Abstract
Drug combinations can improve the control of diseases involving redundant and highly regulated pathways. Validating a multi-target therapy early in drug development remains difficult. Small interfering RNAs (siRNAs) are routinely used to selectively silence a target of interest. Owing to the ease of design and synthesis, siRNAs hold promise for combination therapies. Combining siRNAs against multiple targets remains an attractive approach to interrogating highly regulated pathways. Currently, questions remain regarding how broadly such an approach can be applied, since siRNAs have been shown to compete with one another for binding to Argonaute2 (Ago2), the protein responsible for initiating siRNA-mediated mRNA degradation. Mathematical modeling, coupled with in vitro and in vivo experiments, led us to conclude that endosomal escape kinetics had the highest impact on Ago2 depletion by competing lipid-nanoparticle (LNP)-formulated siRNAs. This, in turn, affected the level of competition observed between them. A future application of this model would be to optimize delivery of desired siRNA combinations in vitro to attenuate competition and maximize the combined therapeutic effect.
Keywords: RNA interference, siRNA, lipid nanoparticles, combination therapy, competition, mathematical modeling
Introduction
Combination therapy has the potential to overcome the redundancy found in complex biological pathways to yield more effective drug treatments.1 It is not surprising that many of the first-line drug treatments have a direct effect on multiple targets, which may be crucial to their efficacy.2 Yet, current drug development focuses on a single target, limiting the likelihood of identifying drugs similar to these in the future. One solution would be a multi-target approach where two or more single-target treatments are developed with the goal of combining them to yield a potent combination therapy.3 However, this approach is hampered by both the cost and difficulty of pursuing multiple small-molecule programs early in the drug development process.
Based on the advancements made in RNAi therapeutics within the past decade,4 now is the right time to leverage the siRNA technology for combination therapy. Small interfering RNAs (siRNAs) are 21–23-nucleotide, double-stranded RNA molecules that negatively regulate gene expression by inducing mRNA cleavage at a complementary site to one of the two siRNA strands.5, 6 This process, referred to as RNA interference (RNAi), occurs through the loading of an siRNA into the RNA-induced silencing complex (RISC), whereby one strand (the passenger) is degraded and the other (the guide) leads RISC to the mRNA cleavage site.7, 8, 9, 10 The Argonaute 2 (Ago2) protein within RISC catalyzes mRNA cleavage of the guide strand/mRNA duplex, leading to its subsequent degradation.11, 12
RNAi has been demonstrated to be a powerful tool for both the identification and validation of genes in cell-based assays and for in vivo applications.13 The advantage of siRNAs over traditional small-molecule drug programs14 places siRNAs in a better position to evaluate the simultaneous inhibition of multiple targets. Moreover, with the advancements in the field of bioinformatics, it has become easy and cost effective to design target-specific siRNAs.15 These siRNAs can inhibit multiple targets in a single metabolic pathway, with high specificity and minimal off-target effects. Recent developments in RNAi formulation and delivery16 have advanced the use of RNAi as a therapeutic itself for targets that would otherwise not be druggable using conventional methods. Recently, the U.S. Food and Drug Administration (FDA) approved the first lipid-nanoparticle (LNP)-based siRNA therapy developed by Alnylam Pharmaceuticals.17 This will indeed be a big boost to the RNAi field going forward.
The delivery vehicles used for siRNA delivery, such as LNPs18 and siRNA conjugates,19, 20 are generic in nature and do not need optimization for every single siRNA. Thus, once we have the siRNAs of interest and the optimized delivery vehicle in hand, we can potentially move directly from the discovery phase to the preclinical phase.21 This saves time and resources when it comes to developing siRNAs as a therapeutic versus small molecules, which require rounds of optimization. Also, because of the enhanced specificity of the individual siRNAs, we can potentially circumvent the issue of drug-drug interactions, which is a concern for small molecules used in combination therapy.22
The strength of utilizing siRNAs for combination therapy is demonstrated by the fact that one of the first clinical trials for systemic RNAi was an siRNA combination targeting the Kinesin Spindle Protein (KSP) and Vascular Endothelial Growth Factor (VEGF).23 We have also explored various combination therapies. For example, by combining Dgat2 siRNA with Mtp siRNA, we were able to rescue steatosis induced by Mtp siRNA treatment alone.24 How broadly such an approach can be applied remains questionable though, given that several groups have shown that siRNAs in vitro can compete with one another for binding to Ago2, leading to reduced activity for one of the two siRNAs within a combination pair.25, 26 Open questions remain regarding the generality of siRNA competition and to what degree siRNA competition translates across delivery platforms and into animals. Particular concerns regarding toxicity remain, given that the introduction of siRNAs to cells has been shown to compete with endogenous miRNAs perturbing these pathways.27 Additionally, the overexpression of short hairpin (sh)RNAs in mice has led to fatalities due to competition between shRNAs for the RNAi machinery.28 We were therefore interested in trying to mechanistically understand the factors that govern siRNA competition and propose ways to alleviate competition in vivo. We chose to use LNPs as our delivery platform for siRNAs, given that this delivery platform is being developed for RNAi-based therapeutics and versions similar to the formulation reported here are already in the clinic.29 In order to understand what dictates competition between multiple siRNAs, and mechanistically explain the differences we observed in the degree of competition between siRNAs when delivered by the commonly used transfection reagent RNAiMax (Lipofectamine) and LNPs, we used a combination of mathematical simulations and in vitro and in vivo experiments. Toward this end, we built upon the recently published mathematical model that was validated for the kinetics of LNP-mediated siRNA delivery.30
Results
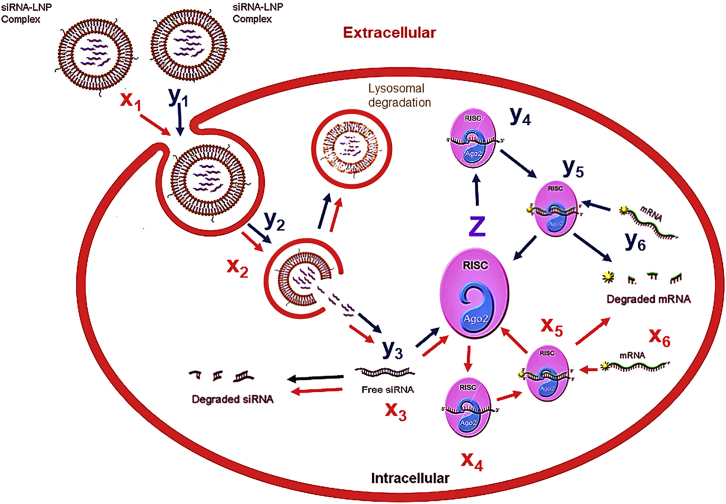
Kinetic Model of LNP-Mediated Delivery of Two siRNAs
We recently published a validated model for LNP-mediated siRNA delivery30 and have further built upon it to simulate the kinetics of a pair of siRNAs, using LNP as the mode of delivery. The extracellular and intracellular volumes are represented as separate modules in a two-compartment model (Figure 1). The various steps are numbered and described in the legend. The units of concentration for the species used in the model are the number of molecules per cell. The model comprises two sets of parallel ordinary differential equations (ODEs) describing the dynamics of the state variables. The input of the model [x1(0), y1(0)] is the extracellular siRNA concentration of two siRNAs (encapsulated in RNAiMax or LNP) at the initial moment of time (t = 0), and the output is the respective mRNA expression x6(T), y6(T), where T is the final time. Both siRNAs compete for Ago2 (z). The system of ODEs we used to simulate the model is as follows:
1.i dx1/dt = x1 − a*x2
1.ii dy1/dt = y1 − a*y2
2.i dx2/dt = a*x1 − (b+c)*x2
2.ii dy2/dt = a*y1 − (b+c)*y2
************************************
3.i dx3/dt = b*x2 − e*x3 − di*x3*z
3.ii dy3/dt = b*y2 − e*y3 − dii*y3*z
************************************
Z. dz/dt = z − di*x3*z − dii*y3*z
************************************
4.i dx4/dt = di*x3*z − f*x4*x6
4.ii dy4/dt = dii*y3*z − f*y4*y6
5.i dx5/dt = f*x4*x6 − g*x5
5.ii dy5/dt = f*y4*y6 − g*y5
6.i dx6/dt = h - i*x6 − k*x5
6.ii dy6/dt = h - i*y6 − k*y5
Figure 1.
ODE Compartment Model of siRNA Combinations
Schematic of the kinetic model with the key steps involved in the LNP-mediated delivery of two siRNAs (x1, y1). Z, Ago2. The various steps involved are as follows: (1) LNP crossing of the plasma membrane, (2) endosomal escape-unpackaging of LNPs, (3) lysosomal degradation of siRNA, (4) loading of siRNA onto the RISC, (5) degradation of siRNA in the cytoplasm, (6) formation of active RISC with target mRNA, (7) cleavage of target mRNA by RISC, (8) transcription rate (of mRNA), and (9) degradation of mRNA.
The initial conditions, the variables and the constant parameters used in the above equations, are described in Table S1. We modeled these differential equations in the Simbiology toolbox developed for MATLAB users (MathWorks, Natick, MA, USA).
A Shift in Potency Was Observed for Target siRNA on the Addition of a Competitor siRNA In Vitro Using RNAiMax as the Transfection Agent
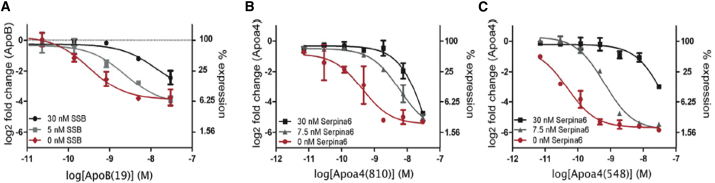
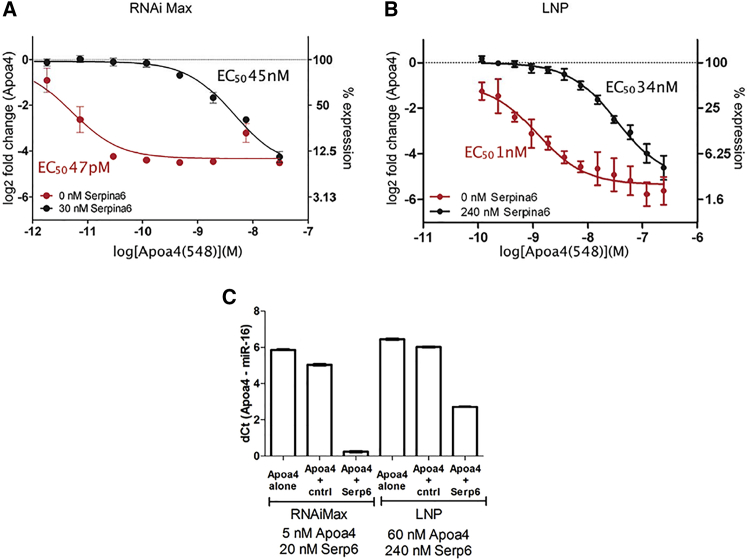
Previous work has demonstrated the potential for two siRNAs to compete with one another for assembly into RISC.31, 32, 33 In these studies, siRNA competition led to an observed decrease in mRNA silencing for the out-competed siRNA at a target-to-competitor siRNA ratio of 1:1 or less. To investigate the extent that competition influences siRNA activity, we first wanted to identify siRNA pairs from our existing siRNA library that exhibit robust competition under previously reported conditions. A total of 48 siRNA combination pairs were evaluated, and significant inhibition of mRNA silencing was observed for 24 siRNA combinations at a 1:6 target-to-competitor siRNA ratio and for 34 siRNA combinations at a 1:30 target-to-competitor siRNA ratio (data not shown). We next examined competition by measuring the change in potency (half-maximal effective concentration [EC50]) caused by the presence of a competitor siRNA for three siRNA pairs exhibiting a range of activity loss due to competition. Here, decreased potency (EC50) was observed for ApoB(19), Apoa4(810), and Apoa4(548) siRNAs by sequentially increasing the concentration of a competitor siRNA: SSB(291) or Serpina6(1011) (Figures 2A–2C).
Figure 2.
Competition between siRNAs’ Shifted siRNA Potency EC50
(A–C) Dose-response curves for (A) ApoB(19) with or without a competitor siRNA (SSB(291), (B) Apoa4(810) with or without a competitor siRNA Serpina6(1011), and (C) Apoa4(548) with or without a competitor siRNA Serpina6(1011) were measured in Hepa 1-6 cells for ApoB(19) or in primary mouse hepatocytes for Apoa4(810) and Apoa4(548). Data were fit to a sigmoidal dose-response curve; error bars, ±SD, n = 2.
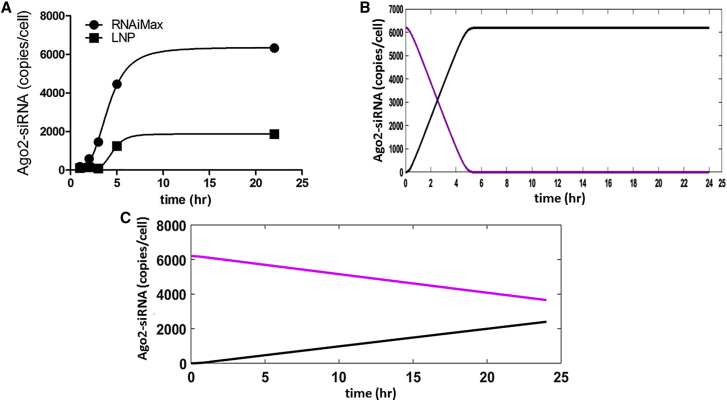
Comparison of Ago2-Loading Kinetics of SSB(291), Using RNAiMax and LNP as the Delivery Vehicle, Derived Experimentally and Through Mathematical Simulations
We encapsulated 10 nM SSB(291) siRNA into RNAiMax and LNP and then used loop RT-PCR of anti-mouse Ago2 immunoprecipitate to measure the Ago2-loading kinetics of the two different delivery vehicles in vitro. The amount of SSB(291) siRNA bound to Ago2 was ∼3-fold lower when LNP was used as the delivery vehicle, as compared with RNAiMax (Figure 3A). These results matched our simulations generated using our previously validated mathematical model (Figures 3B and 3C, RNAiMax and LNP, respectively). Together, these data and our modeling results suggest that transfection with RNAiMax exhibits faster Ago2-loading kinetics relative to LNP-mediated delivery and is thus capable of saturating Ago2 under these conditions.
Figure 3.
Comparison of Experimental and Simulated Kinetics of SSB(291) binding to Ago2, Using RNAiMax- or LNP-Mediated Delivery
(A) A 3-fold greater amount of SSB(291) siRNA bound to Ago2 was observed for RNAiMax-mediated (circles) delivery relative to LNP (squares) delivery at 10 nM siRNA concentration. Data are represented as the absolute number of siRNA copies per cell. The average expression level for each group is shown (±SD = 50, based on n = 3). siRNA was transfected using either RNAiMax or LNP. The assay was performed in Hepa 1-6 cells 4.4 × 106 cells/10 cm plate. Simulation kinetics of SSB(291) siRNA bound to Ago2 using (B) RNAiMax (black line) and corresponding Ago2 depletion (purple line). (C) Simulated kinetics of SSB(291) siRNA bound to Ago2 using LNP (dotted black line) and corresponding Ago2 reduction (dotted purple line).
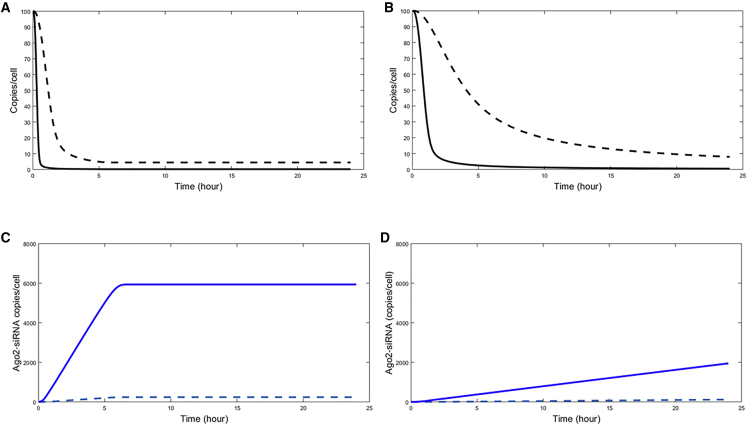
Simulation of the Kinetics of the Target siRNA on Addition of a Competitor siRNA
In order to better understand the impact of the delivery vehicle on the kinetics of various intracellular species (mRNA knockdown and Ago2 loading), we simulated mRNA knockdown of the target siRNA with the competitor siRNA in a 1:6 ratio and without it (Figures 4A and 4B). Our simulations suggest that the difference between the Ago2-loading kinetics of the target siRNA [ApoB(19)] alone versus with a competitor siRNA [SSB(291)] is more profound in the case of RNAiMax- than with LNP-mediated delivery (Figures 4C and 4D).
Figure 4.
Results of Numerical Simulated Kinetics of ApoB(19) +/− Competitor siRNA SSB(291), Using RNAiMax and LNP
(A–D) Simulation of kinetics of mRNA knockdown of target siRNA ApoB(19) without (black solid line) and with (black dotted line) the competitor SSB(291), upon delivery by RNAiMax (A) and LNP (B). Simulation of kinetics of Ago2 loading of target siRNA ApoB(19) without (blue solid line) and with (blue dotted line) the competitor SSB(291), upon delivery by RNAiMax (C) and LNP (D).
Competition between siRNAs Is Substantially Attenuated, and Binding to Ago2 Is Reduced When LNP Is Used as the Delivery Vehicle, as Compared with RNAiMax In Vitro
To determine if the degree of competition between siRNAs is dependent on the mode of delivery, we evaluated competition for the same siRNA pair [Apoa4(548) and Serpina6(1011)], using both LNP- and RNAiMax-mediated delivery. In the case of RNAiMax (Figure 5A), we observed a 1,000-fold reduction in the half-maximal effective concentration (EC50) of the target siRNA Apoa4(548), when a competitor siRNA, Serpina6(1011), was added. On the contrary, in the case of LNP (Figure 5B), we saw only a modest reduction (30-fold) in EC50. In order to see the effect of the mode of delivery of siRNA combinations on Ago2 loading, we determined the relative amount of siRNA bound to Ago2, using both RNAiMax and LNP. We chose a 1:4 ratio of target siRNA, Apoa4(548), to competitor siRNA, Serpina6(1011). In the case of RNAiMax, we observed an ∼6-fold reduction in dCt for Apoa4-Ago2 loading relative to the dCt when no competitor siRNA was added (Figure 5C). Since the efficiency of LNP-mediated delivery is much lower than RNAiMax, we started with 12-fold higher concentrations of the siRNAs while maintaining the same target-to-competitor ratio (1:4). This normalized the relative amount of siRNA bound to Ago2, and thus the relative amount of siRNA bound to Ago2 became comparable between the two delivery methods. Without the addition of any competitor siRNA, the levels of Apoa4-Ago2 loading were comparable for both RNAiMax and LNP. However, when the competitor siRNA was added, we observed only an ∼2-fold reduction in dCt for Apoa4-Ago2 loading in the case of LNP-mediated delivery (Figure 5C)
Figure 5.
Competition between siRNAs Is Substantially Attenuated, and Binding to Ago2 Is Reduced When LNP Is Used as the Delivery Vehicle, as Compared to RNAiMax in Primary Mouse Hepatocytes
(A–C) A dose response for Apoa4(548) siRNA treatment was measured with or without the addition of the Serpina6(1011) competitor siRNA, using either (A) RNAiMax- or (B) LNP-mediated delivery. Data were fit to a sigmoidal dose-response curve; error bars, ±SD, n = 2. (C) The first three bars show the relative amount of siRNA bound to Ago2, measured for cells treated with Apoa4(548) alone, with Apoa4(548) in combination with a non-competing control siRNA (Apoa4+cntrl) and in combination with Serpina6(1011) (Apoa4+Serp6). The latter three bars show the same combination of siRNAs tested using LNP as the delivery vehicle. Data represented the difference in signal (dCt) for Apoa4 relative to miR-16, which served as an internal control; error bars, ±SD, n = 3.
Combination siRNA Delivery by LNP at a Target-to-Competitor siRNA Ratio of 1:5 Attenuates Competition between the siRNAs In Vivo
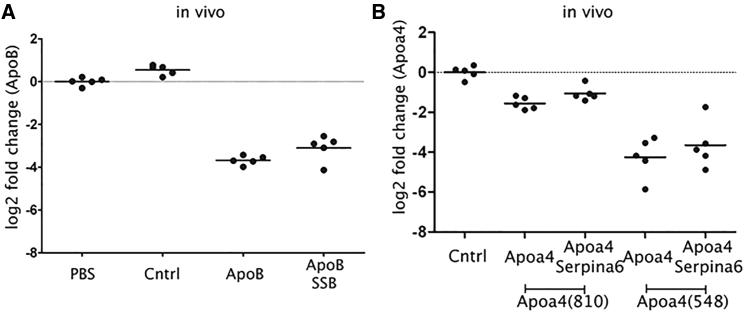
We next attempted to determine to what degree competition between siRNAs translates from an in vitro cell culture system to an in vivo animal model. Toward this end, we evaluated mRNA silencing in mice treated with 1 mg/kg of ApoB(19), Apoa4(810), or Apoa4(548), alone or in combination with 5 mg/kg of the corresponding competitor siRNA (Figure 6). siRNAs were encapsulated within LNPs, and the lipid dose was adjusted by adding additional lipid to the single-target siRNA treatment to ensure that the total lipid dose was equivalent for the single-target siRNA relative to the combination (target plus competitor siRNA) dose. For each target siRNA, similar levels of silencing were observed, whether or not the competitor siRNA was added 1 day after the dose (Figures 6A and 6B). We also performed a comparison study examining if siRNA competition would be higher if the two siRNAs were formulated together into an LNP, and our results revealed no difference in the degree of competition observed for siRNAs formulated together or hand-mixed immediately prior to siRNA administration (data not shown).
Figure 6.
Competition between siRNAs Is Not Observed in Mice
(A and B) A 1 mg/kg dose of (A) ApoB(19) or (B) Apoa4(810) or Apoa4(548) was administered to mice intravenously (i.v.), alone or in combination with 5 mg/kg of the corresponding competitor siRNA using LNP. LNP was added to the single siRNA treatment to ensure that an equivalent amount of lipid was administered in the siRNA dose relative to the combination dose. Data represented as individual animals and group means (bars). Cntrl represents the negative control siRNA treatment group.
Initial Levels of Ago2 Expression Do Not Drive Competition between siRNAs In Vivo at a 1:5 Target-to-Competitor siRNA Ratio
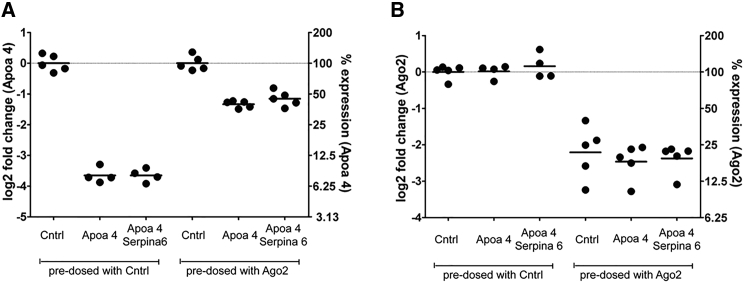
Reducing Ago2 expression levels has been shown to induce greater competition between siRNAs for RNAiMax-mediated delivery in cells by lowering the amount of siRNA needed to reach the threshold for Ago2 saturation.31 To determine if knocking down Ago2 could also lead to greater competition in vivo, we pre-treated mice with a 3 mg/kg dose of an Ago2 or control siRNA on day −1, administered either target siRNA [Apoa4(548)] alone or in combination with a competitor siRNA [Serpina6(1011)] encapsulated within LNPs on day 0, and measured Ago2 expression levels on day 1. Although the expression levels were reduced for the Ago2 pre-treated groups (Figures 7A and 7B), similar levels of Apoa4 silencing were observed, with or without the addition of the competitor siRNA Serpina6 (Figure 7A and 7B).
Figure 7.
Reducing Ago2 Expression Levels Influences siRNA Potency but Not siRNA Competition In Vivo
(A) Apoa4 knockdown 1-day after Apoa4(548) siRNA treatment ± 5-fold excess of a competitor siRNA [Serpina6(1011)]. Apoa4(548) was administered intravenously (i.v.) at 1 mg/kg, with or without the simultaneous injection (i.v.) of 5 mg/kg Apoa4(548). LNP was added to the Apoa4(548)-only siRNA treatment, to ensure that an equivalent amount of lipid was administered for the single siRNA dose relative to the combination dose. Mice were pre-treated with an siRNA targeting Ago2 [Ago2(722)] or negative control siRNA (Cntrl) 2 days prior to the dose of the siRNA combination. (B) Similar levels of Ago2 knockdown were observed across all Ago2 siRNA-treated groups. Individual mice (circles) and the average expression level for each group (bars) are shown.
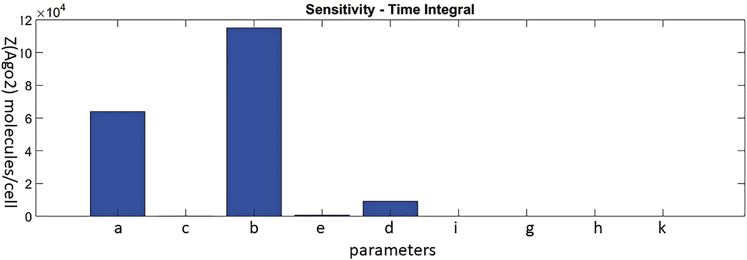
Sensitivity Analysis of the Effect of Various Model Parameters on Ago2 Depletion
The sensitivities of Z (Ago2 depletion) with respect to all model parameters are ranked (Figure 8). The conclusion from these rankings is that endosomal escape and cell entry have the highest impact on Ago2 depletion. These two parameters are in turn dependent on the delivery vehicle. siRNA loading into Ago2 also has an impact, but not as substantial as the other two parameters.
Figure 8.
Parametric Sensitivity Analysis Showing the Influence of Model Parameters on Ago2 Depletion
Sensitivity analysis carried out for all the model parameters, using the Simbiology toolbox in MATLAB. The bar graphs show the consolidated sensitivity metrics of Z (Ago2 depletion) with respect to model parameters. Parameters ranked from highest to lowest impact are as follows: endosomal escape (b), cell entry (a), siRNA-Ago2 loading (d), and degradation of siRNA in the cytoplasm (e). The other parameters did not seem to have an effect on Ago2 loading.
Discussion
The introduction of excess siRNAs into a cell leads to the saturation of Ago2.31 Saturation leads to competition, resulting in a reduction in siRNA potency when siRNAs are combined. This is exactly what we observed in the case of RNAiMax-mediated delivery. To probe further, we systematically studied the effect of a competitor siRNA on the kinetics of a target siRNA. We have previously shown that the key steps involved in the LNP-mediated delivery of siRNA are endosomal escape and Ago2 loading of siRNA into a RISC complex.30 Since Ago2 saturation leads to competition between siRNAs, these two key steps would have a role in determining the level of competition. Toward this goal, we built an ODE model to compare the kinetics of two competing siRNAs delivered into cells by an LNP (Figure 1). Once the initial model was defined through parameters determined by actual biochemical experiments, we systematically changed the model parameters to see the corresponding effect on the kinetics of the target siRNA in the presence of a competitor siRNA. Although others have built similar competition models for small RNAs,34 they do not directly address the issue of competition of siRNAs and the impact on the therapeutic effect. Moreover, the model was not supported by experimental data. The advantage of our model over the other published models is that we validated the critical steps involved in the delivery of siRNA previously and, based on our sensitivity analysis, determined the impact of these key parameters on LNP-mediated delivery of siRNA. For an individual siRNA, we previously showed that endosomal escape had a greater impact on silencing than siRNA-Ago2 loading.30
When siRNAs were administered in vitro with an efficient transfection reagent that was able to saturate Ago2 (RNAiMax), we observed competition in a portion of the siRNA pairs tested, but not all. Our mathematical model provides a possible explanation for these data by emphasizing the importance of Ago2-loading kinetics. The model predicts that, in order to observe competition, a sufficient quantity of the competitor siRNA has to be delivered into the cytoplasm by the delivery vehicle relatively quickly to saturate Ago2 (Figure S1). In parallel, the biochemical assay used in this study for measuring Ago2-loading kinetics can serve as a screening tool for possible combination pairs. We used this biochemical assay to assess the difference in siRNA-loading kinetics between RNAiMax and LNP (Figure 3A). Our simulations showed that Ago2 is saturated in the case of RNAiMax-mediated delivery but not when using LNP (Figures 3B and 3C). These simulations correlate with the in vitro experiments mentioned above. This result recapitulates our understanding of the differences between the kinetics of the two delivery vehicles, which can, in turn, be attributed to the corresponding differences in their biophysical properties and the manner in which the siRNAs are encapsulated within them.30
We demonstrated that competition between siRNAs for Ago2 in vitro is dependent on the delivery vehicle used. LNP-mediated delivery resulted in a reduction in the degree of competition observed for siRNA combinations. As compared with RNAiMax, LNP-mediated delivery resulted in a 20-fold decrease in the potency for the target siRNA when administered alone (Figure 5). This was observed in both Hepa 1-6 cells and in primary mouse hepatocytes, suggesting that these observations are not unique to a particular cell line. When we compared siRNAs across these cells, we did not observe an appreciable difference in the degree of competition that occurs in Hepa 1-6 cells relative to primary mouse hepatocytes (data not shown). Presumably, both the decrease in potency and the decrease in the severity of competition for LNP-mediated delivery can be attributed to less efficient cell entry, endosomal escape from the lipid complex, or both.
We modeled the in vitro kinetics of the target siRNA with and without the competitor siRNA and under RNAiMax- and LNP-mediated delivery (Figure 4). Our simulations indicate, as expected, that mRNA knockdown and siRNA-loading kinetics decrease in the presence of a competitor. With the LNP-mediated delivery, this effect is more pronounced. Ago2 in the case of LNP-mediated delivery is not saturated, and the effect of the competitor is therefore diminished (Figure S1).
In order to test our hypothesis, the target siRNA binding to Ago2 was measured at earlier time points. We observed that less siRNA was bound to Ago2 for LNP-mediated delivery relative to RNAiMax, even though 12-fold more siRNA was added using the LNP formulation (Figure 5C). As a control, we measured the number of copies of miR-16, which remained unchanged upon treatment (data not shown). Less siRNA bound to Ago2 for LNP-mediated delivery at these earlier time points suggests slower Ago2 loading. To further illustrate this point, we matched the siRNA concentration for RNAiMax- and LNP-mediated delivery, and the difference in the amount of siRNA bound to Ago2 was more pronounced (Figure S2). Together, these data suggest that transfection with RNAiMax exhibits faster Ago2-loading kinetics relative to LNP-mediated delivery.
When the LNP is administered in vivo, the ease of release of the siRNA from the complex is no longer the only rate-limiting factor, as is the case in vitro. There are additional barriers that the LNP has to overcome before it can cross the cell membrane and get into the tissue of interest.35, 36 Only a very small percentage of the injected LNPs are able to make it to the target cells,37 following which they fuse with the cell membrane, get internalized via endocytosis, and release their payload, i.e siRNA, into the cytoplasm. As a result of all these barriers, LNP-mediated siRNA delivery is less efficient in vivo, leading to a shift in potency and a reduced level of siRNA competition. We have shown previously that the majority of the siRNA dose administered to mice is cleared within several hours of administration, and only the siRNA bound to Ago2 remains.38 It is the amount of siRNA bound to Ago2 and not the total siRNA dose that correlates with knockdown. Therefore, the intracellular kinetic steps (endosomal escape, Ago2 binding, and crossing the cellular membrane, as shown in our sensitivity analysis) play a much larger role in the competition kinetics and are addressed with our mathematical model. Additionally, our in vivo competition experiments use the same delivery vehicle and different competing siRNAs. Therefore, each siRNA is exposed to the same delivery barriers, since it is encapsulated in the same LNP, and barriers to delivery, such as tissue distribution, were therefore excluded from our model. After the siRNAs are released from the LNP into the endosome, the additional intracellular barriers (Ago2 binding, siRNA degradation) will affect the kinetics based on the siRNA sequence. These kinetic steps are represented in our model (steps 3 and Z).
Previously published in vivo results indicate that 1–6 mg/kg of siRNA does not saturate Ago2.38 Analysis of Ago2 mRNA levels following escalating doses of luciferase or SSB(291) siRNA administration failed to reveal any significant increase in Ago2 mRNA expression in the liver (data not shown). The quantitative Ago2 time-dependent loading analysis indicates that, for the SSB(291) siRNA sequence used in this study, ∼300 copies were loaded per cell, leading to a 50% reduction in mRNA expression in vitro. Interestingly, this estimated value is similar to values reported previously in vitro and in vivo.39 Our simulations also reflect these numbers and recapitulate the importance of siRNA-loading kinetics as the main driver for competition. Although each LNP may contain up to 4,000 siRNAs,40 our data suggest that the large majority does not even reach Ago2, because of the inefficient release of the siRNA from the LNP, which affects its ability to saturate RISC and thereby to induce competition. Whereas in vitro we were able to observe competition at a ratio of 1:6 (target:competitor) with the RNAiMax transfection reagent, that same effect was not observed in vivo. Our attempt to knockdown Ago2 in order to impact competition (Figure 7) illustrates that the initial Ago2 levels alone do not determine the level of competition, as previously hypothesized.31 Our model presents a detailed view and a system-level understanding of the effect of the kinetic steps prior to Ago2 siRNA loading and their relative impact on competition between siRNAs delivered by LNPs.
Interestingly, since competition is attenuated between siRNAs when LNPs are used as the delivery vehicle, it should be plausible to see a combined therapeutic effect with multiple siRNAs using LNP-mediated delivery. The challenge would be to direct the optimal amount of siRNAs released from the LNPs into the cytoplasm for Ago2 loading. Future studies should be directed toward achieving the desired combination effect of multiple siRNAs in vivo by optimizing the ratios of the respective siRNAs, based on the Ago2-loading kinetics of the individual siRNAs, encapsulated within LNPs. As demonstrated, kinetics of LNP-mediated delivery of the individual siRNAs can be simulated from the mathematical model, without having to conduct expensive and time-consuming in vitro-in vivo experiments. Finally, it is worth noting that the FDA is expected to release new guidelines in favor of the development of drug combinations. It is anticipated that the FDA will no longer require proof of efficacy for each active compound within a drug treatment. This should help pave the way for the development of drug combinations that target multiple pathways, which, when inhibited individually, may be only modestly efficacious, but when inhibited together would be robustly efficacious. An advantage of using siRNAs over small molecules as combination therapy is that they can silence or inhibit multiple targets simultaneously without concern for adverse interactions that can occur when two small molecules are combined. Therefore, the new guidelines should favor the use of siRNAs for the development of novel combination therapies.
While more work is needed to understand the underlying mechanism, the development of siRNA combination therapies is well under way. Here, we show that a similar lipid complex used for delivery reduced both the potency of an individual siRNA and the level of competition observed for an siRNA combination relative to RNAiMax. The fact that the same lipid complex exhibited both reduced potency and a decrease in activity loss due to competition suggests that improved potency may come at the risk of increased competition for an siRNA combination. Decreased potency can be a potential advantage in some instances, by enabling targeting of multiple genes simultaneously without concern for activity loss due to competition. Previous work has demonstrated that up to five siRNAs can be co-administered to mice by using a similar lipid complex with ∼10-fold greater potency.41
Both the potency shift and the shift in competition suggest that there are differences in siRNA availability for target silencing between RNAiMax- and LNP-mediated delivery. This difference can be attributed to differences in siRNA uptake, the ease with which the siRNA can escape from the lipid complex, or both. We characterized in depth the biophysical properties of the LNPs used in this study previously by cryo-electron microscopy (cryo-EM; Figure S3) and dynamic light scattering (DLS).42 The nitrogen-to-phosphate ratio (N:P) is 3:3 and the composition of the LNP particle for the delivery vehicle used in this study is as follows: 60% cationic lipid, 38% cholesterol, and 2% polyethylene glycol (PEG). The DLS mean diameter is 96.6 nm, in the optimal range for drug delivery. To characterize the nature of the assembly and interactions that the siRNA has with the two different delivery modalities, we further characterized the two complexes. We measured the amount of siRNA encapsulated within the two modalities in an encapsulation assay and showed that only 30% of the siRNA was encapsulated within the RNAiMax lipoplex, whereas greater than 90% was encapsulated within the LNPs. This result points to the fact that the siRNA was bound to the exterior of the RNAiMax lipoplex rather than being entrapped within the bilayer assembly (as was the case with the LNPs). We next analyzed the differential scanning calorimetric (DSC) profiles for the two complexes. DSC monitors the heat-flow profile continuously and has been used to determine the phase transition point in lipid systems with high accuracy and precision. We measured the transition temperatures (Tm) in the RNAiMax transfection reagent, with and without two independent siRNA cargos (Figures S4A and S4C) and compared the thermograms to the LNP-based system (Figures S4B and S4D). The Apoa4 and Serpina6 siRNAs have measured Tms of 95°C and 82°C, which indicates that the Apoa4 siRNA is a more stable duplex. The RNAiMax transfection reagent does not form measurable phase transitions in the absence or presence of the siRNA cargo, and these data, taken together with our encapsulation measurements, indicate a lack of measurable physical interactions and a lack of self-assembled particles. This result is not surprising, given that the transfection reagent is merely co-administered with the siRNA and does not undergo a rigorous self-assembly process, as in the case of the LNP.43 In contrast, the LNP exhibits three additional phase transitions when assembled with the siRNA cargo and only one phase transition when the LNP is assembled independent of the siRNA cargo. This result, taken together with the encapsulation data, indicates that the siRNA cargo is encapsulated within lipid bilayers. The lipid bilayer undergoes destabilization while the particle is being internalized via endocytosis, and this process results in the displacement of the 21-nucleotide-long siRNA from the lipid complex and its release into the cytoplasm. The multi-vesicular morphology of the LNP, as well as the lamellar-to-inverted hexagonal phase transition temperature, has been shown to play a role in the ability of the LNP to induce endosomal membrane disruption. In the case of the transfection reagent due to the weak interaction with RNAiMax, the siRNA has a greater propensity to escape from the complex and become readily available for subsequent loading onto Ago2. Biophysical characterization of the two delivery vehicles suggests that the tighter the association between the siRNA and the delivery vehicle, the less competition observed.
Based on our in vitro screen, we picked two target siRNAs with different potencies: Apoa4(810) and Apoa4(548). As expected, in the experiments in vitro and in vivo we saw a corresponding difference in silencing based on the potency difference between the siRNAs. However, in the presence of the competitor siRNA Serpina6(1011), we did not observe competition in vivo in both cases. This highlights that there is a key parameter that has a greater impact than the potency of the siRNA in governing the degree of competition (Ago2 depletion by target siRNA). In fact, sensitivity analysis can be used to interrogate the effect of multiple parameters on model output and to shed light on the factors that can affect competition. Based on our sensitivity analysis on the model parameters (Figure 8), our conclusion is that for LNP-mediated siRNA delivery, cell entry and endosomal escape have the largest impact on Ago2 depletion and mRNA knockdown of the target siRNA in the presence of a competitor siRNA. More generally, we may be interested in investigating these parameters further in order to optimize combination therapy, attenuate competition, and maximize therapeutic effect. As a strategy, a delivery vehicle that releases siRNA into the cytoplasm at a slower rate may be more beneficial in order to prevent saturation of Ago2 and thus avoid competition between the siRNAs.
Future application of this model would be to optimize the desired siRNA combinations to attenuate competition and maximize the combined therapeutic effect by focusing on the key parameters that affect competition between siRNAs. Continuing to build on this approach of using a combination of biochemical assays and mathematical modeling, we can devise optimization routines implemented in silico and apply this insight to novel siRNA combinations to attenuate competition and maximize therapeutic effect.
Materials and Methods
siRNA Design
siRNAs were designed to the mRNA transcripts using a previously published design algorithm.44 The siRNA sequences are listed in Table S1 and contained the following chemical modifications added to the 2′ position of the ribose sugar when indicated: deoxy (d), 2′ fluoro (f), or 2′ O-methyl (o). Modification abbreviations are given immediately preceding the base to which they were applied. Passenger strands were capped with an inverted abasic nucleotide on the 5′ and 3′ ends.
siRNA Synthesis
siRNAs were synthesized by methods previously described.45 For each siRNA, the two individual strands were synthesized separately by using solid-phase synthesis, then purified separately by ion-exchange chromatography. The complementary strands were annealed to form the duplex siRNA. The duplex was then ultrafiltered and lyophilized to form the dry siRNA. Duplex purity was tested with liquid chromatography-mass spectrometry (LCMS) and for the presence of endotoxin by standard methods.
Preparation of the siRNA-LNP Complex
LNPs were made using the cationic lipid Octyl CLinDMA (2-{4-[(3b)-cholest-5-en-3-yloxy]-octyl}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol, and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) in a 60:38:2 molar ratio, respectively. siRNAs were incorporated in the LNPs46 with high encapsulation efficiency by mixing siRNA in citrate buffer with an ethanolic solution of the lipid mixture, followed by a stepwise diafiltration process at a lipid-to-siRNA molar ratio of 3.3:1. Cholesterol was purchased from Northern Lipids, PEG-DMG was purchased from NOF, and Octyl CLinDMA was synthesized by Merck. The encapsulation efficiency of the particles was determined, using an SYBR Gold fluorescence assay in the absence and presence of Triton; fluorescence was measured at λex = 495 nm, λem = 535 nm. The particle size measurements were performed with a Wyatt DynaPro plate reader. The siRNA and lipid concentrations in the LNP were quantified by a high-performance liquid chromatography (HPLC) method, developed in-house using a photodiode array (PDA) and charged aerosol detector (CAD).
In Vitro
siRNAs were transfected into Hepa 1-6 cells and primary hepatocytes plated at 3,500 and 16,000 cells per well, respectively, using 0.9 μL RNAiMax (Lipofectamine; Invitrogen). Cells were incubated for 18–24 h prior to isolating RNA, using the Cells to Ct kit (Ambion) according to the product protocol. TaqMan Gene Expression Assays (Applied Biosystems) were performed as described within the product protocol using the following primer probes: Mm00838341_m1 (Ago2), Mm00431814_m1 (Apoa4), Mm01545154_g1 (ApoB), Mm00432327_m1 (Serpina6), Mm00447374_m1 (SSB), and 4352339E (Gapdh). All reactions were performed in duplicate, and data were analyzed using the ddCt method, with GAPDH serving as the internal control.47 The relative amount of siRNA bound to Ago2 was quantitated using stem-loop RT-PCR of anti-mouse Ago2 immunoprecipitate, as described previously.38, 39
In Vivo
All in vivo work was performed according to an approved animal protocol, as set by the Institutional American Association for the Accreditation of Laboratory Animal Care. C57BL/6 male mice (Charles River) ∼8 weeks of age were administered siRNAs by intravenous (i.v.) injection. Animals were euthanized by CO2 inhalation. Immediately after euthanasia, sections from the right medial lobe of each liver were excised, placed in RNALater (for TaqMan Gene Expression Analysis), and stored at 4°C or flash frozen (for siRNA quantification) and stored at −80°C until further use. Quantification of the siRNA concentration in the liver was determined as described previously.38, 39 For TaqMan Gene Expression Analysis, RNA was isolated using QIAGEN’s RNeasy96 Universal Tissue Kit, according to the supplied product protocol. An on-column DNase I treatment was performed, and samples were washed three times prior to elution with 100 μL of RNase-free water. Reverse transcription was performed with the Cells to Ct kit (Ambion) in a 20 μL volume with 350 ng RNA in 1× reverse transcriptase and buffer incubated at 37°C for 1 h. TaqMan Gene Expression Assays (Applied Biosystems) were performed as described within the product protocol, using the primer-probes listed above.
Kinetic Simulations
The mathematical model presented in Figure 1 was simulated, using the Simbiology platform developed by MathWorks and modeled, using a system of differential equations, as shown in the Results section. We solved the equations numerically, using a sundials solver, and the simulations are presented in Figures 3 and 4. We also carried out a sensitivity analysis (Figure 8) using the Simbiology toolbox in MATLAB to determine the critical parameters that control competition. The software uses “complex-step approximation” to calculate derivatives of reaction rates. The relationship between the model input and parameters and the model output is described by a set of differential equations that describe the change in the model variables over time. The aim of the sensitivity analysis is to identify how the output of the model depends on the uncertainty in the parameters.
Author Contributions
B.A., R.M., B.G., A.C., T.S., J.K., S.T., J.W., and R.M.H. performed the experiments. R.M., E.C., M.C., B.A., J.C., W.M.F., N.K., and D.R designed the experiments and analyzed and interpreted the data. B.A., R.M., and D.R. wrote the manuscript.
Conflicts of Interest
All authors were employed by Sirna Therapeutics/Merck & Co., Inc. at the time the experiments were conducted.
Acknowledgments
We thank Duncan Brown for siRNA design and Natalya Dubinina from the in vivo pharmacology group for support with the “in-life” portion of this work.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.03.004.
Supplemental Information
References
- 1.Lopez J.S., Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat. Rev. Clin. Oncol. 2017;14:57–66. doi: 10.1038/nrclinonc.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Csermely P., Agoston V., Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Ramsay R.R., Popovic-Nikolic M.R., Nikolic K., Uliassi E., Bolognesi M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018;7:3. doi: 10.1186/s40169-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorsett Y., Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 6.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Rand T.A., Petersen S., Du F., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 11.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 13.Tsai W.H., Chang W.T. Construction of simple and efficient siRNA validation systems for screening and identification of effective RNAi-targeted sequences from mammalian genes. In: Ochs M., editor. Gene Function Analysis, Methods in Molecular Biology (Methods and Protocols) Volume 1101. Humana Press; 2014. pp. 321–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talevi A. Multi-target pharmacology: possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front. Pharmacol. 2015;6:205. doi: 10.3389/fphar.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tafer H. Bioinformatics of siRNA design. Methods Mol. Biol. 2014;1097:477–490. doi: 10.1007/978-1-62703-709-9_22. [DOI] [PubMed] [Google Scholar]
- 16.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials (Basel) 2017;7:77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhr O.B., Coelho T., Buades J., Pouget J., Conceicao I., Berk J., Schmidt H., Waddington-Cruz M., Campistol J.M., Bettencourt B.R. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study. Orphanet J. Rare Dis. 2015;10:109. doi: 10.1186/s13023-015-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Shum K.-T., Burnett J.C., Rossi J.J. Nanoparticle-Based Delivery of RNAi Therapeutics: Progress and Challenges. Pharmaceuticals (Basel) 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018;9:723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zatsepin T.S., Kotelevtsev Y.V., Koteliansky V. Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int. J. Nanomedicine. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulusu K.C., Guha R., Mason D.J., Lewis R.P.I., Muratov E., Kalantar Motamedi Y., Cokol M., Bender A. Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives. Drug Discov. Today. 2016;21:225–238. doi: 10.1016/j.drudis.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Tabernero J., Shapiro G.I., LoRusso P.M., Cervantes A., Schwartz G.K., Weiss G.J., Paz-Ares L., Cho D.C., Infante J.R., Alsina M. First-in-Humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 24.Tep S., Mihaila R., Freeman A., Pickering V., Huynh F., Tadin-Strapps M., Stracks A., Hubbard B., Caldwell J., Flanagan W.M. Rescue of Mtp siRNA-induced hepatic steatosis by DGAT2 siRNA silencing. J. Lipid Res. 2012;53:859–867. doi: 10.1194/jlr.M021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanudji M., Machalek D., Arndt G.M., Rivory L. Competition between siRNA duplexes: impact of RNA-induced silencing complex loading efficiency and comparison between conventional-21 bp and Dicer-substrate siRNAs. Oligonucleotides. 2010;20:27–32. doi: 10.1089/oli.2009.0195. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Yoo J.W., Lee J.H., Hahn Y., Kim S., Lee D.K. Identification of sequence features that predict competition potency of siRNAs. Biochem. Biophys. Res. Commun. 2010;398:92–97. doi: 10.1016/j.bbrc.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 27.Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 29.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 30.Mihaila R., Ruhela D., Keough E., Cherkaev E., Chang S., Galinski B., Bartz R., Brown D., Howell B., Cunningham J.J. Mathematical modeling: a tool for optimization of lipid nanoparticle-mediated delivery of siRNA. Mol. Ther. Nucleic Acids. 2017;7:246–255. doi: 10.1016/j.omtn.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers T.A., Lima W.F., Nichols J.G., Crooke S.T. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35:6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koller E., Propp S., Murray H., Lima W., Bhat B., Prakash T.P., Allerson C.R., Swayze E.E., Marcusson E.G., Dean N.M. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keck K., Volper E.M., Spengler R.M., Long D.D., Chan C.Y., Ding Y., McCaffrey A.P. Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency. Mol. Ther. 2009;17:538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loinger A., Shemla Y., Simon I., Margalit H., Biham O. Competition between small RNAs: a quantitative view. Biophys. J. 2012;102:1712–1721. doi: 10.1016/j.bpj.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tros de Ilarduya C., Arangoa M.A., Düzgüneş N. Transferrin-lipoplexes with protamine-condensed DNA for serum-resistant gene delivery. Methods Enzymol. 2003;373:342–356. doi: 10.1016/s0076-6879(03)73022-5. [DOI] [PubMed] [Google Scholar]
- 36.Opanasopit P., Nishikawa M., Hashida M. Factors affecting drug and gene delivery: effects of interaction with blood components. Crit. Rev. Ther. Drug Carrier Syst. 2002;19:191–233. doi: 10.1615/critrevtherdrugcarriersyst.v19.i3.10. [DOI] [PubMed] [Google Scholar]
- 37.Barron L.G., Gagné L., Szoka F.C., Jr. Lipoplex-mediated gene delivery to the lung occurs within 60 minutes of intravenous administration. Hum. Gene Ther. 1999;10:1683–1694. doi: 10.1089/10430349950017680. [DOI] [PubMed] [Google Scholar]
- 38.Wei J., Jones J., Kang J., Card A., Krimm M., Hancock P., Pei Y., Ason B., Payson E., Dubinina N. RNA-induced silencing complex-bound small interfering RNA is a determinant of RNA interference-mediated gene silencing in mice. Mol. Pharmacol. 2011;79:953–963. doi: 10.1124/mol.110.070409. [DOI] [PubMed] [Google Scholar]
- 39.Pei Y., Hancock P.J., Zhang H., Bartz R., Cherrin C., Innocent N., Pomerantz C.J., Seitzer J., Koser M.L., Abrams M.T. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010;16:2553–2563. doi: 10.1261/rna.2255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peer D., Park E.J., Morishita Y., Carman C.V., Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love K.T., Mahon K.P., Levins C.G., Whitehead K.A., Querbes W., Dorkin J.R., Qin J., Cantley W., Qin L.L., Racie T. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford R., Dogdas B., Keough E., Haas R.M., Wepukhulu W., Krotzer S., Burke P.A., Sepp-Lorenzino L., Bagchi A., Howell B.J. Analysis of lipid nanoparticles by Cryo-EM for characterizing siRNA delivery vehicles. Int. J. Pharm. 2011;403:237–244. doi: 10.1016/j.ijpharm.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 43.Abrams M.T., Koser M.L., Seitzer J., Williams S.C., DiPietro M.A., Wang W., Shaw A.W., Mao X., Jadhav V., Davide J.P. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majercak J., Ray W.J., Espeseth A., Simon A., Shi X.P., Wolffe C., Getty K., Marine S., Stec E., Ferrer M. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2006;103:17967–17972. doi: 10.1073/pnas.0605461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wincott F., DiRenzo A., Shaffer C., Grimm S., Tracz D., Workman C., Sweedler D., Gonzalez C., Scaringe S., Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mihaila R., Chang S., Wei A.T., Hu Z.Y., Ruhela D., Shadel T.R., Duenwald S., Payson E., Cunningham J.J., Kuklin N., Mathre D.J. Lipid nanoparticle purification by Spin Centrifugation–Dialysis (SCD): A facile and high-throughput approach for small scale preparation of siRNA–lipid complexes. Int. J. Pharm. 2011;420:118–121. doi: 10.1016/j.ijpharm.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.