Abstract
Fatty acid esters of hydroxy fatty acids (FAHFAs) are lipid mediators with promising antidiabetic and anti-inflammatory properties that are formed in white adipose tissue (WAT) via de novo lipogenesis, but their biosynthetic enzymes are unknown. Using a combination of lipidomics in WAT, quantitative trait locus mapping, and correlation analyses in rat BXH/HXB recombinant inbred strains, as well as response to oxidative stress in murine models, we elucidated the potential pathway of biosynthesis of several FAHFAs. Comprehensive analysis of WAT samples identified ∼160 regioisomers, documenting the complexity of this lipid class. The linkage analysis highlighted several members of the nuclear factor, erythroid 2 like 2 (Nrf2)-mediated antioxidant defense system (Prdx6, Mgst1, Mgst3), lipid-handling proteins (Cd36, Scd6, Acnat1, Acnat2, Baat), and the family of flavin containing monooxygenases (Fmo) as the positional candidate genes. Transgenic expression of Nrf2 and deletion of Prdx6 genes resulted in reduction of palmitic acid ester of 9-hydroxystearic acid (9-PAHSA) and 11-PAHSA levels, while oxidative stress induced by an inhibitor of glutathione synthesis increased PAHSA levels nonspecifically. Our results indicate that the synthesis of FAHFAs via carbohydrate-responsive element-binding protein–driven de novo lipogenesis depends on the adaptive antioxidant system and suggest that FAHFAs may link activity of this system with insulin sensitivity in peripheral tissues.
Introduction
White adipose tissue (WAT) metabolism plays an important role in whole-body energy balance. Moreover, dysregulation of both glucose and lipid metabolism in this tissue underlies development of disease associated with obesity and type 2 diabetes. However, the mechanistic links between glucose and lipid metabolism in WAT of obese subjects and systemic insulin resistance are not fully explored. De novo lipogenesis (DNL) converts excess energy from carbohydrates to energy-dense neutral lipids in both the liver and WAT (1–4). Although DNL in the liver is usually associated with systemic insulin resistance, DNL in WAT correlates with insulin sensitivity (5–8). In addition to generating lipid stores, DNL might also serve as a source of signaling molecules. Within the last decade, a number of bioactive lipids produced in WAT (lipokines) via DNL have been described, including palmitoleic acid (PO) (7), alkyl ether lipids (9), and fatty acid esters of hydroxy fatty acids (FAHFAs) (6), which all promote insulin sensitivity and ameliorate insulin resistance. Namely, FAHFAs represent a new immunometabolic link between DNL in WAT and systemic metabolism due to multiple beneficial effects of these extremely potent lipokines on both glucose–insulin homeostasis and inflammation as observed in insulin-resistant mice as well as humans, while their reduced levels may contribute to diabetes risk (6,10–12).
FAHFAs consist of a fatty acid (e.g., palmitic acid [PA]) esterified to the hydroxyl group of a hydroxy fatty acid (e.g., hydroxystearic acid [HSA]), abbreviated as PAHSA, and the position of the branching carbon defines a regioisomer (e.g., 9-PAHSA). There are several regioisomer families derived from PA, PO, stearic acid (SA), oleic acid (OA), linoleic acid (LA), and docosahexaenoic acid (DHA) with tissue-specific distribution documented so far (6,10,11,13–15). WAT represents a major site of carbohydrate-responsive element-binding protein (ChREBP)-dependent FAHFA synthesis (6,11,16). Serine hydrolase carboxyl ester lipase (17,18) and threonine hydrolases (19) were identified as FAHFA-metabolizing enzymes, but the biosynthetic enzymes involved are unknown (18). Given the low susceptibility of saturated acyl chains to oxidation (20), the complexity of the FAHFA family, and their stereospecific metabolism (10,21), a combination of nonenzymatic and enzymatic reactions is probably involved in FAHFA biosynthesis. Although all FAHFAs are structurally related, they are probably biologically as heterogenous as eicosanoids—they are composed of different fatty acids, and even the regioisomers within one family respond differently to physiological stimulations (6,11). Therefore, there could be individual genes/proteins responsible for the differences among regioisomers (substrate specificity toward the branching position and/or acyl chain) as well as genes responsible for major regulation of the whole pathway (e.g., ChREBP [16]).
In this study, the BXH/HXB recombinant inbred (RI) strains, derived from the spontaneously hypertensive rat (SHR) strain and the normotensive Brown Norway (BN) strain (22), were used for linkage and correlational analyses of physiological traits, lipidomics data, and gene expression levels to investigate potential candidate genes involved in FAHFA biosynthesis.
Research Design and Methods
Materials and Reagents
All chemicals were purchased from Sigma-Aldrich (Prague, Czech Republic) unless stated otherwise. FAHFA standards (5-,9-,10-,12-, and 13-PAHSA, OAHSA, and SAHSA as well as 13C4-9-PAHSA) were purchased from Cayman Pharma (Neratovice, Czech Republic), and 5-, 7-, and 9-OAHPA; 7-OAHSA; 5-, 7-, and 9-PAHPA; 13-DHAHLA; and 12-DHAHOA were synthesized as previously described (11,13). PA, PO, SA, OA, LA, and DHA are the acids defined above, AA is arachidonic acid, and the H prefix stands for hydroxy fatty acid.
Animals
BXH/HXB RI strains (n = 30) were derived from SHR/OlaIpcv (referred to as the SHR) and BN-Lx/Cub (referred to as the BN) progenitor strains (22). Nuclear factor, erythroid 2 like 2 (Nfe2l2, also known as Nrf2) transgenic SHR (23) (referred to as the SHR-Nrf2) were derived by microinjecting SHR zygotes with the mix of Sleeping Beauty construct containing mouse Nrf2 cDNA under control of the SV universal promoter and mRNA of the SB100× transposase (23). Genotyping of positive rats was done by PCR with the following primers: mNrf2-413F, GCA ACT CCA GAA GGA ACA GG, and mNrf2-573R, AGG CAT CTT GTT TGG GAA TG (23). All rats were housed in an air-conditioned animal facility at 22°C and allowed free access to food and water. Lipidomic, biochemical, and metabolic phenotypes were assessed in 6-week-old 14-h fasted male rats that were raised on standard laboratory chow (Altromin, Germany). Animals were killed by cervical dislocation in a fasted state (food removed at 6 p.m., animals killed between 8 a.m. and 9 a.m. the next day), and samples of epididymal WAT were stored in liquid nitrogen to prevent degradation until assay, as the samples from specific strains were collected during a period of 6 months. To explore the interaction between metabolic status and Nrf2, SHR-Nrf2 and SHR males (n = 7) were fasted for 24 h (9 a.m. to 9 a.m.) followed or not by refeeding chow for 6 h, and WAT was dissected and stored as described above; eventually, metabolipidomic, proteomic, gene expression, and FAHFA analyses were performed as previously described (11,24). Oral glucose tolerance test (OGTT) was performed using a glucose load of 300 mg/100 g body weight after overnight fasting. Blood was drawn from the tail without anesthesia before the glucose load (0 min time point) and at 30, 60, and 120 min thereafter. To characterize the role of redox status, 16 C57BL/6J adult male mice (The Jackson Laboratory, Bar Harbor, ME) were randomly divided into two groups: untreated mice or mice treated with buthionine sulfoximine (BSO), an inhibitor of glutathione (GSH) synthesis. BSO was supplied in drinking water at the final concentration of 20 mmol/L for 3 days. All mice were fed standard laboratory chow (extruded ssniff R/M-H; ssniff Spezialdiaten, Soest, Germany). Animals were killed in the random-fed state under ether anesthesia, and epididymal WAT was dissected and stored in liquid nitrogen till the analysis. The experiments were conducted in agreement with the Animal Protection Law of the Czech Republic and were approved by the Ethics Committee of the Institute of Physiology, Czech Academy of Sciences, Prague.
Wild-type C57BL/6J mice (The Jackson Laboratory) and peroxiredoxin 6 (Prdx6) null mice on the C57BL/6J background (25) were maintained and killed in a similar manner to that described for the experiments above. Epididymal WAT of adult animals was dissected and stored at −80°C till the analysis. Use of the mice was approved by the University of Pennsylvania Animal Care and Use Committee. Thiobarbituric acid reactive substances and 8-isoprostane were measured using kits from Cayman chemicals (Ann Arbor, MI) (26).
FAHFA Analysis
Analysis of FAHFAs was performed as previously described (10,11) and optimized for a large set of samples. Aliquots of 50–70 mg of WAT were homogenized using a MM400 bead mill (Retsch, Haan, Germany), chilled to −20°C in a mixture of citric acid buffer and ethyl acetate (27), and further extracted with ethyl acetate (1:3 final ratio). An internal standard 13C4-9-PAHSA was added to the homogenate (1 ng/sample). The organic phase was collected, dried in a SpeedVac (Savant SPD121P; Thermo Fisher Scientific), resuspended in dichloromethane, and applied on HyperSep SPE columns (500 mg/10 mL, 40–60 µm, 70Å) (Thermo Fisher Scientific). FAHFAs were eluted from the SPE columns with ethyl acetate, concentrated using a SpeedVac, resuspended in methanol, and immediately analyzed using liquid chromatography–mass spectrometry (LC-MS) as follows.
Chromatographic separation was performed in an ultra performance LC UltiMate 3000 RSLC (Thermo Fisher Scientific) equipped with a YMC-Triart C18 ExRS 1.9-µm, 2.1 × 150 mm column (YMC, Kyoto, Japan). The flow rate was 200 µL/min at 50°C using gradient elution: solvent A (70% water, 30% acetonitrile, 0.01% acetic acid, pH 4) and solvent B (50% acetonitrile, 50% isopropanol) for 1 min (100% solvent A), 5 min (10% solvent A), 23 min (10% solvent A), and 25 min (100% solvent A), while a linear gradient was maintained between the steps. Ultra performance LC was coupled to a QTRAP 5500/SelexION mass spectrometer (Sciex, Framingham, MA). FAHFAs were detected in negative electrospray mode using multiple reaction monitoring (Supplementary Table 1) and MS/MS/MS as previously described (11).
Statistical Analyses
Linkage and correlation analyses of WAT FAHFA levels in BXH/HXB RI strains were performed using Matrix eQTL (28) to identify quantitative trait loci (QTLs) at genome-wide statistical significance at P < 0.05 and false discovery rate (FDR) <0.1. To identify the best candidate genes, we used biological knowledge–driven analysis using databases such as Rat Genome Database (RGD) (rgd.mcw.edu) and PubMed. Specifically, within each identified genomic position (± 3 Mbp), we have also searched for any protein linked to 1) lipid metabolism and an ability to handle acyl chain, 2) peroxidation or oxidation of lipids, and 3) formation of reactive oxygen species (ROS) based on data from RGD (accessed 21 November 2017). Substrate specificity of candidate proteins and their potential relevance to FAHFA metabolism were further explored via PubMed search for primary publications (gene or protein name, accessed 21 November 2017). The GeneNetwork database includes RI strains gene expression data in tissues relevant to the pathogenesis of metabolic syndrome (kidney, left ventricle, epididymal fat, brown fat, hippocampus, and liver) (29). In addition, the database also includes parameters of glucose and lipid metabolism and was used to explore a link of FAHFAs to these traits (30).
The data in transgenic rats and mouse experiments are expressed as means ± SEM. Statistical analysis was performed with SigmaStat. For most experiments, unpaired two-tailed Student t test was used to determine statistical significance among two groups and P < 0.05 was considered significant. Two-way ANOVA was used for the refeeding experiment (Fig. 5).
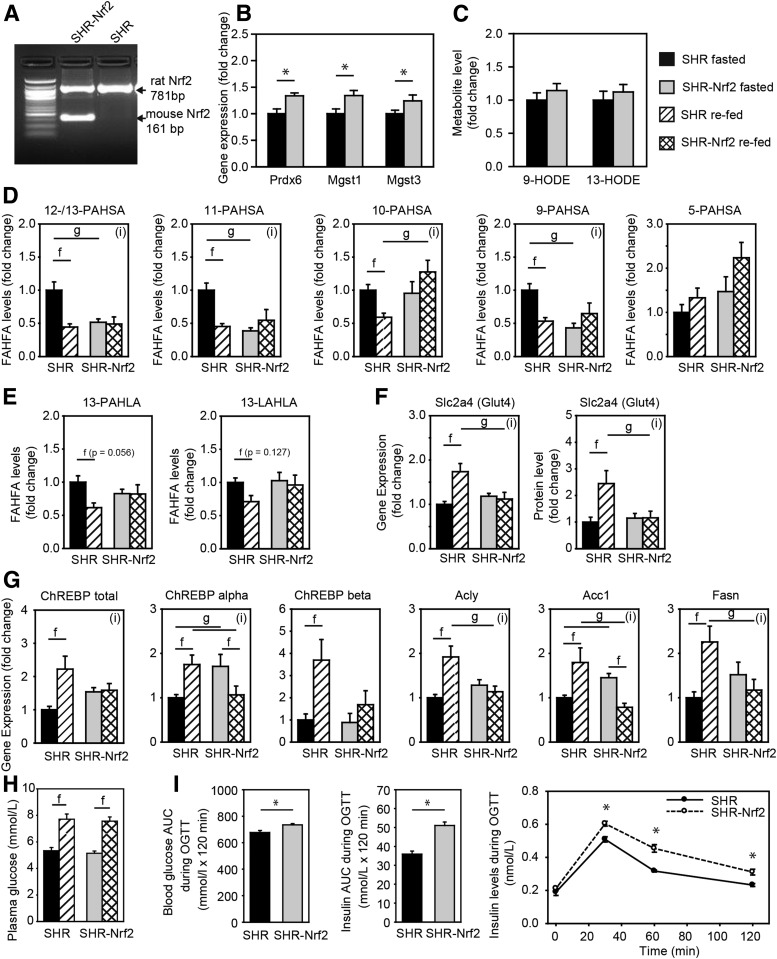
Figure 5.
Effect of Nrf2 overexpression on DNL in WAT. SHR wild-type and SHR-Nrf2 transgenic rats were fasted for 24 h and either sacrificed or refed chow for 6 h. A: Transgenic expression of murine Nrf2 in WAT. B: Expression of positional candidate genes. C: Levels of 9- and 13-HODE (also known as hydroxylinoleic acid [HLA]) in WAT. D: Levels of PAHSA regioisomers. E: Levels of HLA-derived FAHFAs. F: Gene expression and protein levels (MS label-free quantitation) of Glut4 (Slc2a4). G: Gene expression of DNL pathway. Acly, ATP citrate lyase; Acc1, acetyl-CoA carboxylase; Fasn, fatty acid synthase. H: Plasma glucose levels. Data are expressed as means ± SEM, n = 7, fold change over SHR fasted group. I: OGTT. Blood glycemia and insulin levels during the OGTT. AUC, area under the curve. Data are expressed as means ± SEM, n = 10–11. *Student t test: significantly different at P < 0.05. Two-way ANOVA/Holm-Sidak test: (i), statistically significant interaction between genotype and nutritional state at P < 0.05; f, significantly different nutritional state P < 0.05; g, significantly different genotype P < 0.05.
Results
Targeted Lipidomics of FAHFAs
The analysis of FAHFAs from WAT is complicated due to the fact that the two-step lipid extraction is followed by a prolonged chromatography to separate the regioisomers. We tested several lipid extraction techniques to optimize the analysis. Maximal recovery of very hydrophobic FAHFAs was achieved with ethyl acetate and citrate buffer mixture (11,27). The chromatographic separation of FAHFA regioisomers was optimized for PAHSAs using the C18 column with high carbon load, and the final method allowed near baseline peak separation of PAHSA regioisomers as well as regioisomers of other FAHFAs. Using the MS/MS/MS fragmentation patterns (11) and retention times of synthetic standards (11,13), we were able to identify ∼160 regioisomers of 49 FAHFA derived from 7 physiologically relevant fatty acids (Table 1 and Supplementary Fig. 1). There were several other FAHFA regioisomers detected, but their identification was not definitive due to low intensities and coelution.
Table 1.
The number of regioisomers in the corresponding FAHFA family
| HPA | HPO | HSA | HOA | HLA | HAA | HDHA | |
|---|---|---|---|---|---|---|---|
| PA | 6 | 4 | 8 | 4 | 2 | 1 | 2 |
| PO | 6 | 4 | 8 | 4 | 2 | 1 | 2 |
| SA | 4 | 4 | 6 | 4 | 2 | 1 | 2 |
| OA | 6 | 4 | 6 | 4 | 2 | 1 | 2 |
| LA | 6 | 4 | 8 | 4 | 2 | 1 | 2 |
| AA | 6 | 4 | 5 | 4 | 2 | 1 | 2 |
| DHA | 4 | 2 | 3 | 4 | 2 | 1 | 2 |
FAHFA Profiles
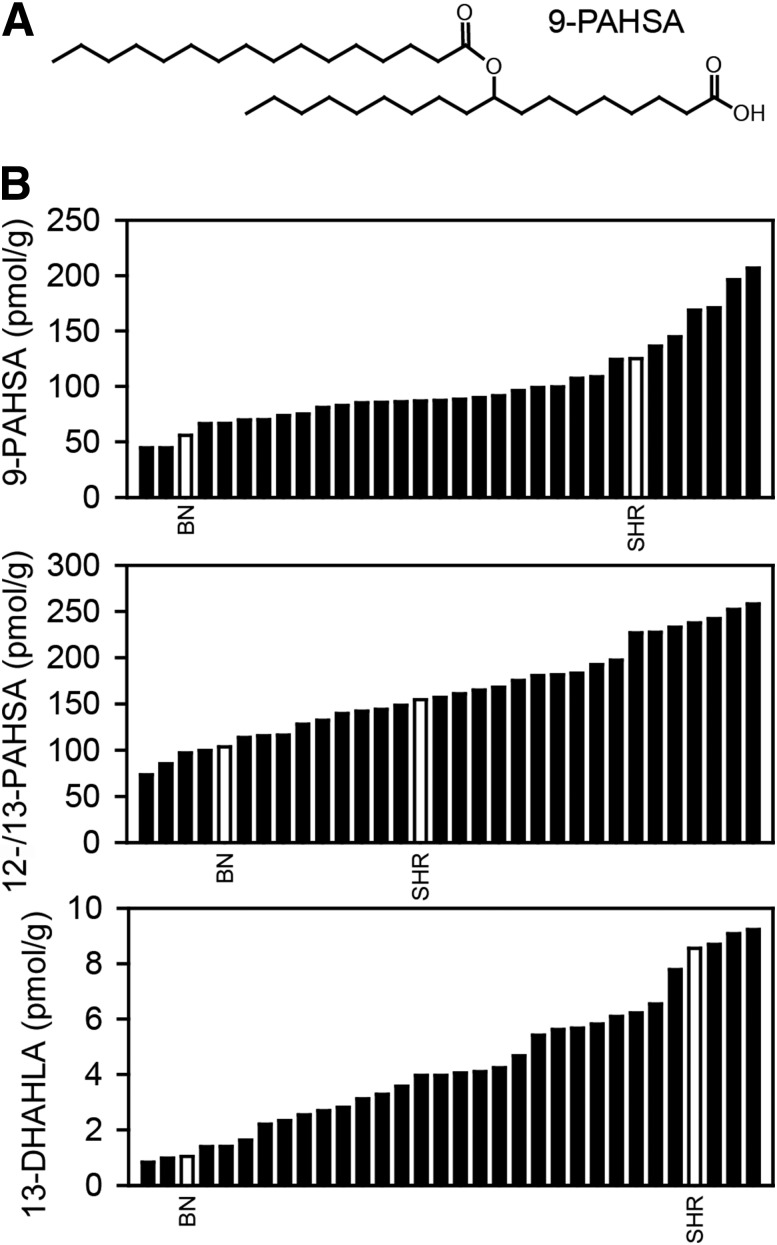
We analyzed WAT levels of FAHFA regioisomers in 30 RI and two progenitor strains. Distribution of 9-PAHSA, 12-/13-PAHSA (6), and 13-DHAHLA (11) among BXH/HXB RI strains was continuous, suggesting a polygenic inheritance. BN rats showed significantly reduced levels of FAHFA regioisomers when compared with SHR rats (Fig. 1).
Figure 1.
Distribution of FAHFAs among RI strains. A: Structure of 9-PAHSA. B: Levels of 9-PAHSA, 12-/13-PAHSA, and 13-DHAHLA in epididymal WAT of RI strains. 12- and 13-PAHSA levels are reported together due to separation issues (6). Values are expressed as medians, and progenitor strains are highlighted as empty bars (n = 5–6).
Relationships of FAHFAs With Parameters of Lipid and Glucose Metabolism
The regioisomer 9-PAHSA was shown to be involved in enhanced glucose uptake and insulin sensitivity of peripheral tissues (6). The correlation between 9-PAHSA trait and physiological parameters of the BXH/HXB RI strains (30) confirmed that 9-PAHSA levels were positively associated with the difference between maximal and basal glucose uptake into collagenase-liberated adipocytes from WAT (Pearson r = 0.466; P = 0.015) (Supplementary Fig. 2), thus supporting the beneficial effect of 9-PAHSA on glucose homeostasis. Interestingly, isoproterenol-induced lipolysis of isolated adipocytes (22) correlated with 9-PAHSA trait (Pearson r = 0.415; P = 0.034), suggesting that 9-PAHSA also augments the capacity of lipid mobilization from WAT.
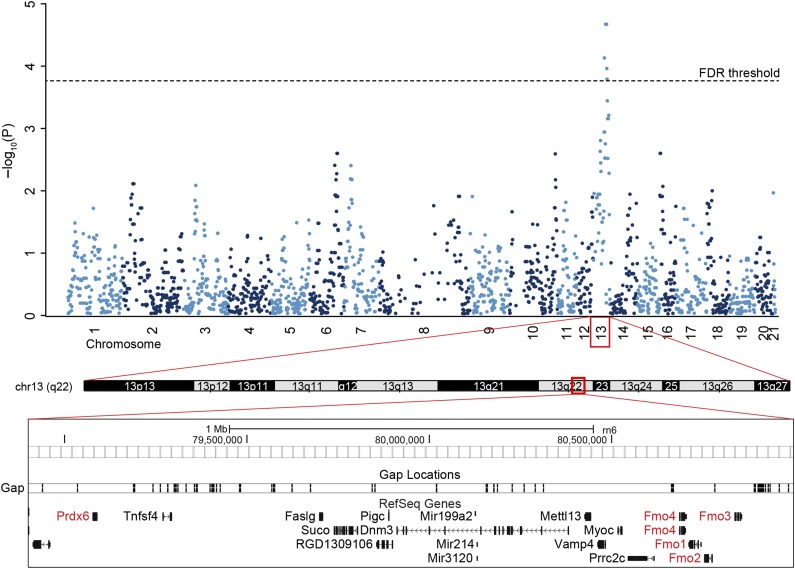
QTL Mapping
Genome-wide linkage analysis revealed several significant QTLs associated with FAHFA regioisomers on chromosomes 4, 5, 10, 13, and 15 at FDR <0.1. These QTLs were divided into four groups based on the chemical structure of associated FAHFAs (Table 2). The first group comprises QTLs associated with PAHSA regioisomers. The strongest association was observed for 12-/13-PAHSA with the peak of QTL linkage in the immediate vicinity of the Prdx6 as a positional candidate gene on chromosome 13 (Fig. 2). Other positional candidate genes relevant to FAHFA metabolism were the Fmo family and Mgst3, as well as Mgst1 in case of 9-PAHSA trait. Additional potential QTL for group 1 was found on chromosome 12 near the marker D12Rat10 in the proximity of the ChREBP-coding gene Mlxipl.
Table 2.
Positional candidate genes in FAHFA metabolism
| Trait | QTL range (Mb) | P value | FDR | RGD ID | Symbol |
|---|---|---|---|---|---|
| Group 1 |
|||||
| 9-PAHSA |
4: 161.8–161.8 |
<0.00003 |
<0.047 |
70927 |
Mgst1 |
| 12-/13-PAHSA |
13: 78.9–85.4 |
<0.00017 |
<0.058 |
71005 |
Prdx6 |
|
Fmo family |
|||||
| 1306373 |
Mgst3 |
||||
| Group 2 |
|||||
| 8-PAHPA |
4: 159.9–167.2 |
<0.00014 |
<0.081 |
620515 |
Olr1 |
| |
70927 |
Mgst1 |
|||
| 11-/12-SAHPA |
4: 181.5–181.5 |
<0.00049 |
<0.083 |
||
| 11-/12-SAHPA |
5: 25.8–66.5 |
<0.00074 |
<0.089 |
1584701 |
Acnat1 |
| |
1359532 |
Acnat2 |
|||
| |
2190 |
Baat |
|||
| 11-/12-SAHPA |
15: 25.8–26.3 |
<0.00068 |
<0.089 |
||
| Group 3 |
|||||
| 12-SAHOA |
3: 136.4–155.3 |
<0.00025 |
<0.086 |
3439 |
Ptgs1 |
| 8-OAHOA |
5: 130.6–130.6 |
<0.00002 |
<0.034 |
1306999 |
Gpx7 |
| 9-PAHOA |
10: 104.7–107.7 |
<0.00003 |
<0.012 |
1311950 |
Fads6 |
| 13-PAHLA |
10: 104.7–107.7 |
<0.00008 |
<0.028 |
1311950 |
Fads6 |
| Group 4 |
|||||
| 13-AAHLA | 4: 0.1–14.6 | <0.00022 | <0.092 | 2301 | Cd36 |
Acnat1, acyl-CoA amino acid N-acyltransferase 1; Acnat2, acyl-CoA amino acid N-acyltransferase 2; Baat, bile acid-CoA:amino acid N-acyltransferase; Cd36, CD36 molecule; Fads6, fatty acid desaturase 6; Fmo, flavin containing monooxygenase; Gpx7, glutathione peroxidase 7; Mgst1, microsomal glutathione S-transferase 1; Mgst3, microsomal glutathione S-transferase 3; Olr1, oxidized low density lipoprotein receptor 1; Ptgs1, prostaglandin-endoperoxide synthase 1.
Figure 2.
Linkage analysis of 12-/13-PAHSA on chromosome 13. Manhattan plot of 12-/13-PAHSA trait. Mapping of 12-/13-PAHSA trait on chromosome 13. Positional candidate genes Prdx6 and Fmo family are highlighted in red.
The second group comprised QTLs associated with SAHPA regioisomers, the inverse combination of parental fatty acids from group 1, and PAHPA, the PA metabolite. Linkage analysis revealed positional candidate genes on chromosome 4 (Olr1, Mgst1) and chromosome 5 (Acnat1, Acnat2, Baat; see Table 2 for the full gene names). The third group included QTLs associated with FAHFAs with one and two double bonds (PAHOA and PAHLA), and QTL linkage revealed Fads6, Ptgs1, and Gpx7 as the positional candidate genes. The fourth group of polyunsaturated FAHFAs included Cd36 as the candidate gene for the chromosome 4 QTL.
Role of Positional Candidate Genes in PAHSA Metabolism
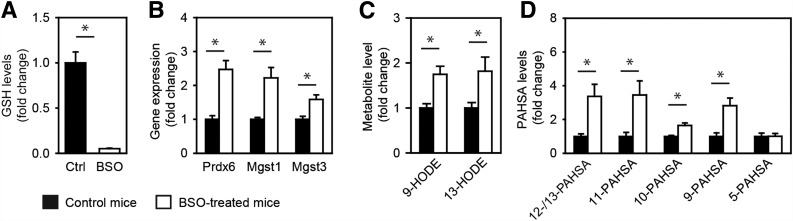
To limit the extent of the investigation, we focused our attention mainly on the possible role of the candidate genes in PAHSA metabolism. Candidate genes from groups 1 and 2 are all target genes for Nrf2 (31,32), a transcription factor that exerts a complex control over the cellular redox status and lipid metabolism. Prdx6 codes for a GSH peroxidase, while Mgst1 and Mgst3 code for GSH S-transferases (31); the activity for all three enzymes is sensitive to GSH depletion caused by BSO, an inhibitor of GSH synthesis. We treated mice by adding BSO to their drinking water (20 mmol/L) for 3 days. Induction of oxidative stress by GSH depletion (Fig. 3A) caused an increase in the levels of all three gene products (Fig. 3B and Supplementary Table 3), increase of the markers for lipid peroxidation (9- and 13-hydroxyoctadecadienoic acids [HODE]) (Fig. 3C), and an increase of 12-/13-, 11-, 10-, and 9-PAHSA (Fig. 3D), suggesting that redox status and ROS scavenging are involved in FAHFA biosynthesis.
Figure 3.
Effect of GSH synthesis inhibition on PAHSAs in epididymal WAT of C57BL/6J control mice and mice treated with BSO. A: GSH levels. B: Gene expression. C: Levels of 9- and 13-HODE (also known as hydroxylinoleic acid [HLA]). D: Levels of PAHSA regioisomers. Data are expressed as means ± SEM, n = 8. *Significantly different at P < 0.05.
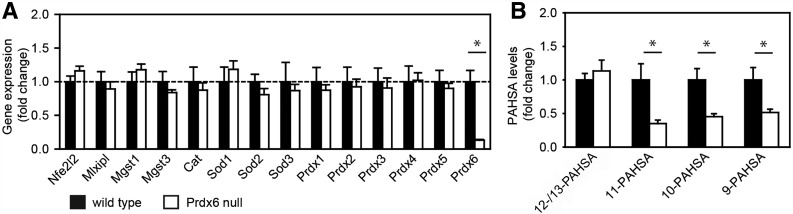
Next, we focused on the involvement of Prdx6, which represents a strong candidate enzyme for PAHSA biosynthesis as it possesses multiple lipid-related enzymatic activities (33), it is a Nrf2 target gene (32), and its role in the pathophysiology of type 2 diabetes has been reported (34). The loss of Prdx6 did not affect gene expression of Nfe2l2, Mlxipl, Mgst1, Mgst3, Cat, Sod1-3, or Prdx1-5 (Fig. 4A), in agreement with previous data showing no compensatory expression of antioxidant enzymes (25) and no increase in lipid peroxidation (Supplementary Fig. 4). In contrast, levels of 11-, 10-, and 9-PAHSA were significantly lower in WAT that had been dissected from Prdx6 null mice as compared with wild-type controls (Fig. 4B), suggesting a direct role of Prdx6 in the synthesis of the regioisomers.
Figure 4.
Effect of Prdx6 knockout on PAHSAs in epididymal WAT. C57BL/6J wild-type mice and Prdx6 null mice. A: Expression of relevant genes in WAT. Nfe2l2, Nrf2; Mlxipl, ChREBP; Mgst, microsomal glutathione S-transferase; Cat, catalase; Sod, superoxide dismutase. B: Levels of PAHSA in WAT. 5-PAHSA levels were not determined. Data are expressed as means ± SEM, n = 5. *Significantly different at P < 0.05.
To directly test the role of Nrf2 in PAHSA synthesis, we have used SHR and SHR-Nrf2 transgenic rats (Fig. 5A). As expected, expression of Prdx6, Mgst1, and Mgst3 in epididymal WAT of fasted rats was significantly higher in the SHR-Nrf2 animals, showing that chronic activation of Nrf2 pathway leads to activation of antioxidant defense system in this model (Fig. 5B), but had no effect on the markers for lipid peroxidation (Fig. 5C). Next, we aimed to characterize the effect of Nrf2 on FAHFA levels in WAT, which is also dependent on the feeding status of the animals. Therefore, epididymal WAT was dissected from SHR and SHR-Nrf2, from fasted animals and from animals refed the chow diet. In the fasted state, levels of 13-/12-, 11-, and 9-PAHSA were reduced in WAT from fasted SHR-Nrf2 as compared with SHR, while levels of 10- and 5-PAHSA were not different in the rats of the two genotypes. In accordance with the previous study in mice (6), in the control SHR, levels of PAHSA decreased in response to the refeeding; however, no effect of the refeeding was observed in SHR-Nrf2 (Fig. 5D). Levels of 13-PAHLA and 13-LAHLA showed that Nrf2 transgenic expression did not affect the synthesis of HLA-derived FAHFAs, while synthesis of HSA-derived FAHFAs and response to refeeding was altered. These results suggested involvement of mutual interactions between Nrf2 and feeding status in the modulation of the PAHSAs levels in WAT.
The refeeding of chow diet to rats should induce DNL in WAT; glucose represents the most common supply of carbon units for DNL, and its availability might limit DNL in WAT. Quantification of the expression of the Glut4 (Slc2a4) gene at both the transcript and the protein level in WAT indicated that Nrf2 transgenic expression counteracted chow diet–mediated induction of the gene and, therefore, limited glucose availability during the refeeding period (Fig. 5F). In fact, the refeeding response of all the genes critical for DNL in WAT that were in focus of our study (ChREBP, Acly, Acc1, Fasn) was dysregulated in SHR-Nrf2 (Fig. 5G). Thus, while chow refeeding induced ChREBP-α and -β expression in SHR, this effect was abrogated in SHR-Nrf2. The expression of the lipogenic genes (Acly, Acc1, Fasn) was significantly upregulated in SHR but not in SHR-Nrf2 in response to the chow refeeding. Levels of plasma glucose in the refeeding experiment (Fig. 5H) and both glucose and insulin levels during OGTT suggested that SHR-Nrf2 animal were insulin resistant.
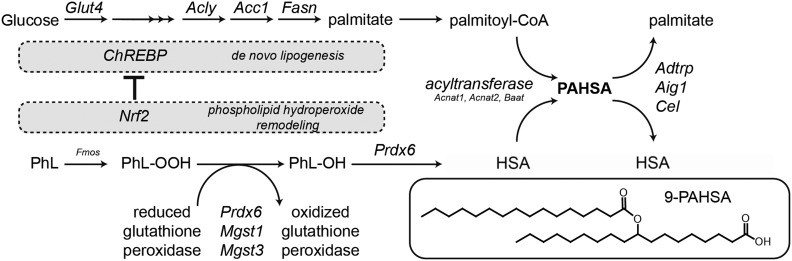
Collectively, these results further document the complex interaction between Nrf2 and feeding status (see above), especially the negative influence of Nrf2 on lipogenesis and the paradoxical dissociation between the changes in activity of the DNL pathway and PAHSA levels in WAT (see also Yore et al. [6], Vijayakumar et al. [16], Fig. 6, and discussion).
Figure 6.
Proposed scheme for PAHSA synthesis. DNL from glucose to palmitate is controlled by ChREBP. Membrane phospholipids (PhL) are oxidized to phospholipid hydroperoxide (PhL-OOH), which is further converted to a phospholipid with the hydroxy fatty acid (PhL-OH) via GSH-dependent peroxidases. The hydroxy fatty acid (HSA) is liberated from the PhL-OH and subsequently coupled to acyl-CoA coming from the DNL pathway (PA), yielding a PAHSA, which can be later degraded by specific hydrolases. While DNL from glucose is an essential prerequisite for PAHSA synthesis, Nrf2 overexpression limits the DNL pathway. Therefore, it seems plausible that fine-tuning of enzymatic and nonenzymatic cellular antioxidant defenses and coordination with the DNL pathway is needed for PAHSA synthesis and that substrate specificity of peroxidases/phospholipases might play a role in the synthesis of individual regioisomers.
Discussion
The principal finding of this study was that biosynthesis of FAHFAs in WAT involved the adaptive Nrf2-regulated antioxidant system. Therefore, our results further document that the defense against oxidative stress is interconnected, in a complex way, with DNL in WAT and suggest that FAHFAs may link activity of the adaptive antioxidant response in WAT with insulin sensitivity in peripheral tissues.
The comprehensive LC-MS analysis of FAHFAs revealed the complexity of this lipid class. We have identified ∼160 regioisomers combined of seven fatty acids and seven hydroxy fatty acids. The distribution of the branching positions among the hydroxy fatty acids suggests that the carbons in the middle of the molecule, and preferentially in the vicinity of a double bond, are the primary targets for oxidation. The chromatography was optimized for separation of PAHSAs; thus, the separation of other FAHFAs was not optimal and there might be other regioisomers that coelute. In general, there are common branching patterns for saturated HPA and HSA as well as monounsaturated HPO and HOA. The hydroxyl position on polyunsaturated HLA and HDHA matched the common enzymatic products (11), while the HAA branching at C 11 was unexpected. Importantly, since the analysis does not recognize cis/trans double bond isomers, R/S enantiomers, or unusual FAs, there might be more than ∼300 FAHFA species. To limit the extent of the investigation, we focused our attention mainly on PAHSAs.
The correlation between 9-PAHSA trait and the parameters of adipocyte metabolism already characterized in the BXH/HXB RI strains confirmed beneficial effects of PAHSAs on the essential metabolic functions of WAT (6). The QTL analysis highlighted a pathway of Nrf2 target genes involved in antioxidative defense. Nrf2 mediates the cross talk between lipid metabolism, DNL, and antioxidant defense mechanisms in the liver and WAT (35,36), demonstrating that defense against oxidative stress is interconnected with lipogenesis (3,37,38). Reduced levels of Nrf2 were associated with the development of oxidative stress, redox status disbalance, and diabetes complications including cardiomyopathy, nephropathy, retinopathy, foot ulceration, and microangiopathy in human patients and in mice (35,39). Transient activation of the Nrf2 system by naturally occurring compounds, such as plant polyphenols, results in raising antioxidant levels and may explain the beneficial metabolic effects of these micronutrients (37,39,40). Moreover, there is synergistic induction of lipid catabolism and anti-inflammatory lipids in WAT and reduction of adiposity in mice with induced obesity, in response to the combined intervention using calorie restriction and n-3 fatty acids that correlates with an increase in WAT levels of 15d-PGJ2, the lipid signaling mediator that stimulates the Nrf2 system (41,42). Therefore, the ChREBP–Nrf2–FAHFA axis may be involved in beneficial effects of various (micro)nutrients on metabolic health and insulin sensitivity and contribute to amelioration of diabetes complications. This important possibility should be explored in the future.
The positional candidate genes related to lipid metabolism include the Fmo family linked to production of ROS (11,43,44), four enzymes with GSH peroxidase activity scavenging phospholipid hydroperoxides (45–48) (different from selenium-containing phospholipid hydroperoxidase Gpx4), and peroxisomal acyltransferases (49). Although the Fmo family preferentially catalyzes oxygenation of heteroatoms (44), Fmo forms a stable 4a-hydroperoxy-FAD intermediate, which can serve as a source of hydrogen peroxide for lipid peroxidation (43). The four identified antioxidative enzymes (Prdx6, Gpx7, Mgst1, Mgst3) exert peroxidase activity with phospholipid hydroperoxides as substrates (45–48), while Prdx6 also possesses phospholipase A2 and lysophosphatidylcholine acyltransferase activities. These enzymes can thus produce a phospholipid with a hydroxy fatty acid or even cleave this hydroxy fatty acid in the case of Prdx6. The family of peroxisomal acyltransferases (Baat, Acnat1, Acnat2) conjugate long-chain and very-long-chain saturated acyl-CoAs to taurine or glycine, but complete substrate specificity has not yet been identified (49). Thus, it might be possible that they can esterify the hydroxy fatty acid with saturated acyl-CoA, as the known acyltransferases are not involved (18). Of note, FAHFAs were identified as the products of DNL in WAT, which relies on NADPH production to drive the lipogenesis. Therefore, NADPH can serve for both fatty acid elongation and GSH-disulfide reductase reaction.
We demonstrated in this study that the inhibition of GSH synthesis by BSO resulted in uniformly elevated PAHSA levels and that deletion of Prdx6 decreased PAHSA levels. This suggests that lipid peroxidation under elevated oxidative stress conditions, GSH peroxidases and a reduction of hydroperoxy phospholipids are involved in formation of HSA. Heterogeneous reduction or elevation of PAHSA regioisomers might be related to the incidence of peroxidation at specific carbon sites (see below), substrate specificity of an acyltransferase, or preferential degradation of some regioisomers (e.g., 12- and 13-PAHSA [19]). A previous report (34) documented that Prdx6 null mice develop a phenotype similar to early-stage type 2 diabetes manifested as increased insulin resistance of peripheral tissues, decreased insulin secretion, and systemic inflammation. The present results with Prdx6 null mice indicating decreased levels of 9-PAHSA complement the original description of PAHSAs (6) as lipids with potent antidiabetic and anti-inflammatory activities.
Unexpectedly, the overexpression of Nrf2 accompanied by the increased expression of cellular antioxidant defense genes led to lower or unchanged PAHSA levels. This discrepancy could be explained by the dysregulation of the ChREBP-dependent DNL pathway. While chronic stimulation of the Nrf2 pathway might supply more hydroxy fatty acids, reduction of de novo synthesized palmitate limits the rate of PAHSA synthesis. Therefore, it seems plausible that the fine-tuning of enzymatic and nonenzymatic cellular antioxidant defenses and coordination with the DNL pathway is needed for FAHFA synthesis and that substrate specificity of peroxidases might play a role in the synthesis of individual regioisomers. We have confirmed the observation by Kahn and colleagues (6) that changes in the activity of DNL due to feeding status do not correlate with the changes of PAHSA levels in WAT, in spite of the essential role of the ChREBP–DNL axis in the formation of PAHSA in WAT (16). Also, genetic activation of Nrf2 in mice led to a repression of both hepatic and WAT lipogenesis (38,50). Therefore, we hypothesize that formation of new fatty acids due to DNL activity is essential for formation of PAHSA, but the rate of PAHSA formation is controlled by other mechanisms that involve remodeling of membrane phospholipid hydroperoxides, as described below (see also Fig. 6). Given the diversity of the FAHFA class, it is plausible that other pathways are also involved in the synthesis of various fatty acid combinations, regioisomers, and enantiomers. The critical step seems to be the synthesis of the parental hydroxy fatty acid. In the case of unsaturated fatty acids, enzymatic and nonenzymatic oxygenation follows the known pathways (e.g., 13-HLA, 12-HOA as major regioisomers). The (poly)unsaturated FAHFAs (13-AAHLA, 13-DHAHLA) are associated with genes related to desaturation (Scd6) and to scavenging of oxidized lipids (Cd36) and might be involved in the immunometabolic cross talk within WAT (1,12).
Based on the current knowledge, we propose a pathway for 9-PAHSA biosynthesis (Fig. 6). Glucose is converted to citrate and further to palmitate in WAT via the DNL pathway, which might be controlled by a cascade of 1) target of rapamycin complex 2 (mTORC2) (3), 2) ChREBP-β (3,5), 3) Nrf2, and 4) Nrf2 targets like Prdx6. Membrane phospholipids can be randomly oxidized by ROS, or phospholipid precursors can be synthesized via DNL (9) and enzymatically oxidized. Although the spontaneous (per)oxidation of saturated stearic acid is less likely compared with polyunsaturated fatty acids, the Fmo family is linked to ROS production and the family substrate specificity has not yet been completely identified (43,44). Also, the peroxidation of palmitate was located preferentially at the center of the molecule (20), similar to PAHSA regioisomer distribution. Phospholipid hydroperoxide can form various types of lipid peroxidation products (malondialdehyde, 4-hydroxynonenal, glyoxal, etc.) or can be reduced by enzymes with GSH peroxidase or transferase activity (Prdx6, Mgst1, Mgst3) to a phospholipid with hydroxy acyl chain. This reaction is dependent on cellular redox state and probably controlled via an Nrf2 master regulator. The hydroxy fatty acid is usually cleaved through an oxidized phospholipid membrane remodeling process (e.g., Prdx6 PLA2 activity) and the free hydroxy fatty acid can be esterified with acyl-CoA by acyltransferase (probably via Baat, Acnat1, or Acnat2 [18]) as was shown in HEK293T cells (21). The final esters are eventually degraded by serine and threonine hydrolases (17,19).
The future challenge will be to validate the identified positional candidate genes to delineate the role for individual FAHFAs in lipid and glucose metabolism, as well as to establish the role of these novel lipokines in the defense against diabetes-induced oxidative stress and associated complications such as diabetic cardiomyopathy and nephropathy. Also, the identification of potential phospholipid intermediates in FAHFA synthesis and detailed structural characterization of FAHFA regioisomers will be needed in order to identify specific receptors and signaling pathways. Elucidation of the involvement of the ChREBP–Nrf2–FAHFA axis in the beneficial health effects of various micronutrients could help in prevention and treatment of obesity-associated diseases.
Supplementary Material
Article Information
Acknowledgments. C.D. and A.B.F. thank Drs. Patrick Seale and Jeff Ishibashi (University of Pennsylvania) for assistance with isolation of WAT from mice. The Proteomic Core Facility at BIOCEV (Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University in Vestec) is acknowledged for the proteomic measurements.
Funding. This work was supported by grants from the Czech Science Foundation (GA16-04859S and GJ17-10088Y) and the Ministry of Education, Youth and Sports of the Czech Republic (LTAUSA17173). This publication is supported by the project BIOCEV (CZ.1.05/1.1.00/02.0109) through the European Regional Development Fund.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. O.K., M.B., J.S., V.L., V.Z., and C.D. performed the animal studies. O.K. and M.B. performed the lipid analysis. O.K., F.K., and N.H. performed the statistical evaluation. F.K., J.K., and M.P. performed the genetic analysis. O.K. and M.V. performed the proteomic study. L.B. and T.D. prepared FAHFA standards. A.B.F., J.K., and M.P. contributed to the discussion. O.K. designed the studies and wrote the manuscript. O.K. and M.P. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the Organ Crosstalk in Obesity and NAFLD (J3) of the Keystone Symposia on Molecular and Cellular Biology, Keystone, CO, 21–25 January 2018.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-1087/-/DC1.
References
- 1.Yilmaz M, Claiborn KC, Hotamisligil GS. De novo lipogenesis products and endogenous lipokines. Diabetes 2016;65:1800–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eissing L, Scherer T, Tödter K, et al. . De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nat Commun 2013;4:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Y, Wallace M, Sanchez-Gurmaches J, et al. . Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nat Commun 2016;7:11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solinas G, Borén J, Dulloo AG. De novo lipogenesis in metabolic homeostasis: more friend than foe? Mol Metab 2015;4:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman MA, Peroni OD, Villoria J, et al. . A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012;484:333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yore MM, Syed I, Moraes-Vieira PM, et al. . Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014;159:318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 2008;134:933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flachs P, Adamcova K, Zouhar P, et al. . Induction of lipogenesis in white fat during cold exposure in mice: link to lean phenotype. Int J Obes 2017;41:372–380 [DOI] [PubMed] [Google Scholar]
- 9.Lodhi IJ, Yin L, Jensen-Urstad AP, et al. . Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab 2012;16:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brezinova M, Kuda O, Hansikova J, et al. Levels of palmitic acid ester of hydroxystearic acid (PAHSA) are reduced in the breast milk of obese mothers. Biochim Biophys Acta 2018;1863:126–131 [DOI] [PubMed]
- 11.Kuda O, Brezinova M, Rombaldova M, et al. . Docosahexaenoic acid-derived fatty acid esters of hydroxy fatty acids (FAHFAs) with anti-inflammatory properties. Diabetes 2016;65:2580–2590 [DOI] [PubMed] [Google Scholar]
- 12.Masoodi M, Kuda O, Rossmeisl M, Flachs P, Kopecky J. Lipid signaling in adipose tissue: connecting inflammation & metabolism. Biochim Biophys Acta 2015;1851:503–518 [DOI] [PubMed]
- 13.Balas L, Bertrand-Michel J, Viars F, et al. . Regiocontrolled syntheses of FAHFAs and LC-MS/MS differentiation of regioisomers. Org Biomol Chem 2016;14:9012–9020 [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Kind T, Vaniya A, Gennity I, Fahrmann JF, Fiehn O. An in silico MS/MS library for automatic annotation of novel FAHFA lipids. J Cheminform 2015;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Moraes-Vieira PM, Castoldi A, et al. . Branched fatty acid esters of hydroxy fatty acids (FAHFAs) protect against colitis by regulating gut innate and adaptive immune responses. J Biol Chem 2016;291:22207–22217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijayakumar A, Aryal P, Wen J, et al. . Absence of carbohydrate response element binding protein in adipocytes causes systemic insulin resistance and impairs glucose transport. Cell Reports 2017;21:1021–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolar MJ, Kamat SS, Parsons WH, et al. . Branched fatty acid esters of hydroxy fatty acids are preferred substrates of the MODY8 protein carboxyl ester lipase. Biochemistry 2016;55:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn B, Herman M, Saghatelian A, Homan E. Lipids that increase insulin sensitivity and methods of using the same. Patent WO2013166431 A1. 3 May 2013
- 19.Parsons WH, Kolar MJ, Kamat SS, et al. . AIG1 and ADTRP are atypical integral membrane hydrolases that degrade bioactive FAHFAs. Nat Chem Biol 2016;12:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodnitz MH, Nawar WW, Fagerson IS. Autoxidation of saturated fatty acids. II. The determination of the site of hydroperoxide groups in autoxidizing methyl palmitate. Lipids 1968;3:65–71 [DOI] [PubMed] [Google Scholar]
- 21.Nelson AT, Kolar MJ, Chu Q, et al. . Stereochemistry of endogenous palmitic acid ester of 9-hydroxystearic acid and relevance of absolute configuration to regulation. J Am Chem Soc 2017;139:4943–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubner N, Wallace CA, Zimdahl H, et al. . Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 2005;37:243–253 [DOI] [PubMed] [Google Scholar]
- 23.Ivics Z, Mátés L, Yau TY, et al. . Germline transgenesis in rodents by pronuclear microinjection of Sleeping Beauty transposons. Nat Protoc 2014;9:773–793 [DOI] [PubMed] [Google Scholar]
- 24.Manakov D, Ujcikova H, Pravenec M, Novotny J. Alterations in the cardiac proteome of the spontaneously hypertensive rat induced by transgenic expression of CD36. J Proteomics 2016;145:177–186 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med 2004;37:1736–1743 [DOI] [PubMed] [Google Scholar]
- 26.Lee I, Dodia C, Chatterjee S, Feinstein SI, Fisher AB. Protection against LPS-induced acute lung injury by a mechanism-based inhibitor of NADPH oxidase (type 2). Am J Physiol Lung Cell Mol Physiol 2014;306:L635–L644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin JH, Liu LY, Yang MH, Lee MH. Ethyl acetate/ethyl alcohol mixtures as an alternative to folch reagent for extracting animal lipids. J Agric Food Chem 2004;52:4984–4986 [DOI] [PubMed] [Google Scholar]
- 28.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012;28:1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics 2003;1:299–308 [DOI] [PubMed] [Google Scholar]
- 30.Pravenec M, Křen V, Landa V, et al. . Recent progress in the genetics of spontaneously hypertensive rats. Physiol Res 2014;63(Suppl. 1):S1–S8 [DOI] [PubMed] [Google Scholar]
- 31.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 2014;39:199–218 [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med 2009;46:146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher AB. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch Biochem Biophys 2017;617:68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacifici F, Arriga R, Sorice GP, et al. . Peroxiredoxin 6, a novel player in the pathogenesis of diabetes. Diabetes 2014;63:3210–3220 [DOI] [PubMed] [Google Scholar]
- 35.Xue P, Hou Y, Chen Y, et al. . Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 2013;62:845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pi J, Leung L, Xue P, et al. . Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem 2010;285:9292–9300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo HA, Lee IK. The role of Nrf2: adipocyte differentiation, obesity, and insulin resistance. Oxid Med Cell Longev 2013;2013:184598 [DOI] [PMC free article] [PubMed]
- 38.Slocum SL, Skoko JJ, Wakabayashi N, et al. . Keap1/Nrf2 pathway activation leads to a repressed hepatic gluconeogenic and lipogenic program in mice on a high-fat diet. Arch Biochem Biophys 2016;591:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiménez-Osorio AS, Picazo A, González-Reyes S, Barrera-Oviedo D, Rodríguez-Arellano ME, Pedraza-Chaverri J. Nrf2 and redox status in prediabetic and diabetic patients. Int J Mol Sci 2014;15:20290–20305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue M, Momiji H, Rabbani N, Bretschneider T, Rand DA, Thornalley PJ. Frequency modulated translocational oscillations of Nrf2, a transcription factor functioning like a wireless sensor. Biochem Soc Trans 2015;43:669–673 [DOI] [PubMed] [Google Scholar]
- 41.Kansanen E, Kivelä AM, Levonen AL. Regulation of Nrf2-dependent gene expression by 15-deoxy-Delta12,14-prostaglandin J2. Free Radic Biol Med 2009;47:1310–1317 [DOI] [PubMed] [Google Scholar]
- 42.Flachs P, Rühl R, Hensler M, et al. . Synergistic induction of lipid catabolism and anti-inflammatory lipids in white fat of dietary obese mice in response to calorie restriction and n-3 fatty acids. Diabetologia 2011;54:2626–2638 [DOI] [PubMed] [Google Scholar]
- 43.Siddens LK, Krueger SK, Henderson MC, Williams DE. Mammalian flavin-containing monooxygenase (FMO) as a source of hydrogen peroxide. Biochem Pharmacol 2014;89:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huijbers MM, Montersino S, Westphal AH, Tischler D, van Berkel WJ. Flavin dependent monooxygenases. Arch Biochem Biophys 2014;544:2–17 [DOI] [PubMed] [Google Scholar]
- 45.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem 2000;275:28421–28427 [DOI] [PubMed] [Google Scholar]
- 46.Jakobsson PJ, Mancini JA, Riendeau D, Ford-Hutchinson AW. Identification and characterization of a novel microsomal enzyme with glutathione-dependent transferase and peroxidase activities. J Biol Chem 1997;272:22934–22939 [DOI] [PubMed] [Google Scholar]
- 47.Mosialou E, Morgenstern R. Activity of rat liver microsomal glutathione transferase toward products of lipid peroxidation and studies of the effect of inhibitors on glutathione-dependent protection against lipid peroxidation. Arch Biochem Biophys 1989;275:289–294 [DOI] [PubMed] [Google Scholar]
- 48.Zhao T, Singhal SS, Piper JT, et al. . The role of human glutathione S-transferases hGSTA1-1 and hGSTA2-2 in protection against oxidative stress. Arch Biochem Biophys 1999;367:216–224 [DOI] [PubMed] [Google Scholar]
- 49.Hunt MC, Siponen MI, Alexson SE. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim Biophys Acta 2012;1822:1397–1410 [DOI] [PubMed]
- 50.Xu J, Kulkarni SR, Donepudi AC, More VR, Slitt AL. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes 2012;61:3208–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.