Abstract
Helicobacter pylori (H. pylori) infection is the strongest recognized risk factor for gastric adenocarcinoma. Since previous observations have shown that polymorphisms in innate immune system genes, as well as vitamin D (VitD) levels, could modify the risk of infection with Helicobacter pylori (H. pylori), we analyzed the relation between single nucleotide polymorphisms (SNPs) in TLRs (TLR1, TLR2, TLR4) CD14, RUNX3 and VitD levels with H. pylori infection. A case-control study on four hundred sixty Lebanese individuals was conducted. Eleven SNPs in total were genotyped and gene expression analysis using real-time PCR was performed in white blood cells of a subsample of eight individuals. A total of 49% of the participants were affected. Although no direct association was found between the SNPs and H. pylori infection, rs4986790G>A and rs4986791T>C in TLR4 were negatively associated with VitD levels (β = −0.371, p = 5 × 10−3 and β = −0.4, p = 2 × 10−3, respectively), which was negatively associated with H. pylori infection (OR = 0.01, p < 1 × 10−3). TLR4 expression was 3× lower in individuals with H. pylori compared with non-infected (p = 0.01). TLR4 polymorphisms, expression, and VitD could be implicated in H. pylori infection and further development of gastric adenocarcinoma.
Keywords: Helicobacter pylori, Toll-like Receptor 4, single nucleotide polymorphisms, gene expression, vitamin D
1. Introduction
Helicobacter pylori (H. pylori) infection has high incidence rates worldwide and is the major cause of several gastrointestinal symptoms, ranging from mild gastritis to gastric adenocarcinoma [1]. It has been estimated that up to half of the world’s population harbor this infection in their stomach [1]. Population-based studies report that developing countries have a higher prevalence rate of H. pylori than their developed counterparts [1]. Using data from a nationally representative, cross-sectional study on Lebanese adults, Naja et al. found that the prevalence of H. pylori infection was 52%, a rate comparable to other developing countries [2].
The host innate immune system plays a key role in the initiation and subsequent progression of H. pylori-associated pathogenesis [3]. The reason for the variable phenotypic expression of the infection is multifactorial and combines the effects of bacterial virulence factors, host genetic constitution, and environmental exposures [4]. Gastric epithelial cells are the primary target and the first point of contact for H. pylori infection, which actively contribute to the innate immune responses by signaling through pattern recognition receptors, such as toll-like receptors (TLRs) [4]. One of the factors that could modify H. pylori infection is genetic predisposition; single nucleotide polymorphisms (SNPs) in TLR genes, such as TLR1, TLR2, TLR4 and TLR10, were shown to alter the bacterial binding, thereby modulating the risk of infection and subsequent cancer [5]. A large genome-wide association study (GWAS) meta-analysis, conducted on 10,938 individuals, identified an association between rs10004195 in TLR1 and H. pylori seroprevalence [6]. In addition, many other small-scale studies revealed that SNPs, in genes such as TLR2, TLR4, CD14 and RUNX3, are associated with H. pylori infection and/or seropositivity [6,7,8,9,10]. Despite their importance, these findings should be investigated among other independent populations [6].
In addition to the innate immune system, serum vitamin D (VitD) levels have been implicated in H. pylori infection in the literature in recent years [11]. Low VitD concentration could be a predisposing factor for severe Th1-type aggression to the stomach epithelium in H. pylori gastritis patients [11]. The proposed mechanism for this link is via VitD-inducing immune modulator properties against H. pylori by decreasing inflammatory chemokine and cytokine levels [12]. In fact, it was shown that VitD production in macrophages is stimulated by TLRs as part of the innate immune response to intracellular bacteria [13,14].
Based on all the above, the association between polymorphisms in TLRs CD14, RUNX3 and VitD with H. pylori infection warrants further investigation. Therefore, we investigated the association between eleven SNPs (Supplementary Data, Table S1) and VitD with H. pylori infection in a Lebanese population composed of four hundred sixty individuals, divided between cases and controls. Furthermore, we assessed the gene expression of TLR4 in white blood cells (WBCs) of a subsample of eight individuals divided according to H. pylori infection status.
2. Methods
2.1. Study Design and Participants
This was a cross-sectional population-based study involving four hundred sixty Lebanese unrelated individuals free of chronic disease (cardiovascular disease or cancer). Recruitment occurred between 2015 and 2016 at the Hospital center of North-Zgharta a major tertiary care hospital in Northern Lebanon. Study participants aged 18 years or above were recruited at the gastroenterological unit after being referred for endoscopic examination of the upper gastrointestinal (GI) tract (gastroscopy) to obtain a biopsy. A retrospective chart review was conducted to determine study eligibility. Only patients with GI (dyspeptic) symptoms, mainly epigastric pain and gastritis, and who were undergoing gastroscopy were included in the study. H. pylori status (positive or negative) was determined after upper GI endoscopy, where gastric biopsy specimens from the antrum, body, and fundus region were collected on a plate containing formalin buffer. A pathologist examined these samples using hematoxylin and eosin (H&E) staining. The semi-quantitative method of scoring according to the Updated Sydney Classification System was applied. Controls were first examined through the same procedure and not found to be carrying H. pylori; in addition, they were matched in terms of age, gender, body mass index (BMI) categories, marital status and alcohol consumption.
2.2. Data Collection
Participants involved in the present study were recruited in accordance with the latest version of the Declaration of Helsinki for Ethical Principles for Medical Research Involving Human Subjects. Ethical approval was obtained from the local institutional review board Clinical Research Ethics Committee at the Lebanese University. Protocols for genetic studies were approved by the local ethics committees for the protection of subjects involved in biomedical research (2182/28). We obtained written informed consent from all the participants.
Complete medical examinations were carried out for all individuals, with data collected including demographic, anthropometric, clinical and measurements. A BMI was considered normal when a value of < 25 kg/m2 was observed. Biochemical measurements including glycemia, lipid profile, and VitD were measured with commercial kits (Roche Diagnostics, Basel, Switzerland). Serum levels of total 25-hydroxyvitamin D [25(OH) D], which is the sum of D2 and D3, were measured using Elecsys™ Vitamin D total assay (Roche Diagnostics, Basel, Switzerland). Vitamin D deficiency and insufficiency were described for levels of 25(OH)D falling below 30 ng/mL and below 20 ng/mL, respectively [15].
2.3. Genomic DNA Extraction and Genotyping Assays
DNA extraction was performed on whole blood samples obtained from all participants, using a DNA extraction kit from Qiagen (QIAamp DNA Mini Kit, Hilden, Germany), according to the manufacturer’s protocol. A Qubit 3.0 fluorometer (Thermo Fisher Scientific, Selangor, Malaysia) was used to quantify the DNA extracts using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Malaysia). We selected eleven SNPs for genotyping (Supplementary Data, Table S1). Genotyping was performed by the LGC group (Berlin, Germany) using KASP as described previously [16].
2.4. TLR4 Expression in White Blood Cells
Total RNA was isolated from WBCs by an automated isolation procedure using an Aurum™ Total RNA Mini Kit (Life Science Research, Bio-Rad, Singapore, Singapore). RNA quality and stability were carefully tested on 1% agarose gel and reverse-transcribed using an iScript™ cDNA Synthesis Kit (Life Science Research, Bio-Rad). We used designed specific primers for TLR4 quantification using Primer3 software, version 0.4.0 (data available upon request). We carried out cDNA amplification in a reaction volume of 20 uL consisting of 70 ng of cDNA, Solis BioDyne 1× HOT FIREPol Blend Master Mix (Tartu, Estonia) and both primers. Quantitative real-time PCR (qRT-PCR) was performed using the CFX Connect™ Real-Time PCR Detection System (Bio-Rad) with an iTaq Universal SYBR Green Supermix Kit (Bio-Rad) for TLR4 transcripts. All experiments were carried out in duplicates in a total reaction volume of 20 µL containing 0.5 mM of each specific primer. Negative and internal controls were included. All mRNA levels were normalized to the mRNA levels of RNA polymerase II subunit A (POLR2A). The specificity of all PCR products was further verified by electrophoresis on 3% polyacrylamide gel.
2.5. Statistical Analyses
Statistical analyses were carried out using SPSS software, version 20.0 (SPSS, Inc., Chicago, IL, USA). Demographic characteristics (age, gender) anthropometric characteristics (height and weight), clinical (systolic and diastolic blood pressure) and biochemical measurements (lipid profile, glucose, VitD) were compared according to H. pylori infection using chi-square tests for categorical variables and t-tests for continuous ones. Normality was tested using the Shapiro–Wilk test and, when needed, continuous variables were log-transformed to improve their normality. SNPs (rs4833095 in TLR2) deviating from the Hardy–Weinberg equilibrium or showing a null MAF (rs5743708 in TLR2) were excluded.
A multivariate logistic regression was performed to first ascertain the association between all SNPs and H. pylori infection in a model adjusted for age, gender, BMI, education level, marital status, alcohol consumption, and VitD status. Next, multivariate linear regression was performed for all SNPs and VitD levels. All regression analyses (using three models of inheritance: Additive, recessive and dominant) were adjusted for age and gender. The significance level was set at p ≤ 0.005 due to multiple testing.
A Mann–Whitney U-test was performed to compare TLR4 expression levels in the WBCs of H. pylori patients (n = 5), compared to controls (n = 3). The significance level was set at p < 0.05.
3. Results
The demographic, clinical, and biochemical characteristics of participants are shown in Table 1. Approximately half of our participants (49%) were found to have H. pylori infection. Whereas the majority of variables matched between cases and controls, some significant differences were noticed (Table 1). The most apparent significant difference between the two groups was serum VitD levels (p = 1 × 10−4, Table 2). Individuals infected with H. pylori were VitD-deficient, whereas non-affected individuals had normal VitD levels (p < 1 × 10−3, Table 1). In addition to VitD, educational level varied significantly between cases and controls, revealing that the highest infection rate is among those with a university degree (Table 1).
Table 1.
Biological characteristics of the study population according to Helicobacter pylori infection.
| Helicobacter pylori | |||
|---|---|---|---|
| Positive (n = 225) | Negative (n = 235) | p | |
| Mean ± SD | Mean ± SD | ||
| Age (years) | 39.28 ± 13.9 | 41.86 ± 14.26 | 0.05 |
| Gender | |||
| Males, n (%) | 88 (39.1%) | 80 (34%) | 0.25 |
| Females, n (%) | 137 (60.9%) | 155 (66%) | |
| BMI Category | |||
| Normal | 109 | 128 | 0.12 |
| Overweight and Obese | 116 | 107 | |
| Education Level | |||
| Primary | 32 | 56 | 6 × 10−3 |
| School | 52 | 57 | |
| University | 141 | 122 | |
| Marital Status | |||
| Not Married | 70 | 69 | 0.38 |
| Married | 155 | 166 | |
| Alcohol consumption | |||
| No | 145 | 153 | 0.48 |
| Yes | 80 | 82 | |
| SBP (mmHg) | 131.03 ± 1.51 | 131.37 ± 1.64 | 0.05 |
| DBP (mmHg) | 6.78 ± 0.88 | 6.78 ± 0.93 | 0.47 |
| Glycemia (mg/dL) | 121 ± 46 | 95 ± 21 | 0.18 |
| Cholesterol (mmol/L) | 1.75 ± 0.41 | 1.87 ± 0.4 | 0.009 |
| TG (mmol/L) | 1.22 ± 0.92 | 1.68 ± 1.45 | 0.001 |
| HDL (mmol/L) | 0.46 ± 0.12 | 0.44 ± 0.16 | 0.13 |
| LDL (mmol/L) | 1.14 ± 0.34 | 1.2 ± 0.32 | 0.24 |
| VitD (ng/mL) | 18.04 ± 7.16 | 30.74 ± 15.66 | <1 × 10−3 |
| Insufficiency | 133 | 12 | <1 × 10−3 |
| Deficiency | 56 | 109 | |
| Normal | 15 | 107 | |
Values are arithmetic mean ± standard deviation (SD) or proportion (%). BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; TG, triglycerides; HDL, high-density lipoproteins; LDL, low-density lipoproteins; VitD, vitamin D.
Table 2.
Multiple logistic regression analysis of different risk factors with Helicobacter pylori infection.
| Helicobacter pylori | |||
|---|---|---|---|
| OR | 95% C.I. | p | |
| Age | |||
| <40 years | 1 | ||
| ≥40 years | 0.73 | (0.33–1.62) | 0.44 |
| Gender | |||
| Male | 1 | ||
| Female | 0.7 | (0.37–1.32) | 0.02 |
| BMI category | |||
| Normal | 1 | ||
| Overweight and Obese | 1.49 | (0.82–2.72) | 0.19 |
| Education Level | |||
| Primary | 1 | ||
| School | 2.46 | (0.96–6.34) | 0.06 |
| University | 4.16 | (1.52–11.4) | 0.01 |
| Marital Status | |||
| Not Married | 1 | ||
| Married | 1.03 | (0.5–2.1) | 0.93 |
| Alcohol Consumption | |||
| No | 1 | ||
| Yes | 1.42 | (0.76–2.67) | 0.27 |
| Vitamin D status | |||
| Insufficiency | 1 | ||
| Deficiency | 0.03 | (0.01–0.06) | <1 × 10−3 |
| Normal | 0.01 | (0–0.02) | <1 × 10−3 |
| Gene SNPs | |||
| rs10004195 in TLR1 | 1.23 | (0.8–1.8) | 0.29 |
| rs10759932 in TLR4 | 1.37 | (0.75–2.48) | 0.30 |
| rs10983755 in TLR4 | 0.51 | (0.05–5.44) | 0.58 |
| rs11536889 in TLR4 | 1.51 | (0.82–2.79) | 0.19 |
| rs1898830 in TLR4 | 0.84 | (0.55–1.29) | 0.44 |
| rs4986790 in TLR4 | 7.09 | (1.12–45) | 0.04 |
| rs4986791 in TLR4 | 0.14 | (0.02–0.86) | 0.03 |
| rs2569190 in CD14 | 0.74 | (0.49–1.12) | 0.15 |
| rs760805 in RUNX3 | 0.78 | (0.53–1.15) | 0.21 |
Reference categories of gene single nucleotide polymorphisms were not shown since no significant association was found. C.I., confidence interval.
Gene polymorphisms were not associated with H. pylori when adjusted for different confounding factors (p > 5 × 10−3, Table 2). Among the dependent variables, we found that, while having a university degree increases the risk of H. pylori (OR = 4.16, p = 0.01), normal VitD levels decrease it remarkably (OR = 0.01, p < 1 × 10−3, Table 2).
On the other hand, rs4986790G>A and rs4986791T>C in TLR4 were negatively associated with VitD levels (Table 3). Thus, we concluded that TLR4 SNPs are associated with decreased VitD levels, which could lead to an increased risk of H. pylori infection.
Table 3.
Association of rs4986790G>A and rs4986791T>C in TLR4 with serum vitamin D levels.
| Vitamin D | |||||||
|---|---|---|---|---|---|---|---|
| SNP ID | Gene | Chr | position | n | β [95% C.I.] | S.E | p |
| rs4986790G>A | TLR4 | 9 | 117713024 | 456 | −0.371 [−0.112, −0.63] | 0.13 | 5 × 10−3 |
| rs4986791T>C | 117713324 | 457 | −0.4 [−0.14, −0.663] | 0.13 | 1 × 10−3 | ||
Chr: Chromosome, n: Sample size, S.E., standard error.
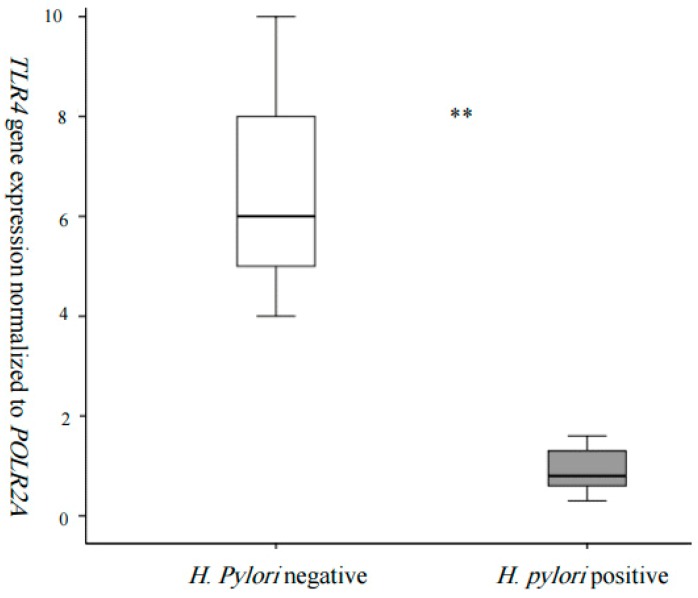
To better assess the relation between TLR4 and H. pylori, we decided to study its expression in the WBCs of a subsample of individuals who were stratified according to their infection status. Gene expression analysis showed that TLR4 was 3 times lower in the WBCs of H. pylori-affected individuals compared to non-affected (Figure 1).
Figure 1.
TLR4 expression in whole blood cells according to H. pylori infection (** p < 0.05).
Quantitative real-time PCR, normalized to the expression of POLR2A, revealed 3× higher expression in the WBCs of with H. pylori (n = 3) when compared to H. pylori (n = 5) (** p < 0.01).
4. Discussion
Herein, we found that rs4986790G>A and rs4986791T>C in TLR4 were associated with VitD, which in turn affected H. pylori infection, implying that those SNPs could have an indirect effect on H. pylori. Specifically, individuals with H. pylori were found to be VitD-deficient. Finally, TLR4 expression was shown to be 3× lower in the WBCs of H. pylori individuals, compared to unaffected ones.
The prevalence of H. pylori infection in this study was found to be 49%, which is comparable to the general Lebanese adult population, according to Naja et al. [2]. This rate is lower than that found in other countries of the Middle East and North Africa region, including Egypt, Libya, Saudi Arabia, Iran, Oman, United Arab Emirates, and Turkey, where the prevalence of H. pylori is higher [14].
We chose to study rs10004195 in TLR1 since it is known to be associated with H. pylori seroprevalence [6]. Despite its importance, we were unable to replicate rs10004195, which can be explained in a number of ways: Compared to Mayerle et al., our sample was highly limited, which makes replication of SNPs with small effects difficult; in contrast to Europeans, Lebanese individuals are multiethnic, originating from less homogeneous descendants, meaning that association studies can be challenging.
The significant negative association between VitD and H. pylori in our study is in concordance with recent findings in previous STUDES [17,18,19]. Yildirim et al. found in their study that VitD deficiency could be a risk factor related to the eradication failure of H. pylori, suggesting the need for the supplementation of VitD before the eradication of H. pylori [19]. In a cross-sectional study, Nasri and Baradaran reported that VitD potentiates the immune response of dialysis patients to H. pylori and could positively affect their chronic inflammatory status [18]. This study suggested that the analogs of VitD, in addition to its antibacterial action against H. pylori, might offer patients on maintenance dialysis a new means by which to control their inflammatory status [18]. Interestingly, previous studies did indicate the antibacterial effects of VitD. According to Hosoda et al., the antibacterial action of VitD against H. pylori is linked to the VitD3 decomposition product, which induces a collapse of cell membrane structures of H. pylori and ultimately lyses the bacterial cells [17]. Recently, Wanibuchi et al. revealed that indene compounds synthetically derived from VitD perform selective antibacterial action against H. pylori [20]. Consequently, VitD3 is suggested as a fundamental structure for the development of new antibacterial substances, providing selective bactericidal action against H. pylori [17].
Since VitD is known for its antineoplastic and antioxidant properties, several studies have explored its relation to cancer progression [21]. In fact, it is involved in cell cycle regulation, cellular proliferation, apoptosis, and angiogenesis [22]. Growing evidence is showing that deficiency in VitD is associated with an increased risk of gastric adenocarcinoma and a poor prognosis [23]. Recently, Singh et al. reported that low serum VitD levels may play a role in the development of incomplete intestinal metaplasia, which is the most frequently observed precancerous change in the gastric mucosa [21].
Existing data show that the total number of WBCs and the numbers of lymphocytes and basophils were significantly increased in H. pylori-positive individuals, compared with unaffected ones [24]. In fact, Kondo et al. demonstrated that the eradication of H. pylori decreases blood neutrophil and monocyte counts [25]. These observations highlight the importance of choosing WBCs as a model to study the relation between TLR4 and H. pylori. Gene expression analysis showed that TLR4 was 3 times lower in the WBCs of H. pylori-affected individuals, compared to controls (Figure 1). Despite the fact that TLR4 expression varied significantly in the WBCs of H. pylori individuals, compared to controls, the difference in expression was modest. This implies that TLR4 is not the sole receptor implicated in H. pylori recognition and the subsequent host response. Evidence for TLR4 implication in H. pylori infection is still contradictory, while Ishara et al. [26] demonstrated that H. pylori infection upregulates the host’s innate immunity through the activation of the TLR4–MD-2 system in the gastric epithelial cells [26]. Smith et al. [27] demonstrated that infection from H. pylori induces responses via TLR2 and TLR5, but surprisingly not TLR4 [27]. This is in contrast with previous studies on mice and guinea pigs, which have provided evidence that TLR4 may be an important contributing factor to the inflammatory response induced by H. pylori [28]. In the current study, we found a modest change in TLR4 expression which may not be detected at the protein levels, because it can be masked by a post-transcriptional control mechanism (such as miRNAs). All the above underlines the necessity of working on adequate cell models to study gene expression when using high-throughput sequencing techniques.
Our findings point out that TLR4 expression in monocytes was altered by the effect of VitD [29]. Supporting our results, in vitro, Do et al. demonstrated that VitD3 was able to modify the protein and mRNA expression of TLR4 [30]. These observations support the hypothesis that VitD may act as an immune modulator. In this regard, it is not surprising to find that TLR4 expression decreased in the WBCs of H. pylori individuals, since they were shown to have low VitD levels, an event that predisposed them to H. pylori infection.
When compared to previous studies, our goal was broader, since we did not only focus on VitD, but we expanded the associations to study the genetics of TLRs and other risk factors in a multiethnic population (Lebanese population). Adding gene expression data in WBCs that are reported to be altered in H. pylori-positive individuals is of interest. Our study has several limitations. The study sample was limited to adult patients at a tertiary healthcare center in one region of Lebanon; therefore, our results cannot be generalized. Our gene expression analysis was done on a quite limited sample size and, thus, it is preliminary, normally, thirty samples are needed to have reasonable and conclusive results.
In conclusion, no direct association was found between TLRs and H. pylori infection; by contrast, SNPs in TLR4 were negatively associated with serum VitD levels, which was negatively associated with H. pylori infection. Finally, TLR4 expression differed according to H. pylori infection. All of the above point to a possible implication in H. pylori infection and further development of gastric adenocarcinoma.
Acknowledgments
The authors would like to thank the participants and their families for their time and effort in the present study.
Supplementary Materials
The following are available online at http://www.mdpi.com/2075-4426/9/1/2/s1, Table S1: List of studies single nucleotide polymorphisms, Table S2: Minor allele frequency of single nucleotide polymorphisms, Table S3: Analysis of deviation from Hardy Weinberg equilibrium.
Author Contributions
S.E.S., C.C. and S.A. conceived of and designed the experiments. L.J. performed the experiments. M.S. and S.E.S. analyzed the data. F.T. contributed reagents/materials/analysis tools. S.E.S., S.A. and C.C. wrote the paper.
Funding
The present work was funded with support from the Lebanese University (SA, SES) Beirut, Lebanon.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Burucoa C., Axon A. Epidemiology of Helicobacter pylori infection. Helicobacter. 2017;22(Suppl. 1) doi: 10.1111/hel.12403. [DOI] [PubMed] [Google Scholar]
- 2.Naja F., Nasreddine L., Hwalla N., Moghames P., Shoaib H., Fatfat M., Sibai A., Gali-Muhtasib H. Association of H. pylori infection with insulin resistance and metabolic syndrome among Lebanese adults. Helicobacter. 2012;17:444–451. doi: 10.1111/j.1523-5378.2012.00970.x. [DOI] [PubMed] [Google Scholar]
- 3.Kusters J.G., van Vliet A.H., Kuipers E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pachathundikandi S.K., Lind J., Tegtmeyer N., El-Omar E.M., Backert S. Interplay of the Gastric Pathogen Helicobacter pylori with Toll-Like Receptors. Biomed. Res. Int. 2015;2015:192420. doi: 10.1155/2015/192420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tongtawee T., Bartpho T., Kaewpitoon S., Kaewpitoon N., Dechsukhum C., Leeanansaksiri W., Loyd R.A., Talabnin K., Matrakool L., Panpimanmas S. Genetic polymorphisms in TLR1, TLR2, TLR4, and TLR10 of Helicobacter pylori-associated gastritis: A prospective cross-sectional study in Thailand. Eur. J. Cancer Prev. 2018;27:118–123. doi: 10.1097/CEJ.0000000000000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayerle J., den Hoed C.M., Schurmann C., Stolk L., Homuth G., Peters M.J., Capelle L.G., Zimmermann K., Rivadeneira F., Gruska S., et al. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 2013;309:1912–1920. doi: 10.1001/jama.2013.4350. [DOI] [PubMed] [Google Scholar]
- 7.Hishida A., Matsuo K., Goto Y., Mitsuda Y., Naito M., Wakai K., Tajima K., Hamajima N. Significant association of RUNX3 T/A polymorphism at intron 3 (rs760805) with the risk of gastric atrophy in Helicobacter pylori seropositive Japanese. J. Gastroenterol. 2009;44:1165–1171. doi: 10.1007/s00535-009-0118-7. [DOI] [PubMed] [Google Scholar]
- 8.Karhukorpi J., Yan Y., Niemela S., Valtonen J., Koistinen P., Joensuu T., Saikku P., Karttunen R. Effect of CD14 promoter polymorphism and H. pylori infection and its clinical outcomes on circulating CD14. Clin. Exp. Immunol. 2002;128:326–332. doi: 10.1046/j.1365-2249.2002.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castano-Rodriguez N., Kaakoush N.O., Goh K.L., Fock K.M., Mitchell H.M. The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: A case-control study and meta-analysis. PLoS ONE. 2013;8:e60327. doi: 10.1371/journal.pone.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang T.J., Chae G.T. Detection of Toll-like receptor 2 (TLR2) mutation in the lepromatous leprosy patients. FEMS Immunol. Med. Microbiol. 2001;31:53–58. doi: 10.1111/j.1574-695X.2001.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 11.Du C., Yang S., Zhao X., Dong H. Pathogenic roles of alterations in vitamin D and vitamin D receptor in gastric tumorigenesis. Oncotarget. 2017;8:29474–29486. doi: 10.18632/oncotarget.15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L., Chen W., Zhu H., Chen Y., Wan X., Yang N., Xu S., Yu C., Chen L. Helicobacter pylori induces increased expression of the vitamin D receptor in immune responses. Helicobacter. 2014;19:37–47. doi: 10.1111/hel.12102. [DOI] [PubMed] [Google Scholar]
- 13.Liu P.T., Stenger S., Li H., Wenzel L., Tan B.H., Krutzik S.R., Ochoa M.T., Schauber J., Wu K., Meinken C., et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 14.Assaad S., Chaaban R., Tannous F., Costanian C. Dietary habits and Helicobacter pylori infection: A cross sectional study at a Lebanese hospital. BMC Gastroenterol. 2018;18:48. doi: 10.1186/s12876-018-0775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alghalyini B., El Shamieh S., Salami A., Visvikis Siest S., Fakhoury H.M., Fakhoury R. Effect of SLCO1B1 gene polymorphisms and vitamin D on statin-induced myopathy. Drug Metab. Pers. Ther. 2018;33:41–47. doi: 10.1515/dmpt-2017-0030. [DOI] [PubMed] [Google Scholar]
- 16.El Shamieh S., Costanian C., Kassir R., Visvkis-Siest S., Bissar-Tadmouri N. Apoe genotypes in lebanon: Distribution and association with hypercholesterolemia and alzheimer’s disease. Per. Med. 2019;16:15–23. doi: 10.2217/pme-2018-0067. [DOI] [PubMed] [Google Scholar]
- 17.Hosoda K., Shimomura H., Wanibuchi K., Masui H., Amgalanbaatar A., Hayashi S., Takahashi T., Hirai Y. Identification and characterization of a vitamin D3 decomposition product bactericidal against Helicobacter pylori. Sci. Rep. 2015;5:8860. doi: 10.1038/srep08860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasri H., Baradaran A. The influence of serum 25-hydroxy vitamin D levels on Helicobacter pylori Infections in patients with end-stage renal failure on regular hemodialysis. Saudi J. Kidney Dis. Transplant. 2007;18:215–219. [PubMed] [Google Scholar]
- 19.Yildirim O., Yildirim T., Seckin Y., Osanmaz P., Bilgic Y., Mete R. The influence of vitamin D deficiency on eradication rates of Helicobacter pylori. Adv. Clin. Exp. Med. 2017;26:1377–1381. doi: 10.17219/acem/65430. [DOI] [PubMed] [Google Scholar]
- 20.Wanibuchi K., Hosoda K., Ihara M., Tajiri K., Sakai Y., Masui H., Takahashi T., Hirai Y., Shimomura H. Indene Compounds Synthetically Derived from Vitamin D Have Selective Antibacterial Action on Helicobacter pylori. Lipids. 2018;53:393–401. doi: 10.1002/lipd.12043. [DOI] [PubMed] [Google Scholar]
- 21.Singh K., Gandhi S., Batool R. A Case-Control Study of the Association between Vitamin D Levels and Gastric Incomplete Intestinal Metaplasia. Nutrients. 2018;10:629. doi: 10.3390/nu10050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleet J.C., DeSmet M., Johnson R., Li Y. Vitamin D and cancer: A review of molecular mechanisms. Biochem. J. 2012;441:61–76. doi: 10.1042/BJ20110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren C., Qiu M.Z., Wang D.S., Luo H.Y., Zhang D.S., Wang Z.Q., Wang F.H., Li Y.H., Zhou Z.W., Xu R.H. Prognostic effects of 25-hydroxyvitamin D levels in gastric cancer. J. Transl. Med. 2012;10:16. doi: 10.1186/1479-5876-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karttunen T.J., Niemela S., Kerola T. Blood leukocyte differential in Helicobacter pylori infection. Dig. Dis. Sci. 1996;41:1332–1336. doi: 10.1007/BF02088556. [DOI] [PubMed] [Google Scholar]
- 25.Kondo Y., Joh T., Sasaki M., Oshima T., Itoh K., Tanida S., Kataoka H., Ohara H., Nomura T., Itoh M. Helicobacter pylori eradication decreases blood neutrophil and monocyte counts. Aliment. Pharmacol. Ther. 2004;20(Suppl. 1):74–79. doi: 10.1111/j.1365-2036.2004.01988.x. [DOI] [PubMed] [Google Scholar]
- 26.Ishihara S., Rumi M.A., Kadowaki Y., Ortega-Cava C.F., Yuki T., Yoshino N., Miyaoka Y., Kazumori H., Ishimura N., Amano Y., et al. Essential role of MD-2 in TLR4-dependent signaling during Helicobacter pylori-associated gastritis. J. Immunol. 2004;173:1406–1416. doi: 10.4049/jimmunol.173.2.1406. [DOI] [PubMed] [Google Scholar]
- 27.Smith M.F., Jr., Mitchell A., Li G., Ding S., Fitzmaurice A.M., Ryan K., Crowe S., Goldberg J.B. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-ĸB activation and chemokine expression by epithelial cells. J. Biol. Chem. 2003;278:32552–32560. doi: 10.1074/jbc.M305536200. [DOI] [PubMed] [Google Scholar]
- 28.Kawahara T., Kuwano Y., Teshima-Kondo S., Kawai T., Nikawa T., Kishi K., Rokutan K. Toll-like receptor 4 regulates gastric pit cell responses to Helicobacter pylori infection. J. Med. Investig. 2001;48:190–197. [PubMed] [Google Scholar]
- 29.Sadeghi K., Wessner B., Laggner U., Ploder M., Tamandl D., Friedl J., Zügel U., Steinmeyer A., Pollak A., Roth E., et al. Vitamin D3 down-regulates monocyte TLR expression and triggers hyporesponsiveness to pathogen-associated molecular patterns. Eur. J. Immunol. 2006;36:361–370. doi: 10.1002/eji.200425995. [DOI] [PubMed] [Google Scholar]
- 30.Do J.E., Kwon S.Y., Park S., Lee E.S. Effects of vitamin D on expression of Toll-like receptors of monocytes from patients with Behcet’s disease. Rheumatology (Oxford) 2008;47:840–848. doi: 10.1093/rheumatology/ken109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.