Abstract
In plants, strict regulation of stomatal pores is critical for modulation of CO2 fixation and transpiration. Under certain abiotic and biotic stressors, pore closure is initiated through anionic flux, with calcium (Ca2+) playing a central role. The aluminum-activated malate transporter 12 (ALMT12) is a malate-activated, voltage-dependent member of the aluminum-activated malate transporter family that has been implicated in anionic flux from guard cells controlling the stomatal aperture. Herein, we report the characterization of the regulatory mechanisms mediating channel activities of an ALMT from the grass Brachypodium distachyon (BdALMT12) that has the highest sequence identity to Arabidopsis thaliana ALMT12. Electrophysiological studies in a heterologous cell system confirmed that this channel is malate- and voltage-dependent. However, this was shown to be true only in the presence of Ca2+. Although a general kinase inhibitor increased the current density of BdALMT12, a calmodulin (CaM) inhibitor reduced the Ca2+-dependent channel activation. We investigated the physiological relevance of the CaM-based regulation in planta, where stomatal closure, induced by exogenous Ca2+ ionophore and malate, was shown to be inhibited by exogenous application of a CaM inhibitor. Subsequent analyses revealed that the double substitutions R335A/R338A and R335A/K342A, within a predicted BdALMT12 CaM-binding domain (CBD), also decreased the channels' ability to activate. Using isothermal titration calorimetry and CBD-mimetic peptides, as well as CaM-agarose affinity pulldown of full-length recombinant BdALMT12, we confirmed the physical interaction between the CBD and CaM. Together, these findings support a co-regulatory mechanism of BdALMT12 activation by malate, and Ca2+/CaM, emphasizing that a complex regulatory network modulates BdALMT12 activity.
Keywords: ion channel, patch clamp, isothermal titration calorimetry (ITC), calmodulin (CaM), calcium, plant physiology, protein–protein interaction, structure–function, stomatal function
Introduction
In plants, the control of CO2 uptake (assimilation) and the loss of water vapor (transpiration) are controlled by pores in the epidermal layer, referred to as stomata. Strict regulation of stomatal function is therefore critical for mediation of plant primary metabolism, growth, and development generally, as well as plant responses to both biotic (pathogenic) and abiotic (environmental) stress.
The opening and closing of the stomata is directly controlled by the release and uptake of ions by guard cells that form the stomatal pore (1, 2). In the case of stomatal closure, upstream signaling is known to mediate the phosphorylation of select anion channels, activating them and leading to Cl− and malate efflux. This anion efflux causes depolarization of the plasma membrane, which stimulates voltage-dependent potassium channels and additional K+ efflux. Overall, this mass ion efflux decreases turgor pressure, reducing swelling of the guard cells and causing the pore to close.
Upstream of the anion channels, signal transduction events stimulated by environmental cues lead to activation of downstream Ca2+-dependent and Ca2+-independent response pathways (3). In the case of the Ca2+-independent pathway, binding of abscisic acid (ABA)3 to its receptor(s) leads to the sequestration and inhibition of type 2C protein phosphatases, such that their target, a sucrose nonfermenting–related kinase (SnRK; e.g. OST1 in Arabidopsis thaliana), becomes activated through autophosphorylation (4). The SnRK subsequently phosphorylates the anion channels (5, 6). In contrast, abiotic stress and ABA are also known to stimulate the production of reactive oxygen species (ROS) through activation of an NADPH oxidase (7). ROS stimulates Ca2+ release into the cytosol, which in turn leads to the activation of Ca2+-dependent kinases (e.g. CPK6 and CPK21/23), which specifically phosphorylate the same anion channels as the Ca2+-independent SnRKs, leading to channel activation and ultimately the same stomatal closure (8–10).
At the channel level, working in concert, two types of anion channels presenting in the plasma membrane of guard cells are known to mediate anion efflux and stomatal closure: the rapid (R-type)- and the slow (S-type)-activating anion channels (11, 12). S-type anion channels are encoded by the slow anion channel 1 (SLAC1) gene in A. thaliana (13, 14) and its homologues (e.g. SLAH3 (13)). Although SLAC1 has been shown to be stimulated by SnRK, CPK, and calcineurin B–like calcium sensors and their calcineurin B–like-interacting protein serine–threonine-type kinases (i.e. Ca2+-independent and Ca2+-dependent pathways (2, 15, 16)), SLAH3, to date, has only been shown to be stimulated by the Ca2+-dependent kinase pathway (16, 17).
In the A. thaliana guard cells the R-type anion channel is encoded by the ALMT12 gene (encoding AtALMT12 (18)). AtALMT12 is one member of a larger family of 14 aluminum-activated malate transporter (ALMT) channels in A. thaliana, better known for releasing malate from the root tip for chelation of aluminum (19, 20). More recently, reports have shown that AtALMT12 is insensitive to Al3+, activated by malate, with very specific voltage-dependent properties, and its deletion selectively impairs stomatal closure (18, 21). As such, it was renamed A. thaliana quick activation anion channel 1 (AtQUAC1) to avoid confusion with other ALMT channels. The secondary structure of AtQUAC1 has been predicted to have six transmembrane segments at its N terminus and a large cytoplasmic C-terminal domain. Similar to SLAC1, AtQUAC1 activation has recently been shown to be controlled by the Ca2+-independent but phosphorylation-dependent SnRK pathway (6). However, that the AtQUAC1 activity was reduced by only 50% with deletion of OST1 (a SnRK) suggests other mechanisms of regulation may also be in play.
Although the ALMT gene family was first identified in wheat (19), the model monocot Brachypodium distachyon ALMT12 has yet to be investigated. A BLAST search yielded seven putative ALMTs in B. distachyon, with one sequence having significant (59%) amino acid identity to AtQUAC1. Using a recombinant expression system, patch-clamp analysis was applied to investigate channel activity and regulation. The observation of Ca2+ sensitivity led to further evaluations of the effect of select kinase and calmodulin (CaM) inhibitors, with results suggesting a regulatory role for CaM in BdALMT12 activity. The relationship between malate, Ca2+, CaM, and stomatal function was investigated in planta. Subsequent electrophysiological evaluation of amino acid substitutions targeting a putative BdALMT12 CaM-binding domain (CBD), along with isothermal titration calorimetry and CaM-agarose affinity pulldown experiments, confirmed the role of the CBD and its ability to mediate a direct interaction between CaM and BdALMT12. The general physiological context and relevance are discussed.
Results
BRADI_3g33980v3 encodes a putative B. distachyon ALMT12 and is expressed in shoot tissue
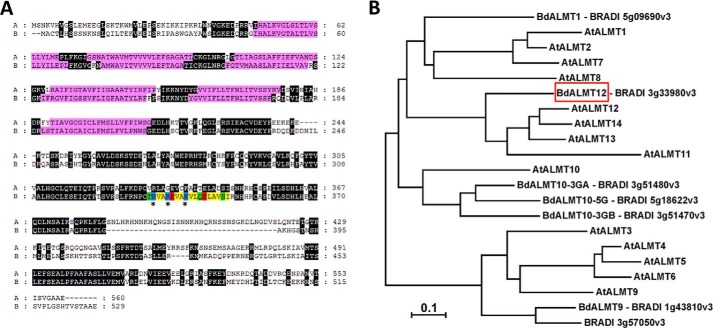
A BLAST search of the Ensembl plants B. distachyon sequence database yielded six unique amino acid sequences with 30–36% identity (BRADI_5g09690v3, BRADI_1g43810v3, BRADI_3g51480v3, BRADI_5g18622v3, BRADI_3g51470v3, and BRADI_3g57050v3) and a single sequence with 59% amino acid identity (BRADI_3g33980v3; NCBI protein accession no. XP_003574370.1; putative BdALMT12) to AtALMT12/ATQUAC1 (Fig. 1A). This latter putative BdALMT12 sequence is 529 amino acids long, with an expected molecular mass of ∼56 kDa. It was found to be ∼90% identical to the Hordeum vulgare (gene id HORVU1Hr1G049820) and Triticum aestivum (gene id TraesCS1D01G194000) closest homologues, and it was 82% identical to Zea mays ALMT12 (GenBankTM accession no. PWZ19427.1). A phylogenetic analysis emphasizes that this particular putative B. distachyon ALMT is the only one of the seven to cluster in clade 3, with A. thaliana ALMTs 11–14 (Fig. 1B). Direct sequence alignments of the A. thaliana clade 3 ALMTs with putative BdALMT12 shows that it does in fact maintain the highest amino acid sequence identity with AtALMT12 (59%), having only 39, 54, and 55% identities, respectively, to AtALMTs 11, 13, and 14. Thus, we refer to this B. dystachyon protein as BdALMT12 going forward.
Figure 1.
Primary structural elements of BdALMT12 and its evolutionary relationships. A, alignment of AtQUAC1 (A) and BdALMT12 (B) amino acid sequences. The alignment was made using Clustal Omega (54) and visualized using GeneDoc (55). Regions highlighted in black are conserved. The location of the six predicted transmembrane helices are highlighted in pink, and a predicted CaM-binding domain is shaded according to Fig. 4G. B, evolutionary relationships of ALMT family members from A. thaliana and B. distachyon. The evolutionary history was inferred using the Neighbor-Joining method (56). The optimal tree with the sum of branch length = 5.50656346 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (57) and are in the units of the number of amino acid substitutions per site. The analysis involved 21 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 105 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (58). Sequence sources (NCBI accession numbers) are as follows: A. thaliana AtALMT1 (AEE28289.1); AtALMT2 (Q9SJE8.2); AtALMT3 (Q9LPQ8.1); AtALMT4 (Q9C6L8.1); AtALMT5 (Q93Z29.1); AtALMT6 (Q9SHM1.1); AtALMT7 (Q9XIN1.1); AtALMT8 (Q9SRM9.1); AtALMT9 (AEE76098.1); AtALMT10 (O23086.2); AtALMT11 (Q3E9Z9.1); AtALMT12 (O49696.1); AtALMT13 (Q9LS23.1); and AtALMT14 (Q9LS22.1). B. distachyon sequence sources are indicated in the figure.
Furthermore, expression analyses showed expression of transcripts arising from the gene BRADI_3g33980v3, encoding BdALMT12, in green leaf tissue taken from both seedlings and adult B. dystachyon plants. Relative expression was found to be 0.313 ± 0.026 (n = 4) and 0.120 ± 0.019 (n = 5), respectively, for young and old plants, compared with the housekeeping UBC18 gene. This is consistent with previous data showing expression of AtALMT12/AtQUAC1 (as well as AtALMT6 and AtALMT9) throughout shoot tissue, compared with AtALMT1 which is expressed in roots (20, 22). Whether BRADI_3g33980v3 is specifically expressed in the guard cells remains to be demonstrated.
Electrophysiological characteristics of a putative BdALMT12 shows co-dependent activation by malate and calcium
To assess the electrophysiological properties of BdALMT12, whole-cell patch clamp of heterologously expressed BdALMT12 in HEK293 cells was performed. Both HEK293 and Xenopus oocytes expression systems have been previously used to assess the electrophysiological properties of plant ion channels as they have relatively few endogenous ion channels, making them relatively “silent” and permitting the expression and evaluation of genes from many different organisms with reproducible outcomes (18, 22–29). Notably, neither system expresses any endogenous malate-sensitive ion channels. Heterologous HEK293 cell expression was chosen for this study as it allows whole-cell electrophysiological recordings, which enable the control of both internal and external cellular buffer solution, whereas the oocyte model does not.
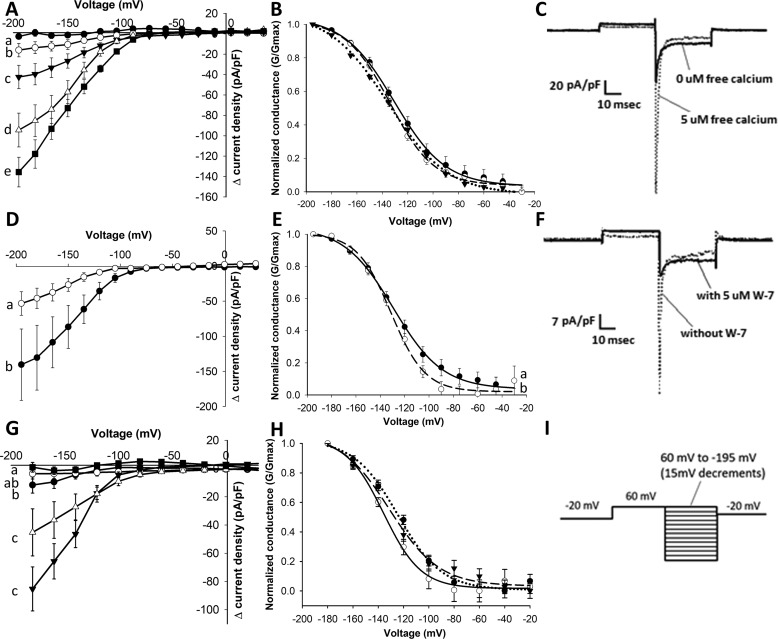
Surprisingly, initial patch-clamp recordings on BdALMT12, in the absence of any Ca2+ or agonist to increase internal Ca2+ (as per the experiments performed previously on AtQUAC1 (18)), yielded no channel activation with or without malate added to the bath solution (Fig. 2, A and C). However, taking advantage of the ability to control the cytosolic Ca2+ concentration via the whole-cell pipette solution during patch-clamp experiments, the malate-dependent activation of BdALMT12 was shown to be dependent on the presence of cytosolic Ca2+. Furthermore, increased cytosolic Ca2+ concentrations were found to increase the current activation, measured at maximum transient activation, in a dose-dependent manner in cells expressing BdALMT12 (Fig. 2, A and C; Fig. S1). The activation is followed by rapid inactivation at prolonged voltage stimulation, where no difference is observed in the inactivation current (at steady state) between different cytosolic Ca2+ concentrations (Fig. S2). No significant increases (regardless of Ca2+ and malate) were observed for the GFP-expressing controls (Fig. S3). Interestingly, there was no voltage shift in peak-normalized conductance between Ca2+ concentrations (Fig. 2B), suggesting cytosolic Ca2+ is regulated only in activation of the channel conductance and not the voltage dependence of the activation.
Figure 2.
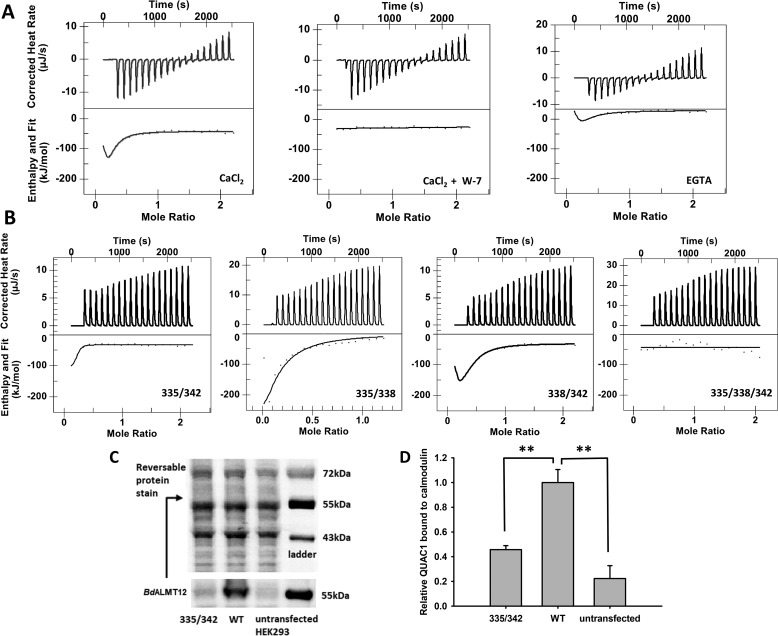
Effect of Ca2+ and pharmacological inhibitors on activity of BdALMT12 after malate activation. Statistical tests and notations are described under “Experimental procedures,” see “Electrophysiology.” A, Ca2+-dependent Δ current density of BdALMT12. HEK cells transfected with BdQUAC1 were patched using different Ca2+ concentrations in the pipette solution including 0 μm (black circle), 0.05 μm (white circle), 0.1 μm (black triangle), 0.5 μm (white triangle), and 5 μm (black square) free Ca2+, n ≥ 8 for each concentration. B, conductance of BdALMT12 channels at different Ca2+ concentrations, including 0.1 μm (solid line), 0.5 μm (broken line), and 5 μm (dotted line) free Ca2+. No statistical differences were detected (p > 0.05). C, representative traces of BdALMT12 current, presented in current density (pA/pF), at −180 mV with 0 and 5 μm free Ca2+ in the pipette solution. D, effect of staurosporine. Pipette solutions contained 0.1 μm free Ca2+ (white circle) or 0.1 μm free Ca2+ and 60 nm staurosporine (black circle) with n = 9 for each treatment. E, conductance of BdQUAC1 channels with addition of staurosporine in the pipette solution. Pipette solutions contained 0.1 μm free Ca2+ (solid line) or 60 nm staurosporine (dotted line). F, representative traces of BdQUAC1 current, presented in current density (pA/pF), at −180 mV with 0.5 μm free Ca2+ and the presence or absence of W-7. G, effect of W-7. Pipette solutions contained 0.5 μm free Ca2+ (black triangle), 0.5 μm free Ca2+ and 1 μm W-7 (white triangle), 0.5 μm free Ca2+ and 5 μm W-7 (black circle), 0.5 μm free Ca2+ and 10 μm W-7 (black circle), and 0 μm free Ca2+ (black square) with n ≥ 6 for each treatment. H, conductance of BdQUAC1 channels with addition of W-7 in the pipette solution. Pipette solutions contained 0.5 μm free Ca2+ (broken line), 0.5 μm free Ca2+ and 1 μm W-7 (dotted line), or 0.5 μm free Ca2+ and 5 μm W-7 (solid line). I, voltage protocol used for the experiments.
Electrophysiological analyses suggest BdALMT12 is regulated by CaM
To determine whether the Ca2+ activation of the BdALMT12 channel is mediated through a Ca2+-activated kinase or CaM mechanism, the effects of applying associated pharmacological inhibitors to the system was assessed. Indeed, HEK293 cells are well-known to maintain expression of many of the canonical eukaryotic regulatory mechanisms, including arrays of protein kinases and CaMs that have been implicated in an array of other biophysical studies (23–29). Herein, the contributions of Ca2+-activated kinases were investigated, based in part on the prediction of putative phosphorylation sites on the C terminus of the channel (e.g. BdALMT12 residues Ser-391, Ser-375, and Ser-394 (30, 31)). In particular, the nonspecific kinase inhibitor staurosporine, which prevents ATP binding to the kinase and thus inhibits any phosphorylation events from occurring, was used (32). Unexpectedly, addition of 60 nm staurosporine to the patch-clamp pipette resulted in an increase in current density (Fig. 2, D and E), rather than a loss of activity as would be expected if the activating Ca2+ effect was mediated by Ca2+-activated kinases.
To examine whether the cytosolic Ca2+ activation effect involves CaM, the activity of BdALMT12 was assessed in the presence of W-7, a naphthalene sulfonamide derivative, which has been shown to inhibit CaM-dependent phosphodiesterase and myosin light chain kinase activities with reported IC50 values of ∼28 and 51 μm, respectively (33). In the case of BdALMT12, the peak currents were observed to decrease significantly when even as little as 1 μm W-7 was used in the pipette solution and were reduced to baseline values with 10 μm W-7 (Fig. 2, F and G), similar to observed channel conductance in the absence of any Ca2+. Only a slight increase in voltage of activation (Fig. 2H) and no change in the rate of inactivation (data not shown) were observed with the addition of W-7. Thus, there does not appear to be much, if any, residual Ca2+-dependent activity that is unaffected by W-7, suggesting that CaM is a primary modulator of the BdALMT12 Ca2+-dependent channel activation under the influence of malate.
Pharmacological studies link BdALMT12 and CaM to calcium + malate-sensitive stomatal regulation in planta
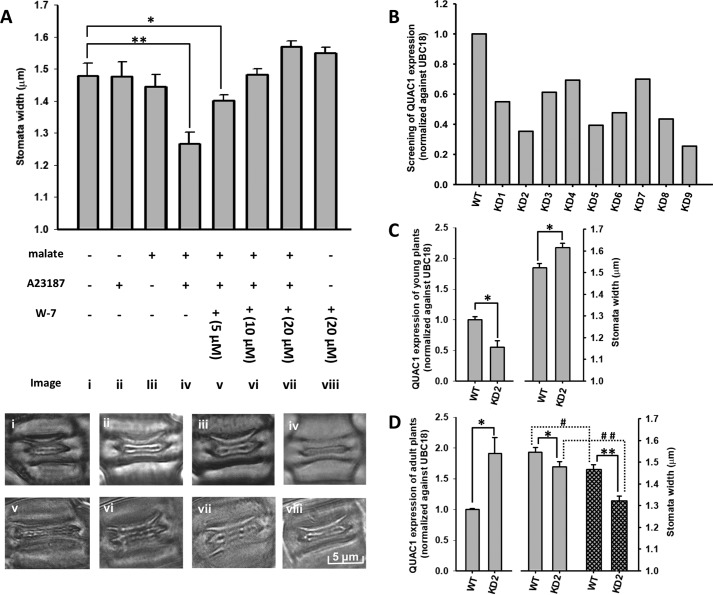
To validate the role of BdALMT12 and CaM in stomatal regulation in planta, the widths of stomatal apertures of B. distachyon leaves were measured after being treated with A23187 (a Ca2+ ionophore that leads to higher Ca2+ in the cytosol) in the presence and absence of malate. Although neither malate nor the ionophore showed any significant effect on stomatal widths on their own, stomatal pores treated with both malate and the ionophore together were significantly smaller, consistent with the combined and very unique role of malate and Ca2+ in activation of BdALMT12 (Fig. 3A). The inclusion of increasing concentrations of W-7 in the malate/A23187 containing buffer negated this effect, yielding concentration-dependent increases in stomatal pore widths, compared with malate/A23187 alone (Fig. 3A). Indeed, by 10 μm W-7, in the presence of malate/A23187, stomatal widths returned to untreated sizes. Notably, the application of W-7 alone did not increase stomatal widths significantly beyond untreated levels, emphasizing that W-7 is most likely reversing the effect of the malate/A23187 and is not simply modulating a different mechanism of stomatal regulation independently, although this possibility cannot be strictly eliminated.
Figure 3.
Effect of malate, a Ca2+ ionophore, and a CaM inhibitor on stomatal aperture in B. dystachyon WT and knockdown plants. Statistical symbols * and # indicate p ≤0.05, and ** and # # indicate p ≤0.01. A, plots of the averages from 100 stomata for each treatment are shown. Significance was determined by a lower-tailed Z test comparing each treatment against the opening control. Representative stomatal images for each treatment are labeled with a lowercase Roman numeral. B, PCR quantification of RNAi knockdown of QUAC1 in B. distachyon T0 generation plants. C, PCR quantification of QUAC1 expression and stomata width of the T1 generation KD2 seedlings (at 3–5 weeks). QUAC1 is significantly knocked down (shown by a one-sample t test, n = 3, with each KD2 paired to a unique WT, with each WT normalized to 1), which causes a small but significant increase in stomatal opening (by a two-sample t test, n = 100 stomata). D, PCR quantification of QUAC1 expression and stomata width of the T1 generation KD2 adult plants (at 8–10 weeks). QUAC1 has rebounded and been overly expressed (by a one-sample t test, n = 19, with WT normalized to 1). Stomata width of KD2 plants is significantly smaller both in the absence (empty pattern) and presence (cross-hatched lines) of malate and A23187 (by a two-sample t test, n = 100 stomata). All error bars represent standard error.
Toward further investigation of the biological function of the BdALMT12 gene, almt12 RNAi knockdown plants were generated in B. distachyon. Initially, nine lines were confirmed with significant knockdown of the gene at the T0 generation (Fig. 3B). Of these, only one line (KD2) still showed a significant knockdown as a young seedling (3–5 weeks old) at the T1 generation (Fig. 3C). Analysis of stomatal pore widths of the KD2 seedlings yielded a small but significant increase in stomatal opening compared with WT plants (Fig. 3C).
Interestingly, by the time the T1 KD2 plants had matured to adults (8–10 weeks old), they had not only lost any knockdown but the expression level of BdALMT12 had rebounded to levels significantly higher than WT adult plants, essentially creating a “BdALMT12-overexpressing condition” (as assessed by PCR; Fig. 3D). Assessment of adult KD2 line stomatal function yielded pore widths significantly smaller than those of WT, consistent with the increased stomatal closure that would be expected from BdALMT12 overexpression (Fig. 3D). The effects that arose due to the changes in BdALMT12 expression levels were further confirmed by investigating the response of the adult “BdALMT12-overexpressing” KD2 T1 generation plants to the exogenous application of the Ca2+ ionophore A23187, along with malate. Consistent with the proposed role of BdALMT12, stomatal pore widths were significantly more responsive to the A23187 and malate in the KD2 line than the WT line, with widths decreasing by ∼11 and 5%, respectively, compared with untreated controls (Fig. 3D; Table 1). Together, these data provide important correlative validation of a possible role for BdALMT12 in mediation of stomatal function, with transient knockdown in KD2 T1 seedlings leading to stomatal opening and the overexpression state of BdALMT12 in adult KD2 T1 plants leading to stomatal closure.
Table 1.
Changes in average stomatal widths induced by BdALMT12 overexpression or exogenous application of a calcium ionophore + malate
Values representing the averages from 100 stomata for each treatment are shown. Significance was determined by a lower-tailed Z test comparing each treatment against the opening control.
| WT stomata width | KD2 stomata width | Δ(WT–KD2) | ||
|---|---|---|---|---|
| μm | μm | |||
| Young, untreated | 1.52 ± 0.02 | 1.62 ± 0.02 | −0.1 ± 0.005 | |
| Adult, untreated | 1.55 ± 0.02 | 1.48 ± 0.02 | 0.06 ± 0.003 | p < 0.001 (2 sample t test) |
| Adult, treated with malate and A23187 | 1.47 ± 0.02 | 1.32 ± 0.02 | 0.15 ± 0.004 | |
| Adult, Δ(untreated − treated) | 0.08 ± 0.005 | 0.16 ± 0.005 | ||
| p < 0.001 (2 sample t test) | ||||
Mutational analyses identify a putative calmodulin-binding domain in BdALMT12
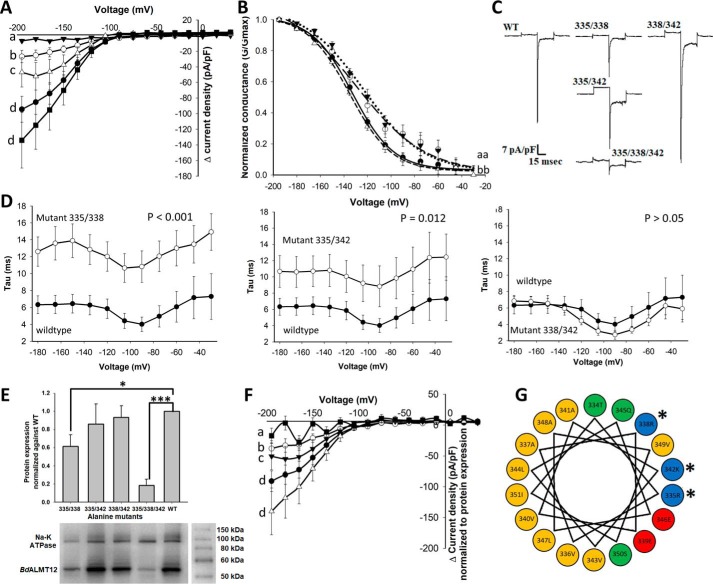
Toward identification of putative CBDs, the BdALMT12 amino acid sequence was analyzed using MI-1 (34), calmodulation meta-analysis (35), and Jpred3 (36) algorithms. Only a single region (BdALMT12 residues 334–351 (Fig. 1A)) met the criteria, yielding both the second highest CBD motif prediction score and displaying a strongly basic and amphiphilic α-helix (Fig. 4G). Toward the evaluation of the biological relevance of this putative CBD region, codons encoding three of the amino acids (Arg-335, Arg-338, and Lys-342 in BdALMT12) included in the basic portion of the helix were mutated to encode Ala in double and triple substitution variants. These residues were selected on the basis that basic residues in the CBD are known to be involved in the primary interactions between the CBD and CaM (37).
Figure 4.
Effect of mutations in the putative BdALMT12 CaM-binding domain on BdALMT12 channel activity. Statistical tests and notations of patch clamp data are described under “Experimental procedures,” see “Electrophysiology.” A, Δ current density of BdALMT12 and BdALMT12 variants. Transfected HEK cells were patched with 0.5 μm Ca2+ in the pipette solution. WT (black circle), variant 335/338 (white circle), variant 335/342 (white triangle), variant 338/342 (black square), and variant 335/338/342 (black triangle) are shown. Each variant was compared with WT only, with n ≥ 8 for each variant or WT. Same letters indicate no significant difference. B, channel conductance of BdALMT12 and BdALMT12 variants, including WT (solid line), variant 335/338 (long broken line), variant 335/342 (dotted line), and variant 338/342 (short broken line). C, representative traces of variant and WT BdALMT12 currents, presented in current density (pA/pF), at −180 mV. D, inactivation time constants of each double variant compared with WT BdALMT12. E, protein expression of variants and WT Myc-tagged BdALMT12 in HEK293 cell membranes. Total isolated cell membrane proteins were analyzed by Western blotting using an anti-Myc tag antibody, with n ≥ 6 for each variant and WT. WT expression was normalized to 1. Significance was determined by one sample t test (*, p = 0.014, ***, p < 0.001). F, Δ current density of BdALMT12 and BdALMT12 variants after normalization to protein expression. Normalization was calculated by subtracting the Δ current density to the GFP Δ current density then dividing by protein expression for WT (black circle), variant 335/338 (white circle), variant 335/342 (black triangle), variant 338/342 (white triangle), and variant 335/338/342 (black square). Significance is denoted as described for A. G, helical wheel depiction of the putative CaM-binding domain in BdALMT12 (amino acids 334–351), highlighting the amphiphilic nature of the helix. Nonpolar, hydrophobic residues are yellow; polar uncharged residues are green; polar acidic residues are red; and polar basic residues are blue. Residues selected for mutagenesis are indicated with an asterisk in both panels. All error bars represent standard error.
Patch-clamp analysis of these variants showed that the triple variant (335/338/342) lacked malate activation altogether and that two double variants, including 335/338 and 335/342, had significantly smaller peak-activated current increases (Fig. 4, A–C). However, no significant difference between the double variant 338/342 and WT was detected, highlighting a particularly notable role for the 335 site in the BdALMT12 putative CBD. To confirm that reduced activities were not caused by low expression of variants on the cell surface, surface biotinylation of transfected cells expressing Myc-tagged BdALMT12 or the Myc-tagged double or triple variants of BdALMT12 was performed to isolate surface proteins, for subsequent Western blot analysis using anti-Myc tag antibody. Although variants with significantly smaller activated current also had somewhat lower protein expression on the membrane surface (Fig. 4E), the overall decrease was not sufficient enough to explain the almost complete lack of current detected for the 335/338 variant in particular. Indeed, following normalization of the activity data to the surface expression data, currents of the deactivated triple variant and the two affected double variants 335/338 and 335/342 were still significantly smaller than WT (Fig. 4F). Additionally, the two partially deactivated double variants 335/338 and 335/342 also demonstrated a significant 10 ± 2-mV depolarizing shift at 50% whole-cell conductance activation increasing the voltage of activation, thereby decreasing the voltage change needed for activation (Fig. 4B). These variants also had a longer inactivation time constant compared with WT (Fig. 4D). Together, these data show that the decrease in peak current densities is not due to the changes in voltage-sensitive activation or inactivation processes, as the observed changes in kinetics should have resulted in larger peak current densities in this event. As such, it would seem that the CaM control over activation supersedes the minor impact on the voltage-gated kinetics of activation and rates of inactivation found with mutational intervention (Fig. 4, B and D, and see “Discussion”).
Interaction analyses show CaM binding to the predicted CBD domain of BdALMT12
With the electrophysiological results highlighting the involvement of the putative CBD domain in regulation of BdALMT12 activity, the physical interaction between the predicted CBD of BdALMT12 and CaM was evaluated using CaM-agarose affinity pulldown, and isothermal titration calorimetry (ITC).
Initial ITC binding studies between a WT BdALMT12 CBD peptide (residues 334–355) and CaM demonstrated a physical interaction as evidenced by the significant release of heat during titration of the peptide into the cell containing CaM, with quantitative analyses highlighting a Kd1 in the low nanomolar range and secondary weaker binding event with a Kd2 in the low micromolar range (Fig. 5A; Table 2). Although elimination of Ca2+ from the titration conditions led to a significant reduction in binding, in fact, some residual interaction was still detected (Fig. 5A). Quantitative analysis of this residual affinity yielded a Kd in the low micromolar range, with complete loss of the low nanomolar range interaction (Table 2), raising the possibility of some Ca2+-independent binding of BdALMT12 to CaM. Notably, this effect of removing Ca2+ is in contrast to the effect of including the CaM inhibitor W-7 in the reaction, which eliminated all release of heat, implying loss of all interactions between CaM and BdALMT12 (Fig. 5A; Table 2).
Figure 5.
Binding of WT and variant CaM-binding peptides to CaM. A, isothermal titration calorimetry analyses of the binding of WT to CaM in the presence of 5 mm Ca2+, 5 mm Ca2+ with 60 μm W-7, or no added Ca2+ with 1.5 mm EGTA for the 0 μm free Ca2+ effect. The amount of W-7 used in the above-mentioned experiment is equal to the amount of CaM presence (60 μm). The 5 mm Ca2+ and no Ca2+ data were fit using the “multiple sites” model, and the 5 mm Ca2+ with W-7 data were fit using the “independent” data. B, isothermal titration calorimetry analyses of the binding of variant CBD peptides to CaM. The binding of all peptides was carried out in the presence of 5 mm Ca2+. Variant 335/342 data were fit using the “multiple sites” model yielding a two-state binding result. The others were fit using the “independent” model and yielded either a single binding event (variant 335/338 and 335/342) or no binding (variant 335/338/342). C, representative Western blotting and reversible protein stain image of full-length WT QUAC1 and variant 335/342 binding to CaM. D, quantification of full-length QUAC1 protein bound to CaM. Wildtype BdALMT12 and variant 335/342 binding to CaM were analyzed by Western blotting using an anti-Myc tag antibody, n = 4. Wildtype and variant QUAC1 were quantified against total proteins eluted from the CaM affinity pulldown, and WT QUAC1 was normalized to 1. Error bars represent standard error. Significance was determined by one sample t test (** indicates p ≤ 0.01).
Table 2.
Binding parameters of wildtype and variant CaM-binding peptide
Unless otherwise indicated, all measurements were made in the presence of 5 mm Ca2+. The “no Ca2+” condition included 1.5 mm EGTA. W-7 concentration was equimolar to CaM concentration. ND means not determined.
| Peptide | Kd1 | ΔH1 | ΔS1 | Kd2 | ΔH2 | ΔS2 |
|---|---|---|---|---|---|---|
| m | kJ/mol | J/mol·K | m | kJ/mol | J/mol·K | |
| Wildtype peptide | 3.452E−8 | 105.4 | 4.962E2 | 3.925E−6 | −174.0 | −4.8E2 |
| Wildtype peptide, no Ca2+ | 9.165E−7 | 69.16 | 3.437E2 | 1.003E−5 | −199.9 | −5.636E2 |
| Variant 338/342 peptide | 4.101E−8 | −48.78 | −2.217E1 | 6.366E−6 | −199.2 | −5.685E2 |
| Variant 335/338 peptide | 3.721E−7 | −75.77 | −131.1 | ND | ND | ND |
| Variant 335/342 peptide | 5.5E−6 | −470.8 | −1478 | ND | ND | ND |
| Variant 335/338/342 peptide | ND | ND | ND | ND | ND | ND |
| Wildtype peptide + W-7 | ND | ND | ND | ND | ND | ND |
Subsequent evaluation of the binding affinities of CaM for double and triple alanine variant CBD peptides, representative of the variants tested by patch clamp, was unsuccessful due to substitution of the charged residues by hydrophobic residues making the peptides prone to formation of aggregates at higher concentrations. Indeed, the Ala-substituted peptide variants were shown to form oligomeric complexes of large particle sizes by dynamic light scattering (data not shown). As such, a new set of more soluble and charge-reversed double and triple variant peptides were synthesized with Asp substitutions at the same representative 335, 338, and 342 sites instead of Ala. Evaluation of the interaction of these Asp-substituted variant peptides showed that the triple mutation peptide 335/338/342 eliminated all binding to CaM (Fig. 5B). Similarly, the two variants 335/338 and 335/342 showed decreased binding affinities with Kd1 values in the low micromolar range, and complete loss of the secondary Kd2 binding event (Fig. 5B and Table 2). The double-variant peptide 338/342 had little impact on interactions, showing similar binding affinities compared with WT. These results are consistent with the electrophysiological analysis of BdALMT12 variants, and together they demonstrate the existence of a functional CaM-binding domain in BdALMT12, and its involvement in the regulation of BdALMT12 activities.
Toward investigating whether these interactions detected by ITC, using BdALMT12 CBD-mimetic peptides, translate to the more biologically relevant full-length BdALMT12, a CaM-agarose affinity pulldown (CAP) experiment was carried out. Western blot analysis of eluted fractions yielded a strong band at the expected molecular weight for WT BdALMT12 compared with samples arising from untransfected cells (Fig. 5C). The intensity of this band was dramatically reduced for cell lysates arising from transfected cells expressing the double Ala variant 335/342 (Fig. 5C). This variant was selected for analysis by CAP based on its low channel activity (Fig. 4A) but WT-like expression surface levels (Fig. 4E). Quantification of the Western blot analysis shows an ∼75% reduction (after subtraction of un-transfected background) in the amount of 335/342 variant eluted, compared with WT BdALMT12 (Fig. 5D), consistent with the significant (100-fold) reduction in binding affinity detected for this variant by ITC (Table 2).
Discussion
Validating prior comments (8), one of the more salient aspects of this study is the demonstration of a requirement for cytosolic Ca2+ for BdALMT12 activation. Further evaluation demonstrated that the effect of Ca2+ depended on CaM. Although such regulation or “calmodulation” of other anion channels has been demonstrated previously (38), this is the first report of the calmodulation of a member of the broader ALMT family.
Following up on initial experiments where BdALMT12 was found to be completely unresponsive to malate activation, the addition of Ca2+ demonstrated that both elevated cytosolic Ca2+ as well as external malate were in fact required for its activation. Although it was subsequently shown that the Ca2+ response was dose-dependent at a fixed concentration of malate, previous studies on AtQUAC1 showed malate activation was itself dose-dependent (18). Thus, how cytosolic Ca2+ concentration would affect the activation of the BdALMT12 channel with respect to malate dose dependence, and vice versa, remains to be determined.
With respect to a more detailed comparison of BdALMT12 and AtQUAC1 electrophysiology, the raw current traces of BdALMT12 (Fig. 2C) are very similar to what has been reported for AtQUAC1 (18). However, BdALMT12 appears to be activated at a lower voltage potential. Indeed, for BdALMT12, a bell-shaped IV curve that starts to approximate that observed for AtQUAC1 would likely only be achieved at considerably more negative voltage pulses. However, such low membrane potential would be nonphysiological and is not realistically achievable by the patch-clamp system. Also worth noting, AtQUAC1 malate-dependent depolarized potentials were found to elicit outward currents (anion uptake), in keeping with AtQUAC1 being activated upon depolarization, whereas malate-dependent hyperpolarized potentials elicited inward currents (anion release), which, as emphasized by the authors thereof, follows with the original hallmarks of R-type channels (18). In contrast, and despite the noted amino acid sequence homology documented herein, malate-dependent BdALMT12 currents are only shown to be activated upon hyperpolarization (eliciting inward currents, representing anion release) in a Ca2+-dependent manner. Thus, it would appear that BdALMT12 becomes more active upon hyperpolarization, emphasizing different voltage-dependent gating behavior between AtQUAC1 and BdALMT12. In this context, the possibility that other factors may be influencing the observed activities cannot be ignored. For example, the lack of observed activation of BdALMT12 at depolarized voltages could be due to the blockage/suppression of outward anionic currents, as reported previously in guard cells (6). In this case, the hyperpolarization-induced instantaneous transient inward currents could then reflect the deactivation, rather than transient activation, of the BdALMT12 anion channels. Alternatively, that the electrophysiological differences highlighted here might be arising due to differences in recombinant hosts (HEK293 cells versus oocytes) should also be kept in mind. Nonetheless, no outward currents (anion uptake) arose from positive potentials in BdALMT12, which is consistent with the primary role of anion channels in stomata closure being the release of anions from guard cells (1).
Interestingly, evaluation of a possible Ca2+-dependent kinase mechanism of activation showed that application of the nonspecific kinase inhibitor (staurosporine) yielded increased whole-cell current density (Fig. 2E). This implies that phosphorylation of the BdALMT12 channel may decrease channel activity and is thus more likely to serve as a feedback inhibition process, rather than a mechanism by which the channel is co-activated by Ca2+. Interestingly, this is in contrast to what was observed previously for the wheat root TaALMT1, where Ca2+-activated kinase inhibition led to loss of activity (30). Although not investigated further here, this observation highlights how tightly regulated BdALMT12 channel activity likely is and how this regulation likely varies with physiological roles within the broader ALMT family.
In the absence of a kinase-associated mechanism for Ca2+-linked activation, a role for CaM, which classically binds Ca2+ to adopt an interacting conformation, was evaluated based on previous reports highlighting a role for CaM in mediating stomatal function more generally (39–42), as well as computational predictions highlighting a putative CBD in the C-terminal domain of the channel (Fig. 4). The observed drastic reduction in activated BdALMT12 currents, upon inclusion of the strong CaM inhibitor W-7 in the pipette solution, validated this hypothesis (Fig. 2D).
Following up on these studies, the pharmacological effect of W-7 on malate + Ca2+-induced closure of stomata in planta was evaluated (Fig. 3). Notably, the Ca2+-signaling events were initiated through the use of a Ca2+ ionophore (A23187), which is known to allow Ca2+ entry into the guard cell cytoplasm and induce stomatal closure (42). Indeed A23187 has also been used to elevate intracellular calcium in many other cellular biological systems as well (43). The use of a Ca2+ ionophore was selected as it circumvents the ABA receptor–signaling pathway, which might otherwise initiate Ca2+-independent events also related to stomatal closure (44). Using this model, W-7 was found to inhibit Ca2+/malate-induced stomatal closure in B. distachyon leaves, demonstrating a role for CaM regulation of stomatal closure in B. distachyon, consistent with previous observations in the Commelina species (39–42). Importantly, the effect of W-7 was to specifically reverse malate + Ca2+-stimulated stomatal closure, showing no significant effect on its own compared with controls, which emphasizes the likelihood of BdALMT12 being a primary player in the CaM-associated stomatal responses documented herein and elsewhere (39–42).
In this context, it is notable that the B. distachyon almt12 RNAi knockdown and re-bound overexpression results do provide some limited evidence of a potential role for BdALMT12 in stomatal closure, with cells expressing less of the gene having larger stomatal openings on average and vice versa (Fig. 3). This is similar to the role demonstrated for AtQUAC1 in A. thaliana (17, 20). Notably, the adult “overexpressing state” was more responsive to malate-induced stomatal closure as is consistent with the proposed model. However, although RNAi knockdown rebound events are not uncommon (45), a detailed understanding of the mechanism leading to such a phenomenon remains enigmatic. As such, any conclusive findings regarding the biological relevance of BdALMT12 await future and more in-depth knockout, localization, and transcriptional expression analyses. In this context of a role for CaM in stomatal regulation, it is notable that the effect was originally documented over 30 years ago. Indeed, the very first report linking CaM to modulation of stomatal function showed significant increases in stomatal aperture widths with the exogenous application of CaM-binding drugs trifluoperazine and compound 48/80 (39). Interestingly, this effect was only relevant at micromolar Ca2+ concentrations, with higher exogenous Ca2+ overwhelming the effects of the CaM inhibitors (39). This is consistent with the low micromolar Ca2+ sensitivity detected for BdALMT12 herein. As well, the pharmacological inhibitor W-7, in particular, has been applied and shown to increase the average aperture width of stomata in Commelina communis L. and Commelina benghalensis species in multiple reports since then, linking CaM to ABA, vanadate, blue-light, and light/dark transition-mediated stomatal closure events (40–42).
With respect to an interaction with CaM, the observation of decreased whole-cell current densities upon variation of the leading CBD basic residue (Arg-335), in combination with other site changes in the basic portion of the predicted helix, further validates this hypothesis (Fig. 4). Interestingly, the R335A variants also demonstrated a small depolarizing shift in the activation voltage of the normalized conductance, such that these required less membrane depolarization to achieve a conductance similar to WT. Additionally, the voltage-dependent process of kinetic inactivation was slowed significantly. These two changes in kinetics suggest a link between the BdALMT12 CBD and voltage dependence, as reported previously in the literature for other channels (38). However, the altered BdALMT12 kinetics did not correlate with a larger current, and thus activation by CaM would seem to be occurring independently of the voltage-gated activation and inactivation processes. Most likely, the structural changes in the R335A-containing variants that affect the CaM interaction also somehow globally affect the structure of the channel, slowing its kinetics. This is supported by absence of any change in inactivation kinetics (data not shown) and only a very minor shift in activation with the pharmacological inhibition of CaM by W-7 on the WT channel (Fig. 2G). Additionally, the Ca2+ dose-response on the WT channel had no effect on inactivation (data not shown) or activation kinetics (Fig. 2B). Together, these findings support a model where mutationally-derived kinetic changes are likely due to modification of the voltage dependence of the channel and not necessarily related to the CaM interaction with the channel. Nonetheless, the decrease in whole-cell current in these variants indicates BdALMT12 activation depends on CaM.
This CaM-linked effect, arising as a result of a direct interaction between the putative BdALMT12 CBD and CaM, was confirmed in vitro by ITC and then for full-length BdALMT12 by CAP (Fig. 5). The ITC-derived two-state binding model, with Kd values of 34 nm and 3.9 μm, respectively, for the WT CBD peptide–CaM interaction, is consistent with other functional CBD–CaM interactions reported in the literature (46). Under these conditions, two of the isolated CBD peptides are expected to bind to a single CaM molecule. Interestingly, a low micromolar affinity interaction was still observed even in the absence of Ca2+, but not in the presence of W-7, which suggests that this low-affinity interaction represents a Ca2+-independent BdALMT12–CaM interaction, similar to that documented previously for a potassium channel (47). The role of a Ca2+-independent interaction would likely be related to keeping signaling pathway members in close proximity to each other, such that upon introduction of Ca2+, activation of the channel is not delayed by belated protein translocation events. With that being said, the possibility of some residual bound Ca2+ contaminating the system cannot be strictly eliminated.
Further investigation demonstrated that the R335D mutation in combination with changes in other sites lowers binding affinities as detected by ITC, and likely results in the loss of a conformational change that normally allows binding of the two CBD peptides to a single CaM molecule. Consistent with the patch-clamp data, mutations at sites not including 335, such as the double variant 338/342, did not affect either binding affinities or functional channel properties. These results are consistent with the CAP results, in which full-length WT BdALMT12 was found to be effectively pulled down by CaM. The full-length double R335A/R342A variant leads to a dramatic reduction in pulldown, emphasizing that the physical interaction is mediated by the identified CBD in full-length BdALMT12. Together, the ITC and CAP show that both Asp and Ala substitutions for Arg-335 are eliciting similar effects related to interfering with the BdALMT12 CBD–CaM interaction.
Together, these results show that BdALMT12 variants that include the substitution at Arg-335 in the putative CBD affect both CaM binding as well as whole cell currents. This lends important support to the ideas that BdALMT12 function is dependent on CaM, the presence of a functional CBD, and a physical interaction between CaM and BdALMT12. All combined, this emphasizes likely biological relevance of the interaction in terms of modifying channel activity.
However, the possibility that CaM is working via alternative or multiple mechanisms that are needed for stomatal closure cannot be ignored. Indeed, a recent report highlighted an interaction between CaM and a novel IQM protein that modulates stomatal function by an as yet unknown albeit Ca2+-independent mechanism (48). This suggests that although CaM can regulate BdALMT12 by direct interaction in a Ca2+-dependent manner, other BdALMT12/Ca2+-independent CaM-linked mechanisms that may be contributing to the overall modulation of stomatal function more broadly cannot be strictly eliminated. Nonetheless, the findings reported herein show that the effect of CaM on BdALMT12 activity in vitro translates to an expected response in planta, suggesting the possibility of physiological relevance.
Toward assessing the potential relevance of this CaM-ALMT12 interaction more broadly, a comparison of sequence motifs (35) and predicted amphipathic helical character (49) of the targeted BdALMTA12 CBD region across an array of monocot and dicot species was completed. In the case of close monocot relatives, such as B. distachyon, Z. mays, Oryza sativa, and Sorghum biocolor, the CBD region appears to be conserved (Table 3). However, the predicted absolute values of the helical hydrophobic moment vectors of this region in more distantly related monocots, such as Aegilops tauschii, Ananas comosus, and Asparagus officinalis, are lower by ∼0.3-fold and the amino acid motif reduced to containing only two basic residues (Table 3). Evaluation of a small cross-section of ALMT12 sequences in dicot plants, including Solanum lycopersicum, Spinacia oleracea, Nicotiana tabacum, Glycine max, Arabidopsis thaliana, and Vitis vinifera all showed an ∼0.3–0.5-fold reduced amphipathic character compared with BdALMT12 in this same region, with the latter three species also reduced to only two basic residues (Table 3). Finally, analysis of TaALMT1, a more distantly related ALMT family member, highlighted ∼0.7-fold reduction in amphipathic character and maintains only a single basic residue in the region. Although there are no strict limits assigned to these values for the prediction of functional CBDs, this analysis highlights that there could be variation in the extent of the relevance of this particular CBD region and CaM-associated activation mechanism across ALMTs and across species. However, this analysis does not preclude the possibility of functional CBDs existing at other locations in the ALMT12 sequences in other species. Indeed, our analysis of the Arabidopsis sequence in particular highlighted a second potential CBD region between residues 475 and 492 with predicted absolute values of the helical hydrophobic moment vectors approximately comparable with the BdALMT12 CBD characterized herein, with several putative conserved basic residues within this region, for future characterization
Table 3.
CaM-binding site prediction comparison for the region homologous to B. distachyon amino acids 334–351
Sequences were obtained from the NCBI Database and aligned using Clustal Omega (54). Regions homologous to the predicted BdALMT12 CBD were evaluated using the calmodulation database and Meta-analysis Predictor software to analyze the full protein sequence (35). The higher the positional prediction score numbers, the higher the likelihood for a binding site, with scoring letters being a continuation of the numbers with A = 10, B = 11, C = 12, etc. Amphipathicity of the helices is expressed as the absolute value of the 3D hydrophobic moment of the helix, calculated using the 3D-HM prediction software, and is expressed as Å·kT/e, where kT/e is the electrostatic potential, and Å is the length of the vector (49).
| Species | Annotation | NCBI accession no. | Motif starting residue no. | Putative CBD motif | CaM motif positional prediction scores | Hydrophobic moment vector (Å·kT/e) |
|---|---|---|---|---|---|---|
| B. distachyon | BdALMT12 | XP_003574370 | 334 | TRVAREVAKVLQELAVSI | 55A999CAAA98898946 | 12.479 |
| Z. mays | ZmALMT12 | XP_008648985 | 338 | TRVAREVAKVLQELAASI | 557666755543343335 | 12.512 |
| O. sativa | OsALMT12 | XP_015614622 | 334 | TRVAREVVKVLQELAVSI | 66BAAADBAA98898946 | 12.405 |
| S. bicolor | SbALMT12 | XP_002437417 | 337 | TRVAREVAKVLQELAVSI | 55A999CAAA98898946 | 12.479 |
| A. tauschii | AgtALMT12 | XP_020161007 | 334 | TRVAREVTKVLQELADSI | 557666755543343335 | 10.084 |
| A. comosus | AcALMT12 | OAY77120 | 309 | IRVAGEVSKVLIELARSI | A8CBBBCAAA87453335 | 8.570 |
| A. officinalis | AoALMT12 | XP_020261302 | 325 | TRVATEVSKVLLELGTSI | 559888977787565557 | 8.206 |
| V. vinifera | AvALMT12 | XP_002278594 | 345 | FQVAAEVSKVLKELANCI | B8A999A88854453335 | 7.002 |
| S. oleracea | SoALMT12 | XP_021854304 | 340 | IRLAAEVSKVVRELANSI | B8A999A88854453335 | 7.840 |
| N. tabacum | NtALMT12 | XP_016493112 | 340 | IRLAGEVTKALKELGDSI | 857666755554453335 | 8.815 |
| G. max | GmALMT12 | XP_003516635 | 344 | IRLAAEVSKVLIELSNSI | B8CBBBCAAA87453335 | 6.646 |
| S. lycopersicum | SlALMT12 | XP_004244701 | 339 | IRLSGEVAKALKELGDSI | 968777866665563335 | 8.329 |
| A. thaliana | AtQUAC1 | O49696 | 331 | VRLAGEVCKALTELADSI | 857666755554453335 | 8.872 |
| T. aestivum | AtALMT1 | Q76LB1 | 324 | GEMSLHSSKVLRDLAMAT | 333342222554476666 | 4.941 |
Overall, this work raises novel hypotheses regarding Ca2+ and CaM-based co-activation mechanisms for the malate-sensitive BdALMT12. It remains to be determined how the Ca2+ dose response might modify malate dose responses and whether CaM's interaction with BdALMT12 is strictly Ca2+-dependent or possibly related to other Ca2+-independent pathways. More broadly, whether ALMT12s from other plant species are also sensitive to Ca2+ and/or CaM remain to be tested experimentally.
Experimental procedures
All chemicals were obtained from Sigma unless otherwise stated.
Plant materials and growth conditions
WT (Bd21) plants were grown in chambers under the following conditions: 60% humidity, with a 16-h photoperiod, at 25 °C in the light and 20 °C in the dark.
Plant transformations were carried out essentially as described previously (50). A 400-bp fragment of the BdALMT12 cDNA (nucleotides 919–1293) was subcloned into pDonor221 (Invitrogen) using Gateway BP Clonase II enzyme mix (Invitrogen). The pDonor221–QUAC1 was then amplified with the universal primer M13 and cloned into the pANIC-8D vector (ABRC (50)) with primer F-RNAi and R-RNAi (Table 4) using LR clonase II enzyme mix (Invitrogen). The vector pANIC-8D carrying a fragment of QUAC1 was then transformed into Agrobacterium strain EHA105. Subsequently, immature Brachypodium seeds were collected. After removal of the lemma, seeds were sterilized by soaking in a solution of 10% bleach and 0.1% Triton X-100 for 4 min and washed three times in sterile water. Embryos were then dissected out of immature seeds and placed on callus initiation media (CIM) (4.43 g of Linsmaier and Skoog medium, 30 g of sucrose, 1 ml of 0.6 mg/ml CuSO4, pH 5.8, with KOH, 2 g of phytagel, 0.5 ml of 5 mg/ml 2.4-D stock solution (2000×) and distilled water to 1 liter) and incubated at 28 °C in the dark. Calluses produced from embryos were spread onto Petri plates every 2 weeks until there were enough calluses for transformation. Transformed Agrobacterium was cultured in 20 ml of CIM at 28 °C to 0.6 OD. Then 200 μl of 10% Synperonic PE/F68 (Sigma) and 20 μl of 200 mm acetoryrinone were added to the Agrobacterium suspension, and 6 g of callus pieces were then added to the suspension and incubated for 5 min. The calluses were then poured onto Petri dishes with filter papers and incubated at room temperature in the dark for 3 days, at which time the calluses were transferred to CIM plates containing 150 mg/liter timentin and 10 mg/liter phosphinothricin (so that only transformed calluses are selected) and incubated at 25 °C. Calluses with green shoots were considered T0 plants and transferred to soil and propagated as per the growth conditions described above.
Table 4.
Primer sequences
F is forward, and R is reverse.
| Name | Primer sequence |
|---|---|
| F-P1 | 5′-CACCGCCGACAAGTTTGTACAAAAAAGCAGGCTTAATAGCTAGCGAGGCCACCATGGCTTGCACTCTACATTCC-3′ |
| R-P1 | 5′-GCCGACCACTTTGTACAAGAAAGCTGGGTACCGCGGTTATTATTCAGCTGCAGTAGAAACTGT-3′ |
| F-R338A | 5′-CCATGCACTAGAGTAGCAGCGGAAGTGGCCAAGGTTTGC-3′ |
| R-R338A | 5′-GCAAACCTTGGCCACTTCCGCTGCTACTCTAGTGCATGG-3′ |
| F-DM1 | 5′-CGAAATCCATGCACTGCAGTAGCAGCGGAAGTGGC-3′, |
| R-DM1 | 5′-GCCACTTCCGCTGCTACTGCAGTGCATGGATTTCG-3′ |
| F-TM | 5′-CGAAATCCATGCACTGCAGTAGCAGCGGAAGTGGCCGCGGTTCTACAAGAGCT-3′ |
| R-TM | 5′-AGCTCTTGTAGAACCGCGGCCACTTCCGCTGCTACTGCAGTGCATGGATTTCG-3′ |
| F-DM2 | 5′-CCATGCACTGCAGTAGCACGAGAAGTGGCCGCGGTTCTA-3′ |
| R-DM2 | 5′-TAGAACCGCGGCCACTTCTCGTGCTACTGCAGTGCATGG-3′ |
| F-DM3 | 5′-CGAAATCCATGCACTAGAGTAGCAGCGGAAGTGGCC-3′ |
| R-DM3 | 5′-GGCCACTTCCGCTGCTACTCTAGTGCATGGATTTCG-3′ |
| F-BdUBC18 | 5′-GGAGGCACCTCAGGTCATTT-3′ |
| R-BdUBC18 | 5′-ATAGCGGTCATTGTCTTGCG-3′ |
| F-BdALMT12 | 5′-ACTGTTGCTGCACTTCATGG-3′ |
| R-BdALMT12 | 5′-CATTGCGTTCTGGTGGTTGA-3′ |
| F-myc | 5′-GATATACCGCGGGCCCGGGATCCGCCCCTCTCC-3′ |
| R-myc | 5′-GATATACCGCGGTTATAGGTCCTCTTCGCTGATTAGCTTTTGTTCTTCAGCTGCAGTAGAAACTGTGTGGCTGCC-3′ |
qPCR expression analysis
BdALMT12 gene expression was assessed by qPCR using the RNeasy plant mini kit (Qiagen). The RNA was reverse-transcribed for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen), according to the manufacturer's protocol. The expression of BdALMT12 was determined with real-time PCR. Each cDNA was amplified using PerfeCTa SYBR Green supermix low ROX (QuantaBio) on the Stratagene Mx3000P qPCR system (Agilent Genomics). The Brachypodium UBC18 gene was used as a control to quantify relative expression levels. Primers for UBC18 (F-UBC18 and R-UBC18) and BdALMT12 (F-BdALMT and R-BdALMT) qPCR are listed in Table 4.
Heterologous BdALMT12 expression plasmid construction, cell culture, and transfection
Total RNA was extracted, and cDNA was synthesized from fresh Brachypodium leaves using the RNeasy plant mini kit (Qiagen) and Superscript III reverse transcriptase (Invitrogen) according to the manufacturers' protocol. Full-length BdALMT12 coding region gene amplification was performed using High Fidelity Phusion polymerase (New England Biolabs). The primers for amplification (P1F and P1R) were designed for Gateway (Invitrogen) cloning or traditional cloning with restriction sites NheI and SacII (Table 4). The amplified PCR products were cloned into pDonor221 using Gateway BP Clonase II enzyme mix according to the manufacturer's protocol. The pDonor221 vector carrying BdALMT12 and pIRES2-eGFP vector (Clontech) was digested with NheI and SacII enzymes (New England Biolabs). The BdALMT12 cDNA was then cloned into pIRES2-eGFP vectors using T4 DNA ligase (New England Biolabs) and transformed the construct into DH5α Escherichia coli cells. The pIRES-eGFP construct allows expression of both the channel and eGFP from the same promoter but unattached in protein construction. This allows for selection of channel-expressing cells during patch-clamp expression via florescence. To detect and determine the expression of BdALMT12 in HEK293 cells, a Myc tag was fused to the C terminus of QUAC1 in the pIRES-eGFP-QUAC1 plasmid. The plasmid was PCR-amplified with primers F-myc and R-myc (Table 4) containing the Myc tag sequence. The PCR products were then treated with DpnI (New England Biolabs), to digest all parental plasmids, and transformed into DH5α E. coli cells. HEK293 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine at 37 °C in 95% air, 5% CO2. Cells were transfected using FuGENE HD transfection reagent (Promega), according to the manufacturer's protocol. Transfected cells were used in patch-clamp experiments the following day.
Electrophysiology
Whole-cell patch clamp was performed with HEKA EPC 10 amplifier, and fluorescing cells were chosen for recording. After capacitance compensation, membrane voltage was clamped from +60 mV to −195 mV with 15-mV decrements. The holding potential was −20 mV. Voltage pulse protocol is showed in Fig. 2I. The standard external solution contained (in mm) 150 NaCl, 1 CaCl2, 1 MgCl2, 10 glucose, 10 mannitol, 10 Hepes, pH 7.3, adjusted with N-methyl-d-glucamine. The external solution with malate containing (in mm) 120 NaCl, 1 CaCl2, 1 MgCl2, 10 glucose, 10 mannitol, 10 Hepes, 30 malic acid, pH 7.3, was added by perfusion. The pipette solution contained (in mm) 130 CsCl, 10 EGTA, 1 MgCl2, 10 Hepes, pH 7.3, and various free Ca2+ concentrations; free Ca2+ concentration was calculated with WEBMAXC software. 60 μm staurosporine (a kinase inhibitor) or 1, 5, or 10 μm W-7 (a CaM inhibitor (Cayman Chem)) was added to the pipette solution in some experiments.
Peak current density of each cell was used to determine Δ current density and conductance. The Δ current density was calculated by subtracting current density recorded before adding malate to the current density recorded after adding malate to the bath solution. Conductance was calculated by division of peak current (after addition of malate) to the driving force and fit with a Boltzmann equation. For clarity, the driving force was the difference of clamped voltages and the chloride equilibrium potential. Chloride equilibrium potential was calculated based on the concentrations of chloride in the pipette and bath solutions (as described), with the following equation: VEq = RT/(zF) ln(Cbath/Cpipette) (where R is the universal gas constant; T was set to 296.15 K, which is equivalent to 23 °C; z is the valence of chloride; F is Faraday's constant; Cbath is the concentration of chloride in the bath solution, and Cpipette is the concentration of chloride in the pipette solution). The fit was a standard four-parameter Boltzmann equation with free running parameters. Specifically, the equation (f = y0 + a/(1 + exp(−(x − x0)/b)) was used: a = max(y) − min(y); b = xwtr(x,y − min(y), 0.5)/4; x0 = x50(x,y − min(y), 0.5); y0 = min(y)). For clarity, although there is no explicit variable of V1/2, one can calculate V1/2 from the ×50 value, which is the potential at which conductance is halfway between max(y) and min(y). The ×50 value was calculated using Sigma Plot from the given sets of data. V1/2 is the value when x = ×50. z is the valance of the ion moving across, which can be back-calculated by the slope of the Boltzmann equation. Slope = RT/(zF); therefore, z = RT/(F·slope) (R is the universal gas constant, T is the temperature in Kelvin, and F is the Faraday's constant.
Only Δ current densities below −100 mV were used in statistical analyses. Overall, Δ current densities were analyzed by two-way repeated measure ANOVA, where voltage points were the repeated factor and either the concentrations of Ca2+, the presence of inhibitors, or the type of QUAC1 (WT, variants) expressed were the second factor. The significance of overall Δ current densities were presented with alphabet letters. Matching notations indicate no significant difference. Conductance was statistically analyzed and presented in the same way as Δ current density.
Time constant (τ) was calculated with the following equation: τ = Cm·(Rss − Ro), where τ (ms) is the time constant; Cm (pF) is the membrane capacitance; Rss (mV/pA) is the resistance at steady state, and Ro is the resistance at peak.
Stomatal aperture measurements
Stomatal assays were performed according to previously reported methods (51, 52). Detached leaves from 4- to 8-week-old plants, or at others times as indicated in the results, were soaked in opening buffer (50 mm KCl, 10 mm Mes, pH 6.1) for 2 h. Leaves were soaked for another 3 h in control buffer (10 mm Mes, 25 μm CaCl2, pH 6.1) or control buffer with addition of either 150 mm malic acid, pH 6.1, 20 μm of a Ca2+ ionophore A23187, or 5, 10, and 20 μm W-7, or two of the three or all three. Leaves were removed from the buffer and immediately dried by gentle blotting with kim-wipe and then painted with a layer of clear nail polish, a process taking less than 30 s. The nail polish peel was used for microscopy using a Zeiss Axiovert 135 microscope, and the obtained images were visualized using ImageJ (53), and ensuring a blind analysis by relabeling of the image files was by a third party. At least 100 stomata were measured, arising from four leaves obtained from four plants for each condition tested. Significance of differences was determined by one-way ANOVA with Fisher's LSD post hoc.
BdALMT12 site-directed mutagenesis
The three basic residues Arg-335, Arg-338, and Lys-342 were changed to alanine using the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol. pIRES-eGFP vector carrying BdALMT12 was used as template with primers F-R338A and R-R338A (Table 4). The plasmid-encoding variant R338A was then used as a template to make the double variant R335A/R338A and triple variant R335A/R338A/K342A with pairs of primers F-DM1 with R-DM1 and F-TM with R-TM, respectively (Table 4). The plasmid encoding the triple variant was used as template to make the double variants R335A/K342A and R338A/K342A with the two pairs of primers F-DM2 with R-DM2 and F-DM3 with R-DM3, respectively (Table 4). Variants were confirmed by sequencing and expressed in HEK293 cells for patch-clamp experiments. Myc tag was also fused to the C terminus of variants, in the same manner as WT, for determining the expression in HEK293 cells by Western blotting. The variant plasmids were PCR with primers F-myc and R-myc (Table 4) containing the Myc sequence. The PCR products were then treated with DpnI (New England Biolabs) and transformed into DH5α E. coli cells.
BdALMT12 protein extraction and Western blotting
WT and variant BdALMT12 fused with Myc tag were expressed in HEK293 cells. All proteins present in the cell membranes of HEK293 cells were extracted with biotinylation using the Pierce cell-surface protein isolation kit (ThermoFisher Scientific), according to the manufacturer's protocol. The extracts were fractionated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes using a transfer apparatus (Bio-Rad). The membranes were blocked with Rapidblock solution 10× (Amresco), according to the manufacturer's protocol. After blocking, the membranes were incubated with horseradish peroxidase–conjugated anti-Myc (1:5000 dilution) and anti-Na,K-ATPase (1:5000 dilution) antibodies for 90 min at room temperature. Blots were washed with TBST buffer (50 mm Tris-Cl, 150 mm NaCl, and 0.1% Tween) three times for 5 min each time, incubated in chemiluminescent substrate (Amersham Biosciences) for 5 min, and developed with the ChemiDoc Imaging System (Bio-Rad). Western bands were quantified with ImageLab software (Bio-Rad).
Isothermal titration calorimetry
The ITC experiments were performed on a Nano-ITC (TA-instrument). BdALMT12 peptides (AA334–AA355) representing the WT CaM-binding domain helix, as well as double variants R335D/R338D, R335D/K342D, R338D/K342D, and triple variants R335D/R338D/K342D with >90% purity were synthesized by Genscript. The peptides and bovine CaM (BioOcean) were dialyzed with 0.1–0.5-kDa MWCO tubing overnight in buffer containing 20 mm Hepes, 100 mm KCl, and 5 mm CaCl2 at pH 7.4. The peptide concentrations were adjusted to 400 μm and titrated into the Nano-ITC cell containing either 60 μm CaM, 60 μm CaM with 60 μm W-7, or just dialysis buffer. Because W-7 inhibits by competing with the peptides and binds to CaM, W-7 was mixed with CaM 30 min prior to the ITC experiment to ensure CaM was fully inhibited. The “no Ca2+” sample was prepared exactly as described above, except that peptide and CaM were dialyzed overnight with three buffer changes into buffer containing 20 mm Hepes, 100 mm KCl, and 1.5 mm EGTA at pH 7.4. The data were processed using the NanoAnalyze software by subtracting the background heat obtained from titrating the peptides into dialysis buffer to the heats obtained from titrating the peptides into the CaM-containing solutions.
CaM-agarose affinity pulldown
WT BdALMT12 and variant 335/342 fused with Myc tag were expressed in HEK293 cells. Cells were rinsed with PBS, pH 7.4 (ThermoFisher Scientific), and gently scraped off the bottom of the flask into PBS. Cell pellets were collected by centrifuging at 500 × g for 3 min and then resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EGTA, 1% Triton X-100, 10% glycerol, and 1× Halt protease inhibitor mixture (ThermoFisher Scientific)) and incubated at 4 °C for 30 min. Total cell lysate was clarified by centrifugation at 10,000 × g for 2 min. Ca2+ was added to the cell lysate to make a final concentration of 8 mm Ca2+. CaM-Sepharose beads (Biovision) were transferred to mini spin columns (VWR) and equilibrated with wash buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.25% Triton X-100, 10% glycerol, 1× Halt protease inhibitor mixture) by centrifuging at 1000 × g for 1 min and repeated two times. The cell lysate was added to the CaM beads and incubated for 8 h at 4 °C. All unbound proteins were removed by centrifuging at 1000 × g for 1 min, and the columns were rinsed four times with wash buffer at 1000 × g for 1 min each time. SDS-PAGE sample buffer containing 20 mm EGTA (62.5 mm Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 20 mm EGTA) was then added to the columns and incubated for 2 h at room temperature. Bound proteins were eluted by centrifuging at 1000 × g for 2 min and loaded onto SDS-polyacrylamide gel for QUAC1 detection by Western blotting as described above. Western bands were quantified by normalizing against total (eluted) protein stain with ImageLab software (Bio-Rad).
Author contributions
K. L. data curation; K. L., N. R., M. C. L., and M. E. L. formal analysis; K. L. and J. C. C. validation; K. L. and N. R. investigation; K. L., N. R., J. C. C., and M. E. L. methodology; K. L., M. C. L., and M. E. L. writing-review and editing; N. R., J. C. C., M. C. L., and M. E. L. supervision; N. R., M. C. L., and M. E. L. visualization; M. C. L. and M. E. L. conceptualization; M. C. L. and M. E. L. funding acquisition; M. C. L. and M. E. L. writing-original draft; M. C. L. project administration.
Supplementary Material
Acknowledgments
We are grateful for training and support in the production of knockdown plants from Joe Hammerlindl at the National Research Council of Canada in Saskatoon, Saskatchewan, Canada. The ITC and dynamic light scattering experiments described here were performed at the Protein Characterization and Crystallization Facility (College of Medicine, University of Saskatchewan, Saskatoon, Saskatchewan, Canada), with help for initial instrument setup from Dr. Michal T. Boniecki.
This work was supported by Natural Sciences and Engineering Discovery Grants 261683-2012 (to M. C. L.) and 371364-2010 (to M. E. L.) and by a grant from the National Research Council of Canada, Wheat Flagship Program to M. C. L. This manuscript is NRCC no. 53630. The authors declare that they have no conflicts of interest with the contents of this article.
This articles contains Figs. S1–S3.
- ABA
- abscisic acid
- ITC
- isothermal titration calorimetry
- SnRK
- sucrose nonfermenting–related kinase
- ROS
- reactive oxygen species
- ALMT
- aluminum-activated malate transporter
- CIM
- callus initiation medium
- qPCR
- quantitative PCR
- pF
- picofarad
- ANOVA
- analysis of variance
- CaM
- calmodulin
- CAP
- CaM-agarose affinity pulldown
- eGFP
- enhanced GFP.
References
- 1. Ward J. M., Mäser P., and Schroeder J. I. (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 71, 59–82 10.1146/annurev.physiol.010908.163204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hedrich R. (2012) Ion channels in plants. Physiol. Rev. 92, 1777–1811 10.1152/physrev.00038.2011 [DOI] [PubMed] [Google Scholar]
- 3. Marten H., Konrad K. R., Dietrich P., Roelfsema M. R., and Hedrich R. (2007) Ca2+-dependent and -independent abscisic acid activation of plasma membrane anion channels in guard cells of Nicotiana tabacum 1. Plant Physiol. 143, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee S. C., Lan W., Buchanan B. B., and Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. 106, 21419–21424 10.1073/pnas.0910601106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreyer I., Gomez-Porras J. L., Riaño-Pachón D. M., Hedrich R., and Geiger D. (2012) Molecular evolution of slow and quick anion channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 3, 263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imes D., Mumm P., Böhm J., Al-Rasheid K. A., Marten I., Geiger D., and Hedrich R. (2013) Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382 10.1111/tpj.12133 [DOI] [PubMed] [Google Scholar]
- 7. Kwak J. M., Mori I. C., Pei Z. M., Leonhardt N., Torres M. A., Dangl J. L., Bloom R. E., Bodde S., Jones J. D., and Schroeder J. I. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633 10.1093/emboj/cdg277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mori I. C., Murata Y., Yang Y., Munemasa S., Wang Y. F., Andreoli S., Tiriac H., Alonso J. M., Harper J. F., Ecker J. R., Kwak J. M., and Schroeder J. I. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLOS Biol. 4, e327 10.1371/journal.pbio.0040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brandt B., Brodsky D. E., Xue S., Negi J., Iba K., Kangasjärvi J., Ghassemian M., Stephan A. B., Hu H., and Schroeder J. I. (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598 10.1073/pnas.1116590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scherzer S., Maierhofer T., Al-Rasheid K. A., Geiger D., and Hedrich R. (2012) Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol. Plant. 5, 1409–1412 10.1093/mp/sss084 [DOI] [PubMed] [Google Scholar]
- 11. Schroeder J. I., and Keller B. U. (1992) Two types of anion channel currents in guard cells with distinct voltage regulation. Proc. Natl. Acad. Sci. U.S.A. 89, 5025–5029 10.1073/pnas.89.11.5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Linder B., and Raschke K. (1992) A slow anion channel in guard cells, activating at large hyperpolarization, may be principal for stomatal closing. FEBS Lett. 313, 27–30 10.1016/0014-5793(92)81176-M [DOI] [PubMed] [Google Scholar]
- 13. Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., and Iba K. (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486 10.1038/nature06720 [DOI] [PubMed] [Google Scholar]
- 14. Vahisalu T., Kollist H., Wang Y. F., Nishimura N., Chan W. Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J. I., and Kangasjärvi J. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491 10.1038/nature06608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K. A., Romeis T., and Hedrich R. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430 10.1073/pnas.0912021106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maierhofer T., Diekmann M., Offenborn J. N., Lind C., Bauer H., Hashimoto K., S Al-Rasheid K. A., Luan S., Kudla J., Geiger D., and Hedrich R. (2014) Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 7, ra86 10.1126/scisignal.2005703 [DOI] [PubMed] [Google Scholar]
- 17. Geiger D., Maierhofer T., Al-Rasheid K. A., Scherzer S., Mumm P., Liese A., Ache P., Wellmann C., Marten I., Grill E., Romeis T., and Hedrich R. (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4, ra32 [DOI] [PubMed] [Google Scholar]
- 18. Meyer S., Mumm P., Imes D., Endler A., Weder B., Al-Rasheid K. A., Geiger D., Marten I., Martinoia E., and Hedrich R. (2010) AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63, 1054–1062 10.1111/j.1365-313X.2010.04302.x [DOI] [PubMed] [Google Scholar]
- 19. Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S. J., Ryan P. R., Delhaize E., and Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653 10.1111/j.1365-313X.2003.01991.x [DOI] [PubMed] [Google Scholar]
- 20. Palmer A. J., Baker A., and Muench S. P. (2016) The varied functions of aluminium-activated malate transporters–much more than aluminium resistance. Biochem. Soc. Trans. 44, 856–862 10.1042/BST20160027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medeiros D. B., Martins S. C., Cavalcanti J. H., Daloso D. M., Martinoia E., Nunes-Nesi A., DaMatta F. M., Fernie A. R., and Araújo W. L. (2016) Enhanced photosynthesis and growth in atquac1 knockout variants are due to altered organic acid accumulation and an increase in both stomatal and mesophyll conductance. Plant Physiol. 170, 86–101 10.1104/pp.15.01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki T., Mori I. C., Furuichi T., Munemasa S., Toyooka K., Matsuoka K., Murata Y., and Yamamoto Y. (2010) Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 51, 354–365 10.1093/pcp/pcq016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schachtman D. P., Schroeder J. I., Lucas W. J., Anderson J. A., and Gaber R. F. (1992) Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258, 1654–1658 10.1126/science.8966547 [DOI] [PubMed] [Google Scholar]
- 24. Cao Y., Ward J. M., Kelly W. B., Ichida A. M., Gaber R. F., Anderson J. A., Uozumi N., Schroeder J. I., and Crawford N. M. (1995) Multiple genes, tissue-specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis thaliana. Plant Physiol. 109, 1093–1106 10.1104/pp.109.3.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leng Q., Mercier R. W., Yao W., and Berkowitz G. A. (1999) Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121, 753–761 10.1104/pp.121.3.753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szabò I., Negro A., Downey P. M., Zoratti M., Lo Schiavo F., and Giacometti G. M. (2000) Temperature-dependent functional expression of a plant K+ channel in mammalian cells. Biochem. Biophys. Res. Commun. 274, 130–135 10.1006/bbrc.2000.3095 [DOI] [PubMed] [Google Scholar]
- 27. Leng Q., Mercier R. W., Hua B.-G., Fromm H., and Berkowitz G. A. (2002) Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128, 400–410 10.1104/pp.010832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pilot G., Lacombe B., Gaymard F., Cherel I., Boucherez J., Thibaud J. B., and Sentenac H. (2001) Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem. 276, 3215–3221 10.1074/jbc.M007303200 [DOI] [PubMed] [Google Scholar]
- 29. Ivashikina N., Deeken R., Fischer S., Ache P., and Hedrich R. (2005) AKT2/3 subunits render guard cell K+ channels Ca2+ sensitive. J. Gen. Physiol. 125, 483–492 10.1085/jgp.200409211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ligaba A., Kochian L., and Piñeros M. (2009) Phosphorylation at S384 regulates the activity of the TaALMT1 malate transporter that underlies aluminum resistance in wheat. Plant J. 60, 411–423 10.1111/j.1365-313X.2009.03964.x [DOI] [PubMed] [Google Scholar]
- 31. Mumm P., Imes D., Martinoia E., Al-Rasheid K. A., Geiger D., Marten I., and Hedrich R. (2013) C terminus-mediated voltage gating of Arabidopsis guard cell anion channel QUAC1. Mol. Plant. 6, 1550–1563 10.1093/mp/sst008 [DOI] [PubMed] [Google Scholar]
- 32. Karaman M. W., Herrgard S., Treiber D. K., Gallant P., Atteridge C. E., Campbell B. T., Chan K. W., Ciceri P., Davis M. I., Edeen P. T., Faraoni R., Floyd M., Hunt J. P., Lockhart D. J., Milanov Z. V., et al. (2008) A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 10.1038/nbt1358 [DOI] [PubMed] [Google Scholar]
- 33. Hidaka H., and Tanaka T. (1983) Naphthalene sulfonamides as calmodulin antagonists. Methods Enzymol. 102, 185–194 10.1016/S0076-6879(83)02019-4 [DOI] [PubMed] [Google Scholar]
- 34. Minhas Fu, and Ben-Hur A. (2012) Multiple instance learning of calmodulin binding sites. Bioinformatics 28, i416–i422 10.1093/bioinformatics/bts416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mruk K., Farley B. M., Ritacco A. W., and Kobertz W. R. (2014) Calmodulation meta-analysis: predicting calmodulin binding via canonical motif clustering. J. Gen. Physiol. 144, 105–114 10.1085/jgp.201311140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cole C., Barber J. D., and Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 10.1093/nar/gkn238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edlund M., Blikstad I., and Obrink B. (1996) Calmodulin binds to specific sequences in the cytoplasmic domain of C-CAM and down-regulates C-CAM self-association. J. Biol. Chem. 271, 1393–1399 10.1074/jbc.271.3.1393 [DOI] [PubMed] [Google Scholar]
- 38. Ben-Johny M., and Yue D. T. (2014) Calmodulin regulation (calmodulation) of voltage-gated calcium channels. J. Gen. Physiol. 143, 679–692 10.1085/jgp.201311153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Donovan N., Martin S., and Donkin M. E. (1985) Calmodulin binding drugs trifluoperazine and compound 48/80 modify stomatal responses of Commelina communis L. J. Plant Physiol. 118, 177–187 10.1016/S0176-1617(85)80146-2 [DOI] [Google Scholar]
- 40. Cousson A., Cotelle V., and Vavasseur A. (1995) Induction of stomatal closure by vanadate or a light/dark transition involves Ca2+-calmodulin–dependent protein phosphorylations. Plant Physiol. 109, 491–497 10.1104/pp.109.2.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimazaki K., Kinoshita T., and Nishimura M. (1992) Involvement of calmodulin and calmodulin-dependent myosin light chain kinase in blue light-dependent H pumping by guard cell protoplasts from Vicia faba L. Plant Physiol. 99, 1416–1421 10.1104/pp.99.4.1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Silva D. L. R., Cox R. C., Hetherington A. M., and Mansfield T. A. (1985) Suggested involvement of calcium and calmodulin in the responses of stomata to abscisic acid. New Phytol. 101, 555–563 10.1111/j.1469-8137.1985.tb02861.x [DOI] [Google Scholar]
- 43. Erdahl W. L., Chapman C. J., Taylor R. W., and Pfeiffer D. R. (1994) Ca2+ transport properties of lonophores A23187, lonomycin, and 4-BrA23187 in a well defined model system. Biophys. J. 66, 1678–1693 10.1016/S0006-3495(94)80959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peterson F. C., Burgie E. S., Park S. Y., Jensen D. R., Weiner J. J., Bingman C. A., Chang C. E., Cutler S. R., Phillips G. N. Jr., and Volkman B. F. (2010) Structural basis for selective activation of ABA receptors. Nat. Struct. Mol. Biol. 17, 1109–1113 10.1038/nsmb.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weiner S. A., Geffre A. G., and Toth A. L. (2018) Functional genomics in the wild: a case study with paper wasps shows challenges and prospects for RNA-interference in ecological systems. Genome 61, 266–272 [DOI] [PubMed] [Google Scholar]
- 46. Reichow S. L., Clemens D. M., Freites J. A., Németh-Cahalan K. L., Heyden M., Tobias D. J., Hall J. E., and Gonen T. (2013) Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat. Struct. Mol. Biol. 20, 1085–1092 10.1038/nsmb.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alaimo A., Nuñez E., Aivar P., Fernández-Orth J., Gomis-Perez C., Bernardo-Seisdedos G., Malo C., and Villarroel A. (2017) Calmodulin confers calcium sensitivity to the stability of the distal intracellular assembly domain of Kv7.2 channels. Sci. Rep. 7, 13425 10.1038/s41598-017-13811-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou Y. P., Duan J., Fujibe T., Yamamoto K. T., and Tian C. E. (2012) AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in Arabidopsis. Plant Mol. Biol. 79, 333–346 10.1007/s11103-012-9915-0 [DOI] [PubMed] [Google Scholar]
- 49. Reisser S., Strandberg E., Steinbrecher T., and Ulrich A. S. (2014) 3D hydrophobic moment vectors as a tool to characterize the surface polarity of amphiphilic peptides. Biophys. J. 106, 2385–2394 10.1016/j.bpj.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mann D. G., Lafayette P. R., Abercrombie L. L., King Z. R., Mazarei M., Halter M. C., Poovaiah C. R., Baxter H., Shen H., Dixon R. A., Parrott W. A., and Neal Stewart C. Jr. (2012) Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236 10.1111/j.1467-7652.2011.00658.x [DOI] [PubMed] [Google Scholar]
- 51. Hilu K. W., and Randall J. L. (1984) Convenient method for studying grass leaf epidermis. Taxon 33, 413–415 10.2307/1220980 [DOI] [Google Scholar]
- 52. Berger D., and Altmann T. (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- 53. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nicholas K. B., Nicholas H. B., and Deerfield D. W. (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News 4, 14 [Google Scholar]
- 56. Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 57. Zuckerkandl E., and Pauling L. (1965) in Evolving Genes and Proteins (Bryson V., and Vogel H. J., eds) pp. 97–166, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 58. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.