Abstract
The time and costs associated with the sequencing of a human genome have decreased significantly in recent years. Many people have chosen to have their genomes sequenced to receive genomics-based personalized healthcare services. To reach the goal of genomics-based precision medicine, health information management (HIM) professionals need to manage and analyze patients' genomic data. Two important pieces of information from the genome sequence are the risk of genetic diseases and the specific medication or pharmacogenomic results for the individual patient, both of which are linked to a patient's genetic variations. In this review article, we introduce genetic variations, including their data types, relevant databases, and some currently available analysis methods and systems. HIM professionals can choose to use these databases, methods, and systems in the management and analysis of patients' genomic data.
Keywords: genetic variation, precision medicine, database
Introduction
In 2015, the Precision Medicine Initiative (PMI) was launched in order to understand how a person's genetics, environment, and lifestyle can help determine the best approach to prevent or treat disease.1 In the past, medical practices followed certain protocols and prescribed medications without considering patients' genetic makeup, environment, or lifestyle, even though healthcare providers have a general understanding of the importance of these factors. One major reason was that accurate data about these factors was not easily accessible. Ten years ago, sequencing a human genome was still very expensive, and no reliable and inexpensive methods of collecting environmental and lifestyle data were available. Even electronic health record (EHR) systems were not widely adopted then. Today, all these factors are easily accessible and reliable because of the availability of high-throughput DNA sequencing technologies, wide adoption of mobile health apps and various types of wearable trackers, and the extensive use of EHR systems in almost all hospitals and clinics in the United States. Therefore, it has become feasible to collect a large amount of health-related data from patients and apply the findings in precision medicine.
Precision medicine will make it possible for physicians to choose the best treatment and prevention strategy for each patient according to their personal health information, including their genetic makeup.2 Some human genetic variations are closely related to certain diseases or individual patient responses to certain medications;3 therefore, physicians can choose specific treatment options to optimize the patient's outcome. For example, in precision medicine, physicians can choose different medications to help their patients quit smoking by examining the patient's speed of nicotine metabolization.4
In the PMI, now renamed All of Us, more than one million Americans will be recruited. Their genomic, EHR, environment, diet, and lifestyle information will be collected and integrated together to investigate the risk for a range of diseases. Individual responses to commonly used drugs, biomarkers associated with common diseases, and targeted therapies will also be investigated. The recruitment of study participants is ongoing. As stated in the executive summary of the PMI Working Group report, “Precision medicine seeks to redefine our understanding of disease onset and progression, treatment response, and health outcomes through the more precise measurement of molecular, environmental, and behavioral factors that contribute to health and disease. This understanding will lead to more accurate diagnoses, more rational disease prevention strategies, better treatment selection, and the development of novel therapies.”5
To successfully reach the goals described in the PMI report, extensive collaboration is needed among researchers and professionals from medicine, public health, genetics/genomics, computer science, and health information management (HIM). These professionals need to work together to recruit study participants, develop methods and tools to collect desired data, analyze and interpret the data, and report the results to the public.
In the desired data items from the PMI, personal genomic information is relatively new for HIM researchers and professionals. In this article we introduce one important type of personal genomic data: genetic variations, including their major types, databases, and some currently available variation analysis methods and systems.
Types of Genetic Variations
A genome is an organism's complete set of DNA sequences. Although people in this world may look different, with different skin color, eye color, hair color, and height, all human genomes are highly similar. More than 99 percent of human genomes from two unrelated individuals are identical.6–8 The differences among these genomes are genetic variations. Most of these genetic variations either do not show noticeable differences or simply make us look different. Some genetic variations can cause diseases.
Several types of genetic variations exist. This article focuses on the following three types of genetic variations:
1. Single nucleotide polymorphisms (SNPs)
2. Short insertions and deletions (INDELs)
3. Copy number variations (CNVs)
Single Nucleotide Polymorphisms
A DNA polymorphism is any difference in the nucleotide sequence between individuals. SNPs are the most common type of genetic variation. They represent a difference in a single nucleotide.9 The difference can be one extra nucleotide (insertion) at the specific location, one missing nucleotide (deletion), or a different nucleotide (substitution), compared with the reference genome sequenced in the Human Genome Project. SNPs represent roughly 90 percent of the human DNA polymorphisms.10 Figure 1 illustrates the concept of SNPs, in which six individuals have either T or C at one specific position in their chromosome 2.
Figure 1.
Illustration of Single Nucleotide Polymorphisms (SNPs). Source: National Human Genome Research Institute. Available at http://www.genome.gov/glossary/index.cfm?p=viewimage&id=185.
SNPs can occur anywhere in a genome. When SNPs occur within a gene or within the regulatory region (which may regulate the expression of a gene or genetic component) near or inside a gene, they may directly affect the gene function or expression and may increase the risk of developing certain diseases,11, 12 especially if the gene is critically important for the normal function of cells. SNPs that occur in intergenic regions (or regions between two genes) are less likely to lead to gene function or expression changes or disease.
Research in pharmacogenomics (the study of how genes affect a person's response to medications) indicates that SNPs may be helpful for predicting an individual's response to certain medications, which is critically important for precision medicine.13 For example, genetic polymorphisms in the CYP450 genes (CYP450 enzymes are responsible for metabolizing drugs14, 15) affect the metabolism of about 25 percent of all medication treatments.16, 17 These genetic polymorphisms can influence medication response, causing it to be normal, enhanced, reduced, or neutralized.18 Additionally, genetic variations in three genes (CYP2C9, VKORC1, and CYP4F2) can result in significant variability in the appropriate dose of warfarin between patients.19
Short Insertions and Deletions
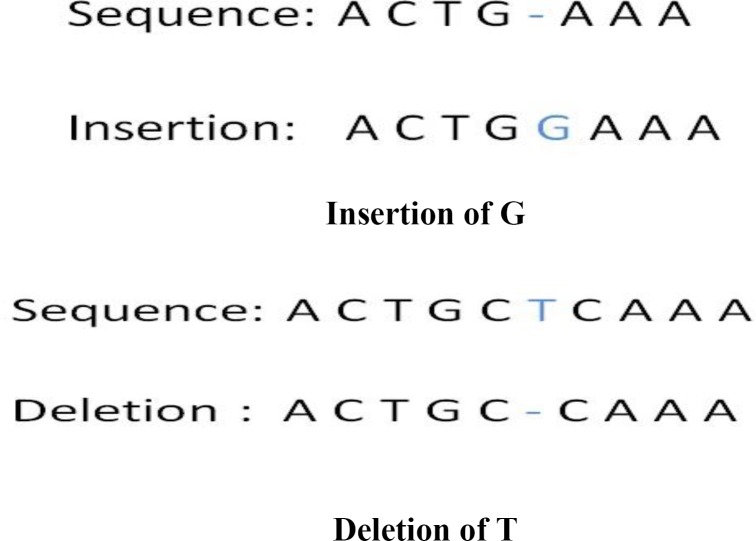
INDELs are insertions or deletions of nucleotides in a DNA sequence. One single nucleotide insertion or deletion is a special case of INDELs. The insertions and deletions can be a long segment (a few hundred or a few thousand nucleotides) of a chromosome.20 Both insertion and deletion of nucleotides will change the number of nucleotides in a DNA sequence. If either of them occur in a gene, then the protein made by the gene may not function properly. For example, the X-linked recessive disorder previously seen in European royal families21 results from the insertion of a large segment of DNA leading to inactivation of a protein required for blood clotting. Figure 2 illustrates the concept of INDELs.
Figure 2.
Illustration of Single-Base Insertions and Deletions (INDELs)
In the human genome, approximately 36 percent of short INDELs are in the genetic region.22 Therefore, some of these INDELs can have an impact on human gene function/expression. Examples of diseases related to short insertions include Huntington disease (CAG repeated more than 35 times in gene HTT) and myotonic dystrophy (a segment of DNA repeated too many times in gene DMPK or CNBP).23 An example of a disease associated with a short deletion is cystic fibrosis (a deletion of three nucleotides at the 508th position on the CFTR gene).24
Copy Number Variations
CNV refers to an intermediate-scale genetic change (in the range from 1,000 nucleotides to several million nucleotides).25, 26 CNVs include additional copies of a DNA sequence segment (duplications) or losses of a DNA segment (deletions).27 For example, A-B-C-D is a long DNA sequence in one chromosome; here, A, B, C, and D are DNA segments longer than 1,000 nucleotides. This chromosome in some people can instead have the following sequence: A-B-B-C-D (a duplication of B) or A-C-D (a deletion of B). The scale of this type of genetic change may affect multiple genes in certain cases.28
There is an increasing concern about the effect of CNVs in the development of complex diseases29 because a number of CNVs overlap with protein coding regions.30 CNVs have been detected in genetic regions associated with complex neurological diseases,31 such as autism,32 Alzheimer's disease,33 and schizophrenia.34
Genetic Variation Databases
Many databases have been created to manage the growing number of genomic data sets. These databases help researchers and healthcare professionals obtain genetic information quickly and conveniently.35 A number of databases contain genetic variations and related information. In this article, we briefly introduce the following two categories of databases:
1. Variation databases that provide information about genetic variations alone.
2. Variations and disease/phenotype databases that provide information about phenotypes and genetic variations. (Phenotype refers to the observable characteristics of an organism, such as its morphology, development, physiological properties, and behavior.)
Variation Databases
The National Center for Biotechnology Information at the National Library of Medicine maintains dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). a database of short genetic variations. This database stores all types of genetic variations of less than 50 base pairs in multiple species.36 Therefore, it includes different types of short genetic variations, such as SNPs, INDELs, and microsatellite markers.
In dbSNP, HIM professionals can find detailed information about each genetic variation, such as the type of the variation, the location of the variation in a chromosome, the relevant genes, and population-specific allele frequencies and genotypes, which are all critically important in precision medicine.37 For instance, according to one recent journal article,38 a SNP with ID rs2981579 is associated with breast cancer. A simple query in dbSNP with this SNP ID can tell us that this genetic variation is in the FGFR2 gene and the specific location of this genetic variation is in chromosome 10, position 121577821; and it occurs in multiple populations, such as European, Asian, and African populations. Therefore, in healthcare practice, HIM professionals can specifically check whether their patients have this genetic variation in this gene (FGFR2) and provide the results to physicians.
The dbVar database (http://www.ncbi.nlm.nih.gov/dbvar) archives large-scale genetic variations for multiple species and is based on various genomic studies. A number of large-scale variations are associated with diseases. Data in dbVar are organized according to the study or publication. Genetic variation types in dbVar include INDELs and CNVs.39 HIM professionals can perform searches in dbVar to determine the type of genetic variation, the location of the variation, the relevant gene, and the clinical relevance of the variation (e.g., pathogenic, likely pathogenic, uncertain significance).
The Database of Genomic Variations (DGV; http://dgv.tcag.ca/dgv/app/home) provides a catalog of human structural variations including INDELs and CNVs. The database is periodically updated according to results reported in peer-reviewed research articles. DGV aims to catalog the highest-quality structural variations in the literature, and it presents the results in a format that is convenient for healthcare professionals and researchers.40 HIM professionals can search the DGV database using keywords, gene symbols, genetic regions, and the specific location on a chromosome. This database can be helpful for HIM professionals to use when collecting data for specific disease registries, such as cancer, stroke, and diabetes registries.
Variations and Disease/Phenotype Databases
Online Mendelian Inheritance in Man (OMIM; http://www.ncbi.nlm.nih.gov/omim) is a comprehensive database that catalogs known diseases and their associated genetic variations. Each OMIM record has a summary of one disease and its relevant genes and variations in human genome as reported in the literature.41 It is particularly useful when HIM professionals want to find all genetic variations associated with one specific disease, or genetic diseases associated with one genetic variation or gene.
The Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php) is the gold-standard resource for comprehensive data on published human inherited disease mutations.42 Each HGMD record includes a reference to the first literature report of a mutation, the associated disease specified in that report, and the gene name, symbol, and chromosomal location. HGMD can be useful to HIM professionals looking for databases to add to a healthcare system data repository. It can also provide useful information on clinical trials, evidence-based best practices, and quality improvement studies.
ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) is a database that provides information about the medical relevance of genetic variations.43 It archives the relationship between medically significant variations and their phenotypes. ClinVar is strongly related to dbSNP and dbVar because it maintains information about the location of variations in human genome assemblies. Unlike dbSNP and dbVar, Clinvar accepts direct submissions of structured details of phenotypes, and it provides interpretation of the functional and clinical significance of genetic variations. ClinVar can be used by HIM professionals when overseeing the clinical registry function within their healthcare organization.
SNPedia (https://www.snpedia.com/) is a wiki-based database.44 Researchers convert information presented in large-scale peer-reviewed genomic studies into a machine-readable format and then store the information in SNPedia so that the information is easily accessible to researchers. SNPedia supports personal genome annotation, interpretation, and analysis. This database links the genetic variations to information about diseases or phenotypic traits published in genomic studies. HIM professionals can refer researchers to the important information provided in this database or use it themselves when compiling annual reports and other business reports that examine linkages of disease.
The use of these databases can be outlined as follows: Short variations can be found in dbSNP. Structural genetic variations can be found in dbVar. Structural variations from healthy people can be searched in DGV. The associations between human SNPs and disease/phenotype can be obtained from SNPedia. OMIM can be used to find the association between human genetic variations and diseases, including an extensive description of relevant genes and phenotypes. HGMD can be used to learn more about the association between human variations and genetic diseases. Table 1 summarizes these genetic variation databases.
Table 1.
Summary of Genetic Variation Databases
| Name | Purpose | Species |
|---|---|---|
| dbSNP | Stores all types of genetic variations less than 50 base pairs | All species |
| dbVar | Archives large-scale genetic variations | All species |
| DGV | Provides a catalog of structural variations, including insertion, deletion, and copy number variations | Human |
| OMIM | Catalogs known diseases with their genetic component | Human |
| HGMD | Includes a reference to the first literature report of a mutation, the associated disease state, and the gene name, symbol, and chromosomal location | Human |
| ClinVar | Archives the medically significant variations and phenotypes | Human |
| SNPedia | Converts the information in large-scale peer-reviewed genomic studies into machine-readable format | Human |
Genetic Variation Analysis
After a genome is sequenced, one critically important step is to analyze the sequence data. Specific to genetic variations, there are four types of analysis:
1. Variation identification,
2. Variation annotation,
3. Variation visualization, and
4. Variation and disease association.
These tasks are typically done by genetics/genomics researchers instead of HIM professionals; therefore, we will only briefly describe them. The more clinically relevant variation analysis task is genetic variation filtering and identification of associated diseases via existing tools, which is described at the end of this section.
Variation Identification
Variation identification is the process of identifying genetic variations from DNA sequence data.45 Usually variation identification can be done by comparing a newly sequenced human genome with the reference human genome and identifying candidate sites at which one or more samples differ from the reference sequence.46 Several tools for identifying variations, such as CRISP, GATK, and SAMtools, are available.47, 48
Variation Annotation
Variation annotation refers to the classification and prediction of the functional impact of variations, followed by filtering and prioritization of the ones that cause diseases.49 Several tools are available for variation annotations; these tools include ANNOVAR,50 VEP,51 and SVA.52
Variation Visualization
Variation visualization refers to the validation and visual representation of genetic variations.53 IGV54 and UCSC Genome Browser55 (https://genome.ucsc.edu) are commonly used variation visualization tools at various levels of detail, from base pair level to chromosome level.
Variation and Disease Association
The study of genetic diseases has a long history.56 However, only after the recent improvements in DNA sequencing,57 researchers can now study genetic variations in the whole genome and their association with diseases. For instance, in recent years, genome-wide association studies (GWAS) have been widely used for the discovery of genetic risk factors associated with human diseases.
Genetic Variation Analysis Tools for Clinical Purposes
In this section, we present several tools that help HIM professionals manage and analyze genetic variations conveniently. After all, once a patient's genomic data is integrated into the patient's EHR, HIM professionals will be the key personnel to use existing software tools to analyze and manage the data and make the analysis results available to physicians for precision medicine practice.
An integrated system of patient genomic information management and analysis for healthcare professionals was created to enable genetic variation analysis and management in the healthcare environment.58 In this Java-based system, HIM professionals can upload the patient's genetic variation data in VCF format (VCF–Variant Call Format, a widely used text file format for describing genetic variations) and obtain reports about the risks of genetic disease and the patient's response to certain medications.
VCF-Explorer is a software tool that can be used to perform analysis for large genetic variation files in the VCF format.59 The software program can run on various computational platforms, from laptops to high-performance computers. HIM professionals can use the graphical user interface (GUI) to filter variations based on specific samples or variant-level annotations.
VCF. Filter is a user-friendly and standalone tool that allows users to search for genetic variations that cause rare diseases.60 The software provides a GUI that helps users design filtering rules and apply them to any VCF file.
The myVCF desktop application allows users to explore, query, visualize, and export genetic variation data.61 The application provides a browser interface that is compatible with Windows, MacOS, and UNIX systems. It can manage multiple VCF files, and it allows users to browse the information contained in the VCF file. The application provides a flexible search engine that allows users to perform queries about region/variant coordinates, gene symbols, or dbSNP IDs.
VariantDB is a web-based annotation and filtering platform that automatically annotates variations with allele frequencies, functional impact, pathogenicity predictions, and pathway information.62
After patients have their genome sequenced, HIM professionals can use variation identification programs to determine genetic variations in patients. They can use the software tools mentioned in this article to filter genetic variations associated with certain diseases, and they can visualize these genetic variations, perform searches in variation-disease databases to learn details about these genetic variations, and explain the results to physicians. Physicians can then design a personalized treatment plan for each patient. In this entire process, HIM professionals do not need to know the detailed implementations of these software programs and databases, but they do need to have basic knowledge of them and know how to search in these databases and use these tools. The obtained results (variations and reports from variation analyses) can be stored in the EHR so that they can be used to support the physician's treatment decisions, clinical coding, and patient outcomes. In other words, working on genomic data is a natural extension of the HIM professional's typical job functions, such as managing data, performing data analysis using software programs, and protecting patients' data security and privacy.63
Conclusion
This article provides a brief introduction to genetic variations and some existing databases and data analysis software programs. Because HIM professionals have the skills for managing and processing large-scale clinical data, they may need only a minimum amount of training in genomics (such as basic concepts, frequently used databases, and data analysis programs) to be able to understand genomic data and extract desired information for clinical practice. In this way, HIM professionals can play a key role in the implementation of precision medicine nationwide.
Contributor Information
Amal Adel Alzu'bi, The Department of Computer Information Systems at Jordan University of Science and Technology in Irbid, Jordan..
Leming Zhou, The Department of Health Information Management at the University of Pittsburgh in Pittsburgh, PA..
Valerie J. M. Watzlaf, The Department of Health Information Management at the University of Pittsburgh in Pittsburgh, PA..
Notes
- 1.Collins, S. F., Varmus H. “A New Initiative on Precision Medicine.”. New England Journal of Medicine. 2015;372(no. 9):793–95. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn R., Adams K., Kowalski, et al R. “The Patient in Precision Medicine: A Systematic Review Examining Evaluations of Patient-Facing Materials.”. Journal of Healthcare Engineering. 2018;2018:9541621. doi: 10.1155/2018/9541621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juengst E., McGowan M. “Why Does the Shift from ‘Personalized Medicine’ to ‘Precision Health’ and ‘Wellness Genomics' Matter?”. AMA Journal of Ethics. 2018;20(no. 9):e881–e890. doi: 10.1001/amajethics.2018.881. [DOI] [PubMed] [Google Scholar]
- 4.Lerman C., A. Schnoll R., W L., Hawk “Use of the Nicotine Metabolite Ratio as a Genetically Informed Biomarker of Response to Nicotine Patch or Varenicline for Smoking Cessation: A Randomised, Double-Blind Placebo-controlled Trial.”. The Lancet Respiratory Medicine. 2015;3(no. 2):131–38. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Precision Medicine Initiative Working Group The Precision Medicine Initiative Cohort Program – Building a Research Foundation for 21st Century Medicine. 2015:p. 1. Available at https://www.nih.gov/sites/default/files/research-training/initiatives/pmi/pmi-working-group-report-20150917-2.pdf. [Google Scholar]
- 6.Shafer A. “Understanding Genetics.”. The Tech, Stanford University. 2006 [Google Scholar]
- 7.Lander, S. E., M. Linton L., Birren B., et al. “Initial Sequencing and Analysis of the Human Genome.”. Nature. 2001;409(no. 6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Venter, C. J., D. Adams M., W. Myers E., et al. “The Sequence of the Human Genome.”. Science. 2001;291(no. 5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 9.Altshuler D., Pollara V.J., Cowles C.R., et al. “An SNP Map of the Human Genome Generated by Reduced Representation Shotgun Sequencing.”. Nature. 2000;407(no. 6803):513–16. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 10.Varela, A. M., Amos W. “Heterogeneous Distribution of SNPs in the Human Genome: Microsatellites as Predictors of Nucleotide Diversity and Divergence.”. Genomics. 2010;95(no. 3):151–59. doi: 10.1016/j.ygeno.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Q., Ancona N., R. Hauser, et al E. “Integrating Genetic and Gene Expression Evidence into Genome-wide Association Analysis of Gene Sets.”. Genome Research. 2012;22(no. 2):386–97. doi: 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talseth-Palmer, A. B., J R., Scott “Genetic Variation and Its Role in Malignancy.”. International Journal of Biomedical Science. 2011;7(no. 3):158–71. [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson B. “SNPs—A Shortcut to Personalized Medicine.”. Genetic Engineering & Biotechnology News. 2008;28(no. 13) [Google Scholar]
- 14.Leon-Cachon, B. R., A. Ascacio-Martinez J., A. Barrera-Saldana H. “Individual Response to Drug Therapy: Bases and Study Approaches.”. Revista de Investigación Clínica. 2012;64(no. 4):364–76. [PubMed] [Google Scholar]
- 15.Ventola, C. L. “Role of Pharmacogenomic Biomarkers in Predicting and Improving Drug Response: Part 1: The Clinical Significance of Pharmacogenetic Variants.”. Pharmacy and Therapeutics. 2013;38(no. 9):545–60. [PMC free article] [PubMed] [Google Scholar]
- 16.Ibid
- 17.Ma, D. J., C. Lee K., M. Kuo G. “Clinical Application of Pharmacogenomics.”. Journal of Pharmacy Practice. 2012;25(no. 4):417–27. doi: 10.1177/0897190012448309. [DOI] [PubMed] [Google Scholar]
- 18.Leon-Cachon, B. R., A. Ascacio-Martinez J., A. Barrera-Saldana H. “Individual Response to Drug Therapy: Bases and Study Approaches.”. Revista de Investigación Clínica. 2012;64(no. 4):364–76. [PubMed] [Google Scholar]
- 19.Salari K., Watkins H., A E., Ashley “Personalized Medicine: Hope or Hype?”. European Heart Journal. 2012;33(no. 13):1564–70. doi: 10.1093/eurheartj/ehs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clancy S. “DNA Deletion and Duplication and the Associated Genetic Disorders.”. Nature Education. 2008;1(no. 1):23. [Google Scholar]
- 21.Rogaev, I. E., P. Grigorenko A., Faskhutdinova G., et al. “Genotype Analysis Identifies the Cause of the ‘Royal Disease.’”. Science. 2009;326(no. 5954):817. doi: 10.1126/science.1180660. [DOI] [PubMed] [Google Scholar]
- 22.Mills, E. R., T. Luttig C., E. Larkins C., et al. “An Initial Map of Insertion and Deletion (INDEL) Variation in the Human Genome.”. Genome Research. 2006;16(no. 9):1182–90. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergeron B. Case Studies in Genes and Disease: A Primer for Clinicians. Philadelphia, PA: American College of Physicians; 2004. [Google Scholar]
- 24.Massie J., Delatycki M. “Cystic Fibrosis Carrier Screening.”. Paediatric Respiratory Reviews. 2013;14(no. 4):270–75. doi: 10.1016/j.prrv.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Stankiewicz P., R J., Lupski “Structural Variation in the Human Genome and Its Role in Disease.”. Annual Review of Medicine. 2010;61:437–55. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 26.Pollex, L. R., A R., Hegele “Copy Number Variation in the Human Genome and Its Implications for Cardiovascular Disease.”. Circulation. 2007;115(no. 24):3130–38. doi: 10.1161/CIRCULATIONAHA.106.677591. [DOI] [PubMed] [Google Scholar]
- 27.Eichler, E. E. “Copy Number Variation and Human Disease.”. Nature Education. 2008;1(no. 3):1. [Google Scholar]
- 28.Check E. “Human Genome: Patchwork People.”. Nature. 2005;437(no. 7062):1084–86. doi: 10.1038/4371084a. [DOI] [PubMed] [Google Scholar]
- 29.Beckmann, S. J., Estivill X., E. Antonarakis S. “Copy Number Variants and Genetic Traits: Closer to the Resolution of Phenotypic to Genotypic Variability.”. Nature Reviews Genetics. 2007;8(no. 8):639–46. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 30.Sebat J., Lakshmi B., Troge, et al J. “Large-Scale Copy Number Polymorphism in the Human Genome.”. Science. 2004;305(no. 5683):525–28. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 31.Girirajan S., D. Campbell C., E E., Eichler “Human Copy Number Variation and Complex Genetic Disease.”. Annual Review of Genetics. 2011;45:203–26. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gai X., M. Xie H., C. Perin, et al J. “Rare Structural Variation of Synapse and Neurotransmission Genes in Autism.”. Molecular Psychiatry. 2012;17(no. 4):402–11. doi: 10.1038/mp.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng X., Y. Demirci F., M.M. Barmada, et al. “Genome-wide Copy-Number Variation Study of Psychosis in Alzheimer's Disease.”. Translational Psychiatry. 2015;5:e574. doi: 10.1038/tp.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, M. S., A. Castellani C., L. O'reilly R. “Copy Number Variation Showers in Schizophrenia: An Emerging Hypothesis.”. Molecular Psychiatry. 2009;14(no. 4):356–58. doi: 10.1038/mp.2008.149. [DOI] [PubMed] [Google Scholar]
- 35.Lathe W., III, Williams J., Mangan M., Karolchik D. “Genomic Data Resources: Challenges and Promises.”. Nature Education. 2008;1(no. 3):2. [Google Scholar]
- 36.Wheeler, L. D., Barrett T., A. Benson D., et al. “Database Resources of the National Center for Biotechnology Information.”. Nucleic Acids Research. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NCBI Resource Coordinators “Database Resources of the National Center for Biotechnology Information.”. Nucleic Acids Research. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michailidou K., Hall P., Gonzalez-Neira, et al A. “Large-Scale Genotyping Identifies 41 New Loci Associated with Breast Cancer Risk.”. Nature Genetics. 2013;45(no. 4):353–61. doi: 10.1038/ng.2563. 361e1–361e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lappalainen I., Lopez J., Skipper, et al L. “DbVar and DGVa: Public Archives for Genomic Structural Variation.”. Nucleic Acids Research. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDonald, R. J., Ziman R., K. Yuen R., et al. “The Database of Genomic Variants: A Curated Collection of Structural Variation in the Human Genome.”. Nucleic Acids Research. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamosh A., F. Scott A., S. Amberger, et al J. “Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders.”. Nucleic Acids Research. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stenson, D. P., Mort M., V. Ball E., et al. “The Human Gene Mutation Database: Building a Comprehensive Mutation Repository for Clinical and Molecular Genetics, Diagnostic Testing and Personalized Genomic Medicine.”. Human Genetics. 2014;133(no. 1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landrum, J. M., M. Lee J., Benson M., et al. “ClinVar: Improving Access to Variant Interpretations and Supporting Evidence.”. Nucleic Acids Research. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cariaso M., Lennon G. “SNPedia: A Wiki Supporting Personal Genome Annotation, Interpretation and Analysis.”. Nucleic Acids Research. 2012;40:D1308–D1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stitziel, O. N., Kiezun A., Sunyaev S. “Computational and Statistical Approaches to Analyzing Variants Identified by Exome Sequencing.”. Genome Biology. 2011;12(no. 9):227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.1000 Genomes Project Consortium “A Map of Human Genome Variation from Population-Scale Sequencing.”. Nature. 2010;467(no. 7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quintánsa B., Ordóñez-Ugaldea A., Cacheiroa P., et al. “Medical Genomics: The Intricate Path from Genetic Variant Identification to Clinical Interpretation.”. Applied & Translational Genomics. 2014;3(no. 3):60–67. doi: 10.1016/j.atg.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altmann A., Weber P., Quast, et al C. “vipR: Variant Identification in Pooled DNA Using R.”. Bioinformatics. 2011;27(no. 13):i77–i84. doi: 10.1093/bioinformatics/btr205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy, J. D., Humburg P., Kanapin A., et al. “Choice of Transcripts and Software Has a Large Effect on Variant Annotation.”. Genome Medicine. 2014;6(no. 3):26. doi: 10.1186/gm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pabinger S., Dander A., Fischer, et al M. “A Survey of Tools for Variant Analysis of Next-Generation Genome Sequencing Data.”. Briefings in Bioinformatics. 2014;15(no. 2):256–78. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLaren W., Pritchard B., Rios, et al D. “Deriving the Consequences of Genomic Variants with the Ensembl API and SNP Effect Predictor.”. Bioinformatics. 2010;26(no. 16):2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge D., K. Ruzzo E., Shianna K.V., et al. “SVA: Software for Annotating and Visualizing Sequenced Human Genomes.”. Bioinformatics. 2011;27(no. 14):1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen, B. C., Cantor M., Dubchak I., et al. “Visualizing Genomes: Techniques and Challenges.” Nature Methods 7no. 3S2010S5–S15. [DOI] [PubMed] [Google Scholar]
- 54.Thorvaldsdottir H., T. Robinson J., P J., Mesirov “Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration.”. Briefings in Bioinformatics. 2013;14(no. 2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raney, J. B., R. Dreszer T., P. Barber G., et al. “Track Data Hubs Enable Visualization of User-defined Genome-wide Annotations on the UCSC Genome Browser.”. Bioinformatics. 2014;30(no. 7):1003–5. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sturtevant “The Linear Arrangement of Six Sex-linked Factors in Drosophila A. as Shown by Their Mode of Association.”. Journal of Experimental Biology. 1913;14:43–59. [Google Scholar]
- 57.Bras J., Guerreiro R., Hardy J. “Use of Next-Generation Sequencing and Other Whole-Genome Strategies to Dissect Neurological Disease.”. Nature Reviews Neuroscience. 2012;13(no. 7):453–64. doi: 10.1038/nrn3271. [DOI] [PubMed] [Google Scholar]
- 58.Alzu'bi A., Zhou L. In 2017 IEEE International Conference on Healthcare Informatics. Los Alamitos, CA: IEEE Computer Society; 2017. “An Integrated Patient Genomic Information Management and Analysis System for Healthcare Professionals.”; pp. 107–13. [Google Scholar]
- 59.Akgün M., Demirci H. “VCF-Explorer: Filtering and Analysing Whole Genome VCF Files.”. Bioinformatics. 2017;33(no. 21):3468–70. doi: 10.1093/bioinformatics/btx422. [DOI] [PubMed] [Google Scholar]
- 60.Müller H., Jimenez-Heredia R., Krolo, et al A.“VCF. Filter: Interactive Prioritization of Disease-linked Genetic Variants from Sequencing Data.” Nucleic Acids Research 45no. W12017W567–W572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pietrelli A., Valenti L. “myVCF: A Desktop Application for High-Throughput Mutations Data Management.”. Bioinformatics. 2017;33(no. 22):3676–78. doi: 10.1093/bioinformatics/btx475. [DOI] [PubMed] [Google Scholar]
- 62.Vandeweyer G., Laer L., Loeys, et al B. “VariantDB: A Flexible Annotation and Filtering Portal for Next Generation Sequencing Data.”. Genome Medicine. 2014;6(no. 10):74. doi: 10.1186/s13073-014-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alzu'bi A., Zhou L., Watzlaf V.“Personal Genomic Information Management and Personalized Medicine: Challenges, Current Solutions, and Roles of HIM Professionals.” Perspectives in Health Information Management (Spring 2014) [PMC free article] [PubMed] [Google Scholar]