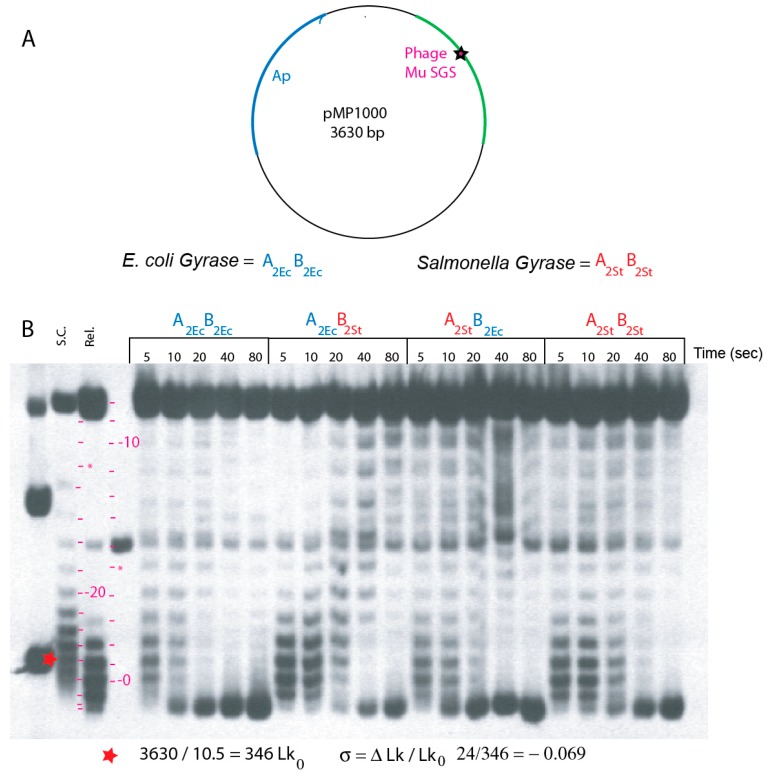
Figure 2.
In vitro assays for supercoiling processivity in enzymes reconstituted from E. coli and S. typhimurium subunits. (A) The map of plasmid pMP1000 includes the nuB101 mutation of the phage Mu SGS that supports processive supercoiling reactions. (B) Southern blot patterns of plasmid DNA incubated with gyrases for different times (sec.) Supercoiled, relaxed, and linear pMP1000 plasmid was loaded in lanes 1–3 as markers. The positions of bands with linking numbers increasing from the relaxed substrate to −12 are shown by red dashes in lane 4; topoisomers higher than −13 reverse and move faster down the gel at positions marked with red dashes between lanes 2 and 3. DNA with higher supercoiling than the in vivo supercoiled species moves beyond the position of −24 *. Blue letters designate E. coli gyrase subunits; red letters designate S. typhimurium gyrase subunits.