Abstract
The variable composition of the chromophore-binding pocket in visual receptors is essential for vision. The visual phototransduction starts with the cis-trans isomerization of the retinal chromophore upon absorption of photons. Despite sharing the common 11-cis-retinal chromophore, rod and cone photoreceptors possess distinct photochemical properties. Thus, a detailed molecular characterization of the chromophore-binding pocket of these receptors is critical to understanding the differences in the photochemistry of vision between rods and cones. Unlike for rhodopsin (Rh), the crystal structures of cone opsins remain to be determined. To obtain insights into the specific chromophore–protein interactions that govern spectral tuning in human visual pigments, here we harnessed the unique binding properties of 11-cis-6-membered-ring-retinal (11-cis-6mr-retinal) with human blue, green, and red cone opsins. To unravel the specificity of the chromophore-binding pocket of cone opsins, we applied 11-cis-6mr-retinal analog-binding analyses to human blue, green, and red cone opsins. Our results revealed that among the three cone opsins, only blue cone opsin can accommodate the 11-cis-6mr-retinal in its chromophore-binding pocket, resulting in the formation of a synthetic blue pigment (B6mr) that absorbs visible light. A combination of primary sequence alignment, molecular modeling, and mutagenesis experiments revealed the specific amino acid residue 6.48 (Tyr-262 in blue cone opsins and Trp-281 in green and red cone opsins) as a selectivity filter in human cone opsins. Altogether, the results of our study uncover the molecular basis underlying the binding selectivity of 11-cis-6mr-retinal to the cone opsins.
Keywords: G protein-coupled receptor (GPCR), membrane protein, photoreceptor, retinoid, rhodopsin, 11-cis-6mr-retinal, protein structure, retinal pigment, vision
Introduction
Both families of visual G protein–coupled receptors, implicated in twilight vision rhodopsin (Rh)4 and responsible for color vision cone opsins, consist of the universal chromophore, 11-cis-retinal, bound to the opsin protein moiety via a protonated Schiff base linkage (1). Besides several structural and functional similarities between Rh and cone opsins, including (i) heptahelical transmembrane architecture, (ii) an ultrafast photoinduced cis-trans isomerization of the chromophore, and (iii) signal transduction through the interaction with heterotrimeric G protein, cone opsins exhibit a relatively faster activation, inactivation, and regeneration than Rh (2, 3). These differences are attributed to the specific interactions between 11-cis-retinal and the chromophore-binding pocket of each pigment. The distinct chromophore–protein interactions also determine the light absorption properties of these receptors (4).
On the molecular level, Rh has been extensively studied by X-ray crystallography (5–9), NMR (10–12), FTIR (13–15), and resonance Raman spectroscopic techniques (16, 17). These structural studies determined that the chromophore-binding pocket of Rh largely comprises hydrophobic residues that enable accommodation of the retinal chromophore molecule and dictate its photochemistry.
Previously, various retinal analogs have been used to probe the specificity of Rh's chromophore-binding pocket and its functional implications (18–20). Recent comprehensive structural and biophysical analyses of the rod pigment regenerated with the 11-cis-6-membered-ring-retinal (11-cis-6mr-retinal) chromophore analog (Rh6mr) showed unique photocyclic behavior of Rh6mr with an atypical cis-dicis retinal photoisomerization, which was entirely mediated by the chemical properties of 11-cis-6mr-retinal featuring a locked C11=C12 cis-trans isomerization (21). In addition, determination of the crystal structure of Rh6mr and molecular mechanics studies led to the conclusion that the 11-cis isomer of 6mr-retinal is the most favorable configuration within the chromophore-binding pocket of Rh.
In contrast to Rh, fewer studies have been conducted to unravel the molecular details of the chromophore-binding pocket of cone opsins, which is in part due to their instability in vitro. Previous studies to delineate the spectral tuning and the G protein activation mechanism of cone opsins focused on regenerating red cone opsin with retinal analogs with varying polyene chain lengths and distinct geometric isomers (22). More recently, the 11-cis-6mr-retinal was utilized to elucidate the basis of the chromophore-binding specificity of green cone opsin (23). Interestingly, green cone opsin was unable to bind 11-cis-6mr-retinal chromophore, suggesting a tighter/rigid chromophore-binding pocket relative to Rh. In this study, we investigated the ability of human blue, green, and red cone opsins to bind the 11-cis-6mr-retinal. We found that among three cone opsins, 11-cis-6mr-retinal formed a Schiff base only with blue cone opsin, resulting in the formation of pigment (B6mr) exhibiting an absorption maximum at 440 nm, and we discerned that the specific amino acid environment of the blue cone opsin chromophore-binding pocket contributed to this property.
Results
Regeneration of rod and cone opsins with 11-cis-6mr-retinal
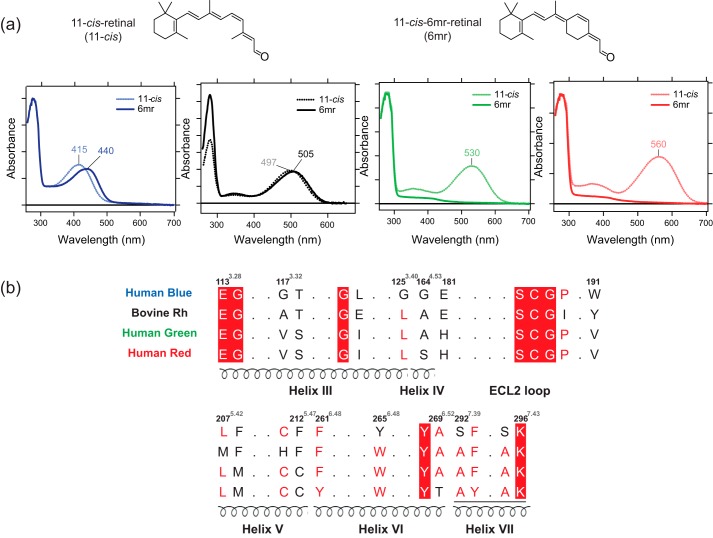
Human blue, green, and red cone opsins were expressed in insect cells, and their regenerative properties with 11-cis-6mr-retinal were tested in the isolated membranes. Rod opsin was prepared in rod outer segment (ROS) membranes isolated from bovine retinas as described under “Experimental procedures.” Opsin membranes were incubated either with 11-cis-retinal or 11-cis-6mr-retinal followed by pigment purification by 1D4 immunoaffinity chromatography. As expected, UV-visible spectroscopy of the samples regenerated with 11-cis-retinal revealed absorption maxima (λmax) of 415, 530, and 560 nm for blue, green, and red cone opsin, respectively. These spectral properties were similar to those obtained previously (24, 25). Interestingly, upon incubation with 11-cis-6mr-retinal, only the blue cone opsin showed formation of light-absorbing pigment with ∼25-nm red-shifted λmax at 440 nm, as compared with blue cone opsin regenerated with 11-cis-retinal. In contrast, both green and red cone opsins were unable to bind 11-cis-6mr-retinal (Fig. 1a). As expected, Rh could be regenerated with 11-cis-6mr-retinal, resulting in a pigment (Rh6mr) with λmax at 505 nm, featuring an 8-nm red shift as compared with Rh.
Figure 1.
Binding of 11-cis-retinal chromophore and its locked 11-cis-6mr-retinal analog to human blue, green, and red cone opsins and bovine rod opsin. a, UV-visible absorption spectra of reconstituted blue cone opsin, Rh, and green and red cone opsins with the native chromophore 11-cis-retinal (blue, black, green, and red dashed lines, respectively) and UV-visible absorption spectra of blue cone opsin, Rh, and green and red cone opsins bound with 11-cis-6mr-retinal analog (blue, black, green, and red solid lines, respectively). b, partial amino acid sequence alignment of human blue, green, and red cone opsins and bovine Rh (black). Conserved residues in the chromophore-binding pocket are shown. The amino acid numbers are based on bovine Rh sequence.
To decipher the basis of chromophore selectivity among the visual receptors, we performed multiple sequence alignment focusing on the amino acid residues in and around the chromophore-binding pocket. The major difference was identified in the primary sequences of cone opsins at amino acid residue 6.48, which plays a critical role in the chromophore entry and spectral tuning. Although green and red cone opsins comprise a Trp at position 6.48 (Trp-281), blue cone opsin features a Tyr at the corresponding position (Tyr-262) (Fig. 1b) (26). The different interactions between residue 6.48 and the β-ionone ring of the chromophore in cone opsins cause a large spectral shift of the λmax in blue as compared with green and red cone opsins (27). Thus, these differences could underlie the binding specificity of 11-cis-6mr-retinal to cone opsins. Moreover, the 6.48 residue is highly conserved among class A G protein–coupled receptors and plays a critical role in the ligand binding and receptor activation (28, 29).
In silico binding analysis of 11-cis-6mr-retinal to cone opsins
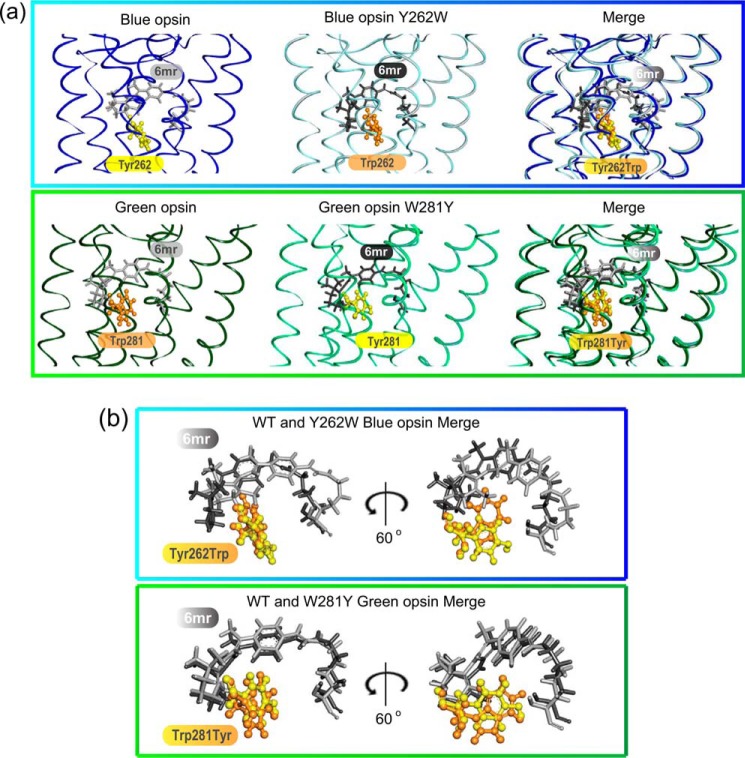
The difference in the binding ability of 11-cis-6mr-retinal to blue and green cone opsins could be attributed to differences in the amino acid composition within their chromophore-binding pocket. To obtain further insights into the potential steric hindrance caused by the 6.48 residue, Rosetta suite was used to calculate the pairwise interaction energies of 11-cis-retinal with Tyr-262 residue of blue cone opsin and Trp-281 residue of green cone opsin. Similarly, the pairwise interaction energies of the swapped mutants (Y262W in blue cone opsin and W281Y in green cone opsin) were also determined. These models were used as templates for calculating the binding free energies and pairwise binding energies between 11-cis-6mr-retinal and residue 6.48 in both wildtype (WT) and the swapped mutants of blue and green cone opsins. Significant differences in the interaction energy profiles of 11-cis-6mr-retinal with all protein models were observed. Binding of 11-cis-6mr-retinal was energetically more favorable in WT blue cone opsin than in its swapped Y262W mutant. Interestingly, the W281Y mutant of green cone opsin displayed a lower binding energy for 11-cis-6mr-retinal as compared with its WT counterpart (Table 1). The molecular modeling studies showed that the orientation of substituted residues did not change, and in both swapped mutants their side chains extended toward the retinal chromophore (Fig. 2, a and b). However, such amino acid substitutions would likely cause global rearrangements in the chromophore-binding pocket, allowing accommodation of 11-cis-6mr-retinal in the larger space of the W281Y green cone opsin as compared with its WT counterpart. Similarly, these spatial changes would also result in the impairment of 11-cis-6mr-retinal fit in the Y262W blue cone opsin mutant due to the exchange of the smaller Tyr into the bulkier Trp residue (Fig. 2). Overall, our molecular modeling analyses suggest a role for residue 6.48 in the chromophore-binding specificity of cone opsins.
Table 1.
Binding energy of 11-cis-6mr-retinal to blue and green cone opsin
The binding free energies and the energies of interaction between 11-cis-6mr-retinal and the 6.48 residue (the Tyr-262 or Trp-281 residues) were calculated in WT or swapped mutants of blue and green cone opsins. Pairwise interaction energies between 11-cis-6mr-retinal and the 6.48 residue are shown in parentheses.
| Binding energy of 11-cis-6mr-retinal |
||
|---|---|---|
| WT | Mutant | |
| kcal/mol | ||
| Blue cone opsin | −9.4 (−2.7) | −8.0 (−2.1) |
| Green cone opsin | −8.4 (−2.0) | −9.8 (−2.6) |
Figure 2.
Comparison of the retinal-binding pockets in WT and mutated cone opsins. a, WT blue cone opsin (dark blue) and the Y262W mutant (light blue) (top panel), WT green cone opsin (dark green) and the W281Y mutant (light green) (bottom panel), and Tyr residues (yellow sticks) and Trp residues (orange sticks) are shown. 11-cis-6mr-Retinal was modeled into the binding pocket of WT and mutated cone opsins and is shown with light gray sticks in WT blue and green cone opsins and with dark gray sticks in mutated blue and green cone opsins. b, magnified view of 11-cis-6mr-retinal conformations in the binding pocket of WT and mutated blue and green cone opsins (top and bottom panels, respectively) relative to the residues of interest, Tyr and Trp, are shown. The same 11-cis-6mr-retinal orientations as in the merged panels of a and after 60° rotation along the y axis are shown.
In vitro binding of 11-cis-6mr-retinal to the swapped mutants of blue and green cone opsins
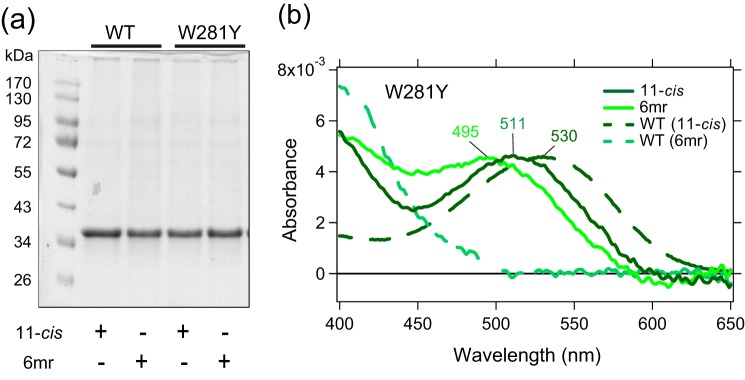
To verify our computational predictions, we prepared the Y262W and W281Y mutants for blue and green cone opsins, respectively, in the pcDNA3.1(+) vector and expressed them in HEK-293 cells. Notably, the expression level of WT green cone opsin was higher as compared with WT blue cone opsin. However, the amino acid substitutions did not significantly affect the expression profiles of the respective cone opsins (Fig. 3a). Opsin pigments were regenerated either with 11-cis-retinal or 11-cis-6mr-retinal and then purified by 1D4 immunoaffinity chromatography. The UV-visible absorption spectra revealed a significant reduction (∼60%) in the ability of Y262W blue cone opsin mutant to bind 11-cis-6mr-retinal, thereby validating our in silico predictions. In contrast, the regeneration of green W281Y cone opsin with 11-cis-6mr-retinal was significantly enhanced as compared with WT green cone opsin (Fig. 3b). However, this mutation also altered the spectral properties of green cone opsin upon binding of natural chromophore 11-cis-retinal. Binding of 11-cis-retinal to W281Y green cone opsin resulted in the formation of pigment with the λmax at 511 nm, ∼19-nm blue-shifted as compared with WT green cone opsin (Figs. 3b and 1a).
Figure 3.
Binding of 11-cis-retinal chromophore and its locked 11-cis-6mr-retinal analog to green cone opsin and the W281Y mutant. a, SDS-polyacrylamide gel of regenerated and purified pigments. Pigment regeneration was performed in HEK-293T cell pellets, which then were purified by 1D4 immunoaffinity chromatography. Proteins were separated on an SDS-polyacrylamide gel (2 μg was loaded in each lane), and the gel was stained with Coomassie Blue. Proteins were deglycosylated with peptide:N-glycosidase F before loading onto the gel. b, UV-visible absorption spectra of reconstituted W281Y green cone opsin regenerated with 11-cis-retinal (11-cis) (dark green), spectra of W281Y green cone opsin regenerated with 11-cis-6mr-retinal (6mr) (light green), and UV-visible absorption spectra of reconstituted WT green cone opsin regenerated with 11-cis-retinal (WT (11-cis)) (dark green dashed line) and with 11-cis-6mr-retinal (WT (6mr)) (light green dashed line) are shown.
Specificity of the chromophore-binding pockets in rod and cone opsins
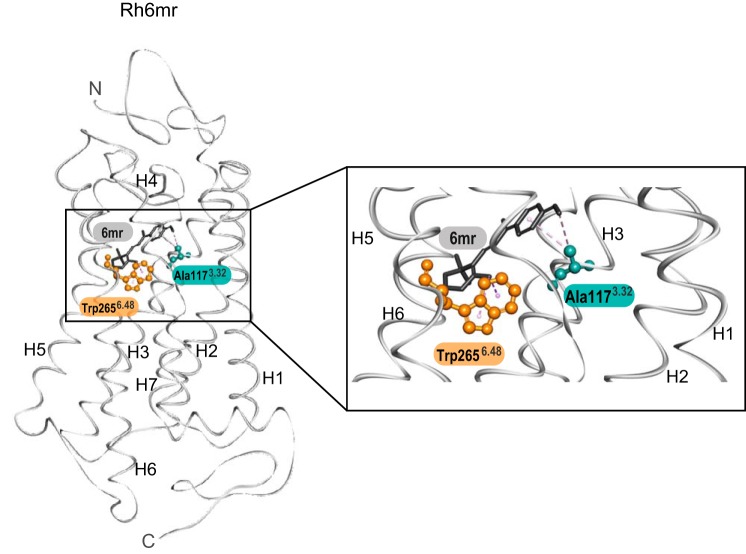
Amino acid residue 6.48 regulates the binding of retinal into the chromophore-binding pocket of opsin pigments. Binding of 11-cis-6mr-retinal is permitted in Tyr6.48-containing blue cone opsin but not in Trp6.48-containing green and red cone opsins. Interestingly, Rh also comprises a Trp in position 6.48 and binds 11-cis-6mr-retinal efficiently (5, 30). This suggests that perhaps Trp-2656.48 is not the only amino acid involved in the chromophore-binding selectivity. Multiple-sequence alignment of cone opsins and Rh identified Val-1323.47 in close vicinity of the retinal-binding pocket of green and red cone opsins. In contrast, blue cone opsin and Rh feature smaller amino acid residues at the corresponding positions. Although Rh contains an alanine (Ala-1173.32) (Fig. 1) (26, 31), blue cone opsin comprises a glycine residue (Gly-1143.29). Conformation of 11-cis-6mr-retinal relative to Trp-2656.48 and Ala-1173.32 is depicted in the crystal structure of Rh6mr (Fig. 4). Although the small Ala residue does not perturb the binding of 11-cis-6mr-retinal to rod opsin, an extra methyl group of Val-1323.47 in green and red cone opsins along with a bulkier Trp6.48 residue could sterically hinder the cyclo-hexyl ring of 11-cis-6mr-retinal and thereby inhibit its entry into the chromophore-binding pocket of these cone opsins.
Figure 4.
The crystal structure of Rh6mr (PDB code 5TE5 (21)) is shown on the left. The magnified region of Rh6mr displaying 11-cis-6mr-retinal in the chromophore-binding pocket is shown on the right. 11-cis-6mr-Retinal is shown with black sticks. Trp-2656.48 and Ala-1173.32 are shown with orange and cyan sticks and balls, respectively. H, helix.
Biophysical properties of B6mr
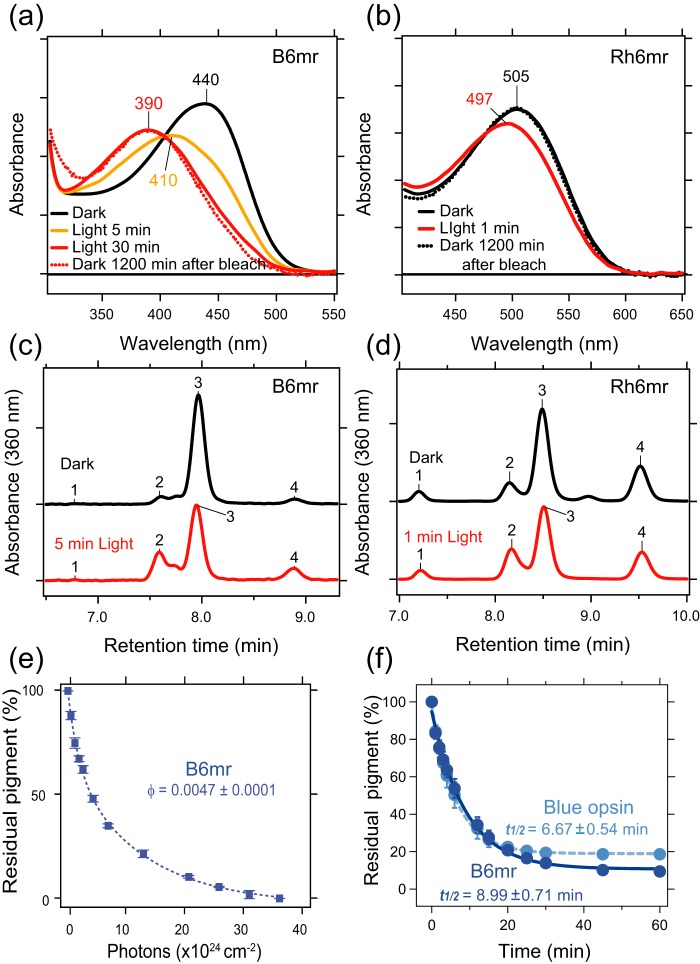
Although Rh6mr features an 8-nm red shift in the λmax as compared with Rh, B6mr displays a 25-nm red shift in its λmax (Fig. 5, a and b, black spectrum) (21). The photobleaching experiments were performed through a 400–440-nm band pass filter for B6mr and through a 480–520-nm band pass filter for Rh6mr. As described previously, Rh6mr requires a prolonged illumination of at least 1 min to reach its photostationary Meta-II–like state absorbing at 497 nm (Fig. 5b, red spectrum), whereas 5-s illumination of Rh in detergent solution at neutral pH is enough to convert 11-cis-retinal to all-trans-retinal and transition to Meta-II state. Rh6mr features a 24-fold lower photosensitivity (quantum yield of 0.027) relative to Rh (quantum yield of 0.65) (32). Interestingly, B6mr required an even longer illumination period of at least 30 min to achieve its photostationary Meta-II–like state with a λmax of 390 nm. The addition of an acid to the sample (pH 2) resulted in a red shift due to the Schiff base protonation, suggesting that the Meta-II–like photoproduct of B6mr possesses an intact, deprotonated Schiff base (Figs. 5a, red spectrum, and 6a, blue spectrum). A shorter illumination of 5 min showed the λmax at 410 nm, corresponding to a mixture of activated and inactive states of B6mr (Fig. 5a, yellow spectrum). Thus, these results indicate that B6mr has even lower photosensitivity than Rh6mr. Indeed, the quantum yield of isomerization of B6mr was calculated as 0.0047, which is about 6-fold lower than that of Rh6mr (0.027) and 138-fold lower than that for Rh (0.65) (Figs. 5e and 7). As shown previously, the Meta-II–like state of Rh6mr does not decay into opsin and free 11-cis-6mr-retinal upon light illumination but instead converts back to its inactive state after 2 h (Figs. 5b, dotted black spectrum, and 7) (21). Interestingly, the Meta-II–like state of B6mr did not revert back to its inactive state within a time range of 20–24 h (Fig. 5a, dotted red spectrum). Comparative retinoid isomeric composition analyses of the inactive and photoactivated Meta-II–like states of Rh6mr and B6mr revealed similar light-stimulated changes in the isomeric composition of 11-cis-6mr-retinal in both pigments. In the absence of the opsin moiety, 11-cis-6mr-retinal exists as a mixture of four isomers: 9,11,13-tricis-, 11,13-dicis-, 11-cis-, and 9,11-dicis-6mr-retinal. These isomers elute as peaks 1, 2, 3, and 4, respectively, from a normal-phase HPLC chromatography column (21, 33). The inactive state of both Rh6mr and B6mr showed a predominant peak that corresponds to 11-cis-6mr-retinal (peak 3). HPLC analyses of light-exposed Rh6mr and B6mr displayed a significant decrease in the intensity of peak 3 and a simultaneous increase of peak 2, which corresponds to the 11,13-dicis-6mr-retinal in both samples, suggesting that 11-cis converts to the 11,13-dicis isomer upon light illumination of B6mr (Fig. 5, c and d), similarly as in Rh6mr.
Figure 5.
Spectral properties of blue cone opsin reconstituted with 11-cis-6mr-retinal compared with rod opsin reconstituted with 11-cis-6mr-retinal. a, UV-visible absorption spectra of blue cone opsin regenerated with 11-cis-6mr-retinal (B6mr) in dark conditions (black spectrum) and after illumination for 5 (orange spectrum) and 30 min (red spectrum). Sample illuminated for 30 min was then kept for 1200 min in the dark (red dotted spectrum). b, UV-visible absorption spectra of rod opsin regenerated with 11-cis-6mr-retinal (Rh6mr) in dark conditions (black spectrum) and after illumination for 1 min (red spectrum). Sample illuminated for 30 min was then kept for 1200 min in the dark (black dotted spectrum). c, HPLC elution profile of retinoid oximes extracted from dark-state B6mr (black line) or from B6mr illuminated for 5 min (red line). d, HPLC elution profile of retinoid oximes extracted from dark-state Rh6mr (black line) or from Rh6mr illuminated for 1 min (red line). e, photosensitivity of B6mr. Samples were illuminated with light from a 150-W Fiber-Lite source delivered through a 400–440-nm band pass interference filter at 20 °C. The percentage of residual pigment was plotted against the incident photon count and fitted with an exponential function. The slope of the fitting line corresponds to the relative photosensitivity of the pigment at the irradiating wavelength. Error bars represent standard deviation (S.D.). f, accessibility of the bulk solvent to the protonated Schiff base in blue cone opsin regenerated with 11-cis-retinal (Blue opsin; light blue) or 11-cis-6mr-retinal (B6mr; dark blue). The UV-visible absorption spectra were recorded every 2 min at 20 °C. The percentage of residual pigment was plotted as a function of time and fitted with an exponential function. Error bars represent S.D. φ, quantum yield.
Figure 6.
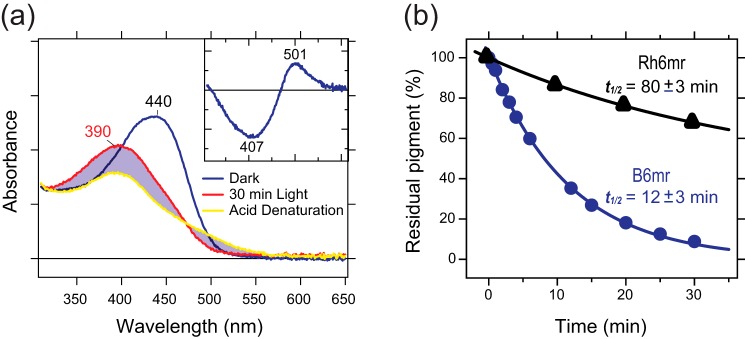
Thermal stability and acidification of blue cone opsin reconstituted with 11-cis-6mr-retinal. a, UV-visible absorption spectra of dark-state B6mr (blue line), light-activated Meta-II–like B6mr (red line), and Meta-II–like B6mr after acid denaturation (yellow line). The pH of the sample was adjusted to 2 with H2SO4 to protonate the Schiff base. Inset in a, the difference spectrum of acidified sample indicates that the Schiff base in Meta-II-like B6mr is in a deprotonated state. b, the thermal stability of purified Rh6mr or B6mr was determined. Samples were incubated at 37 °C in the dark, and their absorbance spectra were recorded every 2 min for 40 min. The change in the absorbance maximum was calculated as a percentage of residual pigment, assuming the absorbance at the initial point as 100%, and plotted as a function of time. These plots were used to calculate the half-lives (t½) of chromophore release. Each experiment was performed in triplicate.
Figure 7.
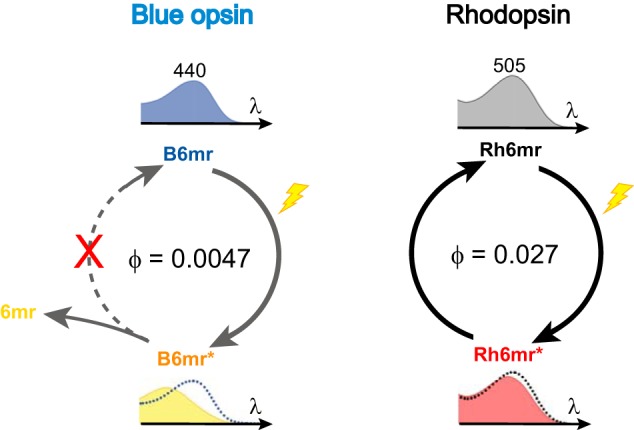
Photochemical modeling of B6mr compared with Rh6mr. B6mr and R6mr feature the red-shifted λmax relative to WT blue cone opsin and Rh (at 440 and 505 nm, respectively). Upon prolonged light illumination, B6mr is converted to the active Meta-II–like state, and 11-cis-6mr-retinal (6mr) is released from the binding pocket, whereas Rh6mr displays reversible photochemical behavior, resulting in back-conversion of its Meta-II–like state to the ground R6mr state. Calculated quantum yields (φ) of B6mr and R6mr were 0.0047 and 0.027, respectively. * indicates the photoactivated (Meta II-like) state of B6mr and Rh6mr.
Previous studies showed that the inactive-state Rh is inaccessible to bulk water; however, binding of 11-cis-6mr-retinal significantly increased solvent accessibility in the inactive state of Rh6mr (21). Thus, we evaluated the accessibility of bulk water to B6mr compared with WT blue cone opsin. Both WT blue cone opsin and B6mr were sensitive to hydroxylamine (NH2OH) in the dark with a half-life of the chromophore release (t½) of 6.67 ± 0.54 and 8.99 ± 0.71 min, respectively. A faster retinal hydrolysis in the presence of NH2OH suggests increased accessibility of bulk water into the chromophore-binding pocket (Fig. 5f). This indicates that binding of 11-cis-6mr-retinal facilitated an additional opening of the cytoplasmic side in B6mr. Next, we assessed the thermal stability of B6mr relative to Rh6mr (Fig. 6b). The results showed that the half-life of the chromophore release in B6mr (t½ = 12 ± 3 min) was significantly shorter than in Rh6mr (t½ = 80 ± 3 min), suggesting a less stable Schiff base formation with Lys7.43 of B6mr as compared with Rh6mr.
Discussion
In addition to the native chromophore 11-cis-retinal, rod opsin can bind retinal geometric isomers and various synthetic retinal analogs (34–36). In this study, we analyzed the binding properties of 11-cis-6mr-retinal to human cone opsin receptors and compared their biochemical and biophysical properties with Rh. As previously reported, rod opsin can efficiently accommodate 11-cis-6mr-retinal in the retinal-binding pocket (4, 22, 30, 34). However, 11-cis-6mr-retinal does not fit very well into the chromophore-binding pocket of green cone opsin (23). Here, we extended this investigation and explored the capability of blue and red cone opsins to bind 11-cis-6mr-retinal. Upon incubation of isolated opsin membranes with 11-cis-6mr-retinal, we found that only blue cone opsin could bind this retinal analog and form a functional pigment absorbing in a blue wavelength range. Similar to Rh6mr, B6mr features a significant change in its spectral properties, displaying a 25-nm red-shifted λmax. This change could be attributed to the differences in the interaction profile of 11-cis-6mr-retinal and 11-cis-retinal with the retinal-binding pocket of blue cone opsin. Additionally, distortion in the C12–C13=C14–C15 dihedral angle of 11-cis-6mr-retinal might contribute to the observed spectral shift. The crystal structure of bovine Rh shows a distortion of the retinal polyene chain at positions C11 and C12, which likely plays a key role in achieving an ultrafast retinal isomerization rate (5). Thus, incorporation of a 6mr-ring between C10 and C13 to the retinal would release the structural constraints and induce depolarization of π-electron on the polyene chain, resulting in the spectral red shift.
Furthermore, the conformation of the C6–C7 single bond is also one of the key factors for spectral tuning. For the bovine Rh, C6–C7 bond is 6-s-cis conformer, resulting in the nonplanar structure from polyene chain to β-ionone ring (37). This nonplanar conformation causes spectral blue shift due to localization of π-electron along the polyene chain. Therefore, the chromophore-binding pocket of B6mr might promote the polyene chain and β-ionone ring planar structure to lead to spectral red shift.
Conversion of Rh to the active Meta-II state in response to light is associated with a 118-nm blue shift in the λmax (498 → 380 nm), occurring as a consequence of the Schiff base deprotonation (4, 30, 38). In Rh6mr, such transition to the active Meta-II–like state is accompanied by a much smaller 8-nm blue shift in the λmax (505 → 497 nm) (21), likely due to the protonated Schiff base, as in Meta-I state of Rh, also featuring a small 8-nm blue shift (4, 30). However, the photostationary Meta-II–like state of B6mr is associated with a much larger 50-nm blue shift in the λmax (440 → 390 nm), which strongly suggests the occurrence of a light-induced deprotonation of the Schiff base. Indeed, acidification of the B6mr Meta-II–like state confirmed deprotonation of the Schiff base upon light illumination (Fig. 6a). In contrast, the Meta-II–like state of Rh6mr features the protonated Schiff base (21). This major difference in the protonation states of Rh6mr and B6mr could explain the lack of photocyclic behavior of B6mr (Fig. 7).
Multiple-sequence alignment of Rh and cone opsins revealed key differences in the amino acid composition of their chromophore-binding pockets. Whereas blue cone opsin features a Tyr residue at position 6.48, green and red cone opsins comprise a Trp residue. This variation in the chromophore environment is likely responsible for the spacial constraints during chromophore binding. To confirm this possibility, in silico calculations of the binding free energies and the pairwise interaction energies between 11-cis-6mr-retinal and the 6.48 residue were performed for WT blue, WT green, Y262W blue, and W281Y green cone opsins. The binding of 11-cis-6mr-retinal to cone opsins was energetically more favorable in the presence of a smaller Tyr residue at position 6.48. Notably, a bulkier Trp resulted in steric hindrance with 11-cis-6mr-retinal, and thus structurally constrained the tolerance of the chromophore-binding pocket for selective binding of 11-cis-6mr-retinal. Indeed, as noted in monkey blue cone opsin, the Y262W mutation resulted in alteration of the chromophore environment and increased the steric constraints along the polyene chain of retinal (39). This caused a planar conformation of the chromophore, which significantly increases the retinal-binding energy. In contrast, a W281Y substitution in green cone opsin reduced the steric clashes, allowing the binding of 11-cis-6mr-retinal. In addition, a W281Y mutation alters the spectral properties of green cone opsin, resulting in the blue shift of the λmax relative to 11-cis-retinal–bound WT green opsin. A similar effect was also reported for another amino acid implicated in the stability of the chromophore-binding pocket in green cone opsin, namely the Pro-2055.40 residue (23, 40). Thus, the environment around the chromophore and/or the specific electrostatic state of the chromophore is essential for the spectral and functional properties of the visual receptors.
Another facet of regulating the spectral tuning in visual pigments involves a network of protein-bound water molecules. It is well known that several water molecules are situated in the vicinity of the retinal chromophore in Rh. These water molecules form an intricate hydrogen bonding network that undergoes significant changes during light-induced activation (41–44). Additionally, some water molecules also facilitate the Schiff base hydrolysis resulting in the chromophore release (31, 45). In cone opsins, an internal water molecule network in the chromophore-binding pocket regulates the spectral properties of cone opsins and has significant effects on the rates of G protein activation (39, 46, 47). Remarkably, a specific water cluster in the chromophore-binding pocket of blue cone opsin was shown to be influenced by the Y262W mutation (39). Therefore, this water-mediated hydrogen bonding network embedded in the chromophore-binding pocket might play a role in providing the structural flexibility required for the binding of 11-cis-6mr-retinal to blue cone opsin.
This study also revealed key structural differences in the chromophore-binding pocket of human cone opsins based on their unique ability to accommodate 11-cis-6mr-retinal. Amino acid residue 6.48 is the key selectivity filter that works in conjunction with surrounding residues to determine binding of 11-cis-6mr-retinal in the chromophore-binding pocket. The presence of smaller Tyr in the retinal-binding pocket instead of bulkier Trp was crucial for the ability to accommodate 11-cis-6mr-retinal to blue cone opsin and W281Y green opsin mutant. Interestingly, this single amino acid substitution in green cone opsin permitted the binding of 11-cis-6mr-retinal. In our previous study (23), we discovered the role of the green cone opsin N terminus for the retinal-binding selectivity. Both green and red cone opsins possess a longer N terminus as compared with blue cone opsin and Rh. Interestingly, this feature is associated with their inability to bind 11-cis-6mr-retinal. Removal of the first 16 N-terminal amino acid residues of green cone opsin enables the binding of 11-cis-6mr-retinal, suggesting that such extended N terminus contributes to protein rigidity. The molecular modeling analysis revealed that the N terminus of green cone opsin forms a short α-helix that interacts with transmembrane helices TM5 and TM6, most likely stabilizing these helices. Interestingly, the interface formed by TM5 and TM6 is the most favorable entrance into the retinal-binding pocket. Removal of the N terminus resulted in increased protein flexibility, allowing 11-cis-6mr-retinal to gain access into the chromophore-binding pocket. Thus, it is likely that among other specific signatures of the chromophore-binding pocket, blue cone opsin and rod opsin are inherently more flexible due to their shorter N terminus as compared with green cone opsin. Trp-281 is directly adjacent to the polyene chain of 11-cis-retinal chromophore, and its substitution to the smaller and more flexible Tyr residue, together with the rearrangement of the associated structural water molecules, might introduce enough conformational flexibility to enable the entry and binding of 11-cis-6mr-retinal even in the presence of the intact N terminus. Overall, both the N terminus and the retinal-binding pocket composition define the selectivity of retinal binding.
Together, these findings provide viable information for designing a new class of retinal-based drug treatment for color blindness. Further investigations will aid in pinpointing all determinants of the chromophore-binding specificity in cone opsins required to advance these developments.
In summary, similar to green cone opsin (23), incubation of red cone opsin with 11-cis-6mr-retinal did not result in the formation of a light-absorbing pigment. Interestingly, 11-cis-6mr-retinal formed a Schiff base with blue cone opsin, resulting in the formation of pigment (B6mr) exhibiting an absorption maximum at 440 nm. These results indicate that the chromophore-binding pocket of blue cone opsin is more flexible as compared with green or red cone opsin and could accommodate the bulkier cyclo-hexyl ring of 11-cis-6mr-retinal. The primary sequence alignment of human Rh and cone opsins in combination with the molecular modeling analyses revealed a highly conserved residue homologous to Trp-265 (residue 6.48) (28, 48) as the key selectivity filter for the chromophore binding in cone opsins. Furthermore, the molecular modeling analyses indicated a more stable interaction between the 11-cis-6mr-retinal and the swapped W281Y green cone opsin mutant with a lower binding energy relative to WT green cone opsin. However, mutation of the Tyr-262 residue to Trp in blue cone opsin completely ablated its binding with 11-cis-6mr-retinal. In agreement with these in silico binding energy calculations, the substitution of Trp-281 residue to Tyr in green cone opsin enabled binding of 11-cis-6mr-retinal. Moreover, unlike Rh6mr, B6mr did not exhibit the photocyclic behavior upon illumination. Altogether, these findings imply that the chromophore-binding pocket of blue cone opsin is optimized to facilitate the Schiff base hydrolysis for faster chromophore release as compared with Rh.
Experimental procedures
Chemicals
n-Dodecyl β-d-maltoside (DDM) was purchased from Affymetrix Inc. (Maumee, OH). 11-cis-Retinal was a generous gift from Dr. Rosaline Crouch (Medical University of South Carolina, Charleston, SC). 11-cis-6mr-Retinal was synthesized as described previously (21, 23) and provided by Novartis (Cambridge, MA).
Constructs
Human blue, green, and red cone opsin cDNAs cloned into a pUC57 vector were synthesized by Genentech (San Francisco, CA). The last 12 amino acids in each cone opsin were replaced by the rod opsin C-terminal amino acid sequence (TETSQVAPA; called 1D4 tag) to enable protein purification. These constructs were subcloned into a pcDNA3.1(+) vector (Invitrogen) according to the manufacturer's protocol. The resulting constructs were used for protein purification and UV-visible spectroscopy experiments, and the sequence of each construct was confirmed by DNA sequencing.
Mutagenesis
The blue cone opsin Y262W and the green cone opsin W281Y mutants were full-length constructs with intact N termini. Mutants of blue and green cone opsin were generated with the Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) according to the manufacturer's procedures.
Expression of cone opsins in insect cells and membrane isolation
Expression constructs of human blue, green, and red cone opsins were prepared in pFastBac HT vectors (Invitrogen) with their N-terminal His tags removed. Constructs were transformed into DH10 Bac to obtain a Bacmid for transfection with X-tremeGene 9 (Roche Applied Science) according to the manufacturer's instructions. Briefly, 3–4 days after transfection with P1 virus, the cell supernatant was collected. P2 and P3 viruses were collected similarly and stored. For larger-scale expression, Sf9 cells at a density of 3.5 × 106 cells/ml of culture were infected with P3 stage virus at a 1:100 volume ratio and cultured for 2 days with shaking at 135 rpm in a 28 °C incubator. Cells were collected 48 h postinfection, and the cell pellet was either kept at −80 °C or used immediately for membrane isolation.
To isolate membranes, first cells were homogenized on ice with a Dounce homogenizer in a hypotonic buffer (25 mm HEPES, pH 7.5, 10 mm MgCl2, 20 mm KCl containing EDTA-free Complete protease inhibitor mixture (Roche Diagnostics GmbH)). The homogenate was then centrifuged at 100,000 × g for 30 min. The supernatant was discarded, and the membrane pellet was homogenized in the same buffer and centrifuged at 100,000 × g for 30 min again. Next, membranes were washed three to four times with 25 mm HEPES, pH 7.5, 1.0 m NaCl, 10 mm MgCl2, 20 mm KCl containing EDTA-free Complete protease inhibitor mixture. Finally, the washed membranes were resuspended in 50% (v/v) glycerol, flash frozen with liquid nitrogen, and stored at −80 °C or used immediately. Pigments were reconstituted with either 11-cis-retinal or 11-cis-6mr-retinal. Retinal was added to the membranes from a DMSO stock solution to a final concentration of 10 μm and then incubated in the dark for 1 h at 4 °C. Regenerated pigments were purified by 1D4 immunoaffinity chromatography.
Expression of cone opsins in HEK-293 cells and pigment reconstitution
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Hyclone, Logan, UT), 5 μg/ml Plasmocin (InvivoGen, San Diego, CA), and 1 unit/ml penicillin with 1 μg/ml streptomycin (Life Technologies) at 37 °C under 5% CO2. Cells were transiently transfected with WT blue, green, or red cone opsin constructs and blue Y262W or green W281Y mutant constructs cloned into pcDNA3.1(+) vectors (Takara Bio USA, Inc., Mountain View, CA) with polyethylenimine (49, 50). Twenty-four hours post-transfection, cells were collected from 40 10-cm plates, and cone opsins were reconstituted with either 11-cis-retinal or 11-cis-6mr-retinal. Alternatively, cell pellets were stored at −80 °C until used. Retinal was added to the cell pellets from a DMSO stock solution to a final concentration of 10 μm and incubated in the dark for 1 h at 4 °C. Regenerated pigments were purified by 1D4 immunoaffinity chromatography.
Preparation of opsin membranes and pigment regeneration
Bovine ROS membranes were isolated from 100 frozen retinas under dim red light as described previously (51). To prepare opsin-containing membranes, ROSs were suspended in 10 mm sodium phosphate, pH 7.0, 50 mm hydroxylamine with Rh concentration of 2 mg/ml; placed on ice; and illuminated with a 150-watt bulb for 30 min at a distance of 15 cm. Then membranes were centrifuged at 16,000 × g for 10 min. The supernatant was discarded, and the pellet was washed four times with 10 mm sodium phosphate, pH 7.0, followed by four washes with 2% BSA and then with four washes with 10 mm sodium phosphate, pH 7.0. The final membrane wash was performed in 20 mm Bistris propane, 100 mm NaCl, pH 7.5, twice. Rh and opsin concentrations were measured with a UV-visible spectrophotometer (Cary 50, Varian, Palo Alto, CA) and quantified using the absorption coefficients ϵ500 nm = 40,600 m−1 cm−1 and ϵ280 nm = 81,200 m−1 cm−1, respectively (52). Regeneration of Rh or Rh6mr with 11-cis-retinal or 11-cis-6mr-retinal, respectively, was accomplished by incubating opsin membranes with the respective retinal (10 μm) overnight at 4 °C. Regenerated pigments were purified by 1D4 immunoaffinity chromatography.
Pigment purification by 1D4 immunoaffinity chromatography
HEK-293T cells transiently transfected with the WT blue, green, or red cone opsin constructs and the blue Y262W or green W281Y mutant constructs were harvested from 40 10-cm plates and centrifuged at 800 × g. Cell pellets were suspended in 50 mm HEPES, 150 mm NaCl, 20 mm DDM, pH 7.0, containing protease inhibitor mixture and incubated for 1 h at 4 °C on a rotating platform. Alternatively, insect cell membranes with reconstituted cone opsins or regenerated bovine ROS membranes were solubilized with the above buffer. The lysate then was centrifuged at 100,000 × g for 1 h at 4 °C, and cone or rod pigments were purified from the supernatant by immunoaffinity chromatography with an anti-Rh C-terminal 1D4 antibody immobilized on cyanogen bromide–activated agarose. Four hundred μl of 6 mg of 1D4/ml of agarose beads were added to the supernatant and incubated for 1 h at 4 °C on the rotating platform. The resin was then transferred to a column and washed with 10 ml of buffer composed of 50 mm HEPES, 150 mm NaCl, and 2 mm DDM, pH 7.0. Pigments were eluted with buffer composed of 150 mm HEPES, 150 mm NaCl, and 2 mm DDM, pH 7.0, supplemented with 0.6 mg/ml TETSQVAPA peptide.
UV-visible spectroscopy of opsin pigments
UV-visible spectra of freshly purified cone or rod pigment samples were recorded in the dark with a Cary 50 UV-visible spectrophotometer. Spectra of the samples were recorded after being exposed to white light delivered from a Fiber-Lite illuminator (150-W lamp) (Dolan-Jenner, Boxborough, MA) at a distance of 10 cm for the indicated time. A band pass 400–440-nm filter was used for illumination of blue cone opsin, and A 480–520-nm filter was used for green cone opsin and Rh. To obtain a difference spectrum of purified cone pigments, the spectrum of the sample recorded in the dark was subtracted from the spectrum of the light-illuminated sample. To check the protonation state of the protein Schiff base, 1 μl of concentrated H2SO4 was added to the sample loaded to the cuvette to decrease the sample pH to 2, and the spectrum was recorded immediately.
HPLC analyses
Samples of either blue cone opsin (B6mr) or bovine Rh (Rh6mr) regenerated with 11-cis-6mr-retinal after their purification by 1D4 immunoaffinity chromatography were denatured for 30 min at room temperature with 50% CH3OH in 100 mm NH2OH. The resulting retinal oximes were extracted with 600 μl of hexane, and their isomeric content was determined by normal-phase HPLC with a Luna 10-μm C18 silica 100-Å, 250 × 4.6-mm column (Beckman). Retinoids were eluted isocratically with 10% ethyl acetate in hexane at a flow rate of 1.4 ml/min. Their signals were detected by absorption at 360 nm (53, 54).
Photosensitivity measurements
Photosensitivity measurements of B6mr and Rh6mr were performed as described previously (21, 32, 55–57). Briefly, B6mr samples maintained at 20 °C were irradiated with light from a 150-W Fiber-Lite source delivered through a 400–440-nm band pass interference filter. The cuvette (width, 4 mm; path length, 1 cm) was placed in a cell holder maintained at 20 °C, and the absorbance change was measured with a UV-visible spectrophotometer. The light intensity was controlled by using a neutral density filter (Thorlabs Inc., Newton, NJ) with a maximum intensity set to either bleach <90% of Rh or deliver 450 μW to B6mr due to its low quantum yield as compared with Rh. B6mr was illuminated for 120 min. UV-visible spectra were recorded at 1.5, 3.5, 5.5, 7.5, 12.5, 20, 37.5, 60, 75, 90, and 120 min. The amount of residual pigment after each irradiation was corrected with the dark spectrum of the pigment. The incident photon flux (s−1 cm−2) was calculated from the power (W) of the incident light measured by a power meter (Thorlabs Inc.). The intensity of the incident light was continuously monitored to correct for any power fluctuations. The percentage of residual pigment was plotted against the incident photon count and fitted with an exponential function. The slope of the fitting line corresponded to the relative photosensitivity of the pigment at the irradiating wavelength (32, 55, 56). Rh photosensitivity was used as a control for all measurements, and the photosensitivity of B6mr was determined as a value relative to that of Rh.
Thermal stability
Purified Rh6mr or B6mr diluted with 50 mm HEPES, 150 mm NaCl, and 2 mm DDM, pH 7.0, to a final volume of 0.4 ml was incubated at 55 °C in the dark. The absorbance spectra were collected every 2 min for 40 min. The absorbance maximum at the initial time point was anticipated to be 100% and used to calculate the percentage of the remaining pigments at specific time points and then plotted as a function of time. These plots were used to calculate the half-lives (t½) of chromophore release upon thermal denaturation.
Modeling
The models of the green and blue cone opsins were generated with the Rosetta software suite (59) as described previously (23). The crystal structure of Rh (Protein Data Bank (PDB) 1U19) (60) and the bovine Rh sequence (UniProtKB accession number P02699) were used as the template for the human green and blue cone opsin protein sequences (UniProtKB accession numbers P04001 and P03999, respectively). The most energetically favorable models were selected.
Retinal was parameterized for use within Rosetta (61). Because the retinal ligand is bound to Lys-312 in green cone opsin and Lys-293 in blue cone opsin by a protonated Schiff base, the ligand–residue complex was treated as a noncanonical amino acid, allowing the structure to be flawlessly used within the Rosetta framework. The coordinates for the retinal–lysine complex were extracted from the 2.2-Å resolution crystal structure of Rh (PDB code 1U19), and a Rosetta noncanonical amino acid parameter file was created. The fixed-backbone design protocol in Rosetta was used to mutate Lys-312 in green cone opsin or Lys-293 in blue cone opsin in the retinal–lysine complex, which was introduced with the conformation present in the crystal structure. All other coordinates in the cone opsin structures were kept constant. To remove any structural inconsistencies or clashes introduced into the cone opsin models, the structure was relaxed with the Rosetta membrane fast relax protocol (58), and the most energetically favorable models were selected. These modeling procedures were repeated to produce a model with 11-cis-6mr-retinal chromophore in the binding site.
Author contributions
K. K., S. G., K. P., and B. J. conceptualization; K. K., S. G., K. P., and B. J. data curation; K. K., S. G., J. T. O., N. S. A., K. P., and B. J. formal analysis; K. K., S. G., and B. J. validation; K. K., S. G., J. T. O., N. S. A., W. S., M. M. S., and B. J. investigation; K. K., S. G., J. T. O., and B. J. visualization; K. K., S. G., J. T. O., N. S. A., W. S., and B. J. methodology; K. K. and B. J. writing-original draft; K. K., S. G., J. T. O., N. S. A., W. S., M. M. S., K. P., and B. J. writing-review and editing; K. P. and B. J. resources; K. P. and B. J. supervision; K. P. and B. J. funding acquisition; K. P. and B. J. project administration.
Acknowledgments
We thank the members of Jastrzebska laboratory for helpful comments on the manuscript. We are also grateful to Dr. Muneto Mogi (Novartis Inc.) for the 11-cis-6mr-retinal.
This work was supported in part by National Institutes of Health Grants EY025214 (to B. J.), EY027283, EY024864, EY009339, EY025451 (all to K. P.), and P30EY011373, CORE grant for Vision Research awarded to the Visual Sciences Research Center (VSRC) at Case Western Reserve University; a Research to Prevent Blindness (RPB) grant (to the Department of Ophthalmology at University of California Irvine); the Canadian Institute for Advanced Research (CIFAR); and Alcon Research Institute (ARI). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Rh
- rhodopsin
- B6mr
- 11-cis-6mr-retinal–bound blue cone opsin
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- DDM
- n-dodecyl β-d-maltopyranoside
- Meta
- metarhodopsin
- ROS
- rod outer segment
- Rh6mr
- 11-cis-6mr-retinal-bound rhodopsin
- TM
- transmembrane
- 11-cis-6mr-retinal
- 11-cis-6-membered-ring-retinal
- W
- watt.
References
- 1. Palczewski K. (2006) G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 10.1146/annurev.biochem.75.103004.142743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ebrey T., and Koutalos Y. (2001) Vertebrate photoreceptors. Prog. Retin. Eye Res. 20, 49–94 10.1016/S1350-9462(00)00014-8 [DOI] [PubMed] [Google Scholar]
- 3. Imamoto Y., Seki I., Yamashita T., and Shichida Y. (2013) Efficiencies of activation of transducin by cone and rod visual pigments. Biochemistry 52, 3010–3018 10.1021/bi3015967 [DOI] [PubMed] [Google Scholar]
- 4. Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., and Kandori H. (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 10.1021/cr4003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., and Miyano M. (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- 6. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., and Ernst O. P. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 10.1038/nature07330 [DOI] [PubMed] [Google Scholar]
- 7. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., and Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 10.1038/nature07063 [DOI] [PubMed] [Google Scholar]
- 8. Salom D., Lodowski D. T., Stenkamp R. E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J. A., and Palczewski K. (2006) Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128 10.1073/pnas.0608022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., and Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 10.1038/nature09789 [DOI] [PubMed] [Google Scholar]
- 10. Mertz B., Struts A. V., Feller S. E., and Brown M. F. (2012) Molecular simulations and solid-state NMR investigate dynamical structure in rhodopsin activation. Biochim. Biophys. Acta 1818, 241–251 10.1016/j.bbamem.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pope A., Eilers M., Reeves P. J., and Smith S. O. (2014) Amino acid conservation and interactions in rhodopsin: probing receptor activation by NMR spectroscopy. Biochim. Biophys. Acta 1837, 683–693 10.1016/j.bbabio.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown M. F., Salgado G. F., and Struts A. V. (2010) Retinal dynamics during light activation of rhodopsin revealed by solid-state NMR spectroscopy. Biochim. Biophys. Acta 1798, 177–193 10.1016/j.bbamem.2009.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heck M., Schädel S. A., Maretzki D., Bartl F. J., Ritter E., Palczewski K., and Hofmann K. P. (2003) Signaling states of rhodopsin. Formation of the storage form, metarhodopsin III, from active metarhodopsin II. J. Biol. Chem. 278, 3162–3169 10.1074/jbc.M209675200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furutani Y., Shichida Y., and Kandori H. (2003) Structural changes of water molecules during the photoactivation processes in bovine rhodopsin. Biochemistry 42, 9619–9625 10.1021/bi034592k [DOI] [PubMed] [Google Scholar]
- 15. Vogel R., Siebert F., Lüdeke S., Hirshfeld A., and Sheves M. (2005) Agonists and partial agonists of rhodopsin: retinals with ring modifications. Biochemistry 44, 11684–11699 10.1021/bi0508587 [DOI] [PubMed] [Google Scholar]
- 16. Yan E. C., Ganim Z., Kazmi M. A., Chang B. S., Sakmar T. P., and Mathies R. A. (2004) Resonance Raman analysis of the mechanism of energy storage and chromophore distortion in the primary visual photoproduct. Biochemistry 43, 10867–10876 10.1021/bi0400148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J. E., Pan D., and Mathies R. A. (2003) Picosecond dynamics of G-protein coupled receptor activation in rhodopsin from time-resolved UV resonance Raman spectroscopy. Biochemistry 42, 5169–5175 10.1021/bi030026d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakanishi K., Yudd A. P., Crouch R. K., Olson G. L., Cheung H. C., Govindjee R., Ebrey T. G., and Patel D. J. (1976) Letter: allenic retinals and visual pigment analogues. J. Am. Chem. Soc. 98, 236–238 10.1021/ja00417a040 [DOI] [PubMed] [Google Scholar]
- 19. Shichida Y., Nakamura K., Yoshizawa T., Trehan A., Denny M., and Liu R. S. (1988) 9,13-dicis-Rhodopsin and its one-photon-one-double-bond isomerization. Biochemistry 27, 6495–6499 10.1021/bi00417a044 [DOI] [PubMed] [Google Scholar]
- 20. Isayama T., Chen Y., Kono M., Degrip W. J., Ma J. X., Crouch R. K., and Makino C. L. (2006) Differences in the pharmacological activation of visual opsins. Vis. Neurosci. 23, 899–908 10.1017/S0952523806230256 [DOI] [PubMed] [Google Scholar]
- 21. Gulati S., Jastrzebska B., Banerjee S., Placeres Á. L., Miszta P., Gao S., Gunderson K., Tochtrop G. P., Filipek S., Katayama K., Kiser P. D., Mogi M., Stewart P. L., and Palczewski K. (2017) Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism. Proc. Natl. Acad. Sci. U.S.A. 114, E2608–E2615 10.1073/pnas.1617446114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kono M., and Crouch R. K. (2011) Probing human red cone opsin activity with retinal analogues. J. Nat. Prod. 74, 391–394 10.1021/np100749j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alexander N. S., Katayama K., Sun W., Salom D., Gulati S., Zhang J., Mogi M., Palczewski K., and Jastrzebska B. (2017) Complex binding pathways determine the regeneration of mammalian green cone opsin with a locked retinal analogue. J. Biol. Chem. 292, 10983–10997 10.1074/jbc.M117.780478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oprian D. D., Asenjo A. B., Lee N., and Pelletier S. L. (1991) Design, chemical synthesis, and expression of genes for the three human color vision pigments. Biochemistry 30, 11367–11372 10.1021/bi00112a002 [DOI] [PubMed] [Google Scholar]
- 25. Ma J., Znoiko S., Othersen K. L., Ryan J. C., Das J., Isayama T., Kono M., Oprian D. D., Corson D. W., Cornwall M. C., Cameron D. A., Harosi F. I., Makino C. L., and Crouch R. K. (2001) A visual pigment expressed in both rod and cone photoreceptors. Neuron 32, 451–461 10.1016/S0896-6273(01)00482-2 [DOI] [PubMed] [Google Scholar]
- 26. Stenkamp R. E., Filipek S., Driessen C. A., Teller D. C., and Palczewski K. (2002) Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim. Biophys. Acta 1565, 168–182 10.1016/S0005-2736(02)00567-9 [DOI] [PubMed] [Google Scholar]
- 27. Fasick J. I., Lee N., and Oprian D. D. (1999) Spectral tuning in the human blue cone pigment. Biochemistry 38, 11593–11596 10.1021/bi991600h [DOI] [PubMed] [Google Scholar]
- 28. Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J. A., and Javitch J. A. (2002) β2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J. Biol. Chem. 277, 40989–40996 10.1074/jbc.M206801200 [DOI] [PubMed] [Google Scholar]
- 29. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., and Elling C. E. (2006) Molecular mechanism of 7TM receptor activation—a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 10.1146/annurev.pharmtox.46.120604.141218 [DOI] [PubMed] [Google Scholar]
- 30. Palczewski K., Hofmann K. P., and Baehr W. (2006) Rhodopsin—advances and perspectives. Vision Res. 46, 4425–4426 10.1016/j.visres.2006.10.009 [DOI] [PubMed] [Google Scholar]
- 31. Stenkamp R. E., Teller D. C., and Palczewski K. (2002) Crystal structure of rhodopsin: a G-protein-coupled receptor. Chembiochem 3, 963–967 [DOI] [PubMed] [Google Scholar]
- 32. Kim J. E., Tauber M. J., and Mathies R. A. (2001) Wavelength dependent cis-trans isomerization in vision. Biochemistry 40, 13774–13778 10.1021/bi0116137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuksa V., Bartl F., Maeda T., Jang G. F., Ritter E., Heck M., Preston Van Hooser J., Liang Y., Filipek S., Gelb M. H., Hofmann K. P., and Palczewski K. (2002) Biochemical and physiological properties of rhodopsin regenerated with 11-cis-6-ring- and 7-ring-retinals. J. Biol. Chem. 277, 42315–42324 10.1074/jbc.M206014200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spalink J. D., Reynolds A. H., Rentzepis P. M., Sperling W., and Applebury M. L. (1983) Bathorhodopsin intermediates from 11-cis-rhodopsin and 9-cis-rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 80, 1887–1891 10.1073/pnas.80.7.1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vogel R., and Siebert F. (2000) Vibrational spectroscopy as a tool for probing protein function. Curr. Opin. Chem. Biol. 4, 518–523 10.1016/S1367-5931(00)00125-3 [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharya S., Ridge K. D., Knox B. E., and Khorana H. G. (1992) Light-stable rhodopsin. I. A rhodopsin analog reconstituted with a nonisomerizable 11-cis retinal derivative. J. Biol. Chem. 267, 6763–6769 [PubMed] [Google Scholar]
- 37. Ahuja S., Eilers M., Hirshfeld A., Yan E. C., Ziliox M., Sakmar T. P., Sheves M., and Smith S. O. (2009) 6-s-cis conformation and polar binding pocket of the retinal chromophore in the photoactivated state of rhodopsin. J. Am. Chem. Soc. 131, 15160–15169 10.1021/ja9034768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jastrzebska B., Orban T., Golczak M., Engel A., and Palczewski K. (2013) Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 27, 1572–1584 10.1096/fj.12-225383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katayama K., Nonaka Y., Tsutsui K., Imai H., and Kandori H. (2017) Spectral tuning mechanism of primate blue-sensitive visual pigment elucidated by FTIR spectroscopy. Sci. Rep. 7, 4904 10.1038/s41598-017-05177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuwayama S., Imai H., Hirano T., Terakita A., and Shichida Y. (2002) Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry 41, 15245–15252 10.1021/bi026444k [DOI] [PubMed] [Google Scholar]
- 41. Yuan S., Filipek S., Palczewski K., and Vogel H. (2014) Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat. Commun. 5, 4733 10.1038/ncomms5733 [DOI] [PubMed] [Google Scholar]
- 42. Okada T., Fujiyoshi Y., Silow M., Navarro J., Landau E. M., and Shichida Y. (2002) Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 5982–5987 10.1073/pnas.082666399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Angel T. E., Gupta S., Jastrzebska B., Palczewski K., and Chance M. R. (2009) Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 10.1073/pnas.0901074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Angel T. E., Chance M. R., and Palczewski K. (2009) Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 10.1073/pnas.0903545106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Janz J. M., and Farrens D. L. (2004) Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J. Biol. Chem. 279, 55886–55894 10.1074/jbc.M408766200 [DOI] [PubMed] [Google Scholar]
- 46. Katayama K., Furutani Y., Imai H., and Kandori H. (2012) Protein-bound water molecules in primate red- and green-sensitive visual pigments. Biochemistry 51, 1126–1133 10.1021/bi201676y [DOI] [PubMed] [Google Scholar]
- 47. Katayama K., Okitsu T., Imai H., Wada A., and Kandori H. (2015) Identical hydrogen-bonding strength of the retinal Schiff base between primate green- and red-sensitive pigments: new insight into color tuning mechanism. J. Phys. Chem. Lett. 6, 1130–1133 10.1021/acs.jpclett.5b00291 [DOI] [PubMed] [Google Scholar]
- 48. Ballesteros J. A., and Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 10.1016/S1043-9471(05)80049-7 [DOI] [Google Scholar]
- 49. Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., and Behr J. P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 10.1073/pnas.92.16.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Y., and Tang H. (2015) High-throughput screening assays to identify small molecules preventing photoreceptor degeneration caused by the rhodopsin P23H mutation. Methods Mol. Biol. 1271, 369–390 10.1007/978-1-4939-2330-4_24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Papermaster D. S. (1982) Preparation of retinal rod outer segments. Methods Enzymol. 81, 48–52 10.1016/S0076-6879(82)81010-0 [DOI] [PubMed] [Google Scholar]
- 52. Wald G., and Brown P. K. (1953) The molecular excitation of rhodopsin. J. Gen. Physiol. 37, 189–200 10.1085/jgp.37.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Van Hooser J. P., Garwin G. G., and Saari J. C. (2000) Analysis of visual cycle in normal and transgenic mice. Methods Enzymol. 316, 565–575 10.1016/S0076-6879(00)16750-3 [DOI] [PubMed] [Google Scholar]
- 54. Garwin G. G., and Saari J. C. (2000) High-performance liquid chromatography analysis of visual cycle retinoids. Methods Enzymol. 316, 313–324 10.1016/S0076-6879(00)16731-X [DOI] [PubMed] [Google Scholar]
- 55. Dartnall H. J. (1968) The photosensitivities of visual pigments in the presence of hydroxylamine. Vision Res. 8, 339–358 10.1016/0042-6989(68)90104-1 [DOI] [PubMed] [Google Scholar]
- 56. Okano T., Fukada Y., Shichida Y., and Yoshizawa T. (1992) Photosensitivities of iodopsin and rhodopsins. Photochem. Photobiol. 56, 995–1001 10.1111/j.1751-1097.1992.tb09722.x [DOI] [PubMed] [Google Scholar]
- 57. Tsutsui K., Imai H., and Shichida Y. (2007) Photoisomerization efficiency in UV-absorbing visual pigments: protein-directed isomerization of an unprotonated retinal Schiff base. Biochemistry 46, 6437–6445 10.1021/bi7003763 [DOI] [PubMed] [Google Scholar]
- 58. Barth P., Schonbrun J., and Baker D. (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U.S.A. 104, 15682–15687 10.1073/pnas.0702515104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee J., Lee D., Park H., Coutsias E. A., and Seok C. (2010) Protein loop modeling by using fragment assembly and analytical loop closure. Proteins 78, 3428–3436 10.1002/prot.22849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., and Buss V. (2004) The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 342, 571–583 10.1016/j.jmb.2004.07.044 [DOI] [PubMed] [Google Scholar]
- 61. Kaufmann K. W., Lemmon G. H., Deluca S. L., Sheehan J. H., and Meiler J. (2010) Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry 49, 2987–2998 10.1021/bi902153g [DOI] [PMC free article] [PubMed] [Google Scholar]