Abstract
Background:
Genome-wide sequencing investigations have identified numerous long noncoding RNAs (lncRNAs) among mammals, many of which exhibit aberrant expression in cancers, including esophageal squamous cell carcinoma (ESCC). Herein, this study elucidates the role and mechanism by which LINC01419 regulates the DNA methylation of glutathione S-transferase pi 1 (GSTP1) in relation to ESCC progression and the sensitivity of ESCC cells to 5-fluorouracil (5-FU).
Methods:
LINC01419 and GSTP1 levels were quantified among 38 paired ESCC and adjacent tissue samples collected from patients with ESCC. To ascertain the contributory role of LINC01419 in the progression of ESCC and identify the interaction between LINC01419 and GSTP1 promoter methylation, LINC01419 was overexpressed or silenced, and the DNA methyltransferase inhibitor 5-Aza-CdR was treated.
Results:
Data from the GEO database (GSE21362) and the Cancer Genome Atlas displayed elevated levels of LINC01419 and downregulated levels of GSTP1 in the ESCC tissues and cells. The silencing of LINC01419 led to decreased proliferation, increased apoptosis, and enhanced sensitivity to 5-FU in ESCC cells. Notably, LINC01419 could bind to the promoter region of the GSTP1 gene, resulting in elevated GSTP1 methylation and reduced GSTP1 levels via the recruitment of DNA methyltransferase among ESCC cells, whereby ESCC progression was stimulated accompanied by reduced ESCC cell sensitivity to 5-FU. GSTP1 demethylation by 5-Aza-CdR was observed to reverse the effects of LINC01419 overexpression in ESCC cells and the response to 5-FU.
Conclusion:
Highly expressed LINC01419 in ESCC promotes GSTP1 methylation, which ultimately acts to promote the event of ESCC and diminish the sensitivity of ESCC cells to 5-FU, highlighting a novel potential strategy to improve 5-FU-based chemotherapy in ESCC.
Keywords: 5-fluorouracil, chemotherapy, DNA methylation, esophageal squamous cell carcinoma, GSTP1, LINC01419
Introduction
Esophageal cancer ranks as the eighth most commonly occurring cancer, while accounting for the sixth most frequent cause of cancer death worldwide.1 Esophageal cancer is generally classified into two main subtypes, namely, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), with the former in recent years exhibiting notably escalating incidences, while the latter is distinctly more common in eastern Africa, Central Asia and China.2 ESCC treatment is generally limited to surgical resection, chemotherapy and radiotherapy methods; however the results are often largely unsatisfactory,3 reflected by the poor 5-year survival rate observed among patients with esophageal cancer.4 The primary factors influencing the low survival rate include rapid progression, local recurrence and distant metastasis.1 A previous study highlighted the role of long noncoding RNAs (lncRNAs) as regulators in the biology of cancer.5 Experimental data previously presented suggest that PlncRNA-1 plays a crucial functional role in ESCC cell proliferation, with studies further highlighting the existence of a relationship between the overexpression of PlncRNA-1 and advanced tumor stage as well as lymph node metastasis, stressing its potential as a promising prognostic marker and therapeutic target for ESCC treatment.6
LncRNAs can be defined as noncoding RNAs with a length above 200 nucleotides, which play important roles in a variety of cellular processes through their interactions with principal components of proteins in the gene regulatory system.7,8 LncRNAs are comprised of various RNA transcripts, including both polyadenylated and nonpolyadenylated lncRNAs, both of which may be sense or antisense, as well as intronic or intergenic in respect to protein-coding genes, with long intervening noncoding RNAs (lincRNAs) belonging to one of the aforementioned types.9 A previous study demonstrated that lincRNA acts to induce the development of ESCC via its interaction with EZH2 through the promotion of POU3F3 methylation.10 Existing literature has highlighted the diagnostic value of LINC01419 in the treatment of primary hepatocellular carcinoma (HCC), emphasizing the overexpression of LINC01419 as a promising strategic target for the treatment of cancer owing to its early diagnostic and prognostic criteria in cases of HCC.11 The application of a dual luciferase reporter gene assay provided verification attesting that LINC01419 could bind to the promoter region of glutathione S-transferase pi 1 (GSTP1), which has been reported to be expressed at a low level in the development of ESCC and cytosine-phosphate-guanine (CpG) island hypermethylation promoter genes, suggesting its potential as a useful biomarker in the early diagnosis of esophageal carcinoma development.12 In addition, 5-fluorouracil (5-FU) treatment has been reported to reduce the rate of cell survival and enhance the chemosensitivity of ESCC cells.13 Therefore, the central objective of the present study was to identify a novel resource for a more comprehensive evaluation on the molecular mechanisms of LINC01419 and GSTP1 in the lesions of esophageal cancer and the sensitivity of ESCC cells to 5-FU.
Materials and methods
Ethics statement
The study was conducted under the approval of the Ethical Committee of Cancer Hospital of Shantou University Medical College, China (2018009). All participating patients signed informed consent documentation. All animals were cared for in strict accordance with the principles of the Institutional Animal Care and Use Committee under a protocol approved by the Medical Animal Care and Welfare Committee of the Shantou University Medical College, China (SUMC2018-012).
Bioinformatics prediction
The GEO database (www.ncbi.nlm.nih.gov/geo) was employed for the bioinformatics prediction. ESCC chip data (GSE100942) and annotate probe files were downloaded, and were obtained from the Affymetrix Human Genome U133 Plus 2.0 array. The Affy installation package of R software was used to conduct the background correction and normalization of each chip data. Next, the Linear Models and Empirical Bayes Methods of the Limma installation package were applied and combined with a traditional Student’s t test in order to conduct a nonspecific filtration process for gene expression profiling, followed by screening of differentially expressed lncRNAs and mRNAs. The ESCC gene expression information was downloaded, and the combination of the Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) database and the R software was employed for data analyses purposes. Differential analysis was performed for the transcriptome profiling data with package edgeR of R. False positive discovery (FDR) correction was applied to the p value using the package, multitest. In order to screen out DEGs (differentially expressed genes), FDR < 0.05 and |log2 (fold change)|>2 was set as the threshold.
Study participants
A total of 38 surgically resected ESCC tissues as well as 38 morphologically normal tissues were initially collected. All collected tissues were previously diagnosed by pathological means. Prior to undergoing operative procedures, the patients were confirmed to have not been treated with radiotherapy or chemotherapy. After collection, the specimens were frozen in liquid nitrogen and stored at −80°C for total RNA extraction from the ESCC tissues. The normal immortalized esophageal epithelial cell line Het-1α and human ESCC cell lines of KYSE70, KYSE450, EC109, and EC9706 were purchased from the Center of Bacterial Species Preservation of the Institute of Cell Biology (Shanghai Institutes for Biological Sciences Chinese Academy of Sciences). The cell lines were subsequently cultured in RPMI1640 medium containing 10% fetal bovine serum (FBS) and incubated in 5% CO2 at a constant temperature of 37°C.
Cell grouping and transfection
The esophageal cancer cells (ECCs) at the logarithmic phase were divided into the LINC01419 oe group (transfected with LINC01419 overexpression plasmid), the negative control (NC) overexpression group (transfected with NC overexpression plasmid), the oe-GSTP1 group (transfected with overexpressing GSTP1 plasmid), the sh-LINC01419 group (transfected with shRNA against LINC01419 plasmid), the sh-NC group (transfected with shRNA against NC plasmid), the 5-Aza-CdR group (treated with demethylating agent, 2.5 umol), the dimethyl sulfoxide (DMSO) group (treated with DMSO), the LINC01419 oe + 5-Aza-CdR group (transfected with LINC01419 overexpression plasmid and treated with demethylating agent, 2.5 µmol), and the LINC01419 oe + DMSO group (transfected with LINC01419 overexpression plasmid and treated with DMSO), the oe-GSTP1 + 5-Aza-CdR group (transfected with GSTP1 overexpression plasmid and treated with demethylating agent, 2.5 umol), the oe-GSTP1 + DMSO group (transfected with GSTP1 overexpression plasmid and treated with DMSO), the LINC01419 oe + NC oe (transfected with LINC01419 overexpression and NC overexpression plasmids), and the LINC01419 oe + oe-GSTP1 group (transfected with LINC01419 overexpression and GSTP1 overexpression plasmids).
The ECC lines EC9706 and KYSE70 at a density of 2 × 105 were inoculated into six-well plates, and then cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco Company, Grand Island, NY, USA) comprised of 10% FBS (Sijiqing Biotechnology, Hangzhou, Zhejiang, China) over a subsequent 24 h period. Next, a 2-µg plasmid of infected cells was added to the plates, and 24 h later, each well was replaced with a fresh selective medium containing 800 g/ml G418 (Calbiochem Company, Darmstadt Germany). Finally, in accordance with the cell growth state, the selective medium was replaced once every 2–3 days. After transfection for 12–15 days, the selective medium (containing 800 μg/ml G418) was replaced once every 4–5 days, until a stable cell population was obtained for subsequent experiments.
Fluorescence in situ hybridization
The specific probe of LINC01419 was synthesized by Bersinbio Company (Guangzhou, Guangdong, China). The ECC line EC9706 was fixed in 4% formalin for 20 min and treated with protease K for 10 min under 37°C conditions. Following two phosphate buffer saline (PBS) washes, the cells were dehydrated with ethanol from a low to high concentration in a successive manner. After 3 min of denaturation at 73°C, 20 µl of hybrid solution (2 µl probe + 18 µl prehybridization solution) was added to the cells for hybridization purposes overnight at 42°C. On the following day, the cells were washed twice with 25% formamide/2 × saline sodium citrate (SSC) at 53°C, after which the cells were rinsed with SSC at a low concentration at 42°C. Finally, 4’, 6-diamidino-2-phenylindole (DAPI) staining and fluorescence detection were conducted.
Dual luciferase reporter gene assay
A base complementary pairing site at the promoter region of the LINC01419 and GSTP1 gene by the Basic Local Alignment Search Tool (BLAST) was verified in connection with the application of a dual luciferase reporter gene assay. The ECC was co-transfected with plasmid DNA [GSTP1-wildtype (WT) or GSTP1-mutant (MUT), LINC01419 or (50 nM) or LINC01419 normal control (LINC01419 NC)]. Following a 24-h period of transfection, the experiment was performed in association with the multifunctional enzyme labeling apparatus based on the instructions of the dual luciferase reporter gene assay kit (BioTek, Winooski, VT, USA). Firefly luciferase and Renilla luciferase were used to assess the relative luciferase activity. The experiment was repeated three times.
Reverse transcription quantitative polymerase chain reaction
Total RNA was extracted using the Trizol method. The cDNA template was synthesized based on the instructions of the high capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). The reverse transcription quantitative polymerase chain reaction (RT-qPCR) experiment was conducted using the Power SYBR® Green Master mix (Applied Biosystems, Carlsbad, CA, USA) and StepOne™ Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The reaction conditions were comprised of pre-denaturation at 95°C for 10 min, with a total of 40 cycles of denaturation at 95°C for 15 s, annealing at an appropriate annealing temperature for 1 min, and extension at 72°C for 30 s, and three wells were set for each pair of primers. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was regarded as the internal reference, while the 2-ΔΔCt method was applied in order to calculate the relative expression. The RT-qPCR primers are displayed in Table 1.
Table 1.
Primer sequences for reverse transcription quantitative polymerase chain reaction.
| Gene | Primer sequence (5’–3’) | Annealing temperatures (°C) |
|---|---|---|
| LINC01419 | F: GAAACTCCGAACACATCTG | 56 |
| R: TTCTCCTGCTGGTTGATT | ||
| GSTP1 | F: CCCTACACCGTGGTCTATTTCC | 56 |
| R: CAGGAGGCTTTGAGTGAGC | ||
| GAPDH | F: TCAAGGCTGAGAACGGGAAG | 56 |
| R: GTGAAGACGCCAGTGGACT |
F, forward; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSTP1, glutathione S-transferase pi 1; R, reverse.
Western blot analysis
Total protein was extracted using a radio-immunoprecipitation assay cell lysis buffer containing phenylmethylsulfonyl fluoride (PMSF), incubated on ice for 30 min at 4°C, and centrifuged for 10 min at 8000 g, after which the supernatant was obtained. The total protein concentration was detected using a bicinchoninic acid (BCA) kit. Next, 50 g of protein was dissolved in 2 × sodium dodecyl sulfate (SDS) loading buffer and boiled for 5 min at 100°C. The above samples then underwent SDS-polyacrylamide gel electrophoresis (PAGE); the proteins were subsequently transferred onto a polyvinylidene fluoride (PVDF) membrane, and mounted in 5% skimmed milk powder at room temperature for 1 h. The PVDF membrane was then cultured with the diluted primary rabbit anti-GSTP1 (ab153949, 1:3000) at 4°C overnight with GAPDH (ab9485, 1:2500) as an internal reference. The membrane was washed three times with tris-buffered saline-tween (TBST) buffer (10 min per wash), and cultured with horseradish peroxidase (HRP)-labeled secondary goat anti-rabbit immunoglobulin (Ig)G (ab97051, 1:2000) for 1 h. The above antibodies were all purchased from Abcam Inc. (Cambridge, MA, USA). After being rinsed in TBST, the membrane was placed on a clean glass plate. The same amount of A fluid and B fluid in the enhanced chemiluminescence (ECL) fluorescence test kit (BB-3501, Amersham, Arlington Heights, IL, USA) were mixed in the darkroom, and added in a dropwise manner onto the membrane and placed into the gel imager. The Bio-Rad image analysis system (Bio-Rad, Inc., Hercules, CA, USA) was used to take photographs, and Quantity One version 4.6.2 software (Bio-Rad, Inc.) was used for analysis. The relative protein level was expressed by the gray value of the corresponding protein band/the gray value of the GAPDH protein band. The experiment was repeated three times in order to obtain the average value.
RNA immunoprecipitation
The cells were collected using cell scrape, washed twice with pre-cooled PBS, and split with the addition of 100 μl of lysis buffer containing proteinase inhibitors and ribonuclease inhibitors on ice for 30 min. After centrifugation at 120,000 g at 4°C for 30 min, the supernatant was subsequently transferred into a centrifuge tube. A small amount of supernatant was taken as the input positive control. Next, 1 μg corresponding antibodies: anti-Dnmt1 (ab13537, mouse antibody), anti-Dnmt3a (ab13888, mouse antibody), anti-Dnmt3b (ab176166, mouse antibody; all from Abcam Inc.) and 10–50 μl protein A/G-beads were added to the remaining supernatant, and incubated overnight at 4°C. After immunoprecipitation, centrifugation at 3000 g at 4°C for 5 min was conducted, the supernatant was discarded. The protein A/G-beads were then washed with 1 ml lysis buffer, and deposited 3–4 times. After each period of washing, the sample was centrifuged with 1000 g at 4°C for 1 min. Finally, 2 × SDS buffer (15 μl) was added, and heated in boiled water for 10 min. Next, the relative RNA was obtained by isolation and purification from the precipitation. The binding effect of LncRNA and DNA methyltransferase was identified using the specific primers of LINC01419 and RT-qPCR.
Chromatin immunoprecipitation
The binding of the GSTP1 gene promoter to the DNA methyltransferase was verified in connection with the instructions of the chromatin immunoprecipitation (ChIP) kit (Millipore, Billerica, MA, USA). Next, 1% formalin was employed to fix the cells for 10 min in order to facilitate DNA and protein cross-linking. Next, DNA was randomly fragmented to 200–800 base pairs (bp) by ultrasonic, and immunoprecipitated with the target protein specific antibodies: anti-Dnmt1 (ab13537, mouse antibody), anti-Dnmt3a (ab13888, mouse antibody), anti-Dnmt3b (ab176166, mouse antibody; all from Abcam Inc.) and IgG in the control group. Finally, 100 µl H2O was used to purify and elute 2.5 µl ChIP DNA which was obtained for RT-qPCR purposes. The binding of the GSTP1 gene promoter and DNA methyltransferase was detected using the GSTP1 primers.
Methylation-specific PCR
The methylation modified DNA (60 ng) was amplified with 25 µl system, including 1 × PCR buffer, 0.4 µl forward and reverse primers respectively (Table 2), 0.2 mmol/l deoxyribonucleoside triphosphates (dNTPs), 5% DMSO, and 1.5 U Taq plus DNA polymerase (BBI Life Sciences Corporation, Canada). The reaction conditions were comprised of pre-denaturation at 95°C for 5 min, followed by the addition of 1.5 U Taq plus DNA polymerase, denaturation at 95°C for 1 min, annealing at 55–65°C for 30 s, extension at 72°C for 1 min, for a total of 10 cycles; denaturation at 95°C for 1 min, annealing at 57°C for 30 s, extension at 72°C for 1 min, for a total of 25 cycles, and extension at 72°C for 10 min.
Table 2.
Primer sequences for MSP of the GSTP1 gene.
| Gene | Primer sequence (5’–3’) | Annealing temperatures (°C) |
|---|---|---|
| Methyl | F: TTC GGG GTG TAG CGG TCG TC | 57 |
| R: GCC CCA ATA CTA AAT CAC GAC G | ||
| Unmethyl | F: GAT GTT TGG GGT GTA GTG GTT GTT | 57 |
| R: CCA CCC CAA TAC TAA ATC ACA ACA |
F, forward; GSTP1, glutathione S-transferase pi 1; MSP, methylation-specific polymerase chain reaction; R, reverse.
Bisulfite sequencing PCR
The modification and purification of hydrogen sulfite in DNA were performed in accordance with the instructions of the DNA vulcanization kit (Epitect Bi.sulfite kit, Qiagen Company, Hilden, Germany). Next, 25 µl of the system was amplified, with the DNA solution (1.0 µl) modified with sodium bisulfite used as a template. The system was comprised of 2 × Taq PCR MasterMix (12.5 μl; containing 0.05 U/µl Taq DNA polymerase, 4 mmol/l MgCl2, 0.4 mmol/l dNTPS), and 0.4 µl forward and reverse primers of the GSTP1 promoter respectively. Next, the water was complemented to the total reaction volume of 25 µl. The reaction conditions included pre-denaturation at 95°C for 15 min, denaturation at 95°C for 30 s, annealing at 52°C for 1 min, and extension at 72°C for 30 s, for a total of 40 cycles, and finally extension at 72°C for 4 min. The sequence of the GSTP1 promoter revealed both the forward primers of 5’-CCCTACACCGTGGTCTATTTCC-3’ with the reverse primer of 5’-CAGGAGGCTTTGAGTGAGC-3’.
5-FU in vitro
ESCC cells at the logarithmic phase were divided into the sh-LINC01419 group (transfected with LINC01419 plasmid), the 5-Aza-CdR group (treated with demethylating agent, 2.5 µmol), DMSO group (treated with DMSO), the LINC01419 oe + 5-Aza-CdR group (treated with overexpression of LINC01419 plasmid, and demethylating agent, 2.5 µmol) and the LINC01419 oe + DMSO group. The cells in each group were treated with 10 μg/ml 5-FU and incubated for 48 h for subsequent experimentation.
CCK-8 assay
Cells at the logarithmic growth phase were inoculated into 96-well plates at a density of 2 × 104 cells/well. Cell viability was detected using a CCK-8 kit every 24 h after inoculation. Each test was conducted following the addition of 10 μl CCK-8 detection solution. The plates were then placed into an incubator for 4 h of incubation, with an absorbance of 450 nm detected using an enzyme labeling instrument.
5-ethynyl-2’-deoxyuridine staining
Cells at a density of 5 × 104 were seeded into 96-well plates for 48 h, incubated and collected. The cells were subsequently incubated for 2 h with the addition of the medium containing 5-ethynyl-2’-deoxyuridine (EdU), washed with PBS, and then fixed in 4% paraformaldehyde for 30 min. Based on the instructions of the EdU kit (Guangzhou RiboBio Co. Ltd., Guangzhou, Guangdong, China), the reagents B, C, D, and E were added for cell incubation purposes. Next, the cells were washed with PBS, incubated with Hoechst33342 staining solution at room temperature for 30 min, washed again with PBS, and observed under a fluorescence microscope. Statistical analyses were performed using ImageJ software (version 1.48u, National Institutes of Health, Bethesda, MD, USA).
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
After 24 h of transfection, the cells were fixed with 2% formaldehyde at room temperature for 1 h, after which they were permeated on the ice for 2 min with 0.1% Triton X - 100 (Beijing Solarbio Science & Technology Co. Ltd., Beijing, China). After three PBS washes, the cells were incubated under a moist and dark environment of 37°C over a period of 50 h with a 50 μl terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling (TUNEL) reaction mixture for 1 h, using the instructions of the TUNEL staining kit. The cells were then observed under an inverted fluorescence microscope (Leica Camera AG Inc., Frankfurt, Germany).
Colony-forming unit
Cells at the logarithmic growth phase were subsequently constructed into single cell suspension and counted, with 1000 cells incubated in 60 mm medium and cultured in an incubator with CO2, with the liquid replaced at regular 3-day intervals. After a 14-day culture process, the culture medium was discarded, followed by three PBS washes, fixation in methanol for 15 min, and stained with crystal violet for 15 min. The number of colonies with 50 cells or more was finally determined under the guidance of a microscope.
Tumorigenicity assay in nude mice
A total of 20 male 6–8 week-old nude mice (weight: 16–20 g), purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China), were recruited for the purposes of the experiment. The ESCC cells (1 × 106) transfected with sh-NC and sh-LINC01419 were dissolved in 200 μl normal saline and subcutaneously injected into the right side of the nude mice. Tumor formation as well as the growth state was observed and recorded at regular intervals. When the tumor was observable, two groups of mice were randomly divided into two subgroups, equal in number. 5-FU (25 mg/kg) or DMSO was injected intraperitoneally into the mice in each subgroup, twice a week (a total of seven times). A Vernier caliper was used to measure the maximum length each tumor every 3 days, and the volume of the tumor was calculated using the formula: V = L × W2 × 0.5 (where L, refers to length and W, refers to the width). Then 28 days later, the xenograft tumors were removed and weighed accordingly.
Statistical analysis
All data analysis was conducted using SPSS version 19.0 software (IBM Corp., Armonk, NY, USA). The measurement data were expressed as the mean ± standard deviation. Comparisons between two groups were analyzed by a Student’s t test, while comparisons among multiple groups were analyzed using one-way analysis of variance. Pairwise comparisons between groups were conducted using a least significant difference test. Double factor variance analysis was applied to analyze the two factor effect. A p value <0.05 was considered to be statistically significant.
Results
LINC01419 expressed at high levels in ESCC tissues and cells
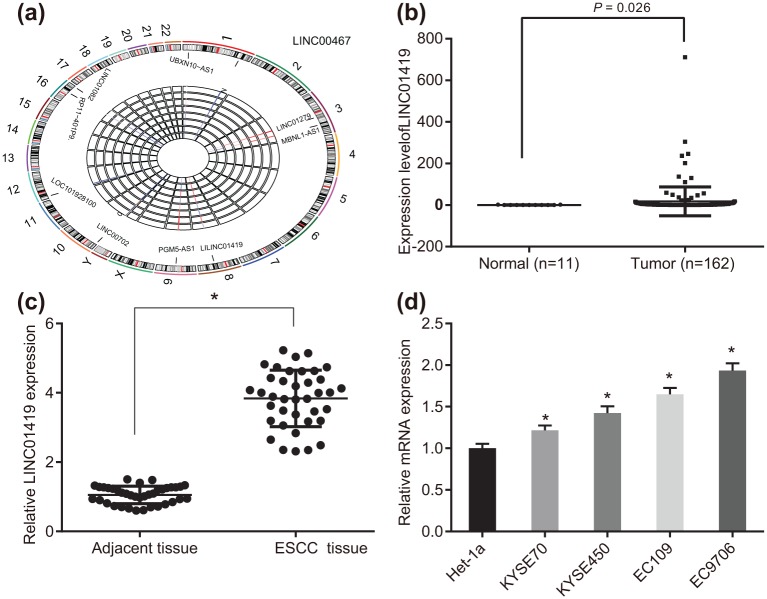
Initially, in order to ascertain whether LINC01419 was an ideal fit for the experiment, bioinformatics prediction was performed. The evaluation of the gene expression profile (GSE100942) of ESCC revealed that LINC01419 was the most drastically upregulated lncRNA in ESCC [Figure 1(a)]. The TCGA database indicated that LINC01419 was overexpressed in ESCC [(Figure 1(b)]. The expression of LINC01419 among the 38 ESCC tissues as well as the 38 normal tissues was subsequently analyzed. The results obtained demonstrated that LINC01419 was highly expressed in ESCC tissues when compared with their adjacent tissues [Figure 1(c)]. Next, we detected how LINC01419 was expressed in the normal immortalized esophageal epithelial cell line Het-1α and human ESCC cell lines KYSE70, KYSE450, EC109 and EC9706. The results indicated that LINC01419 was highly expressed among the KYSE70, KYSE450, EC109 and EC9706 ESCC cells, with the human ESCC cell EC9706 demonstrating the highest levels (1.934 ± 0.087) and KYSE70 demonstrating the lowest levels [Figure 1(d)]. Thus, based on these results we selected the human ESCC cell lines EC9706 and KYSE70 for further experiments.
Figure 1.
LINC01419 was highly expressed in ESCC tissues and cells.
(a) The data analysis of the chip GSE100942; (b) the expression of LINC01419 in TCGA database; (c) RT-qPCR detection results of LINC01419 expression in ESCC cells, the value is measured data, as a measure of the original data, a paired Student’s t test was used to analyze the data, n = 38. *p < 0.05, **p < 0.01, ***p < 0.001 versus the adjacent tissues; (d) RT-qPCR was employed to detect the expression of LINC01419 and GSTP1 in cell lines, the results of which were measurement data, expressed as mean ± standard deviation and analyzed by analysis of variance. The experiment was repeated three times. *p < 0.05, **p < 0.01, ***p < 0.001 versus Het-1α.
ESCC, esophageal squamous cell carcinoma; GSTP1, glutathione S-transferase pi 1; RT-qPCR, reverse transcription quantitative polymerase chain reaction; TGCA, the Cancer Genome Atlas.
Downregulated LINC01419 inhibits ESCC progression and sensitizes ESCC cells to 5-FU in vitro
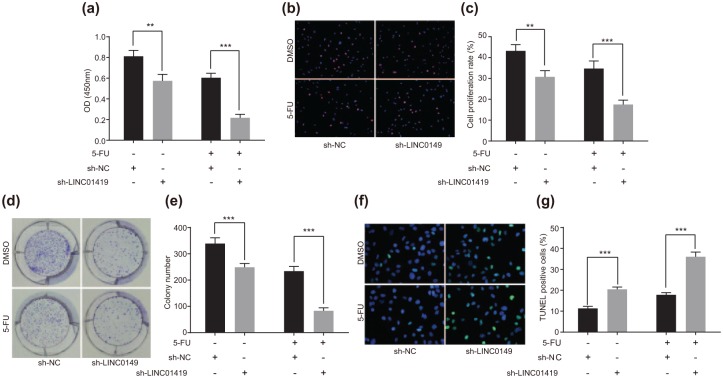
In order to elucidate the effect of LINC01419 on the onset of ESCC, we initially determined the ESCC cell (EC9706) viability after transfection through the application of a CCK-8 assay, the results of which demonstrated that when compared with the sh-NC group, the growth of ESCC cells in the sh-LINC01419 group was markedly declined (p < 0.05); after treatment with 5-FU, compared with the sh-NC + 5-FU group, OD450 value in the sh-LINC01419 + 5-FU group was strikingly decreased [p < 0.05; Figure 2(a)]. At the same time, the cell proliferation experimental results in connection with EdU indicated that cell proliferation in the sh-LINC01419 group was significantly decreased when compared with that in the sh-NC group (p < 0.05); after treatment with 5-FU, comparisons to the sh-NC + 5-FU group exhibited significant decreases in relation to cell proliferation in the sh-LINC01419 + 5-FU group [p < 0.05; Figure 2(b and c)]. A colony-forming unit was subsequently employed in order to determine the colony formation ability of ESCC cells. The results indicated that the colony formation ability of the ESCC cells in the sh-LINC01419 group was significantly diminished when compared with that in the sh-NC group (p < 0.05); after treatment with 5-FU, compared with the sh-NC + 5-FU group, the colony formation ability of ESCC cells was decreased significantly in the sh-LINC01419 + 5-FU group [Figure 2(d and e)]. TUNEL was then applied to measure the transfected ESCC cell apoptosis, the results of which demonstrated that the rate of ESCC cell apoptosis in the sh-LINC01419 group was significantly higher than the sh-NC group (p < 0.05); after treatment with 5-FU, compared with the sh-NC + 5-FU group, cell apoptosis in the sh-LINC01419 + 5-FU group was significantly increased [p < 0.05; Figure 2(f and g)]. Taken together, downregulation of LINC01419 expression was shown to be capable of inhibiting ESCC cell proliferation, promoting cell apoptosis while acting to decelerating the incidence of ESCC. After the transfection of sh-LINC01419, the antiproliferative effect and proapoptotic effect of 5-FU on ESCC cells were observed to have been strengthened, accompanied by augmented sensitivity of the ESCC cells to 5-FU. The results of another ECC line KYSE70 were the same as those described above (Supplementary Figure 1). Based on the results obtained we concluded that LINC01419 deletion could inhibit the occurrence of ESCC and improve the sensitivity of the ESCC cells to 5-FU.
Figure 2.
LINC01419 accelerates the development of ESCC and decreases the sensitivity of ESCC cells to 5-FU.
(a) CCK-8 assay was used to detect ESCC cell viability; (b) EdU staining for cell proliferation; (c) quantitative analysis of cell proliferation; (d) experimental results of colony-forming unit in ESCC cells; (e) quantitative analysis of the experimental data of the colony-forming unit in ESCC cells; (f) apoptosis fluorescence detected by TUNEL staining; (g) quantitative analysis of apoptosis data detected by TUNEL.
*p < 0.05, **p < 0.01, ***p < 0.001. The statistical values of panels a, c, e and g were measurement data, expressed as mean ± standard deviation and compared with two-way analysis of variance. The experiment was repeated three times.
5-FU, 5-fluorouracil; EdU, 5-ethynyl-2’-deoxyuridine; ESCC, esophageal squamous cell carcinoma; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
LINC01419 modulates DNA methylation of GSTP1
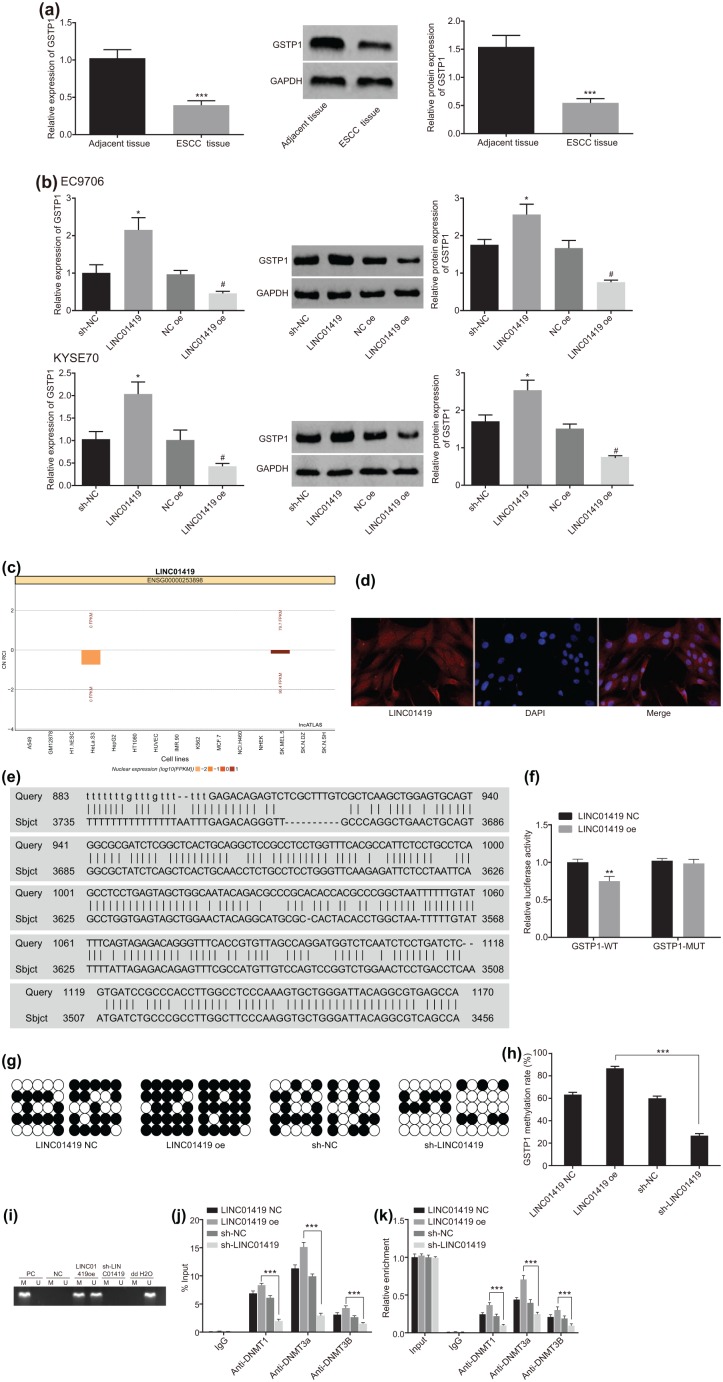
The application of RT-qPCR revealed there to be low levels of GSTP1 expression in the ESCC tissues [Figure 3(a)]. Overexpression of LINC01419 in EC9706 and KYSE70 cells inhibited the expression of GSTP1 [Figure 3(b)]. The lncATLAS website provided evidence indicating the subcellular location of LINC01419, while suggesting that LINC01419 was predominately expressed in the nucleus [Figure 3(c)]. The application of fluorescence in situ hybridization revealed that LINC01419 was mainly expressed in the nucleus [Figure 3(d)]. The results obtained were consistent with the indications of the lncATLAS prediction; the BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) suggested that there were bp binding sites between LINC01419 and the GSTP1 gene promoter [Figure 3(e)]. In order to verify the binding between LINC01419 and GSTP1 gene promoter, a dual luciferase reporter gene assay was performed. Our results demonstrated that compared with the NC group, the luciferase activity in the GSTP1 WT group was significantly decreased, while the GSTP1-mutant (MUT) group exhibited no notable changes [Figure 3(f)], which ultimately suggested that LINC01419 could combine with the promoter region of GSTP1 gene.
Figure 3.
LINC01419 promotes the methylation of GSTP1 gene.
(a) GSTP1 levels in ESCC tissues detected by RT-qPCR and western blot analysis. *p < 0.05, **p < 0.01, ***p < 0.001 versus adjacent tissues. Statistical values were measurement data, presented as mean ± standard deviation and analyzed using a paired Student’s t test; (b) GSTP1 levels in ESCC cells detected by RT-qPCR and western blot analysis. *p < 0.05 versus the sh-NC group, #p < 0.05 versus NC oe group. Statistical values were measurement data, presented as mean ± standard deviation and were analyzed using analysis of variance; (c) prediction of subcellular localization of LINC01419 by lncATLAS website; (d) LINC01419 is mainly distributed in the nucleus, as observed using FISH; (e) Blsat analysis for LINC01419 and the promoter region of GSTP1 gene; (f) the results of dual luciferase reporter gene assay; *p < 0.05, **p < 0.01, ***p < 0.001 versus LINC01419 NC group; (g) detection of methylation of GSTP1 gene by BSP; (h) quantitative analysis of methylation of GSTP1 gene by BSP. *p < 0.05, **p < 0.01, ***p < 0.001. (i) The methylation of the GSTP1 promoter detected with MSP; (j) ChIP verified the binding of GSTP1 promoter region to DNA methyltransferase in patients with ESCC. *p < 0.05, **p < 0.01, ***p < 0.001, versus the normal group; (k) RIP verified the results of LINC01419 binding to DNA methyltransferase, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
Statistical values of luciferase activity of panel (f) were measurement data, presented as the mean ± standard deviation and compared by unpaired Student’s t test. The experiment was repeated three times. Statistical values of panel (h) were measurement data, expressed as mean ± standard deviation and compared by analysis of variance. The experiment was repeated three times. Statistical values of panel (j) and panel (k) were measurement data and were presented as mean ± standard deviation. Comparisons among multiple groups were analyzed using analysis of variance. The experiment was repeated three times.
·, methylation site; º, nonmethylated sites; BSP, bisulfite sequencing polymerase chain reaction; ChIP, chromatin immunoprecipitation; ESCC, esophageal squamous cell carcinoma; FISH, fluorescence in situ hybridization; GSTP1, glutathione S-transferase pi 1; M, methylated; MSP, methylation-specific polymerase chain reaction; NC, negative control; PC, positive control; RIP, RNA immunoprecipitation; RT-qPCR, reverse transcription quantitative polymerase chain reaction; U, unmethylated.
Next, in order to investigate the influence of LINC01419 on the methylation of the GSTP1 gene, we detected the methylation of GSTP1 gene in the ESCC cells transfected with LINC01419 oe and sh-LINC01419 by means of bisulfite sequencing PCR (BSP) and methylation-specific PCR (MSP) experiments. The results obtained revealed that the promoter CpG island of the GSTP1 gene was hypermethylated in the LINC01419 oe group, and the methylation level in the sh-LINC01419 group was lower [Figure 3(g and i)].
Further ChIP experiments on the ESCC cells were conducted. The results demonstrated that compared with the sh-LINC01419 group, binding between the promoter region of GSTP1 and methyltransferase DNMT1, DNMT3A and DNMT3B in the LINC01419 oe group was significantly increased [Figure 3(j)]. At the same time, the results of RIP revealed that when compared with IgG, LINC01419 was able to significantly recruit methyltransferase DNMT1, DNMT3A and DNMT3B. In addition, when compared with the sh-LINC01419 group, binding between LINC01419 and methyltransferase DNMT1, DNMT3A and DNMT3B of the LINC01419 oe group was significantly increased [Figure 3(k)]. The aforementioned results provided evidence demonstrating that LINC01419 could promote the methylation of the GSTP1 gene and then inhibit GSTP1 expression by recruiting DNA methyltransferase into the promoter region of GSTP1.
GSTP1 demethylation inhibits ESCC progression and sensitizes ESCC cells to 5-FU
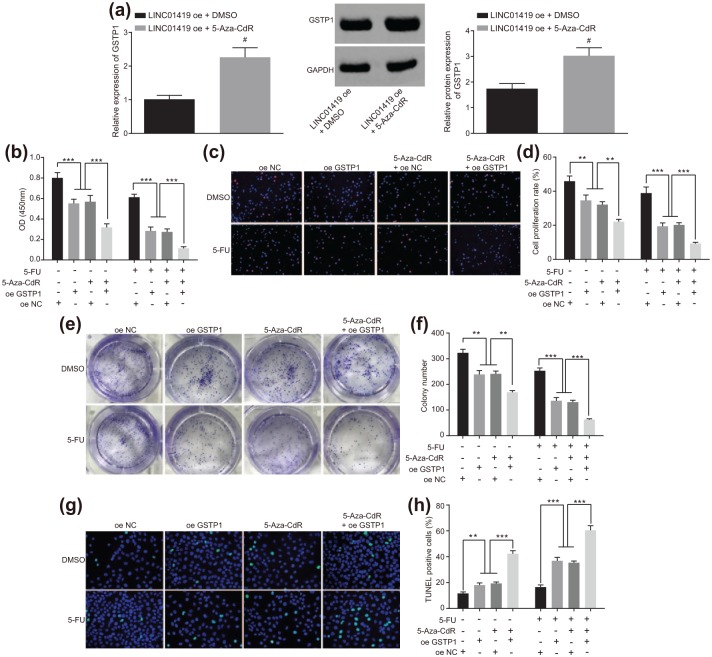
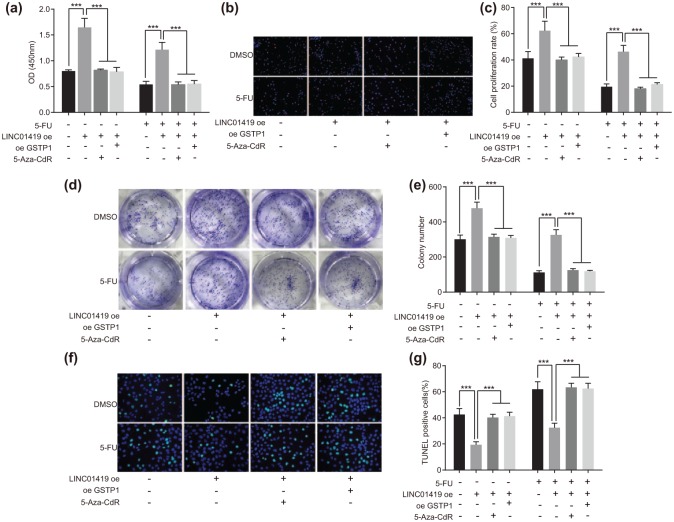
In order to ascertain the effects, associated with GSTP1 gene methylation and the occurrence of ESCC as well as 5-FU sensitivity in ESCC, we initially examined the expression of GSTP1 in the ESCC cells treated with 5-Aza-CdR in the presence of overexpression of LINC01419. The results revealed the expression of GSTP1 in the ESCC cells increased significantly after 5-Aza-CdR treatment. These results suggested that demethylation agent 5-Aza-CdR could reduce the methylation level of GSTP1 and promote the expression of GSTP1. Next, a CCK-8 assay was employed to determine the growth situation of the ESCC cells. The results demonstrated that ESCC cell viability among the cells overexpressing GSTP1 or treated with the 5-Aza-CdR was significantly decreased; after 5-FU treatment, ESCC cell viability in the cells overexpressing GSTP1 or treated with 5-Aza-CdR + 5-FU was significantly decreased [p < 0.05; Figure 4(a)]. Furthermore, the cell proliferation results indicated that ESCC cell proliferation in the cells overexpressing GSTP1 or treated with 5-Aza-CdR was significantly decreased (p < 0.05); after 5-FU treatment, cell proliferation in the cells overexpressing GSTP1 or treated with 5-Aza-CdR exhibited further reductions [p < 0.05; Figure 4(b and c)]. Moreover, the colony-forming experiment results revealed that the colony formation ability of ESCC cells in the cells overexpressing GSTP1 or treated with 5-Aza-CdR was significantly decreased (p < 0.05); after 5-FU treatment, the colony formation ability of ESCC cells of the cells overexpressing GSTP1 or treated with 5-Aza-CdR was further reduced [p < 0.05; Figure 4(d and e)]. Next, TUNEL was applied to detect ESCC cell apoptosis after 5-Aza-CdR treatment, the results of which indicated that ESCC cell apoptosis of the cells overexpressing GSTP1 or treated with 5-Aza-CdR was significantly increased (p < 0.05); following 5-FU treatment, ESCC cell apoptosis of the cells overexpressing GSTP1 or treated with 5-Aza-CdR was significantly elevated [p < 0.05; Figure 4(f and g)]. The above results demonstrated that demethylation of GSTP1 gene decreased ESCC cell proliferation and increased ESCC cell apoptosis, as well as inhibiting the occurrence of ESCC. Following overexpression of GSTP1 or treatment with 5-Aza-CdR, the antiproliferative effect and proapoptotic effect of 5-FU on ESCC cells displayed heightened abilities, indicating that the sensitivity of the ESCC cell to 5-FU was increased after the demethylation of GSTP1 gene. Taken together, the results suggested that GSTP1 gene demethylation could inhibit the occurrence of ESCC and improve the sensitivity of the ESCC cells to 5-FU.
Figure 4.
GSTP1 gene demethylation inhibits the occurrence of ESCC and improves the sensitivity of ESCC cells to 5-FU.
(a) GSTP1 levels in ESCC cells detected by RT-qPCR and western blot analysis, #versus the LINC01419 oe + DMSO group, p < 0.05. (b) ESCC cell viability was evaluated with CCK-8 assay; (c) fluorescence diagram of cell proliferation measured using EdU staining; (d) experimental quantitative analysis of cell proliferation measured using EdU staining. *p < 0.05, **p < 0.01, ***p < 0.001. (e) The results of colony-forming unit of ESCC cell; (f) experimental quantitative analysis of colony-forming unit. *p < 0.05, **p < 0.01, ***p < 0.001. (g) Apoptosis fluorescence detected by TUNEL staining after 5-Aza-CdR treatment; (h) experimental quantitative analysis detected by TUNEL staining after 5-Aza-CdR treatment. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical values were measurement data, presented as mean ± standard deviation and compared by two-way analysis of variance. The experiment was repeated three times.
5-FU, 5-fluorouracil; EdU, 5-ethynyl-2’-deoxyuridine; ESCC, esophageal squamous cell carcinoma; GSTP1, glutathione S-transferase pi 1; RT-qPCR, reverse transcription quantitative polymerase chain reaction; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
DNA methylation of GSTP1 is responsible for the promoting effects of LINC01419 on ESCC progression and the resistance to 5-FU-based chemotherapy
A CCK-8 assay was employed in order to determine whether the relationship between LINC01419 and GSTP1 gene methylation plays a primary role in terms of the sensitivity of the ESCC cells to 5-FU. After the cells (transfected with LINC01419 oe) were simultaneously treated with overexpressed GSTP1 or 5-Aza-CdR, when compared with the cells transfected with LINC01419 oe only, cell viability and proliferation ability were significantly reduced, and the apoptosis was significantly increased (p < 0.05). After treatment with 5-Aza-CdR, compared with the cells transfected with LINC01419 oe only, the ESCC cells (transfected with LINC01419 oe) simultaneously treated with overexpressed GSTP1 or 5-Aza-CdR exhibited significantly decreased cell viability and proliferation ability, and more increased cell apoptosis ability [p < 0.05; Figure 5(a and g)]. The aforementioned findings provided verification that LINC01419 could promote the proliferation of ESCC cells, inhibit cell apoptosis, and facilitate the development of ESCC. After the ESCC cells (transfected with LINC01419 oe) had been treated with overexpressed GSTP1 or 5-Aza-CdR, the stimulatory effect of LINC01419 on the ESCC cells was reversed; the anti-proliferative effect and proapoptotic effect of 5-FU on ESCC cells were strengthened. These results suggested that LINC01419 could promote the development of ESCC by targeting the GSTP1 gene methylation and regulate the sensitivity of ESCC cells to 5-FU.
Figure 5.
LINC01419 decreases the sensitivity of ESCC cells to 5-FU by regulating GSTP1 gene methylation. (a) CCK-8 assay for the detection of cell viability of ESCC cells; (b) fluorescence diagram of EdU cell proliferation, scale bar: 50 µm; (c) experimental quantitative analysis of EdU staining for cell proliferation. *p < 0.05, **p < 0.01, ***p < 0.001. (d) Colony-forming unit results of ESCC cells; (e) experimental quantitative analysis of colony-forming unit. *p < 0.05, **p < 0.01, ***p < 0.001. (f) Apoptosis fluorescence detected by TUNEL staining; (g) experimental quantitative analysis detected by TUNEL staining. *p < 0.05, **p < 0.01, ***p < 0.001. Statistical values of panels (a, c, e and g) were measurement data, presented as mean ± standard deviation and compared by two-way analysis of variance. The experiment was repeated three times.
5-FU, 5-fluorouracil; EdU, 5-ethynyl-2’-deoxyuridine; ESCC, esophageal squamous cell carcinoma; GSTP1, glutathione S-transferase pi 1; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
Downregulated LINC01419 sensitizes ESCC cells to 5-FU in vivo
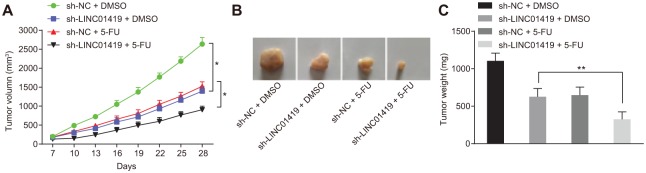
Lastly, in order to verify the effect of LINC01419 on the occurrence of ESCC and assess the sensitivity of the ESCC cells to 5-FU in vivo, we injected the ESCC cells into nude mice in order to observe tumorigenesis in the nude mice, the results of which are illustrated Figure 6. The volume and weight of tumor in sh-LINC01419 + DMSO group were significantly lower than those in sh-NC + DMSO group. After 5-FU treatment, the volume and weight of the tumors among the sh-LINC01419 + 5-FU group were significantly lower than that in the sh-NC + 5-FU group. The results revealed that the downregulation of LINC01419 could significantly inhibit tumor growth and increase the sensitivity of the ESCC cells to 5-FU.
Figure 6.
LINC01419 downregulation prevents ESCC progression and increases the sensitivity of ESCC cells to 5-FU in vivo. (a) the size of xenograft tumor in nude mice. Statistical values were measurement data, presented as mean ± standard deviation and compared by two-way analysis of variance. The experiment was repeated three times, n = 5, *p < 0.05, **p < 0.01, ***p < 0.001. (b) xenograft tumor diagram of nude mice in 28 day; (c) the weight of xenograft tumor in nude mice, n = 5. *p < 0.05, **p < 0.01, ***p < 0.001. The statistical values included measurement data, presented as mean ± standard deviation and compared with analysis of variance. The experiment was repeated three times.
5-FU, 5-fluorouracil; EdU, 5-ethynyl-2’-deoxyuridine; ESCC, esophageal squamous cell carcinoma.
Discussion
In spite of the remarkable continuous improvement of esophageal cancer treatment, patient prognosis remains poor, partly due to the associated high postoperative recurrence rate, with the rate of recurrence reported to be as high as 50% among patients undergoing excision procedures.14 Identi-fication of the entire human genome, DNA methylation pattern and gene expression profile of ESCC postulates a fresh perspective regarding the occurrence and development of ESCC, which highlights the potential of novel therapeutic targets.15 DNA methylation is a key factor in epigenetic regulation of mammalian embryonic development.16 Epigenetic changes, include factors such as, DNA methylation, histone modification, loss and genomic imprinting, all act to play a central role in the occurrence and development of ESCC with genomic variation and inheritance.17 The central objective of the present study was to investigate the molecular mechanism of LINC01419 and its involvement in ESCC as well as the cell sensitivity to 5-FU, in an attempt to identify a fresh theoretical reference in the search for new therapeutic targets for the treatment of esophageal cancer. Collectively, the data of our study demonstrated that LINC01419 was highly expressed in ESCC and promoted the promoter region methylation of the GSTP1 gene, acting to decrease the sensitivity of the ESCC cells to 5-FU.
Based on our findings, LINC01419 was observed to exhibit high levels of expression in ESCC, with downregulated levels of LINC01419 determined to exert an inhibitory effect on ESCC cell proliferation and promote apoptosis, which ultimately confers a protective effect against the occurrence of ESCC. Additionally, when treated with 5-FU, the cells’ sensitivity to 5-FU was elevated, which indicated that the downregulation of LINC01419 could act to inhibit the occurrence of ESCC and improve the sensitivity of ESCC cells to 5-FU. More recently, lncRNAs have been identified as crucial factors in the development of cancer, with certain reports highlighting them as potential biomarker and therapeutic targets in the treatment of various cancers, such as gastric cancer, and ESCC.18,19 Consistently, reports have indicated that the expression of Lincrna-nr_024015 is expressed in the cytoplasm of ESCC cell, while highlighting an association between the overexpression of Lincrna-nr_024015 and rs8506 A alleles and the survival rate of ESCC patients.20 Moreover, another study indicated that Linc00460 was positively related to ESCC tumor node metastasis stage, lymph node metastasis, and forecasted poor prognosis, and suppressed ESCC cell growth via regulating cell proliferation and cell cycle.21 More importantly, Linc00460 can function as an oncogene and could be a valuable prognostic biomarker in the diagnosis and treatment of ESCC.22 Furthermore, existing literature has suggested that LINC01419 transcripts are expressed at a high level at the early stages of HCC which is linked to patient prognosis in connection with its regulation of cell cycle genes.23 Chemotherapy is often combined with 5-FU to treat ESCC, with 5-FU capable of inhibiting ESCC proliferation in a dose-dependent manner.24
The results of the current study demonstrated that LINC01419 promoted the methylation of the GSTP1 gene. More specifically, LINC01419 was observed to promote the methylation of the GSTP1 gene by means of recruiting DNA methyltransferase targets. Glutathione S- transferase is widely known to be a carcinogenic substance related to the metabolism of tobacco, while GSTP1 polymorphism has been correlated with the course of ESCC.25 A previous study demonstrated that GSTP1 Ile105Val polymorphism was an independent risk factor, and GSTT1-WT was an independent protective factor in ESCC, which was suggestive of a contrary role played by GSTP1 and GSTT1 polymorphisms as risk factors in ESCC.26 LincRNA H19 has been reported to various regulate proteins, including GSR, G6PD, GCLC, GCLM, and GSTP1, which are NRF2-target genes.27 Generally, the inactivation of GSTP1 by obtained somatic CpG island promoter has been shown to be hypermethylated in various cancer subtypes.28
Furthermore, our study revealed that following demethylation of GSTP1 gene, cell proliferation was decreased, while apoptosis was increased, with the occurrence of ESCC being inhibited. Following treatment with 5-FU, the sensitivity of cancer cells to 5-FU was increased, suggesting that GSTP1 gene demethylation might inhibit the occurrence of ESCC and improve the sensitivity of ESCC cell to 5-FU. Research has revealed that the combination chemotherapy with 5-FU and cisplatin is an effective treatment for esophageal carcinoma. This being said, the distinguishing factors that affect sensitivity to 5-FU and cisplatin could well prove to be an essential factor in the bid to improve the efficacy of chemotherapy in cases of ESCC.29 Methylation of the GSTP1 gene CpG island promoter is involved in the occurrence and development of multiple tumors, which could be a biomarker for early screen, diagnosis and prognosis.30 Studies have emphasized the ability of GSTP1 expression is an important predictor of the clinical outcome or drug resistance to ESCC. Lymph node metastasis, and overexpression of GSTP1 were all determined to be independent predictors linked to poor prognoses, with this association tightly correlated with the chemotherapeutic efficacy of 5-FU combined with cisplatin in ESCC patients.31
Conclusion
In conclusion, the key findings of the study suggest that LINC01419 is overexpressed in ESCC and promotes the promoter region methylation of the GSTP1 gene, acting to reduce the sensitivity of cancer cells to 5-FU. The detailed regulatory mechanism is depicted in Figure 7. Our results ultimately provide strong evidence that therapeutic strategies should be directed towards the downregulation of LINC01419, which may be a promising and clinically viable target in the treatment of ESCC.
Figure 7.
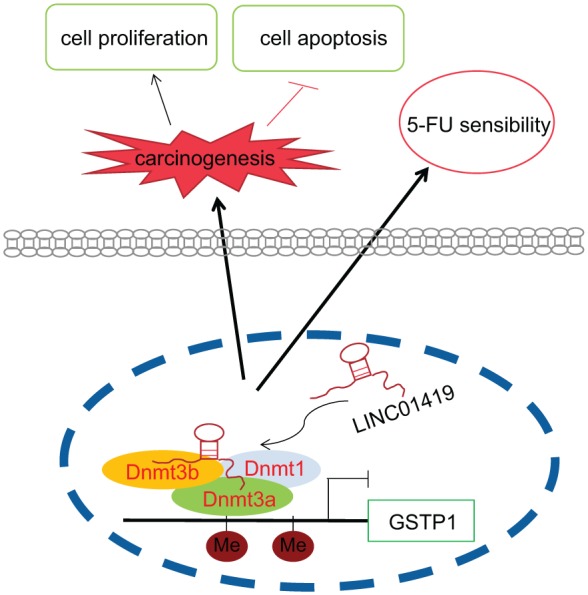
LINC01419 is localized in the nucleus and its overexpression enhances the ability of GSTP1 to recruit methyltransferase DNMT1, DNMT3A and DNMT3B. The overexpression of LINC01419 promotes methylation of the promoter region of the GSTP1 gene, as well as acting to promote ESCC cell growth and reduce the sensitivity of ESCC cells to 5-FU.
5-FU, 5-fluorouracil; ESCC, esophageal squamous cell carcinoma; GSTP1, glutathione S-transferase pi 1.
Supplemental Material
Supplemental material, Supplemental_Figure_1 for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supplementarl_Figure_2 for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supplementary_Material_1_-_Blast_sequence for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supplementary_Material_2_ for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental Material
Supplemental material, Supplementary_Material_3_ for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Acknowledgments
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article. Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin and Yu-Qi She contributed equally to this work as co-first authors.
Footnotes
Funding: This study was supported by surface project of 2018 Clinical Pharmaceutical Research Fund of WU JIEPING Foundation of the Chinese Medical Association. (Project ID: LCVX-M002) and Letter from the Third Batch and Fourth Batch of Medical and Health Science and Technology Program Project Funds in Shantou City in 2018 [Shan Fu Ke letter (2018) 238].
Conflict of interest statement: The authors declare that there is no conflict of interest.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Jian-Liang Chen, Clinical Laboratory, Cancer Hospital of Shantou University Medical College, Shantou, China.
Zhi-Xiong Lin, Radiotherapy Department, Cancer Hospital of Shantou University Medical College, Shantou, China.
Yun-Sheng Qin, Chest Surgery, Cancer Hospital of Shantou University Medical College, Shantou, China.
Yu-Qi She, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, Shantou, China.
Yun Chen, Clinical Pharmacy Research Center, Shantou University Medical College, Shantou, China.
Chen Chen, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, Shantou, China.
Guo-Dong Qiu, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, Shantou, China.
Jie-Ting Zheng, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, Shantou, China.
Zhong-Lin Chen, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, Shantou, China.
Shu-Yao Zhang, Department of Pharmacy, Cancer Hospital of Shantou University Medical College, No. 7, Raoping Road, Shantou 515031, Guangdong Province, China.
References
- 1. Okuda M, Inoue J, Fujiwara N, et al. Subcloning and characterization of highly metastatic cells derived from human esophageal squamous cell carcinoma KYSE150 cells by in vivo selection. Oncotarget 2017; 8: 34670–34677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim JA, Shah PM. Screening and prevention strategies and endoscopic management of early esophageal cancer. Chin Clin Oncol 2017; 6: 50. [DOI] [PubMed] [Google Scholar]
- 3. Nakajima M, Kato H. Treatment options for esophageal squamous cell carcinoma. Expert Opin Pharmacother 2013; 14: 1345–1354. [DOI] [PubMed] [Google Scholar]
- 4. Cao HH, Zheng CP, Wang SH, et al. A molecular prognostic model predicts esophageal squamous cell carcinoma prognosis. PLoS One 2014; 9: e106007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi WH, Wu QQ, Li SQ, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol 2015; 36: 2501–2507. [DOI] [PubMed] [Google Scholar]
- 6. Wang CM, Wu QQ, Li SQ, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci 2014; 59: 591–597. [DOI] [PubMed] [Google Scholar]
- 7. Achawanantakun R, Chen J, Sun Y, et al. LncRNA-ID: Long non-coding RNA IDentification using balanced random forests. Bioinformatics 2015; 31: 3897–3905. [DOI] [PubMed] [Google Scholar]
- 8. Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep 2012; 45: 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brate J, Adamski M, Neumann RS, et al. Regulatory RNA at the root of animals: dynamic expression of developmental lincRNAs in the calcisponge Sycon ciliatum. Proc Biol Sci 2015; 282: 20151746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W, Zheng J, Deng J, et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology 2014; 146: 1714–1726.e1715. [DOI] [PubMed] [Google Scholar]
- 11. Dai M, Chen S, Wei X, et al. Diagnosis, prognosis and bioinformatics analysis of lncRNAs in hepatocellular carcinoma. Oncotarget 2017; 8: 95799–95809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu R, Yin L, Pu Y, et al. Functional alterations in the glutathione S-transferase family associated with enhanced occurrence of esophageal carcinoma in China. J Toxicol Environ Health A 2010; 73: 471–482. [DOI] [PubMed] [Google Scholar]
- 13. Lou P, Sun X, Zhou J, et al. [Effect of RAD18-siRNA on proliferation and chemotherapy sensitivity of human esophageal squamous cell carcinoma ECA-109 cells]. Zhejiang Da Xue Xue Bao Yi Xue Ban 2016; 45: 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu JB, Qiang FL, Dong J, et al. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol 2011; 17: 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen C, Peng H, Huang X, et al. Genome-wide profiling of DNA methylation and gene expression in esophageal squamous cell carcinoma. Oncotarget 2016; 7: 4507–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature 2014; 511: 606–610. [DOI] [PubMed] [Google Scholar]
- 17. Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surg Today 2013; 43: 1355–1364. [DOI] [PubMed] [Google Scholar]
- 18. Gu Y, Chen T, Li G, et al. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol 2015; 11: 2427–2441. [DOI] [PubMed] [Google Scholar]
- 19. Hao Y, Wu W, Shi F, et al. Prediction of long noncoding RNA functions with co-expression network in esophageal squamous cell carcinoma. BMC Cancer 2015; 15: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han L, Liu S, Liang J, et al. A genetic polymorphism at miR-526b binding-site in the lincRNA-NR_024015 exon confers risk of esophageal squamous cell carcinoma in a population of North China. Mol Carcinog 2017; 56: 960–971. [DOI] [PubMed] [Google Scholar]
- 21. Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep 2017; 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang GW, Xue YJ, Wu ZY, et al. A three-lncRNA signature predicts overall survival and disease-free survival in patients with esophageal squamous cell carcinoma. BMC Cancer 2018; 18: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang H, Zhu C, Zhao Y, et al. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget 2015; 6: 43770–43778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hatata T, Higaki K, Nanba E, et al. Inhibition of nuclear factor-kappaB activity by small interfering RNA in esophageal squamous cell carcinoma cell lines. Oncol Rep 2011; 26: 659–664. [DOI] [PubMed] [Google Scholar]
- 25. Zendehdel K, Bahmanyar S, McCarthy S, et al. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer Causes Control 2009; 20: 2031–2038. [DOI] [PubMed] [Google Scholar]
- 26. Song Y, Du Y, Zhou Q, et al. Association of GSTP1 Ile105Val polymorphism with risk of esophageal cancer: a meta-analysis of 21 case-control studies. Int J Clin Exp Med 2014; 7: 3215–3224. [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng ZG, Xu H, Suo SS, et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep 2016; 6: 26093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schnekenburger M, Karius T, Diederich M. Regulation of epigenetic traits of the glutathione S-transferase P1 gene: from detoxification toward cancer prevention and diagnosis. Front Pharmacol 2014; 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minegaki T, Takara K, Hamaguchi R, et al. Factors affecting the sensitivity of human-derived esophageal carcinoma cell lines to 5-fluorouracil and cisplatin. Oncol Lett 2013; 5: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu B, Jiang Y, Zhu F, et al. Demethylation effects of elemene on the GSTP1 gene in HCC cell line QGY7703. Oncol Lett 2016; 11: 2545–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamoto Y, Konishi H, Ichikawa D, et al. Significance of GSTP1 for predicting the prognosis and chemotherapeutic efficacy in esophageal squamous cell carcinoma. Oncol Rep 2013; 30: 1687–1694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_Figure_1 for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental material, Supplementarl_Figure_2 for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental material, Supplementary_Material_1_-_Blast_sequence for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental material, Supplementary_Material_2_ for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology
Supplemental material, Supplementary_Material_3_ for Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation by Jian-Liang Chen, Zhi-Xiong Lin, Yun-Sheng Qin, Yu-Qi She, Yun Chen, Chen Chen, Guo-Dong Qiu, Jie-Ting Zheng, Zhong-Lin Chen and Shu-Yao Zhang in Therapeutic Advances in Medical Oncology