Abstract
The heart plays the vital role of propelling blood to the entire body, which is essential to life. While maintaining heart function is critical, adult mammalian hearts poorly regenerate damaged cardiac tissue upon injury and form scar tissue instead. Unlike adult mammals, adult zebrafish can regenerate injured hearts with no sign of scarring, making zebrafish an ideal model system with which to study the molecular mechanisms underlying heart regeneration. Investigation of heart regeneration in zebrafish together with mice has revealed multiple cardiac regeneration genes that are induced by injury to facilitate heart regeneration. Altered expression of these regeneration genes in adult mammals is one of the main causes of heart regeneration failure. Previous studies have focused on the roles of these regeneration genes, yet the regulatory mechanisms by which the expression of cardiac regeneration genes is precisely controlled are largely unknown. In this review, we will discuss the importance of differential gene expression for heart regeneration, the recent discovery of cardiac injury or regeneration enhancers, and their impact on heart regeneration.
Keywords: heart, zebrafish, regeneration, enhancer, transcription, gene regulation, development
1. Introduction
The ability to regenerate damaged cardiac tissues varies across vertebrate species. While neonatal mice can regenerate injured hearts during the first week of life, adult mammalian hearts regenerate poorly and form scar tissue instead. Scar formation impedes the ability of the heart to propel blood to the whole body, leading to increased morbidity and mortality [1]. Unlike adult mammals, adult nonmammalian species including zebrafish, goldfish (Carassius auratus), giant danio (Devario aequipinnatus), newts, axolotls, and frogs can regenerate injured hearts [2,3,4,5,6,7]. Thus, investigating these models to understand heart regeneration can provide insights into heart repair that may be applied to mammals.
Heart regeneration studies using zebrafish and mice have revealed multiple factors influencing cardiac regeneration. Zebrafish live in an aquatic environment that has a low oxygen level, and this hypoxic condition likely enables the proliferation of cardiac muscle cells, or cardiomyocytes (CMs) [8]. The number of complete sets of chromosomes also affects CM proliferation. While nonregenerative hearts, such as adult mammalian hearts, are composed of CMs that are primarily polyploid (containing more than two sets of homologous chromosomes), almost all zebrafish CMs are diploid (containing two sets of homologous chromosomes) [9]. Notably, the percentage of diploid CMs is highly variable across mouse strains and correlates with the ability to enhance post-injury CM proliferation in adult mice [9]. Interestingly, genetically modified zebrafish in which a majority of CMs are polyploid fail to regenerate, indicating that CM polyploidization acts as a heart regeneration barrier [9,10]. In addition to environmental and cellular structure discrepancies, other main determinants of cardiac regenerative capacity are differentially expressed regeneration-driving genes [11,12,13,14]. Transcriptomic analysis of CMs, fibroblasts, leukocytes, and endothelial cells from infarcted and non-infarcted neonatal (P1) and adult (P56) mice has revealed that adult, but not neonatal, CMs and endothelial cells fail to reactivate proliferation-related genes upon cardiac injury, which contributes to the loss of cardiac regeneration in the adult stage [13]. This study demonstrated that regulatory programs governing neonatal cardiac regeneration may be altered in adults to prevent the reactivation of regeneration gene expression. Thus, transcriptional regulation and underlying regulatory mechanisms are crucial for heart regeneration. In this review, we first describe how the transcription of regeneration genes implicates heart regeneration in multiple species. We then highlight recent discoveries of regulatory elements controlling the transcription of regeneration genes: cardiac regeneration enhancers.
2. Differentially Expressed Genes Underlie Maintenance or Loss of Cardiac Regenerative Capacity
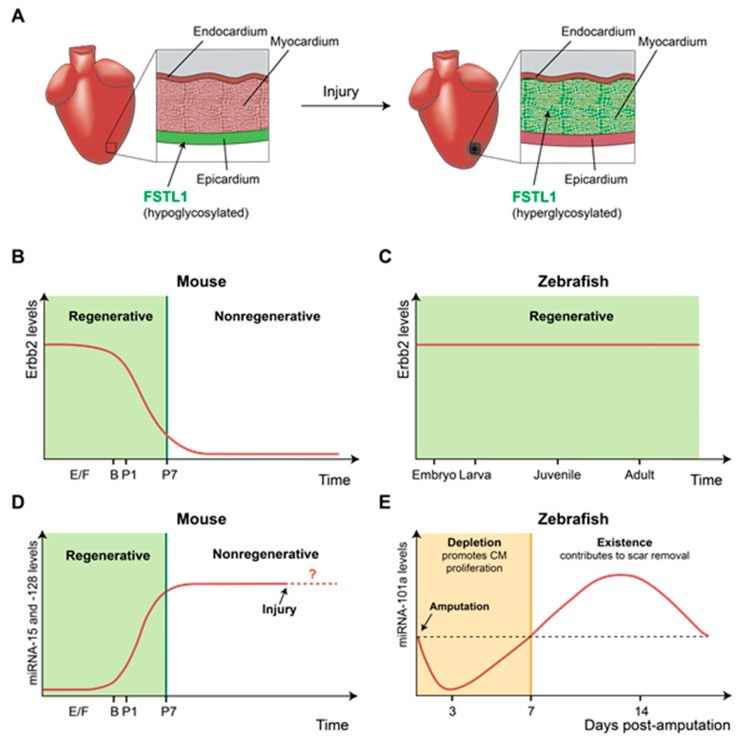
Follistatin-like 1 (Fstl1) was discovered as a potent CM mitogen secreted from an epicardial mesothelial cell line [15]. Delivering Fstl1 protein via an epicardial patch stimulates cardiac regeneration in injured mouse and swine hearts, thereby improving cardiac function. In uninjured mouse hearts, Fstl1 is typically produced by the epicardium, the outermost layer covering the cardiac chambers. However, cardiac injury in mice downregulates Fstl1 expression in the epicardium but strongly induces Fstl1 expression in the myocardium. Interestingly, unlike epicardium-derived Fstl1, CM-derived Fstl1 does not have the potential to promote CM proliferation, possibly due to hyperglycosylation [15]. As shown by this study, cellular sources of regeneration factors that are altered upon injury can affect post-translational modification, resulting in different mitogenic effects from regeneration factors (Figure 1A). An interesting question is how and what regulatory mechanisms control the expression of the same target gene in distinct cell types upon injury.
Figure 1.
Differential gene expression affects regenerative ability. (A) Fstl1 expression in the mouse heart is altered upon injury. In uninjured hearts, the epicardium produces Fstl1, which has the potential to promote cardiomyocyte (CM) proliferation. However, in injured hearts, Fstl1 is mainly produced by the myocardium rather than the epicardium. Fstl1 derived from the myocardium appears to be unable to promote CM proliferation due to glycosylation. (B) The level of Erbb2, a receptor of Nrg1, declines after birth, resulting in poor regenerative capacity in the mouse heart. (C) In zebrafish, Erbb2 expression is maintained throughout life, and a sustained Erbb2 level contributes to retaining cardiac regenerative capacity in the adult stage. (D) The expression level of the miRNA-15 family and miRNA-128 increases after birth in mice to inhibit heart regeneration. However, it is unclear whether expression of these miRNA is maintained upon injury. (E) The expression level of miRNA-101a is dynamic during heart regeneration in adult zebrafish. Depletion of miRNA-101a in the early regenerative stage promotes CM proliferation, while the presence of miRNA-101a in the late regenerative stage contributes to scar tissue removal. The ability to precisely modulate miRNA expression during heart regeneration enables zebrafish to retain remarkable regenerative capacity. In (B,D): E/F, embryonic/fetal period; B, birth; P, postnatal period.
Neuregulin 1 (Nrg1) is another potent CM mitogen in mammals, as demonstrated in neonatal mice and cultured human myocardium, and NRG1 administration has been proposed as a therapeutic strategy to promote cardiac regeneration [16,17,18,19]. The cardiomyogenic effect of NRG1 was tested with neonatal mouse hearts and pediatric patient-derived myocardium [16]. Interestingly, NRG1 therapy is most effective at an early age, such as 3 days in neonatal mice and 6 months in diseased human heart cells; however, the therapeutic effect significantly declines thereafter [16]. Another independent study demonstrated that Nrg1 effectively induced CM proliferation before postnatal day 7 (P7) in mice, but its effects diminished thereafter [18]. The limited time window of Nrg1-induced regeneration is determined by the expression level of Erbb2, a Nrg1 receptor, in CMs, as Erbb2 expression is sharply downregulated in neonatal mice after P7 (Figure 1B). In injured adult mouse hearts, transient induction of active Erbb2 can restore the ability to regenerate CMs, supporting the positive influence of Erbb2 levels on NRG1/ERBB2 signaling-mediated cardiac regeneration.
Nrg1 also has a mitogenic effect in adult zebrafish hearts [20]. Upon cardiac injury, Nrg1 expression in adult zebrafish is upregulated in multiple cells, including regulatory T cells [21] and perivascular cells of ventricular wall [20], to stimulate CM proliferation. In addition, overexpression of Nrg1 in uninjured hearts triggers CM proliferation, causing cardiomegaly through persistent addition to the cardiac muscle wall [20]. Unlike adult mammals, adult zebrafish maintain Erbb2 expression in the heart, which contributes to retaining cardiac regenerative capacity in injured hearts and stimulating CM proliferation in response to ectopic Nrg1 expression in uninjured hearts (Figure 1C). Together with mouse data, these results illustrate that losing the capability to express regeneration genes in aged mammalian hearts may act as a roadblock to heart regeneration. Further experiments to define the regulatory mechanisms suppressing Erbb2 expression in mouse hearts after P7 or those maintaining Erbb2 levels in adult zebrafish hearts will lead to the identification of new molecular targets for heart repair.
MicroRNAs (miRNAs), a class of small noncoding RNAs that play roles in RNA silencing and posttranscriptional regulation [22], are one of the main regulators of cardiac regeneration. In mice, the miRNA-15 family and miR-128 are expressed at low levels shortly after birth, but their expression is robustly increased at P7, a time point at which CMs lose their regenerative capacity [23,24]. The expression pattern of these miRNAs as well as their negative role in heart regeneration was revealed by functional studies, showing that overexpression of these miRNAs in neonatal mice inhibits CM proliferation and that their depletion at the adult stage enhances CM proliferation. Neonatal mouse hearts appear to downregulate the expression of several miR-15 family members in response to cardiac injury, revealing the presence of regulatory mechanisms that may hamper the negative effects of miRNA on heart regeneration (Figure 1D) [23].
Adult zebrafish hearts also express inhibitory miRNAs, such as miR-101a and miR-133, in uninjured hearts [25,26]. Cardiac miR-101a expression is profoundly reduced in response to cardiac injury and steadily declines until 3 days post-amputation (dpa), when CMs dedifferentiate and start to proliferate. Subsequently, miR-101a expression significantly increases above the level of uninjured hearts by 7–14 dpa before returning to uninjured levels at 30 dpa. The dynamic miR-101a level during heart regeneration appears to be essential, as shown by functional analysis; miR-101a depletion at the onset of cardiac injury enhances CM proliferation whereas the sustained reduction in miR-101a after injury prevents scar tissue removal [25]. These studies suggest that adult zebrafish hearts maintain the regulatory mechanisms, by which miR-101a expression is precisely modulated to facilitate heart regeneration (Figure 1E). Due to a lack of data describing the endogenous levels of miRNA-15 and miRNA-128 in adult mouse hearts upon injury, it is unclear whether the absence of the ability to downregulate inhibitory miRNA expression upon injury contributes to nonregenerative hearts. However, possessing a regulatory program controlling dynamic miRNA expression is undoubtedly one factor for maintaining cardiac regenerative capacity.
Immune cells also play important roles during heart regeneration. While inflammation triggers nonregenerative scarring in adult mice after cardiac injury, acute inflammation is required for cardiac regeneration and drives CM proliferation in neonatal mice [27]. These different inflammation effects on heart regeneration may be caused by the differing compositions of macrophage subsets in regenerating adult and neonatal hearts [28,29]. The majority of neonatal cardiac macrophages originate from the embryonic yolk sac and reside in the heart. Upon cardiac injury, these resident heart macrophages mediate angiogenesis and regeneration without scar formation. By contrast, in adult hearts following cardiac injury, the resident macrophages are not proliferative but are instead replaced by infiltrating proinflammatory monocytes and monocyte-derived macrophages, resulting in scar formation. Several studies have demonstrated that macrophages and monocytes produce regenerative factors, including interleukin-6 (IL-6) and myeloid-derived growth factor (MYDGF), which contribute to the regenerative capacity of hearts [30,31]. These studies suggest that distinct gene expression profiles in immune cells also affect heart repair.
Collectively, these studies have provided evidence that preserving regulatory functions to ensure regeneration factor expression constitutes the core mechanism of heart regeneration. Thus, the next critical question is where the regulatory DNA elements are in the genome and which molecular components modulate their activity.
3. Enhancers Are Key Regulatory Elements Controlling Cardiac Gene Expression and Function
Enhancers are key cis-regulatory DNA elements controlling spatiotemporal gene expression [32,33]. The number of enhancers is expected to surpass the number of protein-coding genes, as up to one million putative enhancers have been identified in the human genome [34,35], suggesting that multiple enhancers regulate the expression of a single gene in various circumstances. Enhancers often contain clusters of sequence-specific transcription factor binding motifs, and cooperative interactions of binding factors at the enhancer locus drive target gene expression in a specific context. Active enhancers are characterized by accessible DNA regions, which are devoid of conventional nucleosomes, and the presence of specific histone modifications, including histone H3 lysine 4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) [34]. The most common methods to predict active enhancer candidates include mapping accessible chromatin landscape using DNase I hypersensitive site sequencing (DNase-seq) [34] or assaying for transposase-accessible chromatin using sequencing (ATAC-seq) [36], and analyzing genome-wide profiles of active enhancer markers, such as H3K24me1 and H3K27ac using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) [37,38]. The in vivo functionality of enhancers can be validated by a transgenic assay, in which enhancer sequences coupled with a minimal promoter and a reporter gene are introduced into an animal to test reporter gene expression in a context of interest [39]. In recent years, several groups have also validated enhancer activity by enhancer deletions using genome editing followed by examining the disruption of target gene expression and the phenotype [40,41,42].
Multiple studies have been performed to map cardiac enhancer landscapes in developing and adult mammalian hearts. A multitiered computational analysis, using experimentally validated cardiac enhancers identified a cluster of sequence features shared by cardiac enhancers, with which more putative cardiac enhancers throughout the genome were selected. Further in vivo transgenic enhancer assays using zebrafish and mice verified that significant numbers of putative enhancers displayed activity in hearts [43]. Using ChIP-seq with p300/CREB-binding protein (CBP), transcriptional coactivators associated with enhancers, Blow et al. identified over 3000 cardiac enhancer candidates from embryonic mouse cardiac tissue, and a significant portion of the selected candidates exhibited in vivo cardiac activity [44]. An experimentally defined genome-wide map of human heart enhancers was also generated by ChIP-seq experiments of p300/CBP [45]. Distally localized p300/CBP peaks were markedly enriched near genes associated with cardiovascular development, function, and diseases, suggesting that these sequences may control the expression of cardiovascular genes. Transgenic mouse reporter assays with 65 enhancer candidates revealed that the majority of these cardiac enhancer candidates were authentic in vivo heart enhancers [45], indicating their potential roles in cardiac development and diseases.
Recent work has provided evidence of the importance of cardiac enhancers for cardiac function. Dickel et al. generated a comprehensive genome-wide catalog of cardiac enhancers in mammals by integrating nearly all the available epigenomic profiles of hearts, ranging from prenatal to adult stages of human and mouse heart samples [40]. This cardiac enhancer meta-analysis defined over 80,000 putative cardiac enhancers, and a subsequent retrospective approach matched these cardiac enhancers with the developmental stages in which they are potentially active. In addition to the transgenic enhancer assay, the investigators employed genome editing to delete cardiac enhancer sequences and then assessed the necessity of heart enhancers for cardiac development and function. Two different cardiac enhancers, mm77 and mm771, upstream of Myl2 and Myh7, respectively, were selected because of their association with cardiac diseases in humans. Interestingly, loss of either mm77 or mm771 resulted in decreased expression of Myl2 or Myh7, respectively, and led to abnormal cardiac organization and reduced cardiac function [40]. These results highlight the importance of cardiac enhancers for cardiac health.
Cardiac enhancer studies have shown that enhancers are key regulators that maintain cardiac function. Thus, questions have arisen regarding whether there are regeneration-associated enhancers responsible for cardiac injury and how their activity is regulated.
4. Epicardial Enhancers Drive Epicardial Factor Expression in Developing and Injured Hearts
The epicardium plays crucial roles during cardiac development by providing multipotent progenitor cells for cardiac fibroblasts, vascular smooth muscle cells, adipocytes, pericytes, endothelium, and endocardium, while some results are controversial [46,47,48,49,50]. The epicardium is also a vital tissue that promotes cardiac repair. Following cardiac injuries, epicardial cells contribute to the generation of various cardiac cells, including vascular smooth muscle cells, perivascular cells, and myofibroblasts [49,51,52,53]. In addition, cardiac injuries rapidly activate the epicardium in an organ-wide manner to re-express embryonic developmental genes such as retinaldehyde dehydrogenase 2 (Raldh2), T-box transcription factor 18 (Tbx18), and Wilms tumor 1 (Wt1), and the activated epicardium acts as a paracrine center to stimulate CM proliferation [54,55,56]. Although genetic factors activated in the epicardium are well described, the upstream regulation of epicardial gene activation during development or in response to cardiac injury has just begun to be understood.
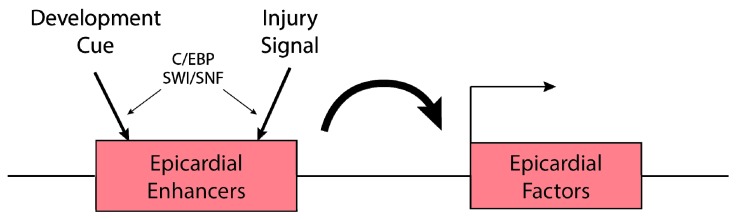
Multiple epicardial genes, such as the Tbx18, transcription factor 21 (Tcf21), Raldh2, and Wt1, are highly enriched in both embryonic and injured adult epicardium [57]. To identify the regulatory elements governing their epicardial expression, Huang et al. surveyed enhancer activity of 39 evolutionarily conserved regions (CRs) associated with the epicardial genes. Through screening with cultured and in vivo mouse hearts, the investigators demonstrated that Raldh2 CR2 and Wt1 CR14 were capable of directing epicardial gene expression [58]. Further analysis demonstrated that CCAAT/enhancer binding protein (C/EBP) transcription factors bound to Raldh2 CR2 and Wt1 CR14 epicardial enhancers, and together with HOX, MEIS, and Grainyhead transcription factors, established a transcriptional network for embryonic gene expression in the epicardium. Notably, Raldh2 CR2 and Wt1 CR14 enhancers are also activated in adult mouse hearts in response to myocardial infarction (MI) [58], suggesting that embryonic epicardial enhancers are reactivated to repurpose the developmental gene program in adult hearts after injury (Figure 2).
Figure 2.
Epicardial enhancers are activated by development cues and cardiac injury. Several evolutionarily conserved regions near epicardial factors are activated during development and upon cardiac injury to drive gene expression in epicardium. C/EBP and the SWI/SNF complex mediate epicardial enhancer activation.
Another independent study revealed the epigenetic regulation of epicardial activation. Brahma-related gene 1 (Brg1) and Brahma (Brm), catalytic subunits of the Switch/Sucrose nonfermentable (SWI/SNF) chromatin-remodeling complex, are expressed in the epicardium of developing and injured adult mouse hearts [59]. BRG1 binds to several evolutionarily conserved regions (ECRs) in the Wt1 locus that have been identified as epicardial enhancers of both developing and injured adult mouse hearts. BRG1 recruitment to Wt1 epicardial enhancers is mediated by C/EBP-beta, and Brg1 and C/EBP-beta are required for Wt1 activation in the epicardium of developing and injured hearts (Figure 2) [58,59]. This work demonstrates that injury cues activate the ATP-dependent SWI/SNF chromatin-remodeling complex, which regulates chromatin accessibility to induce transcription of injury-responsive genes. Collectively, epicardial enhancer studies in mice have provided evidence of the presence of epicardial enhancers in both developing and injured hearts, implying that developmental enhancers can be reactivated to drive injury-induced expression.
5. Cardiac Tissue Regeneration Enhancers in Zebrafish Ensure Regeneration-Specific Expression
An intriguing question is whether cardiac injury activates regeneration-specific enhancers to establish and/or maintain genetic programs supporting regeneration. To address this question, the zebrafish model offers exceptional experimental advantages. Unlike adult mammals, adult zebrafish possess a remarkable capacity to regenerate injured hearts, making zebrafish an ideal model for studying the regulatory mechanisms of adult heart regeneration [2]. Because transgenesis is relatively simple and easy in zebrafish, a growing number of studies have employed the zebrafish system to test whether nonmammalian and mammalian enhancer candidates exhibit activity in various tissues or organs, including hearts [43,60,61]. In addition, various transgenic lines and genetic drivers are available for directing transgene expression in distinct cardiac cell types, such as CMs and epicardium, allowing the identification of cell type-specific enhancers [49,62].
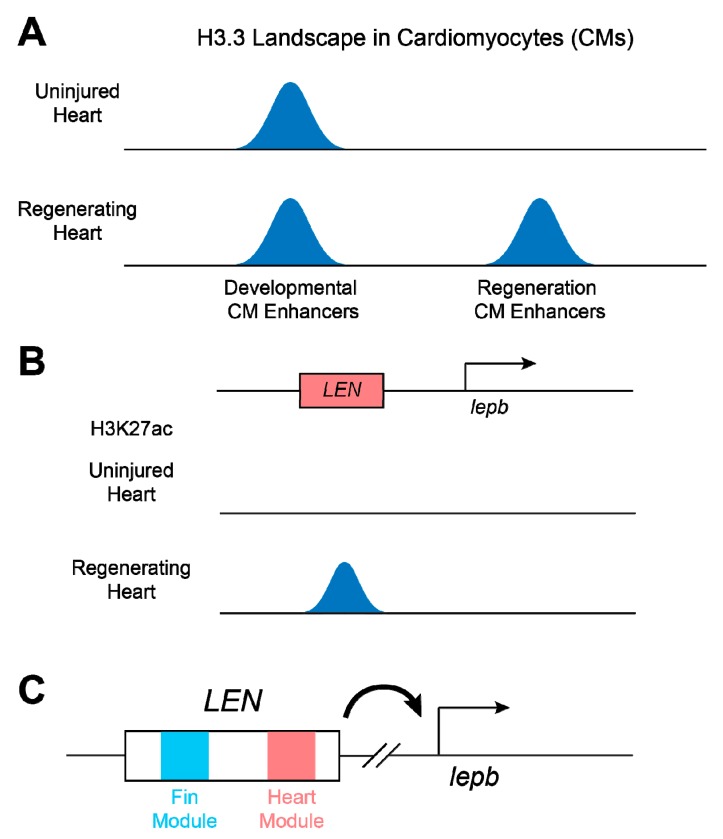
Active enhancers are thought to be accessible for sequence-specific transcription factors to bind enhancer regions. Gaining accessibility results in high turnover of the nucleosome by replacing canonical nucleosomes with unstable nucleosomes containing histone variants H2A.Z and H3.3 [63,64,65]. Because the H3.3 histone variant is selectively enriched at transcriptionally active genes, promoters, insulators, and enhancers, a new way has emerged to identify cis-regulatory elements, including active enhancers, by analyzing H3.3 profiles [62,66]. In a recent study, Goldman et al. utilized the specificity of H3.3 in the zebrafish system to find CM-specific regeneration enhancers. The investigators created transgenic zebrafish in which a biotinylated H3.3 (H3.3-bio) was specifically expressed in CMs to generate CM-specific H3.3 profiles of uninjured and regenerating hearts [62]. Computational analysis of CM-specific H3.3 profiles identified a massive number of de novo H3.3 peaks associated with regeneration. Transgenic assays with zebrafish carrying the cis-regulatory elements marked by H3.3 in regenerating CMs revealed that most of these regeneration-associated H3.3 peaks are cardiac regeneration enhancers directing regeneration-dependent gene expression in myocardium (Figure 3A). Further motif analysis with validated regeneration enhancers predicted potential regeneration modules that may construct core regulatory elements activated in CMs of regenerating hearts [62]. Because the H3.3-bio transgenic system does not require a cell isolation procedure, this approach provides a non-invasive method to obtain cell type-specific enhancer profiles and to reveal cell type-specific regulatory mechanisms in regenerating hearts.
Figure 3.
Cardiac regeneration enhancer elements. (A) Cardiomyocyte (CM)-specific enhancers in zebrafish. CM-specific histone H3.3 profiling of uninjured and regenerating zebrafish hearts captures regulatory elements preferential for heart development and regeneration. (B) ChIP-seq analysis with active enhancer markers, such as H3K27ac, identifies lepb-linked regeneration enhancer (LEN), which can direct regeneration-specific expression in hearts and fins of zebrafish. (C) LEN consists of tissue-specific regeneration modules, each of which mediates gene transcription in their target tissues.
Cardiac regeneration-specific enhancers were also discovered by employing the zebrafish system [67]. Unbiased transcriptomic analysis with regenerating fin and cardiac tissues defined the leptin b (lepb) gene, one of two zebrafish paralogs related to mammalian Leptin [68], as a robustly injury-induced gene in hearts and fins. Interestingly, a lepb:EGFP bacterial artificial chromosome (BAC) reporter line containing a 105-kb DNA sequence upstream of the lepb start codon barely drove EGFP expression in hearts or fins throughout life, but strongly induced EGFP expression in hearts or fins during regeneration. Surveying active enhancer candidates in lepb BAC regions identified two distal fragments that were marked by H3K27ac in regenerating, but not uninjured, hearts. Particularly, a 1.3-kb small fragment among these putative enhancers was capable of directing regeneration-dependent expression without developmental induction, indicating that this lepb-linked element is a regeneration-specific enhancer (Figure 3B). Notably, transgenic lines carrying a series of lepb regeneration enhancer (LEN) sequences revealed that LEN consisted of fin- and cardiac-specific regeneration modules [67], providing evidence that different tissue injuries use distinct gene regulatory networks to drive the expression of the same target gene (Figure 3C).
Enhancers rapidly change during evolution, contributing to physiological and morphological diversity [33,69]. Cardiac regeneration enhancers might also evolve rapidly, suggesting that changes in cardiac regeneration enhancers play important roles in diverse cardiac regeneration abilities across species. Zebrafish LEN is unlikely to be conserved in mammalian genomes at the sequence level, implying that mammals may have lost regeneration enhancers during evolution, and thus have limited regenerative capacity. However, zebrafish LEN can surprisingly drive injury-dependent expression in damaged mouse hearts [67], implying that mammalian gene regulatory networks have the potential to activate zebrafish elements. This result also posits that evolutionary loss of cardiac regeneration enhancers might affect regeneration-driving gene expression and consequently lead to deficient heart regeneration in mammals. This logic can be applied to develop therapeutic strategies to enhance regenerative potential; engineering active cardiac regeneration enhancers in regeneration-deficient animals can awaken dormant expression of regenerative factors and subsequent enhancement of heart regeneration. In zebrafish, coupling LEN with Nrg1 enables regeneration-restricted induction of Nrg1 in the wound area, and the temporal ectopic Nrg1 induction enhances CM proliferation [67]. These data provide a proof of concept that regeneration enhancer engineering presents a promising strategy to precisely deliver the CM mitogen to injured hearts. It will be interesting to examine whether delivery of regenerative factors using cardiac regeneration enhancer engineering will overcome the barriers of regeneration-deficient hearts and augment heart repair.
6. Conclusions
The differential composition of cardiac regeneration enhancers in the genome can lead to distinct gene expression upon cardiac injury, resulting in diverse levels of heart regeneration ability in vertebrates. Recent advances in genomics have enabled us to identify genome-wide landscapes of active enhancers, demonstrating that ambitious efforts will allow the mapping of all regeneration enhancers across species. Such profiles will provide tremendous benefits for potential applications of regenerative medicine. Comparative genomic analysis combined with cross-species transgenic assays will provide insight into how alterations of regeneration enhancers can lead to a decline (or an increase) in regenerative capacity. Biochemical and computational assays will uncover upstream factors and essential motifs, revealing novel gene regulatory networks underlying heart regeneration. More importantly, when applied to gene therapy, regeneration enhancers derived from animals that can naturally regenerate their hearts can be used to drive regeneration factor expression in injured mammalian hearts. This may allow us to unlock the latent healing power of hearts and ultimately to apply these insights to improving heart repair in humans.
Acknowledgments
The authors thank K. Yang (UW-Madison, Madison, WI, USA) for assistance with artwork. We apologize to authors whose important studies cannot be cited due to space constraints.
Author Contributions
I.J.B. and J.K. conceptualized and wrote the manuscript.
Funding
This work was supported by the Vice Chancellor for Research and Graduate Education start-up fund (AAC6429), the UW-Madison Stem Cell and Regenerative Medicine Center start-up fund (AAC8979), the School of Medicine and Public Health start-up fund (AAC8355), the Cell and Regenerative Biology start-up fund, and an American Heart Association grant (AHA16SDG30020001) to J.K.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Richardson W.J., Clarke S.A., Quinn T.A., Holmes J.W. Physiological Implications of Myocardial Scar Structure. Compr. Physiol. 2015;5:1877–1909. doi: 10.1002/cphy.c140067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 3.Grivas J., Haag M., Johnson A., Manalo T., Roell J., Das T.L., Brown E., Burns A.R., Lafontant P.J. Cardiac repair and regenerative potential in the goldfish (Carassius auratus) heart. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2014;163:14–23. doi: 10.1016/j.cbpc.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafontant P.J., Burns A.R., Grivas J.A., Lesch M.A., Lala T.D., Reuter S.P., Field L.J., Frounfelter T.D. The giant danio (D. aequipinnatus) as a model of cardiac remodeling and regeneration. Anat. Rec. 2012;295:234–248. doi: 10.1002/ar.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberpriller J., Oberpriller J.C. Mitosis in adult newt ventricle. J. Cell Biol. 1971;49:560–563. doi: 10.1083/jcb.49.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flink I.L. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: Confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat. Embryol. 2002;205:235–244. doi: 10.1007/s00429-002-0249-6. [DOI] [PubMed] [Google Scholar]
- 7.Rumyantsev P.P. Post-injury DNA synthesis, mitosis and ultrastructural reorganization of adult frog cardiac myocytes. An electron microscopic-autoradiographic study. Z. Zellforsch. Mikrosk. Anat. 1973;139:431–450. doi: 10.1007/BF00306596. [DOI] [PubMed] [Google Scholar]
- 8.Puente B.N., Kimura W., Muralidhar S.A., Moon J., Amatruda J.F., Phelps K.L., Grinsfelder D., Rothermel B.A., Chen R., Garcia J.A., et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson M., Barske L., Van Handel B., Rau C.D., Gan P., Sharma A., Parikh S., Denholtz M., Huang Y., Yamaguchi Y., et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 2017;49:1346–1353. doi: 10.1038/ng.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Rosa J.M., Sharpe M., Field D., Soonpaa M.H., Field L.J., Burns C.E., Burns C.G. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev. Cell. 2018;44:433–446.e437. doi: 10.1016/j.devcel.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassat E., Mutlak Y.E., Genzelinakh A., Shadrin I.Y., Baruch Umansky K., Yifa O., Kain D., Rajchman D., Leach J., Riabov Bassat D., et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morikawa Y., Heallen T., Leach J., Xiao Y., Martin J.F. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547:227–231. doi: 10.1038/nature22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quaife-Ryan G.A., Sim C.B., Ziemann M., Kaspi A., Rafehi H., Ramialison M., El-Osta A., Hudson J.E., Porrello E.R. Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation. 2017;136:1123–1139. doi: 10.1161/CIRCULATIONAHA.117.028252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Meara C.C., Wamstad J.A., Gladstone R.A., Fomovsky G.M., Butty V.L., Shrikumar A., Gannon J.B., Boyer L.A., Lee R.T. Transcriptional reversion of cardiac myocyte fate during mammalian cardiac regeneration. Circ. Res. 2015;116:804–815. doi: 10.1161/CIRCRESAHA.116.304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei K., Serpooshan V., Hurtado C., Diez-Cunado M., Zhao M., Maruyama S., Zhu W., Fajardo G., Noseda M., Nakamura K., et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polizzotti B.D., Ganapathy B., Walsh S., Choudhury S., Ammanamanchi N., Bennett D.G., dos Remedios C.G., Haubner B.J., Penninger J.M., Kuhn B. Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Sci. Transl. Med. 2015;7:281ra45. doi: 10.1126/scitranslmed.aaa5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganapathy B., Nandhagopal N., Polizzotti B.D., Bennett D., Asan A., Wu Y., Kuhn B. Neuregulin-1 Administration Protocols Sufficient for Stimulating Cardiac Regeneration in Young Mice Do Not Induce Somatic, Organ, or Neoplastic Growth. PLoS ONE. 2016;11:e0155456. doi: 10.1371/journal.pone.0155456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Uva G., Aharonov A., Lauriola M., Kain D., Yahalom-Ronen Y., Carvalho S., Weisinger K., Bassat E., Rajchman D., Yifa O., et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 19.Bersell K., Arab S., Haring B., Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 20.Gemberling M., Karra R., Dickson A.L., Poss K.D. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4:e05871. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui S.P., Sheng D.Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D., Kikuchi K. Zebrafish Regulatory T Cells Mediate Organ-Specific Regenerative Programs. Dev. Cell. 2017;43:659–672.e655. doi: 10.1016/j.devcel.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 23.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D., Mammen P.P., Rothermel B.A., Olson E.N., Sadek H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W., Feng Y., Liang J., Yu H., Wang C., Wang B., Wang M., Jiang L., Meng W., Cai W., et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat. Commun. 2018;9:700. doi: 10.1038/s41467-018-03019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beauchemin M., Smith A., Yin V.P. Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development. 2015;142:4026–4037. doi: 10.1242/dev.126649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 26.Yin V.P., Lepilina A., Smith A., Poss K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012;365:319–327. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han C., Nie Y., Lian H., Liu R., He F., Huang H., Hu S. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 2015;25:1137–1151. doi: 10.1038/cr.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aurora A.B., Porrello E.R., Tan W., Mahmoud A.I., Hill J.A., Bassel-Duby R., Sadek H.A., Olson E.N. Macrophages are required for neonatal heart regeneration. J. Clin. Investig. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavine K.J., Epelman S., Uchida K., Weber K.J., Nichols C.G., Schilling J.D., Ornitz D.M., Randolph G.J., Mann D.L. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl. Acad. Sci. USA. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C., Li Y., Wu Y., Wang L., Wang X., Du J. Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J. Biol. Chem. 2013;288:1489–1499. doi: 10.1074/jbc.M112.419788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korf-Klingebiel M., Reboll M.R., Klede S., Brod T., Pich A., Polten F., Napp L.C., Bauersachs J., Ganser A., Brinkmann E., et al. Myeloid-derived growth factor (C19orf10) mediates cardiac repair following myocardial infarction. Nat. Med. 2015;21:140–149. doi: 10.1038/nm.3778. [DOI] [PubMed] [Google Scholar]
- 32.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nord A.S., Blow M.J., Attanasio C., Akiyama J.A., Holt A., Hosseini R., Phouanenavong S., Plajzer-Frick I., Shoukry M., Afzal V., et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 35.Rivera C.M., Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczynski B., Riddell A., Furlong E.E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 39.Kvon E.Z. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106:185–192. doi: 10.1016/j.ygeno.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Dickel D.E., Barozzi I., Zhu Y., Fukuda-Yuzawa Y., Osterwalder M., Mannion B.J., May D., Spurrell C.H., Plajzer-Frick I., Pickle C.S., et al. Genome-wide compendium and functional assessment of in vivo heart enhancers. Nat. Commun. 2016;7:12923. doi: 10.1038/ncomms12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osterwalder M., Barozzi I., Tissieres V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S., et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hewitt K.J., Katsumura K.R., Matson D.R., Devadas P., Tanimura N., Hebert A.S., Coon J.J., Kim J.S., Dewey C.N., Keles S., et al. GATA Factor-Regulated Samd14 Enhancer Confers Red Blood Cell Regeneration and Survival in Severe Anemia. Dev. Cell. 2017;42:213–225.e214. doi: 10.1016/j.devcel.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narlikar L., Sakabe N.J., Blanski A.A., Arimura F.E., Westlund J.M., Nobrega M.A., Ovcharenko I. Genome-wide discovery of human heart enhancers. Genome Res. 2010;20:381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blow M.J., McCulley D.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.May D., Blow M.J., Kaplan T., McCulley D.J., Jensen B.C., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., et al. Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 2011;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acharya A., Baek S.T., Huang G., Eskiocak B., Goetsch S., Sung C.Y., Banfi S., Sauer M.F., Olsen G.S., Duffield J.S., et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grieskamp T., Rudat C., Ludtke T.H., Norden J., Kispert A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ. Res. 2011;108:813–823. doi: 10.1161/CIRCRESAHA.110.228809. [DOI] [PubMed] [Google Scholar]
- 48.Katz T.C., Singh M.K., Degenhardt K., Rivera-Feliciano J., Johnson R.L., Epstein J.A., Tabin C.J. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikuchi K., Gupta V., Wang J., Holdway J.E., Wills A.A., Fang Y., Poss K.D. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi Y., Cavallero S., Patterson M., Shen H., Xu J., Kumar S.R., Sucov H.M. Adipogenesis and epicardial adipose tissue: A novel fate of the epicardium induced by mesenchymal transformation and PPARgamma activation. Proc. Natl. Acad. Sci. USA. 2015;112:2070–2075. doi: 10.1073/pnas.1417232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez-Rosa J.M., Peralta M., Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012;370:173–186. doi: 10.1016/j.ydbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Van Wijk B., Gunst Q.D., Moorman A.F., van den Hoff M.J. Cardiac regeneration from activated epicardium. PLoS ONE. 2012;7:e44692. doi: 10.1371/journal.pone.0044692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dube K.N., Thomas T.M., Munshaw S., Rohling M., Riley P.R., Smart N. Recapitulation of developmental mechanisms to revascularize the ischemic heart. JCI Insight. 2017;2 doi: 10.1172/jci.insight.96800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou B., Honor L.B., He H., Ma Q., Oh J.H., Butterfield C., Lin R.Z., Melero-Martin J.M., Dolmatova E., Duffy H.S., et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Investig. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikuchi K., Holdway J.E., Major R.J., Blum N., Dahn R.D., Begemann G., Poss K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schnabel K., Wu C.C., Kurth T., Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao J., Poss K.D. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 2018 doi: 10.1038/s41569-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang G.N., Thatcher J.E., McAnally J., Kong Y., Qi X., Tan W., DiMaio J.M., Amatruda J.F., Gerard R.D., Hill J.A., et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira J.M., Howard S., Villa Del Campo C., Bollini S., Dube K.N., Masters M., Barnette D.N., Rohling M., Sun X., Hankins L.E., et al. BRG1-SWI/SNF-dependent regulation of the Wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat. Commun. 2017;8:16034. doi: 10.1038/ncomms16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapoor A., Sekar R.B., Hansen N.F., Fox-Talbot K., Morley M., Pihur V., Chatterjee S., Brandimarto J., Moravec C.S., Pulit S.L., et al. An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am. J. Hum. Genet. 2014;94:854–869. doi: 10.1016/j.ajhg.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smemo S., Campos L.C., Moskowitz I.P., Krieger J.E., Pereira A.C., Nobrega M.A. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum. Mol. Genet. 2012;21:3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldman J.A., Kuzu G., Lee N., Karasik J., Gemberling M., Foglia M.J., Karra R., Dickson A.L., Sun F., Tolstorukov M.Y., et al. Resolving Heart Regeneration by Replacement Histone Profiling. Dev. Cell. 2017;40:392–404.e395. doi: 10.1016/j.devcel.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P., Zhao J., Wang Y., Wang M., Long H., Liang D., Huang L., Wen Z., Li W., Li X., et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang J., Hu J., Karra R., Dickson A.L., Tornini V.A., Nachtrab G., Gemberling M., Goldman J.A., Black B.L., Poss K.D. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J.M. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 69.Long H.K., Prescott S.L., Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]