. 10.1097/JTO.0b013e318290868f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Cancer Institute NIoH, US Department of Health and Human Services. Common Terminology Criteria for Adverse Events (CTCAE) (version 5.0). NIH publication 09-7473, Bethesda, MD, National Cancer Institute. 2009. Available online: https://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/CTCAE_v5_Quick_Reference_8.5x11.pdf
- 24.Gupta S, Wallace MJ, Cardella JF, et al. Quality improvement guidelines for percutaneous needle biopsy. J Vasc Interv Radiol 2010;21:969-75. 10.1016/j.jvir.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 25.Tsou MH, Tsai SF, Chan KY, et al. CT-guided needle biopsy: value of on-site cytopathologic evaluation of core specimen touch preparations. J Vasc Interv Radiol 2009;20:71-6. 10.1016/j.jvir.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 26.Yang W, Sun W, Li Q, et al. Diagnostic Accuracy of CT-Guided Transthoracic Needle Biopsy for Solitary Pulmonary Nodules. PLoS One 2015;10:e0131373. 10.1371/journal.pone.0131373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Shi L, Xiao Z, et al. Coaxial technique-promoted diagnostic accuracy of CT-guided percutaneous cutting needle biopsy for small and deep lung lesions. PLoS One 2018;13:e0192920. 10.1371/journal.pone.0192920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winokur RS, Pua BB, Sullivan BW, et al. Percutaneous lung biopsy: technique, efficacy, and complications. Semin Intervent Radiol 2013;30:121-7. 10.1055/s-0033-1342952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon SB, Zakowski MF, Pao W, et al. Core needle lung biopsy specimens: adequacy for EGFR and KRAS mutational analysis. AJR Am J Roentgenol 2010;194:266-9. 10.2214/AJR.09.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferretti GR, Busser B, de Fraipont F, et al. Adequacy of CT-guided biopsies with histomolecular subtyping of pulmonary adenocarcinomas: influence of ATS/ERS/IASLC guidelines. Lung Cancer 2013;82:69-75. 10.1016/j.lungcan.2013.07.010 [DOI] [PubMed] [Google Scholar]
- 31.Heerink WJ, de Bock GH, de Jonge GJ, et al. Complication rates of CT-guided transthoracic lung biopsy: meta-analysis. Eur Radiol 2017;27:138-48. 10.1007/s00330-016-4357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. 10.1148/radiol.2017161659 [DOI] [PubMed] [Google Scholar]
- 33.Kim H, Park CM, Koh JM, et al. Pulmonary subsolid nodules: what radiologists need to know about the imaging features and management strategy. Diagn Interv Radiol 2014;20:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]