Abstract
Oncolytic vaccinia virus (OVV) has demonstrated appropriate safety profiles for clinical development. Although designed to kill cancer cells efficiently, OVV sensitivity varies in individual cancers, and predictive biomarkers of therapeutic responses have not been identified. Here we found that OVV was much more efficient in KFTX paclitaxel-resistant ovarian cancer cells compared to that in KFlow paclitaxel-sensitive cells. Microarray analysis identified long non-coding RNA urothelial carcinoma-associated 1 (UCA1) upregulation, which contributed to both enhanced paclitaxel resistance and OVV spread. In addition, UCA1 expression correlated with efficient OVV spread in other ovarian cell lines and primary cancer cell cultures. When host pathways underlying OVV spread were analyzed, differences were detected in the activation of the Rho GTPase Cdc42, suggesting that filopodia formation enhances OVV cell-to-cell spread and tumor migration. Moreover, we established a clinically relevant mouse model of peritoneal metastasis using KFTX or KFlow cells. Paclitaxel exerted anti-tumor effects on KFlow, but not KFTX, tumors. In mice bearing KFTX cells after paclitaxel failure, OVV treatment induced the regression of residual tumors and improved survival. Our findings demonstrated that UCA1 promotes OVV cell-to-cell spread in ovarian cancer, resulting in enhanced therapeutic outcome.
Keywords: oncolytic vaccinia virus, biomarker, long non-coding RNA, UCA1, chemotherapy
Graphical Abstract
Introduction
Oncolytic viruses have emerged as novel candidates for cancer therapy owing to their inherent potential to selectively replicate and destroy tumor cells.1 With the recent approval of talimogene laherparepvec for advanced melanoma, oncolytic virotherapy has earned its place in clinical settings.2 Furthermore, studies have demonstrated that oncolytic vaccinia virus (OVV) is a promising alternative to existing therapies for the treatment of advanced cancer.3, 4, 5
In a phase 1 clinical trial of intravenous oncolytic pox virus, a Western Reserve OVV strain, two of 11 patients demonstrated prolonged virus replication in tumor tissues.6 Moreover, one patient showed a mixed response with the resolution of some liver metastases. In a phase 1b clinical trial of intravenous Pexa-Vec, a Wyeth OVV strain, 10 of 15 patients developed radiologically stable disease.7 However, there were no complete or partial responses in refractory or heavily pre-treated patients. Therefore, a predictive biomarker to determine patients who are more likely to benefit from treatment or to anticipate therapeutic effects or timing is vital. In addition, studies to elucidate variations in the susceptibility of human malignant cancers to OVV are urgently needed. In this basic and translational research, we address the value of expression of the long non-coding RNA (lncRNA) urothelial carcinoma-associated 1 (UCA1) as a potential predictive biomarker for both OVV and paclitaxel (PTX) chemotherapy in patients with ovarian cancer.
UCA1 was first identified as a sensitive and specific marker for human bladder carcinoma and cancer-upregulated drug-resistant (CUDR) expression.8, 9 Several groups have reported that UCA1 is highly expressed in ovarian cancer, colorectal cancer, and pancreatic cancer, among others, suggesting that it has the potential to be a biomarker for the diagnoses of these cancers.10, 11 UCA1 has been reported to function as a decoy for endogenous RNA, specifically by acting as a sponge for microRNAs (miRNAs), resulting in their downregulation, which has been implicated in enhanced drug resistance.12, 13 Moreover, UCA1 overexpression enhances proliferation and metastasis in several cancer cell lines through ERK, PI3K, Wnt, or Akt signaling.14, 15, 16 However, UCA1 has not previously been reported to be associated with oncolytic virotherapy outcome.
Here we show the possibility of applying UCA1 expression as a predictive biomarker for both OVV and PTX chemotherapy in vitro and in vivo for ovarian cancer. Further functional studies revealed the detailed mechanisms underlying the regulatory effect of UCA1 on OVV spread. Importantly, these results could enable the identification of patients more likely to respond to OVV.
Results
UCA1 Contributes to Enhanced PTX Resistance and Vaccinia Virus-Mediated Oncolysis
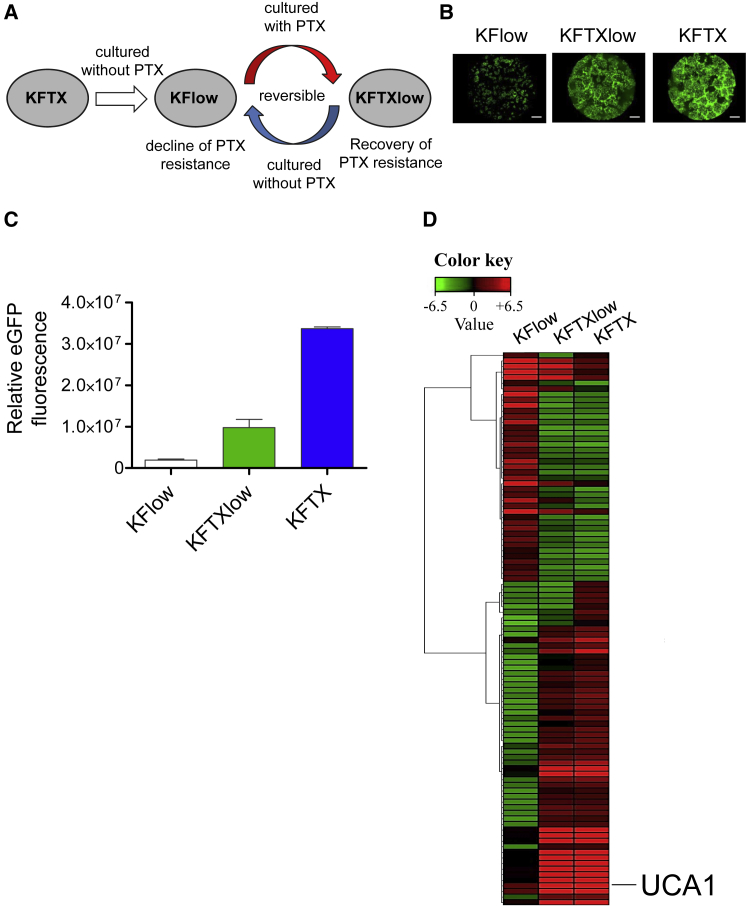
PTX-sensitive KFlow cells were isolated from KFTX cells cultured without the selection pressure of PTX. Further, KFlow cells regained resistance by incubating them with PTX, resulting in PTX-resistant KFTXlow cells (Figure 1A). These cell lines were infected with OVV-LG (LucGFP) at an MOI of 0.01. Interestingly, during KFTX infection, OVV-LG induced massive cytopathic effects (CPEs) after robust viral EGFP expression (Figures 1B and 1C). In contrast, weak CPEs and EGFP expression were induced in KFlow cells, whereas intermediate CPEs and EGFP expression were induced in KFTXlow cells. These results suggest that genes that are modulated according to PTX resistance are potential host factors that are involved in the oncolytic effects of OVV-LG.
Figure 1.
Identification of Candidate Genes Involved in Paclitaxel Resistance and Efficacy of Oncolytic Vaccinia Virus Spread
(A) Schema of KFlow, KFTXlow, and KFTX cells. (B) EGFP images of KFlow, KFTXlow, and KFTX cells after infection with OVV-LG (MOI = 0.01) for 72 h. Scale bar, 1,000 μm. (C) The intensity and area of viral EGFP brightness was measured using a Keyence BZ-X700 fluorescence microscope (n = 3). (D) RNA from KFlow, KFTXlow, and KFTX cells was collected and analyzed by an Agilent Sure Print G3 Human Gene Expression 8 × 60K v.2 Microarray (Takara Bio). The heatmap was constructed using multiExperimental Viewer (MEV) v.4.9 software. Data with error bars represent mean ± SEM.
Cellular gene expression profiles among these cell lines were compared by microarray analysis (Figure 1D). Results of 100 significantly dysregulated genes are shown in Figure 1D. Some candidate gene expression patterns among KFTX, KFTXlow, and KFlow cells were correlated with OVV growth efficacy in these cell lines (Table1). Among candidate genes, UCA1 expression was most dysregulated in KFTX (129.2-fold change) and KFTXlow (51.5-fold change) cells, as compared to that in KFlow cells. Moreover, UCA1 expression patterns among KFTX, KFTXlow, and KFlow cells were in complete accordance with OVV growth efficacy, which differed by more than 10-fold between KFlow and KFTXlow cells and 3-fold between KFTXlow and KFTX cells (Figure 1C). For these reasons, we hypothesized that UCA1 might play an important role in vaccinia virus-mediated oncolysis.
Table 1.
Top 10 Maximally Upregulated and Downregulated Genes Based on Microarray Analysis
| Gene Symbol | GenBank Accession | Scale Signal KFlow | Scale Signal KFTXlow | Scale Signal KFTX | |
|---|---|---|---|---|---|
| Upregulated genes | UCA1 | NR015379 | 233 | 11,996 | 30,100 |
| EEF1A2 | NM001958 | 965 | 31,458 | 40,666 | |
| S100P | NM005980 | 2,792 | 78,119 | 98,929 | |
| STEAP1 | NM012449 | 103 | 3,983 | 3,484 | |
| ABCB1 | NM000927 | 770 | 40,125 | 46,599 | |
| SMARCA1 | NM003069 | 100 | 2,194 | 2,294 | |
| ITGBL1 | NM004791 | 97 | 1,669 | 5,022 | |
| IGF2 | NM000612 | 34 | 306 | 797 | |
| LOXL2 | NM002318 | 70 | 723 | 704 | |
| PADI1 | NM013358 | 201 | 2,610 | 5,596 | |
| Downregulated genes | BATF3 | NM018664 | 2,360 | 88 | 1,025 |
| AKR1B15 | NM001080538 | 54,250 | 16,433 | 2,456 | |
| NEFL | NM006158 | 15,321 | 3,143 | 561 | |
| FSTL1 | NM007085 | 6,418 | 60 | 60 | |
| IRX4 | NM016358 | 32,083 | 418 | 346 | |
| CES1 | NM001266 | 14,067 | 437 | 365 | |
| GJB6 | NM006783 | 3,192 | 242 | 53 | |
| NELL2 | NM006159 | 3,104 | 154 | 149 | |
| SMO | NM005631 | 5,148 | 256 | 81 | |
| PHEX | NM0004444 | 1,041 | 76 | 54 |
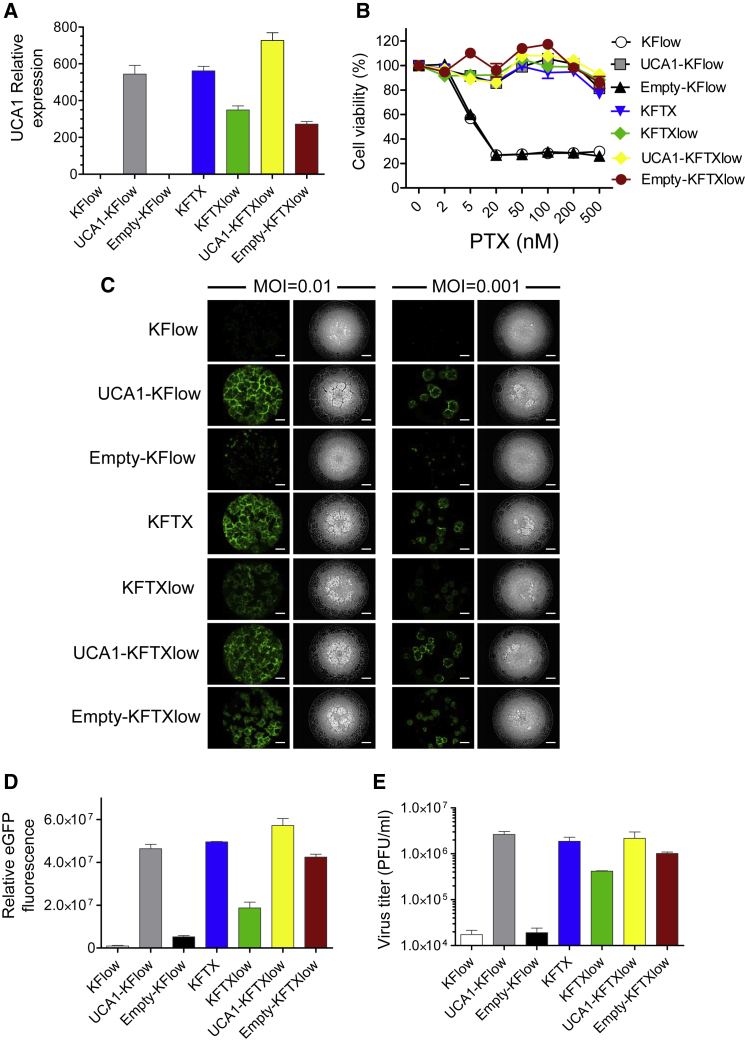
To analyze the effects of UCA1 on PTX resistance and OVV-LG oncolysis, KFlow and KFTXlow cells were transfected with pcDNA3.1/UCA1 (UCA1) or pcDNA3.1 (empty) plasmids (Figure 2A). PTX susceptibility was determined by cell viability assays. Based on viability, the resistance of UCA1-KFlow cells was significantly increased, in a dose-dependent manner, as compared to that of KFTX and KFTXlow cells (Figure 2B). Furthermore, the upregulation of UCA1 induced CPEs and EGFP expression in OVV-LG-treated KFlow and KFTXlow cells, which were equivalent to those observed in KFTX cells at MOIs of 0.01 and 0.001 (Figures 2C and 2D). Moreover, viral progeny was quantified by plaque assays. Consistent with the quantification of EGFP levels, UCA1 upregulation in KFlow and KFTXlow cells enhanced viral yields in KFTXlow cells to levels comparable to those observed in KFTX cells (Figure 2E). However, overexpression of candidate genes except UCA1 did not change OVV growth efficacy (data not shown). In addition, the depletion of UCA1 in KFTX cells decreased viral EGFP expression compared to that in control cells (Figure S1). These results suggested that UCA1 regulates viral replication and spread in addition to PTX resistance.
Figure 2.
Both Paclitaxel Resistance and Oncolytic Vaccinia Virus Spread Are Recovered by the Overexpression of UCA1
(A) Enhanced UCA1 expression was confirmed by qRT-PCR (n = 3). (B) KF cell lines were treated with PTX (0, 2, 5, 20, 50, 100, 200, and 500 nM). Cell viability was determined 48 h after PTX treatment (n = 3). (C) EGFP (left) and bright-field (right) images of KF cell lines are shown after infection with OVV-LG (MOI = 0.01 and 0.001) for 72 h. Scale bar, 1,000 μm. (D) The intensity and area of viral EGFP brightness were measured using a Keyence BZ-X700 fluorescence microscope (n = 3). (E) Plaque-forming units (PFUs) of OVV were determined based on titration using RK-13 cells (n = 3). Data with error bars represent mean ± SEM.
UCA1-Mediated Enhanced Vaccinia Virus-Mediated Oncolysis Is Common in Ovarian Carcinomas
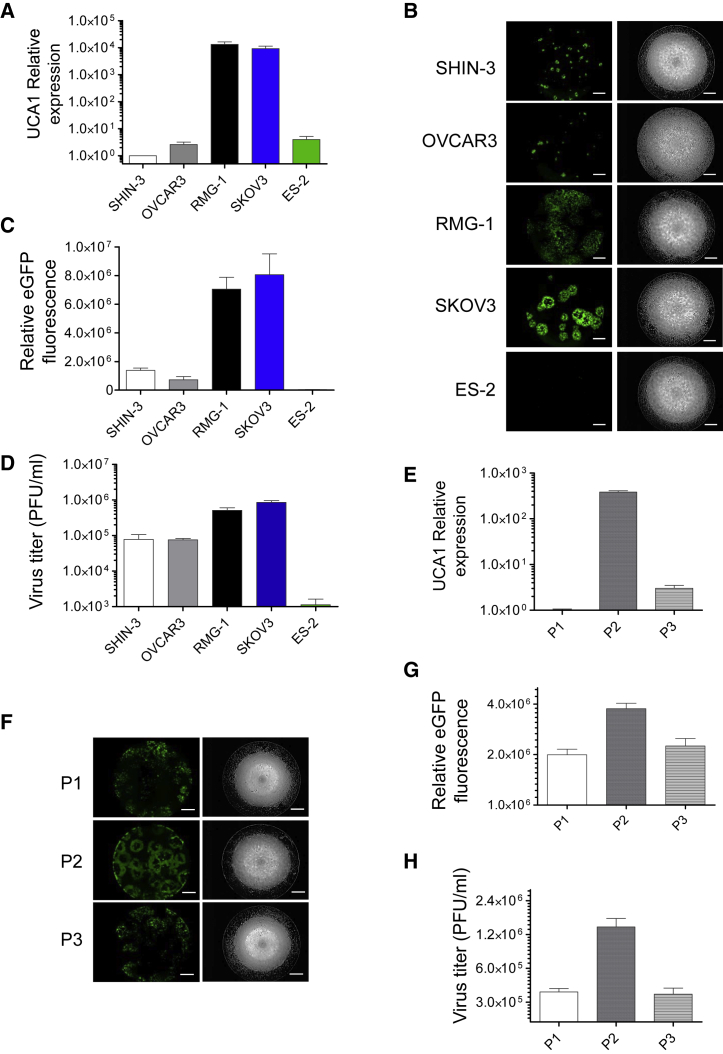
Based on the aforementioned observations, we investigated whether UCA1 expression is involved in the oncolytic effects of OVV-LG in other ovarian cancer cell lines. For this, a comparison of UCA1 expression and oncolytic effects was carried out using SHIN-3, OVCAR3, RMG-1, SKOV3, and ES-2 human ovarian cancer cell lines. We demonstrated that UCA1 expression was obviously inconsistent and that it was overexpressed in SKOV3 and RMG-1 cells compared to expression in SHIN-3, OVCAR3, and ES-2 cells (Figure 3A). These UCA1-high- and UCA1-low-expression cell lines were then infected with OVV-LG at an MOI of 0.001. In SKOV3 and RMG-1 cells, massive CPEs with robust EGFP expression were induced, as compared to those in SHIN-3, OVCAR3, and ES-2 cells (Figures 3B and 3C). In addition, the quantification of EGFP expression and viral progeny yielded consistent results (Figure 3D). Further investigation revealed that UCA1 expression correlated with the oncolytic effects of OVV in primary ovarian cancer cells (Figures 3E–3H). These results suggested that UCA1 regulates the oncolytic effects of OVV and might be considered a promising biomarker to predict oncolytic activity in ovarian cancer.
Figure 3.
Correlation between UCA1 and Oncolytic Vaccinia Virus Spread as Observed in Other Ovarian Cancer Cell Lines
(A and E) UCA1 expression was detected by qRT-PCR in SHIN-3, OVCAR3, RMG-1, SKOV3, and ES-2 cells (A) and primary ovarian cancer cells (E) (n = 3). (B and F) EGFP (left) and bright-field (right) images of SHIN-3, OVCAR3, RMG-1, SKOV3, and ES-2 cells (B) and primary ovarian cancer cells (F) after infection with OVV-LG (MOI = 0.001 or 0.01) for 48 h. Scale bar, 1,000 μm. (C and G) Intensity and area of viral EGFP brightness in SHIN-3, OVCAR3, RMG-1, SKOV3, and ES-2 cells (C) and primary ovarian cancer cells (G) were measured using a Keyence BZ-X700 fluorescence microscope (n = 3). (D and H) Plaque-forming units (PFUs) of OVV in SHIN-3, OVCAR3, RMG-1, SKOV3, and ES-2 cells (D) and primary ovarian cancer cells (H) were determined by titration using RK-13 cells (n = 3). Data with error bars represent mean ± SEM.
UCA1 Enhances Vaccinia Virus Oncolysis by Modulating Viral Cell-to-Cell Spread, but Not Binding, Entry, and Replication
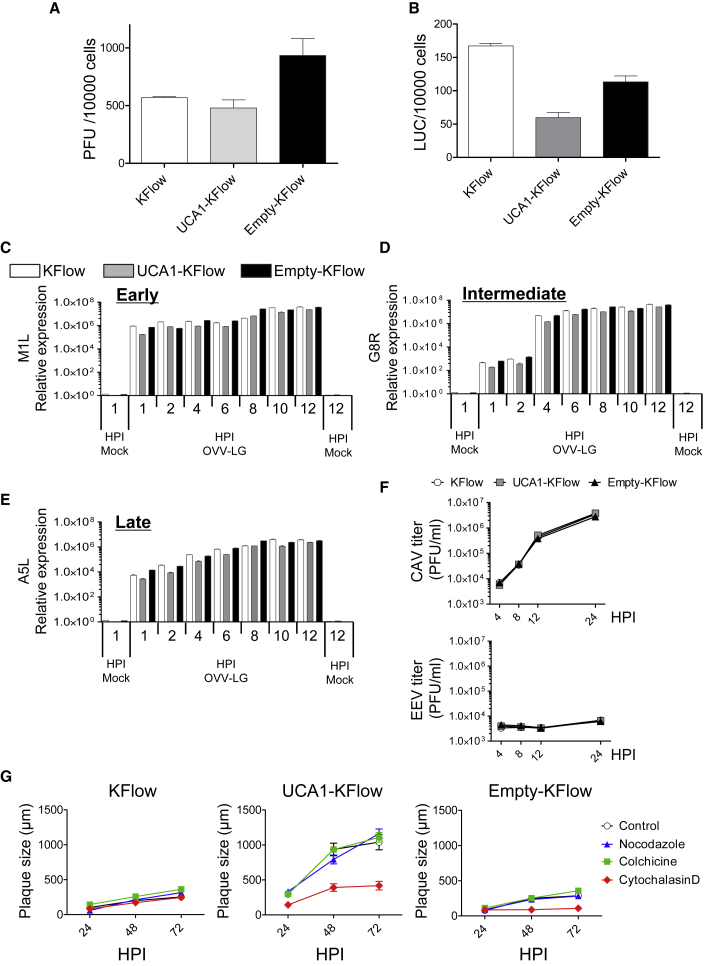
To highlight the mechanism associated with enhanced OVV-LG-mediated oncolysis, we identified which stage of OVV replication is regulated by UCA1. First, we determined if UCA1 affects OVV-LG binding and entry. It was previously reported that inhibiting OVV entry results in decreased replication.17 Based on binding assays, the production of viral progeny by cell-attached viruses was not changed with high UCA1 expression (Figure 4A). Regarding entry assays, viral firefly luciferase activity (Fluc) was not affected by UCA1 expression (Figure 4B).
Figure 4.
UCA1 Expression Specifically Targets Oncolytic Vaccinia Virus Cell-to-Cell Spread and Increases the Oncolytic Effect of OVV-LG
(A and B) KFlow, UCA1-KFlow, and Empty-KFlow cells were infected with OVV-LG (MOI = 10) for 30 min. After ample washes, the number of plaque-forming units (PFUs) of OVV was determined by titration using RK-13 cells (A), and viral luciferase activity was determined (B) (n = 3). (C–E) Gene expression in KFlow, UCA1-KFlow, and Empty-KFlow cells after either mock infection or infection with OVV-LG (MOI = 5). RNA was extracted at 1, 2, 4, 6, 8, 10, and 12 h postinfection (hpi). qRT-PCR was performed with primers specific for M1L (C), G8R (D), A5L (E), and GAPDH (C–E, as an internal control) (n = 3). (F) One-step growth curve. KFlow, UCA1-KFlow, and Empty-KFlow cells were infected with OVV-LG (MOI = 5). Cells (CAV) and supernatant (extracellular enveloped virus [EEV]) were collected, and the number of OVV PFUs was determined by titration using RK-13 cells (n = 3). (G) KFlow, UCA1-KFlow, and Empty-KFlow cells were treated with nocodazole (25 nM), colchicine (2.5 nM), and cytochalasin D (KFlow and Empty-KFlow, 25 nM; UCA1-KFlow, 50 nM) and infected with OV-LG (MOI = 0.001). Representative plaques formed under semisolid medium were photographed and calculated with the analysis application measurement module (Keyence) (n = 10). Data with error bars represent mean ± SEM.
Next, we analyzed the production of early (M1L), intermediate (G8R), and late (A5L) viral transcripts by real-time PCR. In cells infected with OVV-LG, viral genes representing all three phases were not induced by high UCA1 expression (Figures 4C–4E). It was reported that the presence of fewer extracellular enveloped viruses (EEVs), which are required for spread between neighboring cells and distant cells, significantly affects virus production.18, 19 In the early infectious phase, EEV and cell-associated virus (CAV) titers were not stimulated by high UCA1 expression (Figure 4F). Lastly, because it was shown that the efficacy of intracellular transport and cell-to-cell spread is important for the oncolytic effects of OVV,20, 21 selective inhibitors of intracellular transport (nocodazole and colchicine) and cell-to-cell-spread (cytochalasin D) were used, and plaque assays were performed without the induction of cytotoxicity (Figure S2). In UCA1-KFlow cells, inhibitors of intracellular transport had little effect on plaque sizes; however, an inhibitor of cell-to-cell spread completely inhibited plaque formation (Figure 4G). These results indicate that UCA1 specifically affects OVV-LG cell-to-cell spread.
UCA1 Enhances the Cell-to-Cell Spread of OVV via Cdc42 Activation
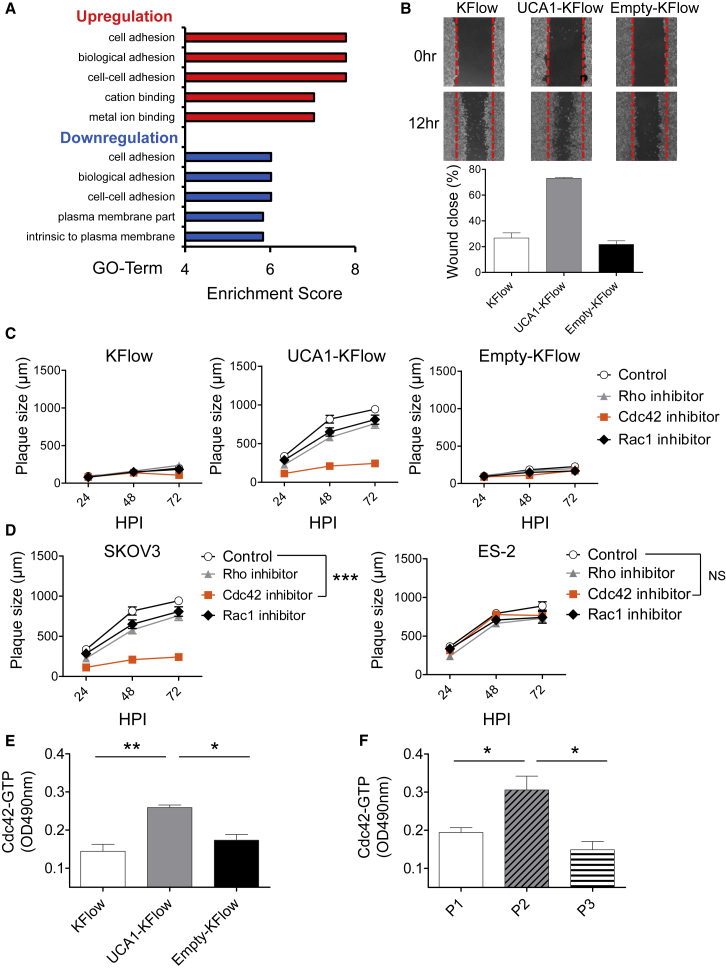
We hypothesized that UCA1 regulates actin polymerization, thereby promoting cell-to-cell spread based on two observations. First, gene ontology analysis based on a microarray comparing KFlow and UCA1-KFlow cells indicated alterations in cell adhesion pathways; however, functional assays were then required (Figure 5A). Second, high UCA1 expression mediated dramatic increases in the numbers and lengths of filopodia, which are actin-rich structures involved in numerous cellular processes like cell migration and cell-cell interactions (Figure S3). In support of this hypothesis, wound-healing assays demonstrated that UCA1 enhances cell migration and adhesion upon serum starvation (Figure 5B).
Figure 5.
Cdc42 Activation Is Critical for the Cell-to-Cell Spread of Oncolytic Vaccinia Virus-LG Induced by UCA1 Expression
(A) Results of gene ontology analysis after microarray analysis. Gene lists correspond to 2-fold changes in KFlow and UCA1-KFlow cells. (B) A pipette tip was used to introduce a scratch in a monolayer of KFlow, UCA1-KFlow, and Empty-KFlow cells in the absence of serum. The percentage of wound closure was estimated using the analysis application measurement module (Keyence) (n = 6). (C and D) KFlow, UCA1-KFlow and Empty-KFlow (C) and SKOV3 and ES-2 cells (D) were treated with Rhosin (targeting Rho; 50 nM), ML-141 (targeting Cdc42; KFlow, UCA1-KFlow, and Empty-KFlow, 20 nM; SKOV3, 18 nM; ES-2, 15 nM), and NSC23766 (targeting Rac1; KFlow, UCA1-KFlow, Empty-KFlow, and SKOV3, 30 nM; ES-2, 15 nM) inhibitors and infected with OV-LG (MOI = 0.001). Representative plaques formed under semisolid medium were photographed and calculated with the analysis application measurement module (Keyence) (n = 10). (E and F) Activation of Cdc42 in KFlow, UCA1-KFlow and Empty-KFlow (E) and primary ovarian cancer cells (F) was analyzed by performing a colorimetric G-LISA assay (n = 3). Data with error bars represent mean ± SEM. One-way ANOVA was used for (C) and (D).*p < 0.05, **p < 0.01, ***p < 0.001.
Since previous studies suggested that cell adhesion and migration are regulated by Rho GTPases, which have an important role in the assembly and disassembly of the actin cytoskeleton,22, 23 we examined whether the suppression of Rho, Rac1, and Cdc42, members of the Rho family of small GTPases, could inhibit plaque growth. The addition of ML141, a Cdc42 inhibitor, resulted in a significant decrease in plaque size, as compared to that with other inhibitors against Rac1 and Rho (Figure 5C; Figure S2). We also examined the effects of Rho, Rac1, and Cdc42 suppression on OVV-LG plaque size in UCA1-high- and UCA1-low-expression cell lines, namely SKOV3 and ES-2 cells, respectively (Figure 5D). As expected, the addition of a Cdc42 inhibitor resulted in inefficient cell-to-cell spread in SKOV3, but not in ES-2, cells. Moreover, Cdc42 expression was not changed between KFlow (scale signal: 41,996) and UCA1 KFlow cells (scale signal: 37,793) based on microarray analysis. Nevertheless, when the expression of active Cdc42 was tested, we observed that UCA1 expression increased Cdc42 activation (Figure 5E); we also noted a correlation between UCA1 and activated Cdc42 expression in primary ovarian cancer cells (Figure 5F). These results suggest that UCA1-mediated Cdc42 regulation enhances OVV-LG cell-to-cell spread through filopodia formation.
OVV Exerts Increased Oncolytic Effects on PTX-Resistant KFTX Cells in a Mouse Model of Peritoneal Metastasis
We previously found that transgene insertion into the hemagglutinin (HA) gene of OVV-LG does not affect native vaccinia virus replication.24 Further, OVV-LG is suitable to elucidate correlations between UCA1 expression and OVV spread. However, OVV-LG maintains viral pathogenicity.24 We thus utilized OVV-VGFΔ/O1LΔ for therapeutic experiments. Since two viral proteins, specifically VGF and O1, contribute to viral spread and replication through activation of the epidermal growth factor receptor (EGFR)-dependent mitogen-activated protein kinase (MAPK)/ERK pathway, deletion of both genes inhibits pathogenic viral replication in normal cells, whereas therapeutic replication is retained in tumor cells with constitutive ERK1/2 activation.25
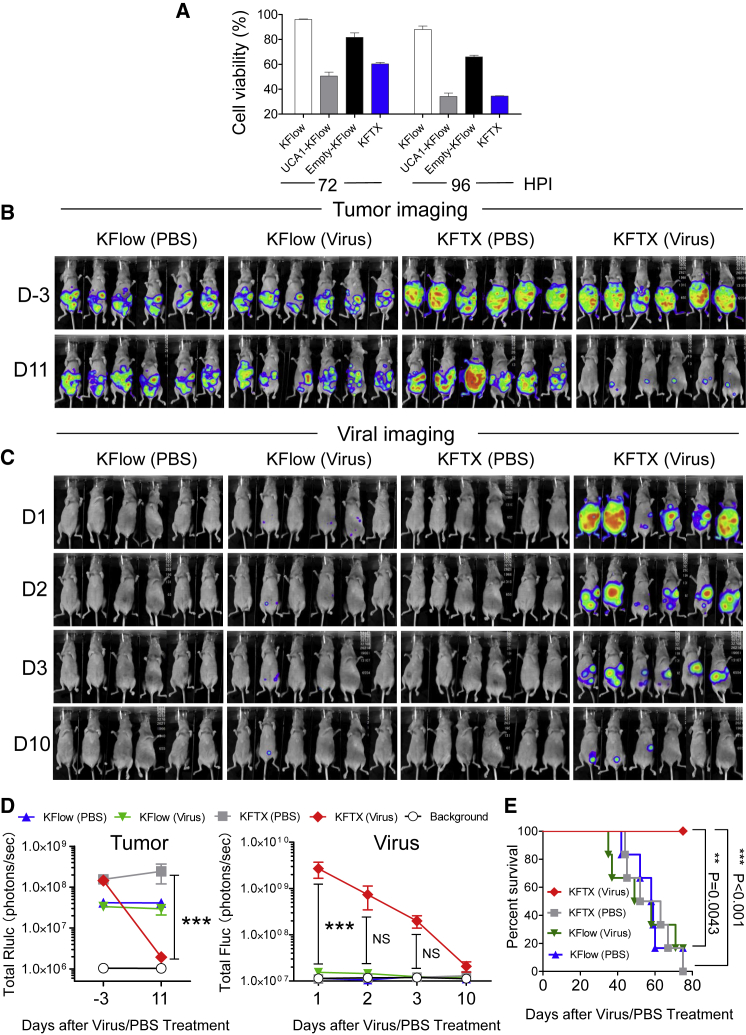
To assess the oncolytic effects of OVV-VGFΔ/O1LΔ, we examined its cytotoxicity using KFlow, UCA1-KFlow, Empty-KFlow, and KFTX cells (Figure 6A). OVV-VGFΔ/O1LΔ mediated enhanced oncolysis in PTX-resistant KFTX and UCA1-KFlow cells compared to that in PTX-sensitive KFlow and Empty-KFlow cells. Therefore, we applied a peritoneal dissemination model using PTX-resistant KFTX or PTX-sensitive KFlow cells expressing Renilla luciferase. Cells were injected into BALB/cAJcl-nu/nu mice, and, after confirming tumor growth based on Renilla luciferase activity, mice were intraperitoneally administered OVV-VGFΔ/O1LΔ or control PBS (Figure 6B). On day 1 after viral injection, mice bearing KFTX cells showed tumor-specific high virus-associated signals, whereas mice bearing KFlow cells exhibited little viral replication (Figures 6C and 6D). On day 10 after viral injection, viral signals in mice bearing KFTX cells disappeared, which was accompanied by a reduction in tumor signals. The treatment of mice harboring KFTX cells with OVV-VGFΔ/O1LΔ resulted in the significant inhibition of tumor growth, by more than two log orders, compared to that in control PBS-treated animals (Figure 6D). In terms of animal survival, treating KFTX-harboring mice with OVV-VGFΔ/O1LΔ led to a significant improvement, but the same treatment in KFlow-bearing mice had no effect (Figure 6E). These data demonstrated that OVV-VGFΔ/O1LΔ is an effective therapy for PTX-resistant ovarian cancer formed from KFTX cells.
Figure 6.
Oncolytic Vaccinia Virus-VGFΔ/O1LΔ Efficiently Exerts Oncolytic Effects against Paclitaxel-Resistant KFTX Cells In Vitro and In Vivo
(A) KFlow, UCA1-KFlow, Empty-KFlow, and KFTX cells were infected with OVV-VGFΔ/O1LΔ (MOI = 0.5). After 72 or 96 h of infection, cell viabilities were determined (n = 3). (B and C) BALB/cAJcl-nu/nu mice were intraperitoneally injected with KFTX or KFlow cells stably expressing Renilla luciferase (n = 6). After confirming tumor growth, mice were administered an intraperitoneal injection of OVV-VGFΔ/O1LΔ (1 × 106 PFUs) or PBS. Tumor imaging (B) and viral imaging (C) are shown. (D) Bioluminescence signals (in photons/s) were calculated from the imaging data of tumor (Rluc) and viral (Fluc) signals. (E) Survival curves of mice are shown after virus injection. A log-rank (Mantel-Cox) test was used to analyze significance. **p < 0.01, ***p < 0.001. Data with error bars represent mean ± SEM. Two-way ANOVA was used for (D).
UCA1 Expression Potentially Predicts the Therapeutic Strategy for Ovarian Cancer Based on Conventional PTX or Novel Oncolytic Virotherapy
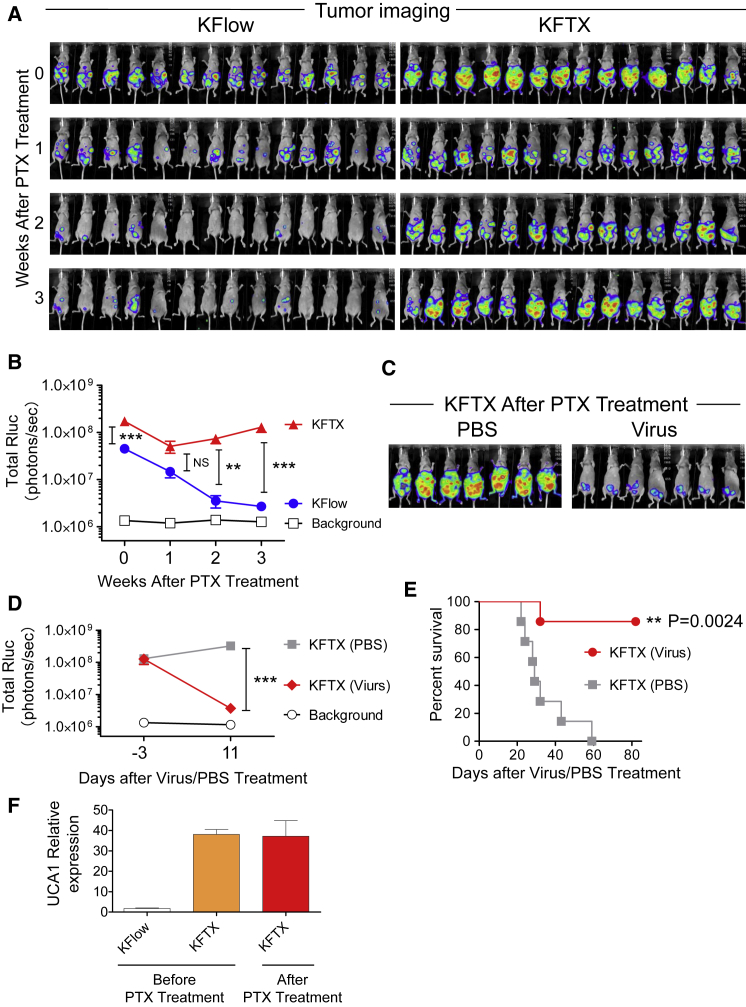
For the development of a clinically relevant mouse tumor model, nude mice bearing tumors derived from intraperitoneally injected KFTX or KFlow cells were injected with PTX. After 3 weeks of administration, PTX efficiently exerted anti-tumor effects on KFlow tumors, whereas KFTX tumors showed only marginal regression for 1 week after treatment and then gradually regrew (Figures 7A and 7B). Importantly, PTX-treated mice bearing KFTX tumors were injected with OVV-VGFΔ/O1LΔ or control PBS, resulting in the dramatic regression of residual tumors. On day 9 after OVV-VGFΔ/O1LΔ injection, a reduction in tumor signals was observed in mice injected with virus (Figures 7C and 7D). In terms of animal survival, treating KFTX-bearing mice with OVV-VGFΔ/O1LΔ led to a significant improvement in survival, compared to that in control PBS-treated animals. After all treatments, the survival of mice bearing KFTX tumors and treated with virus was prolonged compared to that in animals treated with PBS (Figure 7E). Further, UCA1 expression was quantified in excised tumor tissue before and after PTX administration. Consistent with in vitro data, high UCA1 expression was maintained in KFTX tumors (Figure 7F). Taken together, these results indicate that tumors expressing high levels of UCA1 should be treated with OVV rather than PTX, whereas those with low UCA1 expression should receive the opposite therapeutic regimen.
Figure 7.
UCA1 Expression Predicts the Therapeutic Effect of Paclitaxel and Oncolytic Vaccinia Virus-VGFΔ/O1LΔ
BALB/cAJcl-nu/nu mice were intraperitoneally injected with KFTX or KFlow cells stably expressing Renilla luciferase. After confirming tumor growth, PTX (1 mL, 10 mg/kg) in saline was intraperitoneally injected twice per week. Then 3 weeks later, mice were administered a single intraperitoneal injection of OVV-VGFΔ/O1LΔ (1 × 106 PFUs) or PBS. (A) Tumor imaging is shown before and after PTX treatment (n = 14). (B) Bioluminescence signals (in photons/s) were calculated from the tumor imaging data. (C) Tumor imaging is shown after OVV-VGFΔ/O1LΔ injection (n = 7). (D) Bioluminescence signals (in photons/s) were calculated from the tumor imaging data. (E) Survival curves for the mice are shown after virus injection. A log-rank (Mantel-Cox) test was used to analyze significance. (F) At 3 weeks before and after PTX treatment, for the extraction of KFTX and KFlow tumors, mice were sacrificed, and tumors were collected and frozen quickly using liquid nitrogen. Collected tumors were disrupted in lysis buffer and UCA1 expression was detected by qRT-PCR (n = 9–15). ∗∗p<0.01, ∗∗∗p<0.001. Data with error bars represent mean ± SEM. Two-way ANOVA was used for (B) and (D).
Discussion
Various viruses such as adenovirus, herpes simplex virus, Newcastle disease virus, and vaccinia virus have been engineered and clinically investigated as oncolytic viruses.2, 5, 26, 27 OVV has many advantages for oncolytic virotherapy.28 First, the range of tropism for OVV in mammalian cells is broad, and, thus, it could be applied to many types of cancers. Second, the whole life cycle of OVV occurs in the cytoplasm, indicating that it is not incorporated into the host genome, which thereby remains intact. Third, OVV spreads via the bloodstream; it is thus expected that OVV will exert oncolytic effects not only in primary tumors but also in metastatic cancer via remote infection. Finally, the replication cycle of OVV is fast, which should facilitate rapid viral replication and spread, maximizing therapeutic effects. Accordingly, OVV is a promising cancer therapy, and the efficiency of viral replication and spread will ultimately play a crucial role in therapeutic outcome.29
Promising clinical data based on various strains of OVV have indicated both therapeutic efficacy and safety.6, 7, 30 However, targeting appropriate cancer types and grades using OVV remains difficult, and combinations with other cancer therapies is likely to be primarily attempted. In this study, xenograft mouse models of PTX-resistant and PTX-sensitive ovarian cancer were found to be efficiently treated with PTX and OVV, respectively. Although high UCA1 expression was maintained in resected tumors after PTX treatment, UCA1 expression was not detected in normal tissues, such as ovary and urothelial tissues (data not shown). Therefore, this therapeutic strategy could accurately reflect clinical situations with the use of UCA1 as a correlative marker (Figure 7).
First, the expression patterns of some genes, including UCA1 and ITGBL1, were found to be correlated with OVV growth in KFTX, KFTXlow, and KFlow cells (Table 1). However, the overexpression of candidate genes other than UCA1 did not change OVV growth efficacy. For this reason, we selected UCA1. Importantly, multiple studies have shown that it plays an important biological role in tumor growth, metastasis, and malignancy and that it might act as a potential biomarker and therapeutic target for various cancers.9, 31, 32 In clinical specimens, high UCA1 expression was observed in cancer tissue compared to normal tissue.31 Furthermore, the function of UCA1 as an oncogene has gradually become elucidated. For example, elevated expression induces cisplatin resistance via SRPK1 activation in ovarian cancer.33 Further, high UCA1 expression induces PTX resistance through the miR-129/ABCB1 pathway in ovarian cancer.34 Considering that UCA1 regulates chemotherapy sensitivity through multiple pathways, it might modulate PTX resistance through a mechanism other than that required for viral spread.
UCA1 is highly expressed in several cancers and promotes metastasis and progression by inducing ERK signaling.15, 35 Consistent with previous reports, ERK phosphorylation was consistently high and UCA1 was found to further activate ERK in serum-stimulated KFlow and UCA1-KFlow cells (Figure S4A). Furthermore, ERK activation is known to play an important role in the early stages of OVV infection, and it is also crucial for the generation of virus progeny.36 Thus, we examined whether ERK activation plays a key role in promoting OVV spread. The ERK pathway was found to be activated by epidermal growth factor (EGF) stimulation, and the activation of ERK was measured (Figure S4A). Further, KFlow and UCA1-KFlow cells were infected with OVV and treated with EGF stimulation. OVV-mediated EGFP expression in KFlow cells was not enhanced by ERK activation following EGF stimulation (Figure S4B). In addition, we found that OVV replication and progeny were not promoted by high UCA1 expression (Figures 4C–4F). These results suggested that further activation of ERK by EGF stimulation had no effect on OVV replication and progeny in serum-stimulated tumor cells but that another pathway regulated by UCA1 is crucial for OVV spread.
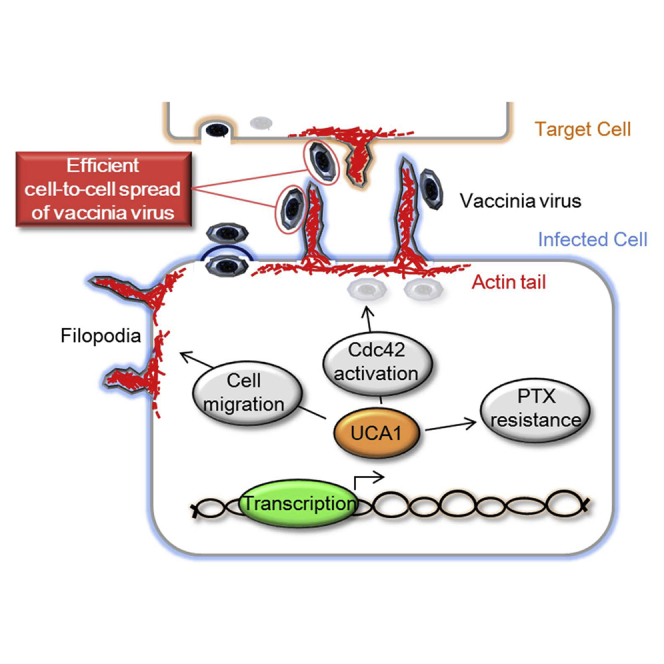
OVV cell-to-cell spread is an important process and is supported by some viral proteins through actin-based motility. During the process of fusion between the extracellular membrane of intracellular enveloped virus and the cell membrane, B5 is exposed at the vaccinia virus surface of cell-associated enveloped virus (CEV).37 The detailed function of B5 is unknown with respect to actin-based motility. However, deletion of B5 dramatically changes vaccinia morphology. A36 localizes to the inner cell membrane after CEV is exposed on the cell surface. Phosphorylated A36 has a binding site for Nck, which mediates the recruitment of neural Wiskott-Aldrich-syndrome protein (N-WASP), resulting in Arp2/3 complex binding to existing actin filaments.38 The activated Arp2/3 complex helps to propel the virus into neighboring cells. However, based on our results, viral gene expression was not changed by UCA1 expression, and host cell-signaling pathways related to actin filament branching had a critical role in viral virulence. The small guanosine triphosphate (GTP)-binding protein Cdc42 has a major role in inducing filopodia formation by regulating actin polymerization.39 A recent report demonstrated that infection by the IHD-J vaccinia virus strain induces Cdc42 activation and filopodia formation.40 ITSN1 is induced by viral infection-activated Cdc42 and associated N-WASP-actin polymerization.38, 41, 42 These results indicate that Cdc42 has an important role in viral spread by inducing filopodia formation. Consistent with this, we found that Cdc42 inhibitors blocked effective viral spread mediated by high UCA1 expression (Figures 5C and 5D).
This research is the first to indicate that host factor-regulated small G proteins mediate efficient OVV cell-to-cell spread; specifically, vaccinia virus proteins directly regulate G protein activation. Recent studies have shown that vaccinia virus encodes F11, a viral protein with a conserved Rho-binding domain that inhibits its downstream signaling.43, 44 The loss of RhoA signaling results in the modulation of cortical actin filaments beneath the plasma membrane and contributes to increased virus release. In this study, inhibition of the Rho GTPases RhoA and Rac1 had no effect on viral cell-to-cell spread at non-cytotoxic concentrations (Figures 5C and 5D). Further, activation of RhoA and Rac1 were consistently low before and after infection (data not shown). These results suggest that actin filaments are not altered and that cell-to-cell spread is triggered by filopodia formation induced by Cdc42 activation in ovarian cancer. In primary ovarian cancer cell lines, a correlation between UCA1 expression and Cdc42 activation was observed. However, low UCA1 expression was observed in the A2780 ovarian cancer cell line, despite highly activated Cdc42 signaling, resulting in efficient cell-to-cell OVV spread (data not shown). Taken together, these results suggest that UCA1 regulates the therapeutic effect of OVV by activating Cdc42 in most ovarian cancer cells and that other pathways might be involved in the activation of Cdc42 in some ovarian cancers and other cancer types. Like UCA1, Cdc42 activation is a potential biomarker to predict the therapeutic effect of OVV in other cancer types.
Intriguingly, the spread of other oncolytic viruses such as adenovirus and measles virus was not found to be induced by UCA1 (data not shown). The life cycle of these oncolytic viruses is independent of actin assembly.45, 46, 47 The OVV LC16mO strain was used for this study. Furthermore, many widely used vaccinia virus strains were shown to exhibit enhanced cell-to-cell spread by inducing Cdc42-dependent actin polymerization.38, 40 Moreover, there are different strains of OVV, such as WR, Copenhagen, LC16mO, and GLV-1h68. LC16mO was found to be highly similar to GLV-1h68 based on phylogenetic analysis, and it shares orthologous open reading frames (ORFs) with WR and Copenhagen based on sequence comparisons.48 These results also suggest that UCA1 does not interact with other pathogenicity factors and that it could predict the therapeutic effects of other vaccinia virus strains like WR, Copenhagen, GLV-1h68, and Wyeth; further efforts are needed to establish UCA1 as a predictive biomarker.
In summary, UCA1 triggers a variety of intracellular mechanisms and independently regulates PTX resistance and Cdc42 activation in ovarian cancer. Cdc42-induced filopodia formation promotes tumor invasion and migration, which is necessary for cell-to-cell spread, and dramatically enhances the therapeutic effect of OVV. Since these mechanisms were proven in primary ovarian cancer cells and in vivo, UCA1 expression has the potential to be a biomarker that could predict responses to both PTX chemotherapy and OVV.
Materials and Methods
Cell Culture
SHIN3,49 SKOV3, RMG-1,50 OVCAR3,51 KFTX,52 KFlow, and KFTXlow cells were cultured in RPMI-1640 with 10% fetal bovine serum (FBS). A2780 cells53 were cultured in DMEM with 10% FBS. ES-2 cells54 were cultured in DMEM/Ham’s F-12 plus 10% FBS. RK13 cells were cultured in E-MEM plus 5% FBS. A549 cells were cultured in Ham’s F-12K plus 10% FBS. SKOV3, RK13, and A549 cells were purchased from the American Type Culture Collection (ATCC). All clinical P1, P2, and P3 samples were taken from recurrent ascites after chemotherapy, including platinum-based drugs and performed at the Saitama Medical University International Medical Center. Ascite cells were cultured in RPMI-1640 with 20% FBS. After reaching confluence, cells were trypsinized and cultured. All cultures were supplemented with 10% FBS and 1% penicillin-streptomycin (Gibco, Grand Island, NY, USA). Approval for this project was obtained from the Ethics Committee of Saitama Medical University International Medical Center (12-096) and Tottori University (2543). All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Microarray
Total RNA from each cell line was extracted using the NucleoSpinRNA kit according to the manufacturer’s protocol (Takara Bio, Shiga, Japan). Using total RNA, microarray experiments were outsourced for Agilent Array expression analysis (Takara Bio). The RNA expression array was performed using the Agilent Sure Print G3 Human Gene Expression 8 × 60K version 2 (v.2) Microarray. These data were deposited (https://www.ncbi.nlm.nih.gov/geo/; GEO: GSE122358).
Hierarchical Clustering Analysis
After extracting the expression values from the gene expression profile, a bidirectional hierarchical clustering heatmap was constructed using multiExperimental Viewer (MEV) v.4.9 software.
Gene Ontology Analysis
Microarray data comparing KFlow and UCA1-KFlow cells were subjected to gene ontology analysis. Biologically relevant features were constructed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) tools (https://david.ncifcrf.gov/). Gene lists corresponding to 2-fold changes between KFlow and UCA1-KFlow cells were uploaded to DAVID for gene ontology analysis.
Cell Viability Assays
PTX resistance was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA). Each cell line was plated at 6 × 103 cells/well in 96-well plates. After 12 h, PTX was added to the media at pre-determined concentrations. After another 48 h, cell viability was measured.
UCA1 Stable Transfectants
The UCA1 gene was amplified using Phusion High-Fidelity Polymerase (New England Biolabs, Ipswich, MA, USA) with primers (5′-CTG GAT CCT GAC ATT CTT CTG GAC AAT GAG-3′ and 5′-CTG CGG CCG CAT ATT AGC TTT AAT GTA GGT GGC-3′), using KFTX cDNA as the template. The PCR product was digested with BamHI and NotI and cloned into the same restriction site of pcDNA3.1. KFlow and KFTXlow cells were transfected with pcDNA3.1-UCA1 or pcDNA3.1 using FuGENE HD Transfection Reagent (Roche, Indianapolis, IN, USA), according to the manufacturer’s instructions. Then, transfected cells were selected with G418 (500 μg/mL) for 3 weeks, resulting in UCA1-KFlow or Empty-KFlow cells, respectively.
OVV Preparation
Recombinant viruses were constructed as described previously.25 Briefly, RK13 cells were infected with LC16mO24 at an MOI of 0.02, and then they were transfected with pVNC110-SP-LucGFP or pUC19-VGF-SP-LucGFP plasmid, resulting in the generation of OVV-LG or OVV-VGFΔ viruses, respectively. After harvesting progeny viruses 2–5 days later, each virus was selected based on LucGFP expression, through three serial plaque purifications. Finally, the insertion of LucGFP was verified by sequencing the modified region. Similarly, OVV-VGFΔO1LΔ was constructed by infecting RK13 cells with OVV-VGFΔ viruses, as described, and then transfecting them with the pUC19-O1-P-DsRed plasmid. All viruses were propagated in RK13 or A549 cells and stored at −80°C. For in vivo experiments, all viruses were purified using OptiPrep (Axis-Shield, Oslo, Norway), according to the manufacturer’s protocol.
OVV Infection
Cells (6–10 × 103/well) were seeded in 96-well plates 36 h before OVV infection. Each cell line was infected with OVV at an MOI of 0.01 or 0.001. After 48–72 h, 96-well plates were photographed under phase-contrast or fluorescence microscopy using a BZ-X700 microscope (Keyence, Osaka, Japan). EGFP fluorescence was quantified based on intensity and area using Hybrid Cell Count (Keyence, Osaka, Japan).
OVV-VGFΔ/O1LΔ Cytotoxicity Assays
Cells (6 × 103/well) were seeded in 96-well plates 36 h before OVV infection. Each cell line was infected with OVV-VGFΔ/O1LΔ at an MOI of 0.5. At 72 and 96 h post-infection, cell viabilities were determined using a CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega), according to the manufacturer’s instructions.
CAV and EEV Titration
Cells were plated at 3 × 104/well in 24-well plates. After 36 h, cells were infected at an MOI of 5 plaque-forming units (PFUs)/cell for 1 h at 37°C and then washed with medium and incubated for the indicated times. Supernatants, as EEV, were collected and centrifuged at 700 × g for 10 min to separate the cells as CAV. Titers of viruses present in the supernatant and cellular fractions were then determined using RK13 cells.
Quantification of UCA1 Expression
Cells were plated at 2 × 105/well in 6-well plates. After 36 h, total RNA from each cell line was extracted using the NucleoSpinRNA kit according to the manufacturer’s protocol (Takara Bio). Each total RNA sample was subjected to reverse transcription using the High-Capacity cDNA Reverse Transcription Kit, according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA). Then, expressions of UCA1 and the endogenous control GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were quantified by real-time PCR using TaqMan probes (Applied Biosystems, Carlsbad, CA, USA). Real-time PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The relative expression of UCA1 was calculated by the comparative threshold cycle method.
Gene Expression of Vaccinia Virus Genome
Cells were plated at 2 × 105/well in 6-well plates. After 36 h, cells were infected with OVV-LG at an MOI of 5. Total RNA was extracted at 1, 2, 4, 6, 8, 10, and 12 h post-infection and converted to cDNA. Real-time PCR was performed using SYBR Green Master Mix with the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Transcripts were detected with the following primer pairs: M1L forward (5′-AAC GGA CCA CAT CCT TCT TC-3′) and M1L reverse (5′-ATC CAA ACG CGT GTGATA AA-3′); G8R forward (5′-GCG GAT CTG TAA ACA TTT GG-3′) and G8R reverse (5′-CCT TGGACA CAG GAA GAT TAA A-3′); and A5L forward (5′-TTT CCA TCC GAT TGT TGT GT-3′) and A5L reverse (5′-AGT TCA CTC CTT CCA GCG TT-3′). GAPDH was used as the internal loading control, and standardization among samples was performed using the primer pair GAPDH forward (5′-CCT GTT CGA CAG TCA GCC G-3′) and GAPDH reverse (5′-GA CCA AAT CCG TTG ACT CC-3′).
Plaque Formation Assays
Cells were plated at 2 × 105/well in 6-well plates. After 36 h, cells were infected with OVV-LG at an MOI of 0.001. After a 1-h incubation, E-MEM containing 0.8% methyl cellulose supplemented with 5% FBS and reagents was added to each well, and plaque sizes were calculated with a BZ-X700 microscope. Reagents used for plaque assays were as follows: nocodazole (Wako, Osaka, Japan), colchicine (Wako), cytochalasin D (Wako), Rhosin (Merck, Boston, MA, USA), ML-141 (Sigma, St. Louis, MO, USA), and NSC23766 (Abcam, Cambridge, UK).
Wound-Healing Assays
Cells were plated at 6 × 105/well in 6-well plates. After 24 h, confluent cells were scratched with a pipette tip in serum-free media. Cells were fixed at 0 and 12 h and processed for microscopic analysis. Phase-contrast images were captured using a BZ-X700 microscope.
Virion-Binding Assays
To analyze viral binding affinity, cells were plated at 1 × 105/well in 12-well plates. After 36 h, cells were infected with OVV-LG at an MOI of 10 at 4°C for 30 min. After ample washes, cell-attached viruses were collected, and virus titers were determined using RK13 cells.
To measure early viral gene expression, cells were plated at 1 × 104/well in 96-well plates. After 36 h, cells were infected with OVV-LG at an MOI of 10 at 4°C for 30 min. After ample washes, cells were incubated for 1 h and the ONE-Glo Luciferase Assay System was used.
GTPase Activation Assays
Cdc42 activation was measured using Cdc42-specific G-LISA activation kits (Cytoskeleton, Denver, CO, USA), according to the manufacturer’s instructions. Briefly, KFlow, UCA1-KFlow, and Empty-KFlow cells, as well as primary ovarian cancer cell cultures, were plated at 1 × 105 cells/well in 6-well plates. After 36 h, G-LISA kits comprising 96-well plates coated with the binding domain of the corresponding GTPase effector protein and GTPase-GTP were detected using specific antibodies, followed by absorbance measurements at 490 nm.
In Vivo Experiments
Protocols for the following animal experiments were approved by the Animal Experiment Committee of the Institute of Medical Science, University of Tottori, Japan.
For the first in vivo experiments to assess the oncolytic effect of OVV-VGFΔ/O1LΔ, 5–10 × 106 cells KFlow and KFTX ovarian cancer cells stably expressing Renilla luciferase in 200 μL PBS were intraperitoneally injected into 5-week-old female BALB/cAJcl-nu/nu mice (Clea, Tokyo, Japan). To confirm tumor growth non-invasively, 150 μL ViviRen In Vivo Renilla Luciferase Substrate (0.925 mg/kg, Promega) was intraperitoneally injected before virus injection. Then, mice were administrated a single intraperitoneal injection of OVV-VGFΔ/O1LΔ in 200 μL PBS or control PBS. After virus injection, 200 μL VivoGlo Luciferin, In Vivo Grade (150 mg/kg, Promega) was injected subcutaneously on days 1, 2, 3, and 10 to monitor viral replication and spread. On day 11, tumor size was measured based on Renilla luciferase activity. Each luminescence image was obtained using the NightSHADE LB985 (Berthold, Tokyo, Japan) imaging system, and, during imaging, mice were anesthetized with isoflurane.
For the second in vivo experiments to develop a clinically relevant mouse tumor model, 5–10 × 106 KFlow and KFTX cells in 200 μL PBS were intraperitoneally injected into 5-week-old female BALB/cAJcl-nu/nu mice (Clea). After confirming tumor growth, PTX (10 mg/kg, Mylan, PA, USA) in 1 mL saline was intraperitoneally injected twice per week. Then 3 weeks later, tumor size was measured based on Renilla luciferase activity. On the following day, mice were administrated a single intraperitoneal injection of OVV-VGFΔ/O1LΔ in 200 μL PBS or control PBS. On day 11 after injection, tumor sizes were measured as stated.
For the isolation of KFTX and KFlow tumors, 5–10 × 106 cells in 200 μL PBS were intraperitoneally injected into 5-week-old female BALB/cAJcl-nu/nu mice. After confirming tumor growth, PTX (10 mg/kg, Mylan) in 1 mL saline was intraperitoneally administered twice per week. Then 3 weeks later, mice were sacrificed, and tumors were collected and frozen quickly using liquid nitrogen. Tumors were disrupted using lysis buffer (Nucleo SpinRNA) and a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan) and centrifuged at 2,000 × g for 30 s.
Statistical Analysis
Differences in cytolytic activity and in vivo viral replication between treatment groups were analyzed for statistical significance by performing one-way or two-way ANOVA, and a Bonferroni test when ANOVA showed overall significance. p values < 0.05 were considered statistically significant. Survival curves were constructed using the Kaplan-Meier method. Survival times were statistically analyzed by performing a log-rank test. Data were analyzed using GraphPad Prism v.5.
Author Contributions
T.N. supervised the entire project. T.N. and K. Horita designed the study and interpreted all results and concluded that UCA1 promotes OVV cell-to-cell spread in ovarian cancer, resulting in enhanced therapeutic outcome. T.N. and K. Horita also wrote the manuscript. K. Horita performed the majority of the experiments. H. Kurosaki, M.N., and N.K. constructed recombinant vaccinia viruses. H.I., T.O., and T.H. isolated and maintained paclitaxel-resistant ovarian cancer cell lines. K. Hasegawa and S.S. established primary cancer cell cultures. H. Kono and M.I. purified and titrated recombinant vaccinia viruses.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Dr. Yasuhiko Kiyozuka (Nara Medical University, Kashihara, Japan) for providing the SHIN-3 cells, Dr. Masahiko Nishiyama (Saitama Medical University, Hidaka, Japan) for providing RMG-1 cells, Dr. Takashi Iwata (Keio University, Shinjuku, Japan) for providing OVCAR3 and ES-2 cells, Dr. Seiji Mabuchi (Osaka University, Suita, Japan) for providing A2780 cells, and Dr. Yoshihiro Kikuchi (National Defense Medical College, Tokorozawa, Japan) for providing KFTX cells. This work was supported by JSPS-KAKENHI (15H04310 and 17K19597 to T.N.) and AMED (JP17lm0203039 to T.N.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.03.003.
Supplemental Information
References
- 1.Russell S.J., Peng K.W., Bell J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poh A. First oncolytic viral therapy for melanoma. Cancer Discov. 2016;6:6. doi: 10.1158/2159-8290.CD-NB2015-158. [DOI] [PubMed] [Google Scholar]
- 3.Fend L., Yamazaki T., Remy C., Fahrner C., Gantzer M., Nourtier V., Préville X., Quéméneur E., Kepp O., Adam J. Immune checkpoint blockade, immunogenic chemotherapy or IFN-α blockade boost the local and abscopal effects of oncolytic virotherapy. Cancer Res. 2017;77:4146–4157. doi: 10.1158/0008-5472.CAN-16-2165. [DOI] [PubMed] [Google Scholar]
- 4.Thorne S.H., Hwang T.H.H., O’Gorman W.E., Bartlett D.L., Sei S., Kanji F., Brown C., Werier J., Cho J.H., Lee D.E. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J. Clin. Invest. 2007;117:3350–3358. doi: 10.1172/JCI32727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downs-Canner S., Guo Z.S., Ravindranathan R., Breitbach C.J., O’Malley M.E., Jones H.L., Moon A., McCart J.A., Shuai Y., Zeh H.J., Bartlett D.L. Phase 1 study of intravenous oncolytic poxvirus (vvDD) in patients with advanced solid cancers. Mol. Ther. 2016;24:1492–1501. doi: 10.1038/mt.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S.H., Breitbach C.J., Lee J., Park J.O., Lim H.Y., Kang W.K., Moon A., Mun J.H., Sommermann E.M., Maruri Avidal L. Phase 1b trial of biweekly intravenous Pexa-Vec (JX-594), an oncolytic and immunotherapeutic vaccinia virus in colorectal cancer. Mol. Ther. 2015;23:1532–1540. doi: 10.1038/mt.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Chen W., Yang C., Wu W., Wu S., Qin X., Li X. Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int. J. Oncol. 2012;41:276–284. doi: 10.3892/ijo.2012.1443. [DOI] [PubMed] [Google Scholar]
- 9.Wang X.S., Zhang Z., Wang H.C., Cai J.L., Xu Q.W., Li M.Q., Chen Y.C., Qian X.P., Lu T.J., Yu L.Z. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 10.Liu F.T., Dong Q., Gao H., Zhu Z.M. The prognostic significance of UCA1 for predicting clinical outcome in patients with digestive system malignancies. Oncotarget. 2017;8:40620–40632. doi: 10.18632/oncotarget.16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong H.H., Hou L.K., Pan X., Wu C.Y., Huang H., Li B., Nie W. Long non-coding RNA UCA1 is a predictive biomarker of cancer. Oncotarget. 2016;7:44442–44447. doi: 10.18632/oncotarget.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian Z., Jin L., Zhang J., Yin Y., Quan C., Hu Y., Feng Y., Liu H., Fei B., Mao Y. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan J., Li X., Wu W., Xue M., Hou H., Zhai W., Chen W. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382:64–76. doi: 10.1016/j.canlet.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Wang G., Yang L., Qu J., Yang Z., Zhou X. Knockdown of long non-coding RNA UCA1 increases the tamoxifen sensitivity of breast cancer cells through inhibition of wnt/β-catenin pathway. PLoS ONE. 2016;11:e0168406. doi: 10.1371/journal.pone.0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Ying H.Q., He B.S., Pan Y.Q., Deng Q.W., Sun H.L., Chen J., Liu X., Wang S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C., Liang G., Yang S., Sui J., Yao W., Shen X., Zhang Y., Peng H., Hong W., Xu S. Dysregulated lncRNA-UCA1 contributes to the progression of gastric cancer through regulation of the PI3K-Akt-mTOR signaling pathway. Oncotarget. 2017;8:93476–93491. doi: 10.18632/oncotarget.19281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsley A.C., Weisberg A.S., Wagenaar T.R., Moss B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 2006;80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison K., Haga I.R., Pechenick Jowers T., Jasim S., Cintrat J.C., Gillet D., Schmitt-John T., Digard P., Beard P.M. Vaccinia virus uses retromer-independent cellular retrograde transport pathways to facilitate the wrapping of intracellular mature virions during virus morphogenesis. J. Virol. 2016;90:10120–10132. doi: 10.1128/JVI.01464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blasco R., Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter G.C., Rodger G., Murphy B.J., Law M., Krauss O., Hollinshead M., Smith G.L. Vaccinia virus cores are transported on microtubules. J. Gen. Virol. 2003;84:2443–2458. doi: 10.1099/vir.0.19271-0. [DOI] [PubMed] [Google Scholar]
- 21.Hollinshead M., Rodger G., Van Eijl H., Law M., Hollinshead R., Vaux D.J.T., Smith G.L. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 2001;154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mack N.A., Georgiou M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 2014;5:10. doi: 10.4161/21541248.2014.973768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobes C.D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 24.Hikichi M., Kidokoro M., Haraguchi T., Iba H., Shida H., Tahara H., Nakamura T. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol. Ther. 2011;19:1107–1115. doi: 10.1038/mt.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura, T. (2017). Mitogen-activated protein kinase-dependent recombinant vaccinia virus (MD-RVV) and use thereof. US Patent US9809803B2, filed November 20, 2014, and granted November 7, 2017.
- 26.Kanerva A., Nokisalmi P., Diaconu I., Koski A., Cerullo V., Liikanen I., Tähtinen S., Oksanen M., Heiskanen R., Pesonen S. Antiviral and antitumor T-cell immunity in patients treated with GM-CSF-coding oncolytic adenovirus. Clin. Cancer Res. 2013;19:2734–2744. doi: 10.1158/1078-0432.CCR-12-2546. [DOI] [PubMed] [Google Scholar]
- 27.Zamarin D., Palese P. Oncolytic Newcastle disease virus for cancer therapy: old challenges and new directions. Future Microbiol. 2012;7:347–367. doi: 10.2217/fmb.12.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirn D.H., Thorne S.H. Targeted and armed oncolytic poxviruses: a novel multi-mechanistic therapeutic class for cancer. Nat. Rev. Cancer. 2009;9:64–71. doi: 10.1038/nrc2545. [DOI] [PubMed] [Google Scholar]
- 29.Zeh H.J., Downs-Canner S., McCart J.A., Guo Z.S., Rao U.N.M., Ramalingam L., Thorne S.H., Jones H.L., Kalinski P., Wieckowski E. First-in-man study of western reserve strain oncolytic vaccinia virus: safety, systemic spread, and antitumor activity. Mol. Ther. 2015;23:202–214. doi: 10.1038/mt.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mell L.K., Brumund K.T., Daniels G.A., Advani S.J., Zakeri K., Wright M.E., Onyeama S.J., Weisman R.A., Sanghvi P.R., Martin P.J., Szalay A.A. Phase I trial of intravenous oncolytic vaccinia virus (GL-ONC1) with cisplatin and radiotherapy in patients with locoregionally advanced head and neck carcinoma. Clin. Cancer Res. 2017;23:5696–5702. doi: 10.1158/1078-0432.CCR-16-3232. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Cao X., Zhang L., Zhang X., Sheng H., Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother. Pharmacol. 2016;77:629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 32.Li J.Y., Ma X., Zhang C.B. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F., Zhou J., Xie X., Hu J., Chen L., Hu Q., Guo H., Yu C. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015;62:432–438. [PubMed] [Google Scholar]
- 34.Wang J., Ye C., Liu J., Hu Y. UCA1 confers paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis. Biochem. Biophys. Res. Commun. 2018;501:1034–1040. doi: 10.1016/j.bbrc.2018.05.104. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z.Q., He C.Y., Hu L., Shi H.P., Li J.F., Gu Q.L., Su L.P., Liu B.Y., Li C., Zhu Z. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi: 10.1016/j.canlet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Andrade A.A., Silva P.N., Pereira A.C., De Sousa L.P., Ferreira P.C., Gazzinelli R.T., Kroon E.G., Ropert C., Bonjardim C.A. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 2004;381:437–446. doi: 10.1042/BJ20031375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breiman A., Smith G.L. Vaccinia virus B5 protein affects the glycosylation, localization and stability of the A34 protein. J. Gen. Virol. 2010;91:1823–1827. doi: 10.1099/vir.0.020677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphries A.C., Donnelly S.K., Way M. Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J. Cell Sci. 2014;127:673–685. doi: 10.1242/jcs.141366. [DOI] [PubMed] [Google Scholar]
- 39.Miki H., Sasaki T., Takai Y., Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 40.Mercer J., Knébel S., Schmidt F.I., Crouse J., Burkard C., Helenius A. Vaccinia virus strains use distinct forms of macropinocytosis for host-cell entry. Proc. Natl. Acad. Sci. USA. 2010;107:9346–9351. doi: 10.1073/pnas.1004618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malacombe M., Ceridono M., Calco V., Chasserot-Golaz S., McPherson P.S., Bader M.F., Gasman S. Intersectin-1L nucleotide exchange factor regulates secretory granule exocytosis by activating Cdc42. EMBO J. 2006;25:3494–3503. doi: 10.1038/sj.emboj.7601247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanderson C.M., Way M., Smith G.L. Virus-induced cell motility. J. Virol. 1998;72:1235–1243. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handa Y., Durkin C.H., Dodding M.P., Way M. Vaccinia virus F11 promotes viral spread by acting as a PDZ-containing scaffolding protein to bind myosin-9A and inhibit RhoA signaling. Cell Host Microbe. 2013;14:51–62. doi: 10.1016/j.chom.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Cordeiro J.V., Guerra S., Arakawa Y., Dodding M.P., Esteban M., Way M. F11-mediated inhibition of RhoA signalling enhances the spread of vaccinia virus in vitro and in vivo in an intranasal mouse model of infection. PLoS ONE. 2009;4:e8506. doi: 10.1371/journal.pone.0008506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stepanenko A.A., Chekhonin V.P. A compendium of adenovirus genetic modifications for enhanced replication, oncolysis, and tumor immunosurveillance in cancer therapy. Gene. 2018;679:11–18. doi: 10.1016/j.gene.2018.08.069. [DOI] [PubMed] [Google Scholar]
- 46.Lapp S., Pfankuche V.M., Baumgärtner W., Puff C. Viral oncolysis - can insights from measles be transferred to canine distemper virus? Viruses. 2014;6:2340–2375. doi: 10.3390/v6062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong P., Agosto L.M., Munro J.B., Mothes W. Cell-to-cell transmission of viruses. Curr. Opin. Virol. 2013;3:44–50. doi: 10.1016/j.coviro.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q., Liang C., Yu Y.A., Chen N., Dandekar T., Szalay A.A. The highly attenuated oncolytic recombinant vaccinia virus GLV-1h68: comparative genomic features and the contribution of F14.5L inactivation. Mol. Genet. Genomics. 2009;282:417–435. doi: 10.1007/s00438-009-0475-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imai S., Maeda H., Kiyozuka Y., Noda T., Moriyama I., Ichijo M. Characterization of the CA125 antigen secreted from a newly established human ovarian cancer cell line (SHIN-3) Acta Pathol. Jpn. 1989;39:43–49. doi: 10.1111/j.1440-1827.1989.tb02401.x. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu M., Hiyama K., Tanimoto K., Yunokawa M., Otani K., Ohtaki M., Hiyama E., Kigawa J., Ohwada M., Suzuki M. Prediction of individual response to platinum/paclitaxel combination using novel marker genes in ovarian cancers. Mol. Cancer Ther. 2006;5:767–775. doi: 10.1158/1535-7163.MCT-05-0408. [DOI] [PubMed] [Google Scholar]
- 51.Nishio H., Yaguchi T., Sugiyama J., Sumimoto H., Umezawa K., Iwata T., Susumu N., Fujii T., Kawamura N., Kobayashi A. Immunosuppression through constitutively activated NF-κB signalling in human ovarian cancer and its reversal by an NF-κB inhibitor. Br. J. Cancer. 2014;110:2965–2974. doi: 10.1038/bjc.2014.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto K., Kikuchi Y., Kudoh K., Nagata I. Modulation of cisplatin sensitivity by taxol in cisplatin-sensitive and -resistant human ovarian carcinoma cell lines. J. Cancer Res. Clin. Oncol. 2000;126:168–172. doi: 10.1007/s004320050027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mabuchi S., Ohmichi M., Nishio Y., Hayasaka T., Kimura A., Ohta T., Saito M., Kawagoe J., Takahashi K., Yada-Hashimoto N. Inhibition of NFkappaB increases the efficacy of cisplatin in in vitro and in vivo ovarian cancer models. J. Biol. Chem. 2004;279:23477–23485. doi: 10.1074/jbc.M313709200. [DOI] [PubMed] [Google Scholar]
- 54.Tamada Y., Takeuchi H., Suzuki N., Susumu N., Aoki D., Irimura T. Biological and therapeutic significance of MUC1 with sialoglycans in clear cell adenocarcinoma of the ovary. Cancer Sci. 2007;98:1586–1591. doi: 10.1111/j.1349-7006.2007.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.