This review summarizes and discusses recent findings on the complex roles of TOR in integrating nutrient, energy, light, and hormone signals to mediate meristem activation and plant growth and development.
Keywords: Energy, glucose, hormone, light, nitrogen, nutrient, phosphorus, signalling, sucrose, sulfur, target of rapamycin
Abstract
The multidomain target of rapamycin (TOR) is an atypical serine/threonine protein kinase resembling phosphatidylinositol lipid kinases, but retains high sequence identity and serves a remarkably conserved role as a master signalling integrator in yeasts, plants, and humans. TOR dynamically orchestrates cell metabolism, biogenesis, organ growth, and development transitions in response to nutrient, energy, hormone, and environmental cues. Here we review recent findings on the versatile and complex roles of TOR in transcriptome reprogramming, seedling, root, and shoot growth, and root hair production activated by sugar and energy signalling. We explore how co-ordination of TOR-mediated light and hormone signalling is involved in root and shoot apical meristem activation, proliferation of leaf primordia, cotyledon/leaf greening, and hypocotyl elongation. We also discuss the emerging TOR functions in response to sulfur assimilation and metabolism and consider potential molecular links and positive feedback loops between TOR, sugar, energy, and other essential macronutrients.
Introduction
Nutrient signalling is the most ancient and fundamental mechanism to regulate and sustain life, and is essential to modulate cellular activities and organismal development by integrating with other intrinsic regulators and environmental cues. In contrast to the previously prevailing notion that nutrients automatically feed into cellular metabolism and growth, nutrient signalling mechanisms are complex for the tailored regulatory networks in diverse cell types, tissues, and organs with specialized physiology, metabolism, and functions. Extensive studies have documented the pivotal role of target of rapamycin (TOR) protein kinase in the regulation of metabolism, translation, and transcription to fuel cellular proliferation and organismal development and growth (Xiong and Sheen, 2014, 2015; Dobrenel et al., 2016a; Saxton and Sabatini, 2017; Schepetilnikov and Ryabova, 2018; Shi et al., 2018). However, the molecular and cellular mechanisms underlying how plant TOR protein kinase transduces, co-ordinates, and integrates multiple nutrient, light, and hormone signals are only emerging.
The evolutionarily conserved TOR proteins in Arabidopsis thaliana and Homo sapiens share a remarkable 73% amino acid sequence identity in the kinase domains (Xiong and Sheen, 2012). At least two structurally and functionally distinct protein complexes (TORCs) with several regulatory partners have been well characterized in eukaryotes. In mammals, mTORC1 (mammalian/mechanistic TOR complex 1) and mTORC2 share a common subunit LST8 (small lethal with SEC13 protein 8). RAPTOR (regulatory-associated protein of mTOR) is a distinct component in mTORC1, whereas RICTOR (rapamycin-insensitive companion of mTOR) is unique in mTORC2 (Saxton and Sabatini, 2017; Tatebe and Shiozaki, 2017). The multidomain RAPTOR protein regulates the stability, catalytic activity, and substrate binding of the dimeric mTORC1. LST8 is a WD40-domain protein positioned next to the ATP-binding active site cleft in mTORC1 for substrate selectivity and delivery (Aylett et al., 2016). In plants, only TOR, LST8 (encoded by LST8-1 and LST8-2 in Arabidopsis), and RAPTOR (encoded by RAPTOR1A and RAPTOR1B in Arabidopsis) orthologues are present in all sequenced plant species, while no RICTOR orthologue could be identified in plant genomes (Anderson et al., 2005; Mahfouz et al., 2006; Moreau et al., 2012; Xiong and Sheen, 2014; Kravchenko et al., 2015; Dobrenel et al., 2016a; Salem et al., 2018).
Although TORC1 is rapamycin sensitive in mammals and yeasts, early research suggested that flowering plants were insensitive to rapamycin, and the FKBP-rapamycin-binding domain (FRB) of Arabidopsis TOR did not form a complex with rapamycin and FKBP12 (FK506-binding protein 12) in yeast two-hybrid and in vitro pull-down analyses. However, using a more sensitive split luciferase protein–protein interaction assay in Arabidopsis mesophyll protoplasts, it was demonstrated that Arabidopsis and human FKBP12 exhibited similar interactions with the FRB domain of Arabidopsis TOR stimulated specifically by rapamycin. In liquid culture of Arabidopsis seedlings, rapamycin rapidly and effectively inhibits Arabidopsis TOR activity based on the conserved and specific phosphorylation of T449 in S6K1, and strongly suppresses the growth of cotyledons, true leaves, petioles, and primary and secondary roots and root hairs, resembling the tor mutant phenotypes. Mesophyll protoplasts and seedlings were carefully cultured with a minimal volume of liquid medium to facilitate chemical uptake, and were monitored with sensitive hypoxia-inducible marker genes to avoid hypoxia stress (Baena-González et al., 2007; Xiong and Sheen, 2012; Xiong et al., 2013; Deng et al., 2016; Li et al., 2017). Importantly, ectopic expression of either human or yeast FKBP12 or overexpression of Arabidopsis FKBP12 can all further enhance rapamycin sensitivity in Arabidopsis. Moreover, two independent alleles of Arabidopsis fkbp12 mutants exhibit reduced rapamycin sensitivity based on phosphorylation of T449 in S6K1 as well as seedling and root hair development (Xiong and Sheen, 2012; Deng et al., 2016). Variable endogenous FKBP12 protein levels and low rapamycin uptake of plants in solid culture medium may account for the varied rapamycin resistance observed in flowering plants especially at low rapamycin concentrations (Xiong and Sheen, 2015). Recent studies have also demonstrated that the next generation of ATP-competitive and TOR-specific chemical inhibitors have significantly empowered the elucidation of diverse TOR functions in plants (Xiong and Sheen, 2015; Shi et al., 2018).
The origins of organic carbon, nitrogen, phosphorus, and sulfur nutrients for animals and humans are mainly derived from plants’ incredible ability to fix CO2 and to take up and assimilate nitrate (Liu et al., 2017; Y.Y. Wang et al., 2018), phosphate (Chien et al., 2018), and sulfate (Takahashi et al., 2011) from the soil, which generate chemical energy, sugars, amino acids, proteins, nucleic acids, lipids, and vitamins essential to support all life forms (Xiong et al., 2013; Li and Sheen, 2016). TOR protein kinase has been demonstrated to be activated by glucose, energy, oxygen, amino acids, hormones, and growth factors in yeast, animal, and plant systems (Xiong and Sheen, 2015; Dobrenel et al., 2016a; Li and Sheen, 2016; Ben-Sahra and Manning, 2017; González and Hall, 2017; Saxton and Sabatini, 2017; Schepetilnikov and Ryabova, 2018; Shi et al., 2018). In this review, we highlight our current understanding of how sugar, energy, light, and hormone signals modulate TOR activity, which governs transcription, translation, metabolism, cell cycle, and autophagy in diverse aspects of developmental processes in the reference plant A. thaliana. We also explore the emerging scenario that TOR may act as a central integrator to sense and relay carbon, sulfur, nitrogen, and phosphorus nutrient signals in positive feedback regulatory loops to orchestrate the complex nutrient uptake, assimilation, and signalling networks central to plant growth, adaptation, and reproduction.
Sugar and energy signalling
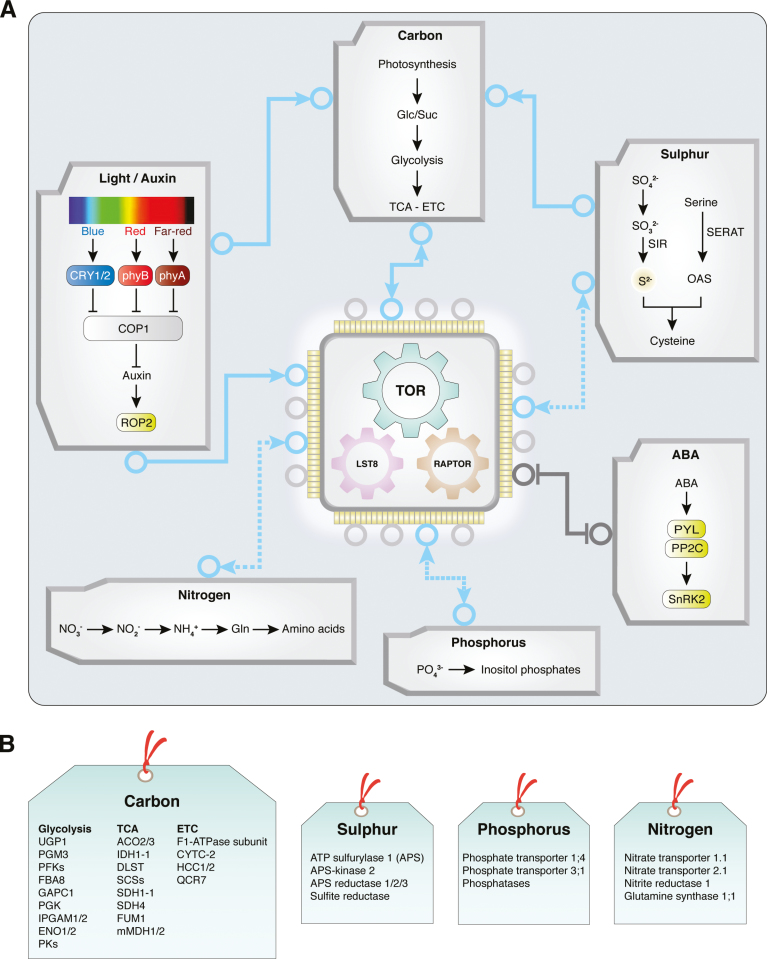
Plant life is centred around the production, transport, utilization, storage, and remobilization of sugars that serve as the primary supplies of energy and building blocks, as well as signalling cues to guide the growth, development, adaptation, defence, survival, and reproduction programmes (Eveland and Jackson, 2012; Ruan, 2014; Sheen, 2014; Eom et al., 2015; Yu et al., 2015; Hulsmans et al., 2016; Li and Sheen, 2016; Wingler, 2018; Wurzinger et al., 2018). Recent findings have illustrated how TOR acts as the central molecular switch to regulate metabolism, cell proliferation, and seedling and adult plant growth in response to sucrose and glucose derived directly or indirectly from photosynthesis. For example, after seed germination at the heterotrophic to photoautotrophic transition checkpoint, physiological levels of glucose and sucrose (15 mM) can replace photosynthesis as the most potent stimuli to promote rapid TOR-dependent root meristem proliferation, root elongation, cotyledon expansion, and root hair production. Other sugars, such as fructose, galactose, and xylose, are much less effective. Neither amino acids nor plant growth hormones can substitute for sugars in the inorganic nutrient medium with nitrate and ammonium. The indispensable role of glucose is explained by the requirement for glycolysis and mitochondrial bioenergetics to activate TOR protein kinase monitored by the phosphorylation of its direct and conserved substrates S6K1/2 at T449 and T455, respectively (Fig. 1A) (Xiong and Sheen, 2012; Xiong et al., 2013).
Fig. 1.
TOR as a central integrator of nutrient, energy, light, and hormone signalling to regulate plant growth. (A) TOR integrates complex signalling pathways in a reciprocal manner. (B) Glucose–TOR target genes are involved in energy and nutrient regulation. The list of genes presented in (B ) was extracted from supplementary table S1 of Xiong et al., 2013. https://media.nature.com/original/nature-assets/nature/journal/v496/n7444/extref/nature12030-s2.xlsx. All microarray data published in Xiong et al., 2013 are available at the Gene Expression Omnibus under accession number GSE40245. TOR, target of rapamycin; LST8, lethal with Sec13 protein 8; RAPTOR, regulatory-associated protein of mTOR; Glc, glucose; Suc, sucrose; Gln, glutamine; SIR, sulfite reductase; SERAT, serine acetyltransferase; OAS, O-acetylserine; ABA, abscisic acid; PYL, pyrabactin resistance 1-like; PP2C, protein phosphatase 2C; SnRK2, SNF1-related protein kinase 2; CRY, cryptochrome photoreceptors; phy, phytochrome; COP1, constitutive photomorphogenesis 1; UGP, UDP-glucose pyrophosphorylase; PGM, phosphoglucomutase; PFK, phosphofructokinase; FBA8, fructose-bisphosphate aldolase 8; GAPC, glyceraldehyde-3-phosphate dehydrogenase C; PGK, phosphoglycerate kinase; IPGAM, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1; ENO, enolase; PK, pyruvate kinase; ACO, ACC oxidase; IDH, isocitrate dehydrogenase; DLST, dihydrolipoamide succinyltransferase; SCS, succinyl coenzyme A synthetase; SDH, succinate dehydrogenase; FUM, fumarase; mMDH, lactate/malate dehydrogenase; CYTC-2, cytochrome c-2; HCC, homologue of the copper chaperone SCO1; QCR7, cytochrome b-c1 complex subunit 7.
The promotion of root and shoot growth in seedlings and adult plants by light and photosynthesis is significantly compromised in various conditional tor-deficient mutants or after treatment with rapamycin or specific ATP-competitive TOR inhibitors. Inhibiting TOR activity leads to transcriptomic and metabolomic reprogramming (Deprost et al., 2007; Ren et al., 2012; Caldana et al., 2013; Montané and Menand, 2013; Xiong et al., 2013; Dong et al., 2015). Moreover, mutations in raptor1b and lst8-1 display a spectrum of related plant growth defects in roots and shoots with delayed flowering and senescence (Anderson et al., 2005; Mahfouz et al., 2006; Moreau et al., 2012; Ren et al., 2012; Kravchenko et al., 2015; Salem et al., 2018). Genetic manipulation of a direct TORC1 phosphorylation substrate TAP46 (Type 2A phosphatase-associated protein of 46 kDa), a regulatory subunit of protein phosphatase 2A, shows that it positively regulates S6K phosphorylation. Transgenic plants overexpressing TAP46 exhibits increased hypocotyl length, enlarged leaves, and enhanced seed size, as well as elevated expression levels of genes associated with ribosome biogenesis, lignin biosynthesis, and nitrogen assimilation. In tap46 RNAi lines, translation and nitrogen assimilation gene expression are reduced, but autophagy is elevated (Ahn et al., 2011, 2015). Defining the TAP46-PP2A substrates will help expand our understanding of TOR signalling in diverse growth processes.
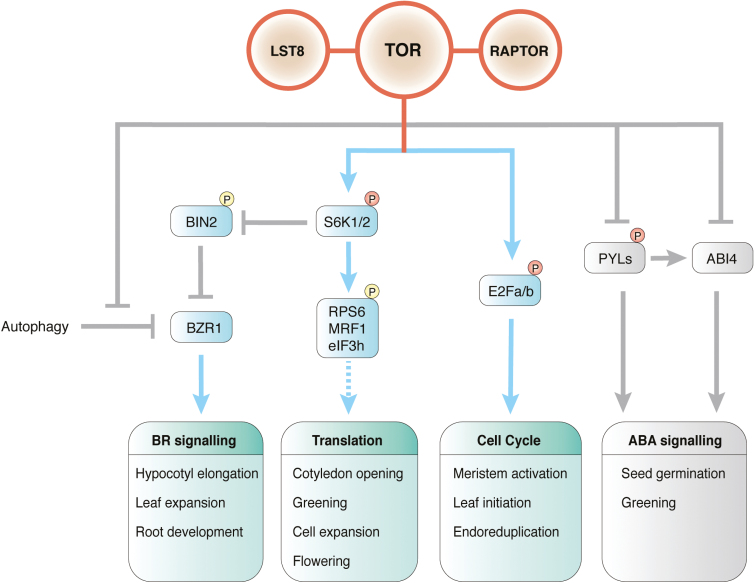
As the conserved direct targets of TOR protein kinase, S6K1/2 relay signalling by phosphorylating RPS6 (the 40S ribosomal subunit S6) and MRF (MA3 domain-containing translation regulatory factor) in response to light, sugar, and energy status in plants (Ren et al., 2012; Xiong et al., 2013; Dobrenel et al., 2016b; Enganti et al., 2017; Lee et al., 2017; Chen et al., 2018). Comprehensive phenotypic analyses of rps6a and rps6b mutants and genetic complementation support downstream roles of RPS6 in light and nutrient-dependent TOR functions in root, leaf, and flowering regulation (Ren et al., 2012), as well as in the control of rRNA synthesis (Kim et al., 2014; Son et al., 2015) (Fig. 2). A recent study shows that TOR plays a critical role in light-dependent RPS6 phosphorylation and protein translation control, but future research will be required to elucidate the precise molecular connection (Chen et al., 2018). Furthermore, MRF1, as a substrate of S6K, was reported to be involved in translation control under dark/starvation conditions, which elevate MRF1 transcripts. Transition from the energy-deficient condition to the light- and glucose-fed condition activates rapid phosphorylation of MRF1 and promotes its association with eIF4A-1 and light polysomal fractions, which may reboot translation (Lee et al., 2017). Intriguingly, the glucose-activated root hair elongation is also suppressed in the oestradiol-inducible tor-es mutant or raptor1b seedlings, or by treatment with rapamycin or ATP-competitive chemical inhibitors in many plant species (Ren et al., 2012; Xiong and Sheen, 2012; Montané and Menand, 2013; Deng et al., 2016; Salem et al., 2018). Although primary auxin-responsive transcription is not affected in the tor-es seedlings (Xiong et al., 2013), a long-term TOR inhibitor treatment for 12 d suggests a link to auxin biosynthesis and signalling in root hair development (Deng et al., 2016). It will require further investigation to determine the role of TOR protein kinase in the epidermal cell fate determination and the morphogenetic plasticity of cell differentiation modulated by complex integration of nutrient, hormone, and stress signals (Salazar-Henao et al., 2016).
Fig. 2.
The TOR signalling network. TOR promotes translation and BR signalling probably through the signalling relay mediated by S6K1/2 and BIN2 substrates. By directly phosphorylating key transcription factors, E2Fa/b, TOR positively regulates the cell cycle. Phosphorylation of ABA-receptor PYLs by TOR represses ABA signalling. TOR, target of rapamycin; LST8, lethal with Sec13 protein 8; RAPTOR, regulatory-associated protein of mTOR; S6K, S6 kinase; E2F, E2 promoter-binding factor; RPS6, ribosomal protein S6; MRF, MA3-containing translation regulatory factor; eIF3h, eukaryotic initiation factor 3h; BIN, brassinosteroid-insensitive; BZR, brassinosteroid signalling positive regulator; TAP46, type 2A-phosphatase-associated protein 46 kDa; PYL, pyrabactin resistance 1-like; PP2C, protein phosphatase 2C; ABI, ABA-insensitive.
In plants growing under light, steady TORC1 mutants or long-term conditional suppression of TOR signalling for days/weeks influence the transcriptome encompassing diverse biological functions in a complex manner (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Dong et al., 2015). The experimental design insuring synchronized glucose stimulation has uncovered transcriptome reprogramming in 3-day-old seedlings with minimal endogenous glucose levels, which maximize the detection sensitivity of primary TOR target genes by dynamic phosphorylation. Within 2 h of treatment at a physiological level of 15 mM glucose, 1318 up- and 1050 down-regulated genes have been identified, which are completely blocked in the tor-es mutant. The sugar- or CO2-regulated transcriptome data sets derived from older seedlings or adult leaves significantly overlap with glucose–TOR target genes detected in young seedlings (Xiong et al., 2013). However, comparative transcriptome analyses indicate that the regulation of some sucrose- or glucose-responsive genes is complex in Arabidopsis plants (Li and Sheen, 2016). It is possible that the precise extent of the regulation of TOR target genes is dictated by cell types, tissues, organs, developmental stages, nutrients, and environmental conditions. Consistently, the sensitivity of young seedlings with relatively abundant meristem tissues and developing cell types facilitates the discovery of previously unknown primary glucose–TOR target genes. Functional categories of cell cycle and DNA synthesis, transcription, and RNA processing are enriched among the novel glucose–TOR-activated genes, whereas those of transcription, protein degradation, and signalling are enriched in the glucose–TOR-repressed genes.
In general, the primary glucose–TOR target genes comprise a myriad of regulatory and metabolic functional categories (Xiong et al., 2013; Xiong and Sheen, 2014). Significantly, rapid glucose–TOR signalling activates evolutionarily conserved bioenergetic and anabolic processes, including genes involved in glycolysis, the tricarboxcyclic acid (TCA) cycle, the mitochondrial electron transport chain (ETC) (Fig. 1B), the oxidative pentose phosphate (OPP) pathway, ribosome assembly, protein synthesis machineries, as well as amino acid, lipid, and nucleotide synthesis. These findings suggest a universal and conserved TOR function in controlling translation, and central carbon and energy metabolism in plants (Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012 ; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015). Glucose–TOR signalling also represses genes mediating catabolism, for example autophagy and the degradation of proteins, amino acids, lipids, and xenobiotics that are critical for survival under starvation (Baena-González et al., 2007; Deprost et al., 2007; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013). Importantly, glucose–TOR signalling plays a pivotal role in regulating plant-specific processes. For example, the metabolic genes for enzymes involved in β-oxidation (triacylglyceride lipase and acyl-CoA oxidase) and the glyoxylate cycle (malate synthase and isocitrate lyase) required in the germination programme of Arabidopsis seeds are repressed (Graham, 2008). Plant-specific genes promoting the synthesis of cell wall polymers/proteins, glutathione, indolic/benzoic/aliphatic glucosinolates, lignins, flavonoids, nitrate transport, phosphate metabolism, as well as nitrogen and sulfur assimilation are activated for plant growth, defence, or communication to promote adaptation, fitness, and survival (Keurentjes et al., 2006; Deprost et al., 2007; Takahashi et al., 2011; Moreau et al., 2012; Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015; Malinovsky et al., 2017).
Although the detailed molecular mechanisms mediating glucose activation of TOR protein kinase remain to be fully elucidated, emerging evidence indicates that functional glycolysis and the mitochondrial ETC are required for glucose–TOR signalling. Blocking hexokinase activities by 2-deoxyglucose at the first step of glycolysis and several chemical inhibitors, antimycin A, 2,4-dinitrophenol, and carbonylcyanide m-chlorophenylhydrazone, targeting different steps of the ETC, prevent TOR activation by glucose. Thus, sugar-mediated TOR activity reflects the cellular metabolic and bioenergetic status to mediate energy signalling in plants (Xiong et al., 2013; Li and Sheen, 2016). Although phosphorylation of RAPTOR1B by the plant energy sensor SnRK1 (Snf1-related protein kinase) has been demonstrated under energy deprivation, the primary target genes of TOR and SnRK1 only partially and antagonistically overlap (Li and Sheen, 2016; Nukarinen et al., 2016). Novel mechanisms for SnRK1-indepednent regulation of TOR probably exist for future research exploration. In recent research, the Rho-like small GTPase ROP2 is shown to bind to and activate TOR in the synergistic action of glucose and auxin signalling (Fig. 1A) (Li et al., 2017; Schepetilnikov et al., 2017). Furthermore, the TEL2–TTI1–TTI2 (TTT)–RUVBL1/2 (RuvB-like ATPase and ATP-dependent DNA helicase) complex in animals is important for TORC1 dimerization and activation in glucose signalling (Kim et al., 2013; David-Morrison et al., 2016). The putative TEL2, TTI1, TTI2, and RUVBL orthologues exist in the Arabidopsis genome. Whether the TTT–RUVBL1/2 complex can be formed and play a similar role in activating the TOR complex by sensing sucrose/glucose requires further investigation. It will also be interesting to determine the role of RAPTOR-bodies in plant TORC1 inactivation under glucose starvation (Hughes Hallett et al., 2015).
Light and hormone signalling
During post-embryonic development after seed germination, the root apical meristem (RAM) and shoot apical meristem (SAM) are the primary reservoir for self-renewable stem cells, which supply new cells to support root, leaf, stem, flower, and fruit organogenesis. TOR expression is enriched in meristems (Menand et al., 2002), and glucose–TOR-mediated energy signalling, which requires glycolysis and mitochondrial relays, plays an essential role in activating the quiescent root meristem under light (Xiong et al., 2013). Based on EdU (thymidine analogue 5-ethynyl-2'-deoxyuridine) staining, systemic glucose derived from photosynthesis or exogenously applied glucose rapidly activates DNA synthesis in the mitotic progenitor cells of the primary root meristem and in the endocycling cells of the root elongation zone. TOR directly phosphorylates the E2Fa transcription factor to activate transcription of S-phase genes involved in DNA synthesis. The stem cells surrounding the quiescent centre (QC) are also activated by glucose–TOR signalling to divide, but at a much lower rate than the progenitor cells (Fig. 2) (Xiong et al., 2013). In the SAM, sucrose and red light additively promote the expression of WUSCHEL (WUS) encoding a homeodomain master transcription factor for stem cell regulation in the organizing centre (OC). Genetic analyses reveal the involvement of the red light photoreceptor phytochrome B (phyB) and blue light photoreceptors cryptochrome 1/2 (CRY1/2) in WUS activation by suppressing the master negative regulator COP1 (constitutive photomorphogenesis 1). Importantly, WUS activation by glucose or sucrose mediating energy signalling in the wild type is prevented by 5 μM AZD-8055 inhibiting TOR protein kinase. WUS expression promoted by red light in the wild type or in cop1 in the dark is reduced by AZD-8055. These results support a role for TOR in integrating energy and light signalling to promote stem cell activation in the SAM (Fig. 1A) (Pfeiffer et al., 2016). However, in the QC of the RAM, the expression of WOX5, encoding a homeodomain transcription factor and functionally related to WUS, is not diminished in the tor-es mutant (Xiong et al., 2013). TOR activation stimulated by sugar, energy, and light signalling may exert differential regulations and functions in the RAM and SAM. It will be interesting to identify the molecular regulators mediating TOR activation of WUS in the SAM.
Recent studies have started to unravel the mechanisms underlying glucose–TOR-mediated energy signalling in promoting cell proliferation in leaf primordia. Although glucose alone is sufficient to active RAM proliferation via TOR signalling, glucose and light are synergistically required for the activation of robust cell proliferation based on the expression of pCYCB1;1::GUS as a mitotic marker in leaf primordia (Xiong et al., 2013; Li et al., 2017). The activation of S6K phosphorylation at T449, pCYCB1;1::GUS expression in leaf primordia, as well as expansion and greening of cotyledons and true leaves stimulated by glucose and light are abolished in tor-es plants or by TOR inhibitors, rapamycin and torin2. Genetic analyses support the roles of white, red, and blue light mediated by phyA/B and CRY1/2 photoreceptors to activate the cell cycle in leaf primordia through the suppression of COP1 (Fig. 1A) (Cai et al., 2017; Li et al., 2017). Both E2Fa and E2Fb are phosphorylated and activated by TOR, and support S-phase gene expression and leaf primordia expansion (Fig. 2).
As light particularly activates auxin biosynthesis genes, YUC2, YUC4, and YUC7, in the shoot apex, the effect of light on TOR activation is probably mediated via stimulating auxin biosynthesis (Li et al., 2017). Consistently, light-enhanced auxin accumulation can be monitored and quantified by the DII-VENUS auxin biosensor, and exogenous auxin can replace light to activate TOR signalling in leaf primordia in darkness. Yucasin as an auxin biosynthesis inhibitor prevents light-stimulated auxin accumulation and TOR signalling in leaf primordia. Significantly, ROP2 interacts with TOR, and constitutively activated ROP2 stimulates S6K phosphorylation, pCYCB1;1::GUS expression, and true leaf expansion in the presence of exogenous glucose without light (Fig. 1A) (Li et al., 2017). These thorough analyses and comprehensive evidence corroborate a new report on TOR-dependent promotion of gene expression, proliferation, and expansion of young leaves by sucrose in older seedlings with access to auxin synthesis and transport, or in cop1 (Mohammed et al., 2018). Another finding suggests that sucrose and light are necessary for organogenesis, and sucrose is essential to promote leaf expansion even in cop1 exhibiting features of constitutive photomorphogenesis with open cotyledons and a short hypocotyl (Pfeiffer et al., 2016). Moreover, independent studies have demonstrated a critical role for ROP2 in mediating the well-characterized auxin activation of TOR–S6K–eIF3h signalling on endosomes contributing to translation regulation of mRNAs via upstream ORFs in the 5'-untranslated regions (Figs 1A, 2) (Schepetilnikov et al., 2013, 2017; Schepetilnikov and Ryabova, 2017, 2018). Although plants lack a homologue of the small GTPase RHEB (Ras homologue enriched in brain), which is the key activator of TORC1 in animal and human cells (Saxton and Sabatini, 2017), ROP2 and related proteins may serve similar functions in activating plant TORC1 ( Roustan et al., 2016; Li et al., 2017; Schepetilnikov et al., 2017).
Light-stimulated photomorphogenesis regulates an array of rapid and long-term responses and biological processes by triggering massive reprogramming of the transcriptome and translatome (Wu, 2014). A new study reports that light-enhanced translation is orchestrated by white, blue, and far-red light perception via phyA and CRY1/2 photoreceptors and a signalling pathway composed of COP1, auxin, TOR, and RPS6 (Fig. 1A) (Chen et al., 2018). In de-etiolated young seedlings, light alone inactivates the negative regulator COP1 within 4 h, which leads to activation of auxin signalling for TOR–S6K-dependent phosphorylation of RPS6 without exogenous sugars. Mutants defective in TOR, RPS6A, or RPS6B exhibit delayed cotyledon opening, a characteristic of the de-etiolating process to ensure timely vegetative development of a young seedling. This finding provides a mechanistic view of light-specific translational enhancement in de-etiolation via TOR activation (Chen et al., 2018). As very young etiolated seedlings grown in the dark were used, it would be important to probe the sugar requirement for TOR–S6K–RPS6 activation in older etiolated seedlings when endogenous sugars are completely depleted. For example, in 12-day-old etiolated seedlings grown without sucrose, RPS6 phosphorylation at S237 and S240 is elevated by light exposure for 2 h, but not for 0.5 h. However, RPS6 phosphorylation is promoted by light exposure of as short as 0.5 h in etiolated seedlings grown with 1% sucrose (Enganti et al., 2017). It is possible that the requirement for sugar, light, and/or hormones to activate TOR signalling can vary in biological contexts and developmental stages.
Chloroplast biogenesis and maturation is a key process dependent on light to establish photosynthesis during cotyledon and leaf development. Several studies suggest that TORC1 plays a crucial role in the biogenesis and maturation of chloroplasts to promote cotyledon and leaf greening (Dong et al., 2015; Li et al., 2015; Deng et al., 2016; Sun et al., 2016; Li et al., 2017; Xiong et al., 2017; Mohammed et al., 2018; Shi et al., 2018; Zhang et al., 2018). Treatment with the TOR chemical inhibitor AZD-8055 for 10 d eliminates greening and cotyledon expansion, which is consistent with a broad repression of photosynthesis genes involved in chlorophyll biosynthesis, light reactions, and CO2 fixation (Dong et al., 2015; Li et al., 2015). The characterization of trin1 (tor-inhibitor insensitive 1) reveals a role for ABI4 [abscisic acid (ABA)-insensitive 4], a chloroplast retrograde regulator, in mediating TOR signalling in the seed to seedling transition. Based on the analysis of TRIN1–GUS (β-glucuronidase) activity, it has been suggested that TOR promotes ABI4 degradation and greening (Li et al., 2015). The direct or indirect molecular connection between TOR and ABI4 regulation remains to be elucidated (Fig. 2).
A large-scale phosphoproteomics experiment has identified a conserved serine in the PYL4 ABA receptor exhibiting ABA-dependent dephosphorylation. Functional analyses of the phosphomimetic version of the ABA receptor PYL1 (S119D) indicates that the specific phosphorylation of the ABA receptor abolishes ABA binding, ABI1 (phosphatase PP2C) interaction, as well as the ability to activate SnRK2.6 for ABF2 transcription factor phosphorylation and RD29-LUC reporter gene activation in protoplast assays. An in vitro protein kinase screen identified TOR for specific phosphorylation of related PYL1 at the conserved S119. Furthermore, raptor1 mutants are hypersensitive to ABA but display decreased ABA synthesis. ABA diminishes S6K1 phosphorylation through ABA-activated SnRK2.6 phosphorylation of RAPTOR in TORC1. These findings reveal reciprocal negative regulations between TOR and ABA signalling to balance plant growth and stress responses, consistent with TOR activation and ABA repression of seedling development and greening of cotyledons and leaves (Figs 1A, 2) (Kravchenko et al., 2015; Li et al., 2015; P. Wang et al., 2018). Genetic and biochemical studies with s6k1, Osraptor2, and tor mutants in rice have provided evidence that TOR–RAPTOR2–S6K signalling regulates thylakoid galactolipid biosynthesis and grana modelling for photosynthesis performance (Sun et al., 2016).
Unexpectedly, in seedlings after extended etiolation, TOR inhibition by AZD-8055 or raptor1b mutants increase greening in the dark to light transition. This paradoxically improved greening response after exposure to light reflects a decrease of the chlorophyll precursor Pchlide (protochlorophyllide), ROS (reactive oxygen species) reduction, enhanced POR (NADPH:protochlorophyllide oxidoreductase) activity, and available metabolites, as the greening difference can be over-ridden by sucrose supply. Furthermore, raptor1b mutants are impaired in GA (gibberellin) signalling, ABA hypersensitive, and epistatic to PIF1/3 (phytochrome-interacting factor 1/3) as negative regulators for greening and ROS (Kravchenko et al., 2015; P. Wang et al., 2018; Zhang et al., 2018). In larger seedlings with more nutrient and hormone access, sucrose-activated plastid biogenesis is promoted by TOR signalling based on AZD-8055 treatment (Mohammed et al., 2018). Thus, it is important to determine the roles of TOR signalling in different physiological, metabolic, and developmental states with comprehensive analyses to provide logical explanations for the observed phenotypes.
In the dark, the elongation of etiolated hypocotyls is strongly enhanced by sucrose and glucose. The sugar promotion of hypocotyl elongation is blocked in the tor-es seedlings or by rapamycin treatment, but enhanced by overexpression of TAP46 (Ren et al., 2012; Ahn et al., 2015; Zhang et al., 2016). A key plant hormone promoting the elongation of etiolated hypocotyl is brassinosteroid (BR), and the expression of several target genes of the BR signalling transcription factor BZR1 involved in cell expansion is suppressed in the tor-es mutant. As increasing the concentration of BR and the gain-of-function bzr1-D mutation partially restore hypocotyl elongation in tor-es, TOR probably activates the BR pathway to promote plant growth. Further experimental evidence suggests that glucose–TOR signalling stabilizes BZR1 in the dark, which is degraded via autophagy suppressed by TOR activation (Fig. 2) (Zhang et al., 2016). Recent findings also uncover a new link between TOR and BR signalling by identifying BIN2 (BR-insensitive 2) as a downstream effector of TOR–S6K2 signalling based on a yeast two-hybrid screen using S6K2 as a bait protein. BIN2 encodes a GSK3β protein kinase, which is a negative regulator of BZR1 via direct phosphorylation and nuclear exclusion in BR signalling (Chaiwanon et al., 2016). S6K2 directly phosphorylates BIN2 at S187 and S203, and presumably inhibits the BIN2 function. BIN2-RNAi plants strongly promote shoot development and are relatively insensitive to multiple TOR-specific chemical inhibitors in seedling growth under light, whereas BIN2-overexpressing plants are hypersensitive (Xiong et al., 2017). These results provide novel molecular mechanisms for dual regulation of BR responses by glucose–TOR signalling through autophagy suppression and BZR1 stabilization, as well as TOR–S6K2-mediated BIN2 inactivation to enhance BZR1 nuclear translocation (Fig. 2) (Chaiwanon et al., 2016; Zhang et al., 2016; Xiong et al., 2017). However, novel molecular mechanisms will be required to explain the proposed function of BIN2 in suppressing photoautotrophic growth, which is also suppressed by BR synthesis and signalling (Fig. 2) (Chory et al., 1991).
Sulfur signalling
Sulfur is an essential nutrient for plant growth, and plant sulfur assimilation is carried out by ATP sulfurylase (APS) and APS reductase (APR) to reduce sulfate to sulfite, and sulfite reductase (SIR) further reduces sulfite to sulfide (Fig. 1A) (Takahashi et al., 2011). The relationship between sulfur signalling and the TOR function has been advanced by investigating the SIR-deficient mutant for sulfide production. TOR signalling monitored by S6K phosphorylation and EdU staining in the root meristem are significantly reduced in sir1-1 (Dong et al., 2017; Speiser et al., 2018). Moreover, limitation of sulfide in sir1-1 leads to severe growth retardation, depletion of TCA cycle intermediates, decreased rRNA, reduced global translation, and induced autophagy, all downstream targets of TOR signalling (Xiong et al., 2013; Dong et al., 2017; Speiser et al., 2018). Significantly, grafting the wild-type shoot to the sir1-1 root or supply of glucose restores S6K phosphorylation and the root growth defect associated with the reduced glucose and sucrose, but enhanced fructose levels in sir1-1. As sulfide fumigation can cause fast and significant up-regulation of glucose levels and TOR activity, TOR may play an integrative role in modulating sulfur nutrient sensing in plants (Dong et al., 2017; Speiser et al., 2018). It will be interesting to determine whether sulfide produced in chloroplasts promotes photosynthesis, sugar production, and nitrogen assimilation, all of which are decreased by sulfur deficiency and in sir1-1 (Khan et al., 2010).
A recent study dissecting the resource allocation between stress response pathways and growth-promoting pathways based on blocking sulfur flux from cysteine to glutathione has also generated new insight into sulfur signalling and TOR regulation (Speiser et al., 2018). It is shown that reducing the glutamate–cysteine ligase activity for glutathione synthesis in the cad2 sir1-1 double mutant partially restores plant growth, rescues meristematic activity, and increases TOR activity in sir1-1. It is suggested that TOR may trigger reallocation of cysteine from glutathione to protein synthesis. Moreover, 3-hydroxypropylglucosinolate, a downstream cysteine metabolite mediating plant defence, reversibly inhibits root elongation and meristem activation similar to TOR inhibitors (Malinovsky et al., 2017). Thus, sulfur and its derived metabolites appear to serve a role in balancing plant development and defence via TOR regulation in response to environmental cues. Interestingly, genome-wide transcript profiling data have provided evidence that glucose–TOR signalling activates genes in the sulfur assimilation pathways and glucosinolate biosynthesis (Xiong et al., 2013). There is a reciprocal positive feedback loop between glucose–TOR and sulfur–TOR signalling (Fig. 1A).
Nitrogen and phosphorus signalling
Although sugars derived from photosynthesis drive plant growth and development, and are the most potent nutrient cues to activate TOR signalling immediately, inorganic nitrogen and phosphorus nutrients may serve as the gatekeeper in integrating carbon–nitrogen and carbon–phosphorus nutrient signalling networks to co-ordinate bidirectional shoot–root nutrient communications, developmental plasticity, and adaptation, and to shape organ biomass and architecture (Gojon et al., 2009; Gruber et al., 2013; Liu et al., 2017; Xuan et al., 2017; Chien et al., 2018; Gutiérrez-Alanís et al., 2018; Y.Y. Wang et al., 2018). Depending on the relationship with glucose or glucose-derived energy and metabolite status, the connection between glucose–TOR signalling and sulfur, nitrogen, or phosphorus availability could be different (Fig. 1A). For instance, sulfur deficiency is tightly linked with low endogenous glucose and sucrose levels, and inhibits photosynthesis (Takahashi et al., 2011; Dong et al., 2017). Enhancing photosynthesis or exogenous glucose supply effectively stimulates TOR–S6K signalling and root meristem activity under a low sulfur status. As sulfur deficiency also decreases nitrogen assimilation, adding nitrogen instead of glucose may not relieve this specific nutrient demand (Takahashi et al., 2011; Dong et al., 2017; Forzani et al., 2018; Speiser et al., 2018). In striking contrast, nitrogen or phosphorus deficiency is often associated with higher levels of sugars, and supplying higher glucose enhances expression of nitrate or phosphate starvation response genes (Moore et al., 2003; Price et al., 2004; Karthikeyan et al., 2007; Chien et al., 2018; Leong et al., 2018; Wagner et al., 2018). As nitrogen- or phosphorus-derived metabolites, such as NADH, NADPH, FADH2, ADP, or phosphate, are essential to support the glucose-stimulated bioenergetic processes (Wagner et al., 2018), we surmise that TOR signalling is probably compromised when the nitrogen or phosphorus nutrient status is low in plants. Despite abundant sugars, we speculate that the metabolic and energy generation processes are suppressed until nitrogen or phosphorus nutrients are replenished. Recent characterization of a mitochondrial ATP synthase mutant overcoming phosphite (Phi) (non-metabolizable but triggers phosphorus signalling) responses lends some support to the connection between ATP synthesis and sugar regulation in phosphorus signalling. This phi1 (phosphite-insensitive 1) mutant displays root growth defects and constitutive mitochondrial impairment, resembling treatment with oligomycin (a specific mitochondrial ATPase inhibitor) (Leong et al., 2018). It will be important to determine whether phi1 exhibits lower TOR activity in this phosphorus signalling relay.
Despite the presence of abundant glucose or sucrose, it is conceivable that the TOR signalling pathway cannot be fully activated when either nitrogen or phosphorus is missing. We propose that nitrogen and phosphorus and/or their metabolic derivatives may serve as important nutrient cues to activate TOR signalling in plants (Fig. 1A). Although multiple amino acid sensors for leucine, arginine, and glutamine have been discovered in mammalian systems in the past decade, none of these sensor genes could be identified in plant genomes (Stracka et al., 2014; González and Hall, 2017; Saxton and Sabatini, 2017). Notably, the molecular mechanism of the glutamine–TOR signalling, which requires a functional mitochondrial TCA and ETC, may exist in plants. However, leucine or arginine signalling is different, activating mTORC1 through different sensors and converging on RAG GTPases on the lysosomal surface in animals or the vacuole membrane in budding yeast (Stracka et al., 2014; Saxton and Sabatini, 2017). As no RAG GTPase homologues have been identified in plants, plant TOR probably relies on novel sensors to perceive amino acids (Roustan et al., 2016).
It has been reported that overexpressing TOR results in hypersensitivity to the root growth inhibited by high nitrate (Deprost et al., 2007), whereas the inhibited root growth in nitrogen-free medium is more obvious than in normal nitrogen-containing medium in TOR RNAi lines (Liu and Bassham, 2010). Very recent studies have shown that nitrate, ammonium, or amino acid rapidly activates TOR–S6K phosphorylation in seedlings starved for nitrogen nutrients. These new findings provide the first clue suggesting possible innovative pathways for diverse nitrogen nutrient sensing and signalling mechanisms in nitrogen-deficient plants (Liu et al., 2018). The puzzling observation of amino acid accumulation upon TOR inhibition is associated with a rapid increase in ammonium uptake and assimilation in Chlamydomonas, which suggests possible regulation by TOR of differential nitrogen usage. 13C- and 15N-isotope labelling experiments show that the imported ammonium is used to support de novo synthesis of amino acids only when carbon and nitrogen sources are available (Mubeen et al., 2018). This finding presents a new carbon–nitrogen connection in the context of TOR signalling. Similar studies have not been conducted to test the activation of phosphate or phosphate-derived metabolites to stimulate TOR signalling in phosphate-deficient plants. However, recent progress in the elucidation of the phosphate sensing and signalling mechanisms will facilitate new discoveries to connect phosphorus signalling to TOR regulation, possibly wired into the elaborate sugar–phosphate metabolic and signalling networks (Péret et al., 2011; Valluru and Van den Ende, 2011; Kuo et al., 2014; Couso et al., 2016; Wild et al., 2016; Chien et al., 2018; Gutiérrez-Alanís et al., 2018; Leong et al., 2018). For example, studies in Chlamydomonas and Arabidopsis have indicated the potential roles of inositol polyphosphates (InsPs) or pyrophosphates (PP-insPs) in phosphorus nutrient sensing and TOR signalling (Fig. 1A) (Kuo et al., 2014; Couso et al., 2016). As the null inositol pentakisphosphate 2-kinase1 (ipk1) plant mutant is lethal, analyses of the weaker ipk1 mutants suggest a link to growth defects in shoots and roots, increased phosphate accumulation in shoots, up-regulation of phosphorus starvation response (PSR) genes with phosphate repletion, and a genetic dependence on PHR1 (PHOSPHATE STARVATION RESPONSE 1) and PHL1 (PHR1-LIKE 1), key transcription factors for PSR gene expression (Kuo et al., 2014). The Chlamydomonas vip1-1 mutant lacking inositol hexakisphosphate kinase exhibits hypersensitivity to TORC1 inhibitors, rapamycin, torin1, and AZD-8055, with reduced InsP7 and InsP8 in the presence of acetate, which is reminiscent of high sugar stimulation of the PSR in plants. The accumulation of triacylglycerol in vip1-1 hints at a potential nutrient signalling link among carbon, nitrogen, and phosphorus sensing converged on TOR regulation (Fig. 1A) (Couso et al., 2016).
How fast TOR signalling could be stimulated by nitrogen, phosphorus, or their metabolites will depend on the timing required to generate the signalling molecules, perhaps through the integration of mitochondrial bioenergetics to activate TOR signalling. Curiously, phosphorus starvation leads to primary root meristem exhaustion, but extensive root hair and lateral root growth. The latter traits are the opposite of the root system responses observed in nitrate-deficient conditions (Ren et al., 2012; Xiong and Sheen, 2012; Liu et al., 2017). It is suggested that the adaption of the unique root system architecture to phosphorus starvation is geared to access phosphate more likely in the surface soil. Nitrogen signalling is adjusted to different levels of availability (starvation, low, and high), and promotes primary and lateral root development to forage for nitrogen nutrients deeper in the soil. Nutrient-stimulated specific adjustment in the plasticity and adaptation of plant root architecture probably serves to maximize the best strategies for a broad range of available nutrient concentrations (Gruber et al., 2013).
Perspectives
By integrating recent research progress and key findings on the molecular mechanisms of sugar, sulfur, nitrogen, and phosphorus sensing and signalling processes, our analyses have suggested the possibility that TOR as the energy signalling master regulator plays a central role as a universal and multifaceted nutrient signalling integrator. Emerging molecular connections to photoreceptors perceiving a distinct spectrum of light as well as hormone biosynthesis and signalling pathways have been discovered as the upstream regulators in modulating TOR activation or repression (Fig. 1A). The sensing and signalling components are molecularly wired to target and relay specific nutrient, hormonal, or environmental cues in different organs, tissues, cells, and subcellular compartments. The sites for sensing and signalling could be local or systemic. One major convergent output from the integrated sensing and signalling of macronutrients, light, and hormones is to activate TOR protein kinase to promote bioenergetics, biogenesis, metabolism, cell proliferation, and cell growth in diverse plant developmental programmes (Fig. 2) (Gojon et al., 2009; Takahashi et al., 2011; Gruber et al., 2013; Xiong et al., 2013; Liu et al., 2017; Chien et al., 2018; Gutiérrez-Alanís et al., 2018; Y.Y. Wang et al., 2018). Future research will identify the precise molecular regulators and elucidate their dynamic actions in these fundamental cellular processes. It will also be fruitful to dissect the relatively unexplored regulatory domains to unravel how TOR signalling represses an equally vast spectrum of primary target genes and pathways in stress and immune responses (Ahn et al., 2011; Moreau et al., 2012; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015). Innovative experimental design and new technologies in genomics, proteomics, phosphoproteomics, and metabolomics, as well as in single-cell imaging and biochemical analyses will complement genetic and natural variation screens to uncover new molecular mechanisms underlying fundamental growth activities in the nutrient–TOR signalling network (Caldana et al., 2013; Gruber et al., 2013; Xiong et al., 2013; Li et al., 2015; Couso et al., 2016; Dobrenel et al., 2016a; Liu et al., 2017; Saxton and Sabatini, 2017; Chien et al., 2018; Schepetilnikov and Ryabova, 2018; Shi et al., 2018; P. Wang et al., 2018).
New TOR chemical inhibitors have been deployed for biological assays and genetic screens to probe the functions and regulations of TOR in diverse signalling pathways. It is critical to interpret the findings in immediate or long-term molecular connections by examining the link to direct TOR protein kinase substrates. Unlike rapamycin, which is suggested specifically to inhibit TORC1, the ATP-competitive inhibitors appear to suppress a broader spectrum of TOR functions. It will be informative to combine and compare the effective range and specificity of these distinct inhibitors in future molecular studies of the plant TOR signalling networks. A better understanding of how nutrient availability is transduced to TOR signalling may allow novel strategies in improving efficient sensing and uptake of diverse essential nutrients and their integration into the central growth, anabolic, and biogenesis networks mediated by TOR. As TOR controls global transcription reprogramming and diverse metabolic, cellular, and developmental processes, it remains possible that additional TOR protein complexes may be discovered to modulate novel substrates and functions in the TOR signalling network in plants (Ahn et al., 2011; Ren et al., 2012; Caldana et al., 2013; Xiong et al., 2013; Dong et al., 2015; Dobrenel et al., 2016a; Schepetilnikov and Ryabova, 2018; Shi et al., 2018).
The plant TOR signalling network has evolved to monitor diverse inorganic and organic nutrient availability, including carbon, nitrogen, phosphorus, and sulfur, to support anabolic activities that require integrated inputs of multiple nutrients to promote ribosome biogenesis, protein translation, cell cycle, cell expansion, and photosynthesis. Transcriptome reprogramming stimulated by glucose–TOR signalling in turn up-regulates gene sets encoding functions for sugar, nitrate, phosphate, and sulfate transport, assimilation, and metabolism (Fig. 1). Future efforts will elucidate the molecular mechanisms underlying nitrogen or phosphorus sensing and signalling connections to the TOR regulatory network. The complex interactions and positive feedback loops that tie multiple essential nutrients and their metabolic derivatives as upstream regulatory signals and downstream targets of the plant TOR signalling network will be further unfolded (Fig. 1A). Developing an integrative view of how cells in different organs co-ordinate the acquisition of diverse nutrients required for growth, adaption, and survival will improve our understanding of the local and systemic nutrient sensing and signalling networks.
Acknowledgements
YW, LS, LL, and JS are supported by the National Institute of General Medical Sciences (R01 GM060493, GM070567, and GM129093). LF, YL, and YX are supported by the National Natural Science Foundation of China (31870269). Deposited in PMC for release after 12 months.
References
- Ahn CS, Ahn HK, Pai HS. 2015. Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. Journal of Experimental Botany 66, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. 2011. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. The Plant Cell 23, 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR. 2005. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biology 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. 2016. Architecture of human mTOR complex 1. Science 351, 48–52. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Manning BD. 2017. mTORC1 signaling and the metabolic control of cell growth. Current Opinion in Cell Biology 45, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Li X, Liu Y, Wang Y, Zhou Y, Xu T, Xiong Y. 2017. COP1 integrates light signals to ROP2 for cell cycle activation. Plant Signaling & Behavior 12, e1363946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. 2013. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. The Plant Journal 73, 897–909. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang W, Zhu JY, Oh E, Wang ZY. 2016. Information integration and communication in plant growth regulation. Cell 164, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH. 2018. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proceedings of the National Academy of Sciences, USA 115, 12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien PS, Chiang CP, Leong SJ, Chiou TJ. 2018. Sensing and signaling of phosphate starvation—from local to long distance. Plant & Cell Physiology 59, 1714–1722. [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. 1991. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. The Plant Cell 3, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso I, Evans BS, Li J, Liu Y, Ma F, Diamond S, Allen DK, Umen JG. 2016. Synergism between inositol polyphosphates and TOR kinase signaling in nutrient sensing, growth control, and lipid metabolism in Chlamydomonas. The Plant Cell 28, 2026–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Morrison G, Xu Z, Rui YN, et al. 2016. WAC regulates mTOR activity by acting as an adaptor for the TTT and Pontin/Reptin complexes. Developmental Cell 36, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Yu L, Zheng X, Zhang K, Wang W, Dong P, Zhang J, Ren M. 2016. Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Frontiers in Plant Science 7, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C. 2007. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports 8, 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. 2016a TOR signaling and nutrient sensing. Annual Review of Plant Biology 67, 261–285. [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Mancera-Martínez E, Forzani C, et al. 2016. b The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Frontiers in Plant Science 7, 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Xiong F, Que Y, Wang K, Yu L, Li Z, Ren M. 2015. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Frontiers in Plant Science 6, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, et al. 2017. Sulfur availability regulates plant growth via glucose–TOR signaling. Nature Communications 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Enganti R, Cho SK, Toperzer JD, Urquidi-Camacho RA, Cakir OS, Ray AP, Abraham PE, Hettich RL, von Arnim AG. 2017. Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Frontiers in Plant Science 8, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB. 2015. SWEETs, transporters for intracellular and intercellular sugar translocation. Current Opinion in Plant Biology 25, 53–62. [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. 2012. Sugars, signalling, and plant development. Journal of Experimental Botany 63, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Forzani C, Turqueto Duarte G, Meyer C. 2018. The plant target of rapamycin kinase: a connecTOR between sulfur and growth. Trends in Plant Science 23, 472–475. [DOI] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian JC. 2009. Root uptake regulation: a central process for NPS homeostasis in plants. Current Opinion in Plant Biology 12, 328–338. [DOI] [PubMed] [Google Scholar]
- González A, Hall MN. 2017. Nutrient sensing and TOR signaling in yeast and mammals. The EMBO Journal 36, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. 2008. Seed storage oil mobilization. Annual Review of Plant Biology 59, 115–142. [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N. 2013. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiology 163, 161–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cárdenas-Torres L, Herrera-Estrella L. 2018. Adaptation to phosphate scarcity: tips from Arabidopsis roots. Trends in Plant Science 23, 721–730. [DOI] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP. 2015. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4, e09181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans S, Rodriguez M, De Coninck B, Rolland F. 2016. The SnRK1 energy sensor in plant biotic interactions. Trends in Plant Science 21, 648–661. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG. 2007. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M. 2006. The genetics of plant metabolism. Nature Genetics 38, 842–849. [DOI] [PubMed] [Google Scholar]
- Khan MS, Haas FH, Samami AA, et al. 2010. Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. The Plant Cell 22, 1216–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, et al. 2013. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT–RUVBL1/2 complex. Molecular Cell 49, 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim S, Shin YJ, Hur YS, Kim WY, Lee MS, Cheon CI, Verma DP. 2014. Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. Journal of Biological Chemistry 289, 3901–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko A, Citerne S, Jéhanno I, Bersimbaev RI, Veit B, Meyer C, Leprince AS. 2015. Mutations in the Arabidopsis Lst8 and Raptor genes encoding partners of the TOR complex, or inhibition of TOR activity decrease abscisic acid (ABA) synthesis. Biochemical and Biophysical Research Communications 467, 992–997. [DOI] [PubMed] [Google Scholar]
- Kuo HF, Chang TY, Chiang SF, Wang WD, Charng YY, Chiou TJ. 2014. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. The Plant Journal 80, 503–515. [DOI] [PubMed] [Google Scholar]
- Lee DH, Park SJ, Ahn CS, Pai HS. 2017. MRF family genes are involved in translation control, especially under energy-deficient conditions, and their expression and functions are modulated by the TOR signaling pathway. The Plant Cell 29, 2895–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SJ, Lu WC, Chiou TJ. 2018. Phosphite-mediated suppression of anthocyanin accumulation regulated by mitochondrial ATP synthesis and sugars in Arabidopsis. Plant & Cell Physiology 59, 1158–1169. [DOI] [PubMed] [Google Scholar]
- Li L, Sheen J. 2016. Dynamic and diverse sugar signaling. Current Opinion in Plant Biology 33, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Song Y, Wang K, Dong P, Zhang X, Li F, Li Z, Ren M. 2015. TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Frontiers in Plant Science 6, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y. 2017. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proceedings of the National Academy of Sciences, USA 114, 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Niu Y, Konishi M, et al. 2017. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2010. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5, e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Xiong Y. 2018. Nitrogen–TOR signaling in shoot apex activation. In: EMBO Workshop. Target of rapamycin (TOR) signaling in photosynthetic organisms. Programme and Abstract Book, 96 EMBO Press. [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP. 2006. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. The Plant Cell 18, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Thomsen MF, Nintemann SJ, Jagd LM, Bourgine B, Burow M, Kliebenstein DJ. 2017. An evolutionarily young defense metabolite influences the root growth of plants via the ancient TOR signaling pathway. eLife 6, e29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA 99, 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed B, Bilooei SF, Dóczi R, Grove E, Railo S, Palme K, Ditengou FA, Bögre L, López-Juez E. 2018. Converging light, energy and hormonal signaling control meristem activity, leaf initiation, and growth. Plant Physiology 176, 1365–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B. 2013. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. Journal of Experimental Botany 64, 4361–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. 2003. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. [DOI] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, et al. 2012. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. The Plant Cell 24, 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubeen U, Jüppner J, Alpers J, Hincha DK, Giavalisco P. 2018. Target of rapamycin inhibition in Chlamydomonas reinhardtii triggers de novo amino acid synthesis by enhancing nitrogen assimilation. The Plant Cell 30, 2240–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, et al. 2016. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Scientific Reports 6, 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16, 442–450. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, et al. 2016. Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife 5, e17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. 2004. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. The Plant Cell 16, 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Venglat P, Qiu S, et al. 2012. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. The Plant Cell 24, 4850–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roustan V, Jain A, Teige M, Ebersberger I, Weckwerth W. 2016. An evolutionary perspective of AMPK–TOR signaling in the three domains of life. Journal of Experimental Botany 67, 3897–3907. [DOI] [PubMed] [Google Scholar]
- Ruan YL. 2014. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annual Review of Plant Biology 65, 33–67. [DOI] [PubMed] [Google Scholar]
- Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W. 2016. The regulation and plasticity of root hair patterning and morphogenesis. Development 143, 1848–1858. [DOI] [PubMed] [Google Scholar]
- Salem MA, Li Y, Bajdzienko K, Fisahn J, Watanabe M, Hoefgen R, Schöttler MA, Giavalisco P. 2018. RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiology 177, 565–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. The EMBO Journal 32, 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA. 2017. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. The EMBO Journal 36, 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Ryabova LA. 2017. Auxin signaling in regulation of plant translation reinitiation. Frontiers in Plant Science 8, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Ryabova LA. 2018. Recent discoveries on the role of TOR (Target of Rapamycin) signaling in translation in plants. Plant Physiology 176, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 2014. Master regulators in plant glucose signaling networks. Journal of Plant Biology 57, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Wu Y, Sheen J. 2018. TOR signaling in plants: conservation and innovation. Development 145, dev160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O, Kim S, Shin YJ, Kim WY, Koh HJ, Cheon CI. 2015. Identification of nucleosome assembly protein 1 (NAP1) as an interacting partner of plant ribosomal protein S6 (RPS6) and a positive regulator of rDNA transcription. Biochemical and Biophysical Research Communications 465, 200–205. [DOI] [PubMed] [Google Scholar]
- Speiser A, Silbermann M, Dong Y, et al. 2018. Sulfur partitioning between glutathione and protein synthesis determines plant growth. Plant Physiology 177, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracka D, Jozefczuk S, Rudroff F, Sauer U, Hall MN. 2014. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. Journal of Biological Chemistry 289, 25010–25020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Y, Hu W, Min Q, Kang H, Li Y, Hong Y, Wang X, Hong Y. 2016. Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochimica et Biophysica Acta 1861, 639–649. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Tatebe H, Shiozaki K. 2017. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru R, Van den Ende W. 2011. Myo-inositol and beyond—emerging networks under stress. Plant Science 181, 387–400. [DOI] [PubMed] [Google Scholar]
- Wagner S, Van Aken O, Elsässer M, Schwarzländer M. 2018. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiology 176, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, et al. 2018. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Molecular Cell 69, 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Cheng YH, Chen KE, Tsay YF. 2018. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology 69, 85–122. [DOI] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, et al. 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. [DOI] [PubMed] [Google Scholar]
- Wingler A. 2018. Transitioning to the next phase: the role of sugar signaling throughout the plant life cycle. Plant Physiology 176, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH. 2014. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annual Review of Plant Biology 65, 311–333. [DOI] [PubMed] [Google Scholar]
- Wurzinger B, Nukarinen E, Nägele T, Weckwerth W, Teige M. 2018. The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiology 176, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Zhang R, Meng Z, Deng K, Que Y, Zhuo F, Feng L, Guo S, Datla R, Ren M. 2017. Brassinosteriod insensitive 2 (BIN2) acts as a downstream effector of the target of rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytologist 213, 233–249. [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. 2013. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2012. Rapamycin and glucose–target of rapamycin (TOR) protein signaling in plants. Journal of Biological Chemistry 287, 2836–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2014. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiology 164, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2015. Novel links in the plant TOR kinase signaling network. Current Opinion in Plant Biology 28, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Beeckman T, Xu G. 2017. Plant nitrogen nutrition: sensing and signaling. Current Opinion in Plant Biology 39, 57–65. [DOI] [PubMed] [Google Scholar]
- Yu S, Lian H, Wang JW. 2015. Plant developmental transitions: the role of microRNAs and sugars. Current Opinion in Plant Biology 27, 1–7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, McFarlane HE, Obata T, Richter AS, Lohse M, Grimm B, Persson S, Fernie AR, Giavalisco P. 2018. Inhibition of TOR represses nutrient consumption, which improves greening after extended periods of etiolation. Plant Physiology 178, 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY. 2016. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Current Biology 26, 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]