Abstract
Optical super-resolution techniques reach unprecedented spatial resolution down to a few nanometers. However, efficient multiplexing strategies for the simultaneous detection of hundreds of molecular species are still elusive. Here, we introduce an entirely new approach to multiplexed super-resolution microscopy by designing the blinking behavior of targets with engineered binding frequency and duration in DNA-PAINT. We assay this kinetic barcoding approach in silico and in vitro using DNA origami structures, show the applicability for multiplexed RNA and protein detection in cells, and finally experimentally demonstrate 124-plex super-resolution imaging within minutes.
Keywords: Super-resolution microscopy, DNA nanotechnology, DNA-PAINT, barcoding, multiplexing
The development of optical super-resolution techniques allows researchers to unravel molecular properties of biological systems with thus far unprecedented detail.1−4 While recent technical advancements propel the achievable spatial resolution to the true molecular scale of only a few nanometers,5−7 current implementations are still limited when it comes to imaging many molecular species simultaneously in single cells and beyond. This so-called multiplexing is traditionally achieved using spectrally distinct, fixed labels (e.g., dye-coupled antibodies or nucleic acid probes) on target molecules of interest such as proteins, DNA, or RNA. While spectral multiplexing approaches are relatively straightforward to implement, the amount of “plex” is inherently limited by the number of distinguishable spectral labels in the detectable emission spectrum, which is in most instances three or four.8 However, in order to fully understand the detailed molecular workings of the complex cellular machinery, one ideally would need to be able to look at hundreds if not thousands of unique components and their molecular interplay.
Some efforts to extend spectral multiplexing capabilities include multiparameter and combinatorial detection,9,10 multispectral acquisition,11 and spectrally resolved detection.12,13 While these approaches increase the number of detectable targets, they are ultimately still limited by the spectral properties of fluorescent molecules used to label target structures.
To overcome the limitation inherent in spectral separation for multiplexed detection, several approaches have recently been devised that employ sequential labeling and imaging of targets using spectrally indistinct probes14−17 (i.e., the same dye molecule). Some of these implementations rely on simultaneous labeling of target molecules with orthogonal DNA-barcoded affinity reagents (e.g., antibodies) followed by sequential imaging using dye-labeled complementary oligos (e.g., Exchange-PAINT14 or Universal DNA Exchange17,18). Others use sequential labeling, imaging, and quenching based on dye-conjugated antibodies, e.g., STORM.15,16 While both implementations differ slightly in the combined time for labeling and imaging of each target, the overall experimental time eventually scales linearly with the number of targets to be acquired. This fact ultimately sets a practical limit to the amount of multiplexing achievable. While it might be reasonable to obtain tens of targets with sequential multiplexing, it will become prohibitively time-intensive (and eventually impossible) for hundreds or even thousands of targets. Thus, current multiplexing approaches are inherently limited in terms of the achievable coding depth, overall acquisition time, and ease-of-use.
To overcome this limitation, we here propose an entirely orthogonal approach to achieve multiplexed detection in single-molecule-based super-resolution experiments that allows hundreds or more targets to be imaged simultaneously. Instead of relying on spectral information or sequential imaging, we engineer targets to blink autonomously with precisely adjustable kinetic signatures (i.e., frequency and duration of blinks), essentially providing a distinct kinetic barcode for hundreds of unique molecular species.
In order to implement and demonstrate the concept of engineered blinking kinetics for simultaneous multiplexed super-resolution imaging, we chose DNA-PAINT7,19 as imaging modality. In DNA-PAINT, short dye-labeled oligonucleotides bind transiently to complementary, target-bound DNA molecules, thus creating an apparent blinking at the target site, which in turn is used for stochastic super-resolution microscopy. Due to the versatile programmability of DNA probes, the binding kinetics such as blinking frequency and duration can be tuned precisely and used downstream as “barcodes” for multiplexed detection.
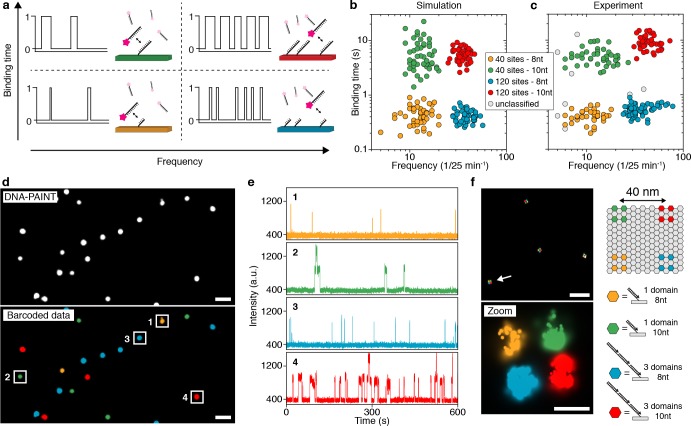
The concept of engineering binding kinetics with DNA-PAINT is schematically shown in Figure 1a. In order to tune the blinking frequency of targets, we label one species with a single DNA-PAINT binding site and an orthogonal species with three binding sites. Assuming a constant influx of imager strands, the blinking frequency will scale linearly with the number of binding sites (e.g., resulting in a blinking frequency of two for the single binding site and a frequency of six for three sites; see Figure 1a). Similarly, we can modulate the blinking duration for a given target molecule by adjusting the length of the docking strand (e.g., 8 nt long docking strands will result in relatively short binding events, while 10 nt long docking sites will result in longer events; see Figure 1a). As binding frequency and duration are independent of each other in DNA-PAINT, these parameters can be combined to perform combinatorial barcoding with just a single imager strand species, thus enabling simultaneous multiplexed imaging.
Figure 1.
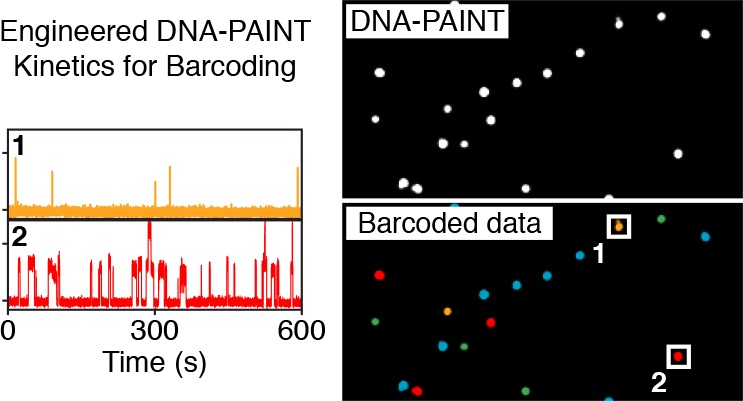
Simultaneous multiplexed super-resolution imaging by engineering blinking kinetics. (a) Engineering blinking kinetics in DNA-PAINT allows the creation of “barcodes” for simultaneous multiplexing, using only a single imager strand species. Frequency can be encoded by designing a certain number of binding sites per target, e.g., a single binding site, leading to a defined blinking frequency. Tripling the number of binding sites triples the blinking frequency (left to right). Similarly, binding duration can be engineered by adjusting the length of the docking strand on a specific target: an 8 nt docking sequence will lead to a “short” binding duration, while a 10 nt docking sequence will result in longer binding (bottom to top). (b) Simulations of four kinetically different structures (40 and 120 binding sites and 8 and 10 nt lengths) show four clearly distinguishable populations corresponding to the engineered frequency and duration levels (see Supplementary Figure 1 for details on cluster detection). (c) Experimental results from DNA origami structures imaged using a single imager strand species show four distinguishable populations in good agreement with in silico data from c (see Supplementary Figure 5 for details on cluster detection). (d) Exemplary overview DNA-PAINT image of the four DNA origami structures (top). Same data set, now color-coded according to identified clusters in c (bottom). (e) Exemplary intensity versus time traces from highlighted regions in d representing each of the four unique DNA origami species. (f) Engineering frequency and duration on DNA origami below the diffraction limit. Each corner of the structure is designed to exhibit a unique kinetic fingerprint. Scale bars: 1 μm (d), 500 nm (f, top), 40 nm (f, bottom). For details regarding simulation parameters and cluster identification, see Methods in Supporting Information.
In order to screen for the optimal conditions to design distinguishable binding kinetics in terms of frequency and duration, we first performed in silico DNA-PAINT experiments. Tuning parameters such as binding time, imager strand concentration, number of binding sites, duration of image acquisition, and others (see online methods for details), we were able to engineer four distinguishable blinking regimes (two blinking frequencies based on 40 and 120 binding sites and two blinking durations based on 400 ms and 5 s) that can now be used for combinatorial barcoding with a single imager strand species only, allowing four-target super-resolution imaging in a relatively short duration of 25 min (Figure 1b and Supplementary Figure 1).
Next, to experimentally validate the in silico results, we turned to DNA origami20 structures to implement the engineered frequency and duration levels, as these structures are exquisitely programmable for super-resolution microscopy.21 We designed four structures carrying 40 and 120 binding sites either with 8 or 10 nt extensions of the same sequence and imaged them simultaneously using a single imager strand species (see also Supplementary Figure 2, Supplementary Tables 2–5, and Supplementary Note 1). In the resulting raw DNA-PAINT data, we performed kinetic analysis for each structure following an initial filtering step (Supplementary Figures 3 and 4) and plotted the obtained binding time and frequency in a 2D plot in the same manner as for the in silico data. Next, we subjected the 2D data set to a clustering analysis (hdbscan;22 for details, see Methods in the Supporting Information), which resulted in four cluster species, in good agreement with the in silico data (Figure 1c and Supplementary Figure 5). This cluster identity now allows us to transform the raw DNA-PAINT image data (Figure 1d, top) to a barcoded pseudocolor image, where each DNA origami structure is assigned to one of the four cluster species (Figure 1d, bottom and Supplementary Figure 6). Examining the intensity versus time traces of four structures that were each assigned to one of the clusters indeed shows the distinct and expected kinetic fingerprints (Figure 1e).
To demonstrate that the kinetic barcoding approach allows satisfactory super-resolution performance, we designed a DNA origami structure with four different “binding spots”, at the four corners of the structure, each with four or 12 binding sites of either 8 or 10 nt length. Again, we performed DNA-PAINT using a single imager strand species and were able to visualize all four corners of the structure, separated by 40 nm. The blinking kinetics of the binding spots were then used to assign each to its correct corner (Figure 1f), demonstrating the application of blinking kinetics for super-resolution microscopy (Supplementary Figures 7 and 8).
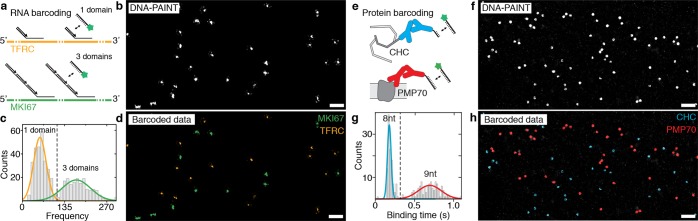
Next, we designed two experiments to demonstrate the general applicability of our simultaneous multiplexing approach in situ in two biologically relevant settings. First, we implemented two-color frequency barcoding for two distinct mRNA species using a combination of DNA-PAINT implemented on a Spinning Disk Confocal microscope23 and smRNA-FISH14,24 (Figure 2a–d). We labeled TFRC and MKI67 mRNA species using two sets of FISH probes displaying 40 and 120 binding sites, respectively (see Figure 2a for probe design and Supplementary Tables 6 and 7 for probe sequences). After image acquisition, the RNA species appear as super-resolved spots in the resulting DNA-PAINT image (Figure 2b, see Methods in the Supporting Information for acquisition details). We then performed kinetic analysis on the individual RNA molecules and obtained a distribution of two populations corresponding to two distinct blinking frequencies, as designed (Figure 2c). Similar to the in vitro case (Figure 1), we assigned a pseudocolor to each of the blinking frequencies and rerendered a barcoded data set (Figure 2d), where we are now able to clearly distinguish TFRC from MKI67 mRNA molecules.
Figure 2.
Engineered binding kinetics allow simultaneous multiplexed super-resolution imaging of RNA and proteins in cells. (a) Scheme showing the implementation of frequency barcoding for smRNA-FISH. Two distinct RNA species (TFRC and MKI67) are labeled with FISH probes featuring 40 binding sites for DNA-PAINT or 120 binding sites, respectively. (b) Resulting DNA-PAINT data after image acquisition shows TFRC and MKI67 mRNA molecules as single spots, which are not yet distinguishable. (c) Plotting the blinking frequency for all detected single mRNA molecules shows a clearly distinguishable distribution of a low and a high frequency, corresponding to the FISH probe set for TFRC (yellow) and MKI67 (green), respectively. (d) Distinct frequencies are used to assign a pseudocolor for each RNA species. (e) Scheme showing the implementation of duration barcoding for protein detection. Two distinct protein species are labeled with DNA-conjugated antibodies featuring an 8 and 9 nt binding site for DNA-PAINT imaging. (f) Resulting DNA-PAINT data after image acquisition shows CHC and PMP70 proteins as clusters, which are not yet distinguishable. (g) Plotting the binding duration for selected protein locations shows a clearly distinguishable distribution of short and long binding species, corresponding to the two proteins. (h) Distinct durations are used to assign a pseudocolor for each protein species. Scale bars: 1 μm.
To demonstrate that binding duration can be used in a similar fashion to barcode biomolecules in cells, we next used DNA-conjugated antibodies targeting the clathrin heavy chain (CHC) and a peroxisomal membrane protein (PMP70) inside HeLa cells, where each secondary antibody species carries a DNA-PAINT binding site of different length (Figure 2e and Supplementary Table 8). Similar to the mRNA experiments, we performed image acquisition and analysis and obtained a DNA-PAINT data set (Figure 2f). This was then transformed using kinetic analysis (Figure 2g) into a pseudocolored, barcoded image, where we were able to clearly distinguish areas of CHC and PMP70 based on the blinking duration (Figure 2h).
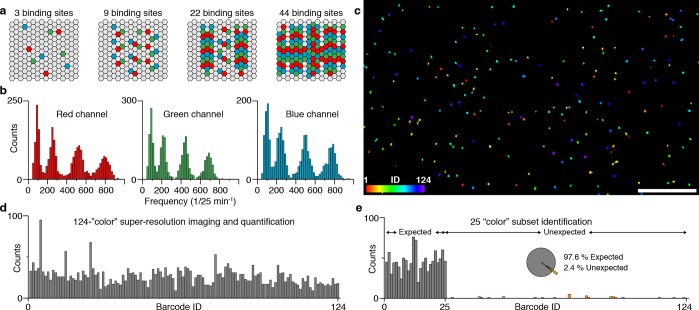
The proof-of-concept experiments for DNA origami, RNA, and protein barcoding underline the general applicability of the kinetic barcoding concept in vitro and in situ. However, to fully demonstrate the power of kinetic barcoding, we turned our attention to the question of how much simultaneous multiplexing can be achieved within a relatively short period of time. By combining four distinguishable binding frequencies with three spectral colors, we should be able to achieve simultaneous, 124-plex super-resolution imaging within a few minutes acquisition time. To demonstrate that this is indeed feasible, we designed and constructed 124 unique DNA origami structures carrying 0, 3, 9, 22, or 44 copies of three orthogonal binding sites each, respectively (Figure 3a and Supplementary Tables 9 and 10). After folding and purification (Supplementary Figure 9), we pooled all 124 distinct DNA origami structures into a single sample and performed 3-color DNA-PAINT imaging using Atto655-, Cy3B-, or Atto488-labeled orthogonal imager strands (Supplementary Table 11). As before, we performed kinetic analysis on each of the structures and plotted the distribution of binding frequencies for each spectral color separately (Figure 3b). We were able to clearly distinguish four distinct frequency populations in each of the three spectral colors. Using these levels, we assigned a unique barcode ID to each of the origami structures in the sample based on color and frequency and were able to render a full 124 pseudocolor super-resolution data set (Figure 3c, see also Methods in the Supporting Information for identification).
Figure 3.
Frequency-based 124-plex super-resolution imaging. (a) DNA origami structures are extended with three unique sequences (red, green, or blue) with 0, 3, 9, 22, or 44 copies, respectively. Using combinatorial labeling, this yields a total of 53 – 1 = 124 unique target structures, achieved by distinguishing five frequency levels and using three spectral colors (i.e., three imager strand species). (b) Binding frequency distribution for all 124 DNA origami structures show four clearly distinguishable frequency levels corresponding to 3, 9, 22, and 44 binding sites for each spectral color (red, green, and blue), respectively. Based on these distributions, a unique barcode ID from a pool of 124 can be assigned to each structure. (c) DNA-PAINT super-resolution image of all 124 DNA origami structures, color-coded according to the assigned binding frequency and spectral color. (d) Quantification of the 124-plex experiment shows that all 124 structures could be identified. In total, 3289 structures were quantified, from which 243 were discarded due to ambiguous frequencies (i.e., overlap of distributions in b). (e) Twenty-five out of 124 structures were imaged in one sample in order to assess identification performance. In total, 1165 structures were quantified, from which 28 were categorized as false-positives (i.e., unexpected) resulting in an accuracy of 97.6%. The ratio between lowest expected and highest unexpected is 20 to 7. Scale bar: 5 μm.
Next, we quantified all DNA origami in the data set and were able to verify that indeed all 124 unique structures could be identified (Figure 3d). Finally, in order to assay the performance in terms of false positive identification of our multiplexing approach, we performed a similar experiment as described above, but now using only a subset of 25 out of the total 124 DNA origami structures (Figure 3e). The resulting quantitative analysis yielded a remarkable accuracy of 97.6%, underlining the robustness of our multiplexing approach based on engineered binding kinetics. We note, however, that this high number of multiplexing and robustness might not be achievable in a straightforward fashion in a cellular setting, e.g., for mRNA barcoding using smRNA-FISH as shown in the examples above, due to the increased complexity in the intracellular environment and suboptimal or target-dependent labeling efficiencies.
In conclusion, we introduced an entirely new barcoding approach for multiplexed detection based on precisely engineering blinking kinetics in stochastic super-resolution microscopy: kinetic barcoding. We demonstrated the implementation using DNA-PAINT both in vitro and in situ, currently reaching 124-plex within minutes. Zooming out, we envision that kinetic barcoding could be applied to ask Systems Biology questions in single cells with super-resolution by simultaneously imaging hundreds of DNA, RNA, and protein targets reaching transcriptomics- and proteomics-style experiments with a simple localization microscopy approach: “Localizomics”. Finally, one could envision its extension to even more multiplexing by implementing readouts such as molecular brightness of imager strands, fluorescence lifetime, and more.25
Acknowledgments
We thank Martin Spitaler and the imaging facility of the MPI of Biochemistry for confocal imaging support. We thank Alexander Auer for microscopy support and help with data acquisition. We thank Julian Bauer and Patrick Schueler for experimental support. We thank Mingjie Dai for help with data processing. We thank William M. Shih for fruitful discussions. O.K.W., S.S., F.St., and J.S. acknowledge support by the QBM graduate school. M.T.S. acknowledges support by the IMPRS-LS graduate school. M.G. acknowledges funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no 796606.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.9b00508.
Materials and methods alongside detailed information about the optical setups, DNA origami self-assembly, RNA-FISH probe design and antibodies, sample preparation and data processing, sequences for DNA origami folding, DNA-PAINT docking and imager sequences, and RNA-FISH probes (PDF)
Author Contributions
⊥ These authors contributed equally to this work. O.K.W. designed and performed experiments and analyzed data. J.B.W. designed and performed experiments and analyzed data. P.C.N. designed and performed DNA origami experiments and analyzed the data. S.S. designed and performed smRNA-FISH and protein barcoding experiments. F.St. and J.S. performed DNA origami and in silico experiments, developed data analysis methods and analyzed data. F.Sc. designed and performed smRNA-FISH experiments, performed DNA origami experiments and analyzed data. M.T.S. developed software and analyzed data. M.G. designed and performed smRNA-FISH experiments and wrote the manuscript. J.S. developed software and analyzed data. H.G. developed software and performed DNA origami experiments. P.Y. supervised the initial phase of the study. P.S. developed data analysis strategies and supervised the study. R.J. conceived and supervised the study, analyzed the data, and wrote the manuscript. All authors reviewed and approved the manuscript.
This research was funded by the German Research Foundation through the Emmy Noether Program (DFG JU 2957/1–1) and the SFB1032 (Project A11), the European Research Council through an ERC Starting Grant (MolMap, Grant agreement number 680241), the Allen Distinguished Investigator Program through the Paul G. Allen Frontiers Group, the Max Planck Society, and the Max Planck Foundation. P.Y. acknowledges support by the NSF (CCF-1317291) and the NIH (1-U01-MH106011 and R01EB018659).
The authors declare the following competing financial interest(s): J.B.W, R.J., and P.Y. filed a provisional patent based on this work. R.J. and P.Y. are co-founders of Ultivue, Inc.
Supplementary Material
References
- Hell S. W.; Sahl S. J.; Bates M.; Zhuang X. W.; Heintzmann R.; Booth M. J.; Bewersdorf J.; Shtengel G.; Hess H.; Tinnefeld P.; Honigmann A.; Jakobs S.; Testa I.; Cognet L.; Lounis B.; Ewers H.; Davis S. J.; Eggeling C.; Klenerman D.; Willig K. I.; Vicidomini G.; Castello M.; Diaspro A.; Cordes T. The 2015 super-resolution microscopy roadmap. J. Phys. D: Appl. Phys. 2015, 48 (44), 443001. 10.1088/0022-3727/48/44/443001. [DOI] [Google Scholar]
- Kanchanawong P.; Shtengel G.; Pasapera A. M.; Ramko E. B.; Davidson M. W.; Hess H. F.; Waterman C. M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468 (7323), 580–4. 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K.; Zhong G.; Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 2013, 339 (6118), 452–6. 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18 (11), 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Dai M. J.; Jungmann R.; Yin P. Optical imaging of individual biomolecules in densely packed clusters. Nat. Nanotechnol. 2016, 11 (9), 798–807. 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarotti F.; Eilers Y.; Gwosch K. C.; Gynna A. H.; Westphal V.; Stefani F. D.; Elf J.; Hell S. W. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 2017, 355 (6325), 606–612. 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- Schnitzbauer J.; Strauss M. T.; Schlichthaerle T.; Schueder F.; Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017, 12, 1198–1228. 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- Dempsey G. T.; Vaughan J. C.; Chen K. H.; Bates M.; Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 2011, 8 (12), 1027–36. 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E.; Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 2012, 9 (7), 743–8. 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M.; Dempsey G. T.; Chen K. H.; Zhuang X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. ChemPhysChem 2012, 13 (1), 99–107. 10.1002/cphc.201100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm A. M.; Cohen S.; Legant W. R.; Melunis J.; Hershberg U.; Wait E.; Cohen A. R.; Davidson M. W.; Betzig E.; Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 2017, 546 (7656), 162–167. 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Kenny S. J.; Hauser M.; Li W.; Xu K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods 2015, 12 (10), 935–8. 10.1038/nmeth.3528. [DOI] [PubMed] [Google Scholar]
- Bongiovanni M. N.; Godet J.; Horrocks M. H.; Tosatto L.; Carr A. R.; Wirthensohn D. C.; Ranasinghe R. T.; Lee J. E.; Ponjavic A.; Fritz J. V.; Dobson C. M.; Klenerman D.; Lee S. F. Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping. Nat. Commun. 2016, 7, 13544. 10.1038/ncomms13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M. J.; Shih W. M.; Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11 (3), 313–U292. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J.; Cordier G. A.; Borbely J. S.; Sandoval Alvarez A.; Lakadamyali M. Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One 2014, 9 (7), e101772 10.1371/journal.pone.0101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley C. C.; Liu S.; Lidke D. S.; Lidke K. A. Sequential superresolution imaging of multiple targets using a single fluorophore. PLoS One 2015, 10 (4), e0123941 10.1371/journal.pone.0123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueder F.; Strauss M. T.; Hoerl D.; Schnitzbauer J.; Schlichthaerle T.; Strauss S.; Yin P.; Harz H.; Leonhardt H.; Jungmann R. Universal Super-Resolution Multiplexing by DNA Exchange. Angew. Chem., Int. Ed. 2017, 56 (14), 4052–4055. 10.1002/anie.201611729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Woehrstein J. B.; Donoghue N.; Dai M.; Avendano M. S.; Schackmann R. C. J.; Zoeller J. J.; Wang S. S. H.; Tillberg P. W.; Park D.; Lapan S. W.; Boyden E. S.; Brugge J. S.; Kaeser P. S.; Church G. M.; Agasti S. S.; Jungmann R.; Yin P. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Lett. 2017, 17 (10), 6131–6139. 10.1021/acs.nanolett.7b02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Steinhauer C.; Scheible M.; Kuzyk A.; Tinnefeld P.; Simmel F. C. Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami. Nano Lett. 2010, 10 (11), 4756–4761. 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440 (7082), 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T.; Strauss M. T.; Schueder F.; Woehrstein J. B.; Jungmann R. DNA nanotechnology and fluorescence applications. Curr. Opin. Biotechnol. 2016, 39, 41–47. 10.1016/j.copbio.2015.12.014. [DOI] [PubMed] [Google Scholar]
- Campello R. J. G. B.; Moulavi D.; Sander J. In Density-Based Clustering Based on Hierarchical Density Estimates; Springer Berlin Heidelberg: Berlin, Heidelberg, 2013; pp 160–172. [Google Scholar]
- Schueder F.; Lara-Gutierrez J.; Beliveau B. J.; Saka S. K.; Sasaki H. M.; Woehrstein J. B.; Strauss M. T.; Grabmayr H.; Yin P.; Jungmann R. Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT. Nat. Commun. 2017, 8 (1), 2090. 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A.; van den Bogaard P.; Rifkin S. A.; van Oudenaarden A.; Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5 (10), 877–879. 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Buck A.; Shih W. M. Single-Molecule Clocks Controlled by Serial Chemical Reactions. Nano Lett. 2017, 17 (12), 7940–7944. 10.1021/acs.nanolett.7b04336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.