The adaptation of plants to their environment requires tight regulation of metabolism and growth processes through central and highly connected signalling pathways. The signalling cascade involving the evolutionarily conserved Target of Rapamycin (TOR) represents just such a central regulatory hub, and research on this protein kinase in plants has progressed significantly during the past decade. TOR is now firmly established as a central player in plant responses to the availability of nutrients such as sugars, stresses including those from pathogens, and hormones. Moreover plant-specific targets and regulators have recently been identified. The reviews in this special issue explore the various facets of regulation exerted by this fascinating kinase as well as its potential for crop improvement.
Eukaryotic cells and organisms need to adjust basic processes such as cell division, growth and metabolism to the available resources and external conditions. These are sensed either directly, as in unicellular eukaryotes, or indirectly, for example through hormonal or nutrient signals, as occurs in multicellular organisms. Central to this regulation is the conserved kinase Target of Rapamycin (TOR), which has been shown to be a critical component of sensing mechanisms.
The discovery of TOR is a perfect example of the serendipitous nature of research. It all started with the identification of a molecule produced by Streptomyces hygroscopicus, a bacterium isolated in the 1970s in a soil sample from the remote and mysterious Easter Island, known as Rapa Nui in Polynesian. This compound was thus named rapamycin (Vezina et al., 1975). Rapamycin was found to inhibit cell proliferation but the mechanism of action was unknown. It was only in the 1990s that Michael Hall’s group in Basel identified mutations in yeast which conferred resistance to rapamycin. This led to the seminal discovery of TOR, inhibition of which by rapamycin leads to arrested growth (Heitman et al., 1991; Montané and Menand, 2019). Michael Hall was later awarded the 2017 Lasker prize for medical research for this paramount discovery.
TOR was subsequently identified in humans, where it was given the name mTOR (mammalian or mechanistic TOR), in various animals including flies and worms (Saxton et al., 2017; Mossmann et al., 2018), flowering plants (Menand et al., 2002) and algae (Perez-Perez et al., 2017). In all eukaryotes, TOR is a very large (around 250 kDa) serine/threonine kinase belonging to the phosphatidylinositol 3-kinase-related kinase (PIKK) family. Indeed, although TOR is a protein kinase, its catalytic domain is unconventional and resembles that of PI lipid kinases. Other members of this family include the conserved DNA damage checkpoint ataxia-telangiectasia mutated (ATM) and ATM-related (ATR) kinases, which are also present in plants.
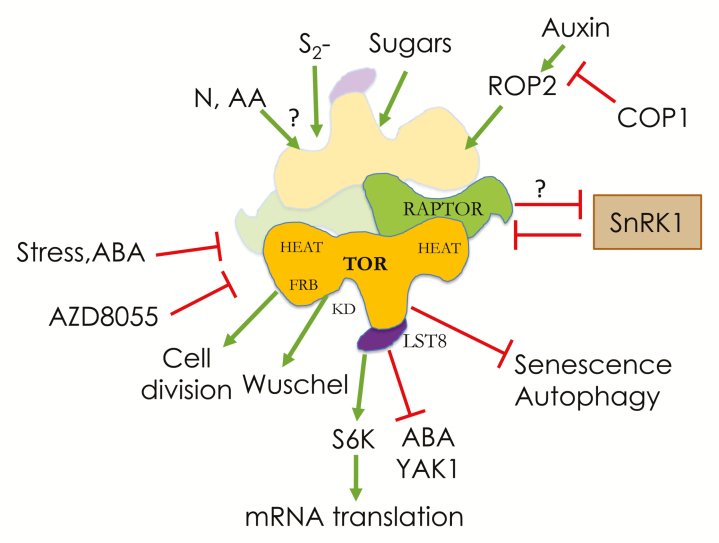
In yeast and animals, where TOR has been studied extensively, various upstream regulators and downstream effectors have been discovered, forming a complex and highly connected signalling pathway (Blenis, 2017). In these organisms ScTOR and mTOR play a major role in the regulation of cell growth and metabolism through the control of gene transcription, cell trafficking, insulin responses as well as protein synthesis and degradation. Therefore, TOR is involved in many human diseases including cancer and diabetes. Thorough biochemical work showed that TOR belongs to two very large complexes, TORC1 and TORC2, containing common and specific companion proteins (Wullschleger et al., 2006; Saxton et al., 2017; Mossmann et al., 2018). Structural models for TOR complexes are now available (Aylett et al., 2016; Karuppasamy et al., 2017). In plants only the TORC1 complex has been described so far which, together with TOR, comprises the evolutionarily conserved LST8 and RAPTOR proteins (Box 1).
Box 1. The basics of plant TOR
The conserved TOR (Target of Rapamycin) protein kinase acts in TORC1 (TOR complex 1) with LST8 and RAPTOR protein partners. This complex is found in all eukaryotes and is an important molecular element connecting nutrient, hormonal and stress signals to metabolism, growth and hormonal responses. TORC1 can be seen as a switch turning on anabolic and growth processes when conditions are favourable and inhibiting catabolism and nutrient recycling by autophagy. TORC1 controls mRNA translation globally but also the translation of specific mRNAs such as those encoding proteins needed for stress or hormone responses. Mounting evidence suggest that TOR and SnRK1/2 (Snf1-related kinases) act in an opposing way and it has been shown that SnRKs inhibit TOR activity by phosphorylating the RAPTOR protein.
In recent years, many targets of TOR have been identified in plants and algae including the ribosomal protein S6 kinase, the PP2A phosphatase partner TAP46, several components of the translation and cell division machinery, and also the PYL ABA receptors.
HEAT, HEAT [Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), TOR1] repeat domain involved in protein–protein interactions; FRB, FKBP12-rapamycin binding domain; KD, kinase domain.
TOR and the control of plant growth
As in yeast and animals, TOR has been implicated in controlling plant growth and cell division (Caldana et al., 2019) and prominent inducers of TOR activity have been shown to be sucrose and glucose (Xiong et al., 2012; Dobrenel et al., 2016a; Shi et al., 2018). Indeed, a strong sugar–TOR growth-controlling axis has emerged from several studies in both roots and shoots (Xiong et al., 2013; Pfeiffer et al., 2016; Wu et al., 2019). Basically, sugars like sucrose and glucose strongly up-regulate TOR activity by a largely unknown mechanism and TOR activates meristems by inducing E2F and Wuschel transcription factors in root and shoot apical meristems, respectively (Xiong et al., 2013; Pfeiffer et al., 2016). Nutrient starvation or defects in nutrient assimilation also affect TOR activity. For example, a decrease in sulfur assimilation due to mutations in the sulfite reductase gene diminishes TOR activity and growth through a reduction is sugar accumulation (Dong et al., 2017). The review by Ahmad et al. (2019) presents further detail on regulation of the plant cell cycle by E2F and other conserved complexes in relation to TOR activity. TOR also influences the cell wall (Leiber et al., 2010), and a new suppressor of lrx1 (leucine-rich repeat extensin 1), a mutation which affects cell wall architecture and is compensated by TOR inhibition, has been identified and found to encode isopropylmalate synthase 1, a protein involved in leucine biosynthesis (Schaufelberger et al., 2019).
Reduction of TOR activity by starvation or other signals increases lifespan for several eukaryotes (Weichhart, 2018). Indeed, inhibition of TOR with rapamycin is currently the only known treatment that increases lifespan in all organisms that have been studied. In plants, however, there is a more complex relationship between TOR activity and aging (Quilichini et al., 2019).
TORC1: master regulator of stress and adaptive responses
In plants, data suggests that activity of the nutrient- and energy-induced TOR kinase is balanced in an antagonistic and probably reciprocal way with one of the starvation-induced kinases of the SnRK (Snf1-related kinase) 1 and 2 families (Box 1; Dobrenel et al., 2016a; Baena-Gonzalez and Hanson, 2017; Jamsheer et al., 2019; Margalha et al., 2019). SnRK1 has been shown to phosphorylate the RAPTOR component of the TORC1 complex and to inactivate TOR (Nukarinen et al., 2016); this function is evolutionarily conserved, since the animal orthologue of SnRK1, the AMP-activated kinase, performs the same post-translational modification to inactivate TOR in response to energy and carbon starvation (Gwinn et al., 2008). Interestingly, changes in subcellular localization of SnRK1 could be involved in inactivating TOR in different compartments (Blanco et al., 2019). SnRK2s have also recently been shown to phosphorylate RAPTOR and to inactivate TOR in response to ABA (Wang et al., 2018). Consistently the same report showed that TOR is able to repress ABA signalling in normal, non-stressed conditions by phosphorylating and inactivating the ABA PYL receptor and that there is a close and antagonistic relationship between TOR and ABA signalling (Box 1; Deprost et al., 2007; Punzo et al., 2018; Forzani et al., 2019, Preprint; Wu et al., 2019). Furthermore, the YAK1 kinase, which is related to ABA signalling (Kim et al., 2016), has been shown to suppress the effects of TOR inhibition (Box 1; Barrada et al., 2019; Forzani et al., 2019, Preprint).
TOR, as a central regulatory module, has connections with many plant hormones apart from ABA (Schepetilnikov and Ryabova, 2018; Jamsheer et al., 2019). For example, brassinosteroids partly suppress the defects in hypocotyl elongation observed in TOR-deficient lines (Zhang et al., 2016) and the ROP2 small Rho protein participates in the auxin-induced stimulation of TOR activity (Li et al., 2017; Schepetilnikov et al., 2017). Light is also an efficient inducer of TOR and recently the stimulation of auxin–ROP2–TOR signalling and mRNA translation observed in the light has been shown to be connected with COP1 (constitutive morphogenesis 1) (Cai et al., 2017; Chen et al., 2018; Ahmad et al., 2019).
Involvement of TOR in the trade-off between growth and defence
Given the central role of the TOR pathway in regulating gene expression, protein synthesis and metabolic adjustments, it could be questioned whether it is also wired to defence responses or targeted during pathogen infection. Growth and defence responses are known to be antagonistic and TOR inhibition was found to prevent growth and activate the salicylic acid-dependent defence pathway in Arabidopsis (Moreau et al., 2012; Dong et al., 2015). Meteignier et al. (2017) found a significant subset of genes translationally regulated by both TOR silencing and activation of the immune response through conditional expression of the Pseudomonas syringae avrRPM1 elicitor. Consistently, TOR-silenced plants display enhanced resistance to P. syringae, leading to the hypothesis that active TOR negatively regulates immunity at the translational level.
In another study, the awr5 effector of Ralstonia solanacearum when expressed in yeast produced similar effects to rapamycin (Popa et al., 2016). TOR inhibition by AZD8055 was also found to block the growth of Xanthomonas citri in Citrus spp. (Soprano et al., 2018). The X. citri effector PthA4 is an interactor of Maf1, a conserved target of TOR, and repressor of Pol III and growth. Maf1 phosphorylation levels decreased upon TOR knockdown and increased following knockdown of the catalytic subunits of PPP4 and PP2A (Ahn et al., 2019).
The role of TOR in the trade-off between growth and immunity was further substantiated in rice; genetic experiments involving overexpression or silencing of OsTOR or OsRAPTOR, combined with rapamycin treatments, showed that TOR negatively regulates several defence-related WRKY and MYB transcription factors, JA and SA signalling and resistance to various pathogens such as Xanthomonas oryzaea (De Vleesschauwer et al., 2018). TOR down-regulation or raptor mutation were also reported to reduce susceptibility to Fusarium graminearum in an Arabidopsis infection model (Aznar et al., 2018). In this work the TOR inhibitor PP242 was also found to reduce leaf infection by Fusarium, although a direct role of the TOR inhibitor on the eukaryotic pathogen itself is probable and cannot be ruled out. Indeed, beside its emerging role in the trade-off between growth and immunity in plant hosts, an active TOR pathway is also involved in the virulence of several eukaryotic pathogens (Shertz and Cardenas, 2011)
Several links between TOR and pathogen infection emerged from virus studies (Margalha et al., 2019). One of the first discoveries was the direct interaction between TOR and the Cauliflower mosaic virus transactivator protein (TAV) which promotes reinitiation of translation along the CaMV multicistronic mRNA (Schepetilnikov et al., 2011). The role of TOR was also investigated in the case of potyviruses, the largest group of plant RNA viruses. It was found that TOR-silenced or AZD8055-treated plants were resistant to Watermelon Mosaic Virus (WMV). Remarkably, AZD8055 treatment was able to eliminate the virus from infected plants (Ouibrahim et al., 2015). More recently, repression of TOR activity by Tombusvirus infection was observed in plants and yeast, and TOR pathway inhibition or genetic repression was shown to reduce virus replication (Inaba and Nagy, 2018). It was further hypothesized that Tombusviruses may recruit the glycolytic pathway, which is regulated by TOR, to furnish energy for virus replication and that TOR repression is part of the plant defence response.
TOR controls mRNA translation
Stress negatively affects plant growth and development, triggering transcriptional, translational and metabolic reprogramming (Bechtold and Field, 2018; Margalha et al., 2019). Indeed, cells can increase or decrease the production of proteins to accommodate changes in response to stress (Bailey-Serres and Juntawong, 2012; Schepetilnikov and Ryabova, 2018).
TOR-related pathways and molecular mechanisms that mediate the impact of stress signals on translation are largely unknown. Low energy stress can drastically decrease global protein synthesis rates, manifested by low levels of active ribosomes (polysomes), but this is reversible in conditions of energy supply (Tomé et al., 2014).
Cold treatment can reduce global polysome levels in rice (Park et al., 2012), while in Arabidopsis they are not significantly affected. Whether TOR is involved in cold stress tolerance in Arabidopsis, as was suggested for AtGCN1 (Wang et al., 2017), would require further research. However, the authors suggest that decreased translation levels in conditions of TOR inhibition might increase the ability of Arabidopsis seedlings to survive under cold stress (Wang et al., 2017).
The most critical TOR upstream input comes from light and photosynthesis; suppression of TOR activity negatively affects light-dependent plant growth (Chen et al., 2018). Although there is a tight link between light, TOR activity, RPS6 (ribosomal protein S6) phosphorylation and global protein synthesis efficiency (Piques et al., 2009; Floris et al., 2013; Enganti et al., 2017), mechanisms used by TOR to impact global protein synthesis have not yet been properly elucidated.
Recent work has identified an MRF (Ma3 Domain-Containing Translation Regulatory Factor) family of proteins that function in translation, especially under energy-deficient conditions (Lee et al., 2017). Strikingly, the human homologue of the MRFs, hPDCD4, inhibits translation initiation by binding eIF4A and preventing its interaction with eIF4G and, thus, formation of the functional eIF4F complex (Loh et al., 2009). In addition, similarly to hPDCD4, which is phosphorylated by S6K1/TOR, MRF1 phosphorylation was regulated positively by S6K1/TOR (Lee et al., 2017).
One efficient way of rapidly altering protein production in response to internal and external stimuli is to regulate mRNA translation selectively. In eukaryotes, TOR is implicated in translational regulation of a specific set of TOP (5’-terminal oligopyrimidine tract)-containing mRNAs via phosphorylation of an La RNA binding protein (LARP1) (Tcherkezian et al., 2014). In plants, heat stress can dramatically induce an mRNA degradation process that is dependent on Arabidopsis LARPs (La-related Proteins) (Deragon and Bousquet-Antonelli, 2015). Although LARP1 was identified as a TOR/S6K1 downstream target in Chlamydomonas reinhardtii and Arabidopsis (Werth et al., 2019; Van Leene et al., 2019), its role in translation of TOP-containing mRNAs (Dobrenel et al., 2016b) remains to be identified.
Active TOR impacts translation of a specific set of mRNAs that harbour upstream open reading frames (uORFs) within their leader regions (Schepetilnikov et al., 2013). Moreover, subunit h of eukaryotic translation initiation factor 3 (eIF3h) was identified as a new TOR/S6K1 target, and its phosphorylation site mapped to Ser178 (Schepetilnikov et al., 2013). The authors proposed that TOR, when activated, promotes phosphorylation of eIF3h and maintains its active phosphorylation status in Arabidopsis to ensure efficient reinitiation events (Schepetilnikov and Ryabova, 2018). However, other players in auxin signalling, such as phospholipase D zeta2 (PLD; required for auxin responses) (Li et al., 2007), which is controlled by auxin, might also participate in TOR activation either directly or via production of phosphatidic acid (PA).
The growing wealth of phosphoproteomic data reveals hundreds of proteins with quantitative changes in their phosphorylation status in response to TOR inhibition in C. reinhardtii (Roustan and Weckwerth, 2018; Werth et al., 2019) and Arabidopsis (Van Leene et al., 2019). Among these TOR signalling-related targets are homologues of an elongation factor 2 kinase (EEF2K), components of the eIF2B complex, eiF4E-binding protein, LARP1 and CTC-interacting domain 4 (CID4). RPS6 Ser240 was clearly identified as a robust and conserved TOR/S6K1-dependent phosphosite in Arabidopsis and Chlamydomonas 40S ribosomal subunit (Dobrenel et al., 2016b; Enganti et al., 2017; Werth et al., 2019). Consistently, an increase in RPS6 phosphorylation was also observed after loss of SnRK1 activity (Nukarinen et al., 2016). However, the role of this evolutionarily conserved TOR-dependent phosphorylation event in the control of mRNA translation is not very well defined and will need further research.
Conclusion and future directions
It is now clear that, as in other eukaryotes, the plant TOR signalling pathway integrates information about external conditions, either directly or through hormonal signals, to mount the necessary physiological, molecular and developmental responses. In recent years the plant TOR field has been blooming. Compared to animals and yeasts, the TOR pathway may be even more critical and integrate specific functions in algae and multicellular plants, which cannot escape from adverse environmental conditions or nutrient scarcity.
A better understanding of the role and regulation of the TOR kinase is still needed but recent phosphoproteomic and interatomic resources will be a great help for deciphering this fascinating regulatory hub (Van Leene et al., 2019). This should open novel routes for increasing crop yield through stimulation of growth, improved pathogen defence and manipulation of metabolite partitioning.
Acknowledgments
Reviews and research articles in this special issue were developed following the EMBO workshop ‘TOR signaling in photosynthetic organisms’ held in Alsace, France in 2018. Journal of Experimental Botany contributed to funding for this meeting. The authors were funded by the DecoraTOR ANR grant (ANR14-CE19-007). CM is also funded by a French State grant (LabEx Saclay Plant Sciences-SPS, ANR-10-LABX-0040-SPS).
References
- Ahmad Z, Magyar Z, Bögre L, Papdi C. 2019. Cell cycle control by the target of Rapamycin signalling pathway. Journal of Experimental Botany 70, 2275–2284. [DOI] [PubMed] [Google Scholar]
- Ahn CS, Lee DH, Pai HS. 2019. Characterization of Maf1 in Arabidopsis: function under stress conditions and regulation by the TOR signaling pathway. Planta 249, 527–542. [DOI] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. 2016. Architecture of human mTOR complex 1. Science 351, 48–52. [DOI] [PubMed] [Google Scholar]
- Aznar NR, Consolo VF, Salerno GL, Martínez-Noël GMA. 2018. TOR signaling downregulation increases resistance to the cereal killer Fusarium graminearum. Plant Signaling & Behavior 13, e1414120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Hanson J. 2017. Shaping plant development through the SnRK1-TOR metabolic regulators. Current Opinion in Plant Biology 35, 152–157. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Juntawong P. 2012. Dynamic light regulation of translation status in Arabidopsis thaliana. Frontiers in Plant Science 3, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrada A, Djendli M, Desnos T, Mercier R, Robaglia C, Montané MH, Menand B. 2019. A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development 146, doi: 10.1242/dev.171298 [DOI] [PubMed] [Google Scholar]
- Bechtold U, Field B. 2018. Molecular mechanisms controlling plant growth during abiotic stress. Journal of Experimental Botany 69, 2753–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco NE, Liebsch D, Guinea DÍaz M, Strand Å, Whelan J. 2019. Dual and dynamic intracellular localization of Arabidopsis thaliana SnRK1.1. Journal of Experimental Botany 70, 2325–2338. [DOI] [PubMed] [Google Scholar]
- Blenis J. 2017. TOR, the gateway to cellular metabolism, cell growth, and disease. Cell 171, 10–13. [DOI] [PubMed] [Google Scholar]
- Cai W, Li X, Liu Y, Wang Y, Zhou Y, Xu T, Xiong Y. 2017. COP1 integrates light signals to ROP2 for cell cycle activation. Plant Signaling & Behavior 12, e1363946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Martins MCM, Mubeen U, Urrea-Castellanos R. 2019. The magic ‘hammer’ of TOR: the multiple faces of a single pathway in the metabolic regulation of plant growth and development. Journal of Experimental Botany 70, 2217–2225. [DOI] [PubMed] [Google Scholar]
- Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH. 2018. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating. Proceedings of the National Academy of Sciences, USA 115, 12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C. 2007. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Reports 8, 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deragon JM, Bousquet-Antonelli C. 2015. The role of LARP1 in translation and beyond. Wiley Interdisciplinary Reviews. RNA 6, 399–417. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D, Filipe O, Hoffman G, et al. 2018. Target of rapamycin signaling orchestrates growth-defense trade-offs in plants. New Phytologist 217, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. 2016a. TOR signaling and nutrient sensing. Annual Review of Plant Biology 67, 261–285. [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Mancera-Martínez E, Forzani C, et al. 2016b. The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Frontiers in Plant Science 7, 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Xiong F, Que Y, Wang K, Yu L, Li Z, Ren M. 2015. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Frontiers in Plant Science 6, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, et al. 2017. Sulfur availability regulates plant growth via glucose-TOR signaling. Nature Communications 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Enganti R, Cho SK, Toperzer JD, Urquidi-Camacho RA, Cakir OS, Ray AP, Abraham PE, Hettich RL, von Arnim AG. 2017. Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Frontiers in Plant Science 8, 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floris M, Bassi R, Robaglia C, Alboresi A, Lanet E. 2013. Post-transcriptional control of light-harvesting genes expression under light stress. Plant Molecular Biology 82, 147–154. [DOI] [PubMed] [Google Scholar]
- Forzani C, Duarte G, Clement G, Huguet S, Paysant-Le-Roux C, Mercier R, Leprince AS, Meyer C. 2019. Mutations of the Atyak1 kinase suppress TOR deficiency in Arabidopsis. Cell Press Sneak Peak. doi: 10.2139/ssrn.3305560 [Preprint]. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell 30, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Inaba JI, Nagy PD. 2018. Tombusvirus RNA replication depends on the TOR pathway in yeast and plants. Virology 519, 207–222. [DOI] [PubMed] [Google Scholar]
- Jamsheer K M, Jindal S, Laxmi A. 2019. Evolution of TOR–SnRK dynamics in green plants and its integration with phytohormone signaling networks. Journal of Experimental Botany 70, 2239–2259. [DOI] [PubMed] [Google Scholar]
- Karuppasamy M, Kusmider B, Oliveira TM, Gaubitz C, Prouteau M, Loewith R, Schaffitzel C. 2017. Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nature Communications 8, 1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Ntui VO, Xiong L. 2016. Arabidopsis YAK1 regulates abscisic acid response and drought resistance. FEBS Letters 590, 2201–2209. [DOI] [PubMed] [Google Scholar]
- Lee DH, Park SJ, Ahn CS, Pai HS. 2017. MRF family genes are involved in translation control, especially under energy-deficient conditions, and their expression and functions are modulated by the TOR signaling pathway. The Plant Cell 29, 2895–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. 2010. The TOR pathway modulates the structure of cell walls in Arabidopsis. The Plant Cell 22, 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y. 2017. Differential TOR activation and cell proliferation in. Proceedings of the National Academy of Sciences, USA 114, 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xue HW. 2007. Arabidopsis PLDzeta2 regulates vesicle trafficking and is required for auxin response. The Plant Cell 19, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, Liu D, Song H. 2009. Structural basis for translational inhibition by the tumour suppressor Pdcd4. The EMBO Journal 28, 274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalha L, Confraria A, Baena E. 2019. SnRK1 and TOR: modulating growth–defense trade-offs in plant stress responses. Journal of Experimental Botany 70, 2261–2274. [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA 99, 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteignier LV, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, Moffett P. 2017. Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. Journal of Experimental Botany 68, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Montané M-H, Menand B. 2019. TOR inhibitors: from mammalian outcomes to pharmacogenetics in plant and algae. Journal of Experimental Botany 70, 2297–2312. [DOI] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, et al. 2012. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. The Plant Cell 24, 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann D, Park S, Hall MN. 2018. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nature reviews. Cancer 18, 744–757. [DOI] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, et al. 2016. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Scientific Reports 6, 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouibrahim L, Rubio AG, Moretti A, Montané MH, Menand B, Meyer C, Robaglia C, Caranta C. 2015. Potyviruses differ in their requirement for TOR signalling. The Journal of General Virology 96, 2898–2903. [DOI] [PubMed] [Google Scholar]
- Park SH, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim YK, Do Choi Y, Kim JK. 2012. Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3’ and 5’ untranslated regions and correlates with differential polysome association in rice. Plant Physiology 159, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Couso I, Crespo JL. 2017. The TOR Signaling Network in the Model Unicellular Green Alga Chlamydomonas reinhardtii. Biomolecules 7, doi: 10.3390/biom7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, et al. 2016. Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. Elife 5, e17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. 2009. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Molecular Systems Biology 5, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa C, Li L, Gil S, Tatjer L, Hashii K, Tabuchi M, Coll NS, Ariño J, Valls M. 2016. The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Scientific Reports 6, 27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo P, Ruggiero A, Grillo S, Batelli G. 2018. TIP41 network analysis and mutant phenotypes predict interactions between the TOR and ABA pathways. Plant Signaling & Behavior 20, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Gao P, Pandey PK, Xiang D, Ren M, Datla R. 2019. Live long and praise TOR: a role for TOR signaling in every stage of plant life. Journal of Experimental Botany 70, 2285–2296. [DOI] [PubMed] [Google Scholar]
- Roustan V, Weckwerth W. 2018. Quantitative phosphoproteomic and system-level analysis of TOR inhibition unravel distinct organellar acclimation in Chlamydomonas reinhardtii. Frontiers in Plant Science 9, 1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schaufelberger M, Galbier F, Herger A, de Brito Francisco R, Roffler S, Clement G, Diet A, Hörtensteiner S, Wicker T, Ringli C. 2019. Mutations in the Arabidopsis ROL17/isopropylmalate synthase 1 locus alter amino acid content, modify the TOR network, and suppress the root hair cell development mutant lrx1. Journal of Experimental Botany 70, 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. The EMBO Journal 32, 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. 2011. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. The EMBO Journal 30, 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA. 2017. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. The EMBO Journal 36, 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Ryabova LA. 2018. Recent discoveries on the role of TOR (Target of Rapamycin) signaling in translation in plants. Plant Physiology 176, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shertz CA, Cardenas ME. 2011. Exploiting and subverting Tor signaling in the pathogenesis of fungi, parasites, and viruses. PLoS Pathogens 7, e1002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Wu Y, Sheen J. 2018. TOR signaling in plants: conservation and innovation. Development 145, doi: 10.1242/dev.160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano AS, Smetana JHC, Benedetti CE. 2018. Regulation of tRNA biogenesis in plants and its link to plant growth and response to pathogens. Biochimica et biophysica acta. Gene Regulatory Mechanisms 1861, 344–353. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. 2014. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes & Development 28, 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco-llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, et al. 2014. The low energy signaling network. Frontiers in Plant Science 5, doi: 10.3389/fpls.2014.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, et al. 2019. Integration of phosphoproteomics and interactomics elucidates substrates of the target of rapamycin kinase in plants. Nature Plants 5, 316–327. [Google Scholar]
- Vézina C, Kudelski A, Sehgal SN. 1975. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. The Journal of Antibiotics 28, 721–726. [DOI] [PubMed] [Google Scholar]
- Wang L, Li H, Zhao C, et al. 2017. The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant, Cell & Environment 40, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, et al. 2018. Reciprocal regulation of the TOR Kinase and ABA receptor balances plant growth and stress response. Molecular Cell 69, 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T. 2018. mTOR as regulator of lifespan, aging, and cellular senescence: a mini-review. Gerontology 64, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth EG, McConnell EW, Couso Lianez I, Perrine Z, Crespo JL, Umen JG, Hicks LM. 2019. Investigating the effect of target of rapamycin kinase inhibition on the Chlamydomonas reinhardtii phosphoproteome: from known homologs to new targets. New Phytologist 221, 247–260. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shi L, Li L, Fu L, Liu Y, Xiong Y, Sheen J. 2019. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. Journal of Experimental Botany 70, 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. 2013. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2012. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. The Journal of Biological Chemistry 287, 2836–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY. 2016. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in arabidopsis. Current Biology 26, 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]