Abstract
A common goal in olfaction research is modeling the link between odorant structure and odor perception. Such modeling efforts require large data sets on olfactory perception, yet only a few of these are publicly and freely available. Given that individual odor perception may be informative on personal makeup and interpersonal relationships, we hypothesized that people would gladly provide olfactory perceptual estimates in the context of an odor-based social network. We developed a web-based infrastructure for such a network we called SmellSpace and distributed 10 000 scratch-and-sniff registration booklets each containing a subset of 12 out of 35 microencapsulated odorants. Within ~100 days, we obtained data from ~1000 participants who rated the odorants along 13 verbal descriptors. To verify that these estimates are comparable to lab-collected estimates we tested 26 participants in a controlled lab setting using the same odorants and descriptors. We observed remarkably high overall group correlations between lab and SmellSpace data, implying that this method provides for credible group-representations of odorants. We further estimated the usability of the data by applying to it two previously published models that used odorant structure alone to predict either odorant pleasantness or pairwise odorant perceptual similarity. We observed statistically significant predictions in both cases, thus further implying that the current data may be helpful toward future efforts of modeling olfactory perception from structure. We conclude that an odor-based social network is a potentially useful instrument for collecting extensive data on olfactory perception and here post the complete raw data set from the first ~1000 participants.
Keywords: odor perception, odorant descriptors, odorant pleasantness, odorant similarity
Introduction
Predicting odorant perception from odorant structure is a major goal in olfaction research. To this end, there is a need for large databases of olfactory perception, yet only few of these are publicly and freely available. The largest is the National Geographic Survey on Smell collected by Wysocki and colleagues (Wysocki and Gilbert 1989). Here ~1.4 million individuals provided detection and identification scores and intensity and pleasantness ratings for 6 odorants. This data has not been publicly posted, but it is available by request. Two additional data sets are available in books. One is the Arctander data (Arctander 1969) that contains perceptual estimates made by one perfumer who rated 3102 odorants and flavor chemicals, and the second is the Atlas of Odor Character Profiles amassed by Andrew Dravnieks and colleagues (Dravnieks 1985). The Dravnieks atlas includes average ratings from ~150 participants who rated 138 monomolecules and 16 mixtures/oils across 146 verbal descriptors. Finally, several recent large-scale efforts orchestrated by Keller, Vosshall, and colleagues, including one that provided ratings from 49 participants who rated 476 odorants across 19 descriptors, have been posted and are available for download (Keller et al. 2012, 2017; Keller and Vosshall 2016). In addition to the above, there are several sources that contain information on many odorants, but at lower resolution, that is, with only one or very few descriptors per odorant and without a numerical scale of applicability. These include the Good Scents Company webpage (http://www.thegoodscentscompany.com/index.html), the Sigma-Aldrich Flavors and Fragrances catalog (Sigma-Aldrich 2016), and various costly software (such as Flavor-Base by Leffingwell & Associates). Each of these describe odorants using one or a handful of primary descriptors but with minimal information on how they were derived. In light of the above, any addition of publicly available large data on olfactory perception may aid in the field effort of modeling perception from structure.
A challenge in generating large perceptual data sets is in how to convince individuals to volunteer. One option is to rely on participants’ scientific curiosity or good will. This was indeed sufficient drive for ~1.4 million participants of the 1986 National Geographic Survey on Smell, but relying on good will alone obviously restricts the extent of time commitment one can expect, or in turn the number of ratings one can obtain. An obvious alternative to good will is financial reward. This has indeed been the path of recent efforts, yet it too has limitations. A known concern associated with the financial reward model is low motivation. Participants who return to lab to provide hundreds upon hundreds of olfactory estimates find it hard to remain motivated, and this can hamper the quality of data. Of course, one can financially incentivize performance, but many tasks, such as rating odorants along verbal descriptors, have no performance element to them (one can estimate consistency, but one cannot be more or less “correct” at subjective ratings). Given these considerations, we put our minds to thinking on how could we convince many people to provide us with their olfactory perceptions, ideally again and again. We identified 2 primary motivations on which we could capitalize: first, individuals gladly invest time in online self-tests, such as personality tests, presumably because this information can serve in building one’s own “narrative psychology” (Dweck 2013). Second, social rewards such as meeting new friends can outweigh monetary rewards (Heyman and Ariely 2004). We therefore hypothesized that if we could satisfy these 2 driving forces using odor perceptions, we would have in hand a powerful tool for data collection. Regarding learning things about oneself, we know that olfactory perception is shaped by both culture (Majid and Levinson 2011) and genetic makeup (Mainland et al. 2014b). Therefore, it is not unlikely that olfactory perception may be linked to individual aspects such as personality. Regarding interactions with others, we know that olfactory perceptions and preferences are linked to human mate selection (Wedekind et al. 1995; Lundström and Jones-Gotman 2009). Therefore, it is not unlikely that olfactory perception may be related to interpersonal interactions. With these hypotheses in mind, we estimated that if we told potential participants that if they give us their olfactory perceptions, we in return could potentially tell them something meaningful about themselves and introduce them to people they may like; we would then obtain quality data. Notably, the current manuscript is not about testing these 2 hypotheses (namely that olfactory preferences predict personal traits and interpersonal relationships), nor do we here make these claims. Here, we merely use these hypotheses as selling points to convince participants to join in a smell-based social network we called SmellSpace (www.smellspace.com). In SmellSpace, participants provided perceptual estimates of odorants and in return received predictions on their personality (with no claims as to the validity of these predictions) and connections with people who smell the world the way they do (with minimal claims as to the value of these matches). This approach allowed us to collect data from ~1000 participants within a relatively short period of time, and this data is made available here in full for use by the community. Such data can be applied to building and/or testing models that link odorant structure to odorant perception.
Materials and methods
All procedures described in this manuscript are consistent with the Declaration of Helsinki for medical research involving human subjects and were approved by the Weizmann Institute of Science Institutional Review Board (IRB). All participants provided online informed consent to participate.
Choosing odorants and descriptors
The combination of the number of odorants and descriptors we used, and their identity, was based on various constraints and ensuing compromises. There is an inherent conflict between the scientific goal of SmellSpace, which is to obtain data on as many odorants and descriptors as possible, versus the participant-related goal of SmellSpace, which is to relate participants to one another. This latter goal relies on our ability to characterize an individual’s olfactory perception or what we call an olfactory perceptual fingerprint (Secundo et al. 2015). Such fingerprints are most efficiently compared across individuals when based on a fixed common set of odorants and descriptors (Secundo et al. 2015). Thus, whereas the scientific goals are to test as many different odorants and descriptors as possible, the social network goals call for a fixed set of odorants and descriptors common across all participants. As a compromise, we opted for a “Fixed Set” (from here on) of odorants and descriptors that will indeed be common to all participants of SmellSpace in order to serve the social network functions and an added “Rotator Set” (from here on) that will each be delivered to a subset of participants and serve to build the growing scientific database. To determine the size of the “Fixed Set,” we go to Figure 4A in Secundo et al. 2015 and observe that 7 ± 3 odorants and 11 descriptors is the smallest number that still allows generation of an olfactory perceptual fingerprint that is individually stable. Thus, we selected 10 odorants that span physicochemical olfactory space (Figure 1A; Table 1) and the 11 descriptors (out of the ~60 tested in Secundo et al. 2015) that provided for the most stable olfactory perceptual fingerprint. In other words, if we generate an individual olfactory perceptual fingerprint for the same person who smells the same odorants day after day, this set of descriptors provides the most stable outcome. The odorants are the first 10 listed in Table 1, and the descriptors are: “Pleasantness”; “Intensity”; “Garlicky”; “Sweet”; “Fruity”; “Chemical”; “Bitter”; “Burnt”; “Spicy”; “Clean-Dirty”; “Fresh-Rotten.” Given that we want to limit participation effort to ~15 min, the Fixed Set takes up most of this time, allowing us to add a Rotator Set of only 2 odorants and 2 descriptors per participant. This is indeed a modest added contribution from each person, but it will add up over time. In this initial run of SmellSpace that included 10 000 printed odorant booklets, we introduced 25 “Rotator” odorants (Table 1; Figure 1A) at 800 copies each (other than 2 that were at 400 copies, see Table 1) and 2 “Rotator” descriptors (“Volatile”; “Develops Over Time”) that were used in the entire run. We would like to reiterate that we are not claiming anything special about these rotating odorants or descriptors. Whereas the fixed sets were determined theoretically (odorants spanning space and descriptors optimizing perceptual fingerprints), the rotators reflect an initial implementation of the goal of SmellSpace, namely to collect new data on descriptors and odorants that have not been matched before. Using SmellSpace, we will be able to estimate the value of such novel combinations.
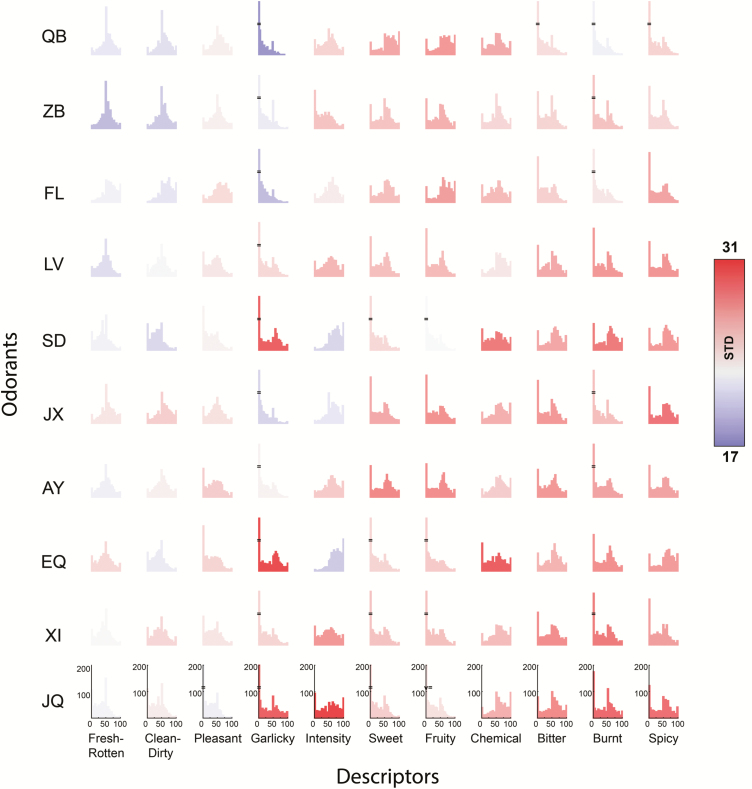
Figure 4.
Descriptors were variably applied across odorants. Histograms for each of the 11 fixed descriptors as they were applied to 10 fixed odorants. Columns arranged according to increasing average column variance (from left to right) and rows according to increasing average row variance in increasing order from top to bottom (the standard deviation is color coded). In other words, “Spicy” and “Fresh-Rotten” were the most and least variably applied descriptors, respectively, and JQ and QB were the most and least variably perceived odorants, respectively.
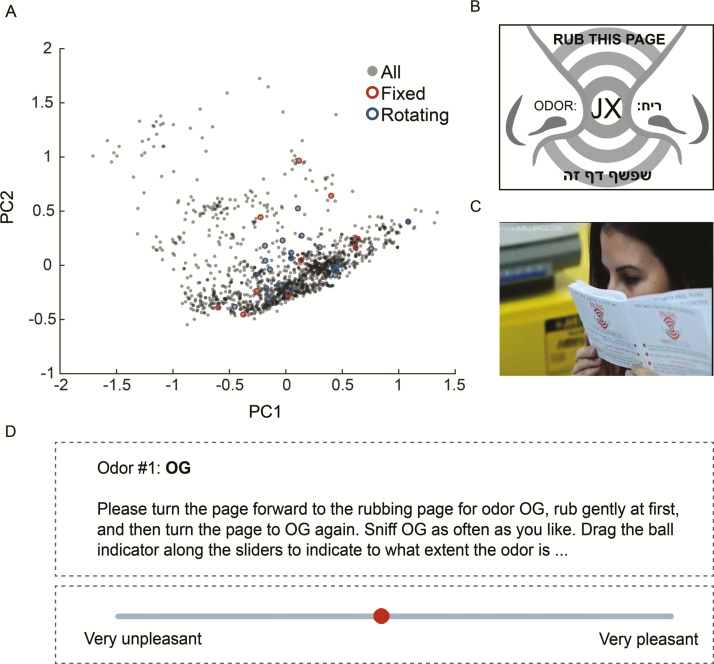
Figure 1.
Odorants were distributed by Scratch and Sniff booklets. (A) The monomolecules used in the study: 10 fixed odorants and 13 rotators (the remaining 12 odorants were mixtures) depicted within the first and second principal components of a representative physicochemical space containing ~1500 odorant molecules. (B) An odorant page from within the booklet. The microencapsulated odorant (in this case; JX) was printed on the entire page. (C) A participant sniffing the booklet. (D) Example of the VAS scale in the webpage.
Table 1.
The odorants used
| Concentration | |||||||
|---|---|---|---|---|---|---|---|
| Odor code | Name | CAS number | (V/V in IPM) | CID | Number of subjects | Intensity ratings | |
| 1 | JX | Laevo-fenchone | 7787-20-4 | 7.60% | 14525 | 985 | 62.6305 |
| 2 | QB | Isoamyl acetate | 123-92-2 | 25.00% | 31276 | 985 | 48.0234 |
| 3 | SD | 3-Propylidene phthalide | 17369-59-4 | 7.90% | 5373603 | 985 | 71.9421 |
| 4 | EQ | Cuminaldehyde | 122-03-2 | 13.60% | 326 | 985 | 74.0538 |
| 5 | AY | Strawberry glycidate 1 (aldehyde C-16) | 77-83-8 | 100.00% | 6501 | 985 | 56.8213 |
| 6 | XI | Nonanal (aldehyde C-9) | 124-19-6 | 100.00% | 31289 | 985 | 48.5157 |
| 7 | FL | Citral | 5392-40-5 | 100.00% | 638011 | 985 | 56.7553 |
| 8 | JQ | Skatole | 83-34-1 | 0.75% | 6736 | 985 | 53.0751 |
| 9 | ZB | Hexanol | 111-27-3 | 30.00% | 8103 | 985 | 33.9107 |
| 10 | LV | 6-Methylquinoline | 91-62-3 | 10.70% | 7059 | 985 | 51.3553 |
| 11 | KO | Musk | 1222-05-5 | 100.00% | 91497 | 65 | 46.3846 |
| 12 | JT | (Mixture) [XI, LV, SD, EQ] | 65 | 78.6154 | |||
| 13 | UV | Damascenone | 23696-85-7 | 10.00% | 5366074 | 53 | 56.8491 |
| 14 | OS | (Mixture) [EQ, ZB, QB, JX] | 53 | 71.6981 | |||
| 15 | SS | ISO E Super | 54464-57-2 | 100.00% | 108242 | 34 | 54.4412 |
| 16 | WZ | (Mixture) [JX, JQ, LV, EQ] | 34 | 75.0294 | |||
| 17 | LB | Iralia pure | 1335-46-2 | 100.00% | 5371084 | 54 | 70.2778 |
| 18 | RZ | (Mixture) [JQ, QB, LV, XI] | 54 | 61.0741 | |||
| 19 | VY | (Mixture) [JX, SD, FL, LV] | 66 | 74.303 | |||
| 20 | MH | Oxane | 59323-76-1 | 10.00% | 101010 | 78 | 82.5641 |
| 21 | SF | (Mixture) [XI, ZB, FL, JX] | 78 | 62.359 | |||
| 22 | ZI | Damscone alpha | 24720-09-0 | 10.00% | 5366077 | 94 | 74.7766 |
| 23 | WV | (Mixture) [JQ, SD, FL, LV, QB, XI, EQ, ZB, JX, AY] | 128 | 78.4922 | |||
| 24 | FE | 4-(2,6,6-trimethylcyclohexen-1-yl)but-3-en-2-one | 8013-90-9 | 26955 | 102 | 64.0294 | |
| 25 | UB | Isoamyl phenyl acetate | 102-19-2 | 100.00% | 7600 | 102 | 68.4804 |
| 26 | NB | Norlimbanol | 70788-30-6 | 25.00% | 116699 | 119 | 76.3697 |
| 27 | TJ | Thiogeraniol | 38237-00-2 | 25.00% | 6365572 | 98 | 79.6224 |
| 28 | FM | Clementine aldehide | 20407-84-5 | 5283361 | 66 | 74.7424 | |
| 29 | IF | Aldehide C7 | 111-71-7 | 10.00% | 8130 | 155 | 48.7806 |
| 30 | HG | Galaxolide | 1222-05-5 | 70.00% | 91497 | 33 | 48.9394 |
| 31 | OA | (Mixture) [ZB, JQ, QB, JX] | 119 | 67.8151 | |||
| 32 | EK | (Mixture) [EQ, SD, QB, ZB] | 98 | 77.2551 | |||
| 33 | AS | (Mixture) [XI, LV, SD, ZB] | 155 | 76.1677 | |||
| 34 | BA | (Mixture) [XI, QB, JQ, EQ] | 33 | 75.9091 | |||
| 35 | NE | IPM pure (the diluent) | 110-27-0 | 34 | 37.9412 |
All odorants used, the first 10 are the fixed odorants and the remaining are rotators, columns as follows: odorant number; 2-letter odorant code; odorant common name; odorant CAS number; odorant dilution volume-by-volume in IPM before encapsulation; compound identification number (CID) identification number; the number of subjects that rated the odorant; the average odorant intensity rating. Note that odorants KO and HG are the same molecular species, KO supplied by Sigma-Aldrich and HG by DreamAir. The mixtures are equal combinations of their components without added dilution. We printed 13 booklet types, each containing the 10 fixed odorants and 2 of the rotators. The 13 rotator pairs were: “KO+JT”; “UV+OS”; NB+OA”; “TJ+EK”; “IF+AS”; “HG+BA”; “SS+WZ”; “LB+RZ”; “FM+VY”; “MH+SF”; “ZI+WV”; “FE+UB”; “NE+WV”. We printed 800 copies of each type but the last, (“NE+WV”) of which we printed 400.
The odorant booklets
In order to deliver uniform odorants to participants at different locations, we opted for scratch-and-sniff (S&S) technology. In brief, S&S uses microencapsulation to deposit encapsulated odorant oils onto printed matter. Once the microcapsules are insulted, for example by scratching, they release the odorant. This method, originally developed by the 3M corporation in the 1960s, has been used in applications related to publishing, advertising, games, toys, and more but is perhaps most known to the field of olfaction research through 2 sources: The first is the already described National Geographic Survey on Smell (Wysocki and Gilbert 1989). The second is the University of Pennsylvania Smell Identification test or UPSIT (Doty et al. 1984). The UPSIT consists of 40 microencapsulated odorants presented to users in the context of a 4-alternative forced-choice discrimination task. The UPSIT became a widely used standard olfactory test, used in clinics to characterize olfactory impairments associated with many diseases and conditions (e.g., Doty et al. 1993). We opted for this method because of its robustness. We printed 10 000 SmellSpace booklets, each containing 12 odorants, 1 on each 10 by 15 cm page (Figure 1B,C; Supplementary Video 1, also at: https://youtu.be/s0IR_cEIpMc). Each odorant page was followed by a blank page that can be used as a pad to scratch the previous page, thus minimizing cross contamination by the user’s hand. Instructions for use were demonstrated in an online video that was made available to users through SmellSpace and can also be seen here in Supplementary Video 1. The odorant oils were prepared in part by us (all monomolecules from Sigma-Aldrich) and in part by a fragrance house (DreamAir). All odorants were diluted using Isopropyl myristate (IPM) (chemical abstracts service [CAS]#: 110-27-0). Dilutions were shipped to a commercial microencapsulation printing company (The Aroma Company) who deposited them onto the printed booklets. In lab, IPM is odorless. To estimate any contaminants associated with the printing process, we included IPM alone as one of the rotator odorants (code NE, Table 1).
The website
We built SmellSpace using Drupal, an open source content management system (www.drupal.org). SmellSpace is currently in Hebrew and English, but the Drupal structure allows translating into additional languages with relative ease. Participants first registered at the login page by providing age and gender alone. Following registration, participants could complete the odorant booklet. This consisted of a process whereby we first verified participant authenticity (human as opposed to bot) by prompting for a unique code that was printed onto the cover of each booklet. Once the code was validated against a list, the participant was guided through the odorant pages and instructed to use a visual analogue scale (VAS) in order to rate each odorant along the 13 verbal descriptors (Figure 1D). In cases where a descriptor has clear separate terms for its opposite extremes, these terms were placed at each end of the VAS. For example, a VAS ranging from “Fresh” to “Rotten” or “Clean” to “Dirty.” Alternatively, other VAS scales had one descriptor, for example, smells like “Garlic” and ranged from “Not at all” to “Very.” After the participant completed rating all odorants, he/she became a member of SmellSpace. This allowed them to navigate through various SmellSpace features such as “Nose Yourself” where the user can observe personality estimates modeled on their olfactory fingerprint (again, with clear disclaimers as to their validity) or “Nose Your Friends” where SmellSpace matches individuals based on their olfactory fingerprints (again, with clear disclaimers as to the value of such matches).
Results
We recruited ~1000 participants in ~100 days
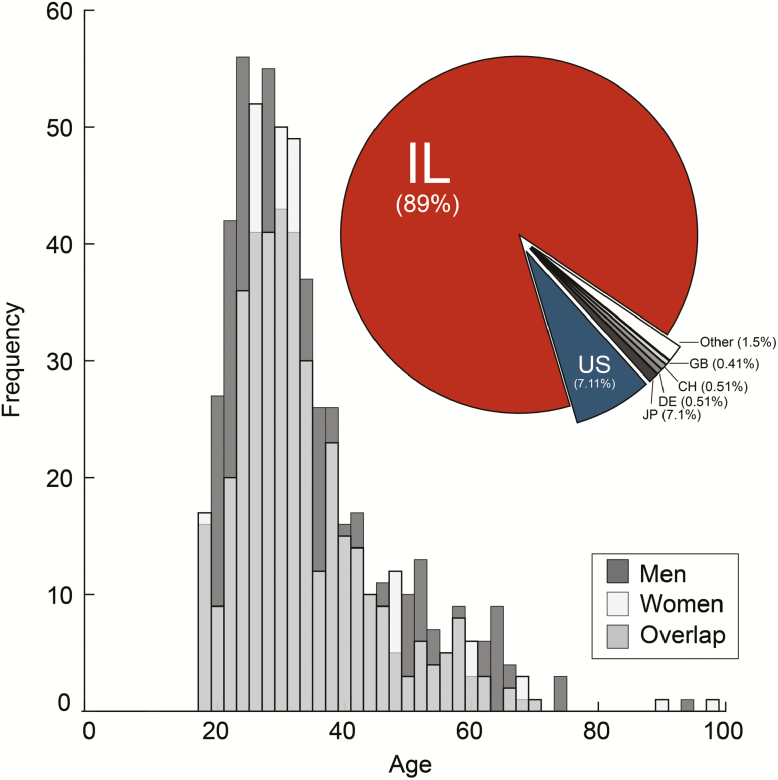
We distributed a first run of 10 000 booklets by various means, including distribution at public lectures and distribution together with a municipal newspaper. Moreover, participants could (and still can) register online and ask that a booklet be mailed to them, thus providing for some international distribution as well. Within ~100 days, 985 participants completed the odorant rating task. The participants were 442 men and 542 women (gender data for one participant was corrupted), their reported age ranged from 10 to 98 with an average of 34.12 (Figure 2), and whereas 849 participants were from Israel, 136 were from 19 other countries (Figure 2 inlay). We refrain from further analyzing data from the 10-year-old participant and one for whom we do not have age data, retaining a total of 983 adult participants for further analysis. All this data is available in Supplementary Data File 1. To gather a sense of the data obtained through SmellSpace, we first provide some descriptive statistics, concentrating on the 10 fixed odorants that were rated by all participants.
Figure 2.
One thousand participants within 100 days. A histogram depicting age and gender of the participants. The insert pie chart denotes country of origin: IL = Israel; US = USA; JP = Japan; DE = Germany; CH = Switzerland; GB = Great Britain.
Odorant intensity varied slightly across 100 days
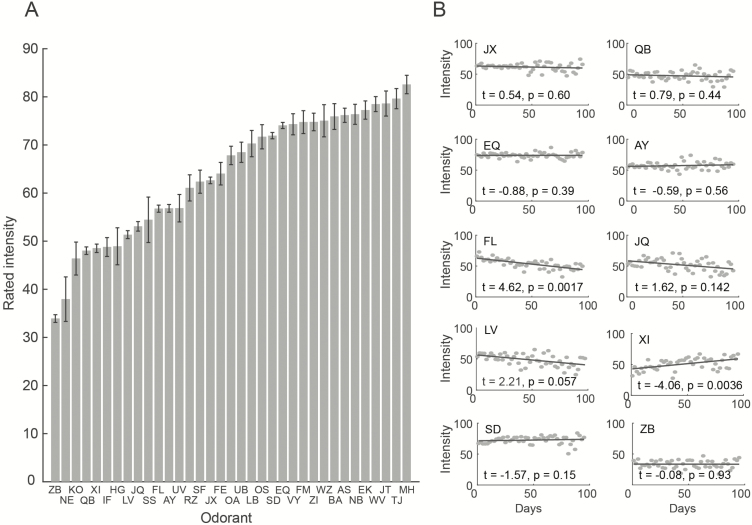
A potential limitation of microencapsulation is that the odorants can leak over time. Thus, we examined both absolute intensity and intensity over time. We plotted the perceived intensity of the fixed odorants over the first 100 days. An analysis of variance (ANOVA) on intensity estimates with conditions of Odorant and Time (binned every 10 days) revealed a main effect of Time (F(9,9829) = 35.93, P = 2.17e-8), a main effect of Odorant (F(9,9) = 173.43, P = 2.7e-306), and a significant interaction (F(81) = 2.79, P = 1.602e-15). The main effect of Odorant reflected odorant-specific intensity differences (Figures 3A and 4; Table 1), whereby odorant ZB stood out as the lowest intensity odorant, not significantly different from the diluent alone (intensity NE [diluent] = 37.94 ± 26.99, ZB = 33.91 ± 25.40, t = −0.9, P = 0.35) (throughout the manuscript reported variability is ±standard deviation in the text and ±standard error in the figures). All other odorants were significantly more intense than the diluent (all t > 2.2, all P < 0.02). The main effect of Time revealed an overall average minor −1.7 ± 9.98% change in perceived intensity from the first to last 10-day time-bin, and the interaction reflected that this change was significant for only 2 odorants, odorant XI with a 17.7% increase (P = 0.0036), and odorant FL with a 16.2% drop (P = 0.0017) but not in any other odorant (Figure 3B). Whereas the reduced intensity of FL over time may reflect leaking, we have no explanation for the increase in perceived intensity of XI. Given that ZB, Hexanol (CAS# 111-27-3), is typically characterized as a perceptible odorant (e.g., “ethereal fusel oil fruity alcoholic sweet green” in the Good Scents data), we speculate that its odd profile in our data (~160 participants didn’t smell it at all) reflects failed encapsulation of this molecule (more on this in the discussion).
Figure 3.
The odorants varied in perceived intensity. (A) Mean and standard error of perceived intensity for all 35 odorants used, rank-ordered by intensity. (B) Perceived intensity of the 10 fixed set odorants across 100 days of distribution (day 0 is the first day of distribution). Each point is the average for that day. The t and p values reflect a two-tailed paired t-test between the first 10 days and last 10 days.
Descriptor application was nonnormally distributed across odorants
Next, we examined the application of each descriptor for each odorant across subjects and observe that none of them were normally distributed (Kolmogorov–Smirnov, all ksstat > 0.0481, all P < 0.02) (Figure 4). We consider this an important observation given that with ~1000 observations in hand, skewed distributions imply meaningful descriptors-to-odorant pairings. Moreover, a 2-way ANOVA with conditions of Odorant (1 through 9) and Descriptor (1 through 11) yielded main effects of Odorant and Descriptor and an interaction of the two, all with F and P values beyond machine precision (i.e., infinite F and P = 0). From this, we conclude that this set of descriptors is indeed variably applied to this set of odorants (Figure 4).
The group estimates obtained through SmellSpace are comparable to group estimates obtained in a controlled lab setting
A potential concern associated with remote testing is the quality of the data. Are such estimates similar to estimates obtained in a typical lab setting? Data quality can be estimated on different fronts: One is data quality per participant, that is, is a participant consistent with him/herself? To estimate this, one needs test–retest measures, which were not part of SmellSpace. An alternative estimate is the quality of data obtained per odorant or per descriptor from the entire group. That is, does a given odorant take on a stable and unique representation and a given descriptor take on a stable and unique pattern of application? This can be estimated by comparing the average ratings across groups of individuals, something that we can do in SmellSpace. With these considerations in mind, we set out on the following path: We first tested and then retested 26 participants (13F, mean age = 29.8 ± 6.19) in a lab experiment using the same web-interface and the same “Fixed Set” of 10 odorants but delivered from sniff-jar rather than scratch-and-sniff source. Here, experiments were conducted in well-controlled rooms specially designed for olfaction psychophysics experiments, subserved by HEPA and Carbon filtration and coated in stainless steel to prevent odor contamination. Each participant completed the entire experiment twice, day after day. Participants were payed for participation. All this data is available in Supplementary Data File 2. This data set will allow us to first determine whether the 26 lab participants are “good” participants (repeatability). Once we verify this, we can then compare the group ratings of these verified participants to equivalent-sized groups from SmellSpace in order to estimate whether the group data we obtain per odorant and per descriptor in SmellSpace is comparable to the group data we obtained from a group of verified participants.
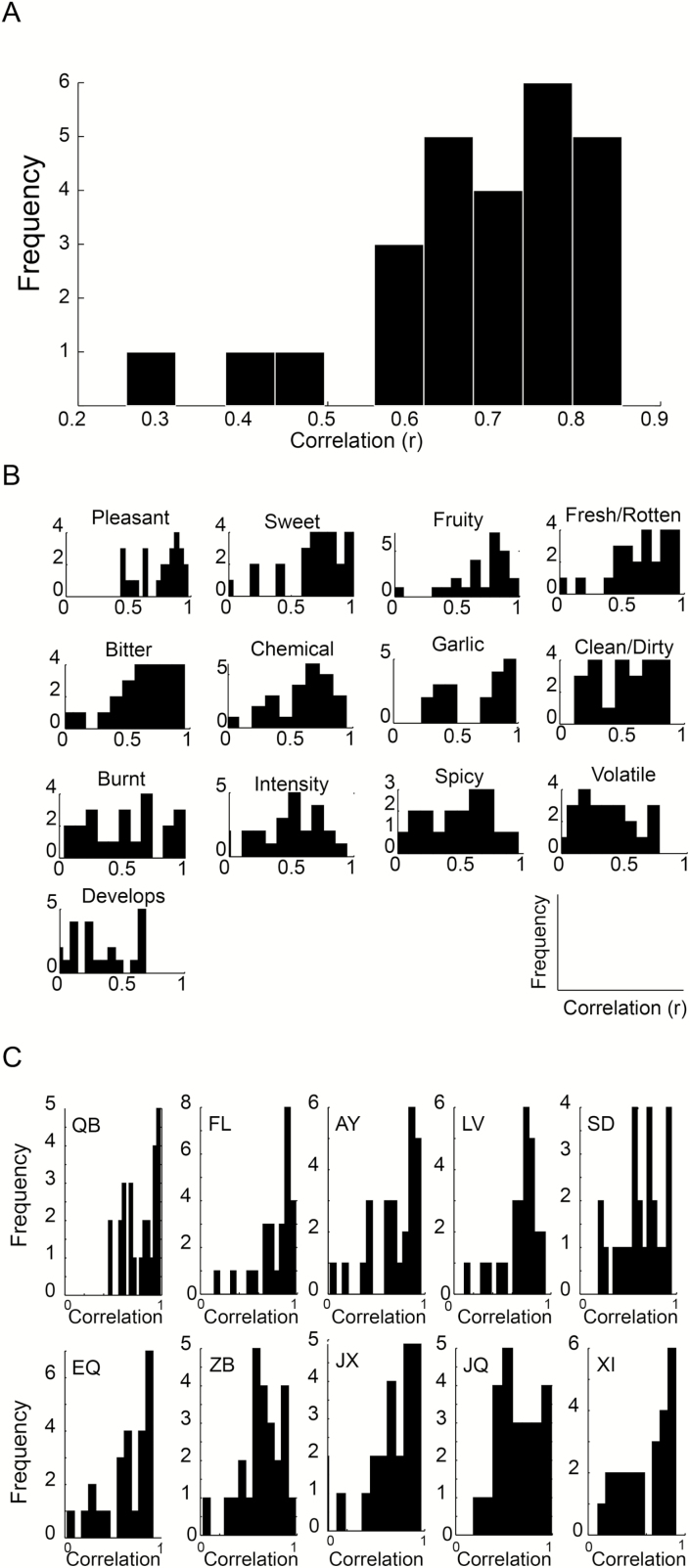
To ask whether the lab participants are “good” participants, we first looked at the entire fixed set (i.e., a vector made of 10 fixed odorants, each rated on 11 fixed descriptors). We observe that the average correlation across days was r = 0.68 ± 0.14, P < 0.0001 (Figure 5A). Looking separately at each descriptor across odorants, we observe the following 11 correlations for fixed descriptors across days: Pleasantness r = 0.75 ± 0.17, P = 0.012; Sweet r = 0.68 ± 0.25, P = 0.03; Fruity r = 0.66 ± 0.27, P = 0.04; Fresh/Rotten r = 0.61 ± 0.29, P = 0.06; Bitter r = 0.56 ± 0.36, P = 0.09; Chemical r = 0.56 ± 0.34, P = 0.09; Garlic r = 0.51 ± 0.42, P = 0.13; Clean/Dirty r = 0.43 ± 0.38, P = 0.21; Burnt r = 0.43 ± 0.37, P = 0.21; Intensity r = 0.41 ± 0.34, P = 0.23; Spicy r = 0.26 ± 0.4, P = 0.46 (Figure 5B) and the following 2 correlations for the rotating descriptors across days: Volatile r = 0.3 ± 0.29, P = 0.4; Develops over time r = 0.24 ± 0.29, P = 0.41 (Figure 5B). Looking separately at each odorant across fixed descriptors, we observe the following 10 correlations across days: odorant QB r = 0.77 ± 0.16, P = 0.002; FL r = 0.77 ± 0.2, P = 0.002; AY r = 0.69 ± 0.25, P = 0.009; LV r = 0.68 ± 0.19, P = 0.010; SD r = 0.63 ± 0.21, P = 0.02; EQ r = 0.61 ± 0.29, P = 0.026; ZB r = 0.60 ± 0.25, P = 0.03; JX r = 0.57 ± 0.37, P = 0.04; JQ r = 0.57 ± 0.34, P = 0.041; XI r = 0.54 ± 0.31, P = 0.056 (Figure 5C). An average overall day-after-day correlation of r = 0.68 and r = 0.75 for pleasantness is consistent with previous observations (Secundo et al. 2015) and allows us to conclude that this is a “good” lab cohort. Indeed, only 3 participants tended to be less consistent than the others (Figure 5A). However, given that even the least consistent participant was still significantly correlated with him/herself day-after-day (r = 0.25, P = 0.0065), we retained these participants.
Figure 5.
Individual lab participants provided consistent ratings. (A) A histogram reflecting the frequencies of day-after-day correlations on the entire fixed set (a 110 unit vector of 11 odorants along 10 descriptors). (B) Histograms reflecting the frequencies of day-after-day correlations on the fixed set of 10 odorants across each of the 13 descriptors. (C) Histograms reflecting the frequencies of day-after-day correlations on the fixed set of 11 descriptors across each of the 10 odorants.
Having observed that these participants are generally “good” participants at the individual level, that is, that individual participants provide similar estimates day after day, we next asked how this converges into group estimates. In other words, we asked whether the mean group rating provided by lab participants is stable day-after-day. Looking at the entire fixed set (i.e., a vector made of 10 fixed odorants, each rated on 11 fixed descriptors), we observe that the correlation of the mean vector in day 1 to the mean vector in day 2 was r = 0.97, P < 0.000001. Moreover, we can look at the correlation in each of the 13 descriptors across each of the 10 odorants. We observed the following correlations for the 11 fixed descriptors: Pleasant r = 0.99, P < 0.000001; Sweet r = 0.99, P < 0.000001; Fruity r = 0.98, P = 0.000001; Spicy r = 0.97, P = 0.000003; Fresh/Rotten r = 0.97, P = 0.000003; Bitter r = 0.94, P = 0.000053; Burnt r = 0.94, P = 0.000053; Garlic r = 0.91, P = 0.00025; Chemical r = 0.88, P = 0.0007; Clean/Dirty r = 0.83, P = 0.0029; Intensity r = 0.83, P = 0.0029 and the following correlations for the 2 rotating descriptors: Volatile r = 0.76, P = 0.01; Develops over time r = 0.54, P = 0.1. We observed the following correlations per odorant: FL r = 0.99, P < 0.00001; LV r = 0.98, P < 0.00001; JX r = 0.98, P < 0.00001; QB r = 0.97, P < 0.00001; JQ r = 0.96, P < 0.00001; ZB r = 0.96, P < 0.00001; AY r = 0.95, P < 0.00001; XI r = 0.94, P < 0.00001; SD r = 0.92, P < 0.00001; EQ r = 0.91, P < 0.00001. These remarkable correlations highlight the power and stability that are gained through group data.
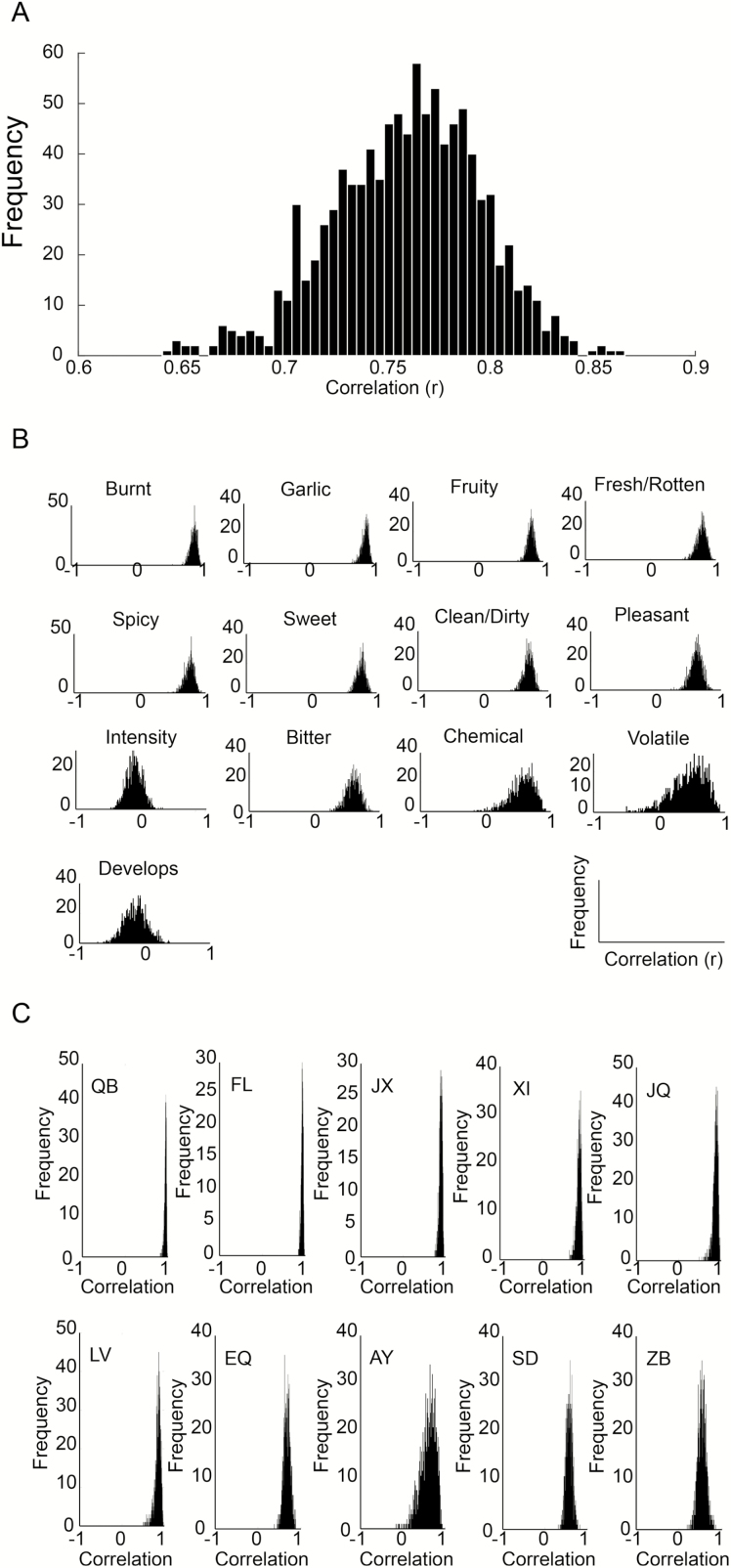
Having observed that this group of lab participants provides consistent odorant group estimates day after day, we next asked whether SmellSpace group estimates are consistent with lab-generated group estimates. We selected from SmellSpace 1000 subgroups of 26 participants each, such that they provided for an exact match in gender and age to the lab cohort (this reflected various recombination of 181 men and 179 women in SmellSpace). We then measured the correlation between lab-generated group estimates (mean across both days) and SmellSpace-generated group estimates. Looking at the entire fixed set (i.e., a vector made of 10 fixed odorants, each rated on 11 fixed descriptors), we observe that the average correlation of the mean lab vector to the mean SmellSpace subgroup vector was r = 0.76 ± 0.033, P < 0.000001 (Figure 6A). Moreover, we can look at the correlation in each of the 13 descriptors across each of the 10 odorants. We observed the following correlations for the 11 fixed descriptors: Burnt r = 0.86 ± 0.05, P = 0.001; Garlic r = 0.86 ± 0.05, P = 0.001; Fruity r = 0.81 ± 0.05, P = 0.004; Fresh/Rotten r = 0.77 ± 0.07, P = 0.009; Spicy r = 0.75 ± 0.07, P = 0.01; Sweet r = 0.75 ± 0.06, P = 0.01; Clean/Dirty r = 0.69 ± 0.07, P = 0.02; Pleasant r = 0.63 ± 0.09, P = 0.05; Bitter r = 0.59 ± 0.11, P = 0.07; Chemical r = 0.58 ± 0.17, P = 0.07; Intensity r = −0.09 ± 0.12, P = 0.8 (Figure 6B) and the following correlations for the 2 rotating descriptors: Volatile r = 0.43 ± 0.26, P = 0.21; Develops over time r = −0.14 ± 0.18, P = 0.69 (Figure 6B). That perceived intensity was uncorrelated is unsurprising given that the lab test was from sniff-jars and SmellSpace was scratch-and-sniff. This lack of correlation renders added value to the remaining perceptual correlations as it implies they reflect genuine odor attributes unrelated to intensity. Without these intensity differences, these correlations may have been even greater. We also observed the following correlations per odorant: QB r = 0.93 ± 0.02, P = 0.000004; FL r = 0.92 ± 0.02, P = 0.000008; JX r = 0.88 ± 0.03, P = 0.00007; XI r = 0.86 ± 0.05, P = 0.00016; JQ r = 0.86 ± 0.05, P = 0.00016; LV r = 0.84 ± 0.07, P = 0.0003; EQ r = 0.677 ± 0.07, P = 0.01; AY r = 0.58 ± 0.17, P = 0.03; SD r = 0.56 ± 0.07, P = 0.04; ZB r = 0.53 ± 0.1, P = 0.062 (Figure 6C). That ZB was the poorest correlated odorant across lab and SmellSpace is consistent with our estimation of failed encapsulation of this odorant.
Figure 6.
SmellSpace group data reflected lab group data. We sampled SmellSpace 1000 times, each time selecting 26 participants age and gender matched to the lab cohort. (A) A histogram reflecting the frequencies of SmellSpace-to-lab correlations on the entire fixed set (a 110 unit vector of 11 odorants along 10 descriptors). (B) Histograms reflecting the frequencies of SmellSpace-to-lab correlations on the fixed set of 10 odorants across each of the 13 descriptors. (C) Histograms reflecting the frequencies of SmellSpace-to-lab correlations on the fixed set of 11 descriptors across each of the 10 odorants.
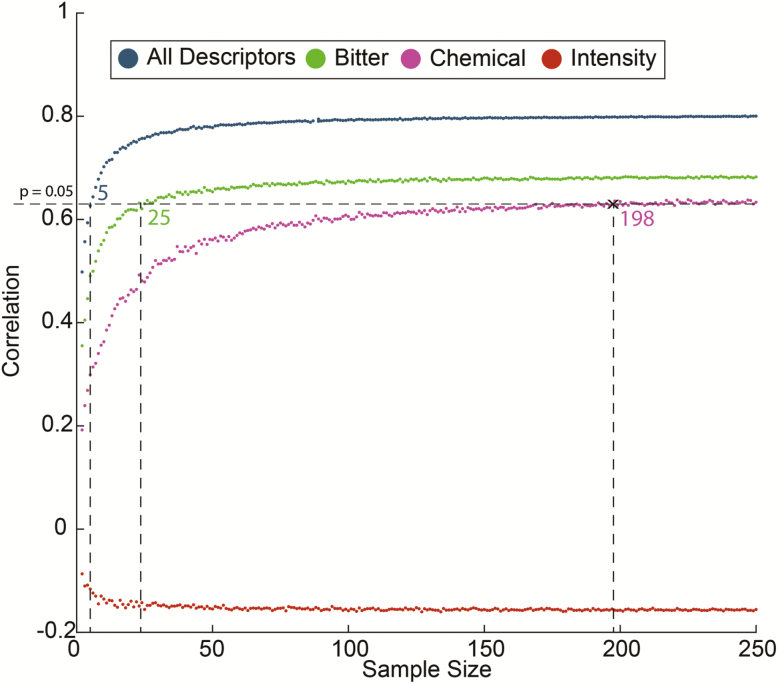
The overall lab vector was highly correlated with the overall SmellSpace vector. However, whereas in lab all 11 fixed descriptors were also each individually highly correlated day-after-day (all r > 0.8, all P < 0.003), 8 of 11 were also significantly correlated between lab and SmellSpace but 3 were not: Intensity r = −0.09 ± 0.12, r = 0.59 ± 0.11, P = 0.07; Bitter r = 0.59 ± 0.11, P = 0.07; Chemical r = 0.58 ± 0.17, P = 0.07. To estimate the dependence of this on the size of the SmellSpace subgroup, we systematically increased the size of the SmellSpace sample. For each sample size, we selected 100 random instances from SmellSpace. Based on all the above observations, we conducted this analysis without odorant ZB. We observe that “Intensity” remains uncorrelated regardless of sample size (Figure 7, red line). For “Bitter,” we observe that merely the removal of ZB was sufficient for obtaining a significant result at 25 participants (Figure 7, green line). For “Chemical,” we observe that only at 198 participants in the group does the SmellSpace rating become significantly correlated with the lab rating (r = 0.64, P < 0.05) (Figure 7, pink line). Thus, we conclude that subgroups of 200 participants from SmellSpace provide for a very strong reflection of what one can expect in a typical lab experiment.
Figure 7.
Correlations with lab data as a function of SmellSpace sample size. We randomly sampled SmellSpace 1000 times for each sample size. For each sample we compared the fixed odorant set vector (without odorant ZB). The dashed line reflects a significant correlation between lab and SmellSpace. At 198 SmellSpace participants, all descriptors but “Intensity” are significantly correlated across lab and SmellSpace.
Predicting odorant pleasantness from odorant structure alone
Having collected the data and verifying that it is comparable to lab-collected data, we next set out to ask whether 2 previous models we generated for linking odorant perception to odorant structure survive testing with this large novel data set.
In Khan et al. (2007), we predicted odorant pleasantness from odorant structure and variations of this prediction have been replicated by others several times (Mandairon et al. 2009; Zarzo 2011; Keller et al. 2017). However, because the manuscript by Khan et al. was focused on the correlation between the principal component of structure and the principal component of perception (pleasantness), its optimized calculation for predicting pleasantness was obscured. Here, we take this opportunity spell out the calculation so that anybody can apply it. Moreover, here, we test the model using novel data.
To predict pleasantness, we first used DRAGON software V6 (Mauri et al. 2006) to obtain 150 previously identified physicochemical features for each odorant (Table 2). Each feature value was multiplied by a previously derived weight (Table 2). These weights also serve to normalize the features that are otherwise of vastly varying scales. To be clear, these 150 features and their weights were previously optimized by us in a previous effort. The 150 values are then summed to produce the pleasantness prediction for each molecule. In other words, we use a weighted linear sum that was previously optimized on different data.
Table 2.
The physicochemical features for predicting pleasantness
| Desc. code | Weight | Desc. code | Weight | Desc. code | Weight |
|---|---|---|---|---|---|
| nSK | 1.262E−04 | SM05_EA(ri) | 6.255E−05 | L3p | −3.57E−06 |
| nCL | −2.980E−06 | SM06_EA(ri) | 4.799E−05 | P2p | −8.92E−06 |
| ZM1 | −9.124E−04 | SM13_EA(ri) | 6.593E−04 | E3p | 1.08E−05 |
| MAXDP | 1.732E−04 | SM11_AEA(bo) | 4.713E−06 | Tu | 2.88E−04 |
| Psi_i_s | 2.826E+00 | SM15_AEA(bo) | −2.647E−04 | Ts | 1.17E−04 |
| SRW08 | 2.014E−04 | SM03_AEA(dm) | −2.160E−04 | Ks | 5.38E−05 |
| SIC0 | 1.247E−05 | Eig15_EA(dm) | 2.592E−05 | Vs | −1.14E−01 |
| CIC5 | −2.021E−05 | Eig06_EA(ri) | −2.532E−04 | HATS0u | 5.42E−05 |
| SpDiam_D | 9.163E−05 | Eig02_AEA(bo) | 6.112E−05 | H7m | 2.78E−07 |
| H_X | −1.603E−04 | Eig06_AEA(bo) | −8.063E−05 | H0v | 5.30E−05 |
| Chi_H2 | 8.301E−05 | Eig11_AEA(bo) | 1.771E−05 | H3v | −9.47E−06 |
| SpDiam_Dt | 7.432E−05 | Mecc | 4.663E−05 | H7v | 1.14E−06 |
| H_D/Dt | −7.089E−02 | SM5_RG | 1.522E−04 | HATS0e | 4.78E−05 |
| EE_D/Dt | 1.570E−04 | SM6_RG | 1.874E−04 | H3p | −1.05E−05 |
| SpAbs_Dz(Z) | −4.421E−04 | VE3_G/D | −6.906E−06 | HATS7p | 8.95E−06 |
| WiA_Dz(e) | 3.562E−05 | VR1_G/D | −2.042E−05 | HATS8p | 7.07E−06 |
| SpAD_Dz(e) | −7.315E−02 | VR3_G/D | −3.026E−05 | HATS2i | 7.90E−05 |
| SpPos_B(m) | 2.513E−04 | TDB08m | −1.927E−06 | R7m+ | −1.08E−07 |
| SpAD_B(m) | 2.149E−04 | TDB07e | 3.715E−05 | R1e | 5.17E−05 |
| SpPosA_B(v) | 5.311E−05 | TDB07p | 5.632E−06 | R8e | 3.85E−05 |
| SM3_B(v) | 1.782E−04 | TDB09s | 6.361E−06 | R8p | 5.84E−06 |
| HyWi_B(p) | 1.113E−04 | TDB02r | 2.296E−05 | R3p+ | 4.05E−07 |
| SpPos_B(p) | 1.357E−04 | RDF155u | −1.171E−05 | R6i | 2.95E−06 |
| SM6_B(p) | 4.491E−04 | RDF035v | 3.212E−05 | DP10 | 6.86E−05 |
| VE1_B(p) | 5.796E−05 | RDF045v | −1.423E−05 | SP15 | −1.90E−04 |
| Chi_B(i) | 2.659E−05 | RDF090v | −1.362E−04 | nCt | −2.12E−04 |
| VE3_B(i) | 7.702E−07 | RDF095e | −6.829E−05 | nR#CH/X | −6.68E−07 |
| ATS5m | 9.451E−05 | RDF085p | −1.584E−04 | nRCHO | 3.14E−05 |
| ATS6v | 9.437E−05 | RDF110p | −2.197E−05 | nArCHO | 4.04E−05 |
| ATS7v | 5.203E−05 | RDF140p | −4.061E−06 | nRC=N | 8.87E−06 |
| ATS6i | 2.640E−04 | RDF015s | 8.325E−01 | nRNR2 | −2.25E−05 |
| ATS8s | 7.139E−05 | RDF080s | 8.029E−05 | nOHp | 8.62E−05 |
| ATSC7i | −9.476E−06 | RDF085s | −3.650E−03 | nHDon | −8.10E−05 |
| MATS3v | 2.435E−05 | Mor13u | 6.520E−06 | C-017 | 1.51E−04 |
| MATS3e | −2.214E−05 | Mor18u | 7.619E−06 | C-026 | −1.85E−05 |
| MATS7s | −1.483E−04 | Mor10m | −6.187E−05 | O-059 | −1.16E−04 |
| GATS4e | 1.629E−05 | Mor17m | 1.195E−05 | SdsCH | 5.53E−04 |
| GATS8i | −3.500E−05 | Mor32m | 9.089E−06 | StsC | −1.69E−06 |
| JGI4 | −1.717E−06 | Mor12v | 6.155E−07 | SdssC | 2.95E−04 |
| JGI5 | 8.027E−06 | Mor27v | −2.076E−06 | NaasC | 6.14E−05 |
| SpMax2_Bh(p) | 1.544E−04 | Mor13e | 1.362E−05 | NsOH | −5.69E−05 |
| SpMax8_Bh(p) | −9.740E−06 | Mor17p | 1.838E−-05 | NaaS | 0.00E+00 |
| SpMin4_Bh(v) | 4.138E−05 | Mor23i | 7.078E−05 | B04[C-S] | −2.11E−05 |
| P_VSA_MR_3 | 3.568E−01 | Mor26i | 2.012E−05 | B10[C-N] | 0.00E+00 |
| P_VSA_m_4 | −4.369E−01 | Mor26s | 5.735E−05 | F03[O-S] | −8.54E−05 |
| P_VSA_v_4 | 5.865E−02 | P1u | 5.349E−05 | F04[C-S] | −2.76E−05 |
| P_VSA_e_3 | −2.517E−01 | L2m | −1.499E−05 | G(O..O) | 2.79E−01 |
| P_VSA_p_2 | −5.653E−01 | G1v | 3.753E−06 | TPSA(NO) | 1.93E−04 |
| P_VSA_p_4 | 5.865E−02 | G2v | 5.278E−05 | Inflammat-80 | −1.96E−06 |
| Eta_betaS | 2.386E−05 | L1e | 2.689E−04 | Infective-80 | −1.16E−04 |
The 150 Dragon features and their associated weighting values that together enable the prediction of odorant pleasantness.
Before applying this calculation, we must restrict our data according to 2 model limitations clearly spelled out in Khan et al: First, the algorithm was tested only for monomolecules and not for mixtures. This restricts us to 23 monomolecules currently in SmellSpace. Second, the pleasantness prediction algorithm works only for odorants equated for perceived intensity. As previously described, the current odorants significantly vary in perceived intensity (Figure 3). To identify a subset of odorants with equal perceived intensity, we applied Hierarchical Clustering to the intensity estimates of the 23 monomolecules we used. This uncovered 3 clusters, 1 containing 1 odorant alone and 2 clusters of 11 odorants each. One of these clusters had a standard deviation of 6.08 and the other had a significantly lower standard deviation of 3.94 and was therefore selected for analysis. This cluster, however, contained odorants HG and KO, which were in fact 2 variants of the same molecule (Table 1). These will obviously yield an identical prediction from the model, so we therefore combined HG and KO, retaining 10 equal intensity monomolecules (Odorants ‘QB’; ‘AY’; ‘XI’; ‘FL’; ‘JQ’; ‘LV’; ‘KOcombined’; ‘UV’; ‘SS’; ‘IF’).
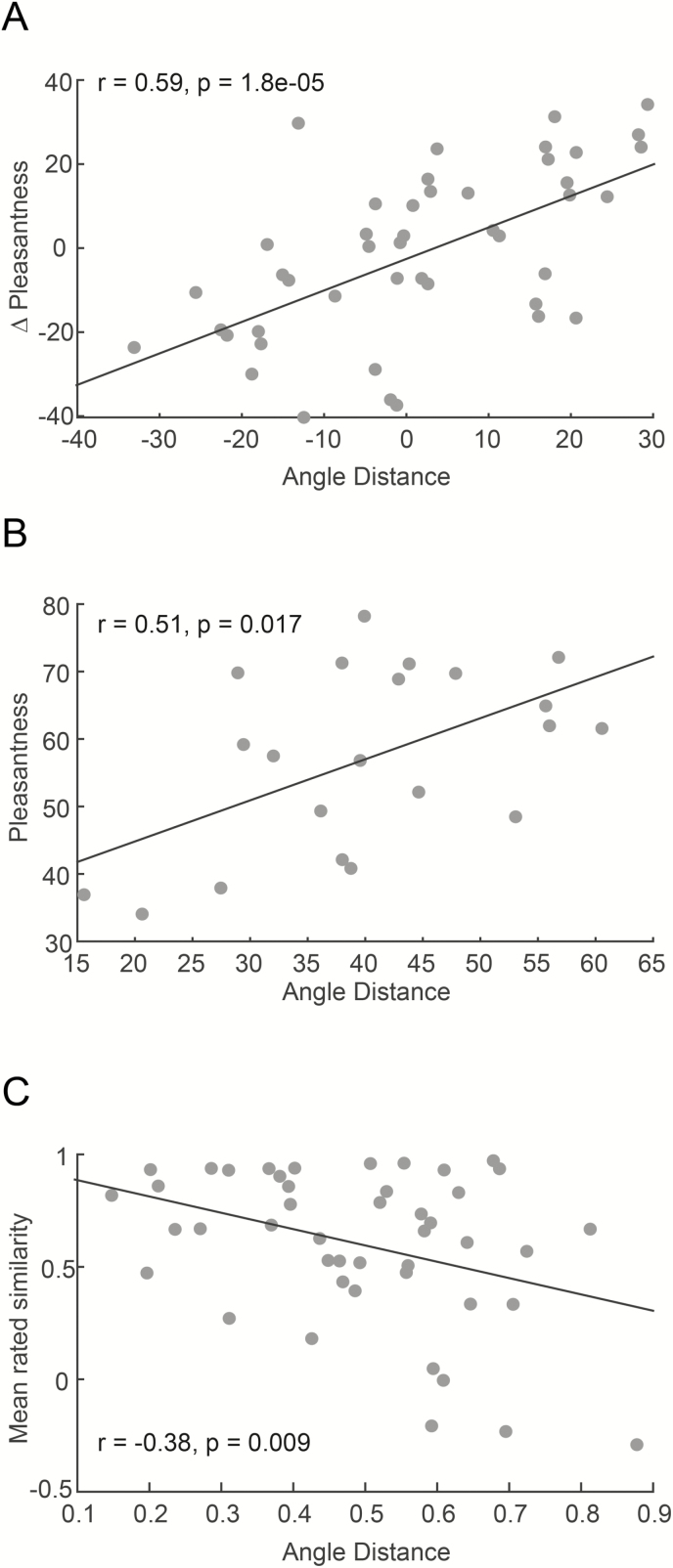
Ten molecules is too small a number for meaningful correlation analysis (it would provide for r = 0.13, P = 0.73). Therefore, initially, rather than looking at the raw pleasantness prediction, we looked at the predicted difference in pleasantness for every pair of molecules. In other words, the predicted versus actual difference in pleasantness between QB and AY, QB and XI, QB and FL, etc. Thus, 10 odorants provide for 45 unique pairwise comparisons. Using this approach we observed a strong correlation between predicted and actual pairwise differences in odorant pleasantness (r = 0.59, P = 1.81e-05) (Figure 8A). Finally, despite the differences in perceived intensity that violate the model assumptions, we also plotted the correlation between predicted and rated absolute pleasantness ratings (not differences) for all 22 monomolecules (excluding ZB) and observed a significant correlation of r = 0.51 (P = 0.017) between predicted and actual odorant pleasantness (Figure 8B).
Figure 8.
Predicting odorant perception from odorant structure in SmellSpace. (A) Predicting pairwise differences in monomolecular odorant pleasantness from odorant structure. Each dot is a comparison of 2 odorants. (B) Predicting odorant pleasantness from odorant structure. Each dot is a monomolecule. (C) Predicting pairwise monomolecular odorant similarity from odorant structure. Each dot is a comparison of 2 odorants.
Predicting pairwise odorant perceptual similarity from odorant structure alone
In Snitz et al. (2013), we predicted odorant perceptual similarity from odorant structure alone. In contrast to the Khan pleasantness model that was optimized for monomolecules but not mixtures, the Snitz similarity model was optimized for mixtures but not monomolecules. However, its clearly acknowledged limitation was that it applied to mixtures where all components were of equal perceived intensity, which again is not the case here. Although the Snitz model was not optimized for monomolecules, it nevertheless provided for reasonable monomolecular similarity estimates, se we once again test the 10 equal intensity monomolecules that provide for 45 pairwise similarity comparisons (e.g., what is the level of perceptual similarity between QB and AY, QB and XI, QB and FL, etc.). For actual pairwise perceived similarity, we derive pairwise odorant similarity from descriptors applied to the odorants. Pairwise perceptual similarity derived from descriptors is very highly correlated with direct similarity ratings (r > 0.85 in Dravnieks et al. 1978 and Callegari et al. 1997). In our method for deriving similarity, each odorant is represented by a vector consisting of its ratings along all descriptors, and these vectors are correlated as a measure of odorant similarity. Using this approach in the past, we observed derived similarity correlated with direct similarity at r = 0.71 (P < 0.0001) (Figure 1C in Khan et al. 2007). As in Snitz et al., to calculate the predicted similarity between any pair of odorants u and v, the distance function between the vector representing odorant and the vector representing odorant was computed as the angle between them in the 21-feature space (Table 3). It was given by: . Where is the dot product between the vectors and and are their Euclidean norms. Using this calculation, we observe a correlation of r = −0.38 (P = 0.009) between predicted distance and actual rated pairwise similarity (Figure 5C) (if we look at all 23 odorants, regardless of perceived intensity, the result is r = 0.18, P = 0.004).
Table 3.
The physicochemical features for predicting pairwise perceptual similarity
| No. | Abbreviation | Description |
|---|---|---|
| 1 | nCIR | Number of circuits (constitutional descriptors) |
| 2 | ZM1 | First Zagreb index M1 (topological descriptors) |
| 3 | GNar | Narumi geometric topological index (topological descriptors) |
| 4 | S1K | 1-path Kier alpha-modified shape index (topological descriptors) |
| 5 | piPC08 | Molecular multiple path count of order 08 (walk and path counts) |
| 6 | MATS1v | Moran autocorrelation—lag 1/weighted by atomic van der Waals volumes (2D autocorrelations) |
| 7 | MATS7v | Moran autocorrelation—lag 7/weighted by atomic van der Waals volumes (2D autocorrelations) |
| 8 | GATS1v | Geary autocorrelation—lag 1/weighted by atomic van der Waals volumes (2D autocorrelations) |
| 9 | EEig05x | Eigenvalue 05 from edge adj. matrix weighted by edge degrees (edge adjacency indices) |
| 10 | ESpm02x | Spectral moment 02 from edge adj. matrix weighted by edge degrees (edge adjacency indices) |
| 11 | ESpm03d | Spectral moment 03 from edge adj. matrix weighted by dipole moments (edge adjacency indices) |
| 12 | ESpm10d | Spectral moment 10 from edge adj. matrix weighted by dipole moments (edge adjacency indices) |
| 13 | ESpm13d | Spectral moment 13 from edge adj. matrix weighted by dipole moments (edge adjacency indices) |
| 14 | BELv3 | Lowest eigenvalue n. 3 of Burden matrix/weighted by atomic van der Waals volumes (Burden eigenvalues) |
| 15 | RDF035v | Radial distribution function—3.5/weighted by atomic van der Waals volumes (RDF descriptors) |
| 16 | G1m | First component symmetry directional WHIM index/weighted by atomic masses (WHIM descriptors) |
| 17 | G1v | First component symmetry directional WHIM index/weighted by atomic van der Waals volumes (WHIM descriptors) |
| 18 | G1e | First component symmetry directional WHIM index/weighted by Sanderson electronegativities (WHIM descriptors) |
| 19 | G3s | Third component symmetry directional WHIM index/weighted by atomic electropological states (WHIM descriptors) |
| 20 | R8u+ | R maximal autocorrelation of lag 8/unweighted (GETAWAY descriptors) |
| 21 | nRCOSR | Number of thioesters (aliphatic) (functional group counts) |
WHIM=weighted holistic invariant molecular descriptors.
The 21 Dragon features that enable predicting perceptual similarity. This table is reproduced from (Snitz et al. 2013).
Discussion
We aimed to develop a novel platform for olfactory perception data collection and to make this data public. In one respect this was a success, namely in that we indeed here post perceptual data from ~1000 self-motivated individuals (Supplementary Data File 1). Moreover, an advantage of SmellSpace is that the subject-specific data will grow over time. We have already launched round 2 of SmellSpace and the existing participants as well as ~400 new participants in round 2 are receiving booklets with 30 additional nonoverlapping odorants, which will bring SmellSpace to 65 odorants. Thus, over time, SmellSpace may provide for a massive data set. Moreover, we have made an effort to functionalize SmellSpace as a research tool. For example, users of SmellSpace can enter the “My Research” tab on the website and order multiple odorant booklets to use in an experiment of their own (free of any charge, pending booklet availability). Any scientist/student can distribute these booklets to different groups (e.g., “Democrats” vs. “Republicans”) and use SmellSpace to test whether these groups differ in terms of odor perception. In turn, an aspect by which we do not consider phase 1 of SmellSpace a success is in the response rate to the initial run. Overall, we were at ~11% response rate. This reflects a very ineffective and expensive path to data collection. That said, we have learned which distribution paths are effective and which are not: Booklets that were ordered by mail yielded effectively 100% response rate, booklets distributed at lectures on olfaction yielded ~20% response rate, yet booklets blindly distributed as attachments to a newspaper, etc., yielded ~1% response rate. In other words, only targeted distribution worked. This will guide our actions in the future development of SmellSpace in an aim to generate the largest possible data set for olfaction research. An additional limitation of SmellSpace is in the perceptual quality of the odorants. Although S&S technology seems to currently be the best way to distribute odorants at massive scale, it is not free of drawbacks. First, some odorants fail to encapsulate, as was likely the case with odorant ZB here. Moreover, we identified an olfactory undertone likely related to the microencapsulation printing process, which slightly contaminated all stimuli. This effect was not dramatic, as evident by the clearly distinct perception of the different odorants (Figure 4) and the high correlation in perception between groups of raters in SmellSpace and groups of raters who smelled nonencapsulated versions of the same odorants in lab (Figure 6). Nevertheless, the perceptual attributes attributed to the diluent (odorant NE) reflect the overall “odor of the booklet” (the paper, the odorant microencapsulation process, etc.) and reflect a limitation of the effort. This highlights that novel methods for odorant dispersion is a need, likely in industry, and clearly in science.
In addition to posting data for the community, we here also conducted 2 analyses aimed at assessing data usability. In the first, we predicted odorant pleasantness from odorant structure alone. A particular strength of this result is that here we tested the model on both molecules and participants who were not part of the model building set. In the past, when we tested the model using novel odorants but the same participants used to build the model, predictive performance was at r = ~0.75. In other words, variance is introduced by both molecules and individuals and here we captured both.
In the second analysis, we predicted odorant pairwise perceptual similarity from odorant structure alone using the approach from Snitz et al. 2013. The Snitz model was designed for odorant mixtures not monomolecules (the correlations for mixtures are r = ~0.7), yet it nevertheless originally provided for correlations of r = ~0.55 between predicted and actual monomolecular pairwise similarity. The Snitz model is applicable to equated intensity odorants alone. Here, we had only 10 equated intensity odorants, providing for 45 pairwise similarity ratings. The correlation between predicted and actual derived similarity for this restricted set was r = 0.38 (P = 0.009). Although a significant result, the power of this link is underwhelming. We hypothesize that this poorer performance than in the 2013 Snitz manuscript likely reflects the small number of odorants and the small number of descriptors available for the current analysis. The latter impacts the quality of the derived similarity estimates. Considering this, an added conclusion from this first phase of SmellSpace is that we intend to increase the number of Fixed descriptors, even at the cost of increased participation time demand.
The results with both models highlight a critical weakness in the world of odor modeling and that is our inability to computationally deal with variation in odorant perceived intensity. Odorant intensity interacts with odorant perception in complex ways (Distel et al. 1999). Effectively modeling and predicting odorant intensity from odorant structure alone will boost performance of models for pleasantness, similarity, and beyond, and is therefore a key future goal in olfactory modeling efforts (Mainland et al. 2014a). Indeed, modeling intensity is one of our ultimate goals for SmellSpace data, but it cannot be addressed at this stage due to the limited number of odorants. We look forward to addressing this once SmellSpace data provides for sufficient scope on this front.
In conclusion, we describe a novel approach to collecting olfactory perceptual data. Analyses of data from SmellSpace provided for big-data replications of 2 previous models for predicting perception from structure: one for predicting odorant pleasantness and another for predicting odorant pairwise perceptual similarity. These replications were presented here primarily as a demonstration for the applicability of the data. The primary contribution of this manuscript is in making the raw data publically available (the complete data set is posted online with this manuscript and at https://www.weizmann.ac.il/neurobiology/worg/materials.html. We look forward to growing this public database over time and further look forward to its use by the community.
Funding
This work was supported by a European Research Council Advanced Grant [670798 SocioSmell to N.S.]. N.S. is also supported by the Rob and Cheryl McEwen Fund for Brain Research.
Conflict of Interest
Given that it is very clear how SmellSpace could be a commercial venture, we would like to very clearly state that we have no commercial interest in SmellSpace and that this effort is for generating scientific data alone.
Supplementary Material
References
- Arctander S. 1969. Perfume and flavor chemicals: (aroma chemicals). Carol Stream (IL): Allured Publishing Corporation. [Google Scholar]
- Callegari P, Rouault J, Laffort P. 1997. Olfactory quality: from descriptor profiles to similarities. Chem Senses. 22:1–8. [DOI] [PubMed] [Google Scholar]
- Distel H, Ayabe-Kanamura S, Martínez-Gómez M, Schicker I, Kobayakawa T, Saito S, Hudson R. 1999. Perception of everyday odors–correlation between intensity, familiarity and strength of hedonic judgement. Chem Senses. 24:191–199. [DOI] [PubMed] [Google Scholar]
- Doty RL, Golbe LI, McKeown DA, Stern MB, Lehrach CM, Crawford D. 1993. Olfactory testing differentiates between progressive supranuclear palsy and idiopathic Parkinson’s disease. Neurology. 43:962–965. [DOI] [PubMed] [Google Scholar]
- Doty RL, Shaman P, Kimmelman CP, Dann MS. 1984. University of Pennsylvania Smell Identification Test: a rapid quantitative olfactory function test for the clinic. Laryngoscope. 94:176–178. [DOI] [PubMed] [Google Scholar]
- Dravnieks A. 1985. Atlas of odor character profiles. Philadelphia: ASTM. [Google Scholar]
- Dravnieks A, Bock F, Powers J, Tibbetts M, Ford M. 1978. Comparison of odors directly and through profiling. Chem Senses. 3:191–225. [Google Scholar]
- Dweck CS. 2013. Self-theories: their role in motivation, personality, and development. London: Psychology Press. [PubMed] [Google Scholar]
- Heyman J, Ariely D. 2004. Effort for payment. A tale of two markets. Psychol Sci. 15:787–793. [DOI] [PubMed] [Google Scholar]
- Keller A, Gerkin RC, Guan Y, Dhurandhar A, Turu G, Szalai B, Mainland JD, Ihara Y, Yu CW, Wolfinger R, et al. ; DREAM Olfaction Prediction Consortium 2017. Predicting human olfactory perception from chemical features of odor molecules. Science. 355:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Hempstead M, Gomez IA, Gilbert AN, Vosshall LB. 2012. An olfactory demography of a diverse metropolitan population. BMC Neurosci. 13:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Vosshall LB. 2016. Olfactory perception of chemically diverse molecules. BMC Neurosci. 17:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan RM, Luk CH, Flinker A, Aggarwal A, Lapid H, Haddad R, Sobel N. 2007. Predicting odor pleasantness from odorant structure: pleasantness as a reflection of the physical world. J Neurosci. 27:10015–10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström JN, Jones-Gotman M. 2009. Romantic love modulates women’s identification of men’s body odors. Horm Behav. 55:280–284. [DOI] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, et al. 2014a. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci. 17:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Lundström JN, Reisert J, Lowe G. 2014b. From molecule to mind: an integrative perspective on odor intensity. Trends Neurosci. 37:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid A, Levinson SC. 2011. The senses in language and culture. Senses Soc. 6:5–18. [Google Scholar]
- Mandairon N, Poncelet J, Bensafi M, Didier A. 2009. Humans and mice express similar olfactory preferences. PLoS One. 4:e4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A, Consonni V, Pavan M, Todeschini R. 2006. Dragon software: an easy approach to molecular descriptor calculations. Match. 56:237–248. [Google Scholar]
- Secundo L, Snitz K, Weissler K, Pinchover L, Shoenfeld Y, Loewenthal R, Agmon-Levin N, Frumin I, Bar-Zvi D, Shushan S, et al. 2015. Individual olfactory perception reveals meaningful nonolfactory genetic information. Proc Natl Acad Sci USA. 112:8750–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigma-Aldrich 2016. Flavors and fragrances products catalog. Darmstadt, Germany: Merck KGaA. [Google Scholar]
- Snitz K, Yablonka A, Weiss T, Frumin I, Khan RM, Sobel N. 2013. Predicting odor perceptual similarity from odor structure. PLoS Comput Biol. 9:e1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind C, Seebeck T, Bettens F, Paepke AJ. 1995. MHC-dependent mate preferences in humans. Proc Biol Sci. 260:245–249. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ, Gilbert AN. 1989. National geographic smell survey. Effects of age are heterogenous. Ann N Y Acad Sci. 561:12–28. [DOI] [PubMed] [Google Scholar]
- Zarzo M. 2011. Hedonic judgments of chemical compounds are correlated with molecular size. Sensors (Basel). 11:3667–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.