Abstract
Objectives
Prior studies have suggested a potential link between nasal microbes and granulomatosis with polyangiitis (GPA; Wegener’s) but these studies relied on culture-dependent methods. This study comprehensively examined the entire community of nasal microbiota (bacteria and fungi) in participants with GPA compared to healthy controls using deep sequencing methods.
Methods
16S rRNA and ITS gene sequencing were performed on nasal microbial DNA isolated from nasal swabs of 60 participants with GPA and 41 healthy controls. Alpha and beta diversity were assessed as well as the relative abundance of the most abundant bacterial and fungal taxa. The effects of co-variates including disease activity and immunosuppressive therapies on microbial composition were evaluated.
Results
Compared to controls, participants with GPA had a significantly different microbial composition (weighted UniFrac p = 0.04) and lower relative abundance of P. acnes and S. epidermidis (for both, FDR-corrected p = 0.02). Disease activity in GPA was associated with a lower abundance of fungal order Malasseziales compared to participants with GPA in remission (p = 0.04) and controls (p = 0.01). Use of non-glucocorticoid immunosuppressive therapy was associated with “healthy” nasal microbiota while participants with GPA who were off immunosuppressive therapy had more dysbiosis (weighted UniFrac p = 0.01). No difference in the relative abundance of S. aureus was observed between GPA and controls.
Conclusions
GPA is associated with an altered nasal microbial composition, at both the bacterial and fungal levels. Use of immunosuppressive therapies and disease remission are associated with healthy microbial communities.
Keywords: granulomatosis with polyangiitis, infections, disease activity
INTRODUCTION
Granulomatosis with polyangiitis (GPA; Wegener’s) is a systemic vasculitis characterized by granulomatous inflammation and up to 90% of participants will develop sinonasal inflammation during the course of disease1. Despite the many therapeutic advances in the management of GPA, relapses remain a significant issue and, in particular, disease activity in the sinuses and nose often persists when a patient is otherwise in clinical remission2. It has been speculated that microbes may be involved in the pathogenesis of GPA. A study using culture data found that chronic nasal carriage of Staphylococcus aureus is associated with a higher risk of relapse3 and two randomized clinical trials showed trimethoprim-sulfamethoxazole prevents relapse in GPA4, 5.
Advances in high-throughput genomic sequencing and the development of new tools for analyzing metagenomic data allow for a better understanding of the dynamic community of microbes (bacteria, fungi, viruses) that inhabit the human body (human microbiome) in a relatively inexpensive and accessible manner. The human microbiota has a large potential to impact many human physiologic functions including immune homeostasis6. Dysbiosis, the imbalance or disruption of microbial communities, has been proposed to contribute to the pathophysiology of several autoimmune diseases including inflammatory bowel disease, rheumatoid arthritis, multiple sclerosis, and type 1 diabetes mellitus7–11. Furthermore, microbial composition varies across different body sites and, while much attention has been given to the gut microbiome, microbial communities in other body habitats, including the nose, have also been implicated in human health and disease12–15.
This is the first study to comprehensively examine the entire community of nasal bacteria and fungi in participants with GPA compared to healthy controls using culture-independent sequencing methods. We chose to focus on the nose due to its distinctive involvement in the natural history of GPA and its role as an active component of the immune system16.
METHODS
Study participants
Participants were recruited through the Penn Vasculitis Center at the University of Pennsylvania. Participants with GPA were eligible if they met the modified American College of Rheumatology classification criteria for GPA17–19. Healthy controls were participants without a systemic vasculitis or other inflammatory disorder. Both participants with GPA and controls were excluded if they had another systemic inflammatory disorder, history of intranasal cocaine use within the prior 3 years, known history of Human Immunodeficiency Virus (HIV) or primary immunodeficiency, lymphoma or other malignancy that mimics AAV. Antibiotic use was not an exclusionary criteria but was incorporated into the analysis. This study was approved by the Institutional Review Board of the University of Pennsylvania and written informed consent was obtained from all participants.
Nasal mucosa was sampled by swabbing the middle meatus with a sterile flocked specimen collection swab (Copan Diagnostics) which was transferred to a −80°C freezer. To control for environmental contamination, negative controls (swab exposed to ambient air) were obtained with each participant sampling and processed in parallel.
Clinical data was collected at time of sampling and included demographics and medication use within prior 6 months (antibiotics, systemic glucocorticoids, and non-glucocorticoid immunosuppressive therapy such as cyclophosphamide, rituximab, azathioprine, methotrexate, etc). For participants with GPA, a detailed disease history was obtained including antineutrophil cytoplasmic antibody (ANCA) type by ELISA (proteinase-3 [PR3], myeloperoxidase [MPO], or negative), disease status determined by the Birmingham Vasculitis Activity Score for Wegener’s Granulomatosis20 (BVAS/WG; BVAS/WG > 0 indicates active disease and BVAS/WG = 0 indicates disease remission), and sinonasal damage according to the ear, nose, and throat items on the Vasculitis Damage Index (VDI)21.
Microbiota profiling using DNA sequencing
DNA was extracted using QIAamp ultraclean production pathogen mini kit. For bacterial profiling, PCR amplification targeting the V1-V2 region of the 16S rRNA gene was performed. For analyses of fungal communities (mycobiome), the internal transcribed spacer (ITS1) region was targeted, using a fungal-specific primer set22. Amplicons were purified and sequenced by the sequencing core of the Penn-CHOP Microbiome program on an Illumina MiSeq instrument, yielding 250bp paired-end sequence reads. Environmental and reagent control samples, consisting of air-exposed swabs, DNA free water, and empty wells were processed alongside participant samples23.
Sequencing data analysis was performed by QIIME 1.9.124. Paired-end reads were quality filtered and joined, clustered into operational taxonomic units (OTUs) with 97% sequence similarity using UCLUST25. Taxonomic assignments were generated by alignment to the Greengenes r13_8 reference databases26. Taking into consideration the various lengths of the ITS1 regions, we used high quality forward reads (R1) for analysis, clustered into OTUs with 95% sequence similarity, and taxonomy assignments generated by comparison to the nt reference database using the consensus method implemented in BROCC software27.
16S reads assigned as Archaea, Mitochondria or Chloroplast were removed. Potential contaminant OTUs showing a strong negative correlation with the amplicon concentration (FDR < 0.05) were also removed28 (Supplementary Table 1). Samples with more than 10,000 read counts for 16S analysis and 4,000 read counts for ITS analysis were retained. To reduce the influence of low-abundant fungi which are potential contaminants, a PicoGreen-corrected OTU abundance was determined by multiplying the relative abundance of fungal taxa by the post-PCR ITS DNA concentration as previously described and validated29.
Taxonomic heatmap was generated at the genus level to depict all taxa with a relative abundance of over 1% in at least one sample. The most abundant OTUs within taxa of interest were also identified for secondary analyses.
Analytical approach
The main comparison of interest was between GPA and controls. Outcome measures included alpha and beta diversity and relative abundance of individual taxa. We measured alpha diversity, or within-sample diversity, using the Shannon diversity index which accounts for evenness and abundance of OTUs within a sample. Beta diversity, which compares dissimilarity of microbial composition between samples, was measured by UniFrac distance which estimates the fraction of a sample’s phylogenetic tree that differs from another sample, with (weighted) or without (unweighted) accounting for the relative abundance of OTUs30, 31. UniFrac distances were visualized using principal coordinate analysis (PCoA). To determine if specific bacterial or fungal taxa were more common or more rare, we evaluated relative abundance (which refers to the percent composition of a taxa relative to the total number of taxa) of the 5 most abundant genera and 10 most abundant OTUs (here on referred to as species). We focused on these relatively abundant bacteria because they are more likely to represent “true” nasal commensals in these low microbial biomass samples. Furthermore, these taxa together represent 92% of all sequences obtained, thus reflecting the dominant microbiota present. In exploratory analyses, differential relative abundance was also investigated between subgroups of GPA according to ANCA type, sinonasal damage, disease activity, and medications.
Detailed profiling of the nasal mycobiota or fungal communities included examination of total fungal abundance and was initially planned to evaluate the relative abundance of the 10 most abundant taxa; however, due to the predominance of the order Malasseziales and the small number of other fungal OTUs detected and their sparse distribution among samples, further investigation of relative abundance focused only on Malasseziales.
Statistical analyses
Categorical variables were compared between groups using chi-square test; continuous variables were compared using the Wilcoxon rank sum test. One-way ANOVA was used to test for differences in alpha diversity (Shannon index) between groups. Beta diversity (UniFrac) was analyzed using permutational analysis of variance (PERMANOVA)32. Relative abundance values were log-transformed and compared using the Wilcoxon rank sum test. Potential fungal-bacterial interactions were assessed by examining associations between abundance of total fungi as well as Malasseziales and the following: 1) UniFrac distance using PERMANOVA, and 2) abundance of 5 most abundant bacterial genera and 10 most abundant species using Wilcoxon rank sum test adjusting for study group (GPA vs control).
Correction for multiple comparisons was performed by the Benjamini-Hochberg false discovery rate (FDR) procedure for all analyses except for exploratory analyses performed within GPA subgroups of interest33. All statistical analyses were conducted with R 3.4.1.
RESULTS
Participant characteristics
There were 101 participants enrolled in this study: 60 participants with GPA and 41 healthy controls. Characteristics at time of sampling are shown in Table 1. Demographics were similar between the 2 groups. Participants with GPA were more likely to have received antibiotics within the prior 6 months (48% vs 29%). Among participants with GPA, 25% had active disease and a majority of participants had received immunosuppressive agents within the prior 6 months.
Table 1:
Participant characteristics at time of nasal sampling
| GPA N = 60 | Control N = 41 | P-value | |
|---|---|---|---|
| Age | 55 (37, 66) | 61 (47, 68) | 0.06 |
| Female | 35 (58%) | 24 (59%) | 0.98 |
| White race | 57 (95%) | 36 (88%) | 0.18 |
| Ever smoker | 19 (32%) | 19 (48%) | 0.11 |
| Current allergies | 14 (23%) | 15 (37%) | 0.15 |
| GPA manifestations | |||
| ANCA type (ever) | N/A | -- | |
| Proteinase-3 | 36 (60%) | ||
| Myeloperoxidase | 15 (25%) | ||
| Negative | 9 (15%) | ||
| Disease duration, years | 4 (2, 7) | N/A | -- |
| Newly diagnosed (within 30 days) | 5 (8%) | N/A | -- |
| Current disease status | N/A | -- | |
| Remission | 45 (75%) | ||
| Severe flare | 2 (3%) | ||
| Limited flare | 10 (17%) | ||
| Persistent disease | 3 (5%) | ||
| Currently active disease of sinonasal area | 10 (10%) | N/A | -- |
| Ever had flare involving sinonasal area | 26 (43%) | N/A | -- |
| Vasculitis Damage Index | |||
| Any sinonasal item | 28 (47%) | N/A | -- |
| Nasal blockade/chronic crusting | 21 (35%) | ||
| Nasal bridge collapse | 11 (18%) | ||
| Chronic sinusitis | 10 (17%) | ||
| Medication use | |||
| Systemic glucocorticoid | |||
| Current | 25 (42%) | 0 | < 0.01 |
| Current or in past 6 months | 31 (52%) | 3 (7%) | < 0.01 |
| Other immunosuppressive drug | |||
| Current | 32 (53%) | 0 | < 0.01 |
| Current or in past 6 months | 42 (70%) | 0 | < 0.01 |
| Current intranasal glucocorticoid | 12 (20%) | 5 (12%) | 0.30 |
| Antibiotic | |||
| Current | 15 (25%) | 6 (15%) | 0.21 |
| Current or in past 6 months | 29 (48%) | 12 (29%) | 0.06 |
| Current trimethoprim-sulfamethoxazole | |||
| Full dose* | 0 | 0 | -- |
| Low dose** | 10 (17%) | 0 | < 0.01 |
Values expressed as median (inter-quartile range) or percentage
Full dose is trimethoprim-sulfamethoxazole 160–800mg twice daily.
Low dose refers to doses used for prophylaxis of Pneumocystis jirovecii pneumonia (trimethoprim-sulfamethoxazole 160–800mg three times a week or trimethoprim-sulfamethoxazole 80–400mg daily).
Bacterial composition and diversity in nasal cavities of participants with GPA and healthy controls
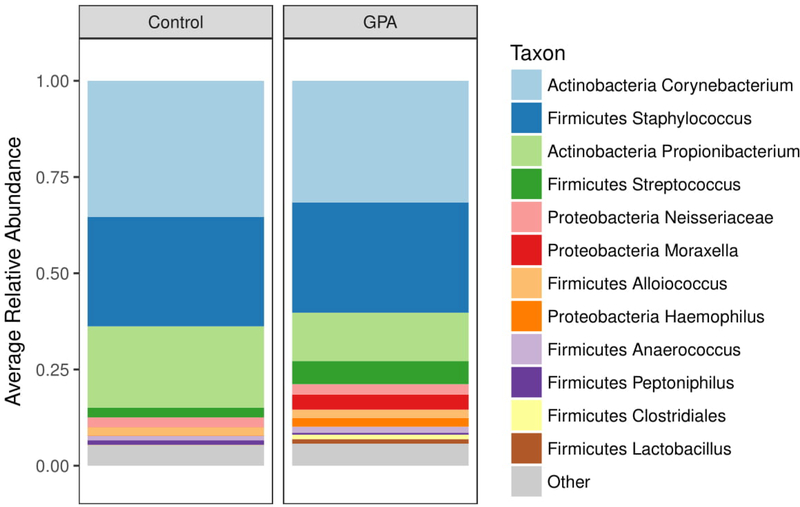
A total of 9,235,460 raw sequence reads were generated from all participant samples and, after quality filtering and removal of contaminants, a total of 9,213,337 high-quality reads were used for analysis (mean reads per sample 91,221 ± 56,533). Corynebacterium, Staphylococcus, and Propionibacterium featured as prominent bacterial taxa in the nasal cavities of the cohort, consistent with previous descriptions of the nasal microbial composition in healthy individuals34 (Figure 1 and Supplementary Figure 1 for heatmap). Within these genera, the most abundant species identified were Corynebacterium tuberculostearicum, Propionibacterium acnes, Staphylococcus aureus, and S. epidermidis.
Figure 1:
The 12 most abundant bacteria that are present in the samples are depicted in a stacked barplot. Corynebacterium, Staphylococcus, and Propionibacterium featured as most abundant genera, similar to previously published studies of nasal microbial composition in healthy individuals.
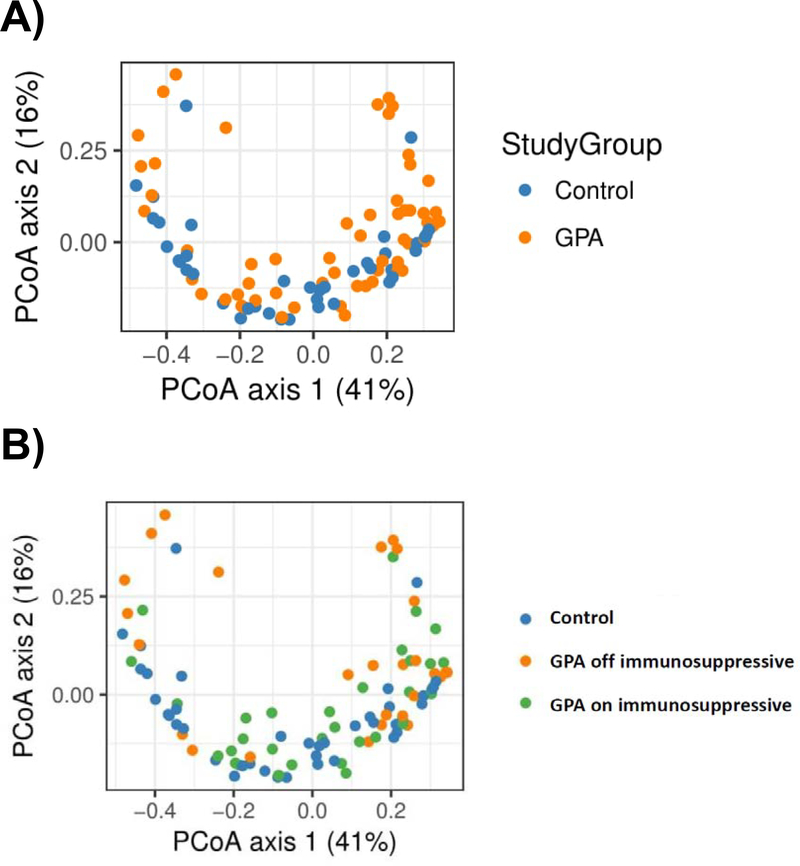
To compare overall composition of bacterial communities, alpha and beta diversity were measured. The Shannon diversity index (alpha diversity) was not significantly different between participants with GPA and healthy controls (p = 0.51). The analysis of beta diversity calculated on UniFrac distances revealed a borderline difference by unweighted UniFrac (i.e., based on the identify of community members) (p=0.05), and a significant difference in weighted UniFrac (i.e., accounting for both community membership and abundances), between participants with GPA and healthy controls, suggesting that microbial composition between the 2 groups is different (p = 0.04) (Figure 2A). We then compared the relative abundances of individual bacterial genera between groups and found that participants with GPA had a lower relative abundance of Propionibacterium (FDR-corrected p = 0.03) compared to healthy controls. When we examined the most abundant species, both P. acnes and S. epidermidis had a significantly lower abundance in GPA compared to controls (for both species, FDR-corrected p = 0.02). No difference was seen in the relative abundance of S. aureus (FDR-corrected p = 0.49).
Figure 2:
Difference in overall microbial composition between (A) controls and granulomatosis with polyangiitis (GPA), and (B) controls and GPA stratified by use of non-glucocorticoid immunosuppressive medications. The principal coordinates analysis (PCoA) plots depict differences in beta diversity based on weighted UniFrac distance, which accounts for phylogenetic differences as well as abundance. (A) There is a significant difference in microbial composition between controls (blue) and participants with GPA (orange) (p = 0.04). (B) Differences in microbial composition are primarily driven by participants with GPA off non-glucocorticoid immunosuppressive therapy (orange) who had a significantly different weighted UniFrac from controls in blue (p = 0.01) while those on immunosuppressive therapies (green) are similar to controls (p = 0.16).
Use of non-glucocorticoid immunosuppressive agents is associated with “healthy” nasal bacteria in GPA
When evaluating the association of co-variates such as medications and disease characteristics with microbial communities in GPA and controls, we found that the nasal microbial communities in participants with GPA receiving non-glucocorticoid immunosuppressive therapies either currently or within the prior 6 months were similar to healthy controls, whereas participants with GPA not receiving non-glucocorticoid immunosuppressive therapies were significantly different from controls. Specifically, there was a significant difference in weighted UniFrac distance between participants with GPA off immunosuppressive therapies compared to controls (p = 0.01) while no difference was observed between GPA on immunosuppressive therapies versus controls (p = 0.16) (Figure 2B). No significant difference in unweighted UniFrac distance was found according to use of immunosuppressive therapies.
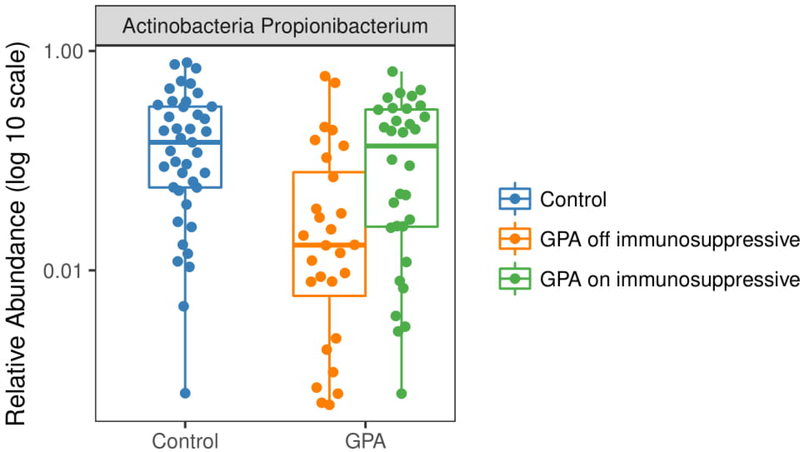
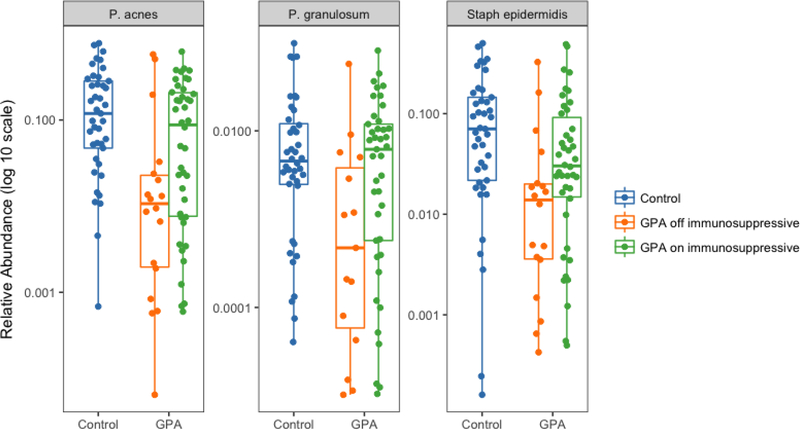
When we evaluated the relative abundance of Propionibacterium and then stratified participants with GPA based on their use of non-glucocorticoid immunosuppressive therapy, we found that data from the participants with GPA who were off immunosuppressive therapy were driving the differences observed between GPA and controls; specifically, participants off immunosuppressive therapy had a significantly lower abundance of Propionibacterium compared to controls (p < 0.01) while participants with GPA on immunosuppression were similar to controls (p = 0.68) (Figure 3). Analysis of the most abundant bacterial species revealed similar differences in relative abundance between groups for P. acnes, P. granulosum, and S. epidermidis (Figure 4). Relative abundances for each sample and OTU are available in Supplementary Table 2.
Figure 3:
Differential abundance of Propionibacterium between controls and participants with granulomatosis with polyangiitis (GPA) stratified by use of non-glucocorticoid immunosuppressive therapy. Participants off immunosuppressive therapy (orange) had a significantly lower abundance of Propionibacterium compared to controls (blue) (FDR-corrected p < 0.01). No difference was observed between participants with GPA on immunosuppression (green) and controls (FDR-corrected p = 0.68) or participants with GPA off immunosuppression (FDR-corrected p = 0.06).
Figure 4:
Differential abundance of Propionibacterium acnes, P. granulosum, and Staphylococcus epidermidis between control and participants with granulomatosis with polyangiitis (GPA) stratified by use of non-glucocorticoid immunosuppressive therapy. Participants with GPA who are off immunosuppressives had a significantly lower abundance of P. acnes, P. granulosum, and S. epidermidis (for all, FDR-corrected p < 0.01) compared to controls and compared to participants on immunosuppression (FDR-corrected p = 0.03, p = 0.01, and p = 0.03, respectively), while participants with GPA on immunosuppression were similar to controls (p > 0.05).
When examining the association between disease activity in GPA and microbial communities, univariate and multivariate analyses found no relationship between disease activity and UniFrac distances, either weighted or unweighted, or relative abundance of bacterial taxa. Similarly, use of antibiotics or prednisone was not associated with UniFrac distance or relative abundance. Additional exploratory analyses within clinically-distinct subgroups of GPA are shown in the Supplementary Text 1.
Participants with active GPA harbor an altered nasal mycobiota
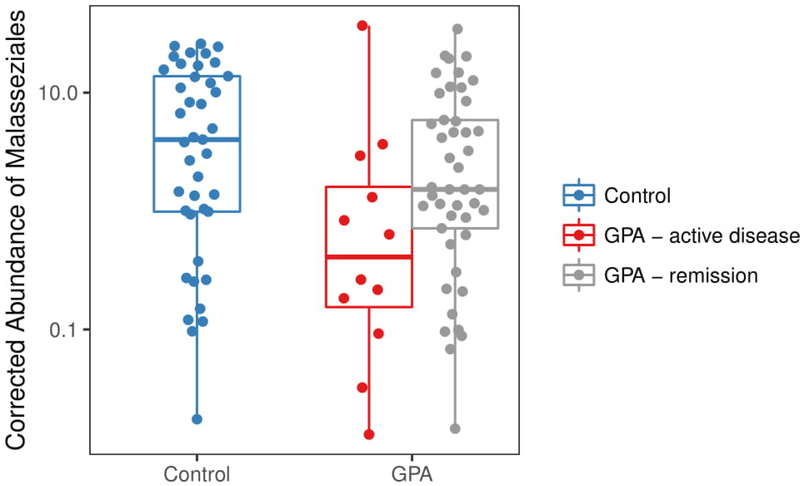
We found a non-significant trend towards lower total fungi abundance in participants with GPA compared to controls (p = 0.06). However, we found that participants receiving prednisone had a lower total abundance of nasal fungi (p = 0.02) compared to healthy controls, and those receiving other immunosuppressive therapies had a significantly higher abundance of fungi (p = 0.03), independent of disease activity, antibiotics, ANCA type, and sinonasal damage. When we investigated the nasal fungal community composition, unclassified Malasseziales was the most abundant fungal taxa present, followed by the genera Penicillium and Aspergillus. Evaluation of Malasseziales demonstrated a significantly lower abundance in GPA compared to controls (p = 0.04). In particular, participants with GPA with active disease appeared to be driving these differences, as they had the lowest abundance compared to participants in remission (p = 0.04) and healthy controls (p = 0.01), while participants with GPA in remission were similar to controls (p = 0.19) (Figure 5). Significant correlations were also found between the 5 most abundant bacterial species and all fungi (Supplementary Figures 2 and 3) as well as just Malasseziales (Supplementary Text 2).
Figure 5:
Differential abundance of Malasseziales between controls and participants with granulomatosis with polyangiitis (GPA) stratified by disease activity. PicoGreen-corrected abundance of Malasseziales was significantly lower in participants with GPA with active disease (red) compared to controls (blue) (p = 0.01) and participants with GPA in remission (gray) (p = 0.04). No difference was seen between participants with GPA in remission and controls (p = 0.19).
DISCUSSION
This study used deep sequencing methods to comprehensively define the entire community of nasal bacteria and fungi in GPA, and found significant differences in nasal microbial composition between participants with GPA and healthy controls. Furthermore, being off immunosuppressive therapy and disease activity were associated with bacterial and fungal dysbiosis, respectively.
For many decades, there has been speculation that infections may have a major influence on disease activity in GPA. With a growing understanding of the human microbiome, a shift occurred within the field of microbiology from focusing only on individual disease-causing organisms to studying the effects of endogenous microbes or commensals as communities. In this study we examined the relationship between microbes and GPA from a different perspective than prior studies, by characterizing the entire community of resident bacteria and fungi in the nasal cavity, an active site of immunity and a known reservoir for pathobionts such as S. aureus. We found that participants with GPA have dysbiosis in the nose and, in particular, a lower abundance of P. acnes and S. epidermidis, both of which have been suggested to be negative competitors of S. aureus14, 35. Most interestingly, differences in microbial composition were primarily driven by participants with GPA who were off non-glucocorticoid immunosuppressive therapy independent of disease activity, suggesting that immunosuppressive therapy is associated with a healthy nasal microbiome in GPA (and conversely, absence of immunosuppressive therapy is associated with nasal dysbiosis). This was an unexpected finding since we had anticipated immunosuppressive therapy would cause more dysbiosis, not less, based on prior studies36. Similar observations have been made in rheumatoid arthritis37.
We postulate there are 2 potential explanations for this association between immunosuppressive use and improvements in nasal dysbiosis: 1) immunosuppressive therapy has direct benefits on the nasal microbiota, or 2) subclinical disease activity, not identified using standard methods of assessment, was present in participants who were off immunosuppressive therapy, which in turn is associated with an altered nasal microbiota. These results suggest either that the manipulation of the nasal microbiome may be a novel therapeutic target and/or that the nasal microbiome may be a more sensitive, non-invasive biomarker of disease activity.
Unlike prior studies, no difference was found in the abundance of S. aureus between GPA and controls, although differences were seen among GPA subgroups (however, these differences were no longer significant after correction for multiple comparisons)38, 39. Several differences in methodology may explain this discrepancy. The current study used sequencing data to identify bacteria which may be more sensitive than culture data for identifying bacteria present; alternatively, sequencing data does not discriminate between viable versus non-viable bacteria. The location within the nasal cavity where swabbing was performed also differed: we sampled the middle meatus to study a mucosal surface whereas prior studies swabbed the anterior nares which is lined with skin-like squamous epithelium. Despite their proximity, spatial variation in microbiota composition (in particular S. aureus colonization) has been found within the nasal cavity, particularly between the middle meatus and anterior nares15.
To our knowledge, only 2 prior studies have examined the nasal mycobiome using culture-independent methods and this is the first to examine the mycobiome in GPA40, 41. Malasseziales, a fungal taxa which has previously been demonstrated to be abundant in the nose, mouth, and skin, was the most abundant fungal taxa in the nasal cavity in this cohort40, 42, 43. Furthermore, Malasseziales was significantly lower in abundance in GPA compared to controls and this difference was most pronounced in participants with active disease. The abundance of Malasseziales correlated with the abundance of several bacteria, suggesting interactions between bacteria and fungi occur in the nose. Prior studies also have suggested that interactions between fungi and bacteria may impact microbial composition and specifically the possibility has been raised that fungi may stabilize microbial community organization44. Future studies further exploring the role of fungi in GPA are warranted.
Limitations to this study include the cross-sectional design which restricts investigations of temporal dynamics and is unable to assess for inter-individual heterogeneity. Given the variability in the microbiome between individuals, a future longitudinal study would have the advantage of controlling for potential confounders through repeated sampling within individuals. Furthermore, causal relationships cannot be determined and confounders such as inflammation itself can perturb the nasal microbiota. Validation of these results in other cohorts along with mechanistic studies are needed to interpret these findings. Although the sample size was adequate to evaluate differences between GPA and controls, the ability to explore differences between GPA subgroups may have been underpowered. Lastly, measurement of disease activity relied on a validated disease activity measure, the BVAS/WG, which may not be sensitive enough to identify subclinical disease activity.
In conclusion, this study found an altered nasal microbial composition associated with GPA, both at the bacterial and fungal levels, and suggests that immunosuppressive therapies and inactive disease status are associated with healthy microbial communities. These findings justify further investigation of host-microbe interactions in GPA, a potentially exciting new dimension to our understanding of the disease, and may be the first step to elucidating critical components of the pathophysiology of disease and identification of novel therapeutics.
Supplementary Material
Acknowledgments:
This work was supported by the CHOP Microbiome Center, the sequencing core of the PennCHOP Microbiome Center. We acknowledge Casey Hofstaedter, Dorothy Kim, Lisa Mattei, and Michael Moraskie who performed DNA extraction and sequencing. We thank the participants for their involvement in the study and Sherry Xu and Marina Fanous for their assistance with recruitment and sample collection.
Funding: This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award K23-AR071514.
Footnotes
Competing interests: None
Availability of data: All sequence data related to this study are available from the United States National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession number SRP149341.
REFERENCES
- 1.Hoffman GS, Kerr GS, Leavitt RY, Hallahan CW, Lebovics RS, Travis WD, et al. Wegener granulomatosis: an analysis of 158 patients. Ann Intern Med. 1992;116(6):488–98. [DOI] [PubMed] [Google Scholar]
- 2.Hogan SL, Falk RJ, Chin H, Cai J, Jennette CE, Jennette JC, et al. Predictors of relapse and treatment resistance in antineutrophil cytoplasmic antibody-associated small-vessel vasculitis. Ann Intern Med. 2005;143(9):621–31. [DOI] [PubMed] [Google Scholar]
- 3.Stegeman CA, Tervaert JW, Sluiter WJ, Manson WL, de Jong PE, Kallenberg CG. Association of chronic nasal carriage of Staphylococcus aureus and higher relapse rates in Wegener granulomatosis. Ann Intern Med. 1994;120(1):12–7. [DOI] [PubMed] [Google Scholar]
- 4.Stegeman CA, Tervaert JW, de Jong PE, Kallenberg CG. Trimethoprim-sulfamethoxazole (co-trimoxazole) for the prevention of relapses of Wegener’s granulomatosis. Dutch Co-Trimoxazole Wegener Study Group. N Engl J Med. 1996;335(1):16–20. [DOI] [PubMed] [Google Scholar]
- 5.Zycinska K, Wardyn KA, Zielonka TM, Krupa R, Lukas W. Co-trimoxazole and prevention of relapses of PR3-ANCA positive vasculitis with pulmonary involvement. Eur J Med Res. 2009;14 Suppl 4:265–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64(10):3083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goverman J, Woods A, Larson L, Weiner LP, Hood L, Zaller DM. Transgenic mice that express a myelin basic protein-specific T cell receptor develop spontaneous autoimmunity. Cell. 1993;72(4):551–60. [DOI] [PubMed] [Google Scholar]
- 11.Marino E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–62. [DOI] [PubMed] [Google Scholar]
- 12.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cope EK, Goldberg AN, Pletcher SD, Lynch SV. Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome. 2017;5(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5(5):e10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14(6):631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi EH, Wormald PJ, Tan LW. Innate immunity in the paranasal sinuses: a review of nasal host defenses. Am J Rhinol. 2008;22(1):13–9. [DOI] [PubMed] [Google Scholar]
- 17.Leavitt RY, Fauci AS, Bloch DA, Michel BA, Hunder GG, Arend WP, et al. The American College of Rheumatology 1990 criteria for the classification of Wegener’s granulomatosis. Arthritis Rheum. 1990;33(8):1101–7. [DOI] [PubMed] [Google Scholar]
- 18.Design of the Wegener’s Granulomatosis Etanercept Trial (WGET). Control Clin Trials. 2002;23(4):450–68. [DOI] [PubMed] [Google Scholar]
- 19.Wegener’s Granulomatosis Etanercept Trial Research G. Etanercept plus standard therapy for Wegener’s granulomatosis. N Engl J Med. 2005;352(4):351–61. [DOI] [PubMed] [Google Scholar]
- 20.Stone JH, Hoffman GS, Merkel PA, Min YI, Uhlfelder ML, Hellmann DB, et al. A disease-specific activity index for Wegener’s granulomatosis: modification of the Birmingham Vasculitis Activity Score. International Network for the Study of the Systemic Vasculitides (INSSYS). Arthritis Rheum. 2001;44(4):912–20. [DOI] [PubMed] [Google Scholar]
- 21.Exley AR, Bacon PA, Luqmani RA, Kitas GD, Gordon C, Savage CO, et al. Development and initial validation of the Vasculitis Damage Index for the standardized clinical assessment of damage in the systemic vasculitides. Arthritis Rheum. 1997;40(2):371–80. [DOI] [PubMed] [Google Scholar]
- 22.Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, et al. Redefining the Chronic-Wound Microbiome: Fungal Communities Are Prevalent, Dynamic, and Associated with Delayed Healing. MBio. 2016;7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. [DOI] [PubMed] [Google Scholar]
- 26.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13(7):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jervis-Bardy J, Leong LE, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 2014;15(10):487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 33.Benjamini Y, Hochberg Y. A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 34.Jervis Bardy J, Psaltis AJ. Next Generation Sequencing and the Microbiome of Chronic Rhinosinusitis: A Primer for Clinicians and Review of Current Research, Its Limitations, and Future Directions. Ann Otol Rhinol Laryngol. 2016;125(8):613–21. [DOI] [PubMed] [Google Scholar]
- 35.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. 2003;69(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tourret J, Willing BP, Dion S, MacPherson J, Denamur E, Finlay BB. Immunosuppressive Treatment Alters Secretion of Ileal Antimicrobial Peptides and Gut Microbiota, and Favors Subsequent Colonization by Uropathogenic Escherichia coli. Transplantation. 2017;101(1):74–82. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. [DOI] [PubMed] [Google Scholar]
- 38.Popa ER, Stegeman CA, Kallenberg CG, Tervaert JW. Staphylococcus aureus and Wegener’s granulomatosis. Arthritis Res. 2002;4(2):77–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tadema H, Abdulahad WH, Stegeman CA, Kallenberg CG, Heeringa P. Increased expression of Toll-like receptors by monocytes and natural killer cells in ANCA-associated vasculitis. PLoS One. 2011;6(9):e24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung WH, Croll D, Cho JH, Kim YR, Lee YW. Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses. 2015;58(3):167–72. [DOI] [PubMed] [Google Scholar]
- 41.Zhao YC, Bassiouni A, Tanjararak K, Vreugde S, Wormald PJ, Psaltis AJ. Role of fungi in chronic rhinosinusitis through ITS sequencing. Laryngoscope. 2018;128(1):16–22. [DOI] [PubMed] [Google Scholar]
- 42.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55(5):494–504. [DOI] [PubMed] [Google Scholar]
- 43.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9(3):e90899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tipton L, Muller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, et al. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome. 2018;6(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.