Abstract
Research with non-human primates (NHP) has been essential and effective in increasing our ability to find cures for a large number of diseases that cause human suffering and death. Extending the availability and use of genetic engineering techniques to NHP will allow the creation and study of NHP models of human disease, as well as broaden our understanding of neural circuits in the primate brain. With the recent development of efficient genetic engineering techniques that can be used for NHP, there’s increased hope that NHP will significantly accelerate our understanding of the etiology of human neurological and neuropsychiatric disorders. In this article, we review the present state of genetic engineering tools used in NHP, from the early efforts to induce exogeneous gene expression in macaques and marmosets, to the latest results in producing germline transmission of different transgenes and the establishment of knockout lines of specific genes. We conclude with future perspectives on the further development and employment of these tools to generate genetically engineered NHP.
Keywords: macaques, marmosets, transgenic animals, gene editing, genome sequencing, transgenesis
Introduction
The ability to modify the mouse genome has given rodents a tremendous importance in biomedical research. Genetic engineering techniques allow both complete or controlled inactivation of endogenous genes, regulated expression of transgenes, and induction of specific mutations. These tools enable the study of neural circuits in the brain. They also allow the investigation of causes for human diseases of genetic origin, including age-related diseases (Bales, 2012; Elder, Gama Sosa, & De Gasperi, 2010; Lampreht Tratar, Horvat, & Cemazar, 2018; Scheikl, Pignolet, Mars, & Liblau, 2010; Wilcock, 2010). However, the significant anatomical, physiological, cognitive and behavioral differences between rodents and humans make it essential to extend the availability of genetic tools to non-human primates (NHP) (Izpisua Belmonte et al., 2015). NHP share the same mammalian order with humans, and both anatomical and functional organization of their brains are homologous to those of humans. In particular, the NHP brain has specialized motor, perceptual and cognitive aptitudes not found in rodents, and neurological and neuropsychiatric disorders are better modeled in NHP than in rodents. Primates are also excellent models of aging, a major co-morbidity factor in many human diseases (Mattison & Vaughan, 2017; Power, Ross, Schulkin, Ziegler, & Tardif, 2013; Riesche et al., 2018; Salmon, 2016; Tardif, Mansfield, Ratnam, Ross, & Ziegler, 2011; Vaughan & Mattison, 2016). Unlike rodent models of aging, which do not develop significant neurodegeneration with aging, aged NHP demonstrate structural and functional brain changes, including an age-dependent decline in cognitive function similar to those reported in humans (Lane, 2000; Mitchell, Scheibye-Knudsen, Longo, & de Cabo, 2015). Extending the availability and use of gene editing techniques to NHP will allow the creation and study of NHP models of human age-related diseases, especially those of genetic origin, as well as broaden our understanding of physiological and pathological status of neural circuits in the primate brain.
In this article, we first review the present state of genetic engineering techniques as applied to NHP, from the early efforts to induce exogeneous gene expression in macaques (Chan, Chong, Martinovich, Simerly, & Schatten, 2001; Chan et al., 2000; Niu et al., 2015; Niu et al., 2010; Yang et al., 2008), to the latest results in producing germline transmission of different transgenes (Liu et al., 2016; Park et al., 2016; Putkhao et al., 2013; Sasaki et al., 2009) and the establishment of knockout lines of specific genes (Y. Chen et al., 2017; Y. Chen, Zheng, et al., 2015; Ke et al., 2016; H. Liu et al., 2014; Z. Liu et al., 2014; Niu et al., 2014; Sato et al., 2016; Wan et al., 2015; Zuo et al., 2017). We then report our own efforts to generate transgenic marmosets expressing genetically-encoded calcium indicators (Park et al., 2016), and talk about our preliminary efforts to generate a marmoset model of CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy). We conclude with some future perspectives on the further development of genetic tools for generation of genetically engineered NHP. Another excellent review of the development of genetically engineered NHP and their potential use in biomedical research can be found in (Chan, 2013).
Generation of transgenic non-human primates: macaques
Several strategies for the delivery of exogenous genes have been explored in the early stages of making transgenic NHP, first in macaques and later in marmosets, as listed in Table 1. The first effort used intracytoplasmic sperm injection (ICSI) into mature rhesus macaque oocytes collected via surgical laparotomy (Chan et al., 2000). The oocytes were injected with plasmid-bound spermatozoa tagged with exogenous DNA encoding the green fluorescent protein (GFP) gene (Chan et al., 2000). The resulting embryos expressed the transgene in vitro and were transferred into surrogate mothers. While integration of the exogenous gene was not detected in the offspring, this study demonstrated the theoretical possibility of transgenesis by ICSI in primates (Chan et al., 2000). In a second attempt to produce transgenic macaques, the same group decided to use a pseudotyped, replication-defective retroviral vector that overexpressed GFP, taking advantage of the well-known ability of retroviruses to efficiently transfer exogenous genes to cell nuclei in a way that allows stable integration of the transgene into the genome (Chan et al., 2001). In 2001, the first transgenic rhesus monkey overexpressing GFP was born following injection of a retroviral vector into the perivitelline space of mature rhesus oocytes, which were later fertilized by ICSI (Chan et al., 2001). Although only one of three offspring was shown to be transgenic, and germline transmission could not be verified, that study clearly demonstrated the feasibility of generating transgenic NHP.
Table 1:
Summary of NHP models overexpressing transgenes
| Transgene | Application | Species | Genetic manipulation | Outcome | Reference |
|---|---|---|---|---|---|
| GFP | Reporter gene | Rhesus macaque | Injection of retroviral vectors into oocytes followed by ICSI | -One live transgenic offspring -Germline transmission not verified -Pregnancies/surrogate: 5/20 (25%) |
Chan et al., 2001 |
| Human mutant huntingtin gene | Huntington’s disease | Rhesus macaque | Injection of lentiviral vectors into oocytes followed by ICSI | -Five live transgenic offspring with clinical features of Huntington’s disease: variable extents of motor dysfunction, chorea and dystonia -Germline transmission confirmed in a follow up study -Pregnancies/surrogate: 6/8 (75%) |
Yang et al., 2008 Moran et al., 2015 |
| GFP | Reporter gene | Common marmoset | Injection of lentiviral vectors into embryos | -Five live transgenic offspring -Germline transmission confirmed in offspring -Pregnancies/surrogate (IVF): 1/13 (7.6%) -Pregnancies/surrogate (natural): 6/37 (16.2%) |
Sasaki et al., 2009 |
| EGFP | Reporter gene | Rhesus macaque | Injection of SIV vectors into embryos | -Two live transgenic offspring with mosaic expression of the transgene -Pregnancies/surrogate: 5/8 (62.5%) |
Niu et al., 2010 |
| Mutant α-synuclein (A53T) | Parkinson’s disease | Rhesus macaque | Injection of lentiviral vectors into oocytes followed by ICSI | -Six live transgenic offspring expressing mutant α-synuclein -Subtle cognitive defects and anxiety like behaviors -Germline transmission not verified -Pregnancies/surrogate: 11/25 (44%) |
Niu et al., 2015 |
| Human MeCP2 | Autism | Cynomolgus macaque | Injection of lentiviral vectors into oocytes followed by ICSI | -Autism-like disorder -Behavioral abnormalities: repetitive circular locomotion, reduction in social interactions, impairment of cognitive functions -Germline transmission confirmed using testicular tissue xenografting -Pregnancies/surrogate: 16/54 (29.6%) |
Liu et al., 2016 |
| GFP | Reporter gene | Cynomolgus macaque | Injection of lentiviral vectors into oocytes or embryos | -Lentivirus injection into oocytes before fertilization achieved homogenous expression of GFP throughout the entire body -Pregnancies/surrogate (post ICSI): 2/3 (66.7%) -Pregnancies/surrogate (before ICSI): 5/17 (29%) |
Seita et al., 2016 |
| GCaMP | Functional reporter gene | Common marmoset | Injection of lentiviral vectors into embryos | -Five live transgenic offspring -Stable and functional GCaMP expression in several different tissues -Germline transmission confirmed -Pregnancies/surrogate (IVF): 13/51 (25.5%) -Pregnancies/surrogate (natural): 9/31 (29%) |
Park et al., 2016 |
| Human mutant ataxin 3-120Q | Polyglutamine diseases | Common marmoset | Injection of lentiviral vectors into embryos | -Seven live transgenic offspring expressing polyQ expanded ataxin 3 with gradual progression of neurological symptoms and motor impairment -Germline transmission confirmed in offspring -Pregnancies/surrogate: 9/18 (50%) |
Tomioka et al., 2017 |
| Human mutant ataxin 3-120Q | Polyglutamine diseases | Common marmoset | Injection of lentiviral vectors into embryos | -Mutant human ataxin 3 gene expression controlled by tet-on system -Germline transmission confirmed in offspring -Pregnancies/surrogate: 14/40 (35%) |
Tomioka et al., 2017 |
Once the first transgenic NHP were generated, attention quickly shifted towards creating NHP models of neurodegenerative diseases. In 2008, the first transgenic rhesus macaque model of Huntington’s disease (HD) was produced by injecting perivitelline space of mature oocytes with high titer lentiviruses expressing both GFP and exon 1 of the human huntingtin (HTT) gene with 84 CAG repeats, which were later fertilized with ICSI and transferred to surrogate mothers (Yang et al., 2008). Integration of both GFP and mutant HTT genes was confirmed in five newborn monkeys, which later developed important clinical features of HD, including variable extents of motor dysfunction and abnormal involuntary movements, such as chorea and dystonia (Yang et al., 2008). To provide an insight of HD cellular pathogenesis, induced pluripotent stem cells (iPSCs) were established from HD monkeys (Chan, Cheng, Neumann, & Yang, 2010). This in vitro model of HD developed distinctive cellular features, including the accumulation of mutant HTT aggregates and the formation of intranuclear inclusions during the course of neural differentiation, holding promise for the development of novel cellular-based therapies that can be tested and validated in HD monkeys prior to being tested in human patients (Carter & Chan, 2012; Y. Chen, Carter, Cho, & Chan, 2014). One example of this promising line of work came in 2014, when stable neural progenitor cell (NPC) lines were derived from HD-iPSCs (Carter et al., 2014). The resulting HD-NPCs were differentiated into neural cells featuring classic HD cellular phenotypes, which were rescued by therapeutic treatments that included RNAi-mediated reduction of HTT transcripts and drug-mediated amelioration of neurotoxicity. These results established this cellular platform of HD as a powerful resource for screening therapeutic strategies aimed at treating HD monkeys (Carter et al., 2014). In parallel with the in vitro studies, longitudinal clinical measurements consisting of morphometric MRI measures of the brain and cognitive behavior evaluation of a single HD transgenic monkey showed reduction in the volume of the striatum and hippocampus with gradual impairment in motor functions and cognitive decline (Chan et al., 2014). These findings were later confirmed in a cohort of HD monkeys which were studied at up to 48 months of age, establishing HD monkeys as a suitable preclinical animal model for HD (Chan et al., 2015).
One of the key justifications for the development of transgenic animals of disease is that the inheritance of the transgene through the germ cells causes inheritance of the disease’s pathogenic phenotype in subsequent generations, allowing researchers to establish a cohort of transgenic animals which mimic human inherited genetic disorders (Chan, 2004; Y. Chen, Niu, & Ji, 2012). Germline transmission of the mutant HTT was confirmed in embryonic stem cells generated from a HD founder (Putkhao et al., 2013) and in second-generation offspring produced via artificial insemination by using intrauterine insemination technique (Moran et al., 2015). The stable germline transmission and expression of the mutant HTT transgene in HD monkeys makes it possible to expand the HD monkey colonies for future translational studies.
A similar strategy was used to produce a transgenic rhesus macaque model of Parkinson’s disease (PD). Oocytes collected from donor females were injected with lentiviral vectors encoding a mutant α-synuclein (A53T) expressed under control of the human ubiquitin promoter (Niu et al., 2015). Six transgenic A53T monkeys were produced, and the mutant A53T was shown to be expressed in their brains. One A53T monkey presented cognitive deficits and anxiety like behaviors compared to an age-matched control, but no obvious motor symptoms were noticed (Niu et al., 2015). However, these transgenic monkeys may develop more robust PD phenotypes as they age, and thus it is important to continue their neurological and behavioral evaluation.
More recently, a lentivirus-based transgenic cynomolgus monkey model of autism was produced via overexpression of the human Methyl-CpG binding protein 2 (MeCP2) (Liu et al., 2016). Overexpression of MeCP2 was confirmed in brain tissues and these transgenic monkeys exhibited an autism-like disorder that was largely characterized by behavioral abnormalities, including repetitive circular locomotion, reduction in social interactions and a mild impairment of cognitive functions. To speed up the generation of F1 offspring, the authors used a testicular tissue xenografting method to rapidly mature sperm cells from a pre-puberty founder (Liu et al., 2016). Oocytes collected from donor females were fertilized via ICSI, and transgenic F1 offspring were born, confirming germline transmission of the transgene (Liu et al., 2016). This method significantly shortened the time required for producing the next generation of transgenic cynomolgus monkeys (~3 years in this study vs. ~5 years under natural conditions).
Since the birth of the first transgenic macaque (Chan et al., 2001), strategies that use different types of lentivirus and optimization of virus injection timing were assessed as a way to improve the methodology for making transgenic NHP. In one study, simian immunodeficiency virus (SIV)-based vector and a protocol for infection of early cleavage-stage embryos were used for making GFP transgenic rhesus monkeys (Niu et al., 2010), showing the usefulness of SIV-based lentiviral vectors for transgenesis in NHP. Early-cleavage stage embryos are known to be more resistant to damages incurred by the microinjection procedure than oocytes, but mosaic expression of the transgene due to delayed lentivirus-mediated gene integration during cell division cycles needs to be further addressed. In another study, transgenic cynomolgus macaques expressing GFP homogenously throughout the whole body were generated by high-titer purified lentivirus injection into matured oocytes before fertilization (Seita et al., 2016). These results can be applied to generate transgenic monkeys that overexpress causative genes for human diseases evenly throughout the entire body.
Generation of transgenic non-human primates: marmosets
Although Old World monkeys, in particular cynomolgus and rhesus macaques, have been traditionally used in biomedical research due to their close relationship to humans, the common marmoset (Callithrix jacchus) has several advantages as an ideal species for developing NHP models of human diseases (Cyranoski, 2014; Kropp, Di Marzo, & Golos, 2017; Mansfield, 2003). The marmoset is a small New World primate that is particularly adapted to the laboratory setting. They are the most prolific of the NHP species, reaching sexual maturity around 15-18 months of age and giving birth twice a year, with litter sizes of 2-3 offspring (Tardif et al., 2003). In 2009, the first GFP transgenic common marmoset was produced via injection of a high titer lentivirus containing an enhanced green fluorescent protein (EGFP) into fertilized embryos (Sasaki et al., 2009). This study revealed that it was beneficial to place the embryos in 0.25 M sucrose solution for making an expanded perivitelline space, allowing delivery of a larger amount of viral particles containing the transgene into preimplantation embryos obtained both from natural mating or from oocytes fertilized in vitro. Owing to the high reproductive efficiency and recent progress in assisted reproductive technologies in this species, germline transmission of transgene was promptly confirmed in GFP-expressing embryos and in offspring derived from transgenic marmosets. This feasibility of rapid expansion of transgenic marmoset colonies has brought more attention to this species (Cyranoski, 2009; Schatten & Mitalipov, 2009) and provided its promising role as a bridge between mouse models of disease and human disorders (Izpisua Belmonte et al., 2015). In particular, marmosets are poised to become a prime NHP aging model, as they are short-lived and yet they show age-related pathologies, including the presence of neurodegeneration, that mirror those seen in humans (Tardif et al., 2011).
After the initial technical breakthrough in making GFP-expressing transgenic marmosets (Sasaki et al., 2009), we reported marmosets expressing functional reporter genes (Park et al., 2016), while another group reported a marmoset model of polyglutamine (polyQ) disease (Tomioka, Ishibashi, et al., 2017; Tomioka, Nogami, et al., 2017). The transgenic marmosets expressing functional reporter genes were generated by our own efforts and the summary of our work and ongoing progress will be discussed later in this review. In an attempt to mimic inherited human neurodegenerative diseases, transgenic marmosets expressing the human ataxin 3 gene with expanded polyQ stretch were developed as a model of polyQ diseases (Tomioka, Ishibashi, et al., 2017). These transgenic marmosets demonstrated no neurological symptoms at birth, but showed slower growth curves, and gradual decreases in spontaneous activity and grip strength. Some hallmarks of the human disease were also demonstrated, including cerebellar atrophy, neurodegeneration and intranuclear polyQ protein inclusions accompanied by gliosis, which recapitulate the neuropathological features of polyQ disease patients. Successful germline transmission was confirmed in F1 offspring derived from a symptomatic F0 transgenic marmoset, allowing for the establishment of a symptomatic marmoset colony that can be used in future translational studies. However, one caveat when trying to establish transgenic NHP models of disease is that transgenic monkeys may carry a high rate of infant mortality, and even juveniles or young adults may acquire early symptoms with rapid disease progression, making it difficult to use these animals to expand the colony. To circumvent this problem, the same group pursued a parallel effort to develop transgenic marmosets expressing the mutant human ataxin 3 gene under control of a tetracyclin-inducible transgene expression (tet-on) system (Tomioka, Nogami, et al., 2017). The offspring harboring the transgene under the control of tet-on system showed increased transgene expression with doxycycline administered in the drinking water and live F1 transgenic marmosets were easily produced from a founder marmoset before the disease onset. Nevertheless, it will be a future challenge in this transgenic model to demonstrate the triggering of disease onset with the relatively modest changes of transgene expression obtained (1.7-1.8 fold) after doxycycline administration.
Thanks to the higher efficiency in transgene delivery to NHP oocytes or embryos via virus-mediated vectors and successful generation of various transgenic NHP models, several additional lines of transgenic marmosets, including Cre expression under cell-specific promoters, Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis models are expected to be available in the near future (Okano & Kishi, 2018). Despite these considerable efforts, however, translational research conducted with transgenic NHP still appears impractical. Technical limitations associated with the use of viral vector-mediated gene delivery range from insert size limitation, to random insertion (integration) site in the genome, to an uncontrollable number of copies of the transgene, to potential silencing of the transgene. These limitations need to be considered when designing and utilizing viral vectors to generate transgenic NHP models.
Targeted gene disruptions in non-human primates
Recent advances in genetic engineering have led to the development of effective tools to introduce site specific double-stranded breaks (DSBs) in DNA, including zinc finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas9 endonucleases (Jinek et al., 2012; Y. G. Kim, Cha, & Chandrasegaran, 1996; T. Li et al., 2011). These programmable site-specific nucleases have already shown their ability to edit the genome in cultured cells and in organisms from plants to human embryos precisely and proficiently (Bak, Gomez-Ospina, & Porteus, 2018; H. Ma et al., 2017). Subsequent to the discovery of the programmable site-specific nucleases, researchers have rapidly applied them to generate NHP knock-out (KO) models of specific genes via mutagenesis mediated by non-homologous end joining (NHEJ) repair mechanisms (Table 2). The relative ease of handling, relatively high efficiency, and multiplexable targeting has made the CRISPR/Cas9 system a preferable option for genomic editing (Doudna & Charpentier, 2014). In 2014, founder cynomolgus macaques harboring Ppar-r, and Rag1 gene modifications were successfully obtained after simultaneous injection of multiple sgRNAs targeting Nr0b1, Ppar-r, and Rag1 with Cas9 mRNA into zygotes (Niu et al., 2014). In a follow-up study, germline transmission of Cas9-mediated genome modifications was examined in gonad and germ cells of Cas9-manipulated monkeys (Y. Chen, Cui, et al., 2015). Results demonstrated extensive gene modifications in various somatic tissues as well as the gonads, providing strong evidence that Cas9-mediated gene modifications can be transmitted through the germline to the next generation. A further assessment of an aborted fetus from this study revealed that the targeted mutations in Nr0b1 were only detected in aborted fetuses, but not in live founders (Kang et al., 2015). Interestingly, the aborted male fetuses carrying Nr0b1 gene mutation displayed similar features to human adrenal hypoplasia congenita and hypogonadotropic hypogonadism, which are known to be caused by mutations in the human homologous gene, providing evidence that genetically engineered monkeys represent more faithful models of human genetic diseases.
Table 2:
Summary of gene edited NHP models
| Edited Gene | Application | Species | Genetic manipulation | Outcome | Reference |
|---|---|---|---|---|---|
| Nr0b1, Ppar-r, and Rag1 | Proof of concept for CRISPR/Cas9 | Cynomolgus macaque | Co-injection of Cas9 mRNA and multiple sgRNAs into single-cell embryos | -Simultaneous disruption of Ppar-r, and Rag1 genes -Germline transmission confirmed in follow-up study -Pregnancies/surrogates: 10/29 (34.5%) |
Niu et al., 2014 Chen et al., 2015 |
| MECP2 | Autism | Rhesus / Cynomolgus macaque | Injection of circular TALEN plasmid into single-cell embryos | -One live MECP2 mutant offspring generated -Male embryonic lethality of MeCP2 mutation shown -Pregnancies/surrogates: 8/26 (30.8%) |
Liu et al., 2014 |
| MECP2 | Autism | Cynomolgus macaque | Injection of TALEN mRNA into single-cell embryos |
-One MECP2 mutant male neonate that did not survive -Pregnancies/surrogates: 3/9 (33.3%) |
Liu et al., 2014 |
| Nr0b1, Ppar-r, and Rag1 | Adrenal hypoplasia congenita (AHC) Hypogonadotropic hypogonadism (HH) |
Cynomolgus macaque | Co-injection of Cas9 mRNA and multiple sgRNAs into single-cell embryos | -Cas9 targeted Nr0b1-deficient male monkey fetus displayed defects in adrenal gland development and abnormal testis morphology | Kang et al., 2015 |
| p53 | Proof of concept for CRISPR/Cas9 | Cynomolgus macaque | Co-injection of Cas9 mRNA and sgRNA into single-cell embryos | -Single-step live p53 biallelic mutant monkeys -Pregnancies/surrogates: 4/13 (30.8%) |
Wan et al., 2015 |
| dystrophin | Duchenne muscular dystrophy (DMD) | Rhesus macaque | Co-injection of Cas9 mRNA and sgRNA into single-cell embryos | -Nine live offspring with multiple mutations of dystrophin gene -Mutant monkeys displayed muscle changes similar to early stage DMD patients -Pregnancies/surrogates: 7/59 (11.9%) |
Chen et al., 2015 |
| MCPH1 | Microcephaly | Cynomolgus macaque | Injection of TALEN mRNA into single-cell embryos | -One live monkey carrying biallelic MCPH1 mutation generated -Microcephaly with hypoplasia of the corpus callosum and upper limb spasticity -Pregnancies/surrogates: 3/9 (33%) |
Ke et al., 2016 |
| IL2RG | Severe Combined Immunodeficiency |
Common marmoset | Injection of ZFN or TALEN mRNA into single-cell embryos | -Nine neonates exhibiting mutations in the IL2RG gene generated -Immunodeficient phenotypes included lack of thymus, reduced T-cell and natural killer cell count in cord blood samples and gradual decrease of B cells in peripheral blood -Germline transmission confirmed in germ cells -Pregnancies/surrogates: 19/113 (16.8%) |
Sato et al., 2016 |
| Prrt2 | Proof of concept for CRISPR/Cas9 | Cynomolgus macaque | Co-injection of Cas9 mRNA and sgRNA into single-cell embryos | -Complete Prrt2 knockout monkey was generated injection of Cas9 mRNA with multiple adjacent sgRNAs that target only a single key exon of the target gene -Pregnancies/surrogates: 1/9 (11.1%) |
Zuo et al., 2017 |
| MECP2 | Autism | Rhesus macaque Cynomolgus macaque |
Injection of circular TALEN plasmid into single-cell embryos | -Four additional live MECP2 mutant offspring generated -Complex behavioral abnormalities, including fragmented sleep, increased stereotypic behavior and reduced social interaction -Pregnancies/surrogates: 14/41 (34.1%) |
Chen et al., 2017 |
In another study, live p53 biallelic mutant cynomolgus monkeys were successfully obtained in a single step without potential off-target mutations after systemic optimization of CRISPR/Cas9 and sgRNA combination (Wan et al., 2015). Although this study provided no descriptions of phenotypes for homozygous mutations in the p53 gene, this result holds great value, as it brings gene editing in NHP closer to practical applications by avoiding the difficulty of producing biallelic monkeys through breeding. Similarly, a CRISPR/Cas9 construct targeting the dystrophin gene was microinjected into fertilized rhesus monkey embryos to generate a monkey model of Duchenne muscular dystrophy (DMD) (Y. Chen, Zheng, et al., 2015). Two stillborn and 9 live offspring had multiple types of dystrophin gene mutations. Despite a high degree of mosaicism, analysis of the stillborn muscle revealed that the CRISPR/Cas9 induced mutations led to depletion of dystrophin and caused muscle changes similar to those found in early stage DMD patients. Since DMD is an age-dependent disorder and live offspring in this study were younger than 6 months at the time of the report, further investigations of their phenotype are yet to be performed.
As CRISPR-Cas9 induced mosaic mutations have drawn concerns about the functional mosaicism in genetically edited animals, approaches for nearly complete knockout of specific genes using multiple sgRNAs were tested in cynomolgus monkeys (Zuo et al., 2017). Zygotic injection of Cas9 mRNA with multiple adjacent sgRNAs that target only a single key exon of the target gene enabled complete gene knockout with high efficiency in a single step. Using this method, cynomolgus macaques with a complete knockout of the Prrt2 gene were successfully generated without significant off-target mutations, offering an efficient one-step process for studying gene function in NHP (Zuo et al., 2017).
The use of TALENs for genomic editing in NHP follows closely behind the use of CRISPR/Cas9. TALEN-mediated gene editing has been an effective tool to induce loss of function mutations in rhesus, cynomolgus monkeys and marmosets (Y. Chen et al., 2017; Ke et al., 2016; H. Liu et al., 2014; Z. Liu et al., 2014; Sato et al., 2016). Knockout of the MECP2 gene was successfully mediated by delivery of three TALENs pairs of exon3 targeting plasmid into single cell rhesus and cynomolgus zygotes together with RAD51, which enhance DNA repair following TALEN mediated DSBs (H. Liu et al., 2014). Cytoplasmic injection of circular plasmid DNA resulted in prolonged episomal form of TALENs expression during early embryonic development without genomic integration and maximized the disruption of the target gene, so that all miscarried and live-born monkeys carried mutations. In addition, when mRNA form of TALENs were injected into cynomolgus embryos in other study (Z. Liu et al., 2014), only one offspring harbored MECP2 mutations out of 5 neonates born, indicating that optimization of targeting TALENs construct, as well as delivery form of the constructs, were important factors for efficient TALEN-mediated mutagenesis approach. Human MECP2 mutations are known to be associated with an autism spectrum disorder (Rett syndrome, RTT) and hemizygous loss of function mutations of MECP2 in males leads to embryonic lethality. In this study, both male rhesus and cynomolgus MECP2 targeted fetuses were miscarried during mid-gestation, confirming its equivalent phenotype to RTT in human males (H. Liu et al., 2014). Later, the same group obtained additional TALEN-mediated MECP2 mutant cynomolgus monkeys (Y. Chen et al., 2017). Similar to previous results, all male mutants were embryonic lethal. Through an imaging, physiological and behavioral analysis of live female mutants, reductions of sub-regional brain volumes were noticed in longitudinal MRI with complicated behavioral abnormalities such as fragmented sleep, increased stereotypic behavior and reduced social interactions in eye tracking experiments. These symptoms are analogous to some of the clinical symptoms of RTT, making the MECP2 deficient mutant NHP valuable for studying the mechanisms of RTT and facilitating the development of new treatments.
Another example of the use of TALEN-mediated gene targeting to model human microcephaly was accomplished in cynomolgus monkeys. The MCPH1 gene was successfully targeted by TALENs mRNA injection into embryos (Ke et al., 2016). One live monkey carrying biallelic MCPH1 mutation was obtained in a single step and reduced MCPH1 expression was confirmed by western blot. Noticeable microcephaly with hypoplasia of the corpus callosum and upper limb spasticity in MCPH1 mutant monkey were similar to symptoms of autosomal recessive primary microcephaly (MCPH) patients, indicating its high potential to be a faithful model of the human disease.
In parallel with gene editing work in rhesus and cynomolgus monkeys, continuous efforts to generate marmoset models of human disease with disrupted expression of targeted genes have been ongoing. Using optimized ZFNs and TALENs targeting the marmoset IL2RG gene, mutant founder marmosets with severe combined immunodeficiency were generated (Sato et al., 2016). Nine of 21 neonates exhibiting mutations in the IL2RG gene were generated after multiple screening of ZFN and TALENs pairs for high targeting efficiency with low mosaic rate. The immunological analysis of IL2RG KO marmoset demonstrated typical immunodeficient phenotypes, including lack of thymus, reduction in the number of T cells and natural killer cells in blood samples collected from the umbilical cord, and gradual decrease of B cells in peripheral blood, all of which mimic clinical symptoms of human X-SCID (X-linked severe combined immunodeficiency) patients. These IL2RG KO marmosets demonstrated the feasibility of employing ZFN/TALEN mediated genome editing in NHP, and can serve as a valuable model for understanding function of the IL2RG gene during the thymic development in primates.
Our own ongoing effort for generation and utilization of genetically modified marmosets
Recently, we generated transgenic marmosets expressing GCaMP under the regulation of either ubiquitous (CMV) or neuron-specific (hSyn1) promoters by lentivirus based transgenesis (Park et al., 2016). The GCaMP protein is one of the GFP-based genetically encoded calcium indicators, which works as a visible marker of cellular calcium dynamics, allowing monitoring of neuronal activity in vitro and in vivo (T. W. Chen et al., 2013). Eight founder transgenic marmosets were born and shown to have stable and functional GCaMP expression in several different tissues. Among these 8 newborns, 3 died of unknown causes within a few hours to 15 days after birth. The remaining 5 founders (3 males, 2 females) developed normally. Successful germline transmission of the transgene was confirmed in F1 marmosets generated from GCaMP transgenic founders of both genders and both CMV and hSyn promoters (Figure 1). We are currently pursuing the establishment of a cohort of GCaMP transgenic marmosets, owing to their short intergeneration time and prolific reproductive capacity (Tardif et al., 2003). Various neuroimaging modalities, including anatomical (MRI) and functional magnetic resonance imaging (fMRI) together with two-photon laser scanning microscopy and electrophysiology have been applied to study the marmoset brain, which will be eventually used to better understanding of corresponding functional network of the human brain (Huang, Merson, & Bourne, 2016; Silva, 2017). Fully grown GCaMP transgenic marmosets have been trained for ongoing awake fMRI studies (Belcher et al., 2016; Hirano et al., 2018; Silva, 2017; Yen, Papoti, & Silva, 2018) and we are currently working on developing multimodal neuroimaging studies to unveil the functional brain networks in marmoset. We believe these transgenic marmosets will be invaluable NHP models in neuroscience, allowing the monitoring of neural activity in awake behaving animals with functional confocal and multi-photon optical microscopy imaging of intracellular calcium dynamics (Santisakultarm et al., 2016).
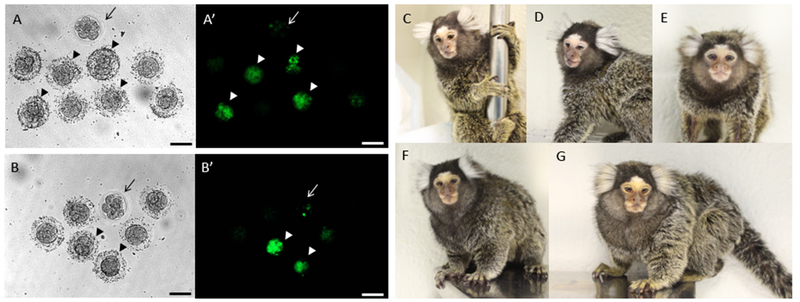
Figure 1:
Confirmation of germline transmission in GCaMP transgenic marmosets. Bright field images (A, B) and corresponding epifluorescence images (A’, B’) of F1 embryos obtained by collecting oocytes from founder marmoset TG-Y (A-A’, CMV-GCaMP5g) or from founder marmoset TG-E (B-B’, CMV-mKO-GCaMP6s) (Park et al., 2016). The oocytes were matured (IVM) and fertilized (IVF) in vitro using sperm from a wild-type marmoset. Some of the resulting embryos expressed GCaMP5g (A’, arrowheads) or GCaMP6s (B’, arrowheads) and were selected for embryo transfer into surrogate females. Wild-type embryos are shown as a control for fluorescence (A-A’, B-B’, arrows). Successful surrogate pregnancies were carried out to term and F1 transgenic marmosets were born. From founder marmoset TG-E (CMV-mKO-GCaMP6s): marmosets TG-R (male, C) and TG-L (female, D). From founder marmoset TG-Y (CMV-GCaMP5g): marmosets TG-F (male, E) and TG-V (male, F). F1 marmoset TG-A (female, G) was obtained by using sperm collected from founder marmoset TG-L (male, hSyn-mKO-GCaMP6s) to fertilize wild-type oocytes.
Concurrently, we are also pursuing development of marmoset models of neurovascular disorders using the advantages of the newest engineered nuclease technologies, in particular the CRISPR/Cas9 system. Successful generation of CRISPR/Cas9 targeted marmoset models have not been reported yet, but marmoset gene targeting with optimized gRNA and CRISPR/Cas9 has been effective in our experience. We are targeting the marmoset notch3 gene to model the CADASIL, a monogenetic cerebral small vessel disease caused by mutations in the notch homolog protein 3 (notch3) (Di Donato et al., 2017; Rutten et al., 2014). Cerebral small vessel disease is an important cause of stroke, cognitive impairment and disability in the elderly and CADASIL provides a unique inheritable model for the study of the most prevalent forms of sporadic small vessel disease (Di Donato et al., 2017). Specific CRISPR gRNAs for targeting the marmoset notch3 gene were designed and gRNA/Cas9 protein complexes were microinjected into the cytoplasm of single cell stage marmoset embryos, resulting in successful mutations in the target region, as confirmed by both T7E1 assay and sanger sequencing of PCR amplicons spanning the targeted exon. We believe that successful development of a notch3 mutant marmoset will lead to a better understanding of the physiological functions of notch3 and give significant insight into the pathogenesis of CADASIL. Development of marmoset model of CADASIL will constitute a useful model for developing novel and effective therapies as well as extending the applicability of CRISPR/Cas9 mediated gene editing for future production of marmoset models of human diseases.
Future perspectives
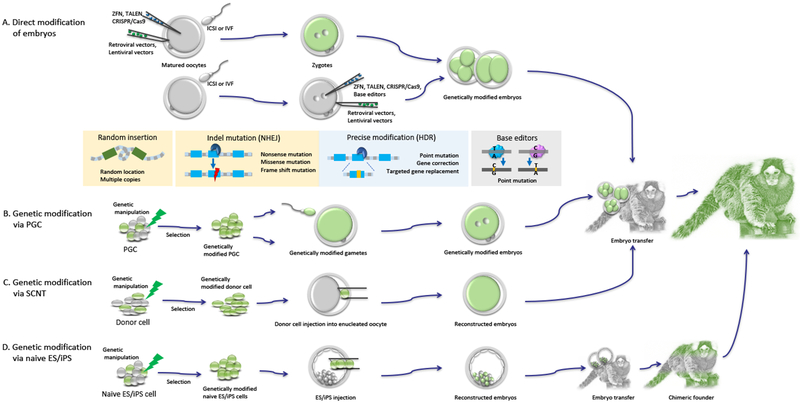
Significant progress has been made in the past 10 years in the development of genetically engineered NHP, bringing great hope to the ability of NHP to bridge the gap between promising, yet incomplete or inadequate rodent-based research, and their use in clinical practice. Figure 2 illustrates current and future approaches to generate genetically modified NHP. However, several challenges still remain before these NHP models can be made widely available.
Figure 2:
Approaches to generate genetically modified NHP: (A) Injection of viral vectors into either oocytes or embryos results in random insertion and integration of multiple copies of the transgene into the genome with high efficiency, but it is not applicable for targeted gene modification. Programmable nucleases such as ZFN, TALEN, and CRISPR/Cas9 are injected into fertilized oocytes to directly modify the embryo genome. (B-D) Precise targeted genetic manipulations of either PGC (B), somatic cells (C) or ES/iPS (D) are performed in cell cultures and the cells carrying the desired mutation are selected for production of genetically engineered NHP. See text for details. PGC: Primordial Germ Cells; SCNT: Somatic Cell Nuclear Transfer; ES: Embryonic Stem cells; IPS: Induced Pluripotent Stem cells; ICSI: Intracytoplasmic Sperm Injection; IVF: In Vitro Fertilization. ZFN: Zinc Finger Nuclease; TALEN: Transcription Activator-Like Effector Nuclease; CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats; Cas9:CRISPR ASsociated protein 9.
The first challenge is the long time necessary to generate genetically engineered NHP. The availability of oocytes or embryos necessary to generate and/or expand genetically modified NHP is significantly limited by the availability of donor females. Second, genetic modifications are inherently inefficient techniques. While viral vector mediated transgenesis (Table 1) and TALENs or CRISPR/Cas9 mediated KO (Table 2) have been successfully demonstrated in NHP, more precise and complicated genome modifications, such as targeted gene knock-in (KI) and making or repairing point mutations without occurrence of genetic mosaicism, remain impractical in NHP, largely due to the low efficiency of these techniques. This can be partially overcome by developing methods to screen and select genetically manipulated embryos prior to implantation. However, embryo biopsy techniques still need to be optimized in NHP. The third challenge is in the number of recipient females needed for implantation of genetically modified NHP. In most reports to date, the number of positive pregnancies per embryo transfer ranges between 7.6% and 75%, depending on the species (Tables 1 and 2). And of those, only a fraction turns into live births. Adding to the above difficulties the long gestation time, low number of offspring and the much longer maturation time of NHP relative to rodents, the production of genetically engineered NHP is a task only possible in few select institutions around the globe.
Another potential approach for making genetically modified animal is to perform gene modification in embryonic stem cells (ESC) or iPSC followed by selection of appropriately modified stem cells via genetic screening which will then develop into chimeric gene edited offspring. Reminiscent of the investigations on conversion of human and monkey ESC into naïve like state (Fang et al., 2014; Hackett & Surani, 2014; Takashima et al., 2014), generation of chimeric cynomolgus fetuses were demonstrated using naïve-like state induced ESCs introduced into host embryos (Y. Chen, Niu, et al., 2015). This approach could pave the way for the production of a new gene edited NHP models of human disease. However, the long time necessary for generation of chimeras, verification of germline transmission and chimeric cleanup by breeding is a major impediment that needs to be overcome. Derivation of germ cells from ESC or iPS holds the potential to yield the gene edited gametes for genetically engineered NHP, but techniques for maintaining stable cultures of NHP germ cell lineages, including primordial germ cells, spermatogonial stem cells and oogonial stem cells, still need to be developed and optimized (Izpisua Belmonte et al., 2015).
The successful generation of cloned cynomolgus macaques by somatic cell nuclear transfer (SCNT) using both fetal fibroblasts and adult cumulus cells has just been reported (Liu et al., 2018). This follows many years of pioneering efforts, including production of rhesus macaques from blastomere transferred embryos (Meng, Ely, Stouffer, & Wolf, 1997), establishment of rhesus ESC derived from SCNT embryos (Byrne et al., 2007), and modification of the SCNT approach for improved nuclear remodeling (Mitalipov et al., 2007; Sparman, Tachibana, & Mitalipov, 2010). In vitro screening of gene-edited donor cells prior to SCNT will circumvent the problems related to off-targeting and mosaicism, as well as open up opportunities for the introduction of predefined genetic modifications that are not feasible in direct genetic manipulation of embryos. Nevertheless, it is important to consider the drawbacks associated with cloning, in particular very low cloning efficiency, which will be challenging to overcome without ready access to a large number of oocytes and recipient females. Other challenges to SCNT include low survival rates, as evidenced by reports of associated birth defects, abortions and early postnatal death (Keefer, 2015; Long, Westhusin, & Golding, 2014; Tan, Proudfoot, Lillico, & Whitelaw, 2016). Further research is necessary to fully develop SCNT techniques to allow them to be effectively and efficiently used to produce genetically modified NHP.
Motivated by the unmet need for the achievement of efficient strategies to introduce or correct point mutations via homology directed repair pathway, new approaches that enable the direct and irreversible conversion of target DNA base are currently under development. The catalytically dead CRISPR/dCas9 was engineered to fuse with a cytidine deaminase (cytidine base editor) or with an adenine deaminase enzyme (adenine base editor), which mediates the targeted base conversion of C to T and T to C, respectively, without requiring a donor template (Gaudelli et al., 2017; Komor, Kim, Packer, Zuris, & Liu, 2016). This system has proven to be an efficient method for generation of targeted point mutations in various species, including plants, mammalian cells, mice and even human embryos (Hess et al., 2016; K. Kim et al., 2017; G. Li et al., 2017; Liang et al., 2017; Y. Ma et al., 2016; Shimatani et al., 2017). The use of base editors could offer an attractive strategy for making various NHP models with single amino acid substitutions and for correcting genetic defects in NHP in the future.
Conclusions
In summary, there’s a bright and promising future for the generation of transgenic and gene-edited NHP. NHP have an essential role in the study of human diseases and for the development of effective therapeutic strategies, particularly in neurological disorders, where aging is a major factor (Chan et al., 2015; Mattison & Vaughan, 2017; Salmon, 2016; Tardif et al., 2011), and in neuropsychiatric disorders that affect cognitive function and behavior (Chan, 2013; Chan et al., 2015; Liu et al., 2016). Much of the initial work to adopt and adapt genetic and gene-editing tools to NHP was done in macaques (Chan, 2013; Chan et al., 2001). However, marmosets offer several advantages as an ideal NHP species for the development of genetically engineered lines (Izpisua Belmonte et al., 2015; Okano & Kishi, 2018; Okano, Miyawaki, & Kasai, 2015), in particular, an intergeneration time and establishment of transgenic lines two-three times faster than in macaques (Park et al., 2016; Sasaki et al., 2009). This is as important a factor as the continued development of efficient genetic tools adapted to NHP. Another factor of major importance in the continued development of genetically modified NHP models of neurological and neuropsychiatric disorders is the support of the general public and of funding agencies, such as the U.S. National Institutes of Health and its Primate Research Centers, the Japanese Consortium BRAIN/Minds – which uses the marmoset as the main animal model for neuroscience research (Okano et al., 2015; Okano et al., 2016), and the recent but very accelerated development of NHP research in China (Liu et al., 2018). While much of the applications of genetically engineered NHP to biomedical research are only now emerging, the relatively few reports to date show that this effort has a tremendous potential to impact our understanding of human disease and to advance the development of effective therapies.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke (Alan P. Koretsky, Scientific Director). This research complies with AJP’s policies on ethical research and treatment of non-human primates. This research adhered to the American Society of Primatologists (ASP) Principles for the Ethical Treatment of Non-Human Primates. This research complied with protocols approved by the NINDS/NIDCD/NCCIH ACUC and adhered to all the legal requirements of the USA.
References
- Bak RO, Gomez-Ospina N, & Porteus MH (2018). Gene Editing on Center Stage. Trends Genet. doi: 10.1016/j.tig.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Bales KR (2012). The value and limitations of transgenic mouse models used in drug discovery for Alzheimer’s disease: an update. Expert Opin Drug Discov, 7(4), 281–297. doi: 10.1517/17460441.2012.666234 [DOI] [PubMed] [Google Scholar]
- Belcher AM, Yen CC, Notardonato L, Ross TJ, Volkow ND, Yang Y, Tomasi D (2016). Functional Connectivity Hubs and Networks in the Awake Marmoset Brain. Front Integr Neurosci, 10, 9. doi: 10.3389/fnint.2016.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Mitalipov SM (2007). Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature, 450(7169), 497–502. doi: 10.1038/nature06357 [DOI] [PubMed] [Google Scholar]
- Carter RL, & Chan AW (2012). Pluripotent stem cells models for Huntington’s disease: prospects and challenges. J Genet Genomics, 39(6), 253–259. doi: 10.1016/j.jgg.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RL, Chen Y, Kunkanjanawan T, Xu Y, Moran SP, Putkhao K, Chan AW (2014). Reversal of cellular phenotypes in neural cells derived from Huntington’s disease monkey-induced pluripotent stem cells. Stem Cell Reports, 3(4), 585–593. doi: 10.1016/j.stemcr.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW (2004). Transgenic nonhuman primates for neurodegenerative diseases. Reprod Biol Endocrinol, 2, 39. doi: 10.1186/1477-7827-2-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW (2013). Progress and prospects for genetic modification of nonhuman primate models in biomedical research. ILAR J, 54(2), 211–223. doi: 10.1093/ilar/ilt035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Cheng PH, Neumann A, & Yang JJ (2010). Reprogramming Huntington monkey skin cells into pluripotent stem cells. Cell Reprogram, 12(5), 509–517. doi: 10.1089/cell.2010.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Chong KY, Martinovich C, Simerly C, & Schatten G (2001). Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science, 291(5502), 309–312. doi: 10.1126/science.291.5502.309 [DOI] [PubMed] [Google Scholar]
- Chan AW, Jiang J, Chen Y, Li C, Prucha MS, Hu Y, Bachevalier J (2015). Progressive cognitive deficit, motor impairment and striatal pathology in a transgenic Huntington disease monkey model from infancy to adulthood. PLoS One, 10(5), e0122335. doi: 10.1371/journal.pone.0122335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Luetjens CM, Dominko T, Ramalho-Santos J, Simerly CR, Hewitson L, & Schatten G (2000). Foreign DNA transmission by ICSI: injection of spermatozoa bound with exogenous DNA results in embryonic GFP expression and live rhesus monkey births. Mol Hum Reprod, 6(1), 26–33. [DOI] [PubMed] [Google Scholar]
- Chan AW, Xu Y, Jiang J, Rahim T, Zhao D, Kocerha J, Bachevalier J (2014). A two years longitudinal study of a transgenic Huntington disease monkey. BMC Neurosci, 15, 36. doi: 10.1186/1471-2202-15-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499(7458), 295–300. doi: 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Carter RL, Cho IK, & Chan AW (2014). Cell-based therapies for Huntington’s disease. Drug Discov Today, 19(7), 980–984. doi: 10.1016/j.drudis.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cui Y, Shen B, Niu Y, Zhao X, Wang L, Huang X (2015). Germline acquisition of Cas9/RNA-mediated gene modifications in monkeys. Cell Res, 25(2), 262–265. doi: 10.1038/cr.2014.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Niu Y, & Ji W (2012). Transgenic nonhuman primate models for human diseases: approaches and contributing factors. J Genet Genomics, 39(6), 247–251. doi: 10.1016/j.jgg.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Chen Y, Niu Y, Li Y, Ai Z, Kang Y, Shi H, Li T (2015). Generation of Cynomolgus Monkey Chimeric Fetuses using Embryonic Stem Cells. Cell Stem Cell, 17(1), 116–124. doi: 10.1016/j.stem.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, Sun YE (2017). Modeling Rett Syndrome Using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys. Cell, 169(5), 945–955 e910. doi: 10.1016/j.cell.2017.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Li XJ (2015). Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet, 24(13), 3764–3774. doi: 10.1093/hmg/ddv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D (2009). Marmoset model takes centre stage. Nature, 459(7246), 492. doi: 10.1038/459492a [DOI] [PubMed] [Google Scholar]
- Cyranoski D (2014). Marmosets are stars of Japan’s ambitious brain project. Nature, 514(7521), 151–152. doi: 10.1038/514151a [DOI] [PubMed] [Google Scholar]
- Di Donato I, Bianchi S, De Stefano N, Dichgans M, Dotti MT, Duering M, Federico A (2017). Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy (CADASIL) as a model of small vessel disease: update on clinical, diagnostic, and management aspects. BMC Med, 15(1), 41. doi: 10.1186/s12916-017-0778-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, & Charpentier E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096. doi: 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Elder GA, Gama Sosa MA, & De Gasperi R (2010). Transgenic mouse models of Alzheimer’s disease. Mt Sinai J Med, 77(1), 69–81. doi: 10.1002/msj.20159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Deng H (2014). Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell, 15(4), 488–497. doi: 10.1016/j.stem.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, & Liu DR (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature, 551(7681), 464–471. doi: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, & Surani MA (2014). Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell, 15(4), 416–430. doi: 10.1016/j.stem.2014.09.015 [DOI] [PubMed] [Google Scholar]
- Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, Bassik MC (2016). Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods, 13(12), 1036–1042. doi: 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Yen CC, Liu JV, Mackel JB, Merkle H, Nascimento GC, Silva AC (2018). Investigation of the BOLD and CBV fMRI responses to somatosensory stimulation in awake marmosets (Callithrix jacchus). NMR Biomed, 31(3). doi: 10.1002/nbm.3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Merson TD, & Bourne JA (2016). In vivo whole brain, cellular and molecular imaging in nonhuman primate models of neuropathology. Neurosci Biobehav Rev, 66, 104–118. doi: 10.1016/j.neubiorev.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Zhang F (2015). Brains, genes, and primates. Neuron, 86(3), 617–631. doi: 10.1016/j.neuron.2015.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, & Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zheng B, Shen B, Chen Y, Wang L, Wang J, Huang X (2015). CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Hum Mol Genet, 24(25), 7255–7264. doi: 10.1093/hmg/ddv425 [DOI] [PubMed] [Google Scholar]
- Ke Q, Li W, Lai X, Chen H, Huang L, Kang Z, Xiang AP (2016). TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res, 26(9), 1048–1061. doi: 10.1038/cr.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer CL (2015). Artificial cloning of domestic animals. Proc Natl Acad Sci U S A, 112(29), 8874–8878. doi: 10.1073/pnas.1501718112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Kim JS (2017). Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol, 35(5), 435–437. doi: 10.1038/nbt.3816 [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, & Chandrasegaran S (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A, 93(3), 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, & Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533(7603), 420–424. doi: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp J, Di Marzo A, & Golos T (2017). Assisted reproductive technologies in the common marmoset: an integral species for developing nonhuman primate models of human diseases. Biol Reprod, 96(2), 277–287. doi: 10.1095/biolreprod.116.146514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampreht Tratar U, Horvat S, & Cemazar M (2018). Transgenic Mouse Models in Cancer Research. Front Oncol, 8, 268. doi: 10.3389/fonc.2018.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MA (2000). Nonhuman primate models in biogerontology. Exp Gerontol, 35(5), 533–541. [DOI] [PubMed] [Google Scholar]
- Li G, Liu Y, Zeng Y, Li J, Wang L, Yang G, Liu J (2017). Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell, 8(10), 776–779. doi: 10.1007/s13238-017-0458-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, & Yang B (2011). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res, 39(1), 359–372. doi: 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Ding C, Sun H, Xie X, Xu Y, Zhang X, Huang J (2017). Correction of beta-thalassemia mutant by base editor in human embryos. Protein Cell, 8(11), 811–822. doi: 10.1007/s13238-017-0475-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Ji W (2014). TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell, 14(3), 323–328. doi: 10.1016/j.stem.2014.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Sun Q (2018). Cloning of Macaque Monkeys by Somatic Cell Nuclear Transfer. Cell, 172(4), 881–887 e887. doi: 10.1016/j.cell.2018.01.020 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, Qiu Z (2016). Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature, 530(7588), 98–102. doi: 10.1038/nature16533 [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou X, Zhu Y, Chen ZF, Yu B, Wang Y, Qiu Z (2014). Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neurosci Bull, 30(3), 381–386. doi: 10.1007/s12264-014-1434-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CR, Westhusin ME, & Golding MC (2014). Reshaping the transcriptional frontier: epigenetics and somatic cell nuclear transfer. Mol Reprod Dev, 81(2), 183–193. doi: 10.1002/mrd.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Mitalipov S (2017). Correction of a pathogenic gene mutation in human embryos. Nature, 548(7668), 413–419. doi: 10.1038/nature23305 [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhang J, Yin W, Zhang Z, Song Y, & Chang X (2016). Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods, 13(12), 1029–1035. doi: 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]
- Mansfield K (2003). Marmoset models commonly used in biomedical research. Comp Med, 53(4), 383–392. [PubMed] [Google Scholar]
- Mattison JA, & Vaughan KL (2017). An overview of nonhuman primates in aging research. Exp Gerontol, 94, 41–45. doi: 10.1016/j.exger.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ely JJ, Stouffer RL, & Wolf DP (1997). Rhesus monkeys produced by nuclear transfer. Biol Reprod, 57(2), 454–459. [DOI] [PubMed] [Google Scholar]
- Mitalipov SM, Zhou Q, Byrne JA, Ji WZ, Norgren RB, & Wolf DP (2007). Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Hum Reprod, 22(8), 2232–2242. doi: 10.1093/humrep/dem136 [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, Scheibye-Knudsen M, Longo DL, & de Cabo R (2015). Animal models of aging research: implications for human aging and age-related diseases. Annu Rev Anim Biosci, 3, 283–303. doi: 10.1146/annurev-animal-022114-110829 [DOI] [PubMed] [Google Scholar]
- Moran S, Chi T, Prucha MS, Ahn KS, Connor-Stroud F, Jean S, Chan AW (2015). Germline transmission in transgenic Huntington’s disease monkeys. Theriogenology, 84(2), 277–285. doi: 10.1016/j.theriogenology.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Guo X, Chen Y, Wang CE, Gao J, Yang W, Li XJ (2015). Early Parkinson’s disease symptoms in alpha-synuclein transgenic monkeys. Hum Mol Genet, 24(8), 2308–2317. doi: 10.1093/hmg/ddu748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Sha J (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156(4), 836–843. doi: 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- Niu Y, Yu Y, Bernat A, Yang S, He X, Guo X, Ji W (2010). Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proc Natl Acad Sci U S A, 107(41), 17663–17667. doi: 10.1073/pnas.1006563107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, & Kishi N (2018). Investigation of brain science and neurological/psychiatric disorders using genetically modified non-human primates. Curr Opin Neurobiol, 50, 1–6. doi: 10.1016/j.conb.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Okano H, Miyawaki A, & Kasai K (2015). Brain/MINDS: brain-mapping project in Japan. Philos Trans R Soc Lond B Biol Sci, 370(1668). doi: 10.1098/rstb.2014.0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Sasaki E, Yamamori T, Iriki A, Shimogori T, Yamaguchi Y, Miyawaki A (2016). Brain/MINDS: A Japanese National Brain Project for Marmoset Neuroscience. Neuron, 92(3), 582–590. doi: 10.1016/j.neuron.2016.10.018 [DOI] [PubMed] [Google Scholar]
- Park JE, Zhang XF, Choi SH, Okahara J, Sasaki E, & Silva AC (2016). Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci Rep, 6, 34931. doi: 10.1038/srep34931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML, Ross CN, Schulkin J, Ziegler TE, & Tardif SD (2013). Metabolic consequences of the early onset of obesity in common marmoset monkeys. Obesity (Silver Spring), 21(12), E592–598. doi: 10.1002/oby.20462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkhao K, Kocerha J, Cho IK, Yang J, Parnpai R, & Chan AW (2013). Pathogenic cellular phenotypes are germline transmissible in a transgenic primate model of Huntington’s disease. Stem Cells Dev, 22(8), 1198–1205. doi: 10.1089/scd.2012.0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesche L, Tardif SD, Ross CN, deMartelly VA, Ziegler T, & Rutherford JN (2018). The common marmoset monkey: avenues for exploring the prenatal, placental, and postnatal mechanisms in developmental programming of pediatric obesity. Am J Physiol Regul Integr Comp Physiol, 314(5), R684–R692. doi: 10.1152/ajpregu.00164.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten JW, Haan J, Terwindt GM, van Duinen SG, Boon EM, & Lesnik Oberstein SA (2014). Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev Mol Diagn, 14(5), 593–603. doi: 10.1586/14737159.2014.922880 [DOI] [PubMed] [Google Scholar]
- Salmon AB (2016). Moving toward ‘common’ use of the marmoset as a non-human primate aging model. Pathobiol Aging Age Relat Dis, 6, 32758. doi: 10.3402/pba.v6.32758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisakultarm TP, Kersbergen CJ, Bandy DK, Ide DC, Choi SH, & Silva AC (2016). Two-photon imaging of cerebral hemodynamics and neural activity in awake and anesthetized marmosets. J Neurosci Methods, 271, 55–64. doi: 10.1016/j.jneumeth.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Nomura T (2009). Generation of transgenic non-human primates with germline transmission. Nature, 459(7246), 523–527. doi: 10.1038/nature08090 [DOI] [PubMed] [Google Scholar]
- Sato K, Oiwa R, Kumita W, Henry R, Sakuma T, Ito R, Sasaki E (2016). Generation of a Nonhuman Primate Model of Severe Combined Immunodeficiency Using Highly Efficient Genome Editing. Cell Stem Cell, 19(1), 127–138. doi: 10.1016/j.stem.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Schatten G, & Mitalipov S (2009). Developmental biology: Transgenic primate offspring. Nature, 459(7246), 515–516. doi: 10.1038/459515a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheikl T, Pignolet B, Mars LT, & Liblau RS (2010). Transgenic mouse models of multiple sclerosis. Cell Mol Life Sci, 67(23), 4011–4034. doi: 10.1007/s00018-010-0481-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita Y, Tsukiyama T, Iwatani C, Tsuchiya H, Matsushita J, Azami T, Ema M (2016). Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci Rep, 6, 24868. doi: 10.1038/srep24868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Kondo A (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol, 35(5), 441–443. doi: 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- Silva AC (2017). Anatomical and functional neuroimaging in awake, behaving marmosets. Dev Neurobiol, 77(3), 373–389. doi: 10.1002/dneu.22456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparman ML, Tachibana M, & Mitalipov SM (2010). Cloning of non-human primates: the road “less traveled by”. Int J Dev Biol, 54(11-12), 1671–1678. doi: 10.1387/ijdb.103196ms [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Smith A (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell, 158(6), 1254–1269. doi: 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Proudfoot C, Lillico SG, & Whitelaw CB (2016). Gene targeting, genome editing: from Dolly to editors. Transgenic Res, 25(3), 273–287. doi: 10.1007/s11248-016-9932-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, & Ziegler TE (2011). The marmoset as a model of aging and age-related diseases. ILAR J, 52(1), 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif SD, Smucny DA, Abbott DH, Mansfield K, Schultz-Darken N, & Yamamoto ME (2003). Reproduction in captive common marmosets (Callithrix jacchus). Comp Med, 53(4), 364–368. [PubMed] [Google Scholar]
- Tomioka I, Ishibashi H, Minakawa EN, Motohashi HH, Takayama O, Saito Y, Seki K (2017). Transgenic Monkey Model of the Polyglutamine Diseases Recapitulating Progressive Neurological Symptoms. eNeuro, 4(2). doi: 10.1523/ENEURO.0250-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka I, Nogami N, Nakatani T, Owari K, Fujita N, Motohashi H, Seki K (2017). Generation of transgenic marmosets using a tetracyclin-inducible transgene expression system as a neurodegenerative disease model. Biol Reprod, 97(5), 772–780. doi: 10.1093/biolre/iox129 [DOI] [PubMed] [Google Scholar]
- Vaughan KL, & Mattison JA (2016). Obesity and Aging in Humans and Nonhuman Primates: A Mini-Review. Gerontology, 62(6), 611–617. doi: 10.1159/000445800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Feng C, Teng F, Yang S, Hu B, Niu Y, Zhou Q (2015). One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res, 25(2), 258–261. doi: 10.1038/cr.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock DM (2010). The usefulness and challenges of transgenic mouse models in the study of Alzheimer’s disease. CNS Neurol Disord Drug Targets, 9(4), 386–394. [DOI] [PubMed] [Google Scholar]
- Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, Chan AW (2008). Towards a transgenic model of Huntington’s disease in a non-human primate. Nature, 453(7197), 921–924. doi: 10.1038/nature06975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CC, Papoti D, & Silva AC (2018). Investigating the spatiotemporal characteristics of the deoxyhemoglobin-related and deoxyhemoglobin-unrelated functional hemodynamic response across cortical layers in awake marmosets. Neuroimage, 164, 121–130. doi: 10.1016/j.neuroimage.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo E, Cai YJ, Li K, Wei Y, Wang BA, Sun Y, Yang H (2017). One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res, 27(7), 933–945. doi: 10.1038/cr.2017.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.