Abstract
Dysregulation of inflammatory cell death is a key driver of many inflammatory diseases. Pyroptosis, a highly inflammatory form of cell death, uses intracellularly generated pores to disrupt electrolyte homeostasis and execute cell death. Gasdermin D, the pore-forming effector protein of pyroptosis, coordinates membrane lysis and the release of highly inflammatory molecules, such as interleukin-1β, which potentiate the overactivation of the innate immune response. However, to date, there is no pharmacologic mechanism to disrupt pyroptosis. Here, we identify necrosulfonamide as a direct chemical inhibitor of gasdermin D, the pyroptotic pore-forming protein, which binds directly to gasdermin D to inhibit pyroptosis. Pharmacologic inhibition of pyroptotic cell death by necrosulfonamide is efficacious in sepsis models and suggests that gasdermin D inhibitors may be efficacious clinically in inflammatory diseases.
INTRODUCTION
Dysregulation of the inflammatory response is a key driver of many debilitating and costly diseases including sepsis and inflammatory bowel disease. Two distinct forms of cell death are important drivers of this inflammatory response. Pyroptosis and necroptosis rely on macromolecular cytosolic complexes to coordinate the formation of destructive membrane pores. These pores accumulate in the cell membrane and disrupt cellular electrochemical potential, causing cell death. Lysis of the cell results in the release of inflammatory molecules that recruit immune effector cells and activate a larger inflammatory response.
In necroptosis, formation of a receptor-interacting protein kinase 1 (RIPK1)/RIPK3/mixed lineage kinase domain-like pseudokinase (MLKL) complex, termed the necroptosome, coordinates RIPK3 phosphorylation of MLKL. This phosphorylation causes MLKL oligomerization and formation of the necroptotic pore (1–3). In contrast, pyroptotic cell death is initiated by cytosolic danger signals that stimulate receptors such as NACHT, LRR, and PYD domains–containing 3 (NLRP3), pyrin, and NLR family CARD domain–containing protein 4 (NLRC4), resulting in the formation of the inflammasome. The inflammasome subsequently coordinates the cleavage and activation of caspase-1. Active caspase-1 cleaves the pore-forming protein, gasdermin D (GSDMD), ultimately resulting in oligomerization of the N-terminal portion of GSDMD (4–10). In the resting state, GSDMD oligomerization is autoinhibited by the intramolecular binding between the N and C termini (11, 12). However, cleavage by an inflammatory caspase relieves this autoinhibition and allows large pyroptotic pores, composed of N-terminal p30-GSDMD oligomers, to form. These N-terminal p30-GSDMD pores then perforate the outer cellular membrane and individual organelle membranes, resulting in outer membrane failure and mitochondrial permeabilization (4, 5, 11, 13, 14). Because GSDMD is a central effector protein of an inflammatory insult, it is essential to inflammatory disease. A small-molecule inhibitor of GSDMD would be important not only for the study of pyroptosis but also to establish GSDMD target validation in inflammatory diseases as diverse as inflammatory bowel disease, inflammatory arthritis, and sepsis.
In this work, we show that necrosulfonamide (NSA) directly binds to GSDMD and inhibits p30-GSDMD oligomerization. NSA blocks pyroptotic cell death and interleukin-1β (IL-1β) release in both primary murine and immortalized human and murine monocytes/ macrophages. NSA does not inhibit other innate immune pathways such as Toll-like receptor (TLR) signaling and gasdermin E (GSDME)–mediated cell death and does not interfere with formation of the inflammasome. Furthermore, NSA treatment ෬reases inflammatory cytokine release and substantially prolongs survival in a murine model of sepsis. Together, our findings demonstrate that NSA directly binds to GSDMD and inhibits p30-GSDMD pore formation, providing a basis for the development of future therapeutics in inflammatory disease.
RESULTS
NSA inhibits pyroptotic cell death in immortalized murine macrophages and human monocytes
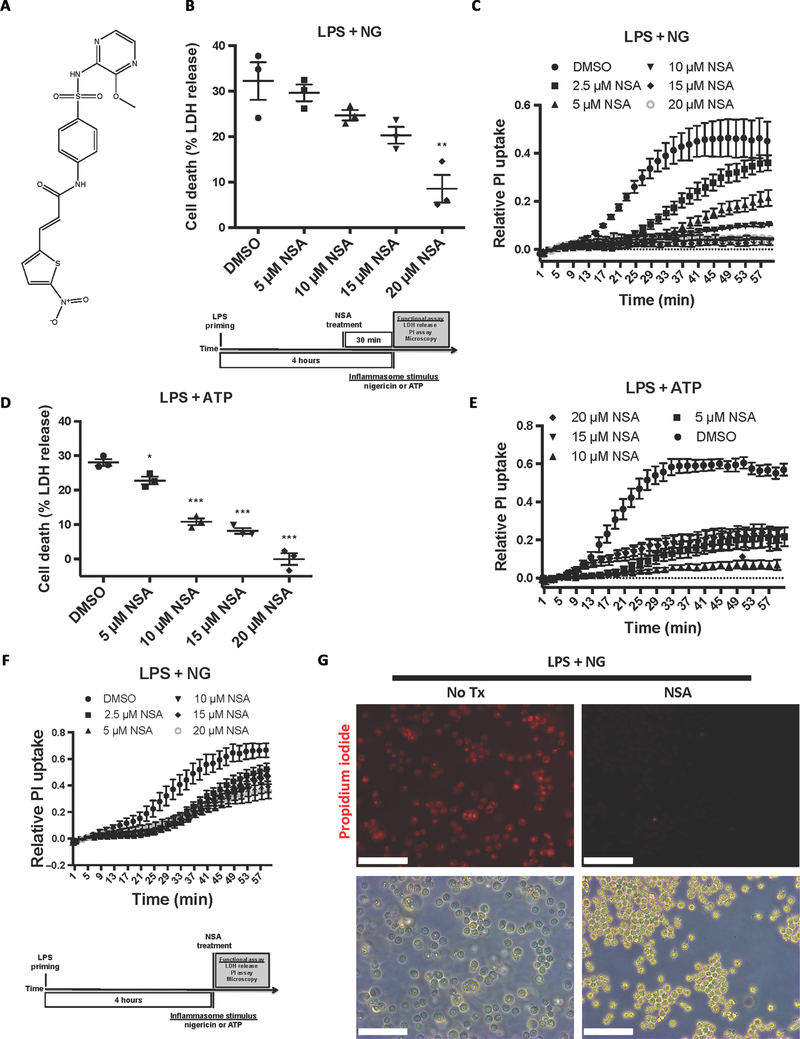
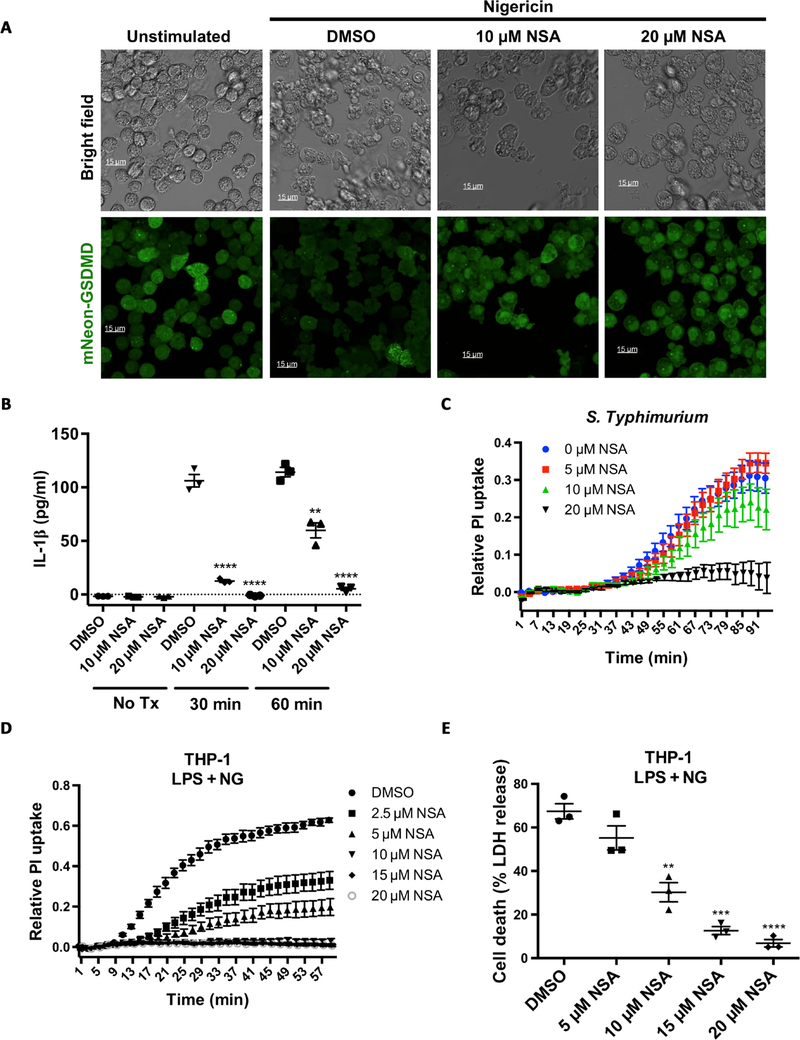
Screening of small-molecule inhibitors of necroptotic cell death in HT29 cells resulted in the discovery of NSA (Fig. 1A) (15). NSA potently inhibits necroptosis through binding to MLKL and disrupting disulfide bonds formed by Cys86 of human MLKL (15). Murine MLKL lacks a cysteine at this location, and thus NSA is unable to inhibit necroptosis in mice. Although distinct from MLKL in some aspects of structure and regulation, disulfide linkages are critical for the oligomerization of p30-GSDMD and the formation of pyroptotic pores (9). Therefore, we asked whether NSA could also inhibit GSDMD oligomerization and pyroptotic cell death. In immortalized bone marrow–derived macrophages (iBMDMs) constitutively expressing NLRP3 and apoptosis-associated speck-like protein containing a CARD (ASC), incubation with NSA after lipopolysaccharide (LPS) priming, but before nigericin addition, inhibited the formation of pyroptotic pores, as measured by propidium iodide (PI) uptake, and cell death, as measured by lactate dehydrogenase (LDH) release (Fig. 1, B and C). Similar suppression of pyroptotic pore formation and cell death by NSA was seen using adenosine 5′-triphosphate (ATP), a separate activator of the NLRP3 inflammasome (Fig. 1, D and E). NSA was also able to inhibit pyroptotic pore formation with simultaneous administration of NSA and nigericin, albeit to a lesser extent than with the short NSA pretreatment. This feature is likely due to the rapid (within minutes) induction of pyroptotic cell death (Fig. 1F). Bright-field and epifluorescent microscopy of iBMDM cells revealed large bubbles resulting from separation of the plasma membrane from the cytoskeleton in the dimethyl sulfoxide (DMSO)–treated cells after treatment with LPS and nigericin as well as the uptake of PI in nearly every cell. Cells treated under the same conditions in the presence of NSA displayed normal morphology and a lack of PI uptake, indicating an absence of pyroptotic cell death (Fig. 1G). Using an internal mNeonGreen tag located in the p30 fragment of GSDMD, Gsdmd−/− iBMDMs were stably reconstituted and the activity of GSDMD was monitored using live cell confocal imaging (16). In cells stimulated with LPS and nigericin, treatment with NSA inhibited aggregation of p30-mNeon-GSDMD, the formation of membrane bubbles, and pyroptotic cell death (Fig. 2A). NSA also inhibited GSDMD-mediated IL-1β release after inflammasome stimulation, demonstrating complete suppression of IL-1β even 60 min after stimulation (Fig. 2B). To then test other inflammasomes, we used Salmonella Typhimurium because S. Typhimurium initiates pyroptosis through activation of the NLRC4 inflammasome. NSA also suppressed pore formation in cells treated with S. Typhimurium (Fig. 2C), thus demonstrating efficacy across multiple inflammasome pathways. To determine whether NSA is also effective at inhibiting pyroptosis in human cells, we next tested NSA for its ability to inhibit pyroptotic cell death in human THP-1 monocytes. NSA inhibited formation of the pyroptotic pore and cell death in a dose-dependent manner in THP-1 cells as measured by PI uptake and LDH release (Fig. 2, D and E). Together, these data suggest that NSA blocks GSDMD pore formation in murine and human cells across multiple inflammasomes, consistent with GSDMD being the target of NSA in pyroptosis.
Fig. 1. NSA inhibits pyroptotic cell death in immortalized murine macrophages.
(A) Chemical structure of NSA. (B to E) Cells were primed with LPS, followed by NSA treatment 30 min before stimulation with nigericin (NG) or ATP. Pyroptotic pore formation was measured kinetically through the uptake of PI, and cell lysis was measured by LDH release. PI uptake is taken from four experiments each with technical duplicates. LDH release is from three independent experiments, each done in technical duplicate. *P < 0.05, **P < 0.01, and ***P < 0.001. (F) Measurement of pyroptotic pore formation assessed by PI uptake with simultaneous administration of NSA with nigericin stimulation. Data represent four experiments each with two technical replicates. (G) Bright-field and epifluorescent microscopy images of iBMDM cells with or without NSA treatment before and after treatment with 10 μM nigericin. Scale bars, 100 μm. All cells were primed with LPS (1 μg/ml) for 4 hours. (B to F) PI and LDH data show means ± SE. Tx, treatment.
Fig. 2. NSA inhibits pyroptotic cell death downstream of multiple inflammasomes in human and murine cells.
(A) mNeon-GSDMD was stably reconstituted into Gsdmd−/− iBMDM cells. Cells were primed with LPS (1 μg/ml), treated with DMSO or NSA, and activated with 10 μM nigericin. Live cell imaging was conducted on an inverted confocal microscope 90 min after stimulation. (B) IL-1β release from iBMDM cells stimulated with LPS and nigericin with DMSO, 10 μM NSA, or 20 μM NSA. IL-1β concentration was determined by sandwich ELISA 30 and 60 min after stimulation. **P < 0.01 and ****P < 0.0001. (C) Pore formation as measured by PI uptake in iBMDM cells treated with S. Typhimurium. iBMDMs were primed for 4 hours followed by activation of the NLRC4 inflammasome with log phase S. Typhimurium. (D and E) THP-1 cells were primed with LPS, followed by NSA treatment 30 min before stimulation with nigericin (NG). Pyroptotic pore formation and cell death were assessed through PI uptake and LDH release, respectively. **P < 0.01, ***P < 0.001, and ****P < 0.0001. (B to E) PI and LDH data are means ± SE and are representative of ≥3 experimental replicates.
NSA inhibits NLRP3- and pyrin-mediated pyroptotic cell death in primary cells
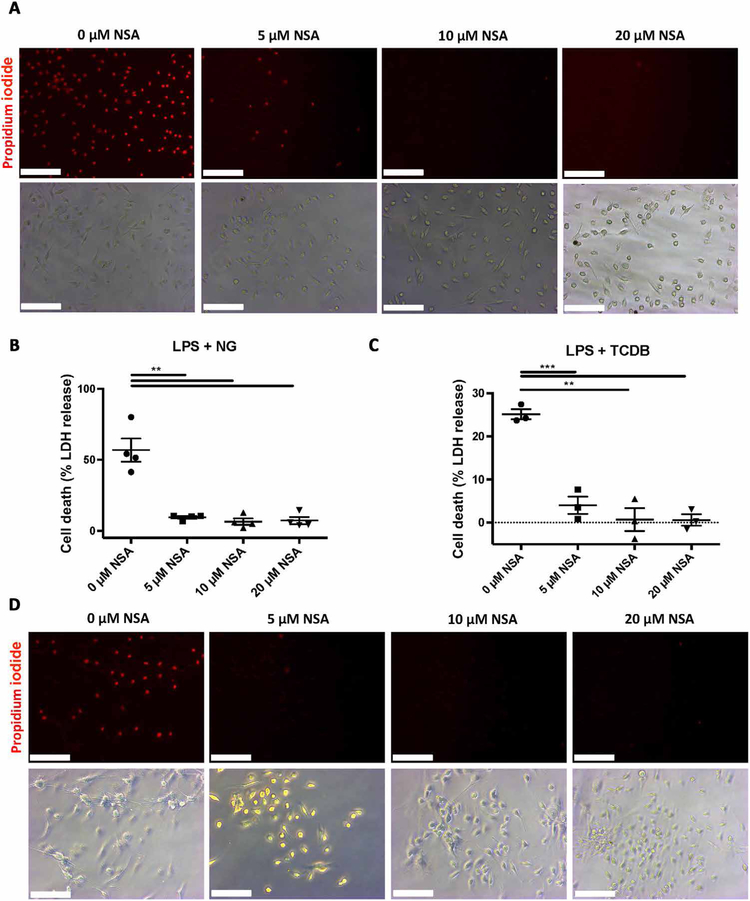
To test whether NSA also inhibited pyroptosis in primary macrophages, we cultured bone marrow–derived macrophages (BMDM) from C57BL/6J mice. After priming with LPS, the NLRP3 inflammasome was activated using nigericin. NSA inhibited PI uptake, as visualized by epifluorescent microscopy, and cell death, as measured by LDH release at 5 μM concentrations (Fig. 3, A and B). Greater suppression was seen in these primary macrophages relative to the immortalized cell line likely due to the fact that these primary cells had not been genetically modified to be highly pyroptotic. To ensure that the inhibition of pyroptotic cell death was not NLRP3-dependent, we tested the ability of NSA to inhibit pyroptosis downstream of activation of the pyrin inflammasome. After priming with LPS, primary macrophages were stimulated with Clostridium difficile toxin B. Once again, NSA inhibited PI uptake and cell death at 5 μM concentrations (Fig. 3, C and D). These findings again suggest that NSA inhibits pyroptosis downstream of multiple inflammasomes and further suggest that as the common pore-forming protein downstream of the pyrin, NLRP3, and NLRC4 inflammasomes, GSDMD is the molecular target of NSA.
Fig. 3. NSA inhibits pyroptosis in primary macrophages.
Primary macrophages were isolated from C57BL/6J mice. (A to D) After priming with LPS (1 μg/ml) for 4 hours, cells were treated with increasing doses of NSA for 30 min before activation of the inflammasome with either 10 μM NG or C. difficile toxin B (TCDB) (0.4 μg/ml). One hour after activation, cells were visualized using bright-field and epifluorescent microscopy. PI was used at 1 μg/ml. Cell death was measured by LDH. (A to D) Data are representative of ≥3 experimental replicates. Scale bars, 100 μm. LDH data show means ± SE. **P < 0.01, ***P < 0.001.
NSA does not inhibit other innate immune pathways or other cell death pathways
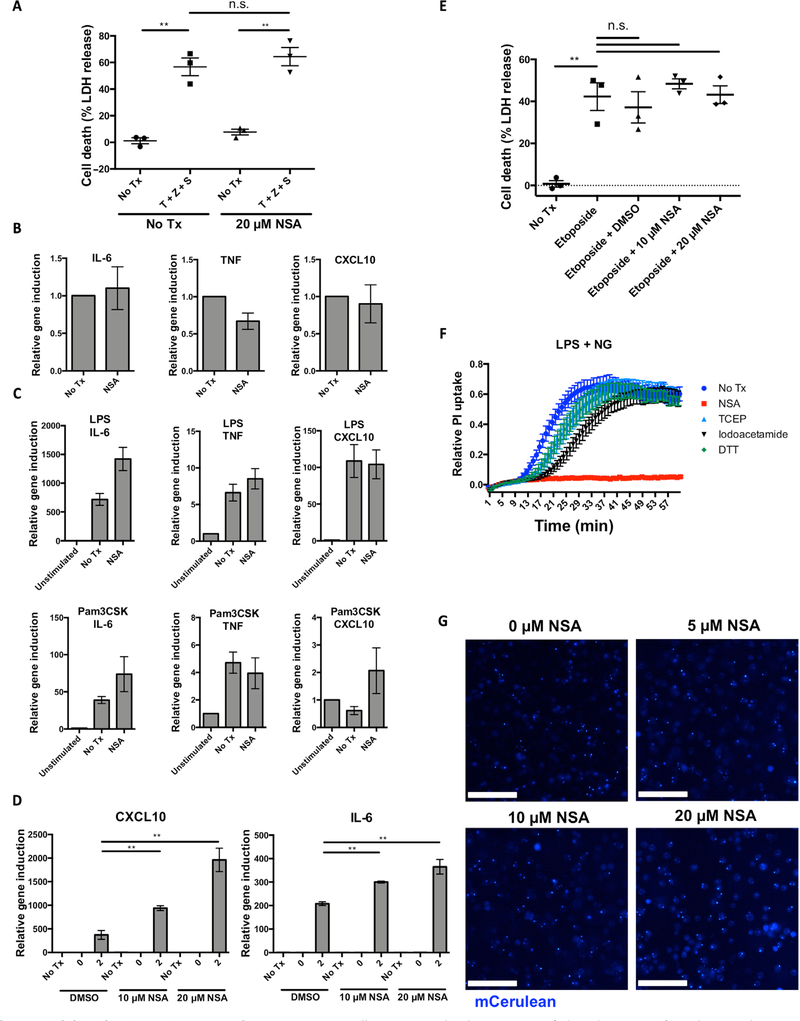
Many known inflammasome inhibitors work by inhibiting upstream of other innate immune pathways or upstream nuclear factor κB signaling. Therefore, it is also important to rule out an effect on other inflammatory cell death pathways like necroptosis. To this end, we tested the effect of NSA on necroptosis in murine cells. Murine iBMDMs, in which MLKL lacks the critical cysteine residue for NSA attachment, were treated with tumor necrosis factor–α (TNF-α), Z-VAD-fmk (zVAD), and the Smac-mimetic GDC-0152 in the presence or absence of NSA. No inhibition of necroptotic cell death was seen with NSA (Fig. 4A), consistent with previous findings that NSA does not inhibit necroptosis in murine cells (15). The impact of NSA on the TLR induction of inflammatory gene transcription was then tested. In unstimulated iBMDM cells, NSA did not alter the basal levels of IL-6, TNF-α, or CXCL10 (Fig. 4B). In response to stimulation with the TLR1/2 agonist, Pam3CSK4, or the TLR4 agonist, LPS, NSA did not inhibit induction of inflammatory genes (Fig. 4C). We further asked whether, by inhibiting pyroptotic cell death, NSA could preserve the inflammasome-independent innate immune response for secondary infection. To determine whether this is the case, iBMDM cells were challenged with S. Typhimurium. As predicted, NSA enhanced the production of other inflammatory signaling molecules as demonstrated by increased levels of CXCL10 and IL-6 through inhibiting the initial pyroptotic cell death (Fig. 4D). The other gasdermin family member known to execute inflammatory cell death is GSDME, which is activated by apoptotic caspase-3 and caspase-7 (17). To test whether NSA was specific to GSDMD-mediated pyroptosis or also inhibited GSDME-mediated necrosis, the ability of NSA to inhibit etoposide-induced cell death was tested. NSA did not inhibit GSDME cleavage or cell death after etoposide treatment (Fig. 4E and fig. S1, A and B). Together, these demonstrate that NSA is selective for GSDMD even among the most similar inflammatory pathways. We then wanted to see whether NSA inhibition of pyroptosis was common to all reducing agents. NSA, but not iodoacetamide, dithiothreitol (DTT), or tris(2-carboxyethyl)phosphine (TCEP), inhibited pyroptotic cell death, demonstrating that NSA is unique in its ability to inhibit pyroptosis (Fig. 4F). We then asked whether NSA inhibited inflammasome assembly. To this end, the ability of mCerulean-ASC–expressing iBMDMs to form ASC specks in response to LPS and nigericin was assessed. ASC oligomers were not inhibited by 5, 10, or 20 μM NSA (Fig. 4G). In all, these results suggest that NSA does not inhibit TLR signaling, GSDME-meditated cell death, murine necroptosis, or inflammasome assembly and further suggest that GSDMD is a direct target of NSA.
Fig. 4. NSA does not inhibit other innate immune pathways.
(A) iBMDM cells were treated with TNF-α, zVAD-fmk, and GDC-0152 for 24 hours in the presence or absence of 20 μM NSA. Cell death was measured by LDH release. (B and C) The impact of NSA on TLR pathways was assessed using RT-PCR of inflammatory gene transcripts after stimulation of TLR1/2 with Pam3CSK or TLR4 with LPS in the presence or absence of 20 μM NSA. (D) qRT-PCR was used to measure CXCL10 and IL-6 expression after infection with S. Typhimurium. Log phase S. Typhimurium was used to infect iBMDM cells at an MOI of 50:1 with induction of IL-6 and CXCL10 genes measured after 0 and 2 hours. Relative induction was based off of GAPDH as the housekeeping reference gene. Data are taken from technical triplicates and are representative of three independent experiments. **P < 0.01. (E) Cell death as measured by LDH assay in macrophages treated for 8 hours with 100 μM etoposide and DMSO, 10 μM NSA, or 20 μM NSA. **P < 0.01. (F) Pyroptotic pore formation as assessed by PI uptake in iBMDM cells after activation of the NLRP3 inflammasome with LPS and nigericin and treatment with 20 μM NSA, TCEP, iodoacetamide, or DTT. (G) iBMDM cells expressing mCerulean-ASC that form specks upon activation of the inflammasome were stimulated with LPS and nigericin in the presence of DMSO or NSA. Formation of specks was visualized using epifluorescent microscopy. Scale bars, 100 μm. (A to F) Data are means ± SE. n.s., not significant.
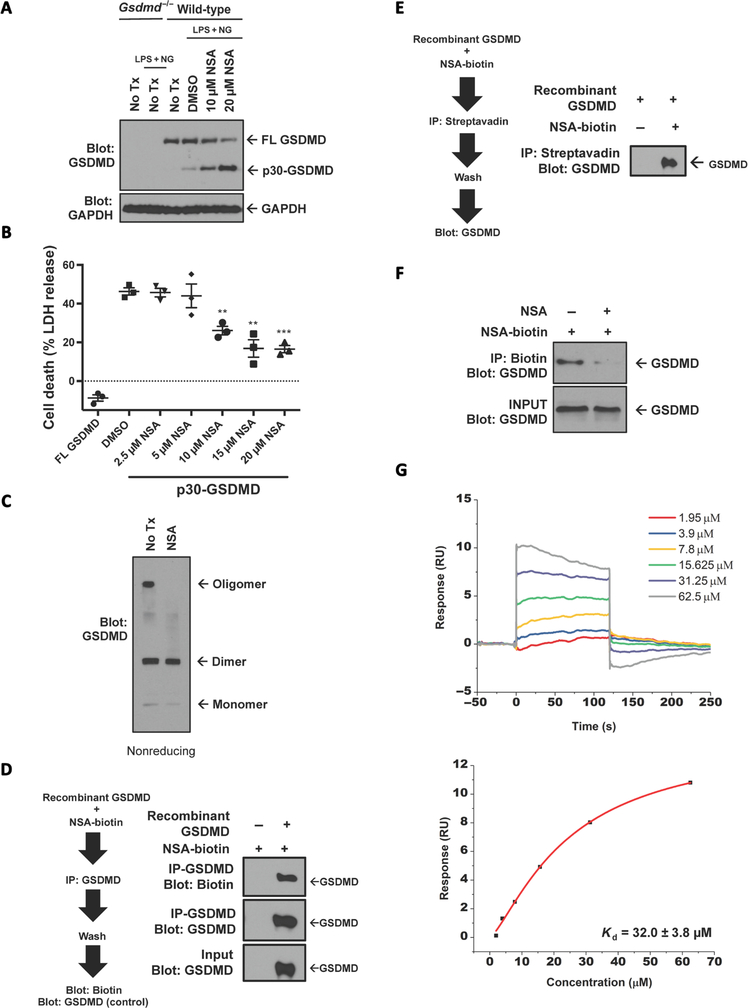
NSA binds directly to GSDMD via Cys191 on GSDMD
While the above results are suggestive of a direct effect of NSA on GSDMD, we aimed to determine the step at which GSDMD was inhibited and whether NSA binds GSDMD directly. As NSA inhibits pyroptotic cell death downstream of the inflammasome (Fig. 4G), the effect of NSA on GSDMD cleavage was tested. Cleavage of GSDMD occurred normally in NSA-treated iBMDMs with higher levels of p30-GSDMD in NSA-treated cells likely due to diminished cell death in the treated cells (Fig. 5A), implying that NSA does not inhibit caspase-1 at these concentrations. Because NSA appeared to be working at the level of p30-GSDMD, the ability of NSA to inhibit GSDMD-mediated cell death downstream of GSDMD cleavage was tested. The cleaved p30 fragment of GSDMD was expressed in human embryonic kidney (HEK)–293T cells with and without exposure to NSA. In this experiment, NSA inhibited cell death induced by expression of the p30 fragment of GSDMD in HEK-293T cells (Fig. 5B), again suggesting that NSA inhibits GSDMD directly. To next determine whether this inhibition was due to the ability of NSA to inhibit oligomerization of p30-GSDMD, we tested the presence of GSDMD oligomers. NSA completely inhibited the formation of oligomers but permitted the formation of p30-GSDMD dimers (Fig. 5C), suggesting that GSDMD inhibits the oligomerization of GSDMD dimers but not the cleavage of GSDMD or the initial dimerization of GSDMD.
Fig. 5. NSA directly interacts with GSDMD.
(A) LPS-primed iBMDM cells were stimulated with 10 μM nigericin in the presence or absence of NSA for 1 hour. Cleavage of full-length (FL)–GSDMD was assessed by Western blot. (B) The ability of NSA to inhibit formation of the pyroptotic pore was tested by transient expression of NTAP–p30-GSDMD in HEK-293T cells using calcium phosphate transfection. Four hours after transfection, medium was changed and DMSO or NSA was used to treat the cells. Twenty-four hours after transfection, LDH release was measured. Data are means ± SE taken from three technical replicates and are representative of three independent experiments. **P < 0.01 and ***P < 0.001. (C) Oligomerization with transient expression of GFP–p30-GSDMD expressed in HEK-293T cells was assessed by Western blot under nonreducing conditions after treatment with DMSO or 20 μM NSA. (D and E) Reciprocal in vitro binding assays of GSDMD to NSA-biotin using pulldown of biotin by streptavidin or pulldown of GSDMD with GSDMD antibody and analysis by Western blot. (F) Competitive binding assay of NSA with NSA-biotin and GSDMD. GFP-GSDMD was expressed in HEK-293T cells using calcium phosphate transfection. The in vitro competitive binding assay between NSA and NSA-biotin at a 5:1 ratio was conducted for 2 hours with analysis of binding by Western blot. (G) Surface plasmon resonance with recombinant GSDMD (immobilized) and NSA demonstrated a binding affinity of 32.0 ± 3.8 μM. IP, immunoprecipitation.
All of these biochemical characteristics of NSA suggest that it binds directly to GSDMD. To test this directly, we purified recombinant, human GSDMD from Escherichia coli (fig. S2A). Immunoprecipitation of recombinant GSDMD demonstrated the direct binding of biotin-conjugated NSA (NSA-biotin) to GSDMD (Fig. 5D and fig. S2B). The same was true of the reciprocal experiment using pulldown with NSA-biotin (Fig. 5E). Competitive binding assay demonstrated that this binding between GSDMD and NSA-biotin could be inhibited by the addition of excess nonbiotinylated NSA (Fig. 5F). Using surface plasmon resonance, the binding affinity of GSDMD to NSA was demonstrated to be 32.0 ± 3.8 μM (Fig. 5G). However, in the cellular context of cleaved p30-GSDMD, with an intact lipid membrane, the affinity may be even higher.
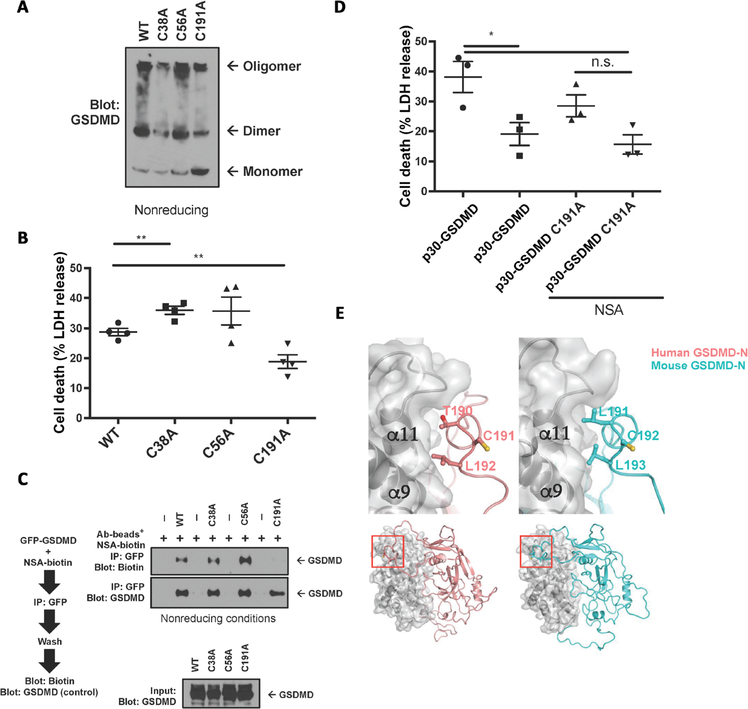
As NSA inhibits MLKL by covalently binding to a unique human cysteine residue and as NSA inhibits p30-GSDMD pore formation in human and murine cells, we then searched for conserved human and murine cysteines within the p30-GSDMD fragment (fig. S3) (15). Cysteine-to-alanine mutations were made at the three conserved cysteines, and oligomerization assays showed that C191A and, to a lesser extent, C38A showed a decreased ability to oligomerize (Fig. 6A). This was especially pronounced in the increased amount of C191A monomer relative to the amount of wild-type (WT), C38A, or C56A monomer, suggesting a less efficient oligomerization process with C191A (Fig. 6A). This feature is in agreement with a previous study that showed that these residues were important for oligomerization (9). Mutation of this cysteine residue on GSDMD was accompanied by a decrease in cell death with expression of the C191A p30-GSDMD mutant relative to WT (Fig. 6B). Given these results and the alkylating nature of NSA, we then tested whether C191A was required for NSA binding. In precipitation experiments, a decrease in binding of the NSA to C191A GSDMD was seen, with no decrease in binding noted with the other cysteine mutants (Fig. 6C). As the C191A mutation decreased oligomerization, cell death, and binding to NSA, we asked whether the C191A mutant altered the ability of NSA to inhibit pyroptotic cell death. WT p30-GSDMD or C191A p30-GSDMD was then expressed in HEK-293 cells with and without NSA addition. While WT p30-GSDMD was able to be inhibited by NSA, C191A showed a statistically insignificant inability to be inhibited by NSA (Fig. 6D). Structural modeling based on the solved crystal structure of GsdmA3 demonstrates that Cys191 lies in close proximity to the second autoinhibitory interface, an area known to be critical for the pyroptotic function of p30-GSDMD (Fig. 6E). Collectively, these results suggest that NSA binds GSDMD directly and that Cys191 is important for this interaction.
Fig. 6. Inhibition by NSA differs between MLKL and GSDMD.
(A) Oligomerization of p30-GSDMD cysteine-to-alanine mutants conserved between mice and humans was tested in nonreducing conditions. (B) The cytotoxicity of p30-GSDMD cysteine-to-alanine mutants, expressed in HEK-293T cells by calcium phosphate transfection, was assessed by LDH release. (C) In vitro binding assays between GFP-GSDMD cysteine-to-alanine mutants and NSA-biotin with analysis by Western blot. (D) HEK-293T cells were transfected with either WT p30-GSDMD or C191A p30-GSDMD and either left untreated or treated with NSA. While NSA decreased WT p30-GSDMD–mediated cell death, the decrease in C191A p30-GSDMD was not statistically significant. (E) Models for the full-length human and murine GSDMDs showing the location of the conserved Cys residues that may engage NSA and are important for oligomerization. The N-terminal domains for human GSDMD and murine GSDMD are shown in ribbons and colored salmon and blue-green, respectively. The C-terminal domains are shown in surface and gray ribbons. Residues Cys191 (human GSDMD) and Cys192 (murine GSDMD) and two surrounding residues are shown as ball-and-stick models, which demonstrate that, while the surrounding residues engage in the N- and C-domain interface, the Cys residues are exposed to the solvent. Approximate locations of the interface (top) in the full-length GSDMD structures are marked in red on the bottom. (B and D) LDH data are means ± SE. Ab, antibody. *P < 0.05, **P < 0.01.
NSA treatment increases survival in murine sepsis
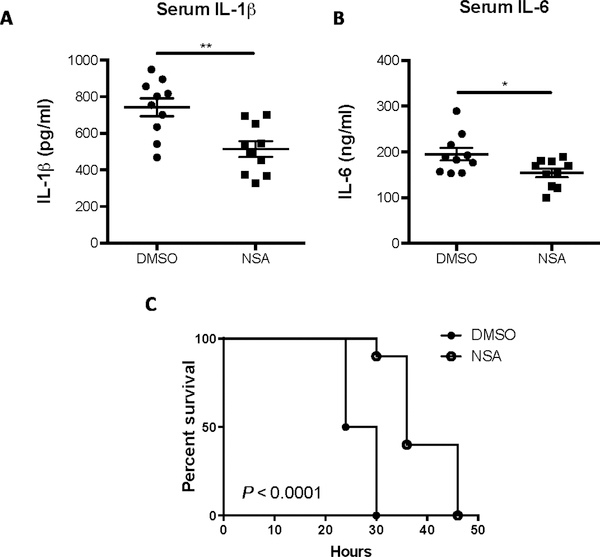
The role of NSA as a therapy in the initial inflammatory response was then tested in vivo in a murine model of sepsis. NSA has not been optimized for pharmacokinetic parameters or for in vivo stability, and after testing, we first determined that the maximal nontoxic dose of NSA was 20 mg/kg. Survival curves in mice given a lethal dose of LPS (25 mg/kg) demonstrated the therapeutic effect of NSA, wherein a single dose of 20 mg/kg NSA administered 30 min before LPS increased median survival by 6 hours (P = 0.009, fig S4). This therapeutic effect was even more pronounced in a second cohort treated with two doses of DMSO or NSA, once 30 min before LPS administration and once 10 hours after LPS administration. Serum taken from these two groups demonstrated a significant decrease in the release of IL-1β and IL-6 in mice treated with NSA (Fig. 7, A and B). In these mice treated with two doses of either NSA or DMSO, NSA increased median survival by 9 hours (P < 0.0001, Fig. 7C). On the basis of our data from the murine sepsis models, it is possible that optimized derivatives of NSA might be of use in the treatment of acute inflammatory diseases.
Fig. 7. NSA treatment extends survival in the LPS model of sepsis.
(A to C) C57BL/6J mice (9 or 10 per group) were injected with LPS (25 mg/kg) and with NSA or DMSO (20 mg/kg) 30 min before and 10 hours after LPS administration. Volume per injection of NSA or DMSO was about 25 μl per mouse. Serum levels of IL-6 and IL-1β, measured by sandwich ELISA, were taken 6 hours after the administration of LPS. Cytokine data are means ± SE. Survival curves were analyzed by log-rank (Mantel-Cox) test demonstrating an increase in median survival of 9 hours in the NSA treatment group (P <0.0001). *P < 0.05, **P < 0.01.
DISCUSSION
Here, the data demonstrate that NSA inhibits formation of the pyroptotic pore in human and murine cells through direct inhibition of the GSDMD pyroptotic pore both in vitro and in vivo (fig. S5). In murine immortalized and primary macrophages, NSA inhibits pyroptotic pore formation, cell death, and release of IL-1β downstream of the pyrin, NLRP3, and NLRC4 inflammasomes. This inhibitory ability translates to human monocytes as well, where NSA inhibits pyroptotic pore formation and cell death. Mechanistically, we found that NSA binds to GSDMD, inhibiting oligomerization of p30-GSDMD, leading to an inability to form the pyroptotic pore and an absence of pyroptotic cell death. NSA does not inhibit dimerization of GSDMD, but rather inhibits the assembly of those dimers into higher-order oligomers. In vitro experiments demonstrated that NSA binds directly to full-length GSDMD with 32 μM binding affinity. This affinity may, in fact, be higher within the cellular context with a lipid membrane and cleaved p30-GSDMD. Through the inhibition of oligomerization, NSA also inhibits release of IL-1β and other pore-mediated signaling molecules, which are responsible for inflammatory disease. NSA specifically inhibits pyroptotic cell death downstream of inflammasome activation while leaving other innate signaling pathways, such as TLR signaling and GSDME-mediated cell death, intact and functional. In vivo, the role of NSA as an inhibitor of cell death was demonstrated with single and double doses of NSA, both of which significantly increased median survival in the LPS model of sepsis.
No inhibitors that directly inhibit GSDMD while conserving inflammasome-independent innate immune function have been reported. Given the role of pyroptotic cell death and IL-1β release in both acute and chronic inflammation and its known role in inflammatory diseases as diverse as atherosclerosis, inflammatory bowel disease, and sepsis, this compound allows us to separate its role in cytokine function from its role in inflammatory cell death (18–22). As an alkylating compound likely targeting cysteine residues, GSDMD and MLKL are not likely to be the only proteins altered by NSA. While our findings do show that Cys191 is crucial for NSA binding and that mutation to alanine disrupts this binding, it is curious that the C191A mutation affects death to a much smaller extent than exposure to NSA. It is possible that binding of a large chemical like NSA is able to disrupt oligomerization to a greater extent than the removal of a single thiol group. On the basis of the fact that NSA targets GSDMD by targeting a cysteine, it is conceivable that NSA might target free cysteines in other molecules at higher concentrations. Further studies comparing pharmacologic and genetic deletion of GSDMD are needed to better understand the specificity of NSA-dependent targeting of GSDMD. Nonetheless, our findings suggest that NSA can directly target GSDMD under pyroptotic conditions, and we propose that NSA could be used as a lead compound to develop more potent inhibitors of GSDMD. This is demonstrated by the identification of the targeted region of GSDMD (surrounding Cys191), the direct binding of GSDMD to NSA as shown by binding assays, surface plasmon resonance assays, and the inhibition of p30-GSDMD toxicity in overexpression assays. In addition, as a chemical probe, NSA allows us to better understand the biophysics of GSDMD pore formation in a manner that cannot be easily measured using in vitro assays. By halting GSDMD pore formation post-GSDMD cleavage and only after GSDMD dimer formation, NSA inhibition shows not only that caspase-1 is functionally intact but also that p30-GSDMD dimers likely form before oligomerization into larger pores. Thus, while the alkylating nature of NSA likely causes off-target effects, its discovery as a GSDMD inhibitor shows that targeting of GSDMD not only is therapeutically feasible but also allows us to better understand the biophysical mechanism of pyroptotic pore formation.
Through a variety of in vitro and in vivo experiments, we demonstrate that NSA binds directly to GSDMD and can ameliorate GSDMD-driven mortality in murine sepsis. The identification of a tool compound in which the biochemistry of pyroptotic pore formation can be studied is important for the field. NSA represents the most distal inhibitor of inflammasome-mediated cell death and IL-1β release yet found. It provides the proof of principle that GSDMD pore formation can be inhibited and validates GSDMD as a viable pharmaceutical target. The therapeutic efficacy of inhibiting the inflammasome-pyroptosis axis has already been demonstrated clinically with the efficacy of IL-1β receptor antagonists to treat inflammatory diseases such as rheumatoid arthritis and the cryopyrin-associated periodic syndromes. Recently, two more studies have supported the importance of inhibiting this cascade as the IL-1β antagonist, anakinra, has been shown to reduce mortality in a subset of sepsis patients, and another IL-1β antagonist, canakinumab, has been shown to significantly reduce cardiovascular events in patients (23, 24). Therefore, there is high therapeutic potential in chemically inhibiting two forms of inflammatory cell death, pyroptosis and necroptosis, in tandem, in human patients during inflammatory disease.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents
HEK-293T cells [American Type Culture Collection (ATCC)] were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 15% SuperCalf Serum (Gemini BioProducts) with 1% antibiotic/antimycotic (Invitrogen). Murine nlrp3−/− iBMDMs constitutively expressing FLAG-NLRP3 and mCerulean-ASC were a gift from E. Latz (University of Bonn). iBMDMs were cultured in 10% SuperCalf Serum with 1% antibiotic/antimycotic and puromycin (3 μg/ml) (InvivoGen). THP-1 cells (ATCC) were cultured in RPMI with 10% fetal bovine serum (Gemini BioProducts) with 1% antibiotic/antimycotic and L-glutamine. Murine macrophages (Alnemri macrophages) used in assays of GSDME cell death were a gift from E. Alnemri (Jefferson College of Biomedical Sciences) and were cultured in DMEM with 10% SuperCalf Serum with 1% antibiotic/antimycotic.
Human GSDMD was cloned into the pNTAP-B or pEGFP vector using Gibson Assembly Cloning with no linking region between GSDMD and epitope tag. NTAP–p30-GSDMD and GFP–p30-GSDMD were created using site-directed mutagenesis (primers designed using PrimerX) introducing a stop codon at Gly276. Cysteine-to-alanine mutants were generated using site-directed mutagenesis on the NTAP–p30-GSDMD vector.
Gibson assembly cloning of the Gsdmd reconstitution vector was conducted using the following primer sets: insert: forward, 5′-AGATGTCGAAGAGaatCCTGGACCGATGGGGTCGGCCTTTGAGC-3′; reverse, 5′-AGAGGTTGATTGTCGACTTAACGCGTCTAGTGGGGCTCCTGGCTC-3′; backbone: forward, 5′-GAGCCAGG AG CCCC AC TA GACGCGTTAAGTCGACAATCAACCTCT-3′; reverse, 5′-GCTCAAAGGCCGACCCCATCGGTCCAGGATTCTCTTCGACATCT-3′. Gibson Assembly cloning of the pNTAP-GSDMD vector was conducted using the following primer sets: insert: forward, 5′-GGGCTGCCCGGGCGGATCCATGGGGTCGGCCTTT-GAGC-3′; reverse, 5′-GAGCCAGGAGCCCCACTAGGGATCCTAAGGTACCAGGTAAGTGTACCCAATTCGC-3′; backbone: forward, 5′-GAGCCAGGAGCCCCACTAGGGATCCTAAGGTACCAGGTAAGTGTACCCAATTCGC-3′; reverse, 5′-GCTCAAAGGCCGACCCCATGGATCCGCCCGGGCAGCCC-3′. The rational design and cloning of mNeonGreen GSDMD (used only in Fig. 2A) were previously described (16). All cloning and mutagenesis were verified using Sanger sequencing. Alignment of human and murine GSDMD amino acid sequences was conducted using Clustal Omega with formatting in ENDscript (ESPript 3.0) (25). Gsdmd−/− iBMDM cells were generated using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) and individually cloned out, with loss of GSDMD expression validated using Western blot as previously described (26, 27).
Assays of cell death
PI assays were conducted as previously described in cells primed with LPS (1 μg/ml) for 4 hours (16, 26, 28). Before the addition of NSA or DMSO, cells were washed with phosphate-buffered saline (PBS) and cultured in a basal salt solution containing PI (1 μg/ml) (Molecular Probes), 120 mM NaCl, 4 mM KCl, 1.5 mM CaCl2, 1 mM magnesium chloride, 25 mM Hepes, 5 mM glucose, and 0.1% bovine serum albumin (BSA) at pH 7.4. Cells were stimulated and read kinetically on a SpectraMax i3x Multimode microplate reader (Molecular Devices) at 533/617 nm (excitation/emission) at 37°C. Maximum fluorescence was obtained using 1% Triton X-100. Relative PI uptake was calculated on the basis of the background and maximum fluorescence in each well and was calculated as relative PI uptake = (sample-background)/ (maximum-background).
Cell death was measured by LDH release assay (Invitrogen). For p30-GSDMD cytotoxicity, HEK-293T cells were transfected with the NTAP–p30-GSDMD construct. Four hours after transfection, medium was changed and NSA or DMSO was added (if applicable). Twenty-four hours after the transfection, LDH release was measured. For cell death in iBMDM, THP-1, or murine macrophages for GSDME experiments, cells were plated at 100,000 cells per well in a 24-well plate. Cells were stimulated with ultrapure LPS (1 μg/ml) (InvivoGen) for 4 hours for GSDMD-mediated cell death. Cells were stimulated with 100 μM etoposide (Sigma) for 8 hours for GSDME-mediated cell death. Cells were then washed once in PBS and the medium was exchanged for the basal salt solution. One hour after stimulation, LDH release was measured.
Necroptotic cell death was induced by treating iBMDM cells with TNF-α (10 ng/ml), 20 μM zVAD-fmk (ApexBio), and GDC-0152 (MedChemExpress) for 24 hours in the presence or absence of 20 μM NSA. Cell death was analyzed by the relative release of LDH. Two-tailed t tests with α set at 0.05 were used for statistical analyses of LDH measurements of cell death.
Cell harvest, SDS–polyacrylamide gel electrophoresis, protein purification, and Western blot
For assessing the cleavage of GSDMD, iBMDM cells were primed with LPS and then stimulated with 10 μM nigericin for 1 hour. For assessing cleavage of GSDME, Alnemri macrophages were treated with etoposide for 8 hours. Cells were harvested in 8 M urea with 5% SDS and protease inhibitor cocktail (Sigma), phenylmethane sulfonyl fluoride, and calyculin (LC Labs). Samples were passaged five times through a 25-gauge needle, combined with SDS containing sample buffer and boiled. Harvesting of samples in nonreducing conditions used 0.5% NP-40 buffer with protease inhibitor cocktail and calyculin. Samples were combined 1:1 with sample buffer in the absence of any reducing agents. Purified protein with molecular weight marker was run on an SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and then fixed and stained with Coomassie brilliant blue in 50% methanol and 5% acetic acid.
E. coli expressing a 6×His-small ubiquitin-like modifier (SUMO)–GSDMD construct were cultured overnight. Bacteria were diluted 1:50 with overnight culture and then induced with isopropyl-β-D-thiogalactopyranoside at an OD600 (optical density at 600 nm) of 0.7. Cells were harvested in tris-HCl and NaCl, sonicated, centrifuged, and purified using a Ni column. On the column, 6×His-SUMO-GSDMD was washed twice and eluted using 500 mM imidazole. The 6×His-SUMO tag was cleaved using ULP-1 and removed using dialysis followed by a Ni column. Purified protein was verified by SDS-PAGE with Coomassie brilliant blue staining. In vitro binding assays with recombinant protein were conducted using 10 μg of recombinant protein incubated with or without biotin-conjugated NSA (Abcam). For pulldown with streptavidin beads (Sigma), beads were blocked in 5% BSA in tris buffer, while protein was precleared with beads that were subsequently discarded. After incubation with 50 μM NSA-biotin, beads were washed five times with tris buffer, combined with sample buffer and β-mercaptoethanol, and boiled. For GSDMD pulldown, recombinant protein G beads (Invitrogen) with mouse anti-GSDMD antibody (64-Y, Santa Cruz Biotechnology) were incubated with 10 μg of recombinant protein. After incubation, beads were washed five times with tris buffer, combined with sample buffer containing β-mercaptoethanol, and boiled. GSDMD was detected using a rabbit anti-GSDMD antibody (HPA044487, Sigma), and biotin was detected using a horseradish peroxidase–conjugated antibiotin antibody (Cell Signaling Technology).
Surface plasmon resonance
NSA binding with GSDMD was measured in PBS buffer with 0.05% (v/v) P20 (pH 7.4) by surface plasmon resonance using a Biacore T200 System (GE Healthcare) at 25°C. The S series sensor chip CM5 (GE Healthcare) was preconditioned at 100 μl/min with successive 20-μl pulses of 50 mM NaOH, 100 mM HCl, 0.1 mM SDS, and 0.085% (v/v) H3PO4. Recombinant GSDMD was immobilized on the sensor chip at high density, and an NSA concentration series ranging from 1.95 to 62.5 μM were injected over the surface. Each concentration was repeated twice, and affinity constant was calculated by BIAevaluation3 software (GE Healthcare). For steady-state analysis, the responses at 5 s after the injection were plotted.
Cytokine assays
The IL-1β enzyme-linked immunosorbent assay (ELISA) was run from supernatants from iBMDMs that were plated at 100,000 cells per well in a 24-well plate. Cells were primed for 4 hours with LPS (1 μg/ml), followed by pretreatment with DMSO, 10 μM NSA, or 20 μM NSA. Cells were activated with 10 μM nigericin for 30 or 60 min. Supernatants were removed and cleared by centrifugation at 13,000 rpm for 1 min. IL-1β in the supernatant was quantified by sandwich ELISA (BioLegend) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on samples following challenge with S. Typhimurium, Pam3CSK (1.0 μg/ml), or LPS (1.0 μg/ml). Overnight cultures of S. Typhimurium were diluted to 1:40 and grown for 2 hours. Cells were then infected at a multiplicity of infection (MOI) of 50:1. qRT-PCR of samples was conducted using primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-AGGCCGGTGCTGAGTATGTC-3′; reverse, 5′-TGCCTGCTTCACCACCTTCT-3′), IL-6 (forward, 5′-GCCTTCTTGGGACTGATGCT-3′; reverse, 5′-TGCCATTGCACAACTCTTTTCT-3′), TNF-α (forward, 5′-GGTGCCTATGTCTCAGCCTCTT-3′; reverse, 5′-GCCATAGAACTGATGAGAGGGAG-3′), and CXCL10 (forward, 5′-TCCTTGTCCTCCCTAGCTCA-3′; reverse, 5′-ATAACCCCTTGGGAAGATGG-3′). Statistical analyses of IL-1β release and gene transcription levels were carried out using two-tailed t tests with α set at 0.05.
LPS-induced model of sepsis in mice
For the LPS murine model of sepsis, mice (n = 10 DMSO control and n = 9 or 10 NSA-treated, C57BL/6J 8-week-old female mice) were injected with LPS (25 mg/kg) (O111:B4; Sigma, catalog # L2630) and either one or two doses of NSA or DMSO (20 mg/kg). Total volume of injected NSA or DMSO was about 25 μl per mouse per dose administered via intraperitoneal injection with an insulin syringe 30 min before the per dose administration of LPS (one dose) or 30 min before LPS administration and 6 hours after administration (two doses). Survival curves were analyzed using a log-rank (Mantel-Cox) test in GraphPad Prism (GraphPad Software Inc.). Serum cytokines were measured by ELISA 6 hours after treatment with LPS.
Imaging
For confocal imaging, eight-well #1.5 chambered coverglasses (Nunc) were coated with collagen I (Corning) at 50 μg/ml in 1% acetic acid for 1 hour in a 37°C incubator, washed with PBS, and then seeded with iBMDM cells at 100,000 per well. Phase contrast and epifluorescent images were acquired on a Leica DMIL light-emitting diode microscope. Live cell confocal imaging was performed on a Leica DMI 6000B inverted microscope within a custom-made environment box, held at 37°C using an AirTherm ATX heater (World Precision Instruments). The objective was a Leica 506375 HC FLUOTAR 25×/0.95 water immersion objective with 0.17-mm glass correction. An argon 488-nm laser line was used for excitation, and 8-bit photomultiplier tubes were used for detection. Deconvolution was performed with Huygens Professional 17.10 using CMLE with SNR 3. Visualization was performed using maximum intensity projections in Imaris (Bitplane). The brightness of the DMSO image was increased relative to the other images for display purposes in Fig. 2A.
Structural modeling
The full-length human GSDMD and murine GSDMD structural models were created using the Multiple Mapping Method (MMM) server (http://manaslu.fiserlab.org/MMM/) with the full-length mGSDMA3 structure (Protein Data Bank code 5b5r) as a template (29).
Supplementary Material
Table S1. Raw data sets.
Fig. S1. Etoposide-induced cell death in macrophages is independent of NSA.
Fig. S2. Purification of human GSDMD.
Fig. S3. Conserved cysteine residues between human and murine GSDMD in the p30-GSDMD fragment.
Fig. S4. Single-dose NSA prolongs survival in the LPS model of sepsis.
Fig. S5. NSA inhibits p30-GSDMD pore formation and cell death.
Acknowledgments:
We thank the Abbott, Li, Xiao, Ramakrishnan, Dubyak, Huang, and Adoro laboratories for thoughtful discussions and scientific insights. We also thank J. Stamler (Case Western Reserve University) for technical advice and critical analysis. We appreciate the help of F. Allen and J. Myers in isolating BMDM cells. Funding: J.K.R., B.L.B., H.C.K., and S.M.C. were supported by NIH grant T32 GM007250. B.L.B. was additionally supported by F31 NS 096857, T32NS077888, and TL1 RR024991. G.R.D. was supported by R01-EY014362 and National Muscular Dystrophy Association grant NMSS RG5130A2/1. X.L. was supported by NIH grant P01HL029582 and a grant from the National Multiple Sclerosis Society (RG-1707-28180). This work was further supported by NIH grants P01091222 and R01086550 and a Harrington Discovery Institute fellowship to D.W.A.
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Huang D, Zheng X, Wang Z.-a., Chen X, He W.-t., Zhang Y, Xu J-G, Zhao H, Shi W, Wang X, Zhu Y, Han J, The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol. Cell Biol 37, e00497–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S, Liu H, Johnston A, Hanna-Addams S, Reynoso E, Xiang Y, Wang Z, MLKL forms disulfide bond-dependent amyloid-like polymers to induce necroptosis. Proc. Natl. Acad. Sci 114, E7450–E7459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, Wang X, Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F, Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM, Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang D-C, Shao F, Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC, GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. U.S.A 113, 7858–7863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Müller DJ, Broz P, Hiller S, GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35, 1766–1778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J, Inflammasomeactivated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latz E, Xiao TS, Stutz A, Activation and regulation of the inflammasomes. Nat. Rev. Immunol 13, 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuang S, Zheng J, Yang H, Li S, Duan S, Shen Y, Ji C, Gan J, Xu X-W, Li J, Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc. Natl. Acad. Sci. U.S.A 114, 10642–10647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Wang C, Rathkey JK, Yang J, Dubyak GR, Abbott DW, Xiao TS, Structures of the Gasdermin D C-terminal domains reveal mechanisms of autoinhibition. Structure 26, 778–784.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He W.-t., Wan H, Hu L, Chen P, Wang X, Huang Z, Yang Z-H, Zhong C-Q, Han J, Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiPeso L, Ji DX, Vance RE, Price JV, Cell death and cell lysis are separable events during pyroptosis. Cell Death Discovery 3, 17070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X, Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Rathkey JK, Benson BL, Chirieleison SM, Yang J, Xiao TS, Dubyak GR, Huang AY, Abbott DW, Live-cell visualization of gasdermin D-driven pyroptotic cell death. J. Biol. Chem 292, 14649–14658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES, Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun 8, 14128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins GR, Wen H, Ting JP-Y, Inflammasomes and metabolic disorders: Old genes in modern diseases. Mol. Cell 54, 297–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doitsh G, Galloway NLK, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Muñoz-Arias I, Greene WC, Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M, Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut 59, 1192–1199 (2010). [DOI] [PubMed] [Google Scholar]
- 21.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE, Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490, 107–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J, Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ; CANTOS Trial Group, Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med 377, 1119–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, Cron RQ, Opal SM, Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of the macrophage activation syndrome: Reanalysis of a prior Phase III trial. Crit. Care Med 44, 275–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, 320–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, Dubyak GR, Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J. Immunol 197, 1353–1367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chirieleison SM, Marsh RA, Kumar P, Rathkey JK, Dubyak GR, Abbott DW, Nucleotide-binding oligomerization domain (NOD) signaling defects and cell death susceptibility cannot be uncoupled in X-linked inhibitor of apoptosis (XIAP)-driven inflammatory disease. J. Biol. Chem 292, 9666–9679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsnelson MA, Rucker LG, Russo HM, Dubyak GR, K+ efflux agonists induce NLRP3 inflammasome activation independently of Ca2+ signaling. J. Immunol 194, 3937–3952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai BK, Fiser A, Multiple mapping method: A novel approach to the sequence-tostructure alignment problem in comparative protein structure modeling. Proteins 63, 644–661 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw data sets.
Fig. S1. Etoposide-induced cell death in macrophages is independent of NSA.
Fig. S2. Purification of human GSDMD.
Fig. S3. Conserved cysteine residues between human and murine GSDMD in the p30-GSDMD fragment.
Fig. S4. Single-dose NSA prolongs survival in the LPS model of sepsis.
Fig. S5. NSA inhibits p30-GSDMD pore formation and cell death.