Abstract
Sucrose non-fermenting 1-related protein kinase 1 (SnRK1) is a central metabolic regulator and the plant orthologue of the mammalian AMP-activated protein kinase (AMPK); both are energy-sensing heterotrimeric enzymes comprising a catalytic α- and regulatory β- and γ-subunits. α-Subunits contain a serine/threonine kinase domain (KD) at their N-terminus that is immediately followed by a small regulatory domain termed the auto-inhibitory domain (AID) in AMPK and the ubiquitin-associated domain (UBA) in SnRK1. Association of the AID with the AMPK KD inhibits activating phosphorylation of the KD by upstream kinases and promotes dephosphorylation, as well as inhibiting AMPK catalytic activity. Despite these mechanistic insights regarding the AMPK AID, the SnRK1 UBA regulatory implications have not been investigated. Using recombinant protein comprising either the KD-only or KD-AID/KD-UBA, we found that the UBA of SnRK1 acts in a distinct regulatory manner to its orthologous AID of AMPK. Firstly, the plant upstream kinase GRIK2 preferentially phosphorylates the SnRK1 KD-UBA. Secondly, the SnRK1 KD in the absence of the UBA shows near identical initial catalytic activity to the KD-UBA, but in comparison a rapid loss of catalytic activity is observed. Our findings indicate that the role of the UBA in SnRK1 regulation may be more akin to that of the UBA in the mammalian AMPK-related kinases rather than its immediate functional orthologue, AMPK. This study adds to a growing body of work demonstrating the divergent regulatory mechanisms of the orthologous plant SnRK1 and mammalian AMPK.
Keywords: SnRK1, AMPK, ubiquitin-associated domain (UBA), auto-inhibitory domain (AID)
Introduction
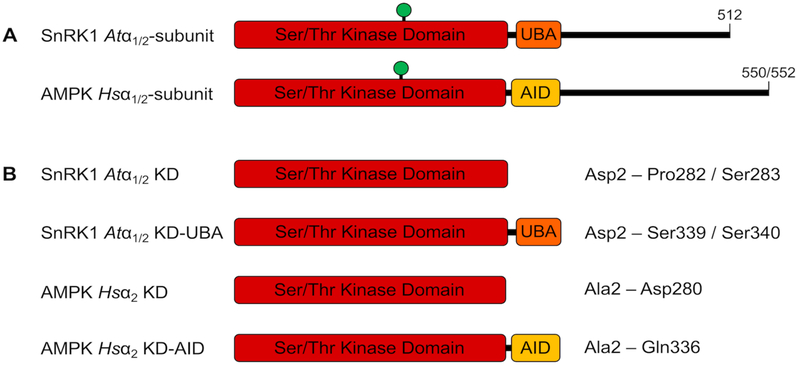
SNF1-related protein kinase 1 (SnRK1) is the plant orthologue of an evolutionarily-conserved family of energy-sensing, central metabolic regulators that include yeast sucrose non-fermenting 1 (SNF1) and mammalian AMP-activated protein kinase (AMPK). Members of the SnRK1/SNF1/AMPK family sense changes in cellular energy status and act at both cellular and systemic levels to restore energy balance [1,2]. These heterotrimeric enzymes comprise a catalytic α- and regulatory β- and γ-subunits, for each of which multiple isoforms exist [3]; for example, there are two α-isoforms for both AMPK and SnRK1. The catalytic subunits comprise an N-terminal serine/threonine kinase domain (KD) that is immediately followed by a three-helix bundle; this is termed the auto-inhibitory domain (AID) for AMPK and the ubiquitin-associated domain (UBA) for SnRK1 (Fig. 1A). The remainder of the α-subunits comprise regions responsible for interaction with β- and γ-subunits and other regulatory elements. Whilst the AMPK and SnRK1 KDs share 63% sequence identity, the respective AID and UBA share only 33–37% sequence identity (Table S1), suggesting their functions in relation to the kinase may differ.
Figure 1.
SnRK1 and AMPK catalytic α-subunits. (A) Both plant and mammalian α-subunits comprise an N-terminal KD followed immediately C-terminally by either a UBA or an AID, respectively. Green circle represents the phosphorylated T-loop threonine. (B) Constructs used in the study, with amino acid boundaries indicated.
Mammalian AMPK is primarily regulated by reversible phosphorylation of the α-subunit T-loop threonine – Thr172 in the α2 isoform – whose phosphorylation is critical for significant activity [4]. Decreasing adenylate charge – i.e. increasing ADP and AMP concentrations relative to ATP – enhances phosphorylation by upstream kinases [5] and protects against dephosphorylation [6]. In addition, AMP further allosterically activates the kinase [6]. The mechanism(s) by which ADP and AMP, which bind to the γ-subunit, causes activation involves conformational change in the heterotrimer whereby the AID dissociates from the hinge region of the KD. This results in a closed, active conformation that renders the KD less susceptible to phosphatases [7] and, as yet inexplicably, a better substrate for its upstream kinases [5,8]. T-loop phosphorylation – at Thr175 in the Arabidopsis thaliana SnRK1 α1 isoform, Thr176 in α2 – is likewise critical for SnRK1 activity [9], and two upstream kinases, Geminivirus Rep-interacting kinase 1 (GRIK1) and GRIK2 [10], have been described, with others likely yet to be identified. Adenylate nucleotides do not, however, play a significant role in the regulation of SnRK1 [2,3,11,12]. Rather, SnRK1 appears to sense energy charge primarily through high-energy sugar-phosphates, most importantly trehalose-6-phosphate, which leads to inhibition of the kinase (through an unresolved mechanism most likely involving a proteinaceous co-factor) [13,14]. The role of reversible phosphorylation in planta is also somewhat unclear, with multiple studies demonstrating that SnRK1 T-loop phosphorylation state does not predictably correlate with SnRK1 activity [9,15–17]. SnRK1 belongs to a larger family of related kinases that include the plant-specific SnRK2 and SnRK3 sub-groups. Whilst these contain regulatory regions C-terminal to their kinase domains that regulate T-loop phosphorylation and catalytic activity, they are unrelated to the UBA of SnRK1 [18]. This then begs the question as to what role the UBA plays in the regulation of SnRK1.
UBAs are present in ten of the twelve mammalian AMPK-related kinases (AMPK-RKs), the only known kinases to possess UBAs in the human genome. Though UBAs in proteins commonly bind ubiquitin, this is not the case for the mammalian AMPK-RKs. Instead, the UBA in these kinases allows liver kinase B1 (LKB1) to phosphorylate their T-loop and thus render them active, though the domain itself is not involved in the LKB1 interaction [19]. LKB1 is one of two AMPK upstream kinases, the other being Ca2+/Calmodulin-dependent kinase kinase β (CAMKKβ) [8], though the AID, in contrast to the UBA of AMPK-RKs, inhibits AMPK T-loop phosphorylation. Despite the observed role in enhancing activating T-loop phosphorylation, investigation of the UBAs of the AMPK-RK microtubule-associated protein/microtubule affinity regulating kinase 1 (MARK1) and MARK2 suggests that they act to reduce catalytic activity [20]. To resolve a seemingly contradictory mechanism of concurrent activation and inhibition, the AMPK-RK UBA has been proposed as a bistable switch element. This hypothesis postulates that the UBA stabilises both the open, inactive and the closed, active conformation depending on phosphorylation state or interaction with co-factors [21].
We sought to define a role for the UBA of the plant SnRK1 α-subunit using recombinantly-expressed A. thaliana (Atα1/2) KD or KD-UBA constructs in comparison to Homo sapiens AMPK α2 isoform (Hsα2) KD and KD-AID equivalents. These constructs comprise only the KD or the KD extended to include the UBA or AID but truncated thereafter (Fig. 1B). We show that the SnRK1 UBA shares sequence similarities with the related but distinct AMPK AID and AMPK-RK UBAs. In an opposite manner to AMPK, the SnRK1 UBA promotes phosphorylation by its upstream kinase GRIK2. Finally, we show that the UBA contributes to the preservation of catalytic activity over time but does not influence initial catalytic activity. The data suggest that the role of the SnRK1 UBA in regulating kinase activity is more akin to that of UBAs from mammalian AMPK-RKs and distinct from that of the AID for AMPK itself. These findings build on a growing body of work showing distinct regulatory mechanisms of SnRK1 in contrast to its opisthokont orthologues.
Materials and Methods
Sequence alignment and statistical analyses
Multiple sequence alignments were performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences were obtained from UniProt (http://www.uniprot.org/). Graphing and statistical analyses were performed using Prism7 (GraphPad).
Constructs
AMPK and SnRK1 (Fig. 1B) and GRIK2 constructs were cloned into and expressed from pGEX-6P-3 plasmid (GE Healthcare Life Sciences) to contain an N-terminal glutathione-S-transferase (GST) tag for purification. pGEX-6P-3 was digested with BamHI and SalI restriction enzymes and digested PCR fragments ligated T4 ligase. CAMKKβ was cloned into and expressed from pET-28xHM plasmid digested with SpeI, modified from pET-28b (Novagen) by incorporation of a linker to contain an N-terminal His6-MYC epitope and SpeI restriction site between NcoI and BamHI sites. Restriction enzymes and T4 ligase from New England BioLabs. Refer to Table S2 for a list of oligonucleotides and restriction enzymes used for PCR fragments. cDNA for PCR amplification was obtained from DNASU at Arizona State University (AMPK, CAMKKβ) or from the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University (SnRK1, GRIK2). All point mutants employed in the study were generated by PCR site-directed mutagenesis using oligonucleotides listed in Table S3 with either Q5 DNA polymerase (New England BioLabs) or Pfu DNA polymerase (Promega), together with T4 ligase.
Protein expression and purification
Constructs were expressed in Escherichia coli BL21(DE3) strain (New England BioLabs) in 2×YT medium: 16 g L−1 tryptone, 10 g L−1 yeast extract, 5 g L−1 NaCl; supplemented with 100 mg L−1 ampicillin and 17 mg L−1 chloramphenicol. Induction was achieved with 400 μM isopropyl-β-D-thiogalactopyranoside for 16 hours at 16 °C upon reaching an OD600 of 0.5. Bacteria were harvested by centrifugation (5,000 g, 15 minutes, 4 °C) and resuspended in homogenisation buffer (20 mM Tris-HCl, pH7.4, 50 mM NaCl, 2 mM dithiothreitol) supplemented with one tablet of protease inhibitor cocktail (Roche Applied Science) per 50 mL buffer. Homogenisation was achieved by incubation with 1 g L−1 lysozyme (Sigma) for 15 minutes at 4 °C with gentle inversion followed by sonication. The homogenisation was cleared by centrifugation (25,000 g, 30 minutes, 4 °C) and the cleared lysate incubated with glutathione-agarose resin (Gold Biotechnology) for 15 minutes at 4 °C with gentle inversion. The agarose was then pelleted by centrifugation (3,000 g, 10 minutes, 4 °C), washed with homogenisation buffer thrice, and the GST-bound recombinant protein eluted with 10 mM reduced glutathione in homogenisation buffer and concentrated using an Amicon Ultra-15 30 kDa MWCO (Millipore). For upstream kinase assays, the GST moiety was removed by incubation with PreScission Protease (GE Healthcare Life Sciences) and KD(-AID/UBA) protein isolated by trapping the free GST on glutathione-agarose. His6-MYC-CAMKKβ was expressed and purified similarly with the following alterations: 50 mg L−1 kanamycin rather than ampicillin, homogenisation buffer lacking dithiothreitol but supplemented with 10 mM imidazole, nickel-agarose resin (Sigma), elution using 300 mM imidazole.
Upstream kinase assay
100 pmol of Hsα2 or Atα1 protein (with GST moiety removed) was incubated with 1 pmol of HsCAMKKβ or AtGRIK2, respectively, in a total volume of 25 μL at 30 °C in 50 mM HEPES, pH7.5, 2 mM MgCl2, 5% (v/v) glycerol, 0.02% (v/v) Brij–35 (Invitrogen), 1 mM dithiothreitol, 200 μM ATP. Reactions were terminated by the addition of 10 μL 4 × SDS sample buffer. The ability of the upstream kinase to phosphorylate its respective T-loop threonine was monitored by Western blotting (following SDS-PAGE) using an anti-AMPKα2-phosphoThr172 antibody (40H9, Cell Signaling Technology) [11].
Kinase activity assay
Enzyme activity assays were performed as previously described [11] using 20 pmol of GST-tagged recombinant protein at 30 °C with SAMS peptide (HMRSAMSGLHLVKRR-amide) as substrate [22,23]. Reactions were measured every two minutes over ten minutes, where the activity for each time point represents the average activity over that particular two-minute period.
Results
Analysis of AID/UBA from AMPK and related kinases
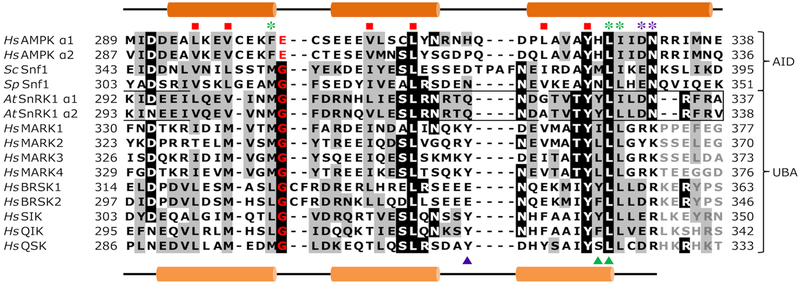
The alignment of the UBA sequences of SnRK1 α-subunits with the AID of AMPK and SNF1 and the UBA from AMPK-RKs reveals several interesting aspects (Fig. 2). Firstly, the residues comprising the hydrophobic core of the AID of AMPK [24] and the UBA of MARKs [20] are conserved in SnRK1, suggesting a conserved three-dimensional fold. Secondly, SnRK1 UBAs share identity or similarity in residues contacting the kinase domain of both AMPK and MARKs. Thirdly, the conserved glycine residue reported by Jaleel et al. as critical for T-loop phosphorylation and catalytic activity [19] is present in SnRK1 but in AMPK is substituted with glutamate. Taken together, we hypothesised that the UBA of SnRK1 may act more like the UBAs of AMPK-RKs to regulate SnRK1.
Figure 2.
Multiple sequence alignment of AMPK, SNF1, SnRK1 and mammalian AMPK-related kinases. Red squares indicate hydrophobic core residues; the conserved UBA glycine residue is highlighted red; black and grey shading indicates residues identical or similar, respectively, to those of the SnRK1; residues involved in hydrophobic (green) and hydrophilic (purple) contacts with the AMPK KD (asterisk) or MARK KD (triangle). Numbering refers to amino acid residue sequence.
The UBA promotes T-loop phosphorylation
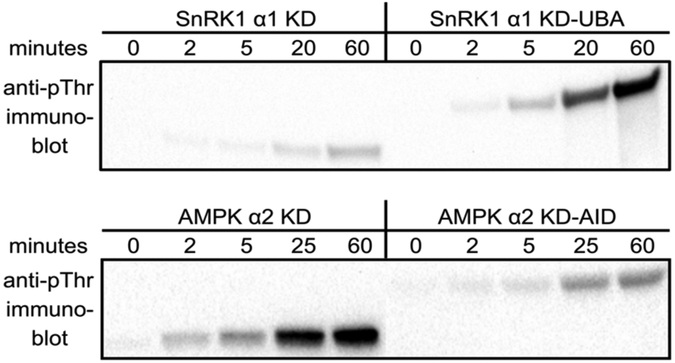
The AMPK AID, when bound to the KD, inhibits activating T-loop phosphorylation by its upstream kinases [5,7,8] whereas the UBA of AMPK-RKs enhances T-loop phosphorylation [19]. Therefore, we sought to determine how the SnRK1 UBA influences T-loop phosphorylation by the plant upstream kinase GRIK2. Upon addition of ATP to start the reaction, Atα1 KD-UBA was more quickly phosphorylated by GRIK2 than was its KD-only counterpart (Fig. 3). This result is in stark contrast to Hsα2 KD-AID, which, as expected, was a vastly inferior substrate for CAMKKβ than its KD-only counterpart (Fig. 3). Clearly, the SnRK1 UBA promotes T-loop phosphorylation, a situation more akin to the role of the UBA in AMPK-RKs [19].
Figure 3.
Immunoblots of SnRK1/AMPK upstream kinase assays ± UBA/AID. The UBA of SnRK1 α1 promotes T-loop Thr phosphorylation by the plant upstream kinase GRIK2. Conversely, the AID of AMPK inhibits T-loop phosphorylation by the mammalian upstream kinase CAMKKβ.
Analysis of catalytic activity
In order to dissect the role of the UBA in KD catalytic activity, the critical T-loop threonine in both SnRK1 and AMPK constructs was mutated to glutamate to mimic phosphorylation so that differences in upstream kinase efficiency towards activating KD and KD-UBA or KD-AID would not influence the results. We used the SAMS peptide as substrate, derived from the AMPK substrate acetyl-CoA carboxylase and commonly used in assays of AMPK and SnRK1 activity [22,23]. Catalytic activities of KD and KD-AID/UBA fragments were assayed over time with activities determined every two minutes for ten minutes. This methodology allows almost-instantaneous activity to be determined at each time point, and thus is a gauge of the change in activity over time.
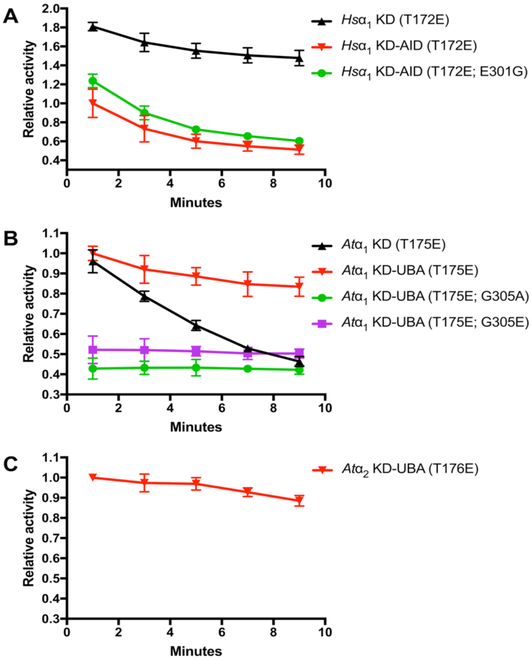
Hsα2 KD displayed an almost two-fold greater initial activity compared to KD-AID (Fig. 4A), demonstrating the auto-inhibitory effect. Interestingly, the activity of the KD-AID decreased over time, such that at 8 to 10 minutes it was roughly half initial (0 to 2 minutes) activity (Fig. 4A). In comparison, the activity of the Hsα2 KD fragment diminished only marginally over the course of the assay. As observed in Fig. 2, the conserved glycine critical for AMPK-RK UBAs to command their LKB1-inducing effect is substituted with glutamate in the AMPK AID. The observed differences in effect on KDs between the AID and UBA may be partly due to this substitution. However, mutation of this residue to glycine had an almost insignificant impact on relieving auto-inhibition (Fig. 4A), indicating that other elements in the AMPK AID may be important in determining effect towards the KD.
Figure 4.
UBA/AID influence on catalytic activity of T-loop threonine-to-glutamate phosphomimetic mutants for Hsα2 (A), Atα1 (B), and Atα2 (C). Relative activity (left y-axis) is given a value 1 that for the respective KD-AID/UBA over the initial (0 to 2 minutes) period. Absolute activity (right y-axis) is indicated in pmol phosphate min−1 nmol−1 enzyme. Activity levels for each point represent the average activity for each two-minute period between which the points are plotted.
Initial (0 to 2 minutes) activities of AtSnRK1 α1 KD and KD-UBA constructs were not significantly different (Fig. 4B), indicating that presence of the UBA does not influence catalytic activity of the SnRK1 KD. However, when tracked over time, whilst KD-UBA retained the majority of its initial activity during the 8 to 10 minutes period, KD-only activity dramatically decreased to less than half its initial (0 to 2 minutes) activity (Fig. 4B). This suggests that the SnRK1 UBA retains the KD in the active conformation, as has been proposed for AMPK-RK UBAs acting as a bistable switch element [21], though in this case the auto-inhibitory effect observed in MARKs [20] was not seen. Mutation of the conserved glycine (G305 in Atα1) to either alanine or glutamate reduced initial catalytic activity to roughly half in both cases (Fig. 4B). However, unlike all other constructs tested, activity of the Atα1 glycine mutants did not change over time. These observations suggest that the conserved glycine is important for the UBA to influence catalytic activity on the KD of SnRK1, though the lack of decrease in activity over time – possibly due to increased locking of the KD in its active state – cannot currently be explained.
We also attempted to perform these assays using the Atα2 isoform. However, despite successfully expressing and purifying the KD and KD-UBA constructs, albeit with much-reduced yield, we only observed activity for Atα2 KD-UBA (Fig. 4C). This fragment, though roughly 4–5 times less active than its Atα1 counterpart, displayed the similar slight diminishment of catalytic activity over time. Notably, similar problems with expression of Atα2 constructs in E. coli have been previously reported [10].
Discussion
SnRK1 is the plant orthologue of mammalian AMPK and fungal SNF1, sharing their conserved role as an energy sensor to mediate central metabolism as well as the heterotrimeric subunit composition. Despite this, it is clear that the mechanism(s) by which SnRK1 senses changes in cellular energy charge and by which these changes regulate catalytic activity are fundamentally distinct from those of its opisthokont orthologues [2,3,11]. The manner by which adenylate nucleotides transmit their effect in AMPK ultimately involves the relative association of the AID with the hinge region of the KD to promote activation or inhibition – respectively, low or high adenylate charge; AID dissociation or association; closed or open KD conformation [7,8]. Whilst AMPK directly senses adenylate charge which regulates both reversible T-loop phosphorylation and allosteric activation, adenine nucleotides likely do not regulate SnRK1 [2,3,11,12], at least directly. An early report demonstrating AMP inhibition of phosphatase-mediated dephosphorylation of the spinach leaf SnRK1 T-loop [25] has not been repeated, though it is possible that the observed regulation by AMP was due to the presence of a co-factor during the SnRK1 purification process. In fact, this is the situation for the regulation of SnRK1 by sugar-phosphates, where the inhibitory effect is observed only in the presence of an unknown intermediary factor seemingly highly dependent on tissue type and developmental stage [13,26,27]. Whatever the case, the lack of (direct) adenylate regulation of SnRK1 necessarily means that the role of the UBA in the regulation of reversible T-loop phosphorylation and catalytic activity be distinct to that of the AMPK AID.
We have shown that the SnRK1 UBA shares sequence similarities with the UBAs of mammalian AMPK-RKs (Fig. 2). AMPK-RKs, whilst having varied binding partners, do not comprise similar β- and γ-subunits to AMPK and SnRK1; adenylate charge is sensed by AMPK through the γ-subunit. Indeed, our observations show that the UBA promotes SnRK1 T-loop phosphorylation by the upstream kinase AtGRIK2 (Fig. 3) in a manner reminiscent of AMPK-RK T-loop phosphorylation by LKB1 [19] and unlike that for AMPK. In a separate study on AMPK-RKs [19], the presence of UBA was essential for LKB1-mediated phosphorylation, whereas in our study the SnRK1 KD-only fragment was still phosphorylated by GRIK2, though to a markedly reduced extent. This perhaps suggests that the qualities imparted by the SnRK1 UBA on the KD are reduced as compared with AMPK-RKs.
The UBA of AMPK-RKs, or at least MARKs, has been shown to possess mild auto-inhibitory properties [20]. However, Jaleel et al. [19] demonstrated that mutation of the conserved glycine to alanine – and thus presumably removal of the function of the UBA domain – drastically reduced catalytic activity of full-length SIK (salt-induced kinase) T-loop threonine-to-glutamate mutant. We find similar reductions in catalytic activity of SnRK1 lacking UBA over time but not for initial catalytic activity (Fig. 4B). Mutation of the conserved glycine (G305) in Atα1 KD-UBA to either alanine or glutamate reduced catalytic activity, indicating its importance to UBA function, though we did not observe the same decrease in activity over time as for KD-only. It is difficult to compare these experimental outcomes, since the studies on the AMPK-RKs measure overall activity at the end-point of the reaction, some 15 to 20 minutes, rather than monitoring activity over time as in the present study. Certainly, at the completion of ten minutes, Atα1 KD activity was strongly reduced compared to KD-UBA; however, the initial activities were indistinguishable (Fig. 4B). Nevertheless, it can be said that the SnRK1 UBA enhances, or rather maintains, catalytic activity over time. A second complication of the MARK study [20] was that catalytic activity was dependent upon T-loop phosphorylation of the various constructs used, which makes direct comparison difficult.
Given the data, the proposal of the UBA of AMPK-RKs as a bistable switch element for the KD [21] appears reasonable to also apply to SnRK1, though whether the SnRK1 UBA could stabilise an inactive KD conformation cannot yet be assumed. Whilst we have demonstrated that the SnRK1 UBA enhances T-loop phosphorylation by GRIK2, maintains catalytic activity over time, and clearly acts in a divergent manner to the AMPK AID, we are yet to understand the significance of the UBA on SnRK1 regulation. How the UBA functions are part of the SnRK1 heterotrimeric complex and how it influences KD conformation and activity in the face of sugar-phosphate inhibition and other regulators and binding partners remains a mystery. Nevertheless, this is the first study to focus on the role of the UBA in SnRK1 regulation and adds to a growing body of evidence demonstrating that the mechanisms of SnRK1 regulation are highly divergent from those of AMPK. Our data suggest that the SnRK1 UBA acts more akin to that of the mammalian AMPK-RKs and therefore that mammalian AMPK itself, in regard to its AID, can be thought of as the kinase divergent from all others in its family.
Supplementary Material
Acknowledgements
This work was supported by Kentucky Science and Engineering Foundation [grants KSEF-2268RDE-014, KSEF-2971-RDE-017], National Science Foundation [grants IIA-1355438, MCB-1252345], Australian Research Council Centre of Excellence in Plant Cell Walls [grant CE1101007] and an Australia Research Council Discovery [grant DP110103161].
References
- [1].Herzig S, Shaw RJ, AMPK: guardian of metabolism and mitochondrial homeostasis, Nat Rev Mol Cell Biol, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Broeckx T, Hulsmans S, Rolland F, The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function, J Exp Bot, 67 (2016) 6215–6252. [DOI] [PubMed] [Google Scholar]
- [3].Emanuelle S, Doblin MS, Stapleton DI, Bacic A, Gooley PR, Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1, Trends Plant Sci, 21 (2016) 341–353. [DOI] [PubMed] [Google Scholar]
- [4].Willows R, Sanders MJ, Xiao B, Patel BR, Martin SR, Read J, Wilson JR, Hubbard J, Gamblin SJ, Carling D, Phosphorylation of AMPK by upstream kinases is required for activity in mammalian cells, Biochem J, 474 (2017) 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE, AMPK is a direct adenylate charge-regulated protein kinase, Science, 332 (2011) 1433–1435. [DOI] [PubMed] [Google Scholar]
- [6].Gowans GJ, Hawley SA, Ross FA, Hardie DG, AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation, Cell Metab, 18 (2013) 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li X, Wang L, Zhou XE, Ke J, de Waal PW, Gu X, Tan MH, Wang D, Wu D, Xu HE, Melcher K, Structural basis of AMPK regulation by adenine nucleotides and glycogen, Cell Res, 25 (2015) 50–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lin SC, Hardie DG, AMPK: Sensing Glucose as well as Cellular Energy Status, Cell Metab, (2017). [DOI] [PubMed] [Google Scholar]
- [9].Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J, A central integrator of transcription networks in plant stress and energy signalling, Nature, 448 (2007) 938–942. [DOI] [PubMed] [Google Scholar]
- [10].Shen W, Reyes MI, Hanley-Bowdoin L, Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop, Plant Physiol, 150 (2009) 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Emanuelle S, Hossain MI, Moller IE, Pedersen HL, van de Meene AM, Doblin MS, Koay A, Oakhill JS, Scott JW, Willats WG, Kemp BE, Bacic A, Gooley PR, Stapleton DI, SnRK1 from Arabidopsis thaliana is an atypical AMPK, Plant J, 82 (2015) 183–192. [DOI] [PubMed] [Google Scholar]
- [12].Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F, The hybrid four-CBS-domain KINβγ subunit functions as the canonical gamma subunit of the plant energy sensor SnRK1, Plant J, 75 (2013) 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ, Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate, Plant Physiol, 149 (2009) 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nunes C, O’Hara LE, Primavesi LF, Delatte TL, Schluepmann H, Somsen GW, Silva AB, Fevereiro PS, Wingler A, Paul MJ, The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation, Plant Physiol, 162 (2013) 1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P, SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation, Plant Physiol, 149 (2009) 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Coello P, Hirano E, Hey SJ, Muttucumaru N, Martinez-Barajas E, Parry MA, Halford NG, Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2, J Exp Bot, 63 (2012) 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A, Lumbreras V, Gonzalez-Guzman M, Antoni R, Rodriguez PL, Baena-Gonzalez E, ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis, Plant Cell, 25 (2013) 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC, The Arabidopsis CDPK-SnRK superfamily of protein kinases, Plant Physiol, 132 (2003) 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jaleel M, Villa F, Deak M, Toth R, Prescott AR, Van Aalten DM, Alessi DR, The ubiquitin-associated domain of AMPK-related kinases regulates conformation and LKB1-mediated phosphorylation and activation, Biochem J, 394 (2006) 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Marx A, Nugoor C, Muller J, Panneerselvam S, Timm T, Bilang M, Mylonas E, Svergun DI, Mandelkow EM, Mandelkow E, Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2, J Biol Chem, 281 (2006) 27586–27599. [DOI] [PubMed] [Google Scholar]
- [21].Marx A, Nugoor C, Panneerselvam S, Mandelkow E, Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases, FASEB J, 24 (2010) 1637–1648. [DOI] [PubMed] [Google Scholar]
- [22].Davies SP, Carling D, Hardie DG, Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay, Eur J Biochem, 186 (1989) 123–128. [DOI] [PubMed] [Google Scholar]
- [23].Dale S, Wilson WA, Edelman AM, Hardie DG, Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I, FEBS Lett, 361 (1995) 191–195. [DOI] [PubMed] [Google Scholar]
- [24].Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW, Structural insight into the autoinhibition mechanism of AMP-activated protein kinase, Nature, 459 (2009) 1146–1149. [DOI] [PubMed] [Google Scholar]
- [25].Sugden C, Crawford RM, Halford NG, Hardie DG, Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5’-AMP, Plant J, 19 (1999) 433–439. [DOI] [PubMed] [Google Scholar]
- [26].Nunes C, Primavesi LF, Patel MK, Martinez-Barajas E, Powers SJ, Sagar R, Fevereiro PS, Davis BG, Paul MJ, Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate, Plant Physiol Biochem, 63 (2013) 89–98. [DOI] [PubMed] [Google Scholar]
- [27].Martinez-Barajas E, Delatte T, Schluepmann H, de Jong GJ, Somsen GW, Nunes C, Primavesi LF, Coello P, Mitchell RA, Paul MJ, Wheat grain development is characterized by remarkable trehalose 6-phosphate accumulation pregrain filling: tissue distribution and relationship to SNF1-related protein kinase1 activity, Plant Physiol, 156 (2011) 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.