INTRODUCTION
Meiotic recombination predominantly occurs at recombination hotspots, discrete kilobase-size regions of the genome where the recombination rate can be thousands of times higher than in adjacent regions1,2. The first human hotspot was described in 19823, and until 2005, only a handful of individual hotspots had been studied in detail. In 2005, the first genome-wide maps of recombination hotspots were created by inferring recombination events from patterns of linkage disequilibrium in human populations4. Over thirty thousand recombination hotspots were identified by this method, opening the door to better understanding the process of meiotic recombination. Building on this work, a major breakthrough was made in 2010, when three labs independently identified the PRDM9 protein as responsible for targeting meiotic DSBs in the human and mouse genomes5–7. PRDM9 directs DSBs to specific hotspots through binding to specific DNA sequences5,8,9 and subsequently trimethylating Lysine 4 of the histone H310. The DNA Double strand break (DSB) formation machinery is recruited to hotspot locations to initiate recombination (reviewed by11–14) and the ends of meiotic DSBs are nucleolitically processed to form long single stranded DNA (ssDNA) overhangs. ssDNA overhangs are bound by recombinases, RAD51 and DMC1, that facilitate the repair of DSBs via homologous recombination. This repair produces either crossover or non-crossover products, depending on the specific mechanism used. Although PRDM9 defines practically all recombination hotspots in mice15 and humans16, many more PRDM9 binding sites are present in the genome than there are hotspots. This indicates that factors other than PRDM9 will shape the recombination landscape and highlights the importance of experimentally determining hotspot locations genome-wide.
Here we describe an approach to map recombination initiation hotspots – the sites of meiotic DSBs17. We first capture DNA bound by DMC1 or RAD51 using chromatin immunoprecipitation (ChIP). Next, we apply a bespoke protocol to enrich for ssDNA and then sequence these ssDNA fragments by high throughput sequencing17 (Fig. 1). Since our approach involves direct physical detection of recombinase-bound ssDNA ends, it is broadly applicable to any sexually reproducing species. The precision of hotspot mapping is independent of polymorphism density (which affects crossover-mapping based methods) and since our method detects the earliest intermediates in DSB repair it can identify hotspots independent of downstream repair pathway decisions. Furthermore, since this method surveys recombination in a population of meiocytes, it yields accurate quantitative estimates of hotspot usage that are not achievable by other techniques. SSDS has been predominantly used in mice15,18–21, but has recently allowed the generation of the first maps of recombination hotspots in individual human genomes16. In addition to mapping meiotic DSBs, SSDS may be useful for detecting DSBs in somatic tissues, or for detecting other single stranded DNA species, such as replication intermediates and certain viruses.
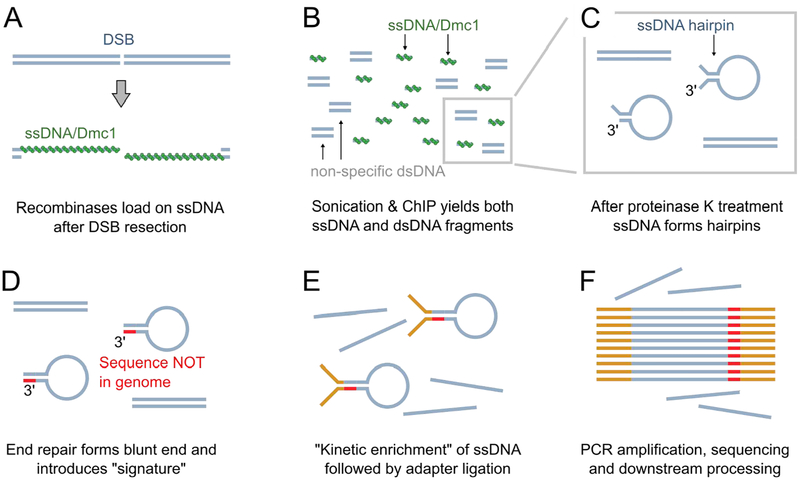
Figure 1. ChIP-SSDS schematic.
(A) ChIP-SSDS captures ssDNA bound by meiotic recombinases to map DSB hotspot locations. (B) After fixation and sonication, DMC1-bound ssDNA is immunoprecipitated using an anti-DMC1 antibody. (C) Deproteinized ssDNA will spontaneously form hairpins, mediated by intra-molecular micro-homologies. (D) The 3’ ends of hairpins are trimmed, then extended, using the 5’ end of the ssDNA molecule as a template. This both introduces a “signature” that allows downstream identification of ssDNA-derived sequencing reads, and ensures that hairpins have double stranded ends. (E) The DNA is denatured, then allowed to return to room temperature. In the reaction timeframe, ssDNA hairpins will re-form because of fast intra-molecular reannealing, however dsDNA will not renature. Sequencing adapters are then ligated to blunt-ended DNA, which will primarily occur at ssDNA-derived hairpins. (F) DNA with ligated adapters is then amplified and sequenced. A computational pipeline then specifically uses the “signature” sequence (D) to identify ssDNA.
CHROMATIN IMMUNOPRECIPITATION (ChIP) AND SINGLE STRANDED DNA SEQUENCING (SSDS)
Reagents:
Paraformaldehyde (PFA) (Sigma, P6148)
5 M NaCl (Sigma, S5150–1L)
SDS (Sigma)
1 M Tris-Cl, pH 6.5 @25 °C (KD Medical)
1 M Tris-Cl, pH 8.0 @25 °C (KD Medical)
KOH (Sigma)
Triton X100 (Sigma)
0.5 M EDTA (Sigma)
0.1 M EGTA (Sigma)
Proteinase Inhibitor cocktail (Roche, 11 836 153 001)
UltraPure DNase/RNase-Free Distilled Water (Invitrogen, 10977–023)
PBS (KD Medical)
TE (KD Medical)
8 M LiCl (Sigma, L7026)
IGEPAL-CA630 (Sigma, 18896)
Sodium deoxycholate monohydrate (Sigma, D5670)
NaHCO3 (Sigma, S5761)
Dynabeads Protein G (Invitrogen, Cat no:10004D)
Proteinase K, 20 mg/ml (Thermofisher # 25530049)
Anti-DMC1 antibody (Abcam, Ab 11054)
MinElute PCR Cleanup Kit (Qiagen, 28004)
TruSeq Nano DNA LT Library Preparation Kit (Illumina)
T4 DNA polymerase (3 μ/μl, New England Biolabs, M0203)
DNA Polymerase I Large (Klenow) Fragment (5 u/μl, New England Biolabs, M0210)
T4 Polynucleotide Kinase (PNK) (10 u/μl, New England Biolabs, M0201)
Klenow Fragment (3’→5’ exo-) (5 u/μl with 10X NEBuffer 2, New England Biolabs, M0212)
Quick ligation kit (New England Biolabs, M2200)
10X T4 ligation Buffer (New England Biolabs, B0202)
dATP and dNTPs (New England Biolabs)
Qubit dsDNA HS Assay Kit (Thermofisher # Q32851) or Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher, P7589)
AMPureXP beads (Beckman Coulter)
Key equipment and consumables:
DNA LoBind 1.5 ml tubes (Eppendorf 022431021) and low retention filtered tips (USA Scientific)
Slide-A-Lyzer G2 dialysis cassette 10k MWCO, 3 ml (Thermo Scientific, 87730)
DynaMagTM-2 Magnet (Invitrogen, 12321D)
15 ml Dounce homogenizer with a tight fit pestle
70 mm Cell Strainer (BD Biosciences, 352350)
Thermomixer (Eppendorf)
PCR machine
Qubit Fluorometer (Thermofisher # Q33216) to use with the Qubit dsDNA HS Assay Kit or any
fluorometer to use with PicoGreen kit.
Bioruptor UCD 200 (Diagenode) and 15ml polystyrene Falcon tubes
Library Quantification Kit (KAPA Biosystems # KK4824)
Buffers:
Note: all buffers are prepared with deionised water and passed through 0.22μm filter
-
10% SDS
1N KOH
-
Quenching buffer, store at 4°C:
1.25 M Glycine
-
Cell wash buffer 1, store at 4°C:
0.25% Triton X-100, 10mM EDTA, 0.5mM EGTA, 10mM Tris-Cl, pH 8.
-
Cell wash buffer 2, store at 4°C:
200mM NaCl, 1mM EDTA, 0.5mM EGTA, 10mM Tris-CL, pH 8
-
Lysis buffer, store at 4°C:
1% SDS, 10mM EDTA, 50mM Tris-Cl, pH 8
Note: Warm to room temperature to allow SDS to re-dissolve before use.
-
Protease Inhibitors 100X, store at −20°C:
1 tablet of Protease Inhibitor Cocktail (Roche, 11 836 153 001) dissolved in 500μl of UltraPure water.
-
ChIP buffer, store at 4°C:
16.7 mM Tris-HCl, pH 8.0, 167mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100.
Note: 1L of this buffer is freshly prepared at Day 1
-
ChIP Wash 1, store at 4°C:
20mM Tris-HCl pH 8, 150mM NaCl, 2mM EDTA, 1% Triton X-100, 0.1% SDS.
-
ChIP Wash 2, stored at 4°C:
20mM Tris-HCl pH 8, 500mM NaCl, 2mM EDTA, 1% Triton X-100, 0.1% SDS.
-
ChIP Wash 3, stored at 4°C:
10mM Tris-HCl pH 8, 0.25M LiCl, 1mM EDTA, 1% Deoxycholic acid, 1% IGEPAL-CA630.
1M NaHCO3, stored at −20 °C:
Note: Warm to room temperature to allow NaHCO3 to re-dissolve before use.
DAY 1: Chromatin preparation and ChIP
Prepare the following:
18.5% PFA (5ml)
Note: Refer to safety data sheet from the supplier while handling PFA and dispose accordingly.
Weigh 0.925g PFA and transfer into a 50ml falcon tube containing 4.8 ml H2O and 35μl 1N KOH
Put the tube into 500 ml glass beaker filled with water to approximately 200 ml. Bring the water in the beaker to boiling using a microwave oven. Mix PFA suspension to dissolve the PFA powder. Repeat if necessary.
Cool down the solution, keep on ice
1% PFA (10ml per sample)
Add 540 μl of the 18.5% PFA to 9.46ml of 1X PBS
Keep at room temperature
Experimental procedure:
Dissect testes, place in a Petri dish with cold 1X PBS, transfer to a dry dish, remove tunica albuginea, gently pull apart testis with fine forceps.
Transfer testes to 10ml of 1% PFA in a 15ml falcon tube, rock for 10 min. at room temperature on a rocker.
Immediately add 1ml of quenching buffer, rock for 5 min. at room temperature and place the sample on ice.
-
Transfer solution with the fixed tissue to a 15ml autoclaved Dounce homogenizer, homogenize the tissue with 10 strokes of the pestle.
Note: Depending on the tissue used, aggregates may remain in the suspension.
Pass the cell suspension through a 70μm cell strainer into a 50ml Falcon tube.
Transfer to a 15ml Falcon tube and spin at 900g for 5 min at 4°C. Remove supernatant.
Resuspend the pellet in 10ml of 1X PBS. Spin at 900g for 5min at 4°C and remove supernatant.
Resuspend in 10ml of Cell Wash Buffer 1. Spin at 900g for 5min at 4°C and remove supernatant.
Resuspend in 10ml of Cell Wash Buffer 2. Spin at 900g for 5min at 4°C and remove supernatant.
Resuspend in 1.5ml of Lysis Buffer and add 15μl of the 100X protease inhibitor stock solution.
-
Transfer supernatant to a 15ml polystyrene Falcon tube
Note: Polystyrene tubes are required for optimal sonication
-
Sonicate chromatin for 15min in the Bioruptor with the following parameters:
15 seconds ON / 45 seconds OFF (Setting: High)
Note: Adjust the time if necessary aiming for a fragment size ~1,000 bp
While waiting for sonication to complete, place a dialysis chamber in 1L of cold ChIP buffer in the cold room. Spin at low speed using a stir bar.
Leave sonicated chromatin at room temperature until SDS is re-dissolved (about 1 min.)
Transfer the chromatin to a 1.5ml Eppendorf tube, spin at 12,000g for 10 minutes at 4°C.
Transfer the supernatant to a fresh 15ml Falcon tube, add 1.5ml of fresh ChIP buffer (total volume = 3ml).
Transfer the 3ml of sonicated chromatin to the dialysis chamber (from step 13) and dialyse for 5 hours at 4°C with constant but slow stirring.
Transfer the dialysed solution into two 1.5 ml tubes and keep 50 μl of the solution to check sonication efficiency.
Add 15 μl of 100X of protease inhibitor stock solution to each tube.
Add 12 μg of DMC1 antibody per tube (24 μg total)
Rotate the tubes slowly overnight at 4°C.
- Check sonication efficiency:
- Add 250 μl H2O to the 50 μl aliquot from step 18
- Add NaCl to 0.2 M
- Boil for 15 minutes to reverse crosslinking
- Cool down to room temperature, add 1 μl of 10 mg/ml RNase A and incubate for 10 minutes at 37°C
- Clean up the DNA with Qiagen MinElute kit using the protocol provided with the kit
- Determine the size of sonicated DNA on 1% agarose gel
DAY 2: ChIP (continued)
Prepare Elution buffer (1ml)
800μl ddH2O
100μl 1M NaHCO3 (bring to room temperature to dissolve particulates)
100μl 10% SDS
Experimental procedure
- Prepare and add the Protein G Dynabeads
- Aliquot 150μl of the bead slurry and separate the beads on the magnet, remove supernatant.
- Wash the beads 3 times with the ChIP buffer.
- Resuspend the beads in ~100 μl of the ChIP sample from Day1, step 21 and transfer back to the rest of the ChIP sample, splitting between the two sample tubes.
- Rotate for 2 hrs at 4°C.
Briefly (1–2 seconds) spin the tubes to collect solution from the lid, separate the beads on the magnet, remove and save the supernatant (store at −20°C).
- Wash the beads/chromatin complex by sequentially suspending the beads in 1.2 ml of each of the cold buffers in the order listed below. After each suspension, rotate for 5 minutes in the cold room, briefly spin, then collect beads on the magnet and carefully dispose of the supernatant.
- ChIP Wash 1
- ChIP Wash 2
- ChIP Wash 3
- TE buffer (2 washes)
Add 150μl of Elution buffer to each tube and mix by gentle flicking.
Incubate in a thermomixer at 65°C, shaking at 800 RPM for 30 minutes.
-
Separate the beads on the magnet and combine the supernatant from both tubes into a fresh 1.5 ml Eppendorf tube for a total volume of 300μl.
Note: Be careful not to discard the supernatant at this step.
To the 300μl sample, add 12μl of 5M NaCl and incubate at 65°C overnight (18–20 hours) to reverse DNA-protein crosslinks.
DAY 3: DNA cleanup
Experimental procedures
-
Digest the protein.
Add 6μl of 0.5M EDTA, 12μl of 1M Tris-HCl pH 6.5 and 5μl of Proteinase K (20mg/ml) to the reverse-crosslinked chromatin and incubate at 45°C for 1 hour.
- Clean up the DNA with Qiagen MinElute kit using the following protocol:
- Transfer the sample to a 15 ml Falcon tube.
- Add 2.4 ml (7 volumes) of Buffer PB and mix well.
- Transfer 0.7 ml of the mixture to a MinElute column, incubate at room temperature for 1 minute and centrifuge for 1 minute.
- Discard flow-through and place the MinElute column back into the same tube.
- Repeat steps c & d until the entire mixture is passed through the column.
- Add 750 μl of Buffer PE to the MinElute column with the bound DNA, incubate at room temperature for 1 minute and centrifuge for 1 minute at full speed.
- Discard flow-through and place the MinElute column back in the same tube.
- Repeat steps f & g once.
- Centrifuge the column for an additional 1 minute at maximum speed.
- Remove residual ethanol around the ring of the column using a pipette.
- Place the MinElute column in a clean 1.5 ml microcentrifuge tube.
- Add 12 μl of Buffer EB to the centre of the membrane, incubate at room temperature for 1 minute and centrifuge for 1 minute at full speed. DNA will be in the eluate.
-
Use Qubit kit (Invitrogen) or PicoGreen kit (ThermoFisher) to measure the resulting DNA concentration of the samples.
Note: typical concentration for anti-DMC1 ChIP samples is within the range of 1–3 ng/μl.
Store samples at −20°C or proceed with library preparation.
DAY 4: Library preparation
The protocol for library preparation is based on Illumina protocols but has some important modifications that ensure optimal library yield from ssDNA. Standard Illumina protocols may not result in a satisfactory library.
All reactions are performed in 1.5 ml Eppendorf tubes. The exception is the PCR reaction, which uses a 0.2 ml tube.
Experimental procedures
-
End-repairEnd-repair pre-mix
10X T4 ligation Buffer 5 μl 10 mM dNTPs 2 μl T4 DNA polymerase (3 U/μl) 1 μl Klenow Fragment (1 U/μl; see Note 1) 1 μl T4 PNK (10 U/μl) 1 μl H2O 30 μl Total 40 μl Note 1: original Klenow concentration is 5 U/μl, prepare a 1/5 dilution every time.
Note 2: Current Illumina protocols perform this step at higher temperatures. This is NOT recommended for SSDS library preparation.- Mix 40 μl of the End-repair pre-mix with 10 μl of the ChIP DNA from Day 3, Step 4.
- Incubate at 20°C in a water bath for 30 minutes.
-
Purification of DNA using Qiagen MinElute PCR cleanup
Add 350 μl (7 reaction volumes) of Buffer PB to the reaction mix and follow the purification protocol provided with the kit. Elute DNA with 12 μl of Buffer EB.
- A-tailingA-tailing pre-mix
10X NEBuffer 2 5 μl Klenow Fragment 3’→5’exo− (5 U/μl) 1 μl 1 mM dATP 10 μl H2O 24 μl Total 40 μl - Mix 40 μl of the A-tailing pre-mix with 10 μl of the end-repaired DNA from step 2.
- Incubate at 37°C for 30 minutes.
-
Purification of DNA using Qiagen MinElute PCR cleanup kit
Add 350 μl (7 reaction volumes) of Buffer PB to the reaction mix and follow the purification protocol provided with the kit. Elute DNA with 12 μl of Buffer EB.
-
Kinetic enrichment for ssDNA
Note: This is the key step in the protocol to enrich for ssDNA.
Incubate the eluted DNA from step 4 at 95°C for 3 minutes in the Thermomixer.
Spin the sample briefly and allow to return to room temperature.
-
Adapter ligationLigation setup
DNA 10 μl Quick DNA ligase Buffer (2X) 15 μl H2O 3 μl Diluted TruSeq Adaptor (see Note 1) 1 μl Quick T4 DNA ligase (see Note 2) 1 μl Total 30 μl Note 1: The stock TruSeq adapter concentration is 15 μM. This should be diluted 1:10 in Buffer EB, and further diluted 1:10. 1 μl of this 1:100 dilution is used for ligation.
Note 2: The T4 DNA Ligase is added last.
Note 3: Current Illumina protocols perform this step at higher temperatures. This is NOT recommended for SSDS library preparation.
Incubate at 20°C for 30 minutes in a water bath.
-
Purification of DNA using Qiagen MinElute PCR cleanup kit
Add 150 μl (5 reaction volumes) of Buffer PB to the reaction mix and follow the purification protocol provided with the kit. Elute DNA with 12 μl of Buffer EB.
- Library amplificationAmplification pre-mix
Illumina TruSeq PCR Master Mix (2X) 25 μl Illumina TruSeq PCR primer cocktail 5 μl H2O 10 μl Total 40 μl - Mix 40 μl of the Amplification pre-mix with the 10 μl of adapter-ligated DNA from step 7.
-
Run the following program on a PCR machine:12–15 cycles of:
- 98 °C
30 sec - 98 °C
10 sec - 60 °C
30 sec - 72 °C
30 sec Followed by:- 72 °C
5 min - 4 °C
hold
- Purification of DNA using AMPureXP beads
-
After PCR, add 0.9X volumes of AMPureXP beads.Note: 0.9X volumes is chosen to optimize the removal of adapter dimers (~125bp) but to keep ssDNA-derived fragments (mean = 205 – 245 bp). This should be carefully calibrated if different beads are used.
- Incubate the PCR tube at room temperature for 10 minutes to bind DNA to the beads.
- Collect the beads on a magnet, discard supernatant.
- With the tube still in the magnet, add 200μl of 80% ethanol.
- Incubate at room temperature for 1 minute.
- Remove and discard ethanol.
- Repeat steps d-f once.
- Dry the beads at room temperature for 3 – 5 minutes or until all the ethanol has evaporated.
- Remove the tube from the magnet and resuspend the beads in 20 μl of Elution buffer (Qiagen EB buffer).
- Incubate at room temperature for at least 2 minutes.
- Collect the beads on a magnet, and transfer supernatant to a new Eppendorf tube.
- Measure DNA concentration using Qubit or PicoGreen kits.
- Store samples at −20°C
-
-
Library quality check
To obtain optimal cluster density for sequencing it is important to accurately quantify the DNA concentration of sequencing libraries.
Perform the following:- Determine the concentration of the library using qPCR (KAPA Library Quantification Kit).
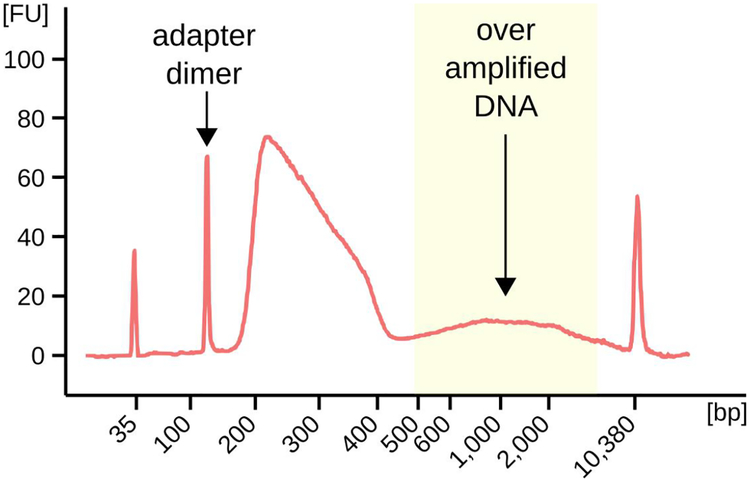
- Check the length distribution of DNA fragments by running an aliquot of the DNA library on the Agilent Technologies 2100 Bioanalyzer using a High Sensitivity DNA chip. Typical bioanalyzer profiles for good SSDS libraries are shown in Fig. 2.
- Calculate the molarity of the library according to concentration and fragment size distribution.
-
Perform paired-end sequencing of SSDS library
Note 1: Computational parsing of ssDNA uses 36bp from read1 and 40bp from read2. Thus, longer sequencing reads are not required.
Note 2: SSDS libraries form secondary structures that appear to adversely affect clustering and sequencing on Illumina platforms. We recommend loading the Illumina flow cell at double the recommended molarity to obtain near optimal cluster density. Running samples in rapid run mode of the Illumina HiSeq 2500 has produced the best results.
Figure 2: Bioanalyser output depicting the fragment size distribution in a typical SSDS library.
The proportions of adapter-dimers and over-amplified DNA may vary considerably between samples. Libraries with a large excess of either of these populations may reduce the yield of ssDNA. This library was not purified using beads (Day 4: step 9) so that the adapter-dimer peak could be clearly seen.
COMPUTATIONAL ANALYSIS: IDENTIFICATION OF DSB HOTSPOTS FROM SSDS DATA.
Most of the software tools for the SSDS data analysis are commonly used for NGS analyses. For more advanced users, it is possible to build a custom pipeline that recapitulates the steps below. The protocol begins with fastq files provided by the sequencing facility. Reads in these fastq files will be aligned to the reference genome, ssDNA will be identified and then DSB hotspots are called.
Materials:
Computer with Ubuntu Linux operating system
Linux user with sudo privileges
-
8 GB of RAM and preferably multiple CPU cores
Note: Instructions are provided for Ubuntu Linux. Installation can be performed with minor modifications on other Linux distributions or Apple Macs. The SSDS pipelines will not run on Windows PCs.
Required programs / packages:
Linux packages:
bioperl
default-jre
git
libxml2-dev
libcurl4-openssl-dev
python-setuptools
python-pip
python-numpy
python-scipy
r-base-core
zlib1g-dev
Perl packages:
Bio::DB::Sam
File::Temp
Getopt::Long
List::Util
Make::Build
Math::Round
Statistics::Descriptive
Switch
R packages
RCurl
XML
R packages (Bioconductor)
Rtracklayer
ShortRead
Other software
macs_2.1.0.20150731
I. SSDS Alignment Pipeline
Introduction:
The following instructions are to be carried out from the linux terminal. In Ubuntu, the terminal can be accessed from the applications menu or by pressing “Ctrl + Alt + T” key combination. In this protocol, terminal commands are displayed in monospaced font and are preceded by the “>” symbol. The “>” should not be included in commands.
For example, the following line:
>apt-get install testPackage
should be interpreted as:
Type “apt-get install testPackage” at the linux terminal, then hit “Enter”
Note: The linux terminal is case-sensitive, therefore all commands must be typed exactly as specified.
Installation:
Install git if required:
>sudo apt-get install git
Use git to clone the SSDS pipeline on your computer:
>git clone https://github.com/kevbrick/SSDSpipeline.git
The easiest way to install the SSDS alignment pipeline is to simply run the configuration script located in the folder downloaded from github:
>cd SSDSpipeline
The configure script will install the SSDS alignment pipeline to /home/$USER/SSDS_pipeline_1.0.0.
To install the pipeline to the default locations:
>sudo./configure.sh
Alternatively, you can specify the installation and / or genomes folders:
>sudo./configure.sh -i /share/SSDS -g /share/SSDSgenomes
The configuration script will install all the required dependencies and test the pipeline.
If the configuration process successfully completes, the pipeline has been installed. Please skip sections 1–3 below. If the configuration process fails, please follow the manual instructions outlined in sections 1–3 below. These instructions also serve as a guide for users of other operating systems.
Manual Installation:
1. Set environment variables:
| SSDSPIPELINEPATH | location of run_SSDS_pipeline binary |
| SSDSPICARDPATH | picard location (recommend: $SSPIPELINEPATH/picard-tools-2.3.0) |
| SSDSFASTXPATH | path to fastx_trimmer binary (recommend: $SSPIPELINEPATH) |
| SSDSSAMTOOLSPATH | path to samtools binary (recommend: $SSPIPELINEPATH) |
| SSDSGENOMESPATH | genomes top level folder |
| SSDSTMPPATH | temporary folder location |
| PERL5LIB | $SSDSPIPELINEPATH must be added to the perl path |
It is best to define these environment variables in your user configuration file (ie. ~/.bashrc). Add the following lines to ~/.bashrc:
export SSDSPIPELINEPATH=/XXX/YYY/SSDSpipeline export SSDSPICARDPATH=/XXX/YYY/SSDSpipeline/picard-tools-2.3.0 export SSDSFASTXPATH=/XXX/YYY/SSDSpipeline export SSDSSAMTOOLSPATH=/XXX/YYY/SSDSpipeline export SSDSGENOMESPATH=/XXX/YYY/SSDSpipeline/genomes export SSDSTMPPATH=/tmp export PERL5LIB=$PERL5LIB:$SSDSPIPELINEPATH
Initialize variables in .bashrc file
>source ~/.bashrc
2. Install dependencies:
The following programs are required for the SSDS pipeline:
Java run-time
Get java runtime (root privileges required):
>sudo apt-get install default-jre
The following Perl packages are required for the SSDS pipeline:
Bio::DB::Sam
File::Temp
Getopt::Long
Math::Round
Statistics::Descriptive
Switch
Make::Build
Install Perl modules as follows (root privileges required):
>export PERL_MM_USE_DEFAULT=1 >sudo cpan File::Temp >sudo cpan Getopt::Long >sudo cpan Make::Build >sudo cpan Math::Round >sudo cpan Statistics::Descriptive >sudo cpan Switch >sudo apt-get install zlib1g-dev >sudo apt-get install bioperl >cd $SSDSPIPELINEPATH/Bio-SamTools-1.43 >perl INSTALL.pl
3. Run the unit tests:
>cd $SSDSPIPELINEPATH/unitTest/ >sh runTest.sh
This will test the addGenome.pl script and the SSDS pipeline using either fastq, fastq.gz or bam files as input. Successful completion of the tests will result in the following output:
Test genome was added and indexed successfully !! SSDS pipeline from FASTQ successful !! SSDS pipeline from FASTQ.GZ successful !!
The SSDS pipeline has been installed successfully and can be run.
Note 1: The SSDS pipeline has been tested and used on UCSC genomes. Therefore, chromosome names in the genome.fa file should follow the “chr##” nomenclature. Chromosome names that lack a “chr” preface or that use letter-based designations may not work.
Note 2: Specific versions of picard, samtools and fastx_trimmer are included in the SSDS pipeline repository. We recommend using these versions as other versions may not be compatible with the SSDS pipeline. Expert users can tweak the pipeline scripts to use different versions of these programs if desired.
Indexing the reference genome with bwa:
The genomes folder is used for both alignment and for sorting BAM files. Each reference genome must be in a unique sub-folder.
The $SSDSPIPELINEPATH/addGenome.pl script will add a genome and generate all the required files. It takes a genome fasta file as input.
>perl $SSDSPIPELINEPATH/addGenome.pl --fa (genome FASTA file) --name (genome name; ie. mm10) --g (genome assembly; ie. mm10) --s (species; ie. mus_musculus)
This will create index the genome and create a folder structure in $SSDSGENOMESPATH. The name provided to the addGenome.pl script is the genome name that will be used for running the pipeline (--g argument).
Alternatively, the genome folder can be populated manually using steps 1–4 below:
- Create the folder. The folder name will be used as the genome name when running the SSDS pipeline:
>mkdir $SSDSGENOMESPATH/genome_name/
- Copy the genome fasta file to the genomes folder:
>cp genome.fa $SSDSGENOMESPATH/genome_name/genome.fa
- Create the genome fasta file index (makes fasta.fai file):
>samtools faidx $SSDSGENOMESPATH/genome_name/genome.fa
-
Index the genome for the bwa version required:
The genome folders structure is as follows (folder names must be exact):>mkdir $SSDSGENOMESPATH/genome_name/BWAIndex >mkdir $SSDSGENOMESPATH/genome_name/BWAIndex/version0.5.x >mkdir $SSDSGENOMESPATH/genome_name/BWAIndex/version0.7.10
Once this structure has been built, index the genome:>cd $SSDSGENOMESPATH/genome2use/BWAIndex/version0.5.x >ln -s ../../genome.fa. >$SSDSPIPELINEPATH/bwa_0.5.x index -a bwtsw genome.fa >cd $SSDSGENOMESPATH/genome2use/BWAIndex/version0.7.10 >ln -s ../../genome.fa. >$SSDSPIPELINEPATH/bwa_0.7.12 index -a bwtsw genome.fa
Create genome dictionary file:>java -jar $SSDSPICARDPATH/picard.jar CreateSequenceDictionary R=$SSDSGENOMESPATH/genome_name/genome.fa O=$SSDSGENOMESPATH/genome_name/genome.dict GENOME_ASSEMBLY=XXXX SPECIES=mySpecies VALIDATION_STRINGENCY=LENIENT
Align reads to reference genome with the SSDS pipeline:
Starting from fastq / fastq.gz files, the pipeline can be run with the following command:
>$SSDSPIPELINEPATH/run_ssDNAPipeline \
--g 22 \
--n {number of threads} \
--fq1 {path to fastq / fastq.gz for read 1} \
--fq2 {path to fastq / fastq.gz for read 2} \
--sample {sample name} \
--lane {lane number; or ANY number} \
--date {date in DDMMYY format} \
--outdir {output folder}
Starting from a paired-end BAM file, the pipeline can be run with the following command:
>$SSDSPIPELINEPATH/run_ssDNAPipeline \
--g {genome name} \
--n {number of threads} \
--bam {path to paired end bam file} \
--sample {sample name} \
--lane {lane number; or ANY number} \
--date {date in DDMMYY format} \
--outdir {output folder}
The arguments for the SSDS pipeline can be accessed from the command line:
>$SSDSPIPELINEPATH/run_ssDNAPipeline -h
Alternatively, they are listed below:
| Argument | Synopsis (* = required) | Detail |
|---|---|---|
| Note: either --bam OR --fq1 & --fq2 arguments are required | ||
| --bam | *Input BAM file | Full path to a paired-end BAM file |
| --fq1 | *Input fastq for read 1 | Full path to a fastq / fastq.gz file for read 1 |
| --fq2 | *Input fastq for read 2 | Full path to a fastq / fastq.gz file for read 2 |
| --g | *Genome name | Name must match a folder in $SSDSGENOMESPATH |
| --n | Number of threads (for alignment step) | Default = 12 |
| --r1BP | Trim size for read 1 (bp) | Integer value (default = 36) |
| --r2BP | Trim size for read 2 (bp) | Integer value (default = 40) |
| --splitSz | Large BAM files are split for ssDNA detection. | A lower value reduces the memory footprint (default = 20000000) |
| --outdir | Output folder | Defaults to BAM / FASTQ folder |
| --sample | Sample name for read group | - |
| --date | Date for read group | Date in DDMMYY format |
| --lane | Lane for read group | - |
| --bwaVers | Bwa version to use | 0.5 or 0.7 [default] |
| --v | Use verbose mode | - |
| --h/help | Show help | - |
Outputs from the SSDS pipeline:
The SSDS pipeline aligns Illumina sequencing reads to a reference genome and then generates output files for each of five sub-categories of DNA types (for more detail, see17):
ssDNA type 1: high confidence ssDNA
ssDNA type 2: low confidence ssDNA
dsDNA: low confidence dsDNA
dsDNA strict: higher confidence dsDNA
unclassified: other ambiguous DNA
Importantly, these designations represent the most likely source of DNA, given the alignment and detectable inverted terminal repeat structure. These designations are not 100% correct: for example, ssDNA with perfect short homology between the start and end of a fragment will be classified as dsDNA. It is very unlikely that dsDNA will ever be classified as ssDNA type 1.
Sequencing reads of each type are output to BAM files. In ssDNA BAM files, the ITRs have been removed, but details of the ITR structure are retained as tag fields:
Microhomology length: uh:i:{number}
Mismatches within microhomology: mm:i:{number}
Distance of microhomology to fragment end (offset): os:i:{number}
ITR length: it:i:{number}
Fragments, representing the entire sequence from the start of the first-end read to the end of the second-end read, are output to BED files. Columns of the BED files correspond to:
Chromosome
Fragment start
Fragment end
Read 1 Quality Score _ Read 2 Quality Score
ITR length _ Microhomology length _ Offset length
Fragment strand
II. Identification of DSB hotspots
Installation:
Install git if required:
>sudo apt-get install git
Use git to clone the callHotspots pipeline on your computer:
>git clone https://github.com/kevbrick/callHotspotsSSDS.git
Configure the pipeline:
The easiest way to configure the SSDS hotspot calling pipeline is to simply run the configuration script located in the folder downloaded from github:
>cd callHotspotsSSDS
The configure script will install the SSDS hotspot calling pipeline to /home/$USER/SSDS_callHotspots_1.0.0. To install the pipeline to the default locations:
>sudo ./configure.sh
Alternatively, you can specify the installation folder:
>sudo ./configure.sh -i /share/SSDS_callHotspots_1.0.0
The configuration script will install all the required dependencies and test the pipeline.
If the configuration process successfully completes, the pipeline has been installed. Please skip sections 1–4 below. If the configuration process fails, please follow the manual instructions outlined in sections 1–4 below. These instructions also serve as a guide for users of other operating systems.
Manual installation:
1. Copy callHotspotsSSDS folder to desired location:
For the callHotspotsSSDS pipeline to be available for all users on a system, it is recommended to copy the entire contents of the callHotspotsSSDS folder to /usr/share:
>cp -r ./callHotspotsSSDS /usr/share
This is a recommended, but NOT a required step.
2. Install dependencies:
The following programs are required to run the callHotspots pipeline:
R (>version 2.10)
MACS (version 2.1.0.20150731)
Pip
First, install bash libraries (root privileges required):
>apt-get install -y r-base-core libxml2-dev libcurl4-openssl-dev python-setuptools python-pip python-numpy python-scipy
To install R packages, start R from the command line:
>R
From the R prompt, run:
>install.packages(“RCurl”, repos=“http://cran.rstudio.com“) >install.packages(“XML”, repos=“http://cran.rstudio.com“) >source(“https://bioconductor.org/biocLite.R”) >biocLite(“rtracklayer”, repos=“http://cran.rstudio.com”) >biocLite(“ShortRead”, repos=“http://cran.rstudio.com“)
MACS can be installed from the command line as follows (root access is required);
>sudo pip install --root=/XXX/YYY/callHotspotsSSDS/macs_2.1.0.20150731 -UMACS2==2.1.0.20150731
The following Perl packages are required for the callHotspots pipeline:
File::Temp
Getopt::Long
List::Util
Math::Round
Statistics::Descriptive
Install Perl modules as follows (root privileges required):
>export PERL_MM_USE_DEFAULT=1 >sudo cpan File::Temp >sudo cpan Getopt::Long >sudo cpan Math::Round >sudo cpan Statistics::Descriptive >sudo cpan List::Util
3. Set environment variables:
| CHSPATH | location of pipeline |
| CHSNCISPATH | path to NCIS R script (recommend: $CHSPATH/NCIS) |
| CHSBEDTOOLSPATH | path to bedtools binaries (recommend: $CHSPATH/bedtools) |
| CHSTMPPATH | temporary folder location |
| PERL5LIB | $CHSPATH must be added to the perl path |
| CHSMACSPATH | path to macs2 binary |
| PYTHONPATH | path to macs2 python libraries must be added |
It is best to define these environment variables in the bash configuration file (~/.bashrc) for each user. /XXX/YYY/ is a placeholder for the installation path of the callHotspotsSSDS folder (i.e. /usr/share/).
Add the following lines:
export CHSPATH=/XXX/YYY/callHotspotsSSDS export CHSNCISPATH=/XXX/YYY/callHotspotsSSDS/NCIS export CHSBEDTOOLSPATH=/XXX/YYY/callHotspotsSSDS/bedtools export CHSTMPPATH=/tmp export PERL5LIB=$PERL5LIB:/XXX/YYY/callHotspotsSSDS export PATH=$PATH:/XXX/YYY/callHotspotsSSDS export CHSMACSPATH=/XXX/YYY/callHotspotsSSDS/macs2.1.0.20150731/usr/local/bin export PYTHONPATH=$PYTHONPATH:/XXX/YYY/callHotspotsSSDS/macs2.1.0.20150731/usr/local/lib/python2.7/dist-packages
Initialize these variables from .bashrc:
>source ~/.bashrc
4. Run the unit tests to ensure that the pipeline works:
This will test the hotspot calling and strength estimation scripts using test data. This test performs peak calling in a 30 Mb region.
>cd $CHSPATH/unitTest/ >sh runTest.sh Successful completion of the tests will result in the following output: Success !! Hotspot calling complete.
The CHS pipeline has been installed successfully and can be run.
Note: Specific versions of MACS, bedtools and NCIS are included in the callHotspots pipeline repository. We recommend using these versions as other versions may not be compatible. Expert users can tweak the pipeline scripts to use different versions of these programs if desired.
Call hotspots:
To identify the locations of DSB hotspots from a DMC1 ChIP-SSDS experiment, we compare treatment and control experiments. This requires:
ssDNA_type_1 fragments BED file from DMC1-SSDS experiment
ssDNA_type_1 fragments BED file from an input or IgG-SSDS experiment
The peak calling pipeline is run using the following syntax:
>$CHSPATH/run_callHotspotsPipeline \
--t {BED file : type 1 ssDNA from dmc1 experiment} \
--c {BED file : type 1 ssDNA from input/IgG experiment} \
--gSz {estimated size of mappable genome} \
--name {prefix for output file names} \
--out {output folder}
The arguments for the hotspot calling pipeline can be accessed from the command line:
>$CHSPATH/run_callHotspotsPipeline -h
Alternatively, they are listed below.
| Argument | Synopsis (* = required) | Detail |
|---|---|---|
| --t | *Treatment BED file | Type 1 ssDNA BED file |
| --c | *Control BED file | Type 1 ssDNA BED file |
| --gSz | *Effective genome size | Estimated size of mappable genome |
| --gName | *Genome/species name | Use pre-computed effective genome size |
| (can be used instead of --gSz) | For human: hg19 | hg38 | hg | hs | human For mouse: mm9 | mm10 | mm | mouse For rat: rn | rat |
|
| --name | *Name prefix for output files | - |
| --out | Output folder | Default = current folder |
| --blist | Blacklist file | BED file of genomic regions with sequencing biases that result in spurious peak calls i.e. for mouse (mm10 genome): $CHSPATH/mm10_hotspot_blacklist.bed |
| --tuniq | Treatment BED file with ONLY unique fragments | Optional, but not recommended |
| --cuniq | Control BED file with ONLY unique fragments | Optional, but not recommended |
| --debug | DEBUG mode | Builds scripts but does not execute. Note: This is a logical argument, so does not take any value (pass as --debug) |
| --q30 | Q30 mode | Use only fragments where both reads have a q-score >= 30. This is useful for removing reads that map with low confidence (i.e. to high copy repeats). Note: This is a logical argument, so does not take any value (pass as --q30) |
| --nc | Do not call peaks | Do not run peak calling. Note: This is a logical argument, so does not take any value (pass as --debug) |
| --h/help | Show help |
Outputs from the hotspot calling pipeline:
The hotspot calling pipeline generates output files of DSB hotspot locations.
| filename | detail |
|---|---|
| $name_peaks.bedgraph | Recentered hotspots with strength estimate (BEDGRAPH file) Column 1: chromosome Column 2: hotspot start Column 3: hotspot end Column 4: hotspot strength Note: Hotspots are recentered as described in 15. Briefly, the hotspot center is defined as the midpoint of the forward and reverse strand ssDNA coverage. |
| $name_peaks.tab | Recentered hotspots table: Column 1: chromosome Column 2: hotspot start Column 3: hotspot end Column 4: hotspot strength Column 5: strength as percentage of total Column 6: strength as rank Column 7: signal fragments Column 8: noise fragments |
The pipeline will also output other files that should only be used for debugging purposes:
| filename | detail |
|---|---|
| $name.NCISout | NCIS output: Treatment:Control ratios |
| $name_model.r | MACS output: R script to visualize MACS peak model for these data |
| $name_peaks.xls | MACS output: raw peak calls (Excel file) |
| $name_summits.bed | MACS output: raw peak summits (BED file) |
| $name_peaks.narrowPeak | MACS output: raw peak calls (BED file) |
| $name_peaks.bed | MACS output: raw peak calls (BED file) |
| $name.macs2callpeak.YYMMDD_NNNNN.OUT | STDOUT from MACS peak calling |
| $name.macs2callpeak.YYMMDD_NNNNN.ERR | STDERR from MACS peak calling |
References
- 1.Arnheim N, Calabrese P & Tiemann-Boege I Mammalian meiotic recombination hot spots. Annual review of genetics 41, 369–399, doi: 10.1146/annurev.genet.41.110306.130301 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Paigen K & Petkov P Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet 11, 221–233, doi: 10.1038/nrg2712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orkin SH et al. Linkage of beta-thalassaemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature 296, 627–631 (1982). [DOI] [PubMed] [Google Scholar]
- 4.Myers S, Bottolo L, Freeman C, McVean G & Donnelly P A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324, doi: 10.1126/science.1117196 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Baudat F et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840, doi: 10.1126/science.1183439 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parvanov ED, Petkov PM & Paigen K Prdm9 controls activation of mammalian recombination hotspots. Science 327, 835, doi: 10.1126/science.1181495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers S et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879, doi: 10.1126/science.1182363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grey C et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 9, e1001176, doi: 10.1371/journal.pbio.1001176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billings T et al. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol 14, R35, doi: 10.1186/gb-2013-14-4-r35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi K, Yoshida K & Matsui Y A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378, doi: 10.1038/nature04112 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Baudat F, Imai Y & de Massy B Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14, 794–806, doi: 10.1038/nrg3573 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Keeney S, Lange J & Mohibullah N Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annual review of genetics 48, 187–214, doi: 10.1146/annurev-genet-120213-092304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter N Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol 7, doi: 10.1101/cshperspect.a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolcun-Filas E & Schimenti JC Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol 298, 179–227, doi: 10.1016/B978-0-12-394309-5.00005-5 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Brick K, Smagulova F, Khil P, Camerini-Otero RD & Petukhova GV Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645, doi: 10.1038/nature11089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratto F et al. DNA recombination. Recombination initiation maps of individual human genomes. Science 346, 1256442, doi: 10.1126/science.1256442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD & Petukhova GV Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res 22, 957–965, doi: 10.1101/gr.130583.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smagulova F, Brick K, Pu Y, Camerini-Otero RD & Petukhova GV The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes & development 30, 266–280, doi: 10.1101/gad.270009.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smagulova F et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378, doi: 10.1038/nature09869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies B et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530, 171–176, doi: 10.1038/nature16931 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grey C et al. In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome Res 27, 580–590, doi: 10.1101/gr.217240.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen L, Kim NK, Mariño-Ramírez L & Landsman D Analysis of biological features associated with meiotic recombination hot and cold spots in Saccharomyces cerevisiae. PLoS One 6, e29711, doi: 10.1371/journal.pone.0029711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]