Highlights
-
•
UCA1 is upregulated in both osteosarcoma tissues and cell lines.
-
•
UCA1 promotes osteosarcoma metastasis both in vitro and in vivo.
-
•
UCA1 increased CREB1 expression by functioning as a ceRNA of CREB1 against miR-582.
-
•
The pro-metastasis role of UCA1 is achieved by promoting EMT.
-
•
UCA1 enhances EMT through CREB1-mediated the PI3K/Akt/mTOR pathway.
Keywords: Osteosarcoma, UCA1, miR-582, CREB1, Metastasis, ceRNA
Abstract
Increasing evidences have demonstrated that Long noncoding RNAs (lncRNAs) are key regulatory RNAs that participate in multiple biological processes. LncRNA urothelial carcinoma-associated 1 (UCA1) is a newly identified lncRNA and functions as a regulator of growth in several cancers. However, the biological function and molecular mechanism of UCA1 in the metastasis of osteosarcoma remain unclear. In this study, we firstly found UCA1 is upregulated in both osteosarcoma tissues and cell lines, and increased UCA1 is associated with higher tumor stage, larger tumor size and poorer prognosis. Then for the first time, we demonstrated that UCA1 promotes the invasion and metastasis of osteosarcoma both in vitro and in vivo. Further mechanistic investigation showed that UCA1 directly interactes with miR-582 and suppresses its expression. Moreover, UCA1 increases CREB1 expression by functioning as a ceRNA against miR-582, thus promoting the EMT process via CREB1-mediated PI3K/AKT/mTOR pathway and finally leading to osteosarcoma metastasis. These findings may extend the function of UCA1 in osteosarcoma progression and provide a promising therapeutic target for osteosarcoma treatment.
1. Introduction
Osteosarcoma is the most frequent primary bone tumor which mainly occurs in adolescents and children [1]. In the past few decades, despite advancements in complete surgical resection and neoadjuvant chemotherapy for osteosarcoma patients, the prognosis is still unfavorable due to the high rate of metastasis. The 5-year survival rate of patients with osteosarcoma having distant metastases is approximately 30% [2]. Therefore, understanding of the molecular mechanisms of osteosarcoma metastasis is important to improve the prognosis of osteosarcoma patients.
Long noncoding RNAs (lncRNAs) are typically longer than 200 nucleotides with little or no protein coding capacity. Accumulating evidences indicate that lncRNAs are aberrantly expressed in many human tumors and play critically important roles in diverse biological processes, such as proliferation, apoptosis and metastasis [3]. LncRNAs regulate gene expressions through multiple mechanisms, including modulating transcription, indirect degradation of RNA transcripts and regulating various post-transcriptional processes [4].
Urothelial carcinoma-associated 1 (UCA1), which is a novelly identified lncRNA, is widely expressed in various tumors and plays key roles in multiple biological processes [5]. Previous studies have demonstrated that UCA1 was highly expressed in ovarian cancer [6], non-small cell lung cancer [7] and prostate cancer [8]. UCA1 was initially found in bladder cancer and could enhance the tumorigenicity of cancer cells both in vitro and in vivo [9]. Bioinformatic analyses showed that UCA1 was highly expressed in tumor tissues from 361 gastric cancer patients and acted as an oncogene to promote cell proliferation. UCA1 also functions as a tumor promoter in colorectal cancer [10]. It was reported that high expression of UCA1 was associated with tumor proliferation and indicated worse prognosis in colorectal cancer [5]. In osteosarcoma, previous study has found that UCA1 was overexpressed in osteosarcoma and promoted cell growth. However, there has been no research focusing on the role of UCA1 in osteosarcoma metastasis. Therefore, to explore how UCA1 participates in osteosarcoma progression, especially its effect on the invasion and migration of osteosarcoma cells, aiming at discovering novel targets for treatment becomes our goal of study.
In this study, we validated that the UCA1 was significantly upregulated in osteosarcoma and, for the first time, we confirmed that UCA1 promotes the metastasis of osteosarcoma both in vitro and in vivo. Also, we are the first to demonstrate that UCA1 interacts with and suppresses miR-582 as a potential ceRNA. The pro-metastatic effect of UCA1 is achieved by inhibiting the epithelial-mesenchymal transition (EMT) via CREB1-mediated PI3K/AKT/mTOR pathway.
2. Materials and methods
2.1. Cell culture and tissue samples
Normal osteoblast cell line hFOB1.19, osteosarcoma cell lines Saos-2, HOS, U2-OS and MG-63 were obtained from the Cell Bank of Chinese Academy Sciences. The hFOB1.19 cells were cultured in d-MEM/F-12, Saos-2 in McCoy's 5A, U2-OS cells in RPMI-1640 and HOS cells in MEM. All of the media and the supplemented 10% fetal bovine serum were from Gibco. All cells were incubated at 37 °C in a 5% CO2 atmosphere.
Human tissue samples of paired normal and primary tumor tissues from 30 osteosarcoma patients were collected from No. 215 Hospital of Shannxi Nuclear Industry. The study was approved by the Medical Ethics Committee and written informed consents were obtained from all patients.
2.2. RNA extraction and quantitative real-time PCR (qPCR)
Total RNA from both tissue samples and cell lines were extracted using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized from 500 ng of total RNA using the PrimeScript RT Reagent Kit (TaKaRa) for detecting mRNA, lncRNA and miRNA levels. Real-time PCR was performed in triplicate using SYBR Premix Ex Taq (TaKaRa) on CFX96 Real-Time PCR Detection System (BioRad). Expression levels of mRNAs, lncRNAs and miRNAs were normalized to GAPDH, 18S rRNA and U6, respectively.
2.3. Oligonucleotides, plasmids construction and cell transfection
The Control/UCA1 shRNA and the full-length human UCA1 sequence were synthesized by Genscript (Nanjing, China) and then sub-cloned into pLKO.1 and pCDNA3.1 vectors, respectively. According to the manufacturer's instructions, cells were transfected with Control/UCA1 shRNA vectors or Control/UCA1 vectors using Lipofectamine® 2000 reagent (Invitrogen) at final concentrations of 2 µg/ml for 48 h.
For stable overexpression of UCA1, the synthesized full-length UCA1 cDNA was cloned into lentiviral expressing vector pLV-puro. After production of lentiviral particles according to the standard protocols, cells were transfected for 24 h with 2 µg/ml polybrene (Sigma) and selected by using 2 µg/ml puromycin for 7 days.
The synthetic miR-NC/miR-582 and inhibitor NC/miR-582 inhibitor purchased from GenePharma (Shanghai, China). All of the oligonucleotides were transfected into cells using Lipofectamine® 2000 reagent (Invitrogen) at final concentrations of 50 nM.
2.4. CCK8 assay and apoptosis assay
Cell proliferation was detected by a Cell Counting Kit-8 (CCK-8) assay (Dojindo) as manufacturer's instructions. Cells were seeded into 96-well plates (2 × 103 cells/well) for 1, 2, 3 and 4 days’ observation. Cell apoptosis was detected by using Apoptosis Detection Kit (KeyGEN) according to the manufacturer's instructions. Cells were stained with florescein isothiocyanate-conjugated Annexin V and PI and analyzed with a FACScan flow cytomete.
2.5. Wound-healing assay and matrigel invasion assay
For the wound-healing assays, wound closures were observed by taking photographs under a microscope 0 and 48 h after scratching. Matrigel invasion assays were performed with Matrigel (BD) and 8 µm, 24-well trans-well chambers (Millipore) following the manufacturer's instructions. Cells (1 × 105) in 200 µl of serum-free medium were added to the upper chamber and cultured for 48 h. Migrated cells were fixed with ethanol and stained with 0.1% crystal violet and photographs were taken of 6 randomly selected fields for each well.
2.6. Western blot analysis
Western blotting was performed according to the standard protocol. In brief, cell lysates quantified with the BCA method were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene floride (PVDF) membrane. The PVDF membrane was incubated with 1:1000 primary antibodies and then 1:5000 goat anti-mouse or goat anti-rabbit IgG-HRP conjugate secondary antibodies (Akea). Blots were detected by ECL system (Alpha Innotech) according to the manufacturer's instructions.
2.7. Animal models
To evaluate the lung metastatic potential of osteosarcoma cells in vivo, 5 × 106 U2-OS/Lenti-Control or U2-OS/Lenti-UCA1 cells in 200 µl of serum-free medium were injected into six-week-old, male nude mice through the tail vein (n = 6 per group). Four weeks later, the mice were sacrificed. Individual organs from the mice were removed, and metastatic tissues were analyzed with H&E staining.
2.8. Luciferase reporter assay
The UCA1 WT/MT 3′-UTR and CREB1 WT/MT 3′-UTR sequences were cloned into pmirGLO plasmid, respectively. The vectors (2 µg/ml) were co-transfected with miR-NC/miR-582 or inhibitor NC/miR-582 inhibitor (50 nM) into cells by Lipofectamine® 2000 reagent (Invitrogen). Luciferase activity was determined by the Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
2.9. RNA immunoprecipitation (RIP) assay
Cells were co-transfected with pLV-MS2 or pLVUCA1-WT-MS2 or pLV-UCA1-MT-MS2 and pMS2-GFP (Addgene). After 48 h of transfection, cells were subjected to a RIP assay by using 5 µg GFP antibody (Sigma-Aldrich) or negative control IgG using RNA Immunoprecipitation Kit (Millipore) according to the manufacturer's instructions. For anti-AGO2 RIP, cells were transfected with miR-NC/miR-582. After 48 h of transfection, cells were used to perform anti-AGO2 RIP assay using 5 µg anti-AGO2 antibody (Millipore) as described above.
2.10. Statistics analysis
Data were analyzed using the SPSS 19.0 software and expressed as the mean ± standard error of the mean (s.e.m) from at least three separate experiments. Two tailed Student's t-tests were used to evaluate statistical significance between two independent groups of samples. The significance of correlations between miR-582 and UCA1 levels were judged via the Pearson product-moment correlation coefficient. Differences were considered significant when *p < 0.05 and ***p < 0.001.
3. Results
3.1. UCA1 is upregulated in osteosarcoma and promotes its growth
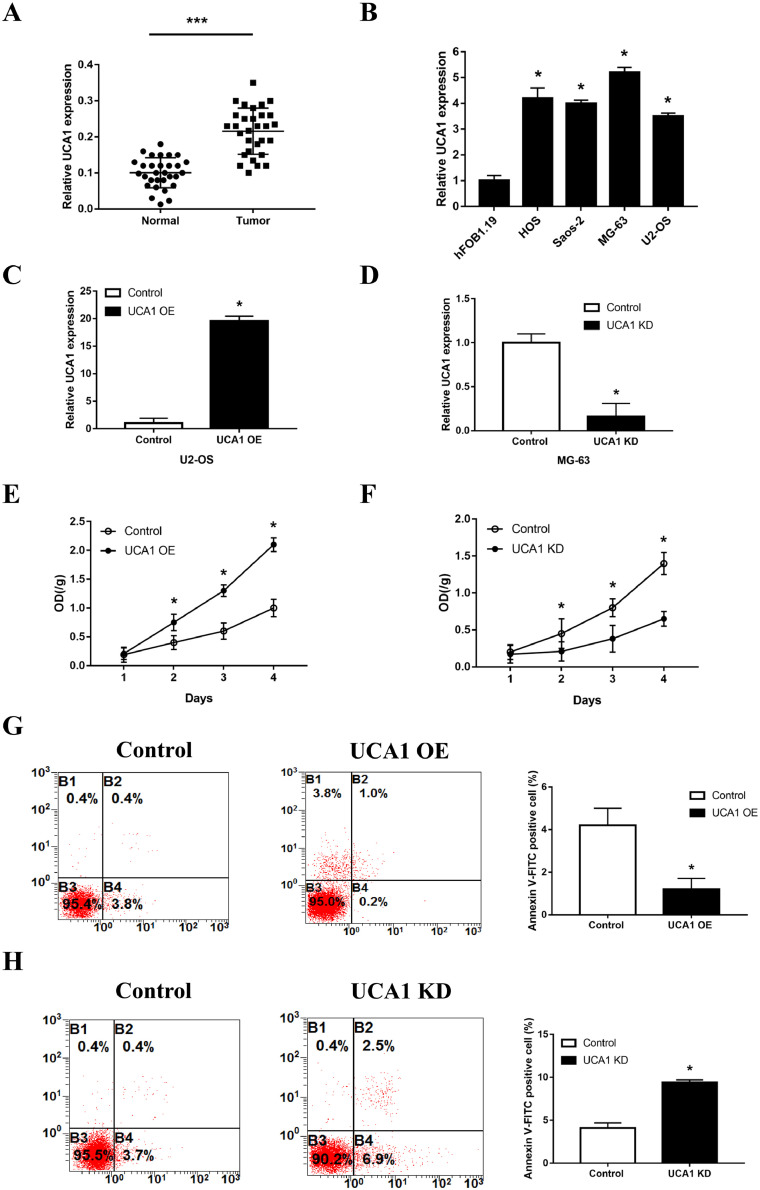
To detect the expression level of UCA1 in osteosarcoma, strictly paired primary tumor tissues and normal tissues were collected from 30 osteosarcoma patients. As displayed in qPCR assays, UCA1 expression was significantly upregulated in osteosarcoma tissues, with the average level over two folds higher than that in normal tissues (Fig. 1A). Similarly, compared with the normal osteoblast cell line hFOB1.19, osteosarcoma cell lines Saos-2, HOS, U2-OS and MG-63 all showed increased level of UCA1 (Fig. 1B). The following clinical analysis showed that increased UCA1 expression was significantly associated with metastasis, larger tumor size and higher clinical stage of osteosarcoma (Table 1). Given the upregulated UCA1 in both the tumor tissues and cell lines, we deduced that UCA1 may promote the growth of osteosarcoma. To clarify this, U2-OS cells, which had the lowest level of UCA1, were transfected with pHBLV Control/UCA1 plasmid to overexpress UCA1 (Fig. 1C) and MG-63 cells, which had the highest level of UCA1, were transfected with Control/UCA1 shRNA to silence the UCA1 expression (Fig. 1D). Results of CCK8 assays showed that UCA1 overexpression significantly promoted cell proliferation (Fig. 1E) and there was a significant repression of growth in UCA1 knockdown group (Fig. 1F) compared with the control groups respectively. Moreover, we found that overexpression of UCA1 markedly inhibited the apoptosis, as shown by an increase of Annexin V-PE positive cells (Fig. 1G), whereas knockdown of UCA1 increased the cell apoptosis (Fig. 1H). These results suggested that UCA1 promotes the growth of osteosarcoma cells by increasing proliferation and decreasing apoptosis.
Fig. 1.
UCA1 is upregulated in osteosarcoma and promotes its growth. (A) Expression levels of UCA1 in paired normal and tumor tissues from 30 osteosarcoma patients detected by qRT-PCT (p<0.0001). (B-D) Expression levels of UCA1 in osteosarcoma cell lines (B, HOS vs hFOB1.19, p = 0.0002; Saos-2 vs hFOB1.19, p<0.0001; MG-63 vs hFOB1.19, p<0.0001; U2-OS vs hFOB1.19, p<0.0001), U2-OS cells transfected with pHBLV Control/UCA1 vectors (C, p<0.0001) and MG-63 cells transfected with Control/UCA1 shRNA (D, p = 0.0013) detected by qRT-PCT. (E-H) CCK8 assays (E, F) and apoptosis assays (G, H) carried out in U2-OS cells transfected with pHBLV Control/UCA1 vectors (E, UCA1 overexpression vs control: day2 p = 0.0303, day3 p = 0.0021; day4 p = 0.0006; G, p = 0.0053) and MG-63 cells transfected with Control/UCA1 shRNA (F, UCA1 knockdown vs control: day2 p = 0.0463, day3 p = 0.0282; day4 p = 0.002; H, p = 0.0002). * P < 0.05 and *** P < 0.001 (Student's t-test).
Table 1.
The relationship between UCA1 expression and clinicopathological features in osteosarcoma patients.
| UCA1 expression levels |
|||
|---|---|---|---|
| Low | High | P | |
| Age (years) | |||
| ≤ 18 | 8 | 9 | 0.713 |
| > 18 | 7 | 6 | |
| Gender | |||
| Female | 9 | 10 | 0.705 |
| Male | 6 | 5 | |
| Location | |||
| Femur/ Tibia | 11 | 10 | 0.69 |
| Elsewhere | 4 | 5 | |
| Tumor size (cm) | |||
| ≤ 5 | 12 | 6 | 0.025 |
| > 5 | 3 | 9 | |
| Stage | |||
| Ⅰ+Ⅱ A | 10 | 2 | 0.003 |
| Ⅱ B/Ⅲ | 5 | 13 | |
| Metastasis | |||
| Yes | 4 | 12 | 0.003 |
| No | 11 | 3 | |
P value was acquired by Pearson chi-square test. The median expression level was used as the cutoff.
3.2. UCA1 promotes the invasion and migration of osteosarcoma both in vitro and in vivo
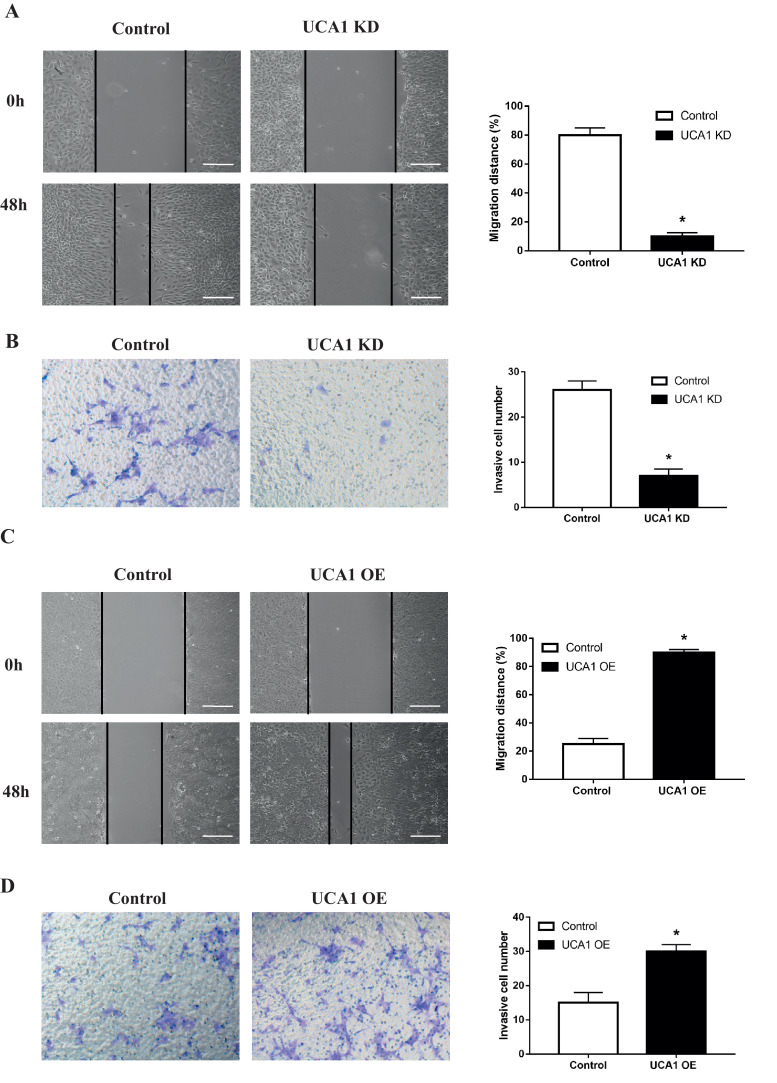
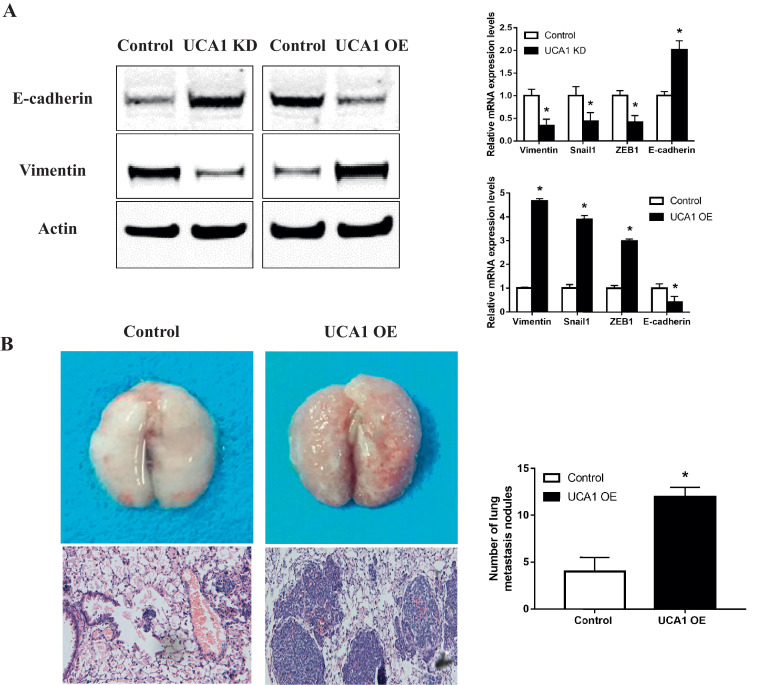
To test the effect of UCA1 on the invasion and migration of osteosarcoma cells, we introduced wound-healing and matrigel invasion assays as well as the Epithelial-Mesenchymal Transition (EMT) markers detection into U2-OS and MG-63 cells treated in the same way as above. We observed that when the endogenous UCA1 was knocked down, MG-63 cells showed decreased capability of migrating to the monolayer of wounded cells (Fig. 2A) and invading through the matrigel treated transwell chambers (Fig. 2B). At the same time, the expression of epithelial marker E-cadherin was upregulated both in mRNA and protein levels, while the mesenchymal markers of Vimentin, Snail1 and ZEB1 were downregulated (Fig. 3A). On the contrary, when the ectopic UCA1 was expressed, U2-OS cells presented increased migration and invasion ability compared with control cells (Fig. 2C and 2D) and the expression pattern of EMT markers were also opposite (Fig. 3A). To further explore the pro-metastasis effect of UCA1 in vivo, we established U2-OS cells that can stably express UCA1 using lentiviral particles and injected these cells into the tail vein of nude mice. Four weeks later, we observed that compared to the control group, U2-OS cells overexpressing UCA1 had stronger capability of invading and migrating to the lung, indicated by more (with the average number of metastasis nodules in UCA1 overexpression group three times higher than that in control group) and larger metastatic nodules in the H&E staining of tissue sections observed under microscope (Fig. 3B). In summary, these data suggested that UCA1 promotes the invasion and migration of osteosarcoma cells both in vitro and in vivo.
Fig. 2.
UCA1 promotes the invasion and migration of osteosarcoma in vitro. Wound-healing assays (A, C) and metrigel invasion assays (B, D) in MG-63 cells transfected with Control/UCA1 shRNA (A, p<0.0001; B, p = 0.0002) and U2-OS cells transfected with pHBLV Control/UCA1 vectors (C, p<0.0001; D, p = 0.0002). * P < 0.05 (Student's t-test).
Fig. 3.
UCA1 promotes the invasion and migration of osteosarcoma in vivo. (A) Western blot and qPCR analyses of EMT markers in MG-63 cells transfected with Control/UCA1 shRNA and U2-OS cells transfected with pHBLV Control/UCA1 vectors (UCA1 knockdown vs control: Vimentin p = 0.0045, Snail1 p = 0.024, ZEB1 p = 0.0054 and E-cadherin p = 0.0013; UCA1 overexpression vs control: Vimentin p<0.0001, Snail1 p<0.0001, ZEB1 p<0.0001 and E-cadherin p = 0.0297). (B) Lungs of nude mice in U2-OS/Lenti-Control and U2-OS/Lenti-UCA1 groups 4 weeks after tail vein injection as labeled by H&E staining. The number of metastatic nodules per nude mouse was counted under a microscope (p = 0.0015). * P < 0.05 (Student's t-test).
3.3. UCA1 directly interacts with miR-582 in osteosarcoma cells
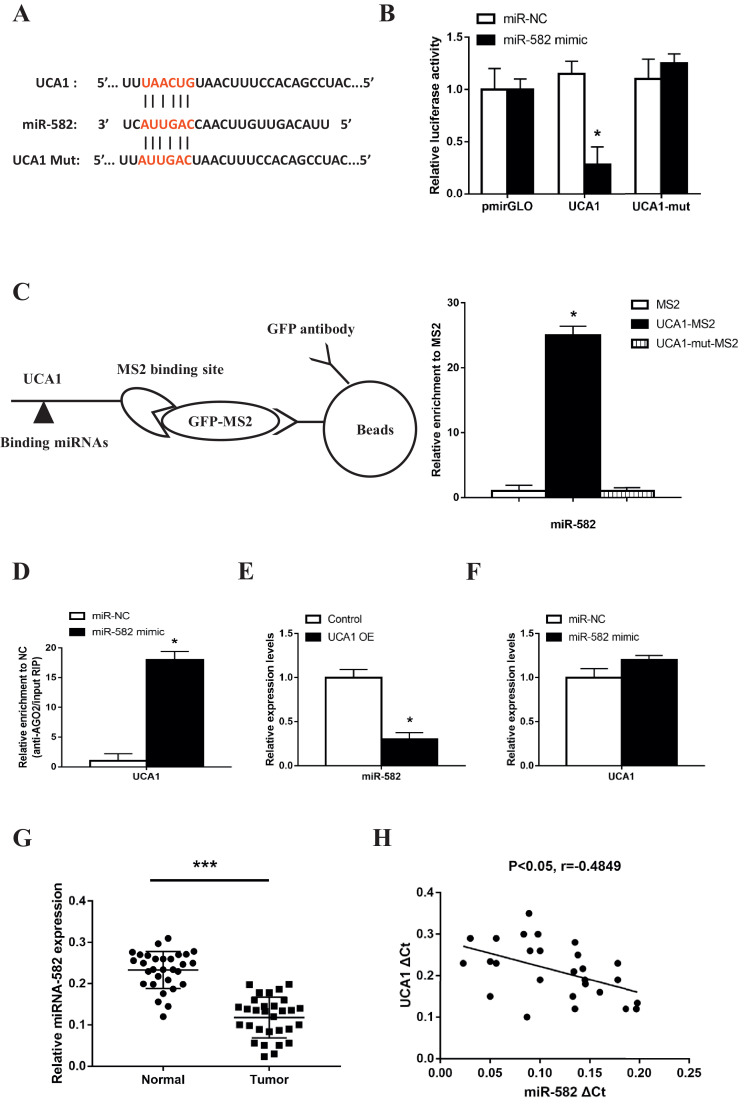
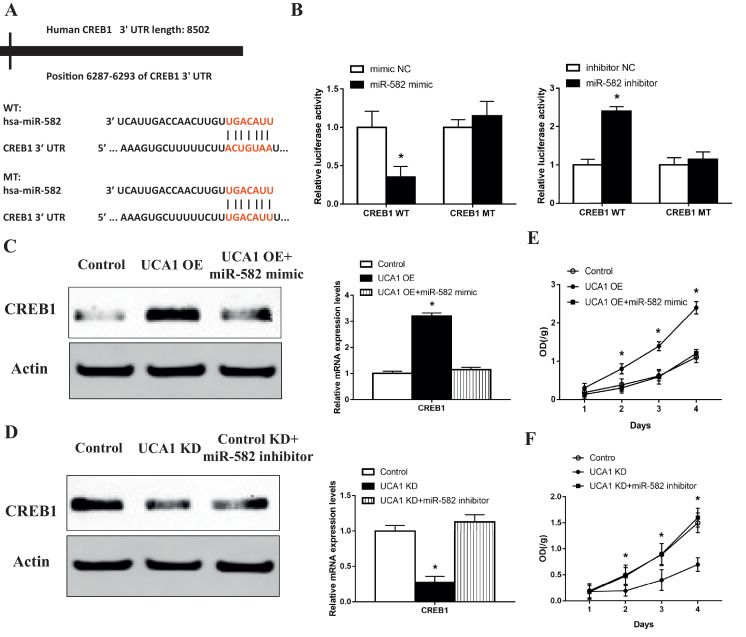
Recent studies have indicated that lncRNAs exert regulatory functions through targeting miRNAs which bind to their 3′-untranslational regions (3′-UTR). After screening and comparison using online software program miRanda (http://mircrorna.org), we found that miR-582 had complementary base pairing with UCA1 (Fig. 4A). We constructed reporter vectors (pmirGLO) containing the wild type (UCA1) and mutated 3′-UTR (UCA1 Mut) of UCA1 where miR-582 is binding. Results of the luciferase reporter assay showed that when we transfected 293T cells with miR-582 mimics together with UCA1 vector, the luciferase activity significantly decreased whereas in the empty vector or UCA1-mut groups, no changes were observed (Fig. 4B). Next we confirmed the direct interaction between UCA1 and miR-582 using RIP assay to pull down the endogenous miRNAs that interact with UCA1 (Fig. 4C). The following qPCR analysis showed that compared to MS2 empty vector and UCA1-mut group, the UCA1 RIP was significantly enriched for miR-582 in U2-OS cells (Fig. 4C). Since it is well defined that miRNAs bind to their targets and induce translational repression and/or RNA degradation through AGO2, a core component of the RNA-induced silencing complex (RISC), we performed anti-AGO2 RIP assay. Results showed high UCA1 level in miR-582-overexpressing MG-63 cells when we use AGO2 to pull down the endogenous lncRNAs, suggesting the direct binding of miR-582 to UCA1 (Fig. 4D). Furthermore, we found that ectopically expressed UCA1 significantly reduced the miR-582 level (Fig. 4E) but overexpression of miR-582 did not alter the UCA1 level (Fig. 4F), indicating that miR-582 could bind to UCA1 but did not induce its degradation. Then we found the miR-582 level was significantly decreased, with the average expression 47.8% lower in tumor tissues than that in the corresponding normal tissues from 30 osteosarcoma patients (Fig. 4G), and the level of miR-582 was negatively correlated with the level of UCA1 in the tumor tissues (Fig. 4H). All of the above data showed that UCA1 directly interacts with UCA1 in osteosarcoma cells.
Fig. 4.
UCA1 directly interacts with miR-582 in osteosarcoma cells. (A) Diagram for the predicted binding site of miR-582 on the WT or Mut 3′-UTR of UCA1. (B) Luciferase reporter assays in 293T cells transfected with miR-NC/miR-582, together with pmirGLO empty vector or reporter vectors containing UCA1/UCA1-mut 3′-UTR (miR-582 vs miR-NC: pmirGLO p>0.99, UCA1 p>0.0019, UCA1 Mut p>0.284). (C) Diagram and the qPCR analysis of the RIP assay performed in U2-OS cells to pull down the miRNAs associated with UCA1 (p<0.0001). (D) Anti-AGO2 RIP assay performed in MG-63 cells transfected with miR-NC/miR-582 followed by qPCR analysis (p<0.0001). (E, F) Expression levels of miR-582 (E) and UCA1 (F) detected by qPCR in UCA1 overexpressing U2-OS cells (E, p = 0.0005) and MG-63 cells transfected with miR-NC/miR-582 (F, p = 0.0663). (G) Expression levels of miR-582 in paired normal and tumor tissues from 30 osteosarcoma patients detected by qRT-PCT (p<0.0001). (H) The correlation between UCA1 and miR-582 expressions in 30 osteosarcoma tissues (p = 0.0104). * P < 0.05 and *** P < 0.001 (Student's t-test).
3.4. UCA1 functions as a ceRNA to upregulate CREB1 and promotes the metastasis of osteosarcoma cells through the AKT pathway
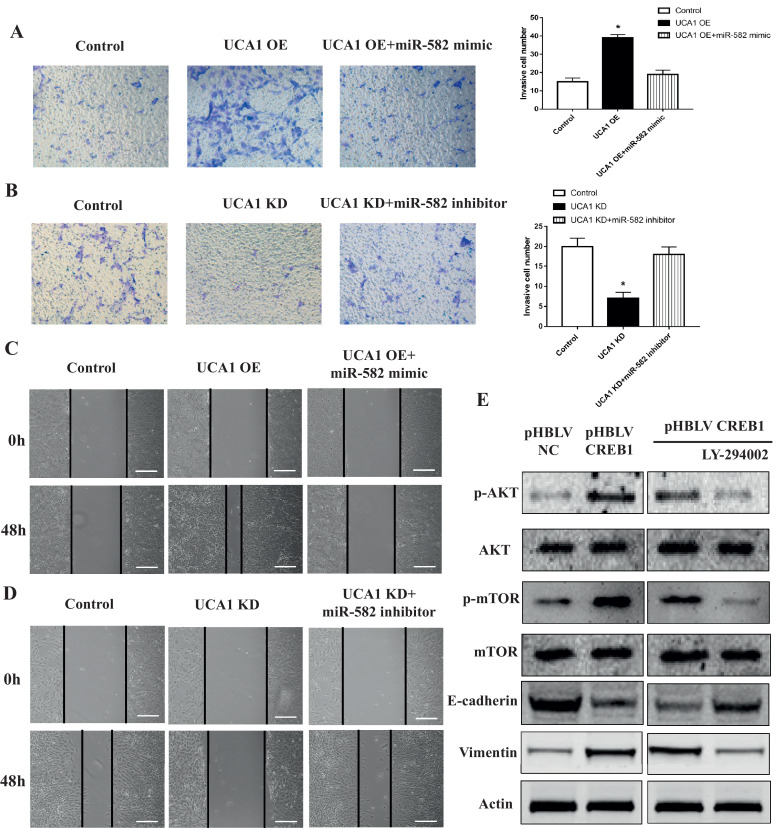
To further explore which gene related to the EMT of osteosarcoma cells is mediating the pro-metastatic effect of UCA1 through interaction with miR-582, we scanned the possible target genes of miR-582 via the publicly available algorithms and focused on the gene CAMP Responsive Element Binding Protein 1 (CREB1). We analyzed the 3′-UTR of CREB1 which includes the putative binding site of miR-582 (WT 3′-UTR) and synthesized the mutated binding site (MT 3′-UTR) (Fig. 5A). After we sub-cloned these two 3′-UTR sequences into the reporter vector, we found the luciferase activity significantly decreased in miR-582 overexpressing MG-63 cells co-transfected with CREB1 WT vector and notably increased in U2-OS cells co-treated with miR-582 inhibitor and CREB1 WT vector compared to control groups (Fig. 5B) compared with the MT 3′-UTR groups, offering a direct evidence of miR-582 targeting CREB1. Then we found that overexpression of UCA1 increased the level of CREB1 in both the protein and mRNA levels, while the co-transfection of miR-582 abrogated this upregulation (Fig. 5C). Meanwhile, we also observed increased proliferation, invasion and migration in UCA1 overexpressing U2-OS cells (Figs. 5E, 6A and C). Similarly, the ectopic expression of miR-582 neutralized the enhancement of these malignent behaviors (Figs. 5E and 6A, C). On the contrary, we found the depletion of UCA1 decreased the expression of CREB1 and the inhibition of miR-582 overcame this suppression (Fig. 5D). Also, the proliferation, invasion and migration of MG-63 cells were decreased with the downregulation of UCA1 and restored with the following transfection of miR-582 inhibitor (Figs. 5F, 6B and D). All of the data suggested that UCA1 functions as a ceRNA to upregulate CREB1 in osteosarcoma cells.
Fig. 5.
UCA1 functions as a ceRNA to upregulate CREB1. (A) Diagram of CREB1 3′-UTR containing the putative conserved target sequence for miR-582. (B) Results of luciferase reporter assays in MG-63 (left) and U2-OS cells (right) co-transfected with CREB1 WT/MT 3′UTR vectors and miR-NC/miR-145 (left) or inhibitor NC/miR-145 inhibitor (right) (miR-582 mimic vs mimic NC: CREB1 WT p = 0.0112, CREB1 MT p = 0.2929; miR-582 inhibitor vs inhibitor NC: CREB1 WT p = 0.0002, CREB1 MT p = 0.3883). (C-F) Expression levels of CREB1 (C, D) CCK8 assays (E, F) in U2-OS cells co-transfected with pHBLV control/pHBLV UCA1 vectors and miR-NC/miR-582 (C, UCA1 overexpression vs control p<0.0001, UCA1 overexpression+miR-582 mimic vs control p = 0.1108; E, day2: UCA1 overexpression vs control p = 0.0239, UCA1 overexpression+miR-582 mimic vs control p = 0.548; day3: UCA1 overexpression vs control p = 0.0016, UCA1 overexpression+miR-582 mimic vs control p = 0.882; day4: UCA1 overexpression vs control p = 0.0004, UCA1 overexpression+miR-582 mimic vs control p = 0.371) and in MG-63 cells co-transfected with Control shRNA/UCA1 shRNA and inhibitor NC/miR-582 inhibitor (D, UCA1 knockdown vs control p = 0.0005, UCA1 knockdown+miR-582 inhibitor vs control p = 0.1535; F, day2: UCA1 knockdown vs control p = 0.0667, UCA1 knockdown+miR-582 inhibitor vs control p = 0.896; day3: UCA1 knockdown vs control p = 0.043, UCA1 knockdown+miR-582 inhibitor vs control p = 0.955; day4: UCA1 knockdown vs control p = 0.0038, UCA1 knockdown+miR-582 inhibitor vs control p = 0.544). * P < 0.05 (Student's t-test).
Fig. 6.
UCA1 functions as a ceRNA to upregulate CREB1 and promotes the metastasis of osteosarcoma cells through the AKT pathway. Metrigel invasion assays (A, B) and wound-healing assays (C, D) in U2-OS cells co-transfected with pHBLV control/pHBLV UCA1 vectors and miR-NC/miR-582 (A, UCA1 overexpression vs control p = 0.0001, UCA1 overexpression+miR-582 mimic vs control p = 0.09; C) and in MG-63 cells co-transfected with Control shRNA/UCA1 shRNA and inhibitor NC/miR-582 inhibitor (B, UCA1 knockdown vs control p = 0.0001, UCA1 knockdown+miR-582 inhibitor vs control p = 0.288; D). (E) Western blot analyses of total/p-AKT, total/p-mTOR, E-cadherin and Vimentin in U2-OS cells co-transfected with pHBLV control/pHBLV CREB1 vectors (left) and CREB1 overexpressing U2-OS cells treated with DMSO (NC)/LY-294002 (right). * P < 0.05 (Student's t-test).
To further study how CREB1 mediates the pro-metastatic effect of its ceRNA UCA1, we tested the activity of AKT/mTOR pathway, which is closely related to the EMT and metastasis of osteosarcoma cells. Western blot analyses showed that when we overexpressed CREB1 in U2-OS cells, both of the phosphorylated AKT and mTOR increased compared to the empty vector group, indicating the activation of this pathway (Fig. 6E, left). Meanwhile, the epithelial marker E-cadherin was downregulated and the mesenchymal marker Vimentin was upregulated (Fig. 6E, left). However, after we treated the CREB1-overexpressing cells with DMSO(NC)/LY-294002, which is an inhibitor of the AKT/mTOR pathway, we found that besides the decreased phosphorylation of AKT and mTOR, the EMT process of U2-OS cells was also overturned (Fig. 6E, right). All of the above data suggested that CREB1 could promote the EMT and metastasis of osteosarcoma cells through activating the AKT/mTOR pathway, thus mediating the pro-metastatic effect of its ceRNA UCA1.
4. Discussion
LncRNAs can function as a vital regulator in a broad range of tumor biological processes, such as proliferation, apoptosis and migration. lncRNA UCA1 is a recently identified noncoding RNA and dysregulation of UCA1 is frequently observed in human cancers. Previous studies have shown that a high LncRNA expression is significantly correlated with poor osteosarcoma prognosis. The increased expressions of LncRNA XIST, SOX2-OT, MALAT1, HNF1A-AS1, PARTICLE, CCAL, 91H, HULC, BCAR4, TUG1, BANCR, MEG3 and HOTTIP indicate poorer prognoses of osteosarcoma patients, while the decreased expressions of LncRNA DANCR and TUSC7 indicate better prognoses of osteosarcoma patients. LncRNA UCA1, BCAR4, HULC, and MALAT1 are independent risk factors for poor overall survival rate in osteosarcoma patients. LncRNA DANCR, PARTICLE, HNF1A-AS1, XIST, CCAL and MEG3 can predict metastasis in osteosarcoma patients [11]. However, the role of UCA1 in regulating osteosarcoma invasion and migration remains unclear. In the present study, for the first time, we revealed that UCA1 promotes the invasion and metastasis of osteosarcoma cells via suppression of miR-582 both in vitro and in vivo.
We firstly confirmed the increased expression of UCA1 in osteosarcoma tissues and cell lines and clarified the positive correlation between UCA1 level and the malignant features of clinical patients. The biological function tests showed that UCA1 promoted the proliferation and inhibited the apoptosis of osteosarcoma cells. Importantly, we found that UCA1 increased the invasive and metastatic capacities of osteosarcoma cells both in vitro and in vivo. Since tumor cells obtain migratory and invasive capabilities by EMT, which causes the cells to lose their intercellular adhesion [12] and to generate an aggressive tumor phenotype that leads to proximal or distant via direct invasion or lymphatic and circulatory systems [13], we detected the alterations of EMT markers following the increase or decrease of UCA1 expression. Results showed that overexpression of UCA1 significantly increased the expression of epithelial marker E-cadherin and decreased expression of the mesenchymal markers Vimentin, Snail and ZEB1, indicating the process of EMT was promoted. Conversely, knockdown of UCA1 suppressed the process of EMT in osteosarcoma cells. For the first time, we demonstrated that UCA1 promotes the process of EMT and thereby induces the invasion and metastasis of osteosarcoma cells.
It has been reported that lncRNAs exert their functions by interacting with different molecules, such as miRNAs, mRNAs and proteins. Among them, the miRNAs can be targeted by binding to the 3′-UTR of lncRNAs and thus making the lncRNAs acting as a miRNA “sponge” or ceRNA [14].Currently, the functional pattern of UCA1 in osteosarcoma cells remains unclear. Recent studies have found that miR-582 acted as a tumor suppressor and could regulate bladder cancer progression both in vitro and in vivo [15]. However, the function of miR-582 in osteosarcoma still remains unclear. In the present study, for the first time, we demonstrated that UCA1 acts as a ceRNA to sequester miR-582. Luciferase reporter and RIP assays demonstrated a direct interaction between UCA1 and miR-582. We also found a negative correlation between UCA1 and miR-582 expressions in osteosarcoma tissues. It is well known that miRNAs are key players in gene regulation and perform functions relying on targeting genes. Using bioinformatic analysis, we found that the 3′-UTR of CREB1 mRNA had a complementary site for miR-582 binding. Luciferase reporter assay, western blot analysis and rescue experiments were performed to further confirm that CREB1 is indeed a target of miR-582 in osteosarcoma cells. Hence, we are the first to demonstrate that UCA1 functions as a ceRNA of CREB1 through interacting with miR-582, thus suppressing its binding to the 3′ UTR of CREB1 mRNA and increasing CREB1 expression in osteosarcoma cells. The PI3K/AKT/mTOR pathway is frequently activated and regulates cell survival, growth and migration in various tumors [16]. Although previous study has found that EMT could be mediated by PI3K/AKT/mTOR pathway [17], the effect of CREB1 on this pathway and the possible role of CREB1 in EMT still remain unknown. For the first time, our results showed that CREB1 induced EMT of osteosarcoma cells by activating the PI3K/AKT/mTOR signaling pathway.
In conclusion, we confirmed that UCA1 is upregulated in osteosarcoma and is related to more advanced clinical features. Importantly, our present study is the first to demonstrate that UCA1 promotes the metastasis of osteosarcoma both in vitro and in vivo. Also, for the first time, we revealed that UCA1, as a ceRNA against miR-582, positively regulates the expression of its target CREB1, thus enhancing the EMT process via CREB1-mediated the PI3K/Akt/mTOR pathway and leading to the invasion and metastasis of osteosarcoma cells. These findings suggested that UCA1 could be a potential biomarker and a promising therapeutic target for osteosarcoma treatment.
Acknowledgments
Acknowledgments
Not applicable
Funding
Not applicable
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Authors' contributions
GS designed the study. RS and HF performed the in vitro experiments. RS and YG performed the animal experimentation. HF and YG prepared the figures. RS, HF and YG collected and analyzed the data. GS wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of No. 215 Hospital of Shannxi Nuclear Industry and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Patient consent for publication
The study was approved by the Medical Ethics Committee and written informed consents were obtained from all patients.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2019.100228.
Appendix. Supplementary materials
References
- 1.Smolle M.A., Pichler M. The role of long non-coding RNAs in Osteosarcoma. Non-coding RNA. 2018;4(1) doi: 10.3390/ncrna4010007. pii:E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed D.R., Hayashi M., Wagner L., Binitie O., Steppan D.A., Brohl A.S., Shinohara E.T., Bridge J.A., Loeb D.M., Borinstein S.C., Isakoff M.S. Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer. 2017;123:2206–2218. doi: 10.1002/cncr.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 4.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M., Mo Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y., Yang Y.N., Yuan H.H., Zhang T.T., Sui H., Wei X.L., Liu L., Huang P., Zhang W.J., Bai Y.X. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L., Cao X., Zhang L., Zhang X., Sheng H., Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother. Pharmacol. 2016;77:629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 7.Nie W., Ge H.J., Yang X.Q., Sun X., Huang H., Tao X., Chen W.S., Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Na X.Y., Liu Z.Y., Ren P.P., Yu R., Shang X.S. Long non-coding RNA UCA1 contributes to the progression of prostate cancer and regulates proliferation through KLF4-KRT6/13 signaling pathway. Int. J. Clin. Exp. Med. 2015;8:12609–12616. [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X.S., Zhang Z., Wang H.C., Cai J.L., Xu Q.W., Li M.Q., Chen Y.C., Qian X.P., Lu T.J., Yu L.Z., Zhang Y., Xin D.Q., Na Y.Q., Chen W.F. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. off. J. Am. Assoc. Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z.Q., Cai Q., Hu L., He C.Y., Li J.F., Quan Z.W., Liu B.Y., Li C., Zhu Z.G. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis. 2017;8:e2839. doi: 10.1038/cddis.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D., Wang H., Zhang M., Jiang S., Zhou C., Fang B., Chen P. Abnormally expressed long non-coding RNAs in prognosis of Osteosarcoma: a systematic review and meta-analysis. J. Bone Oncol. 2018;13:76–90. doi: 10.1016/j.jbo.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acloque H., Adams M.S., Fishwick K., Bronner-Fraser M., Nieto M.A. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J. Clin. Investig. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong J.H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B.M., Sharp P.A., Hopkins N., Yaffe M.B. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. (New York, N.Y.) [DOI] [PubMed] [Google Scholar]
- 14.Xia T., Liao Q., Jiang X., Shao Y., Xiao B., Xi Y., Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchino K., Takeshita F., Takahashi R.U., Kosaka N., Fujiwara K., Naruoka H., Sonoke S., Yano J., Sasaki H., Nozawa S., Yoshiike M., Kitajima K., Chikaraishi T., Ochiya T. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol. Ther. J. Am. Soc. Gene Ther. 2013;21:610–619. doi: 10.1038/mt.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap T.A., Garrett M.D., Walton M.I., Raynaud F., de Bono J.S., Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr. Opin. Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D., Wang S., Chen J., Liu H., Lu J., Jiang H., Huang A., Chen Y. Fibulin-4 promotes osteosarcoma invasion and metastasis by inducing epithelial to mesenchymal transition via the PI3K/Akt/mTOR pathway. Int. J. Oncol. 2017;50:1513–1530. doi: 10.3892/ijo.2017.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding author on reasonable request.