Abstract
To protect against the harmful consequences of viral infections, organisms are equipped with sophisticated antiviral mechanisms, including cell‐intrinsic means to restrict viral replication and propagation. Plant and invertebrate cells utilise mostly RNA interference (RNAi), an RNA‐based mechanism, for cell‐intrinsic immunity to viruses while vertebrates rely on the protein‐based interferon (IFN)‐driven innate immune system for the same purpose. The RNAi machinery is conserved in vertebrate cells, yet whether antiviral RNAi is still active in mammals and functionally relevant to mammalian antiviral defence is intensely debated. Here, we discuss cellular and viral factors that impact on antiviral RNAi and the contexts in which this system might be at play in mammalian resistance to viral infection.
Keywords: antiviral immunity, Dicer, double‐stranded RNA, interferons, RNA interference
Subject Categories: Immunology, RNA Biology
Introduction
Metazoan organisms are constantly exposed to viruses and have evolved diverse mechanisms to combat the invaders. One group of mechanisms operates in a cell‐intrinsic fashion, targeting viral nucleic acids and viral proteins for destruction and/or causing the premature shutdown or demise of infected cells to prevent them from serving as virus producers. Cell‐intrinsic antiviral mechanisms are part of the innate immune system and include RNA interference (RNAi) and the interferon (IFN) system. The two systems operate very differently even though they can both be triggered by virally derived long double‐stranded RNA (dsRNA) or highly base‐paired single‐stranded RNA (ssRNA). DsRNA can derive from the viral genome (in the case of a dsRNA virus) or from annealing of two strands of complementary RNAs, which are generated as RNA virus replication intermediates or DNA virus convergent transcripts. Highly based‐paired ssRNAs are found in hairpins within viral genomes or viral transcripts and are generically referred to as dsRNA, a nomenclature that we retain here even if technically incorrect. Both types of dsRNA are largely absent from uninfected cells and act as hallmarks of viral infection to trigger innate antiviral immune responses.
In RNAi, long dsRNA is cleaved by the type III endoribonuclease Dicer into small interfering RNA (siRNAs) (Bernstein et al, 2001), RNA duplexes of 21–24 nucleotides (nts) in length, with 3′ 2‐nt overhangs and a 5′ mono‐phosphate and a 3′ hydroxyl group on both strands (Fig 1) (Hamilton & Baulcombe, 1999; Zamore et al, 2000; Elbashir et al, 2001b,a). One strand of each siRNA duplex is bound by an Argonaute (Ago) protein, which, together with accessory proteins, forms the RNA‐induced silencing complex (RISC) and mediates the endonucleolytic cleavage (“slicing”) of complementary target RNAs (Hammond et al, 2000; MacRae et al, 2008). Of the four Ago proteins encoded by the mammalian genome, only Ago2 has catalytic activity and is essential for target slicing and RNAi (Liu et al, 2004; Meister et al, 2004; Swarts et al, 2014; Sheu‐Gruttadauria & MacRae, 2017). However, all four Ago proteins are involved in an RNAi‐related process, the microRNA (miRNA)‐mediated gene silencing pathway, which does not involve slicing but translation inhibition and/or mRNA degradation (Bartel, 2009; Jonas & Izaurralde, 2015). Notably, Dicer is also involved in miRNA biogenesis. Vertebrates and nematodes possess a single Dicer that generates both siRNA and miRNAs while most invertebrates express two Dicer proteins. For example, in Drosophila melanogaster, dmDcr‐1 is dedicated to the miRNA pathway while dmDcr‐2 performs antiviral RNAi (Lee et al, 2004).
Figure 1. IFN response and antiviral RNAi triggered by viral dsRNA.
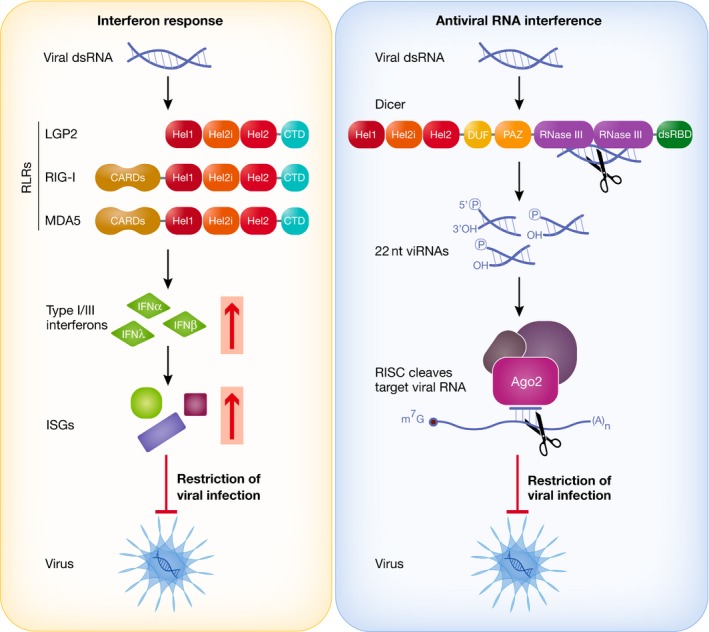
In the cytoplasm of mammalian cells, the RIG‐I‐like receptors (RLRs) RIG‐I and MDA5 detect viral dsRNA and trigger the production of type I interferons, which results in the induction of interferon‐stimulated genes (ISGs) that encode proteins capable of inhibiting viral replication and virus spread. In antiviral RNAi, Dicer cleaves viral dsRNA into viRNAs that are loaded into a RISC complex. As a protein component of this complex, Ago2 degrades viral RNAs with homology to the viRNAs, thereby inhibiting viral replication. RLRs and Dicer share a common DExD/H domain, composed of three helicases (Hel1, Hel2i and Hel2). RIG‐I and MDA5 additionally carry two CARD domains responsible for downstream signalling to MAVS. Dicer possesses two RNase III domains involved in dsRNA dicing.
Three observations indicate that RNAi acts as the major antiviral mechanism of plants and invertebrates (Ding & Voinnet, 2007; Kemp & Imler, 2009; Ding, 2010; Sarkies & Miska, 2013; tenOever, 2016). First, viral infections in these organisms lead to the accumulation of Dicer‐dependent virus‐derived siRNAs (viRNAs) that originate from dsRNA viral replication intermediates and/or RNA hairpins and are homologous to viral RNA sequences (Yoo et al, 2004; Molnar et al, 2005; Galiana‐Arnoux et al, 2006; Ho et al, 2006; van Rij et al, 2006; Wang et al, 2006a; Aliyari et al, 2008; Félix et al, 2011). Second, inactivation of key components of the RNAi pathway results in an increase in viral load in infected cells (Mourrain et al, 2000; Dalmay et al, 2001; Li et al, 2002; Lu et al, 2005; Schott et al, 2005; Wilkins et al, 2005; Deleris et al, 2006; Galiana‐Arnoux et al, 2006; Wang et al, 2006a; Félix et al, 2011). Third, many plant and insect viruses encode viral suppressors of RNAi (VSRs) that interfere with distinct steps of the RNAi pathway, demonstrating the selection pressure imposed by this antiviral system (Pumplin & Voinnet, 2013; Bronkhorst & van Rij, 2014; Csorba et al, 2015).
In contrast, in chordate cells, including mouse and human cells, dsRNA and other nucleic acids associated with viral infection trigger cytosolic innate immune pathways that induce the production of type I IFNs (mainly IFNα and IFNβ) and type III IFNs (IFNλ) (Goubau et al, 2013; Schneider et al, 2014; Wu & Chen, 2014; Schlee & Hartmann, 2016) (Fig 1). These key antiviral cytokines are then secreted and act in an autocrine and paracrine manner by binding to their cognate receptors, i.e. the ubiquitously expressed IFNα/β receptor (IFNAR) and the epithelial cell type‐restricted type III IFN receptor (IL‐28R), which signal to induce hundreds of interferon‐stimulated genes (ISGs) (Schneider et al, 2014). The proteins encoded by these ISGs limit viral replication directly (Schoggins et al, 2011) and serve to enhance adaptive immune responses to the virus (de Veer et al, 2001; Iwasaki & Medzhitov, 2010). For example, the dsRNA‐dependent protein kinase R (PKR) is activated by cytosolic dsRNA and phosphorylates and inactivates the eukaryotic translation initiation factor 2 α (eIF2α), resulting in translational arrest and thwarting the production of both viral and host cell proteins (Pindel & Sadler, 2011). This can ultimately lead to the demise of the infected cell, further undermining the ability of the virus to propagate.
Virally derived long dsRNA or highly based‐paired RNA is detected in the cytosol of mammalian cells by RIG‐I like receptors (RLRs), which include RIG‐I (retinoic acid‐inducible gene I), MDA5 (melanoma differentiation factor 5) and LGP2 (laboratory of genetics and physiology 2) (Fig 1). RIG‐I recognises based‐paired ds or ssRNA with a di‐ or triphosphate (5′PP/5′PPP) at its 5′ extremity (Hornung et al, 2006; Pichlmair et al, 2006; Schlee et al, 2009; Schmidt et al, 2009; Goubau et al, 2014), such as found in the genomes of influenza virus, Sendai virus and reovirus (Baum et al, 2010; Rehwinkel et al, 2010; Weber et al, 2013; Goubau et al, 2014). MDA5 triggers comprise long dsRNAs that accumulate during infection with certain viruses such as picornaviruses and reovirus (Gitlin et al, 2006; Kato et al, 2006; Weber et al, 2006; Pichlmair et al, 2009; Feng et al, 2012). RIG‐I and MDA5 contain two tandem N‐terminal CARDs (caspase activation and recruitment domains) that mediate downstream signalling via the adaptor protein MAVS (mitochondrial antiviral signalling protein), leading to activation of the transcription factors IRF3, IRF7 (interferon regulatory factors 3 and 7) and NFκ‐B (nuclear factor kappa‐light‐chain enhancer of activated B cells). These transcription factors drive the expression of type I and type III IFNs and can directly induce some ISGs. LGP2 lacks CARDs and is unable to induce signalling via MAVS. It is thought to act by modulating responses by the other RLRs (Bruns & Horvath, 2014; Bruns et al, 2014; Parisien et al, 2018).
Thus, plants and invertebrates lack an IFN system and rely on antiviral RNAi to defend against viruses. In contrast, vertebrates have adopted the IFN system for cell‐intrinsic antiviral defence and are thought to have abandoned antiviral RNAi even though they have retained the RNAi machinery and utilise it for miRNA generation and function. Recently, a number of studies have started to question whether the primordial antiviral function of RNAi has truly been abandoned by mammalian cells or whether it can constitute a physiologically relevant antiviral system that complements the IFN pathway. This has become an area of controversy, with some investigators suggesting that RNAi can be a relevant means of cell‐intrinsic restriction to virus infection in mammals while others argue that it is an epiphenomenon with no role in antiviral resistance (Cullen et al, 2013; Cullen, 2014; Ding & Voinnet, 2014; tenOever, 2014, 2017; Jeffrey et al, 2017). In this review, we address this controversy and summarise current understanding of antiviral RNAi pathways in mammals and reflect on the possible contexts in which it might play a role.
Cellular determinants of antiviral RNAi
Detection of dsRNAi in mammalian cells with attenuated IFN responses
To evaluate the possible existence of antiviral RNAi in mammals, it is useful to consider studies that are exempt from virus‐dependent variables such as expression of VSRs. Therefore, we first discuss studies that use synthetic long dsRNA, composed of two perfectly complementary strands, to trigger RNAi, termed here long dsRNA‐mediated RNAi (dsRNAi). This process depends on the successive processing of long dsRNA into a pool of siRNAs and is distinct from RNAi induced experimentally by the introduction of siRNAs (which bypasses the Dicer machinery) (Caplen et al, 2001; Elbashir et al, 2001a) or of short hairpin RNAs (shRNAs, which resemble the structure of pre‐miRNAs) (Brummelkamp et al, 2002; Paddison et al, 2002b; Bartel, 2004).
Long dsRNA‐mediated RNAi was first described in C. elegans (Fire et al, 1998) followed by Drosophila, Trypanosoma brucei, planarians and plants (Kennerdell & Carthew, 1998; Ngô et al, 1998; Waterhouse et al, 1998; Sánchez Alvarado & Newmark, 1999). In mammalian cell lines, long dsRNA had either no effect or displayed a non‐sequence‐specific effect, consistent with activation of the IFN system (Caplen et al, 2000; Elbashir et al, 2001a). Yet, in preimplantation embryos, as well as in oocytes, embryonic stem cells (ESCs) and embryonal carcinoma (EC) cell lines, the introduction of long dsRNA targeting endogenous genes caused a specific reduction in gene expression and induction of phenotypes comparable to those of null mutants, without causing cell death or translational arrest (Svoboda et al, 2000; Wianny & Zernicka‐Goetz, 2000; Billy et al, 2001; Yang et al, 2001; Paddison et al, 2002a). The sequence‐specific silencing induced by long dsRNA in oocytes and ESCs/ECs correlated with a relative inability of these cells to produce and/or respond to IFN (Burke et al, 1978; Francis & Lehman, 1989; Stein et al, 2005; D'Angelo et al, 2016; Wu et al, 2018), which suggested that dsRNAi might be active in mammalian undifferentiated cells but masked or inhibited by the IFN system in differentiated cells.
Antagonism between the IFN response and dsRNAi
Antagonism between the IFN system and dsRNAi was formally tested in somatic cells genetically deficient in MAVS or IFNAR (Maillard et al, 2016). In such cells, introduction of dsRNA resulted in Dicer‐dependent accumulation of siRNAs and Ago2‐dependent sequence‐specific gene silencing (Maillard et al, 2016). A subsequent study showed that the IFN system actively inhibits dsRNAi at least in part through induction of LGP2, which binds Dicer and inhibits processing of long dsRNA into siRNAs (Van der Veen et al, 2018) (Fig 2). In that study (Van der Veen et al, 2018), LGP binding to Dicer did not impact the biogenesis of two household miRNAs although LGP2 has also been reported to interact with the Dicer co‐factor TRBP (HIV TAR RNA‐binding protein) and inhibit the processing of a subset of TRBP‐bound miRNAs (Komuro et al, 2016; Takahashi et al, 2018). Whether LGP2 additionally inhibits dsRNAi via TRBP remains to be addressed.
Figure 2. Impact of viral and cellular determinants on antiviral RNAi.
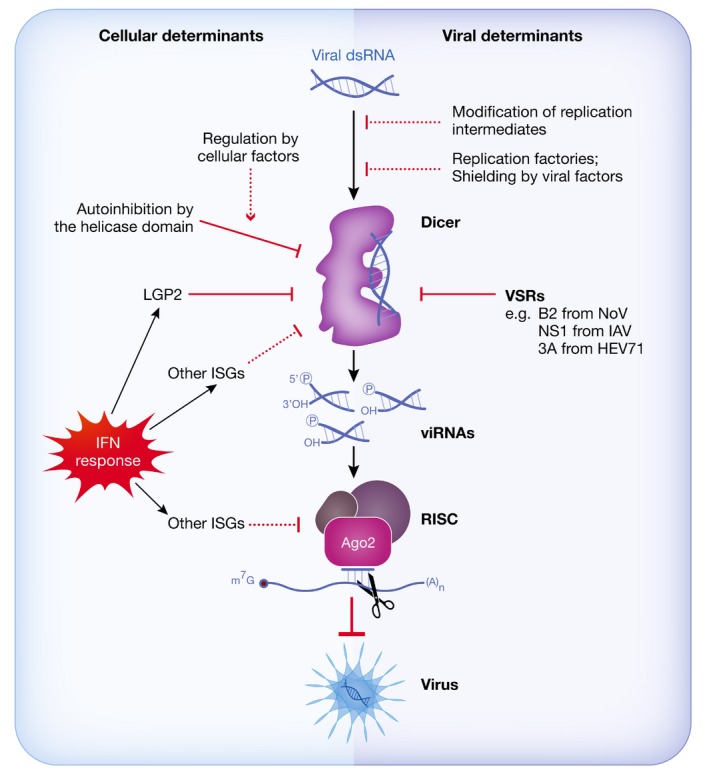
Recognition and dicing of viral dsRNA by Dicer can be influenced by various viral determinants or cellular factors, as described in main text. Known or putative mechanisms that counteract antiviral RNAi are represented with a plain or dashed red line, respectively. Structure of Dicer is based on Liu et al (2018).
It is unclear why somatic cells should inhibit dsRNAi during an IFN response. A clue may come from the observation that mammalian cells stably expressing Drosophila dcr‐2 to artificially boost dsRNAi have impaired induction of IFN upon treatment with poly(I:C), a dsRNA analog (Girardi et al, 2015). Viral infection or treatment with poly(I:C) also induces poly‐ADP‐ribosylation of Ago2 and other RISC components, which inhibits RISC activity and causes a relief in miRNA‐mediated repression of some ISGs (Seo et al, 2013). Perhaps, inhibition of Dicer and RISC is essential for effective stimulation of the IFN pathway, in part by preventing loss of dsRNA substrates for RLR activation. Preservation of dsRNA in infected cells may also ensure that the activity of antiviral proteins encoded by ISGs is not compromised. For example, PKR requires dsRNA of > 30 nts to dimerise and become active for translational repression (Husain et al, 2012). Dicer‐mediated cleavage of long dsRNA could starve the cell of substrates for PKR activation or, more likely, lead to accumulation of 21–22 nt siRNA duplexes that would “quench” PKR monomers, blocking substrate‐dependent dimerisation.
Intrinsic inefficiency of mammalian Dicer in processing long dsRNA
DsRNAi in mammalian cells is further influenced by the molecular properties of its central component: Dicer. This large multi‐domain enzyme comprises an N‐terminal DExD/H helicase domain (containing an ATPase site) followed by a small domain of unknown function (DUF283), a Piwi Argonaute Zwille (PAZ) domain, two tandem RNAse III domains and a C‐terminal dsRNA‐binding domain (dsRBD—Fig 1). The PAZ domain binds the 3′ 2nt‐overhangs found at the extremity of dsRNA substrates, while the RNAse III domains each mediate the cleavage of one strand of the RNA duplex. In vitro studies revealed that human Dicer (hDcr) processes long dsRNA into siRNAs less efficiently than pre‐miRNA into miRNAs (Ma et al, 2008; Chakravarthy et al, 2010). Deletion or partial proteolysis of the helicase domain increases rate of dsRNA cleavage, while only modestly affecting the cleavage of pre‐miRNAs (Provost et al, 2002; Zhang et al, 2002; Ma et al, 2008). Similarly, a deletion mutant of hDcr lacking nearly the entire helicase domain displayed an enhanced ability to process endogenously transcribed long dsRNA and long hairpin RNAs into siRNAs and conferred dsRNAi activity to engineered cells (Kennedy et al, 2015). Finally, mouse oocytes, in which dsRNAi is active, express a shortened isoform of Dicer (DicerO) that lacks the N‐terminal helicase domain and processes endogenous or ectopically expressed long hairpin RNAs more efficiently (Flemr et al, 2013). Together, these data suggest that the helicase domain of Dicer inhibits its catalytic activity for long dsRNA and that its incorporation into the mature enzyme might be regulated by alternative transcription. However, expression of DicerO has not been detected outside mouse germ cells and expression of truncated Dicer isoforms in humans has been reported only in certain cancer cell lines (Potenza et al, 2010; Hinkal et al, 2011; Cantini et al, 2014). An alternative possibility is that modulation of inhibition by the helicase domain could come about not through alternative transcription but through the activity of Dicer‐associated proteins. Structural studies show that the co‐factors TRBP and PACT (Protein Activator of PKR) induce a conformational change in the Dicer helicase domain (Taylor et al, 2013) that could mimic the effect of deletion (Lee et al, 2006; Ma et al, 2008; Chakravarthy et al, 2010; Ota et al, 2013). The exact role and cellular context in which TRBP and PACT might modulate the ability of Dicer to process long dsRNA in vivo remains to be explored.
Viral determinants of antiviral RNAi
Antiviral RNAi is distinct from dsRNAi in (i) the origin of the substrate dsRNA (viral RNA vs exogenous sources) and (ii) the RNAs targeted by the RISC (viral RNA vs host cell mRNA). Antiviral RNAi therefore depends on the efficient production of viRNAs from viral dsRNA and the efficient targeting of viral RNA by the RISC machinery.
Targeting of viral RNA by exogenous small RNAs
Cells transfected with siRNAs or expressing an shRNA targeting viral genomes display sequence‐specific reductions in viral RNA accumulation and virus replication upon challenge with homologous viruses, including human immunodeficiency virus (HIV), hepatitis C virus (HCV), influenza A virus (IAV), West Nile virus (WNV), SARS coronavirus, human papilloma virus and various picornaviruses (Gitlin et al, 2002; Jacque et al, 2002; Lee et al, 2002; Ge et al, 2003; Kapadia et al, 2003; Konishi et al, 2003; Randall et al, 2003; Lu et al, 2004; Phipps et al, 2004; Takigawa et al, 2004; Bitko et al, 2005; Shi et al, 2005; Sim et al, 2005; Werk et al, 2005; Yuan et al, 2005; Kumar et al, 2006; Bousarghin et al, 2009; Qureshi et al, 2018). Similarly, delivery of siRNAs to mice, prior to or concomitant with viral challenge, provides protection against infection with various viruses (Ge et al, 2003; Giladi et al, 2003; McCaffrey et al, 2003; Tompkins et al, 2004; Bitko et al, 2005; Tan et al, 2007; Shah & Schaffer, 2011). RNA viruses engineered to contain perfectly complementary target sites for cellular miRNAs are restricted in cells or tissues expressing the cognate miRNAs (Cawood et al, 2009; Perez et al, 2009; Kelly et al, 2010; Langlois et al, 2013). Finally, in IFN‐defective somatic cells, introduction of long dsRNA conferred sequence‐specific protection from viral challenge dependent on the “slicing” activity of Ago2 (Maillard et al, 2016). Together, these studies show that, if siRNAs are provided directly (bypassing Dicer processing) or are generated from substrates in the absence of an IFN response (shRNAs or dsRNAs in IFN‐defective cells), RISC can access and target viral RNAs to limit viral accumulation. As such, much of the current debate on the role of antiviral RNAi in mammalian cells ultimately centres on the question of whether, during a viral infection, viRNAs are ever produced in sufficient amounts to engage the latent antiviral activity of RISC and exert an antiviral effect.
viRNA production and antiviral RNAi in mammalian cells
viRNAs have key features: (i) a discrete length of ~22 nt, (ii) extremities with 3′ 2nt overhangs and (iii) a strand derived from the positive (+)‐sense viral RNA and a complementary strand commonly derived from the negative (−)‐sense viral RNA or, less frequently, from intramolecular base pairing of viral ssRNA. Initial attempts failed to detect viRNAs in mammalian cells infected with viruses (Pfeffer et al, 2005; Lin & Cullen, 2007), yet the recent emergence of high‐throughput sequencing has allowed the question to be re‐explored more thoroughly. Deep sequencing of differentiated mammalian cells infected with five mammalian viruses [HCV, dengue virus (DENV), WNV, poliovirus and vesicular stomatitis virus (VSV)] revealed the presence of small RNAs derived from the viral genome (viral small RNAs or vsRNAs) (Parameswaran et al, 2010). These vsRNAs, however, did not display size uniformity except in cells carrying a HCV replicon or infected with HCV virions (Parameswaran et al, 2010). Additional deep sequencing experiments of human cells infected with a range of viruses [WNV, DENV, Borna disease virus, IAV, Sindbis virus (SINV)] also reported the detection of vsRNAs but not specifically 22 nt long ones (Girardi et al, 2013; Backes et al, 2014; Bogerd et al, 2014). In addition, deep sequencing of RIG‐I‐ and MDA5‐deficient cells infected with SINV, YFV and the picornavirus coxsackie virus B3 did not reveal viRNA accumulation (Schuster et al, 2017) [although the inhibition of Dicer activity by LGP2 might have dampened the response (Van der Veen et al, 2018)]. Altogether, these experiments argue for limited Dicer‐mediated generation of viRNAs in IFN‐competent differentiated somatic cells.
In contrast, deep sequencing of mESCs infected with the picornavirus encephalomyocarditis virus (EMCV) revealed the accumulation of viral reads with a specific peak at 21‐23 nt and reads that mapped within the first 200‐nt of the EMCV genome and, to a lesser extent, to the 3′end and that, importantly, derived in equal parts from the (+) strand and (−) strand (Maillard et al, 2013). The sequences formed perfectly paired duplexes with 3′ 2‐nt overhangs and were produced in a phase pattern indicative of successive cleavage by Dicer. Finally, the duplexes could be shown to associate with Ago2 and require Dicer for their generation, thereby fulfilling all criteria for bona fide viRNAs (Maillard et al, 2013). Interestingly, production of these viRNAs by mESCs was greatly reduced upon cell differentiation (Maillard et al, 2013), in line with the aforementioned studies reporting little viRNA generation in differentiated somatic cells.
Thus, in some IFN‐deficient cells, including ESCs, infection with viruses allows for viRNA production. Can it elicit a protective RNAi‐dependent response? Early work suggested that knockdown of Dicer in Vero cells, an African green monkey cell line that lacks IFN‐α and IFN‐β genes (Diaz et al, 1988), causes a modest increase in virus production upon IAV infection (Matskevich & Moelling, 2007). In contrast, later reports found that absence of Dicer in HEK 293T cells and/or in mouse embryonic fibroblasts did not impact the accumulation of flaviviruses (DENV, WNV, YFV), alphaviruses (SINV, Venezuelan equine encephalitis virus [VEEV]), IAV, measles virus, HIV and reovirus (Shapiro et al, 2010; Bogerd et al, 2014). Similarly, in engineered cells overexpressing an artificial Dicer that lacks the helicase domain, infection with IAV or poliovirus led to low‐level accumulation of viRNAs, which were loaded onto RISC but had little impact on replication (Kennedy et al, 2015). Finally, expression of a slicing‐deficient Ago2 mutant in Ifnar1 −/− mouse embryonic fibroblasts did not impact infection with Semliki Forest virus (SFV), reovirus or IAV (Maillard et al, 2016). The overall message from those studies is that, even in a context permissive for dsRNAi, viRNA production from viral replication intermediates is too weak to inhibit viral infection. Replication of (+)‐sense RNA viruses often occurs within membranous structures (replication factories), whereas RNAs generated during replication of (−)‐sense RNA viruses rapidly associate with nucleocapsid proteins (Conzelmann, 1998; Romero‐Brey & Bartenschlager, 2014). In addition, the 5′ extremities of certain viral genomes (and replication intermediates) can display various modifications, including a 7‐methylguanosine (Cap) structure, a covalently linked protein (e.g. Vpg, viral protein genome‐linked), highly structured regions or 2–3 phosphates (Fig 2). Whether these features prevent efficient access of Dicer to viral RNA is unknown although it is worth remembering they do not prevent antiviral RNAi in insect cells. A key issue might therefore be antagonism of RNAi by VSRs. This will be discussed in the next section.
VSRs in mammalian viruses
Several proteins encoded by mammalian viruses display VSR activity (1; 2). Expression of influenza virus NS1, vaccinia virus E3L, reovirus σ3 and Nodamura virus (NoV) B2 proteins inhibits RNAi in plants and/or insect cells (Lichner et al, 2003; Bucher et al, 2004; Delgadillo et al, 2004; Li et al, 2004). In mammalian cells, a plethora of mammalian virus‐encoded VSRs, including primate foamy virus type 1 (PFV‐1) Tas, NoV B2, HCV core, IAV NS1, HIV Tat, Ebola VP35, VP30 and VP40, CoV N, SARS‐CoV 7a, YFV capsid, DENV NS4B, human enterovirus 71 (HEV 71) 3A and adenovirus virus‐associated RNA I (VA1), can reduce shRNA/siRNA‐mediated knockdown of reporter genes (Table 1) (Lu & Cullen, 2004; Andersson et al, 2005; Lecellier et al, 2005; Sullivan & Ganem, 2005; Wang et al, 2006b; Haasnoot et al, 2007; Chen et al, 2008; de Vries et al, 2009; Karjee et al, 2010; Fabozzi et al, 2011; Kakumani et al, 2013; Cui et al, 2015; Samuel et al, 2016; Qiu et al, 2017). Most viral proteins identified thus far that display VSR activity share the ability to bind dsRNA and mutations that affect their dsRNA‐binding domain block VSR activity, arguing that their principal mode of action is sequestration of dsRNA from Dicer (Table 1). As dsRNA is a potent inducer of the IFN pathway, most of these VSRs also act as IFN antagonists (García‐Sastre, 2017). It is therefore unclear whether these viral proteins specifically evolved to block RNAi or whether their VSR activity is a byproduct of their role as IFN antagonists (Cullen, 2006). However, some VSRs may function through mechanisms other than dsRNA sequestration (Kakumani et al, 2013), including binding to components of the RNAi pathway: e.g. Ebola virus VP35 and VP30 proteins interact with Dicer co‐factors TRBP and PACT, while HCV core associates with Dicer (Table 1) (Wang et al, 2006b; Chen et al, 2008; Fabozzi et al, 2011). Whether these interactions contribute to VSR activity is unclear. Finally, adenovirus VA1s are small, highly structured RNAs that inhibit shRNA‐mediated RNAi by acting as decoy substrates for Dicer, RISC and exportin 5 (required for nuclear export of pre‐miRNAs and shRNAs) (Lu & Cullen, 2004; Andersson et al, 2005).
Table 1.
List of mammalian virus‐encoded proteins with VSR activity
| Viral genome | Virus name | Virus family | VSR | Properties | Proposed mode of action | References |
|---|---|---|---|---|---|---|
| (+)‐ssRNA | Coronavirus (CoV) | Coronaviridae | N | dsRNA binding | dsRNA sequestration | Cui et al (2015) |
| Severe acute respiratory syndrome coronavirus (SARS‐CoV) | Coronaviridae | 7a | – | – | Karjee et al (2010) | |
| Dengue virus (DENV) | Flaviviridae | NS4B | lack of dsRNA binding | inhibition of Dicer activity | Kakumani et al (2013) | |
| Hepatitis C virus (HCV) | Flaviviridae | capsid | Dicer binding | inhibition of Dicer activity | Wang et al (2006b), Chen et al (2008) | |
| Yellow Fever virus (YFV) | Flaviviridae | capsida | dsRNA binding | dsRNA sequestration | Samuel et al (2016) | |
| Human enterovirus 71 (HEV71) | Picornaviridae | 3A | dsRNA binding | dsRNA sequestration | Qiu et al (2017) | |
| Human immunodeficiency virus 1 (HIV‐1) | Retroviridae | Tat | dsRNA binding | – | Bennasser et al (2005), Triboulet et al (2007), Lin and Cullen (2007)b, Sanghvi & Steel (2011)b | |
| Primate foamy virus type 1 (PFV‐1) | Retroviridae | Tasa | – | – | Lecellier et al (2005) | |
| Nodamura virus (NoV) | Nodaviridae | B2 | dsRNA binding | dsRNA sequestration | Sullivan and Ganem (2005), Aliyari et al (2008), Li et al (2013), Maillard et al (2013) | |
| (−)‐ssRNA | Ebolavirus | Filoviridae | VP30 | Dicer and TRBP binding | inhibition of Dicer activity | Fabozzi et al (2011) |
| VP35 | PACT, TRBP, dsRNA binding | inhibition of Dicer activity | Haasnoot et al (2007), Fabozzi et al (2011) | |||
| VP40 | – | – | Fabozzi et al (2011) | |||
| Marburg virus | Filoviridae | VP35 | dsRNA binding | – | Li et al (2016) | |
| Influenza virus | Orthomyxoviridae | NS1 | dsRNA binding | dsRNA sequestration | Li et al (2004), Bucher et al (2004), Delgadillo et al (2004), Kok and Jin (2006)b, de Vries et al (2009), Kennedy et al (2015), Benitez et al (2015)b, Li et al (2016), Tsai et al (2018) | |
| NP | – | – | Kennedy et al (2015) | |||
| La Crosse virus | Peribunyaviridae | NSs | – | – | Soldan et al (2004) | |
| dsRNA | Reovirus | Reoviridae | σ3a | dsRNA binding | dsRNA sequestration | Lichner et al (2003) |
| dsDNA | Adenovirus | Adenoviridae | VA I, VA II | Dicer binding | Dicer sequestration by acting as decoy RNAs | Lu and Cullen (2004), Andersson et al (2005) |
| Vaccinia virus | Poxviridae | E3L | dsRNA binding | dsRNA sequestration | Li et al (2004), Haasnoot et al (2007) |
VSR activity shown only in non‐mammalian hosts.
Studies questioning VSR activity.
Despite the evidence that many viral proteins from mammalian viruses can act as VSRs in overexpression (i.e. gain‐of‐function) studies, there are relatively few loss‐of‐function studies that show that they actively suppress mammalian antiviral RNAi defence. Persuasive experiments have been done with NoV, a member of the Nodavirus family. Nodaviruses express B2 proteins, which bind long dsRNA and siRNAs in vitro and associate with replication intermediates and viRNAs in infected cells (Chao et al, 2005; Lu et al, 2005; Sullivan & Ganem, 2005; Aliyari et al, 2008). B2 proteins act as potent VSRs in insect cells (Wang et al, 2006a; Aliyari et al, 2008). Notably, B2‐deficient NoV (NoV ΔB2) also replicates less efficiently than parental NoV in mESCs but its accumulation is rescued in mESCs lacking all Ago genes (Maillard et al, 2013). In suckling mice, NoV ΔB2 is highly attenuated and induces accumulation of viRNAs (Li et al, 2013). viRNAs are also detected, although to a lesser degree, upon infection of somatic cells (BHK‐21) with NoV ΔB2, but not with NoV WT (Li et al, 2013). Altogether, these data suggest that the ability of Dicer to process NoV replication intermediates is actively antagonised by the B2 protein.
The NS1 protein from IAV also displays VSR activity when expressed in plants and insect cells (Bucher et al, 2004; Delgadillo et al, 2004; Li et al, 2004). In mammalian cells, NS1 is ineffective against RISC‐loaded siRNAs (Kok & Jin, 2006; Haasnoot et al, 2007; de Vries et al, 2009; Kennedy et al, 2015) but infection of human and African green monkey cells with IAV ΔNS1 but not IAV WT yields readily detectable levels of canonical viRNAs derived from the termini of both strands of the eight viral RNA segments (Li et al, 2016; Tsai et al, 2018). It has been reported that IAV ΔNS1, and to a lesser extent parental wild‐type IAV, VSV and EMCV, replicates more extensively in mouse embryonic fibroblasts expressing a slicing‐deficient Ago2 mutant (Li et al, 2016). However, in other studies, loss of RNAi components did not cause an increase in replication of IAV ΔNS1 (Maillard et al, 2016; Tsai et al, 2018). Interestingly, IAV engineered to express an shRNA or a miRNA targeting a viral gene or a reporter gene integrated in the viral genome, respectively, was attenuated compared to non‐targeting controls (Benitez et al, 2015). This restriction was Dicer‐dependent but independent of NS1 suggesting that, in the context of an infection, NS1′s VSR activity inhibits viRNA production from genome segments but not from short dsRNA hairpins (Benitez et al, 2015; Li et al, 2016; Tsai et al, 2018).
Finally, the HEV71‐encoded protein 3A inhibits shRNA‐mediated silencing in mammalian cells, as well as antiviral RNAi in insect cells, and suppresses Dicer‐mediated biogenesis of siRNAs by binding and sequestering long dsRNA in vitro (Qiu et al, 2017). A point mutation that inactivates 3A's VSR activity reduces viral replication in somatic cells and in suckling mice. Concomitantly, canonical viRNAs derived from both strands of the 5′terminal region of the HEV71 genome are produced, loaded into RISC and able to silence a reporter bearing complementary sites (Qiu et al, 2017). Interestingly, the absence of Dicer increases HEV71 accumulation in infected cells despite the presence of an intact IFN pathway, suggesting that, in this case, antiviral RNAi could function irrespective of the IFN system (Qiu et al, 2017).
Niches for antiviral RNAi?
The generally observed antagonism between the IFN response and dsRNAi suggests that antiviral RNAi may be especially important in cellular niches in which the induction of or the response to IFN is limited. One of those niches might be stem cells. Pluripotent stem cells do not produce IFN upon viral infection or exposure to poly(I:C) and respond poorly to IFN treatment (Chen et al, 2010; Hong & Carmichael, 2013; Wang et al, 2013; Guo et al, 2015; D'Angelo et al, 2016). It is unclear why pluripotent stem cells are refractory to IFN but it may have to do with the fact that self‐renewal is incompatible with the anti‐proliferative effects and pro‐apoptotic effects of the cytokines (Hertzog et al, 1994; de Veer et al, 2001). Furthermore, artificial induction of an IFN response in engineered pluripotent cells compromises differentiation potential (Eggenberger et al, 2019). Thus, pluripotent stem cells may be forced to rely on IFN‐independent mechanisms to combat virus infections. These may include the ability to constitutively express some ISGs that confer an efficient and permanent antiviral state (Wu et al, 2018). In this scenario, antiviral RNAi would constitute an additional mechanism to protect the integrity and function of tissue stem cells in the face of virus infection and thereby contribute to tissue maintenance, repair and regeneration (Xia et al, 2018). Notably, the ability of a virus to infect stem cells might not be needed for its propagation and, therefore, stem cell‐intrinsic antiviral RNAi would benefit the host but not impact on virus transmission.
Future directions in antiviral RNAi
In line with other facets of immunity, it is likely that antiviral RNAi is highly tuneable and that it operates in conjunction with multiple other mechanisms of defence. Further studies are clearly needed to disentangle the complex web that regulates dsRNAi in mammals and to understand its ability to act as a cell‐intrinsic mechanism of antiviral defence. Open questions include what are the cellular factors regulating the activity of Dicer on long dsRNA? Apart from murine germ cells, are there similar truncated Dicer isoforms expressed in other cell types and/or other species? Which viral proteins act as bona fide VSRs in the context of an infection? What are the cell types in which antiviral RNAi is active? Does antiviral RNAi directly impact on viral accumulation upon infection in vivo? These and other questions are likely to enliven the debate on the role of RNAi in mammalian defence from virus attack for years to come.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
CRS is funded by The Francis Crick Institute, which receives core funding from Cancer Research UK (FC001136), the UK Medical Research Council (FC001136), and the Wellcome Trust (FC001136), by an ERC Advanced Investigator Grant (AdG 786674), by a Wellcome Trust Investigator Award (WT106973MA) and by a Prize from the Louis‐Jeantet Foundation. PVM is funded by a Wolfson UCL Excellence Fellowship, and EP is funded by an EMBO Long‐Term Fellowship.
Note added in proof
While this review was in production, a study reported that Zika virus (ZIKV) infection resulted in the production of canonical viRNAs in human neural progenitor cells (hNPCs), the major target cells of ZIKV, but not in neurons differentiated from hNPCs (Xu et al, 2019). This study supports the notion that RNAi can play an antiviral role in certain progenitor cells, including those involved in brain development.
The EMBO Journal (2019) 38: e100941 30872283
Contributor Information
Pierre V Maillard, Email: p.maillard@ucl.ac.uk.
Caetano Reis e Sousa, Email: caetano@crick.ac.uk.
References
- Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW (2008) Mechanism of induction and suppression of antiviral immunity directed by virus‐derived small RNAs in Drosophila . Cell Host Microbe 4: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PCJ, Xu N, Berenjian S, Berkhout B, Akusjarvi G (2005) Suppression of RNA interference by adenovirus virus‐associated RNA. J Virol 79: 9556–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Langlois RA, Schmid S, Varble A, Shim JV, Sachs D, tenOever BR (2014) The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep 8: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, García‐Sastre A (2010) Preference of RIG‐I for short viral RNA molecules in infected cells revealed by next‐generation sequencing. Proc Natl Acad Sci USA 107: 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez AA, Spanko LA, Bouhaddou M, Sachs D, tenOever BR (2015) Engineered Mammalian RNAi can elicit antiviral protection that negates the requirement for the interferon response. Cell Rep 13: 1456–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le S‐Y, Benkirane M, Jeang K‐T (2005) Evidence that HIV‐1 Encodes an siRNA and a Suppressor of RNA Silencing. Immunity 22: 607–619 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double‐stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V, Musiyenko A, Shulyayeva O, Barik S (2005) Inhibition of respiratory viruses by nasally administered siRNA. Nat Med 11: 50–55 [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KLW, Putnam N, Barrows NJ, Sherry B, Scholle F, Garcia‐Blanco MA, Griffin DE, Cullen BR (2014) Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol 88: 8065–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousarghin L, Touze A, Gaud G, Iochmann S, Alvarez E, Reverdiau P, Gaitan J, Jourdan ML, Sizaret PY, Coursaget PL (2009) Inhibition of cervical cancer cell growth by human papillomavirus virus‐like particles packaged with human papillomavirus oncoprotein short hairpin RNAs. Mol Cancer Ther 8: 357–365 [DOI] [PubMed] [Google Scholar]
- Bronkhorst AW, van Rij RP (2014) The long and short of antiviral defense: small RNA‐based immunity in insects. Curr Opin Virol 7: 19–28 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM (2014) Antiviral RNA recognition and assembly by RLR family innate immune sensors. Cytokine Growth Factor Rev 25: 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Leser GP, Lamb RA, Horvath CM (2014) The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5‐RNA interaction and filament assembly. Mol Cell 55: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M (2004) The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 85: 983–991 [DOI] [PubMed] [Google Scholar]
- Burke DC, Graham CF, Lehman JM (1978) Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell 13: 243–248 [DOI] [PubMed] [Google Scholar]
- Cantini LP, Andino LM, Attaway CC, Butler B, Dumitriu A, Blackshaw A, Jakymiw A (2014) Identification and characterization of Dicer1e, a Dicer1 protein variant, in oral cancer cells. Mol Cancer 13: 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen NJ, Fleenor J, Fire A, Morgan RA (2000) dsRNA‐mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252: 95–105 [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA (2001) Specific inhibition of gene expression by small double‐stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA 98: 9742–9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawood R, Chen HH, Carroll F, Bazan‐Peregrino M, Van Rooijen N, Seymour LW (2009) Use of tissue‐specific MicroRNA to control pathology of wild‐type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog 5: e1000440–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA (2010) Substrate‐specific kinetics of Dicer‐catalyzed RNA processing. J Mol Biol 404: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR (2005) Dual modes of RNA‐silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12: 952–957 [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, Huang Y, Cai X, Huang A (2008) HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res 133: 250–258 [DOI] [PubMed] [Google Scholar]
- Chen L‐L, Yang L, Carmichael GG (2010) Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 9: 3552–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann KK (1998) Nonsegmented negative‐strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet 32: 123–162 [DOI] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J (2015) viral silencing suppressors: Tools forged to. Virology 479–480: 85–103 [DOI] [PubMed] [Google Scholar]
- Cui L, Wang H, Ji Y, Yang J, Xu S, Huang X, Wang Z, Qin L, Tien P, Zhou X, Guo D, Chen Y (2015) the nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J Virol 89: 9029–9043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR (2006) Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol 7: 563–567 [DOI] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, tenOever BR (2013) Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 14: 374–378 [DOI] [PubMed] [Google Scholar]
- Cullen BR (2014) Viruses and RNA interference: issues and controversies. J Virol 88: 12934–12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo W, Acharya D, Wang R, Wang J, Gurung C, Chen B, Bai F, Guo Y‐L (2016) Development of antiviral innate immunity during in vitro differentiation of mouse embryonic stem cells. Stem Cells Dev 25: 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC (2001) SDE3 encodes an RNA helicase required for post‐transcriptional gene silencing in Arabidopsis . EMBO J 20: 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego‐Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Delgadillo MO, Sáenz P, Salvador B, García JA, Simón‐Mateo C (2004) Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol 85: 993–999 [DOI] [PubMed] [Google Scholar]
- Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD (1988) Homozygous deletion of the alpha‐ and beta 1‐interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci USA 85: 5259–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW (2010) RNA‐based antiviral immunity. Nat Rev Immunol 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2014) Antiviral RNA silencing in mammals: no news is not good news. Cell Rep 9: 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger J, Blanco‐Melo D, Panis M, Brennand KJ, tenOever BR (2019) Type I interferon response impairs differentiation potential of pluripotent stem cells. Proc Natl Acad Sci USA 172: 201812449–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001a) Duplexes of 21‐nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ (2011) Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol 85: 2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M‐A, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9: e1000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Hato SV, Langereis MA, Zoll J, Virgen‐Slane R, Peisley A, Hur S, Semler BL , van Rij RP, van Kuppeveld FJ (2012) MDA5 detects the double‐stranded RNA replicative form in picornavirus‐infected cells. Cell Rep 2: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P (2013) A retrotransposon‐driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155: 807–816 [DOI] [PubMed] [Google Scholar]
- Francis MK, Lehman JM (1989) Control of beta‐interferon expression in murine embryonal carcinoma F9 cells. Mol Cell Biol 9: 3553–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana‐Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL (2006) Essential function in vivo for Dicer‐2 in host defense against RNA viruses in Drosophila . Nat Immunol 7: 590–597 [DOI] [PubMed] [Google Scholar]
- García‐Sastre A (2017) Ten strategies of interferon evasion by viruses. Cell Host Microbe 22: 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, McManus MT, Nguyen T, Shen C‐H, Sharp PA, Eisen HN, Chen J (2003) RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA 100: 2718–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H, Ketzinel‐Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E (2003) Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther 8: 769–776 [DOI] [PubMed] [Google Scholar]
- Girardi E, Chane‐Woon‐Ming B, Messmer M, Kaukinen P, Pfeffer S (2013) Identification of RNase L‐dependent, 3′‐end‐modified, viral small RNAs in sindbis virus‐infected mammalian cells. MBio 4: e00698–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Lefèvre M, Chane‐Woon‐Ming B, Paro S, Claydon B, Imler J‐L, Meignin C, Pfeffer S (2015) Cross‐species comparative analysis of Dicer proteins during Sindbis virus infection. Sci Rep 5: 10693–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Karelsky S, Andino R (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418: 430–434 [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M (2006) Essential role of mda‐5 in type I IFN responses to polyriboinosinic: polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA 103: 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Deddouche S, Reis e Sousa C (2013) Cytosolic sensing of viruses. Immunity 38: 855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C (2014) Antiviral immunity via RIG‐I‐mediated recognition of RNA bearing 59‐diphosphates. Nature 514: 372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y‐L, Carmichael GG, Wang R, Hong X, Acharya D, Huang F, Bai F (2015) Attenuated innate immunity in embryonic stem cells and its implications in developmental biology and regenerative medicine. Stem Cells 33: 3165–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes E‐J, Prins M, de Haan P, Berkhout B (2007) The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3: e86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA‐directed nuclease mediates post‐transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hertzog PJ, Hwang SY, Kola I (1994) Role of interferons in the regulation of cell proliferation, differentiation, and development. Mol Reprod Dev 39: 226–232 [DOI] [PubMed] [Google Scholar]
- Hinkal GW, Grelier G, Puisieux A, Moyret‐Lalle C (2011) Complexity in the regulation of Dicer expression: Dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br J Cancer 104: 387–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T, Pallett D, Rusholme R, Dalmay T, Wang H (2006) A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J Virol Methods 136: 217–223 [DOI] [PubMed] [Google Scholar]
- Hong XX, Carmichael GG (2013) Innate immunity in pluripotent human cells: attenuated response to interferon‐beta. J Biol Chem 288: 16196–16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G (2006) 5′‐Triphosphate RNA is the ligand for RIG‐I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Husain B, Mukerji I, Cole JL (2012) Analysis of high‐affinity binding of protein kinase R to double‐stranded RNA. Biochemistry 51: 8764–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R (2010) Regulation of adaptive immunity by the innate immune system. Science 327: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque J‐M, Triques K, Stevenson M (2002) Modulation of HIV‐1 replication by RNA interference. Nature 418: 435–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey KL, Li Y, Ding SW (2017) Reply to “questioning antiviral RNAi in mammals”. Nat Microbiol 2: 17053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA‐mediated gene silencing. Nat Rev Genet 16: 421–433 [DOI] [PubMed] [Google Scholar]
- Kakumani PK, Ponia SS, Rajgokul KS, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK (2013) Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 87: 8870–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Brideau‐Andersen A, Chisari FV (2003) Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA 100: 2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karjee S, Minhas A, Sood V, Ponia SS, Banerjea AC, Chow VTK, Mukherjee SK, Lal SK (2010) The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J Virol 84: 10395–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Nace R, Barber GN, Russell SJ (2010) Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J Virol 84: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C, Imler J‐L (2009) Antiviral immunity in Drosophila . Curr Opin Immunol 21: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Whisnant AW, Kornepati AVR, Marshall JB, Bogerd HP, Cullen BR (2015) Production of functional small interfering RNAs by an amino‐terminal deletion mutant of human Dicer. Proc Natl Acad Sci USA 112: E6945–E6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA‐mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Kok K‐H, Jin D‐Y (2006) Influenza A virus NS1 protein does not suppress RNA interference in mammalian cells. J Gen Virol 87: 2639–2644 [DOI] [PubMed] [Google Scholar]
- Komuro A, Homma Y, Negoro T, Barber GN, Horvath CM (2016) The TAR‐RNA binding protein is required for immunoresponses triggered by Cardiovirus infection. Biochem Biophys Res Commun 480: 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Wu CH, Wu GY (2003) Inhibition of HBV replication by siRNA in a stable HBV‐producing cell line. Hepatology 38: 842–850 [DOI] [PubMed] [Google Scholar]
- Kumar P, Lee SK, Shankar P, Manjunath N (2006) A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med 3: e96–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Albrecht RA, Kimble B, Sutton T, Shapiro JS, Finch C, Angel M, Chua MA, Gonzalez‐Reiche AS, Xu K, Perez D, Garcia‐Sastre A, tenOever BR (2013) MicroRNA‐based strategy to mitigate the risk of gain‐of‐function influenza studies. Nat Biotechnol 31: 844–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann‐Che J, Eyquem S, Himber C, Saib A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560 [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li M‐J, Ehsani A, Salvaterra P, Rossi J (2002) Expression of small interfering RNAs targeted against HIV‐1 rev transcripts in human cells. Nat Biotechnol 20: 500–505 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004) Distinct roles for Drosophila Dicer‐1 and Dicer‐2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia‐Sastre A, Ball LA, Palese P, Ding SW (2004) Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 101: 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, Ding SW (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, Reinecker H‐C, Hertzog P, Han Y, Li W‐X, Cheloufi S, Karginov FV, Ding SW, Jeffrey KL (2016) Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol 2: 16250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichner Z, Silhavy D, Burgyan J (2003) Double‐stranded RNA‐binding proteins could suppress RNA interference‐mediated antiviral defences. J Gen Virol 84: 975–980 [DOI] [PubMed] [Google Scholar]
- Lin J, Cullen BR (2007) Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 81: 12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J‐J, Hammond SM, Joshua‐Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang J, Cheng H, Ke X, Sun L, Zhang QC, Wang H‐W (2018) Cryo‐EM structure of human dicer and its complexes with a pre‐miRNA substrate. Cell 173: 1191–1203.e12 [DOI] [PubMed] [Google Scholar]
- Lu S, Cullen BR (2004) Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 78: 12868–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W‐W, Hsu Y‐Y, Yang J‐Y, Kung S‐H (2004) Selective inhibition of enterovirus 71 replication by short hairpin RNAs. Biochem Biophys Res Commun 325: 494–499 [DOI] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman‐Maduro G, Li WX, Ding SW (2005) Animal virus replication and RNAi‐mediated antiviral silencing in Caenorhabditis elegans . Nature 436: 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA (2008) Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA (2008) In vitro reconstitution of the human RISC‐loading complex. Proc Natl Acad Sci USA 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O (2013) Antiviral RNA interference in mammalian cells. Science 342: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Van der Veen AG, Deddouche Grass S, Rogers NC, Merits A, Reis e Sousa C (2016) Inactivation of the type I interferon pathway reveals long double‐stranded RNA‐mediated RNA interference in mammalian cells. EMBO J 35: 2505–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matskevich AA, Moelling K (2007) Dicer is involved in protection against influenza A virus infection. J Gen Virol 88: 2627–2635 [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA (2003) Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol 21: 639–644 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J (2005) Plant virus‐derived small interfering RNAs originate predominantly from highly structured single‐stranded viral RNAs. J Virol 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Rémoué K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Ngô H, Tschudi C, Gull K, Ullu E (1998) Double‐stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA 95: 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K (2013) ADAR1 forms a complex with Dicer to promote microRNA processing and RNA‐induced gene silencing. Cell 153: 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ (2002a) Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA 99: 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS (2002b) Short hairpin RNAs (shRNAs) induce sequence‐specific silencing in mammalian cells. Genes Dev 16: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, Doukas T, Paranjape S, Polacek C, dos Santos FB, Jalili R, Babrzadeh F, Gharizadeh B, Grimm D, Kay M, Koike S, et al (2010) Six RNA viruses and forty‐one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 6: e1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien JP, Lenoir JJ, Mandhana R, Rodriguez KR, Qian K, Bruns AM, Horvath CM (2018) RNA sensor LGP2 inhibits TRAF ubiquitin ligase to negatively regulate innate immune signaling. EMBO Rep 19: e45176–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR (2009) MicroRNA‐mediated species‐specific attenuation of influenza A virus. Nat Biotechnol 27: 572–576 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos‐Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Phipps KM, Martinez A, Lu J, Heinz BA, Zhao G (2004) Small interfering RNA molecules as potential anti‐human rhinovirus agents: in vitro potency, specificity, and mechanism. Antiviral Res 61: 49–55 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C (2006) RIG‐I‐mediated antiviral responses to single‐stranded RNA bearing 5′‐phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C (2009) Activation of MDA5 requires higher‐order RNA structures generated during virus infection. J Virol 83: 10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindel A, Sadler A (2011) The role of protein kinase R in the interferon response. J Interferon Cytokine Res 31: 59–70 [DOI] [PubMed] [Google Scholar]
- Potenza N, Papa U, Scaruffi P, Mosca N, Tonini GP, Russo A (2010) A novel splice variant of the human dicer gene is expressed in neuroblastoma cells. FEBS Lett 584: 3452–3457 [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter‐defence and counter‐counter‐defence. Nat Rev Micro 11: 745–760 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xu Y, Zhang Y, Zhou H, Deng Y‐Q, Li X‐F, Miao M, Zhang Q, Zhong B, Hu Y, Zhang F‐C, Wu L, Qin C‐F, Zhou X (2017) Human virus‐derived small RNAs can confer antiviral immunity in mammals. Immunity 46: 992–1004.e5 [DOI] [PubMed] [Google Scholar]
- Qureshi A, Tantray VG, Kirmani AR, Ahangar AG (2018) A review on current status of antiviral siRNA. Rev Med Virol 9: e1976–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Grakoui A, Rice CM (2003) Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA 100: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C (2010) RIG‐I detects viral genomic RNA during negative‐strand RNA virus infection. Cell 140: 397–408 [DOI] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes Dev 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero‐Brey I, Bartenschlager R (2014) Membranous replication factories induced by plus‐strand RNA viruses. Viruses 6: 2826–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM (2016) Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double‐stranded RNA. Proc Natl Acad Sci USA 113: 13863–13868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA (1999) Double‐stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci USA 96: 5049–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghvi VR, Steel LF (2011) A re‐examination of global suppression of RNA interference by HIV‐1. PLoS One 6: e17246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA (2013) RNAi pathways in the recognition of foreign RNA: antiviral responses and host‐parasite interactions in nematodes. Biochem Soc Trans 41: 876–880 [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, et al (2009) Recognition of 5′ triphosphate by RIG‐I helicase requires short blunt double‐stranded RNA as contained in panhandle of negative‐strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann G (2016) Discriminating self from non‐self in nucleic acid sensing. Nat Rev Immunol 16: 566–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S (2009) 5′‐triphosphate RNA requires base‐paired structures to activate antiviral signaling via RIG‐I. Proc Natl Acad Sci USA 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM (2014) Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Cureton DK, Whelan SP, Hunter CP (2005) An antiviral role for the RNA interference machinery in Caenorhabditis elegans . Proc Natl Acad Sci USA 102: 18420–18424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Tholen LE, Overheul GJ, van Kuppeveld FJM, van Rij RP (2017) Deletion of cytoplasmic double‐stranded rna sensors does not uncover viral small interfering RNA production in human cells. mSphere 2: e00333–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang T‐Y, Krug RM, Sullivan CS (2013) Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PS, Schaffer DV (2011) Antiviral RNAi: translating science towards therapeutic success. Pharm Res 28: 2966–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Varble A, Pham AM, tenOever BR (2010) Noncanonical cytoplasmic processing of viral microRNAs. RNA 16: 2068–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu‐Gruttadauria J, MacRae IJ (2017) Structural foundations of RNA silencing by argonaute. J Mol Biol 429: 2619–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yang DH, Xiong J, Jia J, Huang B, Jin YX (2005) Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res 15: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim ACN, Luhur A, Tan TMC, Chow VTK, Poh CL (2005) RNA interference against Enterovirus 71 infection. Virology 341: 72–79 [DOI] [PubMed] [Google Scholar]
- Soldan SS, Plassmeyer ML, Matukonis MK, Gonzalez‐Scarano F (2004) La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J Virol 79: 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein P, Zeng F, Pan H, Schultz RM (2005) Absence of non‐specific effects of RNA interference triggered by long double‐stranded RNA in mouse oocytes. Dev Biol 286: 464–471 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D (2005) A virus‐encoded inhibitor that blocks RNA interference in mammalian cells. J Virol 79: 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127: 4147–4156 [DOI] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J (2014) The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 21: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nakano Y, Onomoto K, Murakami F, Komori C, Suzuki Y, Yoneyama M, Ui‐Tei K (2018) LGP2 virus sensor regulates gene expression network mediated by TRBP‐bound microRNAs. Nucleic Acids Res 42: D68–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigawa Y, Nagano‐Fujii M, Deng L, Hidajat R, Tanaka M, Mizuta H, Hotta H (2004) Suppression of hepatitis C virus replicon by RNA interference directed against the NS3 and NS5B regions of the viral genome. Microbiol Immunol 48: 591–598 [DOI] [PubMed] [Google Scholar]
- Tan EL, Tan TMC, Tak Kwong Chow V, Poh CL (2007) Inhibition of enterovirus 71 in virus‐infected mice by RNA interference. Mol Ther 15: 1931–1938 [DOI] [PubMed] [Google Scholar]
- Taylor DW, Ma E, Shigematsu H, Cianfrocco MA, Noland CL, Nagayama K, Nogales E, Doudna JA, Wang H‐W (2013) Substrate‐specific structural rearrangements of human Dicer. Nat Struct Mol Biol 20: 662–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR (2014) Response to Voinnet etal . Cell Rep 9: 798–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR (2016) The evolution of antiviral defense systems. Cell Host Microbe 19: 142–149 [DOI] [PubMed] [Google Scholar]
- tenOever BR (2017) Questioning antiviral RNAi in mammals. Nat Microbiol 2: 17052 [DOI] [PubMed] [Google Scholar]
- Tompkins SM, Lo C‐Y, Tumpey TM, Epstein SL (2004) Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA 101: 8682–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable‐Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M (2007) Suppression of microRNA‐silencing pathway by HIV‐1 during virus replication. Science 315: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Kennedy EM, Cullen BR (2018) Influenza A virus‐derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. RNA 24: 1172–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche Grass S, Borg A, Kjær S, Snijders AP, Reis e Sousa C (2018) The RIG‐I‐like receptor LGP2 inhibits Dicer‐dependent processing of long double‐stranded RNA and blocks RNA interference in mammalian cells. EMBO J 37: e97479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR (2001) Functional classification of interferon‐stimulated genes identified using microarrays. J Leukoc Biol 69: 912–920 [PubMed] [Google Scholar]
- de Vries W, Haasnoot J, Fouchier R, de Haan P, Berkhout B (2009) Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J Gen Virol 90: 1916–1922 [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW (2006a) RNA interference directs innate immunity against viruses in adult Drosophila . Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M (2006b) Hepatitis C virus core protein is a potent inhibitor of RNA silencing‐based antiviral response. Gastroenterology 130: 883–892 [DOI] [PubMed] [Google Scholar]
- Wang R, Wang J, Paul AM, Acharya D, Bai F, Huang F, Guo YL (2013) Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double‐stranded RNA. J Biol Chem 288: 15926–15936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR (2006) Double‐stranded RNA is produced by positive‐strand RNA viruses and DNA viruses but not in detectable amounts by negative‐strand RNA viruses. J Virol 80: 5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S , Jacob R, Devignot S, Kochs G, García‐Sastre A, Weber F (2013) Incoming RNA virus nucleocapsids containing a 5′‐triphosphorylated genome activate RIG‐I and antiviral signaling. Cell Host Microbe 13: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werk D, Schubert S, Lindig V, Grunert H‐P, Zeichhardt H, Erdmann VA, Kurreck J (2005) Developing an effective RNA interference strategy against a plus‐strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus‐adenovirus receptor. Biol Chem 386: 857–863 [DOI] [PubMed] [Google Scholar]
- Wianny F, Zernicka‐Goetz M (2000) Specific interference with gene function by double‐stranded RNA in early mouse development. Nat Cell Biol 2: 70–75 [DOI] [PubMed] [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K (2005) RNA interference is an antiviral defence mechanism in Caenorhabditis elegans . Nature 436: 1044–1047 [DOI] [PubMed] [Google Scholar]
- Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32: 461–488 [DOI] [PubMed] [Google Scholar]
- Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann H‐H, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM (2018) Intrinsic immunity shapes viral resistance of stem cells. Cell 172: 423–438.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Li X, Gao W, Fu X, Fang RH, Zhang L, Zhang K (2018) Tissue repair and regeneration with endogenous stem cells. Nat Rev Mater 3: 174–193 [Google Scholar]
- Xu Y‐P, Qiu Y, Zhang B, Chen G, Chen Q, Wang M, Mo F, Xu J, Wu J, Zhang R‐R, Cheng M‐L, Zhang N‐N, Lyu B, Zhu W‐L, Wu M‐H, Ye Q, Zhang D, Man J‐H, Li X‐F, Cui J et al (2019) Zika virus infection induces RNAi‐mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 46: 509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K (2001) Specific double‐stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol 21: 7807–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi‐Gasic E, Haywood V, Archer‐Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Cheung PKM, Zhang HM, Chau D, Yang D (2005) Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J Virol 79: 2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double‐stranded RNA directs the ATP‐dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]


