Abstract
Classical DNA and RNA polymerase (pol) enzymes have defined roles with their respective substrates, but several pols have been found to have multiple functions. We reported previously that purified human DNA pol η (hpol η) can incorporate both deoxyribonucleoside triphosphates (dNTPs) and ribonucleoside triphosphates (rNTPs) and can use both DNA and RNA as substrates. X-ray crystal structures revealed that two pol η residues, Phe-18 and Tyr-92, behave as steric gates to influence sugar selectivity. However, the physiological relevance of these phenomena has not been established. Here, we show that purified hpol η adds rNTPs to DNA primers at physiological rNTP concentrations and in the presence of competing dNTPs. When two rATPs were inserted opposite a cyclobutane pyrimidine dimer, the substrate was less efficiently cleaved by human RNase H2. Human XP-V fibroblast extracts, devoid of hpol η, could not add rNTPs to a DNA primer, but the expression of transfected hpol η in the cells restored this ability. XP-V cell extracts did not add dNTPs to DNA primers hybridized to RNA, but could when hpol η was expressed in the cells. HEK293T cell extracts could add dNTPs to DNA primers hybridized to RNA, but lost this ability if hpol η was deleted. Interestingly, a similar phenomenon was not observed when other translesion synthesis (TLS) DNA polymerases—hpol ι, κ, or ζ—were individually deleted. These results suggest that hpol η is one of the major reverse transcriptases involved in physiological processes in human cells.
Keywords: DNA polymerase, RNA polymerase, reverse transcription, DNA transcription, DNA replication, DNA enzyme, DNA damage, DNA pol eta, translesion synthesis (TLS) enzyme
Introduction
Genetic integrity and stability of DNA are essential for the maintenance of cell homeostasis and function. The genetic material is well-preserved because of the coordination between replication, repair, and cell cycle progression processes, and the DNA polymerases (pols)3 are faithful key players in DNA replication as well as in the DNA repair process. Exposure to exogenous and endogenous agents can result in DNA lesions (1). The so-called translesion synthesis (TLS) DNA polymerases (2, 3) have been characterized over the past 20 years and have been of considerable interest because of their abilities to catalyze DNA polymerization past many DNA adducts, which can be can be an error-prone process.
Historically the DNA and RNA “worlds” were generally considered to be separate until the discovery of viral reverse transcriptase (RT) in the 1970s (4). HIV-1 RT could copy RNA or DNA (5), adding dNTPs. It has been shown that HIV-RT has RNase H as well as DNA polymerase activity (6). Short RNAs are also used in priming DNA synthesis opposite DNA templates, and eukaryotic pol α is one of the DNA polymerases that plays this role (7). In addition, RNA can act as a template in DNA double-strand repair of chromosomal DNA in yeast (8).
Ribonucleotides are the most common DNA “lesions,” even more prevalent than abasic sites, and their presence leads to genomic instability (9–12). The Kunkel laboratory reported that some replicative yeast and human DNA polymerases (α, δ, ϵ) and yeast polymerases β, λ, and ζ occasionally insert rNTPs while copying DNA (9, 13–18). Another possibility for the retention of the backbone sugars of RNA within DNA is the incomplete removal of stretches of RNA from Okazaki fragments during genome duplication (12). In Escherichia coli, ribonucleotides can increase the leading strand fragmentation during the replication process (19). The presence of ribonucleotides in DNA has been linked to systemic autoimmunity (20).
Embedded ribonucleotides in DNA can disturb the normal biological processes such as transcription (21). Ribonucleotides within DNA may interfere with DNA repair and TLS processes. Ribonucleotides are also responsible for chromosomal instability (22). The presence of (ribosyl) adenosine has been shown to affect the repair of 8-oxo-7,8-dihydrodeoxyguanosine (8-oxodG) by DNA glycosylases (23). Many of these inserted ribonucleotides are removed by RNase H2, employing the ribonucleotide excision repair (RER) pathway (10, 15, 24), and even some DNA polymerases (e.g. pol δ) (25). RNase H2 plays an important role in maintaining genomic stability (26), and mutations in any of the three subunits of human RNase H2 cause Aicardi-Goutières syndrome, a human neurological disorder with debilitating consequences (27, 28). RNase H is also necessary for the removal of RNA/DNA hybrids such as R-loop, which is responsible for replication fork collapse that ultimately affects the DNA replication process (29–31). RNase H–deficient systems are involved in the quasi-palindrome–associated mutations (32). Toward this end, the effect of cations on RNase H2 activity has been studied extensively (33).
It has been reported that only making two amino acid substitutions can add transcription (RNA polymerase) activity to Thermococcus gorgonarius Tgo DNA polymerase, indicating the delicate balance of these activities in polymerases (34). However, another archaebacterial Family D DNA polymerase is highly discriminating in selecting for dNTPs more than rNTPs (35).
hpol η, a rather typical TLS DNA polymerase that we have studied extensively (36–40), is able to perform a variety of functions we did not expect (41). The enzyme is capable of using both RNA and DNA templates (and primers) to insert dNTPs or rNTPs. The reverse transcription efficiency was almost as high as DNA polymerase activity (40). hpol ι has also been reported to have “ribo” behavior (42), as does hpol κ (40). These results with ribonucleotides were unexpected and have led to a series of hypotheses and questions about hpol η and other TLS polymerases, extending our own previous biochemical and structural studies on the TLS polymerases (43).
We previously showed that hpol η, as well as another Y family DNA polymerase, hpol κ, can surprisingly use an entire RNA strand as the primer during strand synthesis opposite a DNA template without sacrificing base selectivity, even with a cyclobutane pyrimidine dimer (CPD) or 8-oxodG in the template (43). In addition, isolated hpol η was able to reverse transcribe an RNA strand. Our new results show that hpol η adds rNTPs to DNA primers at physiological rNTP concentrations, even in the presence of dNTPs. We investigated whether CPD lesions affect the removal of ribonucleotides by RNase H2. Our results with cell extracts suggest that hpol η is one of the major reverse transcriptases in human cells.
Results
dATP and rATP compete for incorporation by hpol η at the ends of DNA primers opposite the unmodified base dT and a CPD lesion
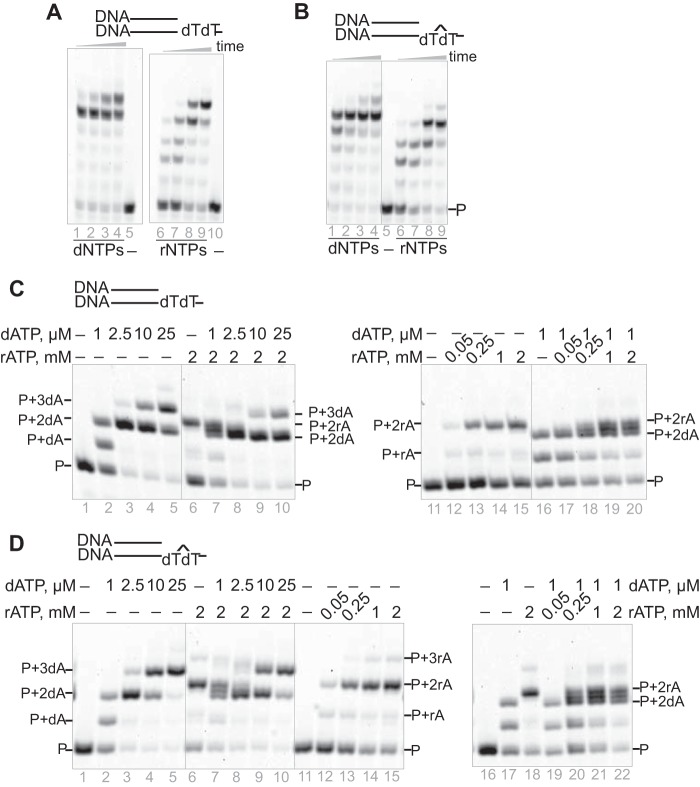
We previously demonstrated that purified human pol η could incorporate rNTPs to extend primers, even beyond CPD lesions (41). However, those studies were done at high concentrations of dNTPs and rNTPs (1 mm). In presence of physiological concentrations of dNTPs or rNTPs (23, 44), hpol η also extended the primers to full-length products (Fig. 1, A and B).
Figure 1.
hpol η can extend a DNA primer opposite an undamaged base or CPD by incorporation of dNTPs or rNTPs. hpol η (500 nm) and the annealed duplex (5 μm) were incubated with physiological concentrations of dNTPs (dATP, 25 μm; dCTP, 30 μm; dGTP, 90 μm; and dTTP, 40 μm) or rNTPs (ATP, 2 mm; CTP, 0.25 mm; GTP, 0.5 mm; and UTP, 0.5 mm) (44) for 5, 10, 30, and 60 min. A, DNA/DNA (dT). B, DNA/DNA (CPD). dATP and rATP competed for incorporation by hpol η (100 nm) in the presence of different concentrations of dATP, rATP, or mixtures of the two. Incubations were done for 5 min. C and D, the annealed substrates were (C) DNA/DNA (dT) (1 μm) and (D) DNA/DNA (CPD) (1 μm). See “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used.
The incorporation depended on the concentration of the dNTP or rNTP. With the rATP concentration fixed at 2 mm and increasing the dATP concentration, the product bands were shifted from primer + 2rA (i.e. two rNTPs incorporated) to primer + 3dA (i.e. three dNTPs incorporated) and primer + 2dA regardless of whether incorporation occurred opposite dT or the DNA lesion CPD (Fig. 1, C and D). Similar product band shifts were also observed when the dATP concentration was 1 μm and the concentration of rATP was varied (Fig. 1, C and D). Thus, dNTPs and rNTPs compete for incorporation by hpol η.
RNase H2 recognizes and incises rA positioned opposite dT or the CPD lesion
RNase H2 cleaves ribonucleotides from dsDNA or DNA/RNA hybrids (24, 28). We examined the effect of incorporating two ribonucleotides (rA) opposite a CPD lesion, and we previously observed that hpol η was capable of generating such a structure (Fig. 1D).
The results showed that RNase H2 could cleave a DNA strand containing two adjacent rAs, whether positioned opposite dTdT or CPD. The activity for cleavage of rArA opposite CPD was slightly lower (Fig. 2, A–C) as compared with the dTdT-containing template (which showed two cleavage products, P1 and P2).
Figure 2.
RNase H2 incises both dTdT:rArA and CPD:rArA. A, for RNase H2 assays the enzyme (11 nm) was mixed with 1 μm oligonucleotides containing dTdT:rArA or CPD:rArA for 2.5, 5, 7.5, 10, 15, 20, and 30 min. B, dTdT:rArA or CPD:rArA (1 μm) was incubated with 1.1, 2.7, 5.5, 8.2, 11, 14, 16, 22, 55, or 109 nm RNase H2. C, quantitation of part B. D, comparison of pre–steady-state kinetics of RNase H2 processing of dTdT:rArA and CPD:rArA. The inset is an extension of the time frame, showing different steady-state rates. Results are presented as means of duplicate assays ± SD (range). S represents the uncut substrate, and P1 and P2 are the products. See “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used.
We measured initial burst rates of cleavage in pre–steady-state kinetic assays (Fig. 2D). In line with the enzyme concentration dependence study in Fig. 2C, the effect of linking the two dT bases (i.e. CPD) was an ∼2-fold attenuation. The burst rate was 33−1 opposite dTdT and 15−1 opposite CPD, and the steady-state rate also reflected inhibition (Fig. 2D, inset).
hpol η in fibroblast cell extracts catalyze incorporation of dNTPs and rNTPs onto a DNA/DNA duplex
To test our hypothesis that hpol η tolerates ribonucleotides during strand extension in a physiological context, extracts from hpol η–deficient (XP-V, cell line XP30RO) and corrected (XP-V + lentivirus-transfected hpol η, cell line XP30RO + Pol η) fibroblast cells were prepared (45, 46) and examined with a DNA/DNA duplex containing either an unmodified dT or a CPD in the template. As expected, considerable primer degradation occurred because of the multiple nucleases in the cells (data not presented). To minimize degradation, a nonlabeled and unrelated ssDNA was added to the reaction mixture at a relatively high concentration as a competitor for the degradation by the nucleases (Fig. 3). In the presence of dNTPs, the elongation patterns for DNA/DNA (dTdT) with the XP-V (cell line XP30RO) and the corrected (cell line XP30RO + Pol η) cell extracts were almost identical, indicating that other human DNA polymerases, instead of hpol η, played the major roles in extending the primer (Fig. 3, B and C).
Figure 3.
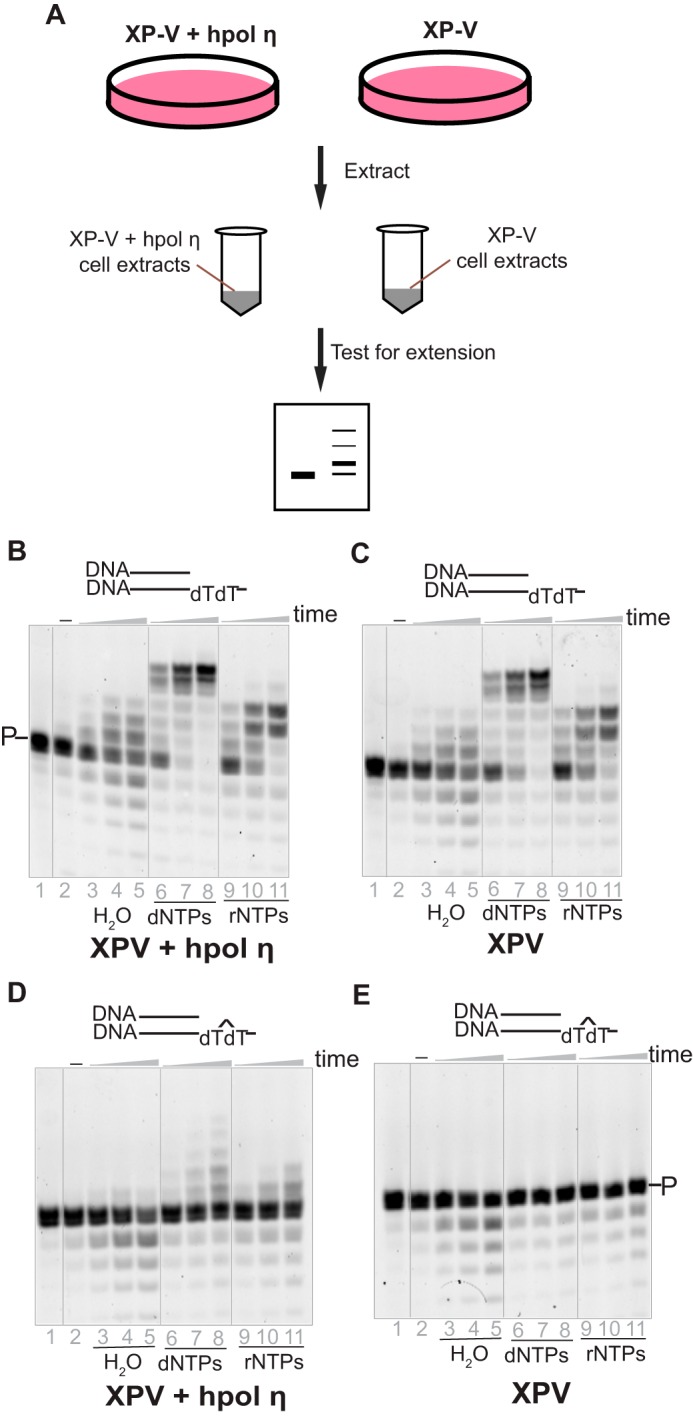
hpol η incorporates rNTPs opposite CPD lesion within human cell extracts. A, hpol η–deficient (XP-V, XP30RO) and corrected cell (XP30RO + Pol η) extracts were prepared, and each cell extract (1 mg protein/ml) was tested with 1 μm native or CPD-containing primer/template DNA substrates. B, the corrected cell extracts with DNA/DNA(dT). C, the XP-V cell extracts with DNA/DNA(dT). D, the corrected cell extracts with DNA/DNA(CPD). E, the XP-V cell extracts with DNA/DNA(CPD). To decrease degradation, a ssDNA, poly dA (100 μm) was added in each reaction. The reactions were conducted in the presence of H2O (negative control), dNTPs, or rNTPs (1 mm each nucleotide) for 10, 30, and 60 min. P indicates the primer and the bands below the primer are degradation products. See “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used.
When a CPD-containing template (paired with a DNA primer) was incubated with XP-V cell (cell line XP30RO) extracts, neither dNTPs nor rNTPs were incorporated (Fig. 3E), consistent with our knowledge that pol η is necessary to bypass the CPD lesion (1, 47, 48). However, with the corrected (XP-V + transfected hpol η, cell line XP30RO + Pol η) cell extracts, not only dNTP but also rNTP incorporation was observed (Fig. 3D, lanes 6–11), although with a limited amount of extension.
hpol η is the major reverse transcriptase in human fibroblasts
Similar experiments were conducted with RNA/DNA and DNA/RNA hybrid duplexes with the two types of fibroblast extracts (Fig. 4). The presence of RNA strands caused the substrates to be degraded more easily, probably by RNase H1 and RNase H2 in the cell extracts, compared with the DNA/DNA substrate (Fig. 4). To prevent degradation of targeted hybrids, limited amounts of cell extracts and large amounts of competitor oligomers (nonlabeled unrelated dsRNA/DNA hybrids and ssDNA) were used in each reaction. Under these experimental conditions, no rNTP insertion was observed for any of the substrates (Fig. 4, A–D).
Figure 4.
Lack of hpol η leads to loss of reverse transcription activity in human cell extracts. A–F, extracts (0.48 mg protein/ml) of hpol η–deficient (XP-V) and corrected cells were tested with DNA/DNA (A and B), RNA/DNA (C and D), and DNA/RNA (E and F), respectively (1 μm each substrate), in the presence of H2O (negative control) and dNTPs (1 mm each nucleotide). An unrelated annealed dsDNA/RNA hybrid (poly dA/poly rU) and ssDNA (poly dA) (100 μm) were added to prevent degradation. The reactions were conducted for 10, 30, and 60 min. See “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used.
Surprisingly, one dNTP insertion was observed for the DNA/RNA substrate by the hpol η–corrected cell extracts (Fig. 4E, lanes 5–7), and in the incubations with the XP-V cell extracts no primer was extended (Fig. 4F). These results demonstrate the critical role of hpol η in reverse transcription and indicate that hpol η is a key reverse transcriptase in human cells.
hpol η is the major reverse transcriptase in HEK293T cells
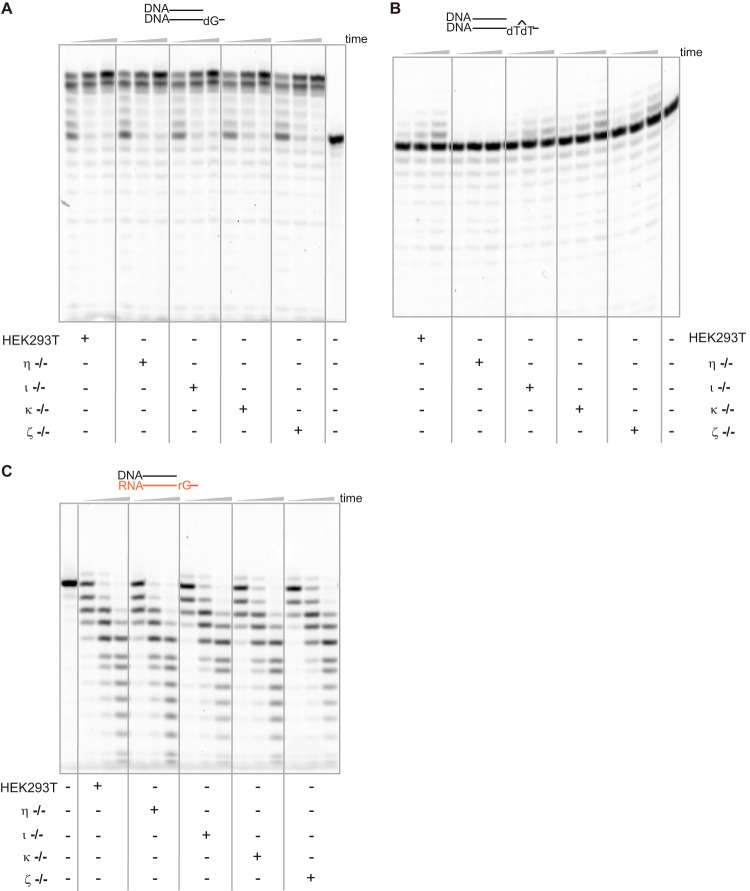
To test the hypothesis that hpol η is the major reverse transcriptase in other human cells, extracts from HEK293T cells were also examined. TLS DNA polymerase-deficient cell lines, deficient in hpols η, ι, κ, and ζ, respectively, were generated and extracts were prepared (49). These cell extracts were capable of extending a DNA primer opposite an undamaged DNA template, although significant degradation was observed (Fig. 5A).
Figure 5.
hpol η, instead of other TLS polymerases, plays a key role in reverse transcription in human cell extracts. Extracts of CRISPR-Cas9 knock-out cell lines were made, and each cell extract (1.4 mg protein/ml) was incubated with 1 μm DNA/DNA (dG), DNA/DNA(CPD), or DNA/RNA (rG), respectively, in the presence of a dNTP mixture (the concentration of each dNTP was 450 μm). An unrelated annealed dsDNA-RNA hybrid and ssDNA (100 μm concentrations) were used to prevent degradation. The reactions were conducted for 10, 30, and 60 min. See “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used.
However, when a CPD lesion was present in the template, no incorporation was observed in the cell extracts of hpol η−/− cells (Fig. 5B), as expected from previous findings on the critical role of hpol η in CPD bypass (1, 50). This result provides confirmation of the phenotype of these cells with regard to the status of hpol η.
Extensive DNA degradation resulted when a DNA primer was positioned opposite an RNA template. However, one-base extension of the primer was observed for all the cell extracts except hpol η−/− (Fig. 5C), confirming the important role of hpol η in its reverse transcriptase mode in human cells.
Discussion
The concentration of nucleotides plays an important role in cell regulation. It is very important to understand the biological consequences of the physiological levels of free rNTPs. In general, the concentrations of rNTPs are much higher than of dNTPs (13) and lead to the incorporation of rNTPs during replication. In the genome, the embedded rNTPs are processed either by RER (RNase H2, topoisomerase I) or postreplication repair mechanisms (51–53). Ribonucleosides in DNA can also be tolerated by multiple DNA polymerases, including the ones belonging to the TLS polymerase family (15, 25).
Although we previously demonstrated that hpol η could incorporate rNTPs into DNA (41, 43), we now show that this occurs under conditions in which dNTPs and rNTPs are present at their physiological concentrations. We tested the possible process and consequence of rNTP incorporation opposite CPD lesions by hpol η. As expected, the incorporation of rNTPs was observed, and hpol η can perform TLS opposite a CPD lesion, utilizing rATP (Fig. 1). rNTPs and dNTPs compete for incorporation by hpol η, and rNTPs can be incorporated in the presence of dNTPs. These results suggest that the low sugar selectivity of the hpol η can promote the incorporation of rATP opposite a CPD lesion into the genome, and it can possibly lead to the accumulation of rNTPs in RNase H–deficient cells.
Our studies were done with purified hpol η (actually the hpol η used is a short version containing the catalytic portion) and cell extracts. Cell extracts have been utilized in studying a variety of functions of DNA processing enzymes, in that all accessory proteins should be present, and what we have observed is a reflection of what is happening in the cells (and by extension, what is happening in human tissues). Carrying out the extension experiments in cells is not feasible, in terms of recovering products to analyze.
Regarding the physiological relevance of our results, we have already speculated on some of the possibilities but do not have proof of any yet (43). One possibility is that hpol η can replace hpol α or PrimPol to conduct translesion synthesis past CPD lesions by extending an RNA primer at the origin of replication or an Okazaki fragment. Another possibility is that hpol η could use a transcript strand as a template to synthesize DNA at a DNA double-strand break (43). However, we can only speculate on those possible functions at this time.
The majority of human DNA pols have now been shown to incorporate rNTPs (43). The selectivity varies considerably and depends on the rNTP concentrations under cellular conditions. Mitochondrial DNA (mtDNA) is known to contain ribonucleotides, which cannot be repaired by RER and may lead to the mtDNA depletion syndrome (MDS) (54). Recent studies have shown that the presence of ribonucleotides slows the speed of mitochondrial DNA replication by pol γ (55); ribonucleotides in the DNA template do not slow replication but incorporation of rNTPs slows replication. PrimPol, a primase-polymerase, is well-known for its two distinct functions as a primase and a TLS polymerase. PrimPol is present in mitochondria, as well as in the nucleus, and may play a crucial role in tolerance of rNTPs during the replication of mtDNA (56). In the archaebacterium Pyrococcus abyssi, all three known DNA polymerases incorporate rNTPs, to varying extents (57). P. abyssi also contains an RNase H2 to repair such lesions.
The RNase H2 activity was affected by the presence of the DNA lesion CPD, either the concentrations of RNase H2 needed for cleavage (Fig. 2C) or the pre–steady-state rate (Fig. 2D). These results indicate that the incorporation of rArA opposite CPD lesions can decrease the efficiency of their removal by RNase H2. One possibility which we cannot exclude is that these circumstances might delay RER in cellular events and can lead to replication stress and genomic instability.
Reverse transcriptases are important factors in the central dogma of molecular biology. In eukaryotes, the self-replicating stretches of genomes use reverse transcriptase for DNA replication, employing RNA as an intermediate. Joyce and Samanta (58) have recently discovered that the RNA polymerase ribozyme can catalyze reverse transcriptase activity. Apart from rNTP incorporation, it is of interest to investigate the detailed role of hpol η as a reverse transcriptase which catalyzes the RNA-dependent polymerization of DNA and is responsible for maintaining genetic information and translation machinery. In this regard, we focused on the roles of hpol η under cellular conditions. Our studies with human fibroblasts and HEK293T cells indicate that hpol η is the major human reverse transcriptase (Fig. 3). Our results suggest that there is a loss of reverse transcriptase activity in an hpol η–deficient system (Fig. 4). However, we observed incorporation of one nucleotide opposite an RNA template in cell lines deficient in other TLS DNA polymerases, pols ι, κ, and ζ (Fig. 5C), whereas hpol η−/− cells did not incorporate any nucleotide opposite an RNA template. These results clearly indicate the importance of hpol η as a reverse transcriptase (Fig. 5C).
Human DNA pol μ (hpol μ) and terminal deoxynucleoside transferase (TdT) both incorporate rNTPs to a large extent, and this activity has been implicated in the repair of chromosome breaks by nonhomologous end joining (59). We have previously postulated that hpol η could utilize RNA primers at origins of replication or facilitate DNA double-stranded break repair by using RNA transcripts as primers (43).
In summary, hpol η can catalyze numerous reactions with ribose-based molecules under cellular conditions. These findings with human cell extracts provide potential directions for understanding the consequences of rNTPs incorporation as well as role of hpol η as a reverse transcriptase.
Experimental procedures
Oligonucleotide substrates
The oligonucleotide containing the CPD lesion was purchased from TriLink BioTechnologies (San Diego, CA) and the others were from Integrated DNA Technologies (Coralville, IA). All of these oligonucleotides were purified by HPLC (reversed phase) by the manufacturers. The primers 5′-FAM-CGG GCT CGT AAG CGT CAT-3′ and 5′-FAM-rCrGrG rGrCrU rCrGrU rArArG rCrGrU rCrArU-3′ were annealed with template oligonucleotides in a 1:1 molar ratio of i) 5′-TCA (CPD)A TGA CGC TTA CGA GCC CG-3′ (CPD indicates cis-syn thymine dimer), ii) 5′-TCA TTA TGA CGC TTA CGA GCC CG-3′, and iii) 5′-TCA TGA TGA CGC TTA CGA GCC CG-3′. In addition, 5′-rUrCrA rUrGrA rUrGrA rCrGrC rUrUrA rCrGrA rGrCrC rCrG-3′ were annealed with 5′-FAM-CGG GCT CGT AAG CGT CAT-3′. For the RNase H2 incision assay, 5′-TCA (CPD)A TGA CGC TTA CGA GCC CG-3′ and 5′-TCA TTA TGA CGC TTA CGA GCC CG-3′ were annealed with 5′-FAM-CGG GCT CGT AAG CGT CAT rArAT GA-3′ to form dTdT:rArA and CPD:rArA complexes.
Cell lines
hPol η–deficient XP-V (XP30RO) human skin fibroblasts and the corrected cells (XP30RO + Pol η) were kindly provided by Prof. James E. Cleaver (University of California, San Francisco) (45, 46). HEK293T cells deficient in the POLH, POLI, POLK, and REV3L genes, which encode DNA pols η, ι, and κ and the catalytic subunit of hpol ζ, respectively, were produced previously using a CRISPR-Cas9 genome editing method and characterized (49, 60).
Protein expression and purification
The catalytic core of WT hpol η (1–432 amino acids) was expressed in E. coli and purified as reported previously (40, 50).
Full-length RNase H2 was expressed in E. coli and purified as described (61, 62). After chromatography on a HisTrap column (GE Healthcare), dialysis, and chromatography on a heparin column (GE Healthcare), the protein was eluted with a 40 mm potassium phosphate buffer (pH 7.0) containing 350 mm NaCl, 1 mm dithiothreitol (DTT), 5% glycerol (v/v), and 0.5 mm EDTA. The final product was aliquoted, flash frozen with liquid N2, and stored at −80 °C.
Extension assays
hpol η was incubated with annealed 5′-FAM–labeled primer-template substrates in the reaction buffer (40 mm Tris-HCl (pH 7.5) containing 10 mm DTT, 0.1 mg/ml BSA, 5% glycerol (v/v), 5 mm MgCl2, and 100 mm KCl) at 37 °C for 5 min before adding the mixtures of nucleotides or a single nucleotide. Reactions were conducted for the indicated times and stopped by addition of a quench solution (95% formamide (v/v) and 10 mm EDTA). The products were loaded onto 18% (w/v) denaturing polyacrylamide gels, separated, and visualized using a Typhoon system (GE Healthcare) (63, 64).
RNase H2 incision assay
A FAM-labeled dTdT:rArA or CPD:rArA duplex (1 μm; see “Oligonucleotide substrates” under “Experimental procedures” for the oligonucleotide sequences used) was incubated with RNase H2 for varying times at 37 °C in the “incision buffer”: 40 mm Tris-HCl (pH 7.5) containing 1 mm DTT, 0.1 mg/ml BSA, 5% glycerol (v/v), 10 mm MgCl2, and 50 mm NaCl. Reactions were stopped by the addition of 95% formamide (v/v) and 10 mm EDTA. For the pre–steady-state kinetic assays, 2 μm dTdT:rArA or CPD:rArA complex and 437 nm RNase H2 (both in incision buffer) were mixed in a KinTek RP-3 instrument (KinTek Corporation, Austin, TX), allowing the reaction to proceed for only a very short period time. The products were separated on 18% (w/v) denaturing polyacrylamide gels and visualized with a Typhoon system (GE Healthcare) (63, 64). The program ImageJ was used for quantitation, and the pre–steady-state kinetic assay data were fit to a burst equation: y = A(1-e−kpt) + kssE0t, using GraphPad Prism software (GraphPad, San Diego, CA), where A represents the apparent concentration of the active form of the enzyme, kp is the burst rate, kss is the steady-state rate, t is time, and E0 is the total enzyme concentration (63, 64).
Extension assays with cell extracts
Xeroderma pigmentosum variant (XP-V) cells, the corrected cell lines (XP-V + transfected hpol η–EGFP), HEK293T cells, HEK293T cell-deficient cells (knocking out hpols η, ι, κ, and ζ, respectively, by CRISPR-Cas9 genome editing method) were as previously reported (49, 60). The cells were grown in Dulbecco's modified Eagle medium (plus 4.5 g/liter d-glucose and l-glutamine) (Life Technologies) plus 10% FBS (v/v) (Life Technologies), penicillin, and streptomycin at 37 °C (5% CO2, v/v).
For the preparation of cell extracts, 75% fluent cells were trypsinized and washed with cold PBS solution. After resuspending cell pellets in hypotonic lysis buffer (10 mm Tris-HCl (pH 7.5) containing 1 mm EDTA, 10% sucrose (w/v), 10% glycerol (v/v), 1 mm DTT, and a protease tablet (Mini cOmplete EDTA-free)), cells were swelled for 20 min at 4 °C and lysed with a Dounce homogenizer. NaCl was added to a final concentration of 0.15 m. Samples were incubated for 10 min at 4 °C, sonicated, and centrifuged at 15,000 × g at 4 °C for 20 min. The supernatants were flash frozen in liquid N2 and stored at −80 °C.
Cell extracts were mixed with annealed DNA substrates, or DNA/RNA hybrids (1 μm) in 40 mm Tris-HCl buffer (pH 7.5) containing 1 mm DTT, 0.1 mg/ml BSA, 5% glycerol (v/v), 5 mm MgCl2, and 100 mm KCl. Single-stranded poly dA (100 μm) was added to the reaction with DNA substrates and both 100 μm poly dA and 100 μm hybrid poly dA/poly rU were added to the reactions using DNA/RNA substrates. The reaction times were 10, 30, and 60 min. After quenching, the final products were separated by denaturing polyacrylamide gels (63, 64).
Author contributions
Y. S. and F. P. G. conceptualization; Y. S. data curation; Y. S. formal analysis; Y. S. validation; Y. S. investigation; Y. S. and F. P. G. methodology; Y. S., P. P. G., and F. P. G. writing-original draft; Y. S., P. P. G., M. E., and F. P. G. writing-review and editing; P. P. G. and F. P. G. visualization; M. E. project administration; L. L., Y. W., and F. P. G. resources; F. P. G. supervision; F. P. G. funding acquisition.
Acknowledgments
We thank Dr. Robert Crouch for the expression vector for RNase H2, Dr. James Cleaver for the cell lines, and K. Trisler for assistance in preparation of the manuscript.
Note added in proof
Following acceptance of our manuscript, our attention was called to a paper (Franklin, A., Milburn, P. J., Blanden, R. V., and Steele, E. J. (2004) Human DNA polymerase-η, an A-T mutator in somatic hypermutation of rearranged immunoglobulin genes, is a reverse transcriptase. Immunol. Cell Biol. 82, 219–225) by one of the authors, in which pol η had been reported to show activity in a product-enhanced reverse transcriptase assay using a 4 mM mixture of dNTPs, and a mechanism had been proposed. Reference to this work had been included in an earlier draft of our manuscript but had been inadvertently deleted in the final version. We believe that our biochemical and cell extract work in Ref. 43 and here in this paper goes beyond this early study in demonstrating the significance of pol η as a reverse transcriptase, but apologize for the oversight.
This work was supported by National Institutes of Health Grants R01 ES026955 and R01 ES010546 (to F. P. G.) and R01 ES025121 (to Y. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
- pol
- (DNA) polymerase
- 8-oxodG
- 8-oxo-7,8-dihydrodeoxyguanosine
- CPD(dTdT)
- cyclobutane pyrimidine dimer (cis-syn)
- dNTP
- deoxyribonucleoside triphosphate
- FAM
- 6-carboxyfluorescein
- h
- human
- mtDNA
- mitochondrial DNA
- RER
- ribonucleotide excision repair
- rNTP
- ribonucleoside triphosphate
- TLS
- translesion synthesis
- XP-V
- xeroderma pigmentosum variant.
References
- 1. Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., and Ellenberger T. (2006) DNA Repair and Mutagenesis, 2nd ed., ASM Press, Washington, DC [Google Scholar]
- 2. Goodman M. F., and Woodgate R. (2013) Translesion DNA Polymerases. Cold Spring Harbor Perspect. Biol. 5, a010363 10.1101/cshperspect.a010363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang W., and Gao Y. (2018) Translesion and repair DNA polymerases: Diverse structure and mechanism. Annu. Rev. Biochem. 87, 239–261 10.1146/annurev-biochem-062917-012405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baltimore D., and Smoler D. (1971) Primer requirement and template specificity of the DNA polymerase of RNA tumor viruses. Proc. Natl. Acad. Sci. U.S.A. 68, 1507–1511 10.1073/pnas.68.7.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kati W. M., Johnson K. A., Jerva L. F., and Anderson K. S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988–25997 [PubMed] [Google Scholar]
- 6. Tian L., Kim M. S., Li H., Wang J., and Yang W. (2018) Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc. Natl. Acad. Sci. U.S.A. 115, 507–512 10.1073/pnas.1719746115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kornberg A., and Baker T. A. (1992) DNA Replication, 2nd ed., W. H. Freeman, New York [Google Scholar]
- 8. Storici F., Bebenek K., Kunkel T. A., Gordenin D. A., and Resnick M. A. (2007) RNA-templated DNA repair. Nature 447, 338–341 10.1038/nature05720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams J. S., and Kunkel T. A. (2014) Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair 19, 27–37 10.1016/j.dnarep.2014.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., Lundström E. B., Johansson E., Chabes A., and Kunkel T. A. (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6, 774–781 10.1038/nchembio.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao N. Y., Schroeder J. W., Yurieva O., Simmons L. A., and O'Donnell M. E. (2013) Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U.S.A. 110, 12942–12947 10.1073/pnas.1309506110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caldecott K. W. (2014) Ribose—An internal threat to DNA. Science 343, 260–261 10.1126/science.1248234 [DOI] [PubMed] [Google Scholar]
- 13. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., and Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 10.1073/pnas.0914857107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watt D. L., Johansson E., Burgers P. M., and Kunkel T. A. (2011) Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair 10, 897–902 10.1016/j.dnarep.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clausen A. R., Zhang S., Burgers P. M., Lee M. Y., and Kunkel T. A. (2013) Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair 12, 121–127 10.1016/j.dnarep.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clausen A. R., Murray M. S., Passer A. R., Pedersen L. C., and Kunkel T. A. (2013) Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. U.S.A. 110, 16802–16807 10.1073/pnas.1309119110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Makarova A. V., Nick McElhinny S. A., Watts B. E., Kunkel T. A., and Burgers P. M. (2014) Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Repair 18, 63–67 10.1016/j.dnarep.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gosavi R. A., Moon A. F., Kunkel T. A., Pedersen L. C., and Bebenek K. (2012) The catalytic cycle for ribonucleotide incorporation by human DNA pol λ. Nucleic Acids Res. 40, 7518–7527 10.1093/nar/gks413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cronan G. E., Kouzminova E. A., and Kuzminov A. (2019) Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision. Proc. Natl. Acad. Sci. U.S.A. 116, 1251–1260 10.1073/pnas.1814512116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Günther C., Kind B., Reijns M. A., Berndt N., Martinez-Bueno M., Wolf C., Tüngler V., Chara O., Lee Y. A., Hübner N., Bicknell L., Blum S., Krug C., Schmidt F., Kretschmer S., et al. (2015) Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 125, 413–424 10.1172/JCI78001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potenski C. J., and Klein H. L. (2014) How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 42, 10226–10234 10.1093/nar/gku773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conover H. N., Lujan S. A., Chapman M. J., Cornelio D. A., Sharif R., Williams J. S., Clark A. B., Camilo F., Kunkel T. A., and Argueso J. L. (2015) Stimulation of chromosomal rearrangements by ribonucleotides. Genetics 201, 951–961 10.1534/genetics.115.181149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crespan E., Furrer A., Rösinger M., Bertoletti F., Mentegari E., Chiapparini G., Imhof R., Ziegler N., Sturla S. J., Hübscher U., van Loon B., and Maga G. (2016) Impact of ribonucleotide incorporation by DNA polymerases β and λ on oxidative base excision repair. Nat. Commun. 7, 10805 10.1038/ncomms10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sparks J. L., Chon H., Cerritelli S. M., Kunkel T. A., Johansson E., Crouch R. J., and Burgers P. M. (2012) RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47, 980–986 10.1016/j.molcel.2012.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams J. S., Clausen A. R., Nick McElhinny S. A., Watts B. E., Johansson E., and Kunkel T. A. (2012) Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ϵ. DNA Repair 11, 649–656 10.1016/j.dnarep.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hiller B., Hoppe A., Haase C., Hiller C., Schubert N., Müller W., Reijns M. A. M., Jackson A. P., Kunkel T. A., Wenzel J., Behrendt R., and Roers A. (2018) Ribonucleotide excision repair is essential to prevent squamous cell carcinoma of the skin. Cancer Res. 78, 5917–5926 10.1158/1538-7445.AM2018-5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cerritelli S. M., and Crouch R. J. (2009) Ribonuclease H: The enzymes in eukaryotes. FEBS J. 276, 1494–1505 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uehara R., Cerritelli S. M., Hasin N., Sakhuja K., London M., Iranzo J., Chon H., Grinberg A., and Crouch R. J. (2018) Two RNase H2 mutants with differential rNMP processing activity reveal a threshold of ribonucleotide tolerance for embryonic development. Cell Rep. 25, 1135–1145.e5 10.1016/j.celrep.2018.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao H., Zhu M., Limbo O., and Russell P. (2018) RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep. 19, e45335 10.15252/embr.201745335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Costantino L., and Koshland D. (2018) Genome-wide map of R-loop-induced damage reveals how a subset of R-loops contributes to genomic instability. Mol. Cell 71, 487–497.e3 10.1016/j.molcel.2018.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belotserkovskii B. P., Tornaletti S., D'Souza A. D., and Hanawalt P. C. (2018) R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair 71, 69–81 10.1016/j.dnarep.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim N., Cho J.-E., Li Y. C., and Jinks-Robertson S. (2013) RNA:DNA hybrids initiate quasi-palindrome-associated mutations in highly transcribed yeast DNA. PLoS Genet. 9, e1003924 10.1371/journal.pgen.1003924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samara N. L., and Yang W. (2018) Cation trafficking propels RNA hydrolysis. Nat. Struct. Mol. Biol. 25, 715–721 10.1038/s41594-018-0099-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cozens C., Pinheiro V. B., Vaisman A., Woodgate R., and Holliger P. (2012) A short adaptive path from DNA to RNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 109, 8067–8072 10.1073/pnas.1120964109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schermerhorn K. M., and Gardner A. F. (2015) Pre-steady-state kinetic analysis of a Family D DNA polymerase from Thermococcus sp. 9°N reveals mechanisms for archaeal genomic replication and maintenance. J. Biol. Chem. 290, 21800–21810 10.1074/jbc.M115.662841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patra A., Su Y., Zhang Q., Johnson K. M., Guengerich F. P., and Egli M. (2016) Structural and kinetic analysis of miscoding opposite the DNA adduct 1, N6-ethenodeoxyadenosine by human translesion DNA polymerase η. J. Biol. Chem. 291, 14134–14145 10.1074/jbc.M116.732487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi J.-Y., and Guengerich F. P. (2005) Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 352, 72–90 10.1016/j.jmb.2005.06.079 [DOI] [PubMed] [Google Scholar]
- 38. Patra A., Nagy L. D., Zhang Q., Su Y., Müller L., Guengerich F. P., and Egli M. (2014) Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 289, 16867–16882 10.1074/jbc.M114.551820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patra A., Zhang Q., Lei L., Su Y., Egli M., and Guengerich F. P. (2015) Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase η. J. Biol. Chem. 290, 8028–8038 10.1074/jbc.M115.637561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su Y., Patra A., Harp J. M., Egli M., and Guengerich F. P. (2015) Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-dihydro-8-oxo-2′-deoxyguanosine. J. Biol. Chem. 290, 15921–15933 10.1074/jbc.M115.653691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su Y., Egli M., and Guengerich F. P. (2016) Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 291, 3747–3756 10.1074/jbc.M115.706226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Donigan K. A., McLenigan M. P., Yang W., Goodman M. F., and Woodgate R. (2014) The steric gate of DNA polymerase ι regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J. Biol. Chem. 289, 9136–9145 10.1074/jbc.M113.545442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su Y., Egli M., and Guengerich F. P. (2017) Human DNA polymerase η accommodates RNA for strand extension. J. Biol. Chem. 292, 18044–18051 10.1074/jbc.M117.809723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]
- 45. Thakur M., Wernick M., Collins C., Limoli C. L., Crowley E., and Cleaver J. E. (2001) DNA polymerase η undergoes alternative splicing, protects against UV sensitivity and apoptosis, and suppresses Mre11-dependent recombination. Genes Chromosomes Cancer 32, 222–235 10.1002/gcc.1186 [DOI] [PubMed] [Google Scholar]
- 46. de Feraudy S., Limoli C. L., Giedzinski E., Karentz D., Marti T. M., Feeney L., and Cleaver J. E. (2007) Pol η is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene 26, 5713–5721 10.1038/sj.onc.1210385 [DOI] [PubMed] [Google Scholar]
- 47. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., and Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 10.1038/21447 [DOI] [PubMed] [Google Scholar]
- 48. Johnson R. E., Kondratick C. M., Prakash S., and Prakash L. (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285, 263–265 10.1126/science.285.5425.263 [DOI] [PubMed] [Google Scholar]
- 49. Wu J., Li L., Wang P. C., You C. J., Williams N. L., and Wang Y. S. (2016) Translesion synthesis of O4-alkylthymidine lesions in human cells. Nucleic Acids Res. 44, 9256–9265 10.1093/nar/gkw662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., and Yang W. (2010) Structure and mechanism of human DNA polymerase η. Nature 465, 1044–1048 10.1038/nature09196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim N., Huang S.-Y. N., Williams J. S., Li Y. C., Clark A. B., Cho J.-E., Kunkel T. A., Pommier Y., and Jinks-Robertson S. (2011) Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564 10.1126/science.1205016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lazzaro F., Novarina D., Amara F., Watt D. L., Stone J. E., Costanzo V., Burgers P. M., Kunkel T. A., Plevani P., and Muzi-Falconi M. (2012) RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell 45, 99–110 10.1016/j.molcel.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Williams J. S., Smith D. J., Marjavaara L., Lujan S. A., Chabes A., and Kunkel T. A. (2013) Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 49, 1010–1015 10.1016/j.molcel.2012.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wanrooij P. H., Engqvist M. K. M., Forslund J. M. E., Navarrete C., Nilsson A. K., Sedman J., Wanrooij S., Clausen A. R., and Chabes A. (2017) Ribonucleotides incorporated by the yeast mitochondrial DNA polymerase are not repaired. Proc. Natl. Acad. Sci. U.S.A. 114, 12466–12471 10.1073/pnas.1713085114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Forslund J. M. E., Pfeiffer A., Stojkovič G., Wanrooij P. H., and Wanrooij S. (2018) The presence of rNTPs decreases the speed of mitochondrial DNA replication. PLoS Genet. 14, e1007315 10.1371/journal.pgen.1007315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bailey L. J., and Doherty A. J. (2017) Mitochondrial DNA replication: A PrimPol perspective. Biochem. Soc. Trans. 45, 513–529 10.1042/BST20160162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lemor M., Kong Z. Q., Henry E., Brizard R., Laurent S., Bossé A., and Henneke G. (2018) Differential activities of DNA polymerases in processing ribonucleotides during DNA synthesis in archaea. J. Mol. Biol. 430, 4908–4924 10.1016/j.jmb.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 58. Samanta B., and Joyce G. F. (2017) A reverse transcriptase ribozyme. Elife 6, e31153 10.7554/eLife.31153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pryor J. M., Conlin M. P., Carvajal-Garcia J., Luedeman M. E., Luthman A. J., Small G. W., and Ramsden D. A. (2018) Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science 361, 1126–1129 10.1126/science.aat2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Du H., Leng J., Wang P., Li L., and Wang Y. (2018) Impact of tobacco-specific nitrosamine-derived DNA adducts on the efficiency and fidelity of DNA replication in human cells. J. Biol. Chem. 293, 11100–11108 10.1074/jbc.RA118.003477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Figiel M., Chon H., Cerritelli S. M., Cybulska M., Crouch R. J., and Nowotny M. (2011) The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières syndrome defects. J. Biol. Chem. 286, 10540–10550 10.1074/jbc.M110.181974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chon H., Vassilev A., DePamphilis M. L., Zhao Y., Zhang J., Burgers P. M., Crouch R. J., and Cerritelli S. M. (2009) Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 37, 96–110 10.1093/nar/gkn913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Su Y., and Guengerich F. P. (2016) Pre-steady-state kinetic analysis of single-nucleotide incorporation by DNA polymerases. Curr. Protoc. Nucleic Acid Chem. 65, 7.23.1–7.23.10 10.1002/cpnc.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. O'Flaherty D. K., and Guengerich F. P. (2014) Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products. Curr. Protoc. Nucleic Acid Chem. 59, 7.21.1–7.21.13 10.1002/0471142700.nc0721s59 [DOI] [PMC free article] [PubMed] [Google Scholar]