Abstract
Focal adhesions (FAs) are specialized sites where intracellular cytoskeleton elements connect to the extracellular matrix and thereby control cell motility. FA assembly depends on various scaffold proteins, including the G protein–coupled receptor kinase-interacting protein 1 (GIT1), paxillin, and liprin-α. Although liprin-α and paxillin are known to competitively interact with GIT1, the molecular basis governing these interactions remains elusive. To uncover the underlying mechanisms of how GIT1 is involved in FA assembly by alternatively binding to liprin-α and paxillin, here we solved the crystal structures of GIT1 in complex with liprin-α and paxillin at 1.8 and 2.6 Å resolutions, respectively. These structures revealed that the paxillin-binding domain (PBD) of GIT1 employs distinct binding modes to recognize a single α-helix of liprin-α and the LD4 motif of paxillin. Structure-based design of protein variants produced two binding-deficient GIT1 variants; specifically, these variants lost the ability to interact with liprin-α only or with both liprin-α and paxillin. Expressing the GIT1 variants in COS7 cells, we discovered that the two PBD-meditated interactions play different roles in either recruiting GIT1 to FA or facilitating FA assembly. Additionally, we demonstrate that, unlike for the known binding mode of the FAT domain to LD motifs, the PBD of GIT1 uses different surface patches to achieve high selectivity in LD motif recognition. In summary, our results have uncovered the mechanisms by which GIT1's PBD recognizes cognate paxillin and liprin-α structures, information we anticipate will be useful for future investigations of GIT1–protein interactions in cells.
Keywords: focal adhesion, crystal structure, structure-function, protein-protein interaction, protein complex, structural biology, cell migration, GTPase activating protein (GAP), complex structure, paxillin (PXN), PTPRF-interacting protein alpha 2 (PPFIA2), single alpha-helix
Introduction
Focal adhesions (FAs)4 are protein-rich, highly specified subcellular structures in mediating the connection between cell and extracellular matrix, which are essential in various cellular processes, including cell spreading, migration, cancer invasion, and neuronal growth (1–4). These processes are closely associated with FA dynamics, which are regulated by FA-associated adaptor and scaffold proteins (5–8). Among these FA-associated proteins, G protein–coupled receptor kinase–interacting proteins, GIT1 and GIT2 serve as GTPase-activating proteins (GAPs) for ADP-ribosylation factors (Arfs) to affect the assembly of integrin adhesion complexes to FAs (9, 10).
The GIT proteins share a conserved domain architecture, which from N terminus to C terminus includes an Arf-GAP domain, three ankyrin repeats, a Spa homology domain, a coiled-coil region, and a paxillin-binding domain (PBD) (Fig. 1A). GITs are involved in regulating FA dynamics by interacting with many FA proteins (11, 12). Among these interactions, the bindings of liprin-α, paxillin, and HIC5, a paxillin-like protein, to GIT1 are mediated by the PBD, folded as a four-helix bundle (13–17). A similar fold is also found in focal adhesion kinase (FAK), termed the FAK-targeting (FAT) domain. Previous structural studies indicated that the FAT-like domains, including PBD, employ a similar way to recognize leucine-aspartic acid (LD) motifs, short helical interacting motifs (18–20).
Figure 1.
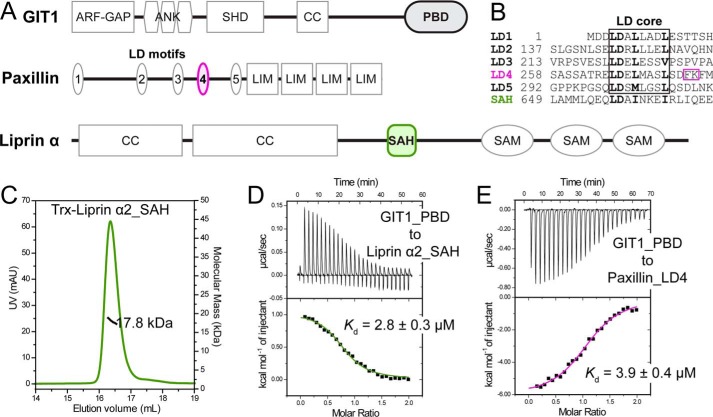
Biochemical characterization of the GIT1/liprin-α and GIT1/paxillin interactions. A, cartoon diagrams of domain organizations for GIT1, paxillin, and liprin-α. The color-coding of the regions is applied throughout the entire paper except as otherwise indicated. B, sequence alignment of five LD motifs in paxillin and the SAH sequence in liprin-α2 showing the consensus sequence in the LD core region. The unique FK sequence found in the LD4 motif is highlighted by a purple box. C, the molecular weight of liprin-α2_SAH was measured using analytical gel filtration chromatography–coupled multi-angle static light scattering. The theoretical molecular mass of monomeric Trx-tagged liprin-α2_SAH is 18.2 kDa. D and E, ITC-based measurement of the bindings of GIT1_PBD to liprin-α2_SAH (D) and paxillin_LD4 (E), respectively. The thermodynamic parameters of the ITC titrations as shown in this and following figures are summarized in Table S1.
The liprin-α family is composed of four highly conserved members (liprin-α1/2/3/4) in mammals; each contains the N-terminal coiled-coils and the three C-terminal sterile α motif repeats (21, 22) (Fig. 1A). As the synaptic scaffold, liprin-α was identified as the GIT1-binding partner in neurons to mediate AMPA receptor targeting (14). In addition to the well-known roles of liprin-α in neurons, emerging evidence has indicated that liprin-α plays an important role in nonneuronal cells by mediating the FA turnover during integrin-mediated migration (23, 24).
Paxillin acts as a scaffold to recruit different proteins in FAs via its five LD motifs and four LIM (Lin11, Isl-1, and Mec-3) domains (18) (Fig. 1A). By connecting integrin to F-actin and other FA-associated proteins, paxillin plays a critical role in assembly and disassembly of FAs (25). Paxillin was suggested to recruit GIT1 to FA by the binding of the LD4 motif of paxillin to the PBD of GIT1, as the deletion of the LD4 motif or the PBD resulted in the disruption of localization of GIT1 to FA (16, 26, 27).
Although the PBD is crucial for the subcellular localization and function of GIT1, the molecular mechanisms underlying the PBD-mediated interactions remain elusive. A recent study indicated that the bindings of liprin-α and paxillin to GIT1 are mutually exclusive (13). However, without detailed interaction information, it would be difficult to understand how these competitive interactions are coordinated and regulated in FA.
Here, we determined crystal structures of GIT1_PBD in complex with a single α-helix (SAH) of liprin-α2 and the LD4 motif of paxillin. Although liprin-α2_SAH and paxillin_LD4 associate with the same binding groove on GIT1_PBD, surprisingly, these two helical peptides adopt binding orientations reversed to each other. Cellular analysis using the designed GIT1 mutations indicated that the GIT1/paxillin interaction controls the GIT1's localization to FA and the GIT1/liprin-α interaction promotes FA assembly. The structure of the GIT1_PBD/paxillin_LD4 complex reveals a novel LD recognition mode that involves a new surface patch on GIT1_PBD in addition to the canonical LD binding surface found in the FAT domain of FAK.
Results
Both liprin-α and paxillin bind with GIT1_PBD via a single α-helix
To understand the assembly mechanism of GIT1, liprin-α, and paxillin, we investigated the binding of GIT1 to liprin-α and paxillin, which is mediated by the paxillin-binding domain of GIT1 (GIT1_PBD). To narrow down the minimal PBD-binding region in liprin-α2, various fragments were expressed in Escherichia coli and purified with high quality (Fig. 1A). As indicated by an isothermal titration calorimetry (ITC)-based assay, a small fragment containing residues 642–671 in liprin-α2 interacts with GIT1_PBD with a binding affinity of ∼3 μm (Fig. 1D). Neither an extension at the N-terminal nor at the C-terminal part of the fragment could significantly increase the binding affinity (Fig. S1A), suggesting that the 30-residue fragment contains essential and sufficient elements for the binding of liprin-α2 to GIT1_PBD. Although this fragment was predicted as a coiled-coil (14), the molecular weight of the fragment measured by multiangle static light scattering matches the theoretical molecular weight of its monomeric state (Fig. 1C and Fig. S1B). Combining the CD spectrum of the fragment (Fig. S1C), we concluded that the fragment folds as a single α-helix. Therefore, the fragment in liprin-α2 was named hereafter as SAH. Similar to liprin-α2_SAH, the LD4 motif of paxillin (paxillin_LD4), also a single α-helix, interacts with GIT1_PBD with a binding affinity of ∼4 μm (Fig. 1E), consistent with the previous reports (15, 16).
Overall structures of GIT1_PBD in complex with liprin-α2_SAH and paxillin_LD4
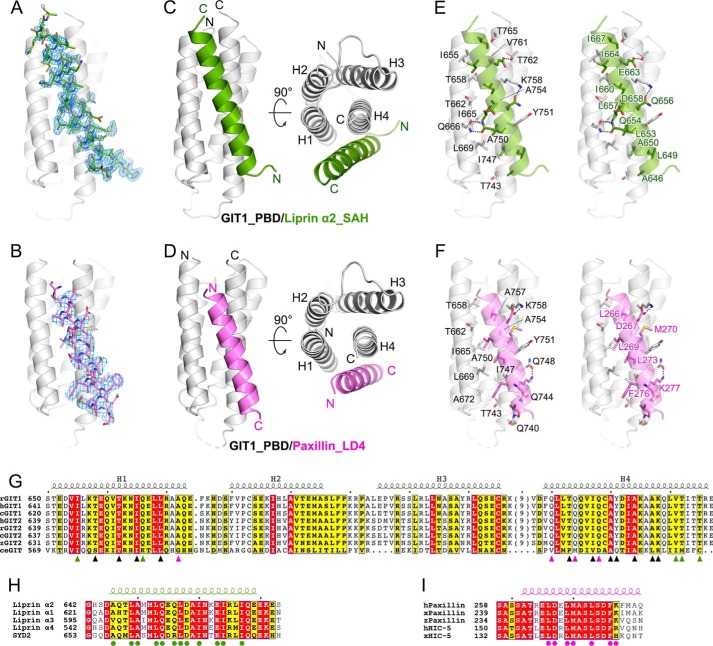
As the bindings of liprin-α and paxillin to GIT1_PBD are mutually exclusive (13) and liprin-α contains a LD-like motif in the SAH region (Fig. 1B), do liprin-α and paxillin share a similar LD-binding model, interacting with GIT1? To address this question, we aimed to solve the complex structures of GIT1_PBD/liprin-α2_SAH and GIT1_PBD/paxillin_LD4 by using X-ray crystallography. Gel filtration analysis showed that GIT1_PBD forms a stable complex with either liprin-α2_SAH or paxillin_LD4 in solution (Fig. S2, A and B). Crystallization trails for the two complex samples yielded high-quality crystals. The crystal structures of the two complexes, GIT1_PBD/liprin-α2_SAH and GIT1_PBD/paxillin_LD4, were successfully determined at 1.8 and 2.6 Å resolutions, respectively, by using the molecular replacement method (Table 1). Most residues of liprin-α2_SAH (29 of the 30 total residues) and paxillin_LD4 (21 of the 24 total residues) in these two structures were clearly assigned (Fig. 2, A and B). As a FAT-like domain, GIT1_PBD adopts the four-helix-bundle conformation. The overall structures of GIT1_PBD in the two complexes are essentially same as its apo-form structure (overall root mean square deviations of 0.9 and 1.0 Å, respectively), indicating that GIT1_PBD does not undergo conformational change upon target binding. Consistent with our biochemical analysis, both liprin-α2_SAH and paxillin_LD4 form single helices to interact with GIT1_PBD on the same surface composed of the first helix (H1) and the last helix (H4) (Fig. 2, C and D), well explaining the competitive binding of liprin-α and paxillin to GIT1_PBD (13).
Table 1.
Statistics of data collection and model refinement
Numbers in parentheses represent the value for the highest-resolution shell.
| Data collection | ||
| Data set | GIT1_PBD/liprin-α2_SAH | GIT1_PBD/paxillin_LD4 |
| Space group | P212121 | I4 |
| Unit cell parameters | ||
| a, b, c (Å) | 88.2, 38.6, 99.1 | 128.8, 128.8, 47.6 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) | 50–1.8 (1.83–1.8) | 50–2.6 (2.64–2.6) |
| No. of unique reflections | 32,467 (1606) | 12,341 (621) |
| Redundancy | 5.3 (5.6) | 6.7 (7.0) |
| I/σ | 25.5 (2.0) | 35.1 (1.5) |
| Completeness (%) | 99.2 (100) | 99.9 (100) |
| Rmerge (%)a | 9.8 (84.2) | 5.4 (98.6) |
| Structure refinement | ||
| Resolution (Å) | 50–1.8 (1.85–1.8) | 50–2.6 (2.86–2.6) |
| Rcryst/Rfree (%)b | 22.7 (32.5)/26.7 (33.4) | 19.4 (25.8)/22.6 (30.1) |
| Root mean square deviations | ||
| Bonds (Å) | 0.003 | 0.002 |
| Angles (degrees) | 0.6 | 0.5 |
| Average B factor | 39.4 | 131.0 |
| No. of atoms | ||
| Protein | 2438 | 2070 |
| Ligand/ion | 3 | 0 |
| Water molecules | 141 | 0 |
| Ramachandran plot | ||
| Favored regions (%) | 99.7 | 98.9 |
| Allowed regions (%) | 0.3 | 1.1 |
| Outliner (%) | 0 | 0 |
a Rmerge = Σ|Ii − Im|/ΣIi, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
b R cryst = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are observed and calculated structure factors. Rfree = ΣT‖Fo| − |Fc‖/ΣT|Fo|, where T is a test data set of about 5% of the total reflections randomly chosen and set aside prior to refinement.
Figure 2.
The crystal structures of GIT1_PBD in complex with liprin-α2_SAH and paxillin_LD4. A and B, Fo − Fc omit maps (contoured at 2.5σ) of the bound SAH (A) and LD4 (B) peptides. The maps are shown with the final model of the corresponding complex superimposed. C and D, ribbon representations of the overall structures of the GIT1_PBD/liprin-α2_SAH (C) and GIT1_PBD/paxillin-LD4 (D) complexes. E and F, the atomic details of the GIT1_PBD/liprin-α2_SAH (E) and GIT1_PBD/paxillin-LD4 (F) interfaces. Hydrogen bonds and salt bridges are indicated by dashed lines. G, sequence alignment of the GIT family members from different species. Residues involved in SAH-specific, LD4-specific, and overlapped binding are marked with green, purple, and black triangles, respectively. The species are indicated as follows: human (h), rat (r), Xenopus tropicalis (x), for Danio rerio (z), Gallus gallus (c), and Caenorhabditis elegans (ce). The secondary structural elements are indicated above the alignment. H, sequence alignment of four liprin-α isoforms in humans and SYD2 in C. elegans. Residues involved in the GIT1_PBD/liprin-α_SAH interaction are marked with green circles. I, sequence alignment of paxillins and HIC-5s. Residues involved in the GIT1_PBD/paxillin-LD4 interaction are marked with purple circles.
Paxillin_LD4 interacts with GIT1_PBD using the similar LD-binding mode for FAT-like domains, in which the LD4 helix is parallel to the H1 helix (19, 28–30) (Fig. 2D and Fig. S3). However, to our surprise, despite containing an LD-like motif, liprin-α2_SAH binds to GIT1_PBD with the reverse orientation, in which the SAH helix is antiparallel to the H1 helix (Fig. 2C). Although the interacting orientations are different for the two PBD-binding peptides, both liprin-α2_SAH and paxillin_LD4 use their hydrophobic sides in their amphipathic helices to tightly pack with the H1/H4 groove of GIT1_PBD through hydrophobic interactions, which are further strengthened by hydrogen bondings (Fig. 2, E and F). In support of our findings, previous structural studies reported that hydrophobic interactions involving the GIT1_PBD/paxillin_LD4 interaction are critical for complex formation (15, 16). Additionally, Asp-267 in paxillin_LD4 forms a salt bridge with Lys-758 in GIT1_PBD (Fig. 2F), which is a characteristic interaction in the LD-binding mode (15, 16, 20). Sequence analysis further shows that the interface residues in GIT1, liprin-α2, and paxillin are highly conserved across species (Fig. 2, G–I), suggesting that the two different GIT1-binding modes observed for liprin-α2_SAH and paxillin_LD4 are likely to be shared by other members of the liprin-α family and HIC-5, respectively.
Structural comparison of the LD4 and SAH-binding modes of GIT1_PBD
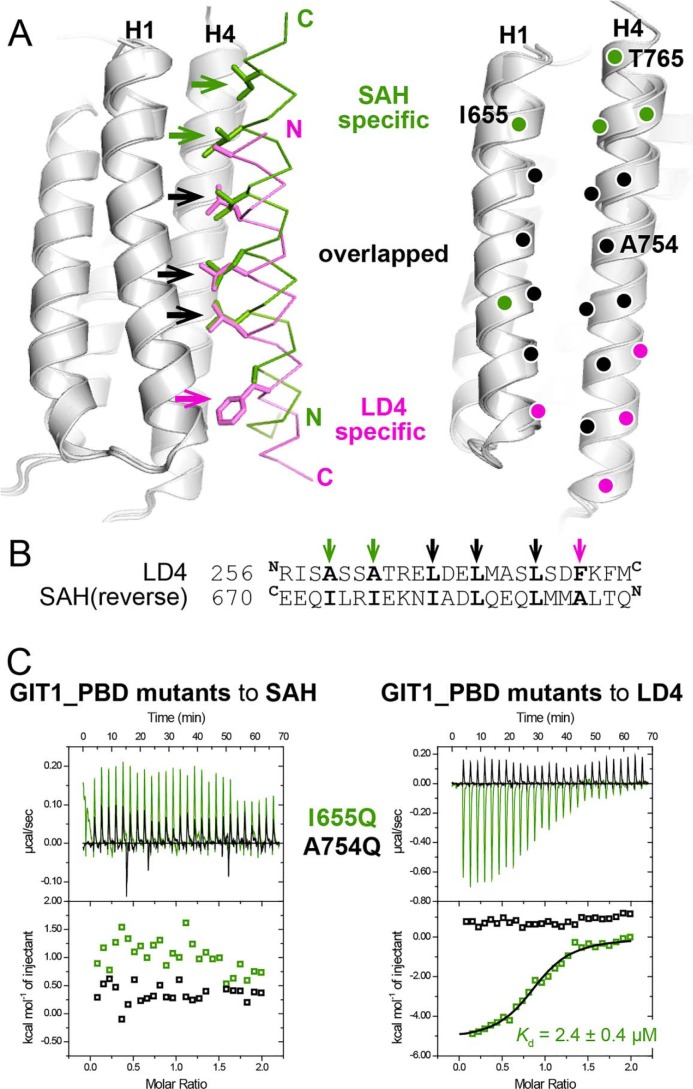
As the bindings of liprin-α2_SAH and paxillin_LD4 to GIT1_PBD are mainly mediated by hydrophobic interactions (Fig. 2, E and F), we carefully analyzed the interfaces by comparing the buried hydrophobic residues of the two bound peptides. In the LD4 peptide, Leu-266LD4, Leu-269LD4, and Leu-273LD4, the conserved leucine residues in the LD core (Fig. 1B), interact with the hydrophobic groove formed by the H1 and H4 helices of GIT1_PBD, which is a general LD-binding feature identified in other FAT-like domains (Fig. S3). Interestingly, by aligning the two peptide-bound GIT1_PBD structures together, we found that the side chains of Ile-660SAH, Leu-657SAH, and Leu-653SAH in the SAH peptide could be overlapped precisely with those of the three leucine residues in the LD4 peptide (Fig. 3A), despite the different helical orientations between the two peptides. Likewise, the reversed amino acid sequence of the SAH peptide can be aligned to the LD4 sequence with the hydrophobic residues matched (Fig. 3B). This suggests the central roles of these hydrophobic residues in the binding of liprin-α2_SAH and paxillin_LD4 to GIT1_PBD.
Figure 3.
The different binding modes of liprin-α2_SAH and paxillin-LD4 to GIT1_PBD. A, the superposition of the two complex structures revealing the different binding modes of GIT1_PBD. The hydrophobic residues of the LD4 and SAH peptide that are involved in the SAH-specific, LD4-specific, and overlapped interactions with the H1/H4 groove of GIT1_PBD are shown as a stick model and indicated by green, purple, and black arrows, respectively, in the left panel. The corresponding interacting residues of GIT1_PBD are indicated by purple, green, and black circles in the right panel. The residues that were analyzed by mutagenesis in this study are labeled with one-letter codes and position numbers. B, pairwise sequence alignment of the LD4 motif and the reversed sequence of the SAH peptide. C, ITC curves of the bindings of two GIT1_PBD mutants to liprin-α2_SAH and paxillin_LD4.
In addition to the three overlapped residues, a few C-terminal residues of both of the two peptides significantly contribute to the intermolecular hydrophobic interactions. As shown in Fig. 3A, Ile-664SAH and Ile-667SAH in the SAH peptide interact with the upper part of the H1/H4 groove, formed by the N terminus of H1 and the C terminus of H4, whereas the bulky side chain of Phe-276LD4 in the LD4 peptide occupies the H1/H4 groove in the lower part. In contrast, the corresponding positions of Ile-664SAH and Ile-667SAH in the LD4 peptide (Ala-262LD4 and Ala-259LD4) and of Phe-276LD4 in the SAH peptide (Ala-650SAH) are all alanine residues (Fig. 3, A and B), of which small side chains barely touch the H1/H4 groove, thereby contributing little to the bindings of either paxillin_LD4 or liprin-α2_SAH to GIT1_PBD. Thus, replacing Ala-650SAH with a phenylalanine in the SAH peptide may artificially create an additional PBD-binding site by mimicking the binding of Phe-276LD4 to GIT1_PBD. In line with this hypothesis, the A650F mutation of liprin-α2_SAH largely enhances the binding (Fig. S4). Based on the above analysis, we conclude that GIT1_PBD can specifically recognize liprin-α2_SAH and paxillin_LD4 by combinatorial usage of different surface patches in the H1/H4 groove, in which the upper and lower parts of the groove provide the binding specificities for the SAH and LD4 peptides, respectively, whereas the middle part of the groove creates the common binding surface for both of the two peptides (Fig. 3A).
The binding mode differences between paxillin_LD4 and liprin-α2_SAH allow us to design GIT1 mutations, A754Q and I655Q, for disruption of specific target interactions with GIT1_PBD. The A754Q mutation largely decreases the hydrophobicity of the overlapped binding surface and imposes steric hindrance for interactions, thereby disrupting the bindings of GIT1_PBD to both liprin-α2_SAH and paxillin_LD4. By contrast, as Ile-655 locates on the SAH-binding surface on GIT1_PBD, the I655Q mutation presumably prevents GIT1_PBD from binding to liprin-α2_SAH while it retains the ability of GIT1_PBD to bind with paxillin_LD4. Fully consistent with our design, neither liprin-α2_SAH nor paxillin_LD4 shows detectable binding to the A754Q mutant, whereas the I655Q mutant interacts with paxillin_LD4 but not liprin-α2_SAH, as indicated by ITC-based measurements (Fig. 3C) and analytical gel filtration analysis (Fig. S2, C–F).
Paxillin and liprin-α play distinct roles in GIT1-mediated FA dynamics
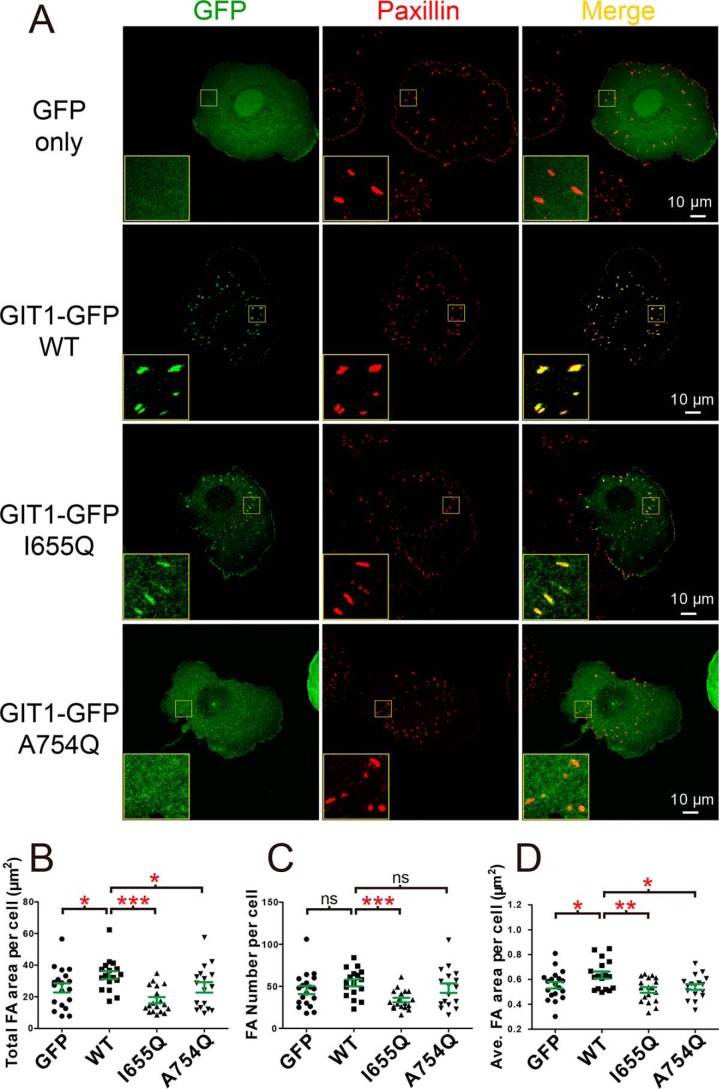
As the core component of FA, paxillin is indispensable for FA assembly (25, 31). Considering the localization of GIT1 and liprin-α to FA for controlling FA dynamics (23, 24, 32, 33), the competitive bindings of liprin-α and paxillin to GIT1 raise a question of how GIT1 coordinates with liprin-α and paxillin to fulfill their functions in FA. To approach this question, the subcellular localization of GIT1 was analyzed by overexpressing the GFP-tagged GIT1 in COS7 cells. The GFP tag was fused to the C terminus of GIT1 to avoid a potential inhibitive effect of N-terminal fused GFP on GIT1's activity (11, 34). As reported previously (16), GIT1-GFP is localized to paxillin-positive FA, whereas GFP alone shows a diffused distribution in cells (Fig. 4A). Consistent with our structural and biochemical analysis, the I665Q mutant of GIT1 remains largely co-localization with paxillin in FA, suggesting that liprin-α is unlikely to play an important role in the FA localization of GIT1. Unlike the I665Q mutant, the A754Q mutant of GIT1, presumably abolishing its binding to paxillin, completely lost its localization to FA (Fig. 4A). These observations indicate that paxillin is required for recruiting GIT1 to FA via the PBD-mediated interaction.
Figure 4.
The cellular effects of the paxillin and liprin-α binding deficient mutations in GIT1. A, cell imaging of exogenous expressed GFP and GFP-tagged GIT1 and its variants. The C-terminal tagged GIT1 constructs were transiently expressed in COS7 cells grown on coverslips. FAs were indicated by endogenous paxillin. Scale bar, 10 μm. B–D, paxillin-labeled FA areas and numbers were measured using the cell-imaging data. Paxillin-positive puncta at cell edge were excluded in quantifications. An average of 18 cells transfected with GFP only, GIT1 WT, and mutants as indicated were analyzed with total FA area per cell (B), the total FA numbers per cell (C), and average area per FA per cell (D). Error bars, S.D. The unpaired Student's t test analysis was used to define a statistically significant difference (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant).
Next, we measured the potential changes of FA in the GIT1-overexpressed COS7 cells by quantification of areas for matured FAs (Fig. 4, B–D). Compared with cells transfected with GFP alone, cells transfected with GIT1-GFP have enlarged FA area, suggesting that the recruitment of GIT1 to FA by paxillin promotes FA assembly. Consistently, as the A754Q mutant of GIT1 fails to localize to FA, neither FA area nor FA number in cells expressing this mutant shows significant changes compared with cells expressing GFP alone. However, GIT1 carrying the liprin-α binding–deficient mutation, I665Q, although remaining localized to FA, shows severely decreased FA numbers in COS7 cells (Fig. 4C), indicating the critical roles of liprin-α in regulating FA dynamics. Because the averaged FA area was reduced in cells expressing the I665Q mutant (Fig. 4D), it is likely that the I665Q mutation impairs the GIT1-promoted FA assembly. Taken together, the above cellular data revealed the distinct roles of GIT1 in the two competitive interactions, in which GIT1 is recruited to FA by binding to paxillin and promotes FA assembly by binding to liprin-α.
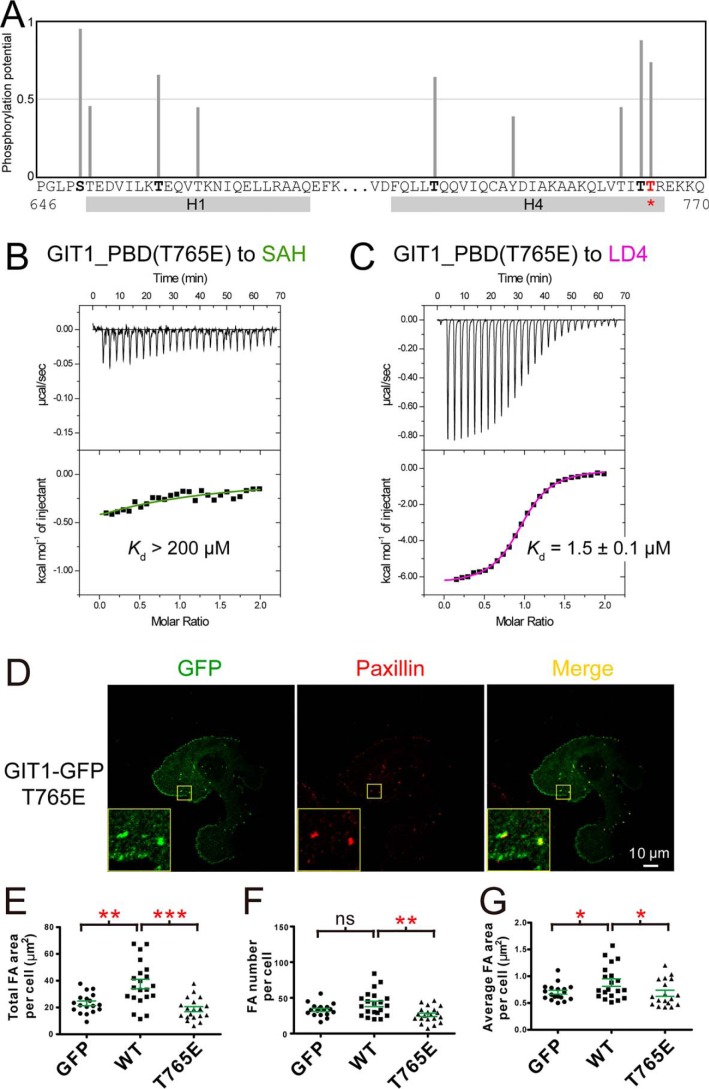
Considering the similar binding affinities of GIT1/liprin-α and GIT1/paxillin (Fig. 1, D and E), it would be interesting to know how these two competitive interactions are switched from each other. Protein phosphorylation is a common mechanism in regulation of protein–protein interactions (35, 36). By analyzing the GIT1_PBD sequence, we found that Thr-765, located on the SAH-specific binding surface (Fig. 3A), is a potential phosphorylation site (Fig. 5A). It is likely that the phosphorylation of Thr-765 specifically blocks the binding of GIT1 to liprin-α by introducing a highly negatively charged phosphoryl group to the hydrophobic interface. Indeed, mutating Thr-765 to a negatively charged glutamate, mimicking its phosphorylated state, almost eliminates the GIT1_PBD/liprin-α2_SAH interaction while maintaining the binding of GIT1_PBD to paxillin_LD4 (Fig. 5, B and C). We further tested the effects of this phosphorylation-mimicking mutation on the FA dynamics. Consistent with our biochemical analysis, the Thr-765 mutation does not affect the localization of GIT1 to FA (Fig. 5D), but it significantly decreases the FA numbers and areas of matured FAs (Fig. 5, E–G), presumably due to the disruption of the GIT1/liprin-α interaction. Thus, the Thr-765 phosphorylation of GIT1 may act as a switch to regulate the two PBD-mediated interactions in vivo.
Figure 5.
The potential phosphorylation regulation of the binding of GIT1_PBD to liprin-α. A, prediction of phosphorylation sites in the H1/H4 groove of GIT1_PBD. Predicted phosphorylation potential value of each serine, threonine, and tyrosine in the sequence is indicated as a bar. The regions of the H1 and H4 helices are indicated. B and C, ITC-based measurements of the affinities for the bindings of the phosphorylation-mimicking mutant T765E of GIT1_PBD to liprin-α2_SAH (B) and paxillin_LD4 (C). D, cell imaging of exogenous expressed GFP-tagged GIT1 T765E mutation in COS7 cells. E–G, FA areas and numbers were analyzed using the same method as described in the legend to Fig. 4.
The unique LD motif recognition mechanism revealed by the GIT1_PBD/paxillin_LD4 complex structure
The typical binding of the helical LD motif to the FAT domains is mainly determined by the characteristic sequence of LDXLXXXL (where X denotes any residue of the 20 common amino acids) (Fig. 1B). Upon binding to FAT domains, the three leucine residues buried in the H1/H4 groove and the aspartate residue form a salt bridge with a highly conserved lysine in the FAT domains (20). Such interactions are also observed in the GIT1_PBD/paxillin_LD4 complex structure (Fig. 2F). However, unlike the FAT domains in FAK and PYK, which interact with the LD2 and LD4 motifs of paxillin in the same fashion (Fig. S3), GIT1_PBD shows a much higher binding selectivity to LD4 than to LD2 (15) (Fig. S5). It has been largely unclear where the selectivity comes from, as the core regions of LD2 and LD4 are nearly identical.
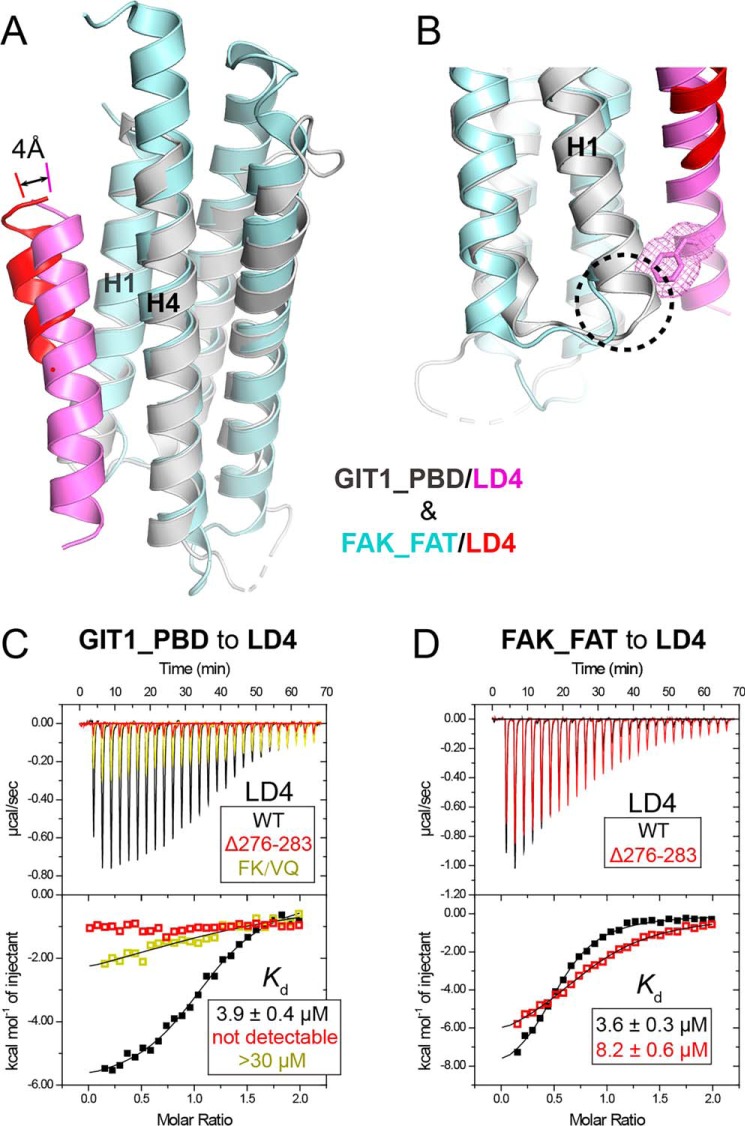
To decode the LD recognition mechanism by GIT1_PBD, the GIT1_PBD/LD4 structure was overlapped with the FAK_FAT/LD4 structure. Compared with the short helix of FAK_FAT-bound LD4, the GIT1_PBD bound LD4 forms a longer helix, in which the C-terminal flanking residues of the LD core are also involved in the helix formation and attach to the H1/H4 groove with a closer distance (Fig. 6A). The helix extension is mainly stabilized by the interaction of Phe-276LD4 and Lys-277LD4 with GIT1_PBD via hydrophobic interaction and hydrogen bonding (Fig. 2F). The involvement of the LD core flanking sequence in the binding was not found in other FAT/LD structures (Fig. S3). Therefore, we ask why FAK_FAT cannot interact with the LD core flanking sequence. Detailed analysis showed that the Phe-276LD4–packing site, formed by the C-terminal part of H1 and the N-terminal part of H4 in GIT1_PBD, is distorted in FAK_FAT by replacing the last turn of the H1 helix in GIT1_PBD with a short loop (Fig. 6B). Such a conformational distortion in FAK_FAT prevents Phe-276LD4 from binding to FAK_FAT.
Figure 6.
Structural comparison of the GIT1-PBD/LD4 complex with the FAK-FAT/LD4 complex. A, structural alignment of the LD4 bound GIT1_PBD and FAK_FAT (PBD code 1OW7). B, the structural determinant of the binding of Phe-276LD4 to the H1/H4 groove of GIT1_PBD. Compared with FAK_FAT, the H1 helix of GIT1-PBD has an additional helical turn, highlighted by a dashed circle, to form the hydrophobic surface patch serving for the recognition of Phe-276LD4 upon LD4 binding. The bulky side chain of Phe-276LD4 is indicated as sticks and meshes. C, ITC titrations of GIT1_PBD to paxillin_LD4 and its mutations. D, ITC titrations of FAK_FAT to paxillin_LD4 and its mutation.
Consistent with our structural findings, the binding of GIT1_PBD to the LD4 motif is either abolished by deleting the LD4 C-terminal sequence (residues 276–283) or largely diminished by mutating Phe-276LD4 and Lys-277LD4 to the corresponding residues (a valine and a glutamine) in the LD2 motif (Fig. 6C), confirming the essential role of Phe-276LD4 in the GIT1_PBD/paxillin_LD4 interaction. Even the replacement of Phe-276LD4 with a smaller hydrophobic residue may interfere with the interaction. As Phe-276LD4 is not conserved in other LD motifs of paxillin, it is very likely that the C-terminal sequences of the LD motifs determine the binding selectivity for GIT1_PBD. In contrast to GIT1_PBD, FAK_FAT lacks an appropriate pocket to hook Phe-276LD4. Indeed, the binding of FAK_FAT to the LD4 motif is only mildly decreased by the deletion mutation (Fig. 6D), indicating that the LD core flanking sequence contributes little to the selectivity for FAK_FAT.
Discussion
FA dynamics are crucial in regulating various cellular processes (5, 7, 8). The involvement of GIT1, liprin-α, and paxillin in FA dynamics has been studied extensively (9, 24, 32, 37, 38). Due to the lack of structural information, the molecular mechanism of the coordinated bindings of GIT1 to liprin-α and paxillin has been largely unknown. In this study, we solved the crystal structures of GIT1_PBD in complex with the short PBD-binding sequences in liprin-α and paxillin, respectively, revealing that GIT1_PBD utilizes the H1/H4 groove to recognize its targets by different modes. These high-resolution structures allow us to design specific binding-deficient mutations of GIT1, providing an amenable way to differentiate the cellular effects of these two competitive interactions. Considering the importance of GIT1 and liprin-α in neuronal development (14, 33, 39–42), our study provides structural information for future investigations of the GIT1/liprin-α interaction in neurons.
Given the presence of both paxillin and liprin-α in FAs, it remains puzzling how GIT1 switches the binding from liprin-α to paxillin or vice versa. Because phosphorylation plays an important role in regulation of GIT1's function (43, 44), it is possible that phosphorylated GIT1 selectively blocks one of the two bindings. In support of this hypothesis, we identified Thr-765 in GIT1_PBD as a potential phosphorylation site, and the phosphorylation-mimicking mutation T765E disrupts the binding of GIT1 to liprin-α but not paxillin. Such phosphorylation may control GIT1 to alternatively interact with the two PBD-mediated interactions. Notably, many protein–protein interactions in FAs are mediated by LD motifs. For example, GIT1_PBD interacts with some LD motifs of paxillin, HIC5, and leupaxin (16, 17), whereas paxillin_LD4 binds with FAK (45), PYK2 (28), and parvin (46, 47). All of these interactions need to be coordinated to control the focal adhesion dynamics. Thus, it is possible that the potential phosphorylation of Thr-765 may also regulate the binding of GIT1 to other FA proteins.
The previous studies (15, 19, 28–30) have indicated that the LD core sequence determines the binding of LD motifs to FAT-like domains (Fig. S3). Nevertheless, the GIT1_PBD/paxillin_LD4 structure, for the first time, reveals that the sequence flanking the LD core contributes significantly to the FAT-like domain/LD interaction (Fig. 6). Although the LD core sequences of different LD motifs are highly similar, their flanking sequences vary a lot (Fig. 1B). Considering that different FAT-like domains show certain selectivity to bind with different LD motifs (29, 30, 45, 49), it is tempting to think that the flanking sequences may participate in the bindings of other FAT-like domains. Therefore, by demonstrating that a FAT-like domain can develop an additional surface to recognize the sequences near the LD core region in LD motifs, this study provides a plausible mechanism for the selective binding of different LD motifs. Despite the weak binding of LD2 to GIT1 (Fig. S5), the multisite binding of paxillin to GIT1 through LD2 and LD4 may enhance the recruitment of GIT1 to FAs.
Although the SAH sequence of liprin-α contains the LD-like motif, surprisingly, it binds with GIT1_PBD using a different mode by adopting the reverse binding orientation (Fig. 3, A and B). By aligning the LD-like sequence of SAH to LD4, we found that the corresponding positions of 276FK277LD4 that play a critical role in the binding of the LD4 motif to GIT1_PBD are replaced by “IQ” in SAH (Fig. 1B), implying that even if SAH bound with GIT1_PBD using the LD4 orientation, the binding affinity would be much lower than in the reverse orientation. Our observation of the reverse binding orientation of the LD-like motif indicates that other FAT-like domain may also recognize the different LD-like motif using different binding modes. In support of this possibility, CD4 was found to interact with FAK_FAT by a reverse orientation (50), despite the fact that the poor quality of the electron density of the binding helix provides few hints to determine which amino acid sequence(s) determines the binding orientation.
Materials and methods
Expression constructs and site-directed mutagenesis
DNA encoding sequences of rat GIT1_PBD (residues 639–770), human liprin-α2_SAH (residues 642–671), human paxillin_LD2 (residues 140–161), and LD4 (residues 252–283) was subcloned into a modified pET-32a vector with an N-terminal thioredoxin (Trx)-His6 tag. Human FAK_FAT (residues 915–1052) was subcloned into modified pET-28a vector with an N-terminal His6 tag. The GFP-tagged full-length GIT1 constructs for cellular assays were PCR-amplified using plasmids as template and cloned into mammalian expression vectors containing GFP. All point mutations were created using a site-directed mutagenesis kit and confirmed by DNA sequencing.
Protein expression and purification
All of the proteins were expressed in BL21(DE3) E. coli cells at 16 °C with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside induced in LB medium. The protein samples were purified using nickel-affinity chromatography followed by size-exclusion chromatography. For isothermal titration calorimetry and analytical gel filtration chromatography, the protein samples contained an N-terminal tag. For crystallization, the N-terminal Trx-His6 tag was cleaved by HRV 3C protease and then separated by size-exclusion chromatography.
Crystallization and data collection
The complex samples were prepared by mixing GIT1_PBD with either liprin-α2_SAH or paxillin_LD4 and were further purified by size-exclusion chromatography. The complexes were concentrated to ∼30 mg/ml. Crystals were obtained by the sitting-drop vapor-diffusion method at 16 °C. To set up a sitting drop, 1 μl of concentrated protein solution was mixed with 1 μl of crystallization solution with 0.2 m potassium iodide, 0.1 m MES, pH 6.5, and 25% (w/v) PEG 4000 (for the GIT1_PBD/liprin-α2_SAH complex) or with 0.1 m HEPES, pH 7.5 and 30% (w/v) PEG 1000 (for the GIT1_PBD/paxillin_LD4 complex). Crystals with better qualities were obtained by removing six N-terminal flexible residues in GIT1-PBD and eight N-terminal flexible residues in paxillin_LD4. Before X-ray diffraction experiments, crystals were soaked in the crystallization solutions containing additional 20% (v/v) PEG 400 or 30% (w/v) PEG 1000 for cryoprotection. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility beamlines BL17U1 and BL19U1. Data were processed and scaled using HKL3000 software.
Structure determination and analysis
The initial phase of the complex structure was determined by molecular replacement in PHASER (51) using the apo-structure of GIT1_PDB (PDB code 2JX0) as the search model. The liprin-SAH or paxillin-LD4 peptide was further built into the corresponding model. The model was refined in PHENIX (52). COOT was used for model rebuilding and adjustments (53). In the final stage, an additional TLS refinement was performed in PHENIX. The model quality was checked by MolProbity (54). The final refinement statistics are listed in Table 1. All structure figures were prepared by using PyMOL (Schroedinger, LLC, New York). The phosphorylation site prediction was performed on the NetPhos 3.1 server (http://www.cbs.dtu.dk/services/NetPhos/)5 (48).
ITC analysis
ITC experiments were carried out on a VP-ITC Microcal calorimeter (Malvern) at 25 °C. All proteins were dissolved in 50 mm Tris buffer containing 100 mm NaCl, 1 mm EDTA, 2 mm DTT at pH 7.5. Each titration point typically consists of injecting 10-μl aliquots of the GIT1_PBD, its mutants, or FAK_FAT at concentration of 400 μm into the solution of containing liprin-SAH, paxillin-LD2/4, or their mutants at a concentration of 40 μm. A time interval of 150 s between two titration points was used to ensure the complete equilibrium of each titration reaction. The titration data were analyzed using the program Origin version 7.0 and fitted by a one-site binding model.
Analytical gel filtration chromatography
Analytical gel filtration chromatography was carried out on an ÄKTA pure system (GE Healthcare). Protein samples at a concentration of 50 μm were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated with 50 mm Tris-HCl buffer, pH 7.5, containing 100 mm NaCl, 1 mm EDTA, and 2 mm DTT.
Multi-angle light-scattering analysis
A miniDAWN TREOS (Wyatt Technology Corp.) coupled with an ÄKTA pure system (GE Healthcare) was used for molar mass measurement. The procedure follows the protocol used for analytical gel filtration analysis.
Cell culture, transfection, and fluorescence imaging
COS7 cells were cultured in Dulbecco's modified Eagle's medium (Corning, 10-013-CVR) supplemented with 10% fetal bovine serum (Pan Biotech) and 50 units/ml penicillin and streptomycin. Transfections of either WT or mutant GIT1/EGFP-expressing plasmids were performed with polyethyleneimine-25,000 (Polyscience) according to the manufacturer's instructions. One day after transfection, the cells were trypsinized, replated on ∼20 μg/ml fibronectin (Millipore)-coated coverslips, and cultured for 1 h. After fixation with 4% paraformaldehyde, the cells were stained with anti-paxillin (BD Biosciences) followed by Alexa 594–conjugated anti-mouse IgG Ab (Invitrogen) and observed under a Nikon A1R confocal microscope. Images were analyzed using ImageJ software (National Institutes of Health).
CD spectroscopy
CD spectra were recorded using a Chirascan spectrometer (Applied Photophysics, Leatherhead, UK). The buffer used contained 20 mm Na2HPO4/KH2PO4 (pH 7.0), 50 mm NaCl. Protein sample was concentrated to 50 μm and loaded into a quartz cell with a 1-mm path length. The measurement was performed every 1 nm with a 0.5-s time per point and averaged from four repeated scans.
Author contributions
M. L. and X. X. data curation; M. L., X. X., J. P., and Z. W. formal analysis; M. L., X. X., G. J., and C. Y. validation; M. L., X. X., J. P., and G. J. investigation; M. L. and X. X. methodology; M. L., X. X., C. Y., and Z. W. writing-original draft; M. L., C. Y., and Z. W. writing-review and editing; X. X., C. Y., and Z. W. funding acquisition; X. X. and Z. W. visualization; C. Y. resources; C. Y. and Z. W. supervision; C. Y. and Z. W. project administration; Z. W. conceptualization.
Supplementary Material
Acknowledgments
We thank the Life Science Research Center, Southern University of Science and Technology (SUSTech), for providing facilities. We thank the staffs from the BL17U, BL18U, and BL19U1 beamlines of the Shanghai Synchrotron Radiation Facility for assistance during data collection.
This work was supported by National Natural Science Foundation of China Grants 31770791 and 31570741 (to Z. W.) and 31870757 (to C. Y.); Natural Science Foundation of Guangdong Province Grant 2016A030312016; Science and Technology Planning Project of Guangdong Province Grant 2017B030301018; and Shenzhen Science and Technology Innovation Commission Grants JCYJ20160229153100269 (to Z. W.), JCYJ20160301112450474 (to X. X.), and ZDSYS20140509142721429 (to C. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 6IUH and 6IUI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- FA
- focal adhesion
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- Arf
- ADP-ribosylation factor
- FAK
- focal adhesion kinase
- FAT
- focal adhesion kinase targeting
- GAP
- GTPase-activating protein
- GIT1
- G-protein-coupled receptor kinase-interacting protein 1
- ITC
- isothermal titration calorimetry
- LD
- leucine-aspartic acid motif
- PAK
- p21-activated kinase
- PBD
- paxillin-binding domain
- PDB
- Protein Data Bank
- SAH
- single α-helix
- Trx
- thioredoxin.
References
- 1. Geiger B., Bershadsky A., Pankov R., and Yamada K. M. (2001) Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2, 793–805 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- 2. Eva R., and Fawcett J. (2014) Integrin signalling and traffic during axon growth and regeneration. Curr. Opin. Neurobiol. 27, 179–185 10.1016/j.conb.2014.03.018 [DOI] [PubMed] [Google Scholar]
- 3. Burridge K., Fath K., Kelly T., Nuckolls G., and Turner C. (1988) Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu. Rev. Cell Biol. 4, 487–525 10.1146/annurev.cb.04.110188.002415 [DOI] [PubMed] [Google Scholar]
- 4. Parsons J. T., Horwitz A. R., and Schwartz M. A. (2010) Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burridge K. (2017) Focal adhesions: a personal perspective on a half century of progress. FEBS J. 284, 3355–3361 10.1111/febs.14195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitra S. K., Hanson D. A., and Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- 7. Wehrle-Haller B. (2012) Structure and function of focal adhesions. Curr. Opin. Cell Biol. 24, 116–124 10.1016/j.ceb.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 8. Haase K., Al-Rekabi Z., and Pelling A. E. (2014) Mechanical cues direct focal adhesion dynamics. Prog. Mol. Biol. Transl. Sci. 126, 103–134 10.1016/B978-0-12-394624-9.00005-1 [DOI] [PubMed] [Google Scholar]
- 9. Vitali T., Girald-Berlingeri S., Randazzo P. A., and Chen P. W. (2017) Arf GAPs: a family of proteins with disparate functions that converge on a common structure, the integrin adhesion complex. Small GTPases, 1–9 10.1080/21541248.2017.1299271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank S. R., Adelstein M. R., and Hansen S. H. (2006) GIT2 represses Crk- and Rac1-regulated cell spreading and Cdc42-mediated focal adhesion turnover. EMBO J. 25, 1848–1859 10.1038/sj.emboj.7601092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou W., Li X., and Premont R. T. (2016) Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 129, 1963–1974 10.1242/jcs.179465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoo S. M., Cerione R. A., and Antonyak M. A. (2017) The Arf-GAP and protein scaffold Cat1/Git1 as a multifaceted regulator of cancer progression. Small GTPases, 1–9 10.1080/21541248.2017.1362496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Asperti C., Astro V., Pettinato E., Paris S., Bachi A., and de Curtis I. (2011) Biochemical and functional characterization of the interaction between liprin-α1 and GIT1: implications for the regulation of cell motility. PLoS One 6, e20757 10.1371/journal.pone.0020757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ko J., Kim S., Valtschanoff J. G., Shin H., Lee J. R., Sheng M., Premont R. T., Weinberg R. J., and Kim E. (2003) Interaction between liprin-α and GIT1 is required for AMPA receptor targeting. J. Neurosci. 23, 1667–1677 10.1523/JNEUROSCI.23-05-01667.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z. M., Simmerman J. A., Guibao C. D., and Zheng J. J. (2008) GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs. J. Biol. Chem. 283, 18685–18693 10.1074/jbc.M801274200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmalzigaug R., Garron M. L., Roseman J. T., Xing Y., Davidson C. E., Arold S. T., and Premont R. T. (2007) GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell. Signal. 19, 1733–1744 10.1016/j.cellsig.2007.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nishiya N., Shirai T., Suzuki W., and Nose K. (2002) Hic-5 interacts with GIT1 with a different binding mode from paxillin. J. Biochem. 132, 279–289 10.1093/oxfordjournals.jbchem.a003222 [DOI] [PubMed] [Google Scholar]
- 18. Brown M. C., Curtis M. S., and Turner C. E. (1998) Paxillin LD motifs may define a new family of protein recognition domains. Nat. Struct. Biol. 5, 677–678 10.1038/1370 [DOI] [PubMed] [Google Scholar]
- 19. Hoellerer M. K., Noble M. E. M., Labesse G., Campbell I. D., Werner J. M., and Arold S. T. (2003) Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 11, 1207–1217 10.1016/j.str.2003.08.010 [DOI] [PubMed] [Google Scholar]
- 20. Alam T., Alazmi M., Gao X., and Arold S. T. (2014) How to find a leucine in a haystack? Structure, ligand recognition and regulation of leucine-aspartic acid (LD) motifs. Biochem. J. 460, 317–329 10.1042/BJ20140298 [DOI] [PubMed] [Google Scholar]
- 21. Serra-Pagès C., Medley Q. G., Tang M., Hart A., and Streuli M. (1998) Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 273, 15611–15620 10.1074/jbc.273.25.15611 [DOI] [PubMed] [Google Scholar]
- 22. Zürner M., and Schoch S. (2009) The mouse and human Liprin-α family of scaffolding proteins: genomic organization, expression profiling and regulation by alternative splicing. Genomics 93, 243–253 10.1016/j.ygeno.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 23. Asperti C., Astro V., Totaro A., Paris S., and de Curtis I. (2009) Liprin-α1 promotes cell spreading on the extracellular matrix by affecting the distribution of activated integrins. J. Cell Sci. 122, 3225–3232 10.1242/jcs.054155 [DOI] [PubMed] [Google Scholar]
- 24. Astro V., Tonoli D., Chiaretti S., Badanai S., Sala K., Zerial M., and de Curtis I. (2016) Liprin-α1 and ERC1 control cell edge dynamics by promoting focal adhesion turnover. Sci. Rep. 6, 33653 10.1038/srep33653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown M. C., and Turner C. E. (2004) Paxillin: adapting to change. Physiol. Rev. 84, 1315–1339 10.1152/physrev.00002.2004 [DOI] [PubMed] [Google Scholar]
- 26. West K. A., Zhang H. Y., Brown M. C., Nikolopoulos S. N., Riedy M. C., Horwitz A. F., and Turner C. E. (2001) The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL). J. Cell Biol. 154, 161–176 10.1083/jcb.200101039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manabe R., Kovalenko M., Webb D. J., and Horwitz A. R. (2002) GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 115, 1497–1510 [DOI] [PubMed] [Google Scholar]
- 28. Lulo J., Yuzawa S., and Schlessinger J. (2009) Crystal structures of free and ligand-bound focal adhesion targeting domain of Pyk2. Biochem. Biophys. Res. Commun. 383, 347–352 10.1016/j.bbrc.2009.04.011 [DOI] [PubMed] [Google Scholar]
- 29. Vanarotti M. S., Finkelstein D. B., Guibao C. D., Nourse A., Miller D. J., and Zheng J. J. (2016) Structural basis for the interaction between Pyk2-FAT domain and leupaxin LD repeats. Biochemistry 55, 1332–1345 10.1021/acs.biochem.5b01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanarotti M. S., Miller D. J., Guibao C. D., Nourse A., and Zheng J. J. (2014) Structural and mechanistic insights into the interaction between Pyk2 and paxillin LD motifs. J. Mol. Biol. 426, 3985–4001 10.1016/j.jmb.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanteti R., Batra S. K., Lennon F. E., and Salgia R. (2016) FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget 7, 31586–31601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Z. S., Manser E., Loo T. H., and Lim L. (2000) Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell. Biol. 20, 6354–6363 10.1128/MCB.20.17.6354-6363.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoefen R. J., and Berk B. C. (2006) The multifunctional GIT family of proteins. J. Cell Sci. 119, 1469–1475 10.1242/jcs.02925 [DOI] [PubMed] [Google Scholar]
- 34. Vitale N., Patton W. A., Moss J., Vaughan M., Lefkowitz R. J., and Premont R. T. (2000) GIT proteins, A novel family of phosphatidylinositol 3,4,5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 275, 13901–13906 10.1074/jbc.275.18.13901 [DOI] [PubMed] [Google Scholar]
- 35. Pawson T., and Nash P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 10.1126/science.1083653 [DOI] [PubMed] [Google Scholar]
- 36. Hunter T. (2000) Signaling—2000 and Beyond. Cell 100, 113–127 10.1016/S0092-8674(00)81688-8 [DOI] [PubMed] [Google Scholar]
- 37. Turner C. E., Glenney J. R. Jr., and Burridge K. (1990) Paxillin–a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 10.1083/jcb.111.3.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asperti C., Pettinato E., and de Curtis I. (2010) Liprin-α1 affects the distribution of low-affinity β1 integrins and stabilizes their permanence at the cell surface. Exp. Cell Res. 316, 915–926 10.1016/j.yexcr.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 39. Webb D. J., Zhang H., Majumdar D., and Horwitz A. F. (2007) α5 integrin signaling regulates the formation of spines and synapses in hippocampal neurons. J. Biol. Chem. 282, 6929–6935 10.1074/jbc.M610981200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong S. T., and Mah W. (2015) A critical role of GIT1 in vertebrate and invertebrate brain development. Exp. Neurobiol. 24, 8–16 10.5607/en.2015.24.1.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhen M., and Jin Y. (1999) The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature 401, 371–375 10.1038/43886 [DOI] [PubMed] [Google Scholar]
- 42. Wong M. Y., Liu C. L., Wang S. S. H., Roquas A. C. F., Fowler S. C., and Kaeser P. S. (2018) Liprin-α3 controls vesicle docking and exocytosis at the active zone of hippocampal synapses. Proc. Natl. Acad. Sci. U.S.A. 115, 2234–2239 10.1073/pnas.1719012115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ĉernohorská M., Sulimenko V., Hajkova Z., Sulimenko T., Sládková V., Vinopal S., Dráberová E., and Dráber P. (2016) GIT1/βPIX signaling proteins and PAK1 kinase regulate microtubule nucleation. Biochim. Biophys. Acta 1863, 1282–1297 10.1016/j.bbamcr.2016.03.016 [DOI] [PubMed] [Google Scholar]
- 44. Totaro A., Astro V., Tonoli D., and de Curtis I. (2014) Identification of two tyrosine residues required for the intramolecular mechanism implicated in GIT1 activation. PLoS One 9, e93199 10.1371/journal.pone.0093199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown M. C., Perrotta J. A., and Turner C. E. (1996) Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135, 1109–1123 10.1083/jcb.135.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lorenz S., Vakonakis I., Lowe E. D., Campbell I. D., Noble M. E., and Hoellerer M. K. (2008) Structural analysis of the interactions between paxillin LD motifs and α-parvin. Structure 16, 1521–1531 10.1016/j.str.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stiegler A. L., Draheim K. M., Li X., Chayen N. E., Calderwood D. A., and Boggon T. J. (2012) Structural basis for paxillin binding and focal adhesion targeting of β-parvin. J. Biol. Chem. 287, 32566–32577 10.1074/jbc.M112.367342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blom N., Gammeltoft S., and Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 10.1006/jmbi.1999.3310 [DOI] [PubMed] [Google Scholar]
- 49. Turner C. E., Brown M. C., Perrotta J. A., Riedy M. C., Nikolopoulos S. N., McDonald A. R., Bagrodia S., Thomas S., and Leventhal P. S. (1999) Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 145, 851–863 10.1083/jcb.145.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garron M. L., Arthos J., Guichou J. F., McNally J., Cicala C., and Arold S. T. (2008) Structural basis for the interaction between focal adhesion kinase and CD4. J. Mol. Biol. 375, 1320–1328 10.1016/j.jmb.2007.11.040 [DOI] [PubMed] [Google Scholar]
- 51. Storoni L. C., McCoy A. J., and Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 10.1107/S0907444903028956 [DOI] [PubMed] [Google Scholar]
- 52. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 54. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.