Abstract
The PTEN/PI3K/mTOR signal transduction pathway is involved in the regulation of biological processes such as metabolism, cell growth, cell proliferation, and apoptosis. This pathway has been extensively studied in mammals, leading to the conclusion that PTEN is a major tumor suppressor gene. PTEN orthologues have been characterized in a variety of organisms, both vertebrates and non-vertebrates, and studies of the associated PTEN/PI3K/mTOR pathway indicate that it is widely conserved. Studies in fungal systems indicated a role of PTEN in fungal defense mechanisms in Candida albicans, and in the developmental process of sporulation in Saccharomyces cerevisiae. The present study was aimed at investigating the role of the PTEN ortholog, ptn1, in Ustilago maydis, the pathogen of maize. U. maydis ptn1 mutant strains where ptn1 gene is deleted or overexpressed were examined for phenotypes associate with mating, virulence and spore formation. While the overexpression of ptn1 had no substantial effects on virulence, ptn1 deletion strains showed slight reductions in mating efficiency and significant reductions in virulence; tumor formation on stem and/or leaves were severely reduced. Moreover, tumors, when present, had significantly lower levels of mature teliospores, and the percent germination of such spores was similarly reduced. Thus, ptn1 is required for these important aspects of virulence in this fungus.
Keywords: Ustilago maydis, PTEN, teliospore development
1. Introduction
Ustilago maydis, is a plant pathogen responsible for smut disease in Zea mays [1]. In the past few decades, U. maydis has developed into a model system for a variety of research areas, including but not limited to, cellular transport processes, plant–pathogen interaction and DNA repair mechanisms [2]. Due to the high level of genetic similarity shared between U. maydis and metazoans, it may serve as a viable model for the study of essential cellular processes of higher eukaryotes [2,3]. Several homologues of human proteins that are involved in DNA repair and genomic stability are present in U. maydis, such as the BRCA2 orthologue, Brh2 [4].
PTEN (phosphatase deleted on chromosome 10; also known as MMAC1 or TEP1 (tensin-like phosphatase) is a human tumor suppressor gene mapping onto chromosome 10 at 10q23.3 [5]. The PTEN gene is either deleted or inactivated in several tumors, including those associated with breast, endometrial, brain, and prostate cancers [6]. The sequence of the phosphatase domain of PTEN is most similar to those of dual specificity protein phosphatases such as cell division cycle 14 (CDC14), baculovirus phosphatase (BVP), DUSP3, and DUSP4. However, the amino-terminal region of PTEN containing the phosphatase catalytic domain, also has homology to proteins such as actin-binding protein, tensin-1 (TNS1), and auxilin, the cofactor of ATPase heat shock protein HSC70, though such proteins are unrelated to PTEN catalytic functions [7]. The PTEN gene codes for a protein that is a major phosphatase enzyme that dephosphorylates PtdIns(3,4,5)P3 (PIP3) to PtdIns(4,5)P2 (PIP2), thereby counteracting the signaling by phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3Ks) and playing a major role in the regulation of the PTEN/AKT/mTOR signaling pathway [8]. Studies conducted in mouse models and humans suggest that PTEN is highly regulated and very critical for cellular functions. It is evident from mouse model studies that a 20% reduction in PTEN expression level is enough to develop a range of tumors [9].
PTEN is mostly known for its tumor suppressor activity; however, it has also been shown to be involved in a variety of other functions, including the regulation of cell size, metabolism, ageing, and the development of neurons [10]. Orthologs of PTEN have been identified in Drosophila melanogaster [11], Caenorhabditis elegans [12,13], Candida albicans [14], and budding yeast [15]. PTEN is essential for Drosophila, where it regulates cell size, cell proliferation, and apoptosis [11]. In C. elegans, mutations in PTEN may cause disruptions in the developmental process, characterized by constitutive dauer (larval morphology) formation [12,13]. The deletion of the PTEN orthologue, TEP1, in the budding yeast, S. cerevisiae, revealed that it has a role in sporulation. TEP1 mutants had defects in the trafficking or deposition of dityrosine, a major component of yeast spore walls [15]. In another yeast species, C. albicans, PTEN is involved in the defense mechanism, where it activates cofilin-1, an actin depolymerization factor during PGE2 (prostaglandin E2)-mediated inhibition of fungal phagocytosis by macrophages [14].
In this study, we have begun the functional characterization of the sole PTEN ortholog, ptn1 (UMAG_03760), in U. maydis. We examined the phenotypes of U. maydis cells overexpressing or deleted for ptn1. In the absence of a functional ptn1 gene, U. maydis haploid cells grow normally in axenic culture, but displayed a reduced capability to tolerate cell wall stress. More strikingly, such mutants, when mixed with compatible mating partners, were defective in tumor formation, teliospore development, and the rate of teliospore germination.
2. Materials and Methods
2.1. Strains and Growth Conditions
The U. maydis strains used in this study are listed in Table 1. Escherichia coli strains TOP10 and DH5α (Invitrogen/Thermo Fisher) were used for all cloning and plasmid maintenance. U. maydis strains were grown at 26 °C in solid or liquid yeast extract-peptone-sucrose medium (YEPS), potato dextrose agar (PDA), Holliday salt solution (HSS) media [16], or Holliday Salt media, as described previously [17]. Strains grown in liquid media were shaken overnight at 250 rpm. Mating media was made with solid PDA and 1% activated charcoal, while stress media was made with solid YEPS and either Congo Red (Fisher Scientific, Fairlawn, NJ, 15 µg/mL), sodium chloride (1 M), calcofluor white (50 µM; Sigma-Aldrich, St. Louis, MO, USA) or sorbitol (1 M), as described in [18].
Table 1.
List of strains.
| Ustilago Strain | Genotype a | References |
|---|---|---|
| 1/2 | a1b1 | [19] |
| 2/9 | a2b2 | [19] |
| 1/2 Δptn1 | a1b1 ptn1::cbxR | This study |
| 1/2 Δptn1J | a1b1 ptn1::hygR | This study |
| 2/9 Δptn1 | a2b2 ptn1::cbxR | This study |
| 2/9 Δptn1J | a2b2 ptn1::hygR | This study |
| FB1 Δptn1J | a1b1 ptn1::hygR | This study |
| 1/2 ptn1otef | a1b1 Potef-ump2, cbxR | This study |
| 2/9 ptn1otef | a2b2 Potef-ump2, cbxR | This study |
| SG200 | a1mfa2 bE1bW2 | [20] |
| SG200 Δptn1 | a1mfa2 bE1bW2 ptn1::hygR | This study |
| d132 | a1a2b1b2 | [21] |
| d132 Δptn1 | a1a2b1b2 ptn1:: hygR | This study |
| 1/2 Δptn1c | a1b1 ptn1::cbxR Potef-ptn1, hygR | This study |
| 2/9 Δptn1c | a2b2 ptn1::cbxR Potef-ptn1, hygR | This study |
| FBD11-7 | a1a1 b1b2 | [16] |
| FBD12-17 | a2a2 b1b2 | [16] |
| Plasmids | Genotype | References |
| DelsGate ptn1 | kanR ptn1::cbxR | [22] |
| Otef-ptn1-cbx | bla(ampR) Potef-ptn1 cbxR | This study |
| Otef-ptn1-hyg | bla(ampR) Potef-ptn1 hygR | This study |
acbxR, carboxin resistance; hygR, hygromycin resistance; the otef promoter is a modified tef promoter in which two direct repeats of a synthetic fragment containing seven tetracycline-responsive elements precede the promoter.
2.2. Vector Construction and Nucleic Acid Manipulations
The ptn1 deletion strains in U. maydis were made, using homologous recombination by the DelsGate protocol, as described previously [22], using primers (Table 2) designated UPTN-K1-K4 to amplify the flanks of ptn1. The strains 1/2 Δptn1 and 2/9 Δptn1 were created by replacing the ptn1 open reading frame (ORF) with the DelsGate deletion construct plasmid containing a carboxin resistance marker. Confirmed Δptn1 mutants were complemented with a plasmid containing a hygromycin resistance gene, and a wild type copy of the gene under the control of the constitutive otef promoter (Potef), to yield 1/2 Δptn1c and 2/9 Δptn1c complemented strains. The overexpression strains of ptn1 were generated by expressing the ptn1 ORF under the control of Potef, using a derivative of p123 bearing a carboxin resistance marker [23]. Potef-based vectors were linearized using SspI digestion prior to transforming U. maydis protoplasts [23]. Additional ptn1 deletion strains 1/2 Δptn1J, 2/9 Δptn1J, FB1 Δptn1J, SG200 Δptn1, and d132 Δptn1 were generated via replacing the ptn1 ORF with a hygromycin resistance cassette. The hygromycin replacement construct was prepared through overlap-PCR [24,25], using primers PTENUPLT, PTENUPRT, PTENDNLT, CPDNRT, HYGPTENLT, HYGPTENRT (Table 2) as described previously [24], with minor modifications.
Table 2.
List of primers.
| Primers | Sequence (5’-3’) |
|---|---|
| UPTN-K1 | TAGGGATAACAGGGTAATCAACAAAATGACGCAACCAC |
| UPTN-K3 | GGGACAAGTTTGTACAAAAAAGCAGGCTAAGAACGATGTTTGCGTGTCAG |
| UPTN-K4 | GGGGACCACTTTGTACAAGAAAGCTGGGTACGTTAGACAAAGCGCAATCA |
| UPTN-K2 | GGAAGGTGATCGTGAAGGAA |
| PTENUPLT | GCAACAAAATGACGCAACC |
| PTENUPRT | GTAGTTACCACGTTCGGCCATAAGGTGGTGGGACTGCTTT |
| PTENDNLT | GCTTTGCTCTTTGCTCTTCTG |
| CPDNRT | GCCTTAATTAATATACGCCCGATTGCTTACC |
| HYGPTENLT | AAAGCAGTCCCACCACCTTATGGCCGAACGTGGTAACTAC |
| HYGPTENRT | AGAAGAGCAAAGAGCAAAGCCTCAGGCCTCATGTTTGACA |
| PTEN5 | TGAGGCCTGAGTGGCCTATATACCCCCTGCCCCTGT |
| PTEN3 | CGAATTCTTGCGCTTTGTCTAACGAAC |
| HYGSPH1RTNEW | TTTGCATGCCTCAGGCCTCATGTTTGACA |
| HYGMFE1 | TTTCAATTGTGGCCGAACGTGGTAACTAC |
Transformed U. maydis strains were selected on YEPS agar plates containing either 3 μg carboxin per mL (Santa Cruz Biotechnology, Dallas, TX, USA) or 150 μg/mL hygromycin B (Gold Biotech). Transformation was confirmed using PCR with primers that would be diagnostic for true transformants, and distinct from bands for wild type (e.g., PTEN5/PTEN3). Also, for Δptn1 derivatives, qPCR was used to confirm the loss of gene expression relative to the wild type. Primers were designed using the Primer3 software (http://bioinfo.ut.ee/primer3/) based on the genome sequence available at JGI (http://genome.jgi.doe.gov/Ustma1/Ustma1.home.html). PCR was carried out in a T100 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). For PCR amplification reactions, either Hot Start ExTaq DNA Polymerase, PrimeSTAR HS DNA Polymerase (Takara Bio, USA), Platinum Taq Green Hot Start DNA Polymerase (Life Technologies Corporation), or EconoTaq DNA Polymerase (Lucigen Corporation) were used.
2.3. RNA Isolation and Expression Analysis
RNA isolation from U. maydis strains and quantitative real-time PCR were carried out, as described in [26]. The RNA expression data were analyzed in GraphPad Prism by using unpaired T-tests after normalization to wild-type expression under high ammonium conditions; a probability value of p < 0.05 was considered as a significant difference.
2.4. Cell Growth, Mating, and Plant Infection
For cell growth rate and mating assays, liquid cultures of the strains grown overnight in 4 mL YEPS liquid media shaken at 260 rpm were used at an absorbance at 600 nm (OD600) of between 0.6–0.8. To determine growth rates, cells grown overnight to reach exponential growth phase were collected, washed, and resuspended in fresh YEPS liquid media. Subsequently, 4 × 106 cells were transferred into 10 mL of fresh YEPS liquid media, and optical density was determined every 2 h for the first 12 h and every 6 hr afterwards, up to 30 h [23,27]. Data were analyzed using Graphpad Prism. Mating assays were performed using U. maydis cells grown overnight in YEPS broth to an OD600 of 0.8, and cells were resuspended in water to an OD600 of 1.6 prior to spotting. A volume of 10 µL of opposite mating-type strains was co-spotted onto PDA charcoal plates, and the mating reaction was observed after incubation at 26 °C for 24–48 h [19].
For plant infection assay, 4 mL overnight cultures were diluted using 50 mL fresh YEPS liquid media and grown overnight to an OD600 of 1.0, harvested and resuspended in water to an OD600 of 3 prior to infection [20]. Plant infection was performed by inoculating 7–9 days old maize seedlings (Golden Bantam seeds, Bunton Seed Co., Louisville, KY and W. Atlee Burpee & Co., 62 Warminster, PA). Haploid strains of opposite mating types were mixed prior to plant infection, while diploid d132 and the solopathogenic SG200 strain were used directly, as previously described [19,28]. The virulence of each treatment group was scored by a disease index (DI) on a scale of 0 to 5, where 0 = No Symptoms, 1 = Chlorosis, 2 = Small leaf tumors, 3 = Large leaf tumors or small stem tumors, 4 = Large stem tumors with bending, and 5 = Death [19,29]. Thirty plants were used in each experiment, and the trials were repeated three times, with plants scored for disease symptoms on 14 and 21 days post-infection (dpi). A haploid a1b1 background mutant strain crossed with a haploid a2b2 mutant strain was compared in each case to the corresponding wild type cross. Additional measures of virulence included estimations of biomass, numbers of leaves, plant length, and chlorophyll content of infected plants. The dry biomass index of the infected plants was measured as previously described [30]. The above-ground parts of the infected plants where both partners were wild type, or both were mutant (i.e., Δptn1 × Δptn1 or ptn1otef × ptn1otef) were collected eight days post-infection (dpi), and washed once with tap water and twice with distilled water. Plants were blotted dry, and the samples were stored in labeled paper bags, and dried at 65 ± 5 °C in an incubator until a constant weight and dry biomass was recorded. Chlorophyll extraction was performed by the DMSO method, and contents were determined spectrophotometrically, as described previously [31,32] Plant infections were also performed on a dwarf variety of maize (Tom Thumb popcorn, High Mowing Organic Seeds, Walcott, VT) mainly to facilitate maize cob infections. For maize cob infection, maize plants were de-tasseled to increase the prospect of cob infection by preventing the pollination of ovules. Cobs were injected, as described previously [29,33]. In separate experiments, dwarf maize plants were infected with wild type (WT) and mutant strains on the leaf, following the protocol described [22].
2.5. Statistical Analysis
Statistical analysis of the disease index data from plant infection was conducted using Kruskal–Wallis Test with a Multiple Comparison Test using R software [34,35]. The data of the plant morphological characteristics such plant length, leaf number, dry biomass, chlorophyll content, spore germination rate. and spore count were analyzed using analysis of variance (ANOVA) followed by Tukey’s test [36] using GraphPad Prism software, and the probability values of p < 0.05 were considered as being significant.
3. Results
3.1. In Silico Analysis of ptn1
Initial analysis of the U. maydis genome sequence revealed the presence of a hypothetical-like gene (UMAG_03760), hereafter known as ptn1, predicted to encode a protein of 848 amino acids. The nucleotide sequence analysis (BLASTn) of the ptn1 gene found the greatest similarity to the PTENB gene (XM_012330711.1) of Pseudozyma hubeiensis, a fungus also belonging to the order Ustilaginales. The U. maydis Ptn1 protein is characterized by the conserved protein tyrosine phosphatase catalytic (PTPc) domain motif (more specifically, the CDC14 subgroup of PTP proteins) present in other PTEN orthologues, as well as a phosphatase tensin-type domain, which is characteristic of all PTEN proteins (Figure 1, Figure S1). In silico prediction tools such as SignalP (http://www.cbs.dtu.dk/services/SignalP) and TargetP (http://www.cbs.dtu.dk/services/TargetP) suggested that Ptn1 is a cytoplasmic protein (Figures S2 and S3).
Figure 1.
Sequence alignment of U. maydis Ptn1 and other PTEN proteins. Ptn1 protein was compared to selected proteins from Basidiomycete fungi Pseudozyma hubeiensis and Sporisorium scitamineum; Ascomycete fungi, Saccharomycetes cerevisiae; and the mammals, Mus musculus and Homo sapiens. The red color shows a high degree of sequence similarity in amino acids at the N-terminal regions between Ptn1 and the other identified proteins.
3.2. Deletion of ptn1 Led to the Reduced Production of the Aerial Hyphae Characteristics of the Mating Reaction on Charcoal Media
Wild type, Δptn1, and overexpression strains (ptn1otef) were spotted onto PDA charcoal plates to assess any defects in mating or post-mating filamentation (Figure 2). Compatible wild type haploids (1/2 a1b1 x 2/9 a2b2) produced dikaryotic hyphae that appeared as white ‘fuzzy’ growth when co-spotted. However, compatible ptn1 deletion strains when co-spotted, showed a reduction in ‘fuzz’, especially when examined after the first 24 h. However, overexpression strains did not show a major change in ability to produce aerial hyphae. The complemented mutants, Δptn1c, were able to partially recover the wild type phenotype, indicative of successful mating.
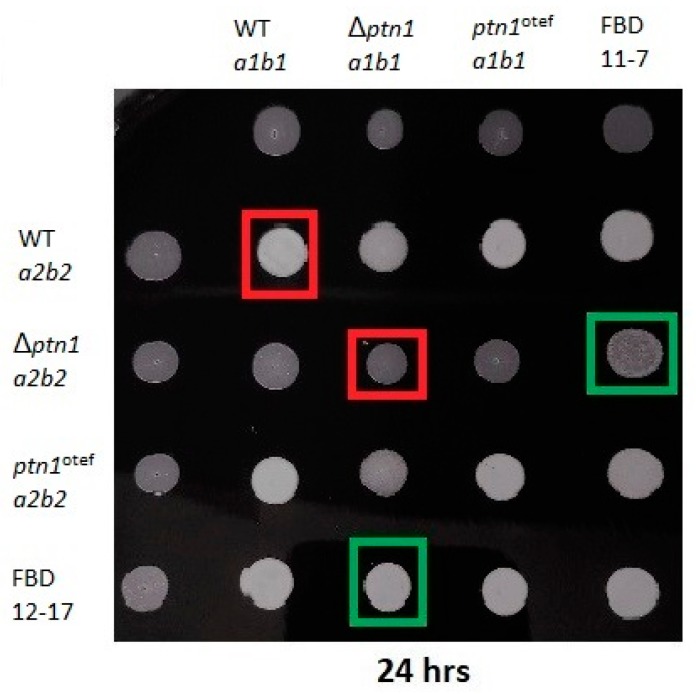
Figure 2.
Mating assay and testing for pheromone production. Charcoal plate mating assay with wild type, Δptn1, Δptn1c, and FBD-tester strains. A positive mating reaction is indicated by a white “fuzz” phenotype of aerial hyphae production. The reduction in filamentation was noticeable at 24 h, while such differences disappeared by 48 h (not shown). Red boxes compare the ‘fuzz’ produced between wild type mating and Δptn1 mating. Green boxes indicate the ‘fuzz’ produced when Δptn1 strains were mixed with tester strains.
To determine whether Δptn1 cells produce pheromone, mating was performed using U. maydis tester strains, FBD11-7 and FBD12-17 (Figure 2). Such diploid strains are homozygous at the a mating locus, and they do not produce aerial hyphae when spotted alone on charcoal, but will become filamentous when they are plated together with cells that produce a different pheromone. The ‘fuzzy’ appearance appeared in the case of both the wild type a1b1 and a2b2 partners, together with their respective tester strains; on the other hand, there was a slight reduction in the production of aerial hyphae for the ptn1 deletion strain in the a2b2 background when paired with tester strain FBD11-7, suggesting reduced production of the a2 pheromone by this mutant.
As an additional level of confirmation for the observed defects in the ‘fuzzy’ phenotype, the mating assay was performed using deletion strains in an additional genetic background (FB1). When mating of the wild type (1/2 × 2/9) was compared with that of ptn1 deletion strains (1/2 Δptn1 × 2/9 Δptn1, 1/2 Δptn1J × 2/9 Δptn1J, FB1 Δptn1J × 2/9 Δptn1J), similar results were obtained as in Figure 1, i.e., there was reduction in mating at 24 h for deletion strains, irrespective of the construct or the background (Figure 3A).
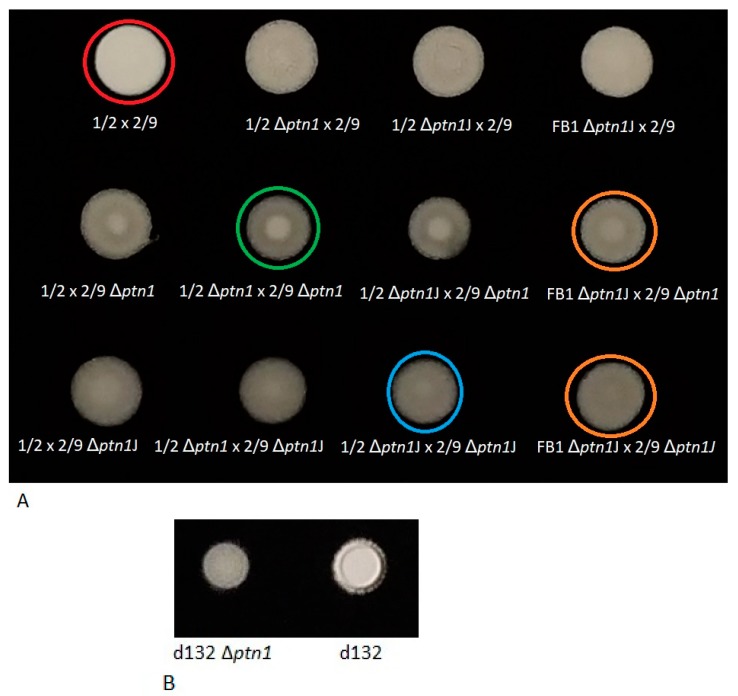
Figure 3.
(A) Mating compared between deletion strains using two different constructs and different genetic backgrounds. Compared to wild type mating (1/2 × 2/9) denoted by the red circles, mating of deletion strains (1/2 Δptn1 × 2/9 Δptn1, green circle, as well as 1/2 Δptn1J × 2/9 Δptn1J, blue circle) showed reduction in mating at 24 h. This result matched the observation made when a haploid deletion strain Δptn1J in the FB1 background was mated with a haploid Δptn1 or Δptn1J strain in the 2/9 background (indicated by orange circles). (B) Formation of aerial hyphae for wild type ptn1ptn1 or mutant ptn1Δptn1 diploid d132 background at 24 h.
Formation of aerial hyphae on charcoal was also examined for ptn1 deletion mutants in the diploid background (d132 Δptn1) and in the solopathogenic SG200 background (SG200 Δptn1). The deletion strain in the SG200 background did not show any change in the ‘fuzz’ intensity (Figure S4). However, the d132Δptn1 strain showed reduced formation of aerial hyphae (Figure 3B), compared to the wild type. This result indicates that ptn1 gene does show some level of haplo-insufficiency in the d132 diploid background, as has been observed for other genes in this species [28]. In contrast, the loss of ptn1 appears at least partially compensated by the presence of the wild type allele in dikaryons as demonstrated by 2/9 x 1/2 Δptn1.
3.3. prf1 Is Upregulated in Δptn1 under Low Ammonium Conditions
The lower than expected formation of aerial hyphae during the mating of the 2/9 Δptn1 a2b2 with the tester strain, FBD 11-7 strain led us to speculate whether there was an alteration in the expression of mating genes affecting pheromone production. The fusion of haploid cells during mating leads to the differentiation of U. maydis into subsequent filamentous dikaryon [20]. Also, environmental conditions such as acidic pH, lipids, and low ammonium conditions can trigger the filamentous response in U. maydis [37,38,39], while acetate inhibits filamentation, due to mating or to exposure to oleic acid [40]. Hence, a qRT-PCR analysis was conducted for several genes involved in the mating pathway, e.g., mfa2, encoding a2 pheromone precursor, pra2, encoding pheromone receptor, and prf1, encoding a transcription factor [41]. The analysis involved comparing the expression levels between wild type 2/9 and 2/9 Δptn1 under high and low ammonium conditions. The expression of all the strains were compared to the wild type 2/9 strain with high ammonium (Figure S5). The data indicated that the prf1 levels decreased between high and low ammonium conditions in wild type, whereas it significantly increased in the Δptn1 mutant from high to low ammonium. There was a significant difference between the relative expression of prf1 between wild type under low ammonium, and Δptn1 mutants under low ammonium. Even though there was no significant difference in the expression of mfa2 and pra2 under similar conditions, the trend showed that while the mfa2 and pra2 genes tended to be less well expressed under low ammonium conditions in wild type, for the Δptn1 mutant, their expression appeared to be in the opposite direction.
3.4. Cell Growth of ptn1 Mutants Is Largely Unaffected, while Stress Tolerance Is Reduced under Certain Conditions
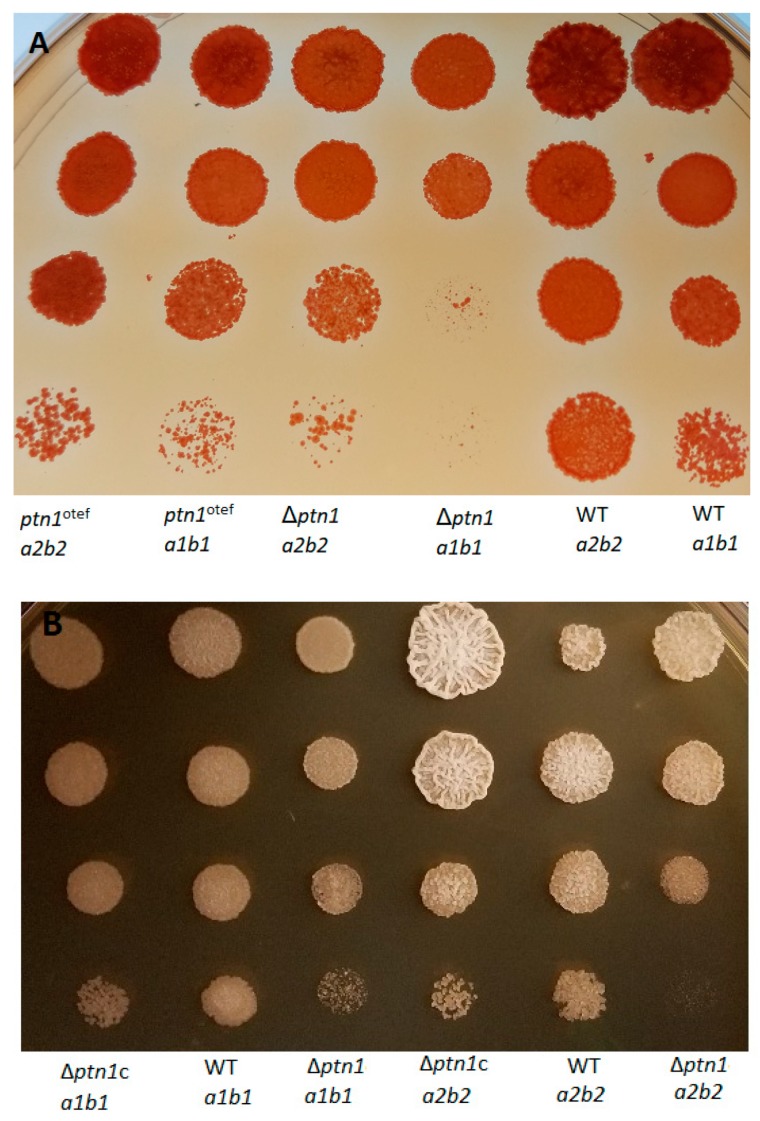
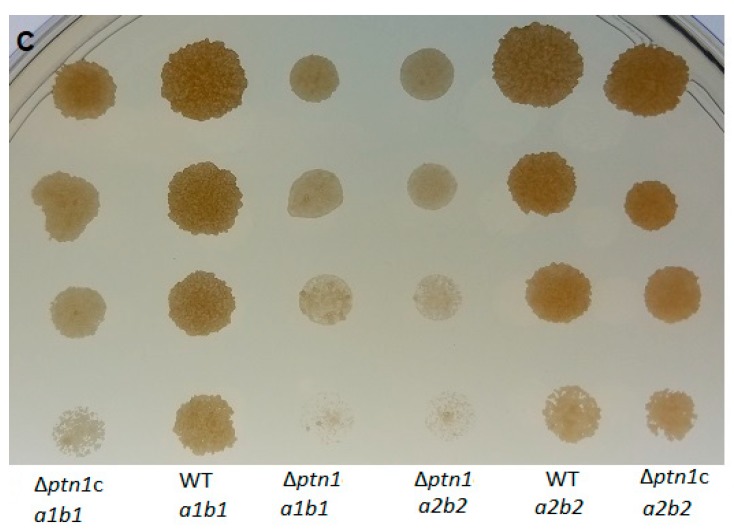
In standard growth media such as yeast extract-peptone-sucrose (YEPS) or potato dextrose broth (PDB), ptn1 mutants were able to grow and showed no significant differences from wild type cells, in terms of growth rate (Figure S6). The effects of stressors, including high osmotic medium, i.e., YEPS supplemented with 1 M sorbitol or 1 M NaCl was tested, but not found to affect growth (data not shown). However, ptn1 mutants showed a difference from wild type strains on YEPS media supplemented with Congo Red (CR) or calcofluor white (CFW). Both ptn1 deletion and overexpression strains showed reduced tolerance when compared to the respective wild type strains (Figure 4A), while the complemented strains were able to partially recover the wild type growth (Figure 4B) in CR media, as well as on CFW media (Figure 4C). Overall, these experiments suggested that the mutation of ptn1 has an impact on the integrity of the cell wall.
Figure 4.
Response of mutants to cell wall stress. (A) 10-fold serial dilutions of the 1/2 a1b1, 2/9 a2b2, Δptn1 a1b1, Δptn1 a2b2, ptn1otef a1b1, and ptn1otef a2b2 strains were spotted onto media containing 1 mM Congo Red, and observations were made at 48 h. Congo Red is a cell wall stressor which prevents glycan microfibril assembly by binding to β-glucans. The 1/2 a1b1 strains were compared with a1b1 mutant strains, while a2b2 background strains were compared amongst themselves. Growth defects were observed in the Δptn1 strains, as well as ptn1otef, as reductions in colony numbers at higher dilutions compared to comparable wild type, for both the a1b1 and a2b2 backgrounds. The overexpression of ptn1 ectopically in deletion mutants was able to partially rescue the growth defect in (B) Congo Red media and (C) calcofluor white media.
3.5. Pathogenicity Is Reduced by ptn1 Deletion
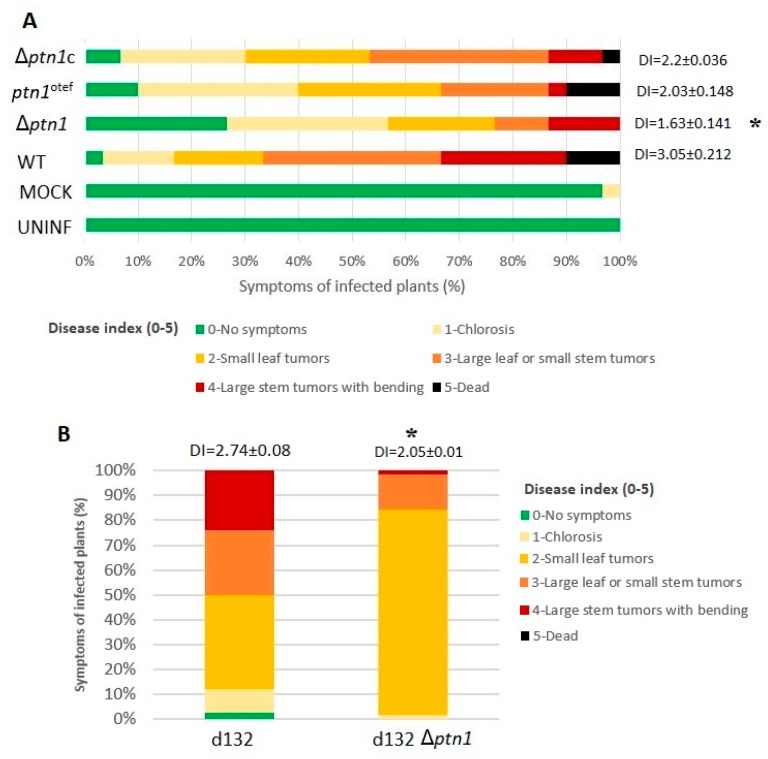
Compatible opposite mating type strains were mixed and injected into maize seedlings to assess the effect of deletion of ptn1 on U. maydis virulence. The results at 14 dpi are presented in the graph as a percentage of symptom formation in infected plants. Statistical analysis was performed on the disease index score (mean ± standard deviation) with a Kruskal–Wallis test and a multiple-comparison test. The results (Figure 5) indicate that deletion of ptn1 results in reduced virulence compared to the wild type, and this reduction was statistically significant compared to the other infections. There were significantly more healthy plants surviving at the end of the study among the plants infected with ptn1 deletion mutants. On the other hand, overexpression of ptn1 did not yield significant changes in the virulence. For infections where both partners carried the ptn1 deletion, the ability of the fungus to form tumors was severely reduced. While in infections with both wild type partners approximately 82% of infected plants had tumors, only roughly 30% of infections had tumors when both mating partners carried the Δptn1 lesion. The complemented Δptn1c mating partners were partially able to rescue this defect. More plants (~90%) showed signs of infection in the Δptn1c infected group when compared to the Δptn1-infected plants (~68%).
Figure 5.
Plant infection. (A) One-week-old maize seedlings were infected with indicated strains of U. maydis. Disease symptoms were scored 14 days post-infection (dpi), and compared to those where both partners were wild type (WT) strains (1/2 a1b1 × 2/9 a2b2). Plants were inoculated with one paired background, indicated on the Y-axis. Disease symptom categories are depicted at the bottom. Numbers to the right of each bar represent mean (±standard deviation) for disease indices (DI). The graphs display the percentage of plants with specific symptoms of infection. The Δptn1 (a1b1 × a2b2) and ptn1otef (a1b1 × a2b2) infections; here, both partners contained the indicated mutations/modifications, displayed reduced pathogenicity. The reduction in Δptn1 strains was statistically significant (p < 0.05) compared to the wild type used in this study. (B) Experiments conducted using diploid strains d132 WT and d132 Δptn1 showed that the deletion of one copy of ptn1 reduces pathogenicity. Numbers above each bar represent mean (± standard deviation) for disease indices (DI). The data were analyzed using Kruskal–Wallis test, followed by a post hoc comparison, and an asterisk (*) indicates significant difference (p < 0.05).
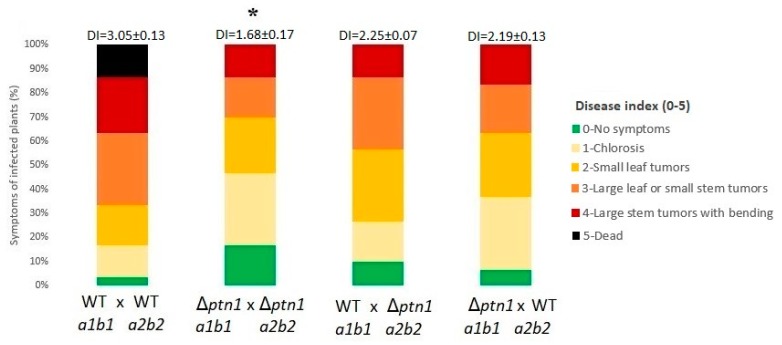
In a separate experiment, deletion strain d132 Δptn1 also showed a reduced severity of infection compared to its wild type progenitor, indicating that one functional copy of ptn1 was insufficient for full pathogenicity (Figure 5B). The phenotypic trend of reduced pathogenicity was confirmed by using deletion mutant in a SG200 background, though the difference (p = 0.05048) was only marginally significant (Figure S7). In addition to the above-mentioned infections using d132 background strains, the ptn1 deletion strain in the a1b1 background mixed with the WT strain in a2b2 background was examined for the effect of virulence when one wild-type copy of the gene was present; i.e., the 1/2 a1b1 Δptn1 strain was crossed with the 2/9 a2b2 strain, and vice versa. This was compared with wild type (1/2 × 2/9) infection and the deletion mutant infection (1/2 Δptn1 × 2/9 Δptn1). The results are shown in Figure 6, and they indicate that the presence of one wild-type copy can rescue the phenotype to some extent.
Figure 6.
Are functional copies of the ptn1 gene in both partners required for pathogenicity? The Δptn1 strains were mated with the wild type strain of the opposite mating-type and compared with infections due to either both wild type or both mutant pairings. The graphs display the percentage of plants with specific symptoms of infection. The combinations of 1/2 Δptn1 a1b1 × 2/9 a2b2 and 1/2 a1b1 × 2/9 Δptn1 a2b2 were more virulent when compared to Δptn1 (a1b1 × a2b2), but less that when compared to 1/2 × 2/9. The numbers above each bar represent the mean (±standard deviation) for disease indices (DI). The data were analyzed using the Kruskal–Wallis test, followed by a post hoc comparison, and an asterisk (*) indicates significant difference (p < 0.05).
3.6. Pathogenicity Calculated by Additional Plant Growth Indicators
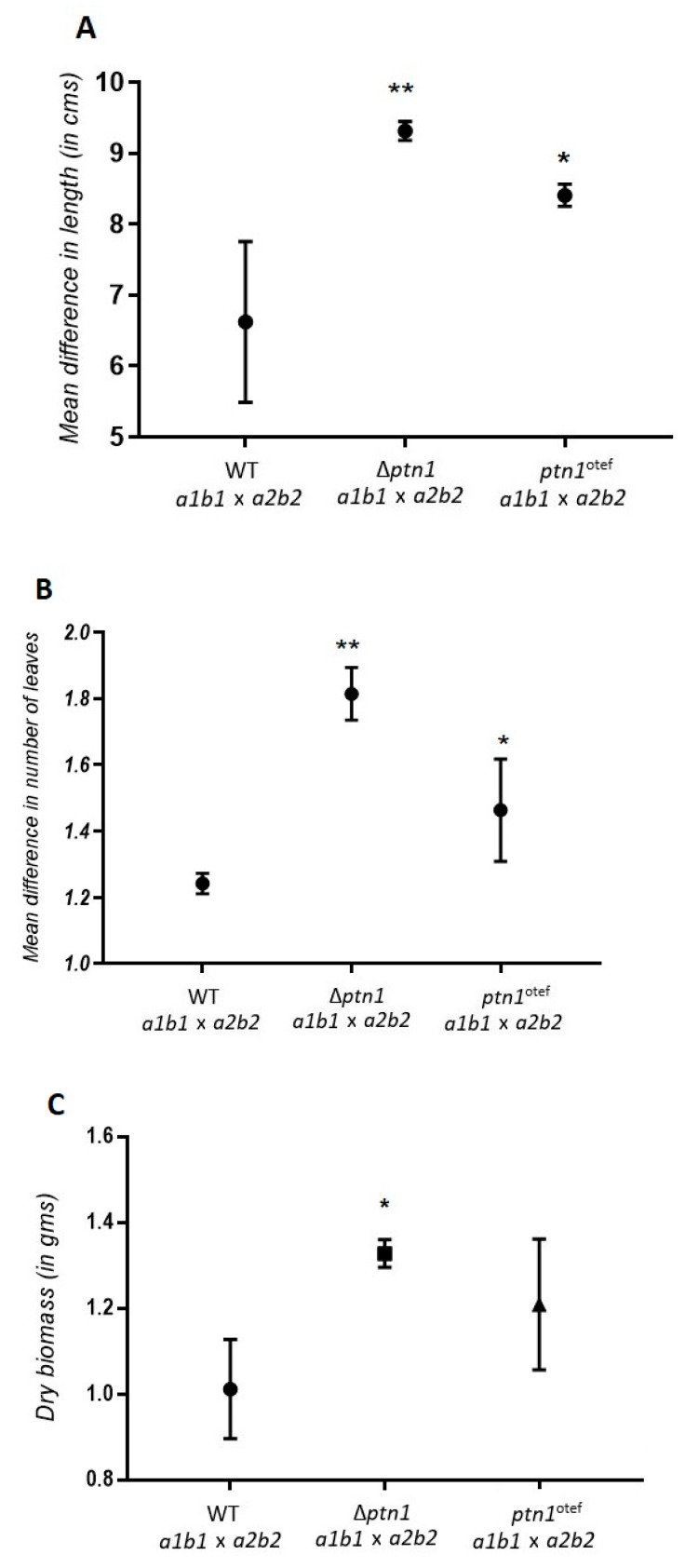
The effect of the deletion of ptn1 on the virulence of the fungus was measured by a range of plant growth indicators. This provided a more comprehensive view of the extent of the effect on pathogenicity. In this experiment, plants infected with wild-type cross (1/2 × 2/9) strains were compared with those infected with deletion cross and overexpression cross strains. A total of 28 plants were used per treatment group, and the experiment was repeated a total of three times. The total number of leaves and the length of the plants were measured on the day of infection, as well as on 4 dpi (days post-infection) and 8 dpi. Measurements were not taken after 8 dpi, as many plants were drooping and unable to stand erect due to infection, thereby posing difficulties in making accurate length measurements. Both the Δptn1 and ptn1 overexpression infections yielded significantly taller plants (i.e., reduced infection) compared to the wild type (Figure 7A). The average number of leaves produced were significantly lower in plants infected with wild-type partners, compared to plants infected with overexpression strains or deletion strains, with the latter showing the highest average number of leaves (Figure 7B). The dry biomass index of the plants infected with wild type strains was significantly lower when compared to Δptn1 infections. The ptn1otef-infected plants had a lower biomass index than the Δptn1-infected plants, but this was higher than for wild-type infections. However, the differences in biomass, plant height, or leaf length of the plants infected with the overexpression strain were not statistically significant when compared to wild type infection, which indirectly indicates that only the deletion of ptn1 results in a significant alteration in the virulence of the fungus.
Figure 7.
Plant growth parameters. Effect on (A) plant length, (B) leaf count, and (C) plant dry weight. The plants were infected with 1/2 a1b1 × 2/9 a2b2, Δptn1 (a1b1 × a2b2) and ptn1otef (a1b1 × a2b2) and the length and the number of leaves produced were measured on dpi-0 (0 days post-infection), dpi-4 and dpi-8. The Δptn1 strains showed a significant reduction in their effect on plant length and leaf production when compared to WT. Means (± standard deviation) of plant lengths and the number of leaves of Δptn1 marked by an asterisk (*) were significantly different (p ≤ 0.05) or (**) were significantly different (p ≤ 0.01) from those of WT; the mean length and number of leaves of ptn1otef was significantly different (p ≤ 0.05) from WT and Δptn1, respectively. The dry biomass (± standard deviation) of the plants infected with WT (a1b1 × a2b2), Δptn1 (a1b1 × a2b2) and ptn1otef (a1b1 × a2b2) were measured. The Δptn1 strains showed an increase in plant biomass when compared to WT, indicating a reduction in virulence. Mean of dry mass of plants infected by Δptn1 was significantly different (p ≤ 0.05) from those of wild type infections. Statistical analysis was done by ANOVA followed by Tukey’s test.
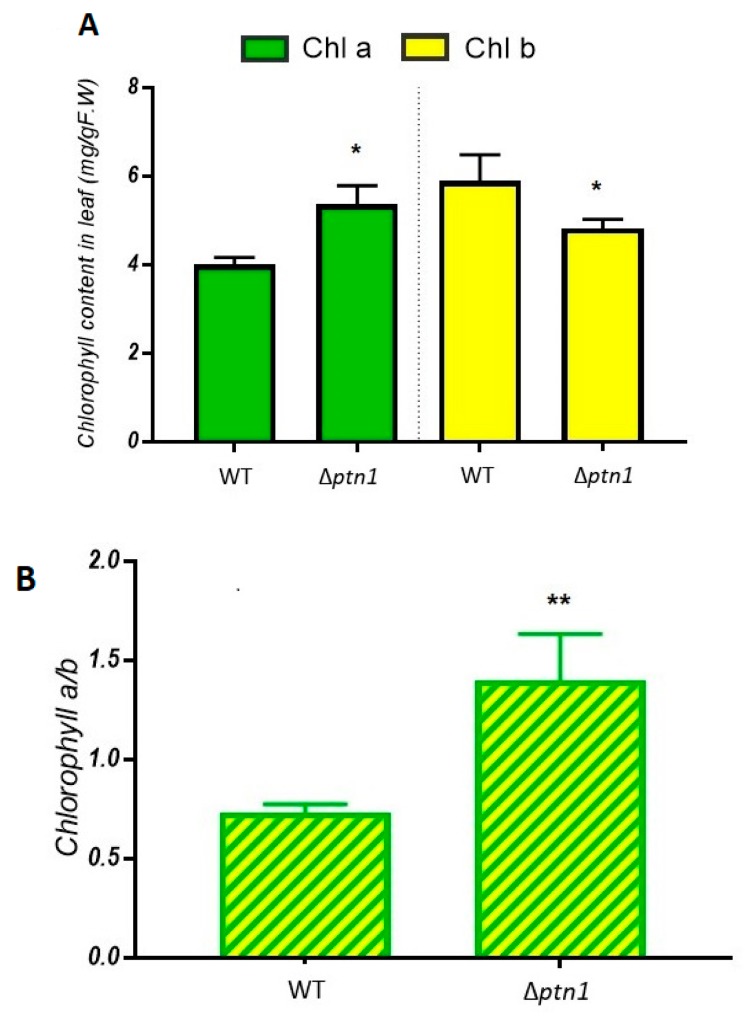
During infection there was normally a color change in the leaves of infected plants, due to chlorosis. The wild-type infected plant leaves had more yellow coloration/chlorosis, compared to Δptn1-infected plants. Several papers have shown, for wild-type U. maydis infections, an increase in chlorosis and a decrease in the chlorophyll content of infected leaves [42,43]. Thus, chlorophyll estimation was performed to examine whether there was a corresponding change in the level of photosynthetic pigments such as chlorophyll a and chlorophyll b between the different treatment groups. The concentrations of both chlorophyll a and chlorophyll b pigments were significantly reduced in wild-type infected plants, when compared to comparable infections, using the deletion strain partners (Figure 8). The chlorophyll a and chlorophyll b data were used to calculate chlorophyll a/b ratio which is indicative of the level of stress in plants [44,45]. The chlorophyll a/b ratio for plants from the Δptn1 infection was significantly higher compared to that for wild type.
Figure 8.
Chlorophyll estimation. (A) Chlorophyll count of leaves of plants infected with WT (1/2 a1b1 × 2/9 a2b2), Δptn1 (a1b1 × a2b2) and ptn1otef (a1b1 × a2b2) were measured using the DMSO method [31]. (B) The plants infected with the Δptn1 strains had higher a/b ratios, and such plants had chlorophyll levels that were more comparable to “mock” infections. Means of chlorophyll a, chlorophyll b, and chlorophyll a/b ratio were marked with an asterisk (*) or two asterisks (**) were significantly different (p ≤ 0.05) and (p ≤ 0.01), respectively, by Tukey’s test.
3.7. Impact of ptn1 Mutations on Spore Formation and Germination
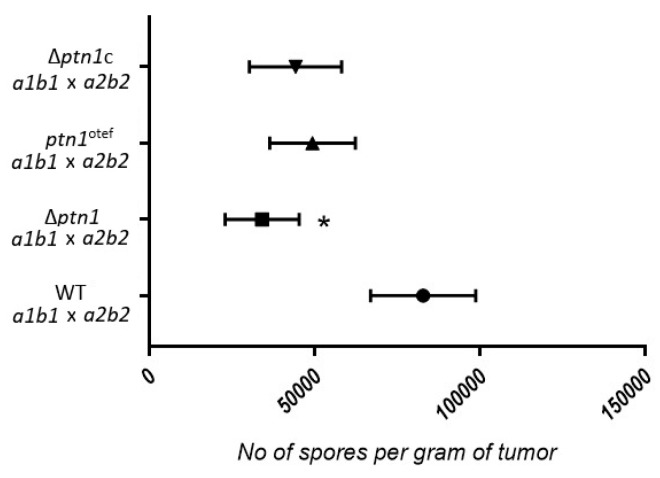
The plants infected with ptn1 deletion strains showed visibly reduced production in tumors in terms of size and number (Figure S9). Tumors were collected from all three treatment groups, 1/2 × 2/9, Δptn1 × Δptn1 and ptn1otef × ptn1otef, and weighed. The experiment was repeated three times. The data indicated that the WT (1/2 × 2/9) fungus produced far more tumors in plants when compared to the deletion strain or the overexpression strain. In the case of deletion strain, tumors produced were significantly fewer than for the WT or overexpression strain infections (p = 0.011, Tukey’s test). Microscopic analyses of tumors from plants infected with compatible wild type strains or compatible Δptn1 strains, showed that the number of mature spores were relatively low in deletion strains (Figure S10). The teliospores from wild-type infections overall were spherical and darkly pigmented, and the number of mature teliospores increased in quantity with the age/size of tumor, becoming clearly visible without magnification. However, while the Δptn1 × Δptn1 infected plants produced some tumors, the quantity of darkly pigmented teliospores was less (Figure 9), and in most cases, they were not obviously visible. It was difficult to quantify the presence of immature spores, due to the inconsistency in the stages of infection between tumors, and even within different parts of the same tumor. However, observations made from multiple independent infections indicated that the number of spores matched the intensity of infection. Crosses between Δptn1 derivatives constitutively expressing an ectopic copy of ptn1 saw an improvement in tumor number and size, as well as spore number, but did not show a complete return to the wild type levels for these traits. Rather, the spore count from Δptn1c was closer to that of ptn1otef than Δptn1, but it was still less compared to wild type (Figure 9).
Figure 9.
Tumors were collected from plants infected with 1/2 a1b1 × 2/9 a2b2, Δptn1 (a1b1 × a2b2), ptn1otef (a1b1 × a2b2) and Δptn1c (a1b1 × a2b2). From each type of infection (e.g., 1/2 a1b1 × 2/9 a2b2) tumors were pooled and used to extract spores. The spore count was obtained using a hemocytometer for each pooled extraction. Presented here are the mean spore count (± standard deviation) for three biological replicates of each infection experiment. There was a significant reduction in spore production in infections by the Δptn1 strains, indicated by * (p < 0.05, by Tukey’s Test).
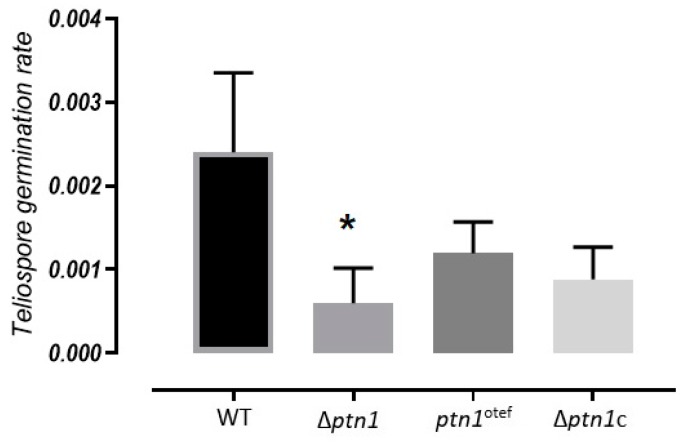
The teliospores were extracted from tumors and allowed to germinate using previously published methods [46]. Tumors from maize plants inoculated with wild type cross (1/2 × 2/9), deletion strain cross (Δptn1 × Δptn1), and the cross of overexpression strains (ptn1otef × ptn1otef) were used for germination studies. Since spore production was reduced in infections with the deletion strain cross, we examined whether there were also defects in spores from such infections. The Δptn1 strains showed a significantly lower rate of germination when compared to that for wild type infections (p < 0.05, Tukey’s test); the complemented Δptn1 derivatives could only partially overcome the defect in spore germination rates (Figure 10).
Figure 10.
Spore germination assay. Spores were extracted from the plant tumors produced from the infection of 1/2 a1b1 × 2/9 a2b2, Δptn1 a1b1 × Δptn1 a2b2, ptn1otef a1b1 × ptn1otef a2b2, and Δptn1c a1b1 × Δptn1c a2b2, and plated onto water agar plates. The plates were incubated at 30 °C for 72 h, and observed for the presence of colonies formed from the germination of spores. Spore germination was reduced in Δptn1 a1b1 × Δptn1 a2b2, and ptn1otef a1b1 × ptn1otef a2b2 combinations, with few colonies being observed after germination. The difference in germination rate (percentage germination per spores plated (± standard deviation) for Δptn1 was statistically significant, indicated by * (p < 0.05) by Tukey’s Test; the differences for the overexpression strain were not statistically significant.
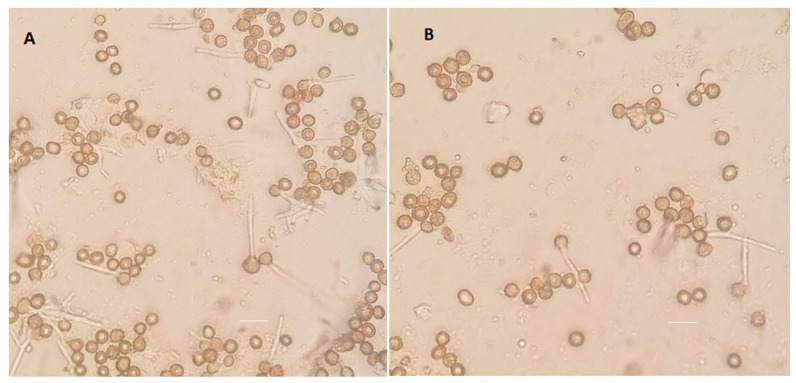
The formation of germination tubes from the spores (Figure 11) was examined to see if such defects could explain the reduced germination rates observed. Spores collected from tumors of plants infected with 1/2 a1b1 × 2/9 a2b2 and Δptn1 a1b1 × Δptn1 a2b2 strains were plated onto 2% water agar plates. The formation of germination tubes was observed microscopically after 24 to 48 h.
Figure 11.
Spore germination tube formation. Microscopic analysis of spores collected from tumors of plants infected with (A) 1/2 a1b1 × 2/9 a2b2 and (B) Δptn1 a1b1 × Δptn1 a2b2 strains to examine possible defects in the formation of germination tube. Spore germination on water agar plates incubated at 29 °C for 30 h was examined using differential interference contrast (DIC) microscopy. Scale bars, 20 μm.
In a pilot experiment to examine additional possible defects of the ptn1 deletion crosses, plant infections were performed on maize ears of a dwarf variety of maize (n = 2) (Figure S11). In this way, we investigated whether the deletion of ptn1 affected teliospore production when inoculated into the developing embryos (ears) of plants with mixtures of compatible wild type or mutant strains. Only the wild-type inoculation produced tumors, and these were filled with black teliospores (Figure S11A); in contrast, the deletion strains did not show ear infection (Figure S11B). As a control for differences in the infection rates on different varieties of maize, the same dwarf variety was infected on the stems of plants; deletion strains failed to yield infection progression beyond chlorosis, and over time, infected plants appeared to be as healthy as uninfected plants. However, the wild-type infection produced severe infection symptoms, with the death of most infected plants (not shown).
4. Discussion
In this study we examined the role of the PTEN orthologue of U. maydis, ptn1, in the pathogenic program of the fungus and in teliospore formation. The predicted Ptn1 protein of U. maydis has a conserved protein tyrosine phosphatase catalytic motif (PTPc) in its N-terminal region, a motif present in the PTEN proteins of most other organisms (Figure S1, Table S1). This is consistent with the finding that the PTEN orthologue of Arabidopsis thaliana, AtPTEN1, shares similarity with higher eukaryotes only in the N-terminal region where the catalytic domain is present [47]. However, the lack of conservation outside of the catalytic PTPc motif suggests that ptn1 may respond mostly to signals that are unique to U. maydis. In our study, it was observed that a mutation in ptn1 may lead to a decrease in tolerance to stress conditions, especially in media supplemented with cell wall stress agents such as Congo Red and calcofluor white. It has been observed in S. cerevisiae that dual-specificity protein tyrosine phosphatases such as Ptp2 and Ptp3 play a regulatory role in cell integrity pathways [48]. It has also been reported in mouse model studies that PTEN is involved in stress response pathways that are connected to oxidative stress [49].
In U. maydis, successful mating is indispensable for the formation of the dikaryon, which is the infectious form of the fungus [50]. Mating assays on charcoal plates and subsequent aerial hyphae indicative of infectious filament formation were used to measure this prerequisite for successful infection. The ptn1 deletion strains had reduced fuzz formation during the initial 24 hr. Since the difference was unnoticeable after 48 h, this indicates that the deletion caused delay in the onset of mating, and thus, only a minor defect in the mating capability. The RNA expression pattern of mfa2 and pra2 under low ammonium conditions could not explain the reduced aerial hyphal production in matings involving 2/9 Δptn1. On the other hand, we did observe the altered expression of prf1 in the Δptn1 a2b2 mutant. Since Prf1 is a transcription factor, its dysregulation could alter the expression of some unidentified downstream targets, leading to translational or post-translation effects on mating-specific targets. It has been reported before that overexpression of ras1(UMAG_11476) resulted in elevated levels of mfa (a pheromone precursor) in U. maydis [51]. A bioinformatic analysis looking for potential binding partners of Ptn1 has indicated that UMAG_11476 is a potential binding partner (Figure S12). Thus, it is plausible that UMAG_11476 is a potential downstream target for Ptn1. In humans, the dosage of PTEN is important for tumor suppression, as is evident from the data that monoallelic mutations (i.e., heterozygous) at the PTEN locus have been found in various cancer types, including prostate cancer, breast cancer, colon cancer, and lung cancer [52]. Our data from the mating and plant infection experiments using ptn1 deletion strains in the d132 diploid background are in accord with this observation, i.e., the loss of one copy of the ptn1 gene is only partially compensated by the presence of the wild-type allele in dikaryons and diploids. It has been observed that deletion strains of biz1 gene in the SG200 background could filament normally, while showing reduced virulence in plant infection [53]. We observed a similar phenotype with SG200 Δptn1.
Infection of maize plants with compatible wild type strains (1/2 a1b1 × 2/9 a2b2) and mutant strains (Δptn1 a1b1 × Δptn1 a2b2 or ptn1otef × ptn1otef) showed that the ptn1 deletion mutants were less virulent than wild type strains. Though the Δptn1 mutants were able to infect maize plants, they showed reduced symptoms, especially in tumor number and size (Figure S9); ptn1 overexpression did not significantly alter virulence. We were able to rescue the virulence partially by complementing the deletion strains with a wild type copy of the ptn1 gene. Although deletion strains constitutively expressing ptn1 ectopically were only partially rescued, the fact that independently-isolated ptn1 deletion mutants, produced by different means and in different genetic backgrounds, had similar levels of defects in measures of pathogenicity (Figure S8), strongly supports the role of the ptn1 gene in virulence for U. maydis. A study of the pathogenic fungus, Thielaviopsis basicola, causing black root rot disease of cotton plants, has shown that plant growth factors such as plant height, dry biomass, etc., are valid indicators of pathogen infection [54]. Similarly, biochemical indicators like chlorophyll content or chlorophyll (a/b) ratio can be used as indicators of level of fungal infection in plants, as shown with Crinipellis perniciosa, causing witches broom disease in cocoa plants [55] and Colletotrichum lindemuthianum infecting Phaseolus vulgaris (common bean) plants [56]. In oxeye daisy (Leucanthemum vulgare) plants infected with the fungus Rhizoctonia solani, an 81% reduction in shoot and a 70% reduction in root biomass were observed [57]. In our study, the reduction in pathogenicity was confirmed in measures of plant height, numbers of leaves, and levels of chlorophyll content. The plants infected with deletion strains were significantly healthier compared to the wild-type infected plants.
The dissection and microscopic analysis of the tumors from plants infected with Δptn1 mutants showed that while tumors were produced, a large proportion of those tumors lacked black teliospores. Moreover, tumors from infections with the deletion strains had significantly fewer spores than tumors from wild-type infections. The spore germination assay found a significantly lower rate of germination of spores from infections by the Δptn1 mutants. However, there was no obvious difference between the wild-type and mutant strains in germination tube formation.
Effects of the ptn1 deletion in vitro, especially in haploid or diploid sporidia, were subtle (Figure 2 and Figure 3). A slight reduction in aerial hyphae production on charcoal, and an increased sensitivity to cell wall stressors were seen for such mutants. A similar cell wall defect was reported for ptn1 (PTEN orthologue) mutants in fission yeast, Schizosaccharomyces pombe [58]. Our study indicates that while appressoria formation and penetration (not shown) were not impaired for the ptn1 mutants, pathogenic development was obstructed at stages after penetration.
It has been noted in the case of some signaling proteins that aberrant expression in either direction can end up in same phenotypes. For example, when the tumor suppressor SRPK1 (serine/arginine-rich splicing factor kinase), which mediates the recruitment of the phosphatase PHLPP1 to Akt (besides its role in regulated splicing) was deleted or overexpressed, it resulted in the promotion of cancer [59]. This is especially a possibility when the same protein has multiple roles in interconnected signal transduction pathways. This may be a reasonable explanation for our observation that overexpression strains (ptn1otef × ptn1otef) also displayed a reduction in virulence, and the haploid mutant cells were also more susceptible to cell wall stressors than the wild type.
In U. maydis, infection of maize leads to the formation of galls or tumors that are filled with fungal hyphae that ultimately develop into teliospores [50]. Several individual processes inside the tumor lead to the formation of teliospores. These include the rounding and swelling of hyphal tips of fungal mycelia, for which functional rum1 and hda1 are necessary [60,61]. It has been shown that subsequent hyphal fragmentation would need fuz1 [50]. Other studies have shown that several genes, including cru1, hgl1, unh1, ust1, and ros1 influence the formation and maturation of teliospores, and some of the phenotypes of mutants in these genes resemble those shown by ptn1 mutants. For instance, cru1 mutants produced tumors that were considerably smaller and that contained very few teliospores [62]. On a similar note, hgl1 mutants produced tumors, but failed to produce teliospores [63]. The deletion of unh1 showed a defect in the formation, maturation, and pigmentation of teliospores [29]. A recent study indicated that the transcription factor Ros1 is a master regulator of sporogenesis in U. maydis. Although ros1 is not required for tumor formation, it is essential during late stages of infection for fungal karyogamy, the proliferation of fungal hyphae, and spore formation [64]. The data from most of these studies indicate that the process of tumor formation is separate from teliospore development, as is also evident in the case of ptn1 mutants. This makes ptn1 part of the growing number of genes that are vital to the complex process of teliospore formation and germination.
In Arabidopsis thaliana, AtPTEN1 (PTEN orthologue) is expressed exclusively in pollen grains during the late stage of development; when expression of AtPTEN1 was suppressed by RNA interference, it resulted in pollen cell death after mitosis [47]. This is interesting, considering the observation that the ptn1 gene in U. maydis is involved in the sexual development of the fungus. Similarly, in S. cerevisiae, TEP1 (PTEN orthologue) mutants showed a defect in the developmental process of sporulation. Moreover, FgTep1p (PTEN orthologue) deletion mutants in the wheat pathogen, Fusarium graminearum, produced fewer conidia and showed an attenuation in virulence on host plants [65].
In conclusion, the results reported in this study illustrate that ptn1 has a role in virulence, especially in tumor production and in the development of teliospores in U. maydis. A growing number of studies show that the process of teliospore formation and germination is intricate, requiring exquisite control of hyphal proliferation inside the plant, the formation of teliospores, the formation of a thick cell wall, and germination involving the completion of meiosis, to produce haploid cells. The cells must make appropriate developmental decisions at each of these steps in response to the prevailing conditions. A defect in the genes responsible for any of these steps can result in failure to produce mature and functional teliospores. Hence, further investigation into the proteins that interact with Ptn1 during spore formation will likely contribute to a deeper understanding of how the complex signaling networks regulate pathogenic development and teliospore development in U. maydis.
Acknowledgments
The authors thank Shahab Taheri for help with plant infections and chlorophyll measurements, and we are extremely grateful to Margaret Wallen and Hector Mendoza for help with RNA extractions and qRT-PCR measurements. We also appreciate the constructive criticism of two anonymous reviewers whose suggestions improved the final manuscript.
Supplementary Materials
The following are available online at http://www.mdpi.com/2309-608X/5/1/1/s1, Figure S1: Motif scan analysis of Ptn1; Figure S2: SignalP 4.1 prediction of the absence of the signal peptide; Figure S3: TargetP prediction of the sub-cellular localization of Ptn1; Figure S4: The charcoal assay for fuzz production, performed with SG200 and SG200 Δptn1 strains; Figure S5: The relative expression analysis of mfa2, pra2, and prf1 (U. maydis mating pathway genes) in wild type and Δptn1 under high and low ammonium conditions; Figure S6: Growth rates of 1/2, 2/9, 1/2 Δptn1, and 2/9 Δptn1 strains grown in YEPS medium; Figure S7: Infection of plants with Δptn1 deletion strains in the SG200 background; Figure S8: Infection of plants with Δptn1 deletion strains in different backgrounds, created through two different methods; Figure S9: Teliospores from infected maize plants; Figure S10: Dissection of tumors under the microscope showing teliospores; Figure S11: Deletion of ptn1 alters the ability to produce tumors in maize cobs; Figure S12: Interaction network analysis of the Ptn1 protein sequence using STRING; Table S1: Protein prediction analysis of Ptn1 using PredictProtein (https://www.predictprotein.org).
Author Contributions
Conceptualization, L.M.K.V. and M.H.P.; Data curation, L.M.K.V. and M.H.P.; Formal analysis, M.H.P.; Funding acquisition, M.H.P.; Investigation, L.M.K.V., J.S.D. and M.N.G.; Methodology, L.M.K.V. and M.H.P.; Project administration, M.H.P.; Resources, M.H.P.; Validation, M.H.P.; Visualization, L.M.K.V.; Writing—original draft, L.M.K.V.; Writing—review & editing, L.M.K.V. and M.H.P.
Funding
This work was supported by the National Institutes of Health (NIH) [sub-awards #OGMB131493C1 and #OGMB131493D1] to [MHP] from [P20GM103436] to [Nigel Cooper, PI]. The contents of this work are solely the responsibility of the authors and do not represent the official views of the NIH. No funds were received for covering the cost to publish in open access. Additional support was from a Research Initiation Grant from the Office of the Vice President for Research to MHP and a GNAS grant to LMKV from the College of Arts and Sciences at the University of Louisville.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. Ustilago maydis as a Pathogen. Annu. Rev. Phytopathol. 2009;47:423–445. doi: 10.1146/annurev-phyto-080508-081923. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg G., Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Nadales E., Nogueira M.F., Baldin C., Castanheira S., El Ghalid M., Grund E., Lengeler K., Marchegiani E., Mehrotra P.V., Moretti M., et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollmeister E., Schipper K., Baumann S., Haag C., Pohlmann T., Stock J., Feldbrugge M. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 2012;36:59–77. doi: 10.1111/j.1574-6976.2011.00296.x. [DOI] [PubMed] [Google Scholar]
- 5.Steck P.A., Pershouse M.A., Jasser S.A., Yung W.K.A., Lin H., Ligon A.H., Langford L.A., Baumgard M.L., Hattier T., Davis T., et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 6.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. A Putative Protein Tyrosine Phosphatase Gene Mutated in Human Brain, Breast, and Prostate Cancer. Science. 1997;275:1943. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 7.Song M.S., Salmena L., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 8.Leslie N.R., Downes C.P. PTEN function: How normal cells control it and tumour cells lose it. Biochem. J. 2004;382:1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E., et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010;42:454. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Chin-Sang I.D. C. elegans as a model to study PTEN’s regulation and function. Methods. 2015;77–78:180–190. doi: 10.1016/j.ymeth.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Huang H., Potter C.J., Tao W., Li D.M., Brogiolo W., Hafen E., Sun H., Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 12.Gil E.B., Malone Link E., Liu L.X., Johnson C.D., Lees J.A. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. USA. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solari F., Bourbon-Piffaut A., Masse I., Payrastre B., Chan A.M.L., Billaud M. The human tumour suppressor PTEN regulates longevity and dauer formation in Caenorhabditis elegans. Oncogene. 2005;24:20. doi: 10.1038/sj.onc.1207978. [DOI] [PubMed] [Google Scholar]
- 14.Serezani C.H., Kane S., Medeiros A.I., Cornett A.M., Kim S.H., Marques M.M., Lee S.P., Lewis C., Bourdonnay E., Ballinger M.N., et al. PTEN directly activates the actin depolymerization factor cofilin-1 during PGE2-mediated inhibition of phagocytosis of fungi. Sci. Signal. 2012;5:ra12. doi: 10.1126/scisignal.2002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heymont J., Berenfeld L., Collins J., Kaganovich A., Maynes B., Moulin A., Ratskovskaya I., Poon P.P., Johnston G.C., Kamenetsky M., et al. TEP1, the yeast homolog of the human tumor suppressor gene PTEN/MMAC1/TEP1, is linked to the phosphatidylinositol pathway and plays a role in the developmental process of sporulation. Proc. Natl. Acad. Sci. USA. 2000;97:12672–12677. doi: 10.1073/pnas.97.23.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brachmann A., König J., Julius C., Feldbrügge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genom. 2004;272:216–226. doi: 10.1007/s00438-004-1067-8. [DOI] [PubMed] [Google Scholar]
- 17.Holliday R. Ustilago maydis. In: King R.C., editor. Bacteria, Bacteriophages, and Fungi: Volume 1. Springer US; Boston, MA, USA: 1974. [Google Scholar]
- 18.Cervantes-Chávez J.A., Ali S., Bakkeren G. Response to environmental stresses, cell-wall integrity, and virulence are orchestrated through the calcineurin pathway in Ustilago hordei. Mol. Plant-Microb. Interact. 2011;24:219–232. doi: 10.1094/MPMI-09-10-0202. [DOI] [PubMed] [Google Scholar]
- 19.Gold S.E., Brogdon S.M., Mayorga M.E., Kronstad J.W. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell. 1997;9:1585–1594. doi: 10.1105/tpc.9.9.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller P., Aichinger C., Feldbrugge M., Kahmann R. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 1999;34:1007–1017. doi: 10.1046/j.1365-2958.1999.01661.x. [DOI] [PubMed] [Google Scholar]
- 21.Kronstad J.W., Leong S.A. Isolation of two alleles of the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA. 1989;86:978. doi: 10.1073/pnas.86.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Pedrajas M.D., Nadal M., Kapa L.B., Perlin M.H., Andrews D.L., Gold S.E. DelsGate, a robust and rapid gene deletion construction method. Fungal Genet. Biol. 2008;45:379–388. doi: 10.1016/j.fgb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Pham C.D., Yu Z., Sandrock B., Bölker M., Gold S.E., Perlin M.H. Ustilago maydis Rho1 and 14-3-3 homologues participate in pathways controlling cell separation and cell polarity. Eukaryot Cell. 2009;8:977–989. doi: 10.1128/EC.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison N., Cavinder B., Townsend J.P., Trail F. Optimized primers and other critical conditions for efficient fusion PCR to generate knockout vectors in filamentous fungi. Fungal Genet. Rep. 2013;60:1–10. doi: 10.4148/1941-4765.1006. [DOI] [Google Scholar]
- 25.Kamper J. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genom. 2004;271:103–110. doi: 10.1007/s00438-003-0962-8. [DOI] [PubMed] [Google Scholar]
- 26.Paul J.A., Wallen R.M., Zhao C., Shi T., Perlin M.H. Coordinate regulation of Ustilago maydis ammonium transporters and genes involved in mating and pathogenicity. Fungal Biol. 2018;122:639–650. doi: 10.1016/j.funbio.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Ho E.C.H., Cahill M.J., Saville B.J. Gene discovery and transcript analyses in the corn smut pathogen Ustilago maydis: Expressed sequence tag and genome sequence comparison. BMC Genom. 2007;8:334. doi: 10.1186/1471-2164-8-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham C.D., Yu Z., Ben Lovely C., Agarwal C., Myers D.A., Paul J.A., Cooper M., Barati M., Perlin M.H. Haplo-insufficiency for different genes differentially reduces pathogenicity and virulence in a fungal phytopathogen. Fungal Genet. Biol. 2012;49:21–29. doi: 10.1016/j.fgb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Doyle C.E., Kitty Cheung H.Y., Spence K.L., Saville B.J. Unh1, an Ustilago maydis Ndt80-like protein, controls completion of tumor maturation, teliospore development, and meiosis. Fungal Genet. Biol. 2016;94:54–68. doi: 10.1016/j.fgb.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Akhtar T., Zia-Ur-Rehman M., Naeem A., Nawaz R., Ali S., Murtaza G., Maqsood M.A., Azhar M., Khalid H., Rizwan M. Photosynthesis and growth response of maize (Zea mays L.) hybrids exposed to cadmium stress. Environ. Sci. Pollut. Res. Int. 2017;24:5521–5529. doi: 10.1007/s11356-016-8246-0. [DOI] [PubMed] [Google Scholar]
- 31.Manolopoulou E., Varzakas T., Petsalaki A. Chlorophyll Determination in Green Pepper Using two Different Extraction Methods. Curr. Res. Nutr. Food Sci. J. 2018;4:52–60. doi: 10.12944/CRNFSJ.4.Special-Issue1.05. [DOI] [Google Scholar]
- 32.Hiscox J.D., Israelstam G.F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979;57:1332–1334. doi: 10.1139/b79-163. [DOI] [Google Scholar]
- 33.Morrison E.N., Emery R.J., Saville B.J. Phytohormone Involvement in the Ustilago maydis-Zea mays Pathosystem: Relationships between Abscisic Acid and Cytokinin Levels and Strain Virulence in Infected Cob Tissue. PLoS ONE. 2015;10:e0130945. doi: 10.1371/journal.pone.0130945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conover W.J. Practical Nonparametric Statistics. 3rd ed. John Wiley; New York, NY, USA: Chichester, UK: 1999. [Google Scholar]
- 35.Redkar A., Hoser R., Schilling L., Zechmann B., Krzymowska M., Walbot V., Doehlemann G. A Secreted Effector Protein of Ustilago maydis Guides Maize Leaf Cells to Form Tumors. Plant Cell. 2015;27:1332–1351. doi: 10.1105/tpc.114.131086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torró I., Bretó P., García-Yzaguirre A. The influence of plant spacing in the early stages of selection of rice (Oryza sativa L.) varieties. Span. J. Agric. Res. 2016;14:e0704. doi: 10.5424/sjar/2016142-8130. [DOI] [Google Scholar]
- 37.Klose J., de Sa M.M., Kronstad J.W. Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 2004;52:823–835. doi: 10.1111/j.1365-2958.2004.04019.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith D.G., Garcia-Pedrajas M.D., Gold S.E., Perlin M.H. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 2003;50:259–275. doi: 10.1046/j.1365-2958.2003.03680.x. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Herrera J., Leon C.G., Guevara-Olvera L., Carabez-Trejo A. Yeast-mycelial dimorphism of haploid and diploid strains of Ustilago maydis. Microbiology. 1995;141:695–703. doi: 10.1099/13500872-141-3-695. [DOI] [Google Scholar]
- 40.Kretschmer M., Lambie S., Croll D., Kronstad J.W. Acetate provokes mitochondrial stress and cell death in Ustilago maydis. Mol. Microbiol. 2018;107:488–507. doi: 10.1111/mmi.13894. [DOI] [PubMed] [Google Scholar]
- 41.Kaffarnik F., Muller P., Leibundgut M., Kahmann R., Feldbrugge M. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 2003;22:5817–5826. doi: 10.1093/emboj/cdg554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kühnemann J., Sonnewald U., Kahmann R., et al. Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008;56:181–195. doi: 10.1111/j.1365-313X.2008.03590.x. [DOI] [PubMed] [Google Scholar]
- 43.Horst R.J., Engelsdorf T., Sonnewald U., Voll L.M. Infection of maize leaves with Ustilago maydis prevents establishment of C4 photosynthesis. J. Plant Physiol. 2008;165:19–28. doi: 10.1016/j.jplph.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Funayama-Noguchi S., Terashima I. Effects of Eupatorium yellow vein virus infection on photosynthetic rate, chlorophyll content and chloroplast structure in leaves of Eupatorium makinoi during leaf development. J. Funct. Plant Biol. 2006;33:165–175. doi: 10.1071/FP05172. [DOI] [PubMed] [Google Scholar]
- 45.Pavlovic D., Nikolic B., Djurovic S., Waisi H., Andjelkovic A., Marisavljevic D. Chlorophyll as a measure of plant health: Agroecological aspects. Pestic. Fitomed. 2014;29:21–34. doi: 10.2298/PIF1401021P. [DOI] [Google Scholar]
- 46.Boyce K.J., Chang H., D’Souza C.A., Kronstad J.W. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryot Cell. 2005;4:2044–2056. doi: 10.1128/EC.4.12.2044-2056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta R., Ting J.T.L., Sokolov L.N., Johnson S.A., Luan S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell. 2002;14:2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattison C.P., Spencer S.S., Kresge K.A., Lee J., Ota I.M. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 1999;19:7651–7660. doi: 10.1128/MCB.19.11.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bankoglu E.E., Tschopp O., Schmitt J., Burkard P., Jahn D., Geier A., Stopper H. Role of PTEN in Oxidative Stress and DNA Damage in the Liver of Whole-Body Pten Haplodeficient Mice. PLoS ONE. 2016;11:e0166956. doi: 10.1371/journal.pone.0166956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banuett F., Herskowitz I. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development. 1996;122:2965–2976. doi: 10.1242/dev.122.10.2965. [DOI] [PubMed] [Google Scholar]
- 51.Muller P., Katzenberger J.D., Loubradou G., Kahmann R. Guanyl nucleotide exchange factor Sql2 and Ras2 regulate filamentous growth in Ustilago maydis. Eukaryot Cell. 2003;2:609–617. doi: 10.1128/EC.2.3.609-617.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmena L., Carracedo A., Pandolfi P.P. Tenets of PTEN Tumor Suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Flor-Parra I., Vranes M., Kamper J., Perez-Martin J. Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell. 2006;18:2369–2387. doi: 10.1105/tpc.106.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma J., Jaraba J., Kirkpatrick T.L., Rothrock C.S. Effects of Meloidogyne incognita and Thielaviopsis basicola on Cotton Growth and Root Morphology. Phytopathology. 2013;104:507–512. doi: 10.1094/PHYTO-06-12-0120-R. [DOI] [PubMed] [Google Scholar]
- 55.Scarpari L.M., Meinhardt L.W., Mazzafera P., Pomella A.W., Schiavinato M.A., Cascardo J.C., Pereira G.A. Biochemical changes during the development of witches’ broom: The most important disease of cocoa in Brazil caused by Crinipellis perniciosa. J. Exp. Bot. 2005;56:865–877. doi: 10.1093/jxb/eri079. [DOI] [PubMed] [Google Scholar]
- 56.Lobato A.K.S., Gonçalves-Vidigal M.C., Vidigal Filho P.S., Andrade C.A.B., Kvitschal M.V., Bonato C.M. Relationships between leaf pigments and photosynthesis in common bean plants infected by anthracnose. N. Z. J. Crop Horticult. Sci. 2010;38:29–37. doi: 10.1080/01140671003619308. [DOI] [Google Scholar]
- 57.Lewandowski T.J., Dunfield K.E., Antunes P.M. Isolate Identity Determines Plant Tolerance to Pathogen Attack in Assembled Mycorrhizal Communities. PLoS ONE. 2013;8:e61329. doi: 10.1371/journal.pone.0061329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra P., Zhang Y., Rameh L.E., Ivshina M.P., McCollum D., Nunnari J.J., Hendricks G.M., Kerr M.L., Field S.J., Cantley L.C., et al. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 2004;166:205–211. doi: 10.1083/jcb.200404150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P., Zhou Z., Hu A., Ponte de Albuquerque C., Zhou Y., Hong L., Sierecki E., Ajiro M., Kruhlak M., Harris C., et al. Both Decreased and Increased SRPK1 Levels Promote Cancer by Interfering with PHLPP-Mediated Dephosphorylation of Akt. Mol. Cell. 2014;54:378–391. doi: 10.1016/j.molcel.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichmann M., Jamnischek A., Weinzierl G., Ladendorf O., Huber S., Kahmann R., Kamper J. The histone deacetylase Hda1 from Ustilago maydis is essential for teliospore development. Mol. Microbiol. 2002;46:1169–1182. doi: 10.1046/j.1365-2958.2002.03238.x. [DOI] [PubMed] [Google Scholar]
- 61.Quadbeck-Seeger C., Wanner G., Huber S., Kahmann R., Kamper J. A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 2000;38:154–166. doi: 10.1046/j.1365-2958.2000.02128.x. [DOI] [PubMed] [Google Scholar]
- 62.Flor-Parra I., Castillo-Lluva S., Pérez-Martín J. Polar growth in the infectious hyphae of the phytopathogen Ustilago maydis depends on a virulence-specific cyclin. Plant Cell. 2007;19:3280–3296. doi: 10.1105/tpc.107.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durrenberger F., Laidlaw R.D., Kronstad J.W. The hgl1 gene is required for dimorphism and teliospore formation in the fungal pathogen Ustilago maydis. Mol. Microbiol. 2001;41:337–348. doi: 10.1046/j.1365-2958.2001.02528.x. [DOI] [PubMed] [Google Scholar]
- 64.Tollot M., Assmann D., Becker C., Altmüller J., Dutheil J.Y., Wegner C.E., Kahmann R. The WOPR Protein Ros1 Is a Master Regulator of Sporogenesis and Late Effector Gene Expression in the Maize Pathogen Ustilago maydis. PLoS Pathog. 2016;12:e1005697. doi: 10.1371/journal.ppat.1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang D., Fan F., Yang J., Wang X., Qiu D., Jiang L. FgTep1p is linked to the phosphatidylinositol-3 kinase signalling pathway and plays a role in the virulence of Fusarium graminearum on wheat. Mol. Plant Pathol. 2010;11:495–502. doi: 10.1111/j.1364-3703.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.