Abstract
T cell activation is tightly regulated by both stimulatory and inhibitory co-receptors and has been a focus in the development of interventions for managing cancer or autoimmune diseases. Targeting the inhibitory co-receptors programmed cell death 1 (PD-1) and cytotoxic T lymphocyte–associated protein 4 (CTLA-4) has successfully eradicated tumors but induced immune-related adverse events in humans and mice. The beneficial and adverse effects of targeting these co-receptors highlight their importance in cancer immunity and also autoimmunity. Although the therapeutic potencies of other inhibitory co-receptors are under extensive investigation, their inhibitory mechanisms and their functional differences are not well understood. Here we analyzed the inhibitory mechanisms of lymphocyte activation gene-3 (LAG-3), another inhibitory co-receptor, by using an in vitro T cell activation system and a high-affinity anti-LAG-3 antibody that strongly interferes with the binding of LAG-3 to its ligand. We found that the expression level of LAG-3 strongly correlates with the inhibitory function of LAG-3, suggesting that LAG-3 functions as a rheostat rather than as a breaker of T cell activation. By evaluating the inhibitory capacities of various LAG-3 variants relative to their expression levels, we found that LAG-3 transduces two independent inhibitory signals through an FXXL motif in the membrane-proximal region and the C-terminal EX repeat. These motifs have not been reported previously for inhibitory co-receptors, suggesting that LAG-3 inhibits T cell activation through a nonredundant inhibitory mechanisms along with the other inhibitory co-receptors. Our findings provide a rationale for combinatorial targeting of LAG-3 and the other inhibitory co-receptors to improve cancer immunotherapy.
Keywords: immunology, T cell, T cell receptor (TCR), inhibition mechanism, immunotherapy, monoclonal antibody, cytokine induction, co-receptor, immune checkpoint, LAG-3
Introduction
T cell activation is tightly regulated by stimulatory and inhibitory co-receptors that modulate the T cell receptor (TCR)3 signal (1–3). Because of the recent success of tumor immunotherapy targeting inhibitory co-receptors, cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and programmed cell death 1 (PD-1), the therapeutic potencies of other inhibitory co-receptors are investigated extensively (4). However, their inhibitory mechanisms as well as their functional differences or the molecular coordination among inhibitory and stimulatory co-receptors are still poorly understood.
Lymphocyte activation gene-3 (LAG-3), an inhibitory co-receptor with structural similarities to CD4, has been shown to negatively regulate autoimmunity, cancer immunity, and infectious immunity by itself or in collaboration with other inhibitory co-receptors, including PD-1 (5–10). LAG-3 deficiency exacerbates type I diabetes in non-obese diabetic mice (5, 8). Mice deficient in both LAG-3 and PD-1 develop lethal autoimmune myocarditis, and simultaneous blockade of PD-1 and LAG-3 efficiently eradicates tumors and clears pathogens in mice (7, 8, 10). Therefore, LAG-3 is a potent therapeutic target of immunotherapies for cancer as well as other diseases (11).
LAG-3 has four Ig-like domains, called domain 1 (D1) to D4, in its extracellular (EC) region. LAG-3 has been reported to associate with major histocompatibility complex class II (MHCII), using its D1 with higher affinity than CD4, and inhibit T cell activation by interfering with the engagement of CD4 by MHCII (12, 13). However, LAG-3 can inhibit T cell activation independently of CD4, and the molecular mechanism of inhibition by LAG-3 has been enigmatic (8). We have recently found that LAG-3 does not bind to MHCII universally but binds selectively to a stable complex of peptides and MHCII (pMHCII) and preferentially inhibits the activation of T cells reactive to stable pMHCII (14). We have also demonstrated that LAG-3 does not interfere with CD4–pMHCII and TCR–pMHCII interactions. Instead, the inhibitory function of LAG-3 requires its intracellular (IC) region, suggesting that LAG-3 transduces the inhibitory signal via its IC region (14).
The IC region of LAG-3, consisting of about 60 amino acid residues, lacks a typical signaling motif with a known signaling mechanism such as the immunoreceptor tyrosine–based inhibitory motif and immunoreceptor tyrosine–based switch motif. However, amino acid sequences from different species show substantial similarity. Workman et al. (15) reported that the KIEELE sequence in the middle of the IC region is conserved among species and is required for LAG-3 to inhibit antigen-dependent activation of 3A9 hybridoma T cells. They also demonstrated that the amino acid substitution of Lys in the KIEELE sequence to Ala was enough to abrogate the inhibitory capacity of LAG-3 (15). However, the other regions have not been extensively analyzed, and how this Lys transduces the inhibitory signal is currently unknown. Iouzalen et al. (16) identified LAG-3–associated protein (LAP) as a molecule that binds to the C-terminal region of LAG-3 with 10 to 15 repeats of Glu and favoring but not limited to Pro (EX repeat). Currently, the function of LAP as well as the EX repeat is unknown. Thus, the molecular basis of the inhibitory signal by LAG-3 remains elusive.
In this study, we used an in vitro T cell activation system and high-affinity anti-LAG-3 antibody (Ab) that strongly interfered with the interaction of LAG-3 and pMHCII to analyze the inhibitory function of LAG-3. We demonstrated that the expression level of LAG-3 strongly correlated with the inhibitory function of LAG-3. Intriguingly, deletion of the KIEELE sequence did not abrogate the inhibitory function of LAG-3. Instead, we found that LAG-3 transduces two independent inhibitory signals through an FXXL motif in the membrane-proximal region (PR) and the C-terminal EX repeat. To date, these motifs have not been reported for the other inhibitory co-receptors, suggesting that LAG-3 inhibits T cell activation using nonredundant inhibitory mechanisms with the other inhibitory co-receptors.
Results
Evaluation of anti-mouse LAG-3 monoclonal Abs
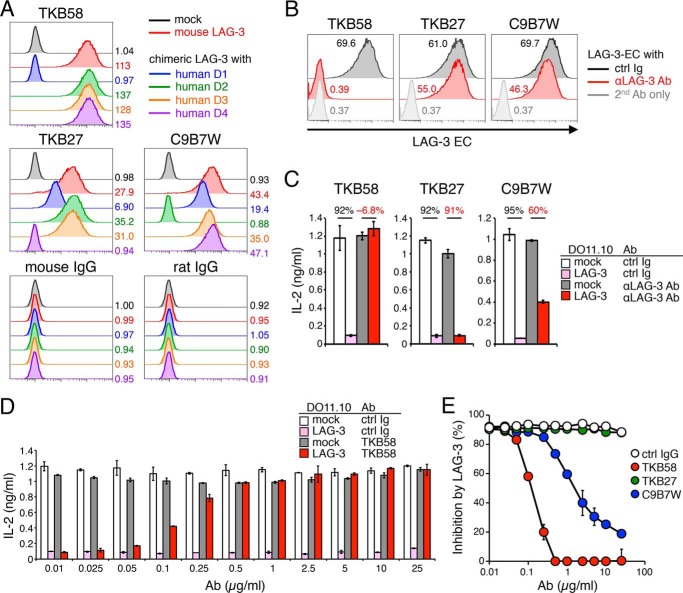
Monoclonal Abs are useful and essential for analyzing the physical and functional properties of molecules. Although the C9B7W clone of the anti-mouse LAG-3 Ab is widely used as a blocking Ab of mouse LAG-3, C9B7W has been reported not to interfere with the binding of LAG-3 to MHCII but postulated to induce conformational changes of LAG-3 to attenuate its function (13). We have recently developed the TKB58 clone of the anti-moue LAG-3 Ab that can strongly block binding of LAG-3 to pMHCII (14). We first characterized TKB58 and C9B7W as well as the TKB27 clone of the anti-mouse LAG-3 Ab, which we developed by immunizing LAG-3–deficient mice with the recombinant protein of the mouse LAG-3 EC region. We generated chimeric molecules of mouse and human LAG-3 in which one of the four Ig-like domains of mouse LAG-3 is swapped with the corresponding Ig-like domain of human LAG-3 and tested their reactivity to TKB58, TKB27, and C9B7W. In accordance with the former report, C9B7W recognized D2 of mouse LAG-3 because it failed to recognize the chimeric molecule with human D2 (Fig. 1A). TBK58 and TBK27 recognized D1 and D4 of mouse LAG-3, respectively (Fig. 1A). Next we tested the capacity of TKB58, TKB27, and C9B7W to block the binding of soluble LAG-3 protein (LAG-3–EC) to stable pMHCII on IIA1.6 B lymphoma cells (14). As shown in Fig. 1B, TKB58 but not TKB27 blocked LAG-3–EC binding to IIA1.6 cells. On the other hand, C9B7W slightly reduced the staining intensity of LAG-3–EC in IIA1.6 cells, suggesting that the conformational change induced by C9B7W might result in weak attenuation of the interaction between LAG-3 and pMHCII.
Figure 1.
Characterization of anti-mouse LAG-3 Abs. A, recognition of mouse LAG-3 D1, D4, and D2 by TKB58, TKB27, and C9B7W clones of anti-mouse LAG-3 Abs. DO11.10 T cells expressing chimeric LAG-3 with the indicated human-derived Ig-like domain were stained with the indicated Abs. Mean fluorescence intensity (MFI) is indicated. B, capacity of anti-mouse LAG-3 Abs to block binding of LAG-3–EC to stable pMHCII on IIA1.6 cells. IIA1.6 cells were stained with LAG-3–EC (5 μg/ml) in the presence or absence of TKB58, TKB27, or C9B7W (5 μg/ml), and bound LAG-3–EC was detected with the secondary Ab (anti-DYKDDDDK tag Ab). MFI is indicated. ctrl, control. C, capacity of anti-mouse LAG-3 Abs to block LAG-3 function. DO11.10 T cells with or without LAG-3 were stimulated with pOVA-pulsed (1 μm) IIA1.6 cells in the presence or absence of TKB58, TKB27, and C9B7W (1 μg/ml). The concentration of IL-2 in the culture supernatant was determined by ELISA. Numbers denote the percentage of LAG-3–dependent inhibition of IL-2 production in the presence of the indicated Ab. D, titration of the capacity of TKB58 to block LAG-3 function. DO11.10 T cells with or without LAG-3 were stimulated as in C in the presence of the indicated amount of TKB58 or its isotype control. The concentration of IL-2 in the culture supernatant was determined by ELISA. E, comparison of the capacity of anti-mouse LAG-3 Abs to block LAG-3 function. The percentage of LAG-3–dependent inhibition of IL-2 production in the presence of the indicated amount of each Ab is shown. Data are the mean ± S.D. of technical duplicates in one experiment. Data are representative of more than three independent experiments.
To examine the capacities of TKB58, TKB27, and C9B7W to block the inhibitory function of LAG-3, we used DO11.10 hybridoma T cells, which recognize the 323–339 segment of chicken ovalbumin (pOVA323–339) in the context of I-Ad. As we have reported before, LAG-3 efficiently inhibited the activation and the secretion of IL-2 from DO11.10 T cells upon co-culture with pOVA323–339–pulsed IIA1.6 cells (Fig. 1C) (8). TKB58 completely blocked the inhibitory effects of LAG-3 at as low as 0.5 μg/ml, whereas TKB27 did not block the inhibitory effects of LAG-3 even at 25 μg/ml (Fig. 1, D and E). In agreement with the former report, we could observe the blocking effect by C9B7W, albeit to a lesser extent than TKB58 (13). However, C9B7W did not completely block the inhibitory effect of LAG-3, even at 25 μg/ml, probably because C9B7W attenuates LAG-3 function by inducing the conformational change of LAG-3 but not by masking the binding surface of LAG-3 to pMHCII (Fig. 1E).
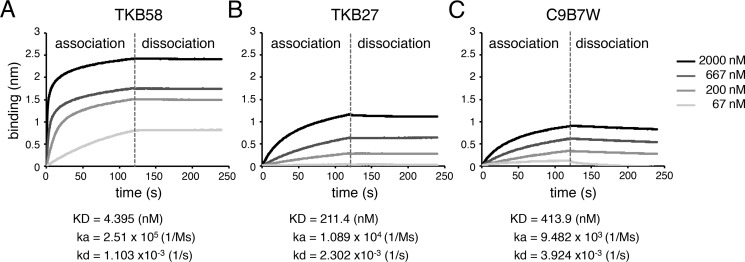
Then we measured binding affinities of TKB58, TKB27, and C9B7W to LAG-3 using biolayer interferometry (Fig. 2). The association rate of TKB58 was 26 and 23 times faster than that of C9B7W and TKB27, whereas the dissociation rate of TKB58 was 2.1 and 3.6 times slower than that of TKB27 and C9B7W. Accordingly, the KD value of TKB58 was 48 and 94 times smaller than that of TKB27 and C9B7W, indicating that TKB58 binds to mouse LAG-3 much more strongly than TKB27 and C9B7W (Fig. 2).
Figure 2.
Binding affinities of TKB58, TKB27, and C9B7W to mouse LAG-3. A–C, binding affinities of anti-mouse LAG-3 Abs to mouse LAG-3 protein. LAG-3–EC was immobilized on the biosensor chip, and the association of TKB58 (A), TKB27 (B), and C9B7W (C) at the indicated concentration was monitored by biolayer interferometry. Chips were washed with PBS to analyze the dissociation kinetics; ka, kd, and kD are shown. Data are representative of more than two independent experiments.
Correlation of the cell surface amount of LAG-3 and its inhibitory effect
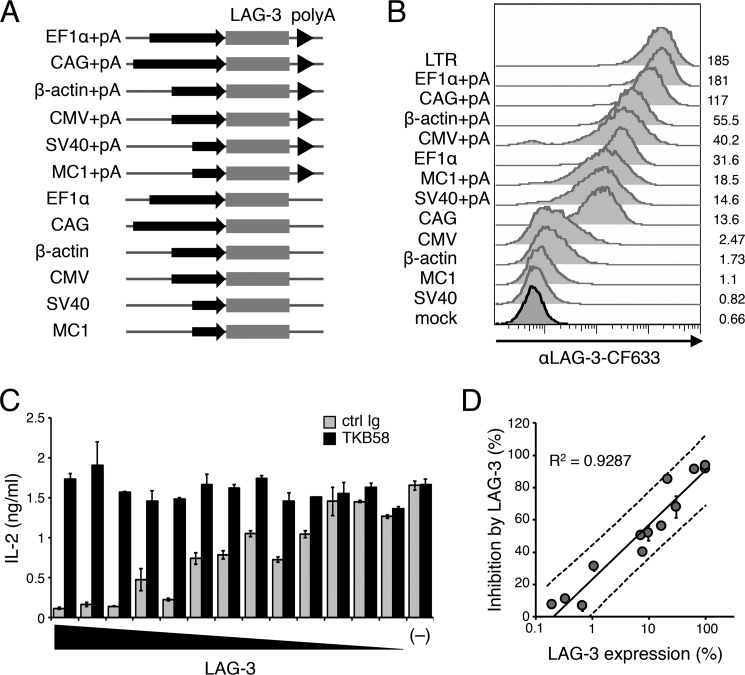
LAG-3 is not expressed on naive T cells but inducibly expressed on T cells upon activation (17). Although the stronger TCR signal induces higher LAG-3 expression, the correlation of the cell surface amount of LAG-3 and its inhibitory effect has not been elucidated. Therefore, we overexpressed LAG-3 on DO11.10 T cells to various levels using seven different promoters (pLTR, pEF1α, pCAG, pβ-actin, pCMV, pSV40, and pMC1) with or without a polyadenylation (poly(A)) signal, which generally prolongs the longevity of transcripts (Fig. 3, A and B). The amount of cell surface LAG-3 was calculated relative to that of DO11.10 T cells expressing LAG-3 under pLTR. LAG-3–dependent inhibitory effects were calculated by comparing the amount of IL-2 in the presence or absence of an anti-LAG-3–blocking Ab (TKB58). As shown in Fig. 3, C and D, the inhibitory effect of LAG-3 showed a strong positive correlation with the cell surface amount of LAG-3 (R2 = 0.9287). Intriguingly, the inhibitory effect of LAG-3 was not saturated, even when LAG-3 was expressed with the strongest promoter tested (pLTR). These results indicate that LAG-3 functions as a rheostat rather than as a breaker of T cell activation.
Figure 3.
Correlation between the amount of LAG-3 and its inhibitory effect. A, schematic of retroviral expression vectors. Mouse LAG-3 cDNA was overexpressed in DO11.10 T cells using the indicated promoters with or without a poly(A) signal. B, DO11.10 T cells with various expression levels of LAG-3. The expression levels of LAG-3 were evaluated by flow cytometry using an anti-LAG-3 Ab (TKB58). MFI is indicated. C, LAG-3–dependent inhibition of IL-2 production in DO11.10 T cells with various LAG-3 expression levels. DO11.10 T cells overexpressing LAG-3 under the indicated promoter with or without a poly(A) signal were stimulated with pOVA-pulsed (1 μm) IIA1.6 cells in the presence of anti-LAG-3–blocking Ab (TKB58) or its isotype control (ctrl Ig). The concentration of IL-2 in the culture supernatant was determined by ELISA. D, the correlation between the percentage of LAG-3–dependent inhibition and the expression level of LAG-3. The amount of LAG-3 was calculated relative to that of DO11.10 T cells expressing LAG-3 under the LTR promoter. The solid line represents the regression line, and dashed lines represent 95% prediction intervals. The correlation coefficient is shown. Data are the mean ± S.D. of technical duplicates in one experiment. Data are representative of more than three independent experiments.
Requirement of the PR for the inhibitory function of LAG-3
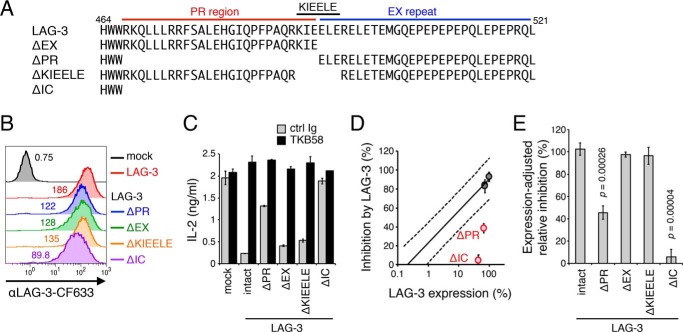
Although the KIEELE sequence in the middle of the IC region of LAG-3 has been reported to be essential for LAG-3 function (15), we could not reproduce the result in our experimental system using DO11.10 T cells (Fig. 4, A–C). Therefore, we examined the requirements of the PR (amino acids 467–492) and the EX repeat (amino acids 493–521) of LAG-3 for its inhibitory function. We generated deletion mutants of LAG-3 and introduced them into DO11.10 T cells (Fig. 4, A and B). Because deletion mutations affected the cell surface expression level of LAG-3 to variable degrees, and we observed a strong correlation between the cell surface amount of LAG-3 and its inhibitory effect (Fig. 3D), we evaluated the inhibitory capacity of each LAG-3 mutant by comparing its expression level and inhibitory effect with the reference values obtained from the panel of DO11.10 T cells expressing LAG-3 to variable degrees (Fig. 3D). In accordance with the former observation, LAG-3ΔIC failed to inhibit IL-2 secretion from DO11.10 T cells upon antigen stimulation, suggesting that LAG-3 inhibits T cell activation by transducing an inhibitory signal via its IC region (Fig. 4, D and E) (14, 15). Intriguingly, LAG-3ΔPR, but not LAG-3ΔEX or LAG-3ΔKIEELE, showed a substantially reduced inhibitory capacity, indicating that the PR, but not the KIEELE sequence, is required for the inhibitory function of LAG-3 (Fig. 4, D and E).
Figure 4.
Requirement of the PR region for the inhibitory function of LAG-3. A, amino acid sequences of the IC region of LAG-3 and its deletion mutants. The PR, KIEELE sequence, and EX repeat are indicated by red, black, and blue lines, respectively. Numbers denote the position of the amino acid from the translation initiation site. B, expression levels of LAG-3 mutants in DO11.10 T cells. DO11.10 T cells expressing LAG-3 and its indicated mutants were stained with TKB58 and analyzed by flow cytometry. C–E, defective inhibition of IL-2 production from DO11.10 T cells by LAG-3 lacking its PR and IC region. DO11.10 T cells expressing LAG-3 and its mutants were stimulated with pOVA-pulsed (1 μm) IIA1.6 cells in the presence of anti-LAG-3–blocking Ab (TKB58) or its isotype control (ctrl Ig). C, the concentration of IL-2 in the culture supernatant was determined by ELISA. The relative expression level and the inhibitory effect of LAG-3 mutants are plotted in the standard plot shown in Fig. 3D. D, LAG-3 mutants with weaker function are highlighted in red. E, the inhibitory effect relative to intact LAG-3 with the same expression level is shown for the indicated LAG-3 mutants. Data are the mean ± S.D. of technical duplicates in one representative experiment from three independent experiments (C) or the mean ± S.D. of three independent experiments (D and E). p values comparing the expression-adjusted relative inhibition of the indicated LAG-3 mutants with that of intact LAG-3 are shown (Student's t test, E).
Requirement of the FXXL motif in the PR for the inhibitory function of LAG-3
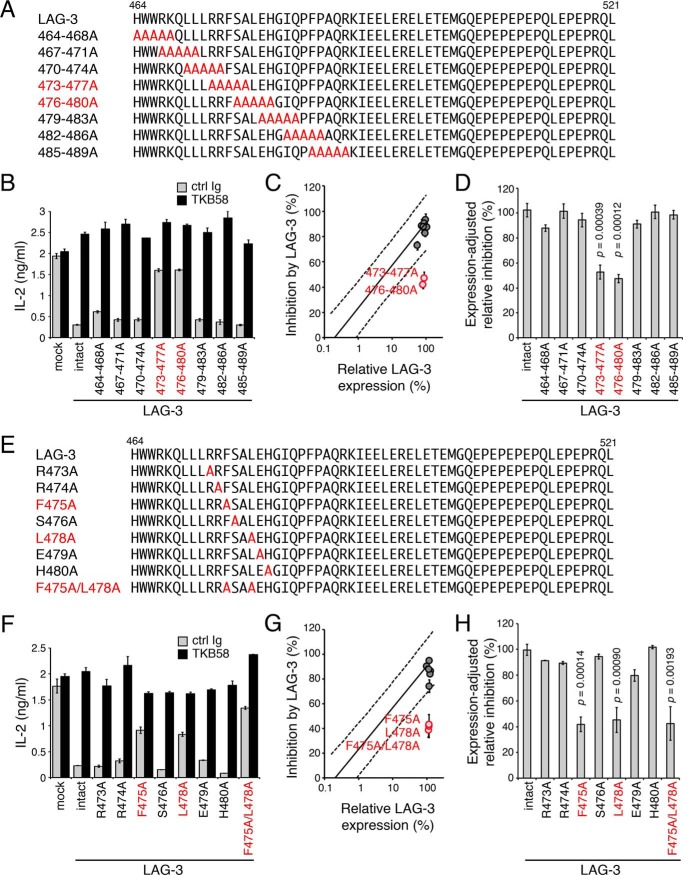
To further characterize the PR, a series of mutants with alanine substitutions in five consecutive amino acid residues in the PR were generated and tested for their expression levels on DO11.10 T cells and inhibitory function (Fig. 5A). LAG-3 mutants with 473–477A and 476–480A substitutions showed reduced inhibitory capacity to a similar level as LAG-3ΔPR, indicating that the responsible amino acid residues are located between Arg-473 to His-480 (Fig. 5, B–D). Then we tested single alanine substitution mutants and found that LAG-3–F475A and LAG-3–L478A showed reduced inhibitory capacity to a similar level as LAG-3ΔPR, suggesting that Phe-475 and Leu-478 play pivotal roles in the PR-dependent inhibitory function of LAG-3 (Fig. 5, E–H). LAG-3 with both F475A and L478A mutations also showed reduced inhibitory capacities to a similar level as LAG-3ΔPR, LAG-3–473–477A, and LAG-3–476–480A (Fig. 5, F–H). Therefore, Phe-475 and Leu-478 likely function in the same inhibitory process, and we named this motif the FXXL motif.
Figure 5.
Requirement of the FXXL motif in the PR for the inhibitory function of LAG-3. A–D, defective inhibition of IL-2 production by LAG-3 with alanine substitutions at 473–477 and 476–480. A, amino acid sequences of the IC regions of LAG-3 and its mutants are shown. Substituted alanine residues and mutants with weaker function are colored in red. B, DO11.10 T cells expressing LAG-3 and its mutants were stimulated with pOVA-pulsed (1 μm) IIA1.6 cells in the presence of anti-LAG-3–blocking Ab (TKB58) or its isotype control (ctrl Ig). The concentration of IL-2 in the culture supernatant was determined by ELISA. C, the relative expression level and the inhibitory effect of LAG-3 mutants are plotted on the standard plot shown in Fig. 3D. LAG-3 mutants with weaker function are highlighted in red. D, the inhibitory effect relative to intact LAG-3 with the same expression level is shown for the indicated LAG-3 mutants. E–H, defective inhibition of IL-2 production by LAG-3 with alanine substitutions at 475 and 480. Amino acid sequences (E) and inhibitory function (F–H) of the indicated LAG-3 mutants are shown as in A–D. Data are the mean ± S.D. of technical duplicates in one representative experiment from three independent experiments (B and F) or the mean ± S.D. of three independent experiments (C, D, G, and H). p values comparing the expression-adjusted relative inhibition of indicated LAG-3 mutants with that of intact LAG-3 are shown (Student's t test, D and H).
Weak EX repeat–dependent inhibition of T cell activation by LAG-3
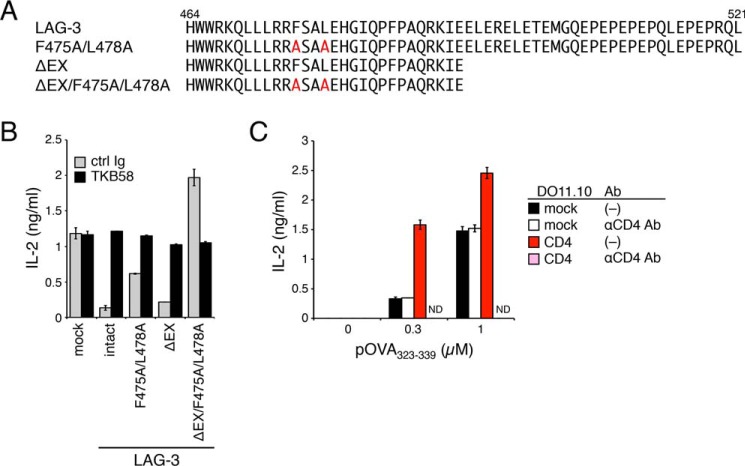
Although the inhibitory capacity of LAG-3ΔPR and F475A/L478A was substantially attenuated, they still weakly inhibited IL-2 production from DO11.10 T cells upon activation. Because LAG-3ΔIC completely lacked the inhibitory function, this residual inhibitory effect is likely mediated by the EX repeat. As expected, by deleting the EX repeat of LAG-3–F475A/L478A, the residual inhibitory effect was completely abrogated (Fig. 6, A and B). Therefore, LAG-3 likely inhibits T cell activation by using two distinct mechanisms that are dependent on the FXXL motif and the EX repeat.
Figure 6.
EX repeat–dependent inhibition of T cell activation by LAG-3. A and B, EX repeat–dependent partial inhibitory effect of the LAG-3 mutant lacking the FXXL motif. A, amino acid sequences of the IC region of LAG-3 and its mutants. B, DO11.10 cells with LAG-3 and its mutants were stimulated with pOVA-pulsed (1 μm) IIA1.6 cells in the presence of anti-LAG-3–blocking Ab (TKB58) or its isotype control (ctrl Ig). The concentration of IL-2 in the culture supernatant was determined by ELISA. C, different manners of co-stimulation by CD4 and the LAG-3 mutant lacking both the FXXL motif and EX repeat. DO11.10 T cells overexpressing mouse CD4 were stimulated with IIA1.6 cells pulsed with the indicated amount of pOVA in the presence or absence of anti-CD4–blocking Ab (GK1.5). The concentration of IL-2 in the culture supernatant was determined by ELISA. Data are the mean ± S.D. of technical duplicates in one experiment. Data are representative of more than three independent experiments. ND, not detected.
The LAG-3 mutant that was defective in both FXXL motif– and EX repeat–dependent inhibitory mechanisms (LAG-3ΔEX/F475A/L478A) rather increased IL-2 production, indicating that LAG-3ΔEX/F475A/L478A functioned as a co-stimulator (Fig. 6, A and B). The co-stimulatory effect depended on the interaction of LAG-3ΔEX/F475A/L478A with pMHCII because the anti-LAG-3–blocking Ab (TKB58) completely canceled the additive effect. As mentioned above, LAG-3 has a structural similarity to CD4 in the EC but not IC region. CD4 is known to associate with Lck, which is essential for initiation of the TCR signal. In agreement with previous reports (18, 19), overexpression of CD4 not only augmented the production of IL-2 upon activation but also rendered DO11.10 T cells dependent on CD4 for activation, as the anti-CD4–blocking Ab (GK1.5) completely abrogated IL-2 production rather than merely canceling the additive effect. These results suggest that the PR of LAG-3 may associate with a signaling molecule that additively augments the strength of the TCR signal (Fig. 6C).
Discussion
In this study, by using an in vitro T cell activation system and high-affinity anti-LAG-3 Ab that interferes with the interaction of LAG-3 and stable pMHCII, we analyzed the inhibitory function of LAG-3 against T cell activation. First we demonstrated that the cell surface amount of LAG-3 strongly correlated with its inhibitory function, which indicates that LAG-3 functions as a rheostat rather than as a breaker of T cell activation. LAG-3 is not expressed on naive T cells but inducibly expressed on T cells upon activation (17). The protein kinase C signaling pathway has been reported to regulate the expression level of LAG-3 by controlling its trafficking from the lysosomal compartment to the cell surface (20). The cell surface expression level of LAG-3 is also regulated by shedding of the EC region by two transmembrane metalloproteases, ADAM10 and ADAM17 (21). These findings clearly indicate that the inhibitory effect of LAG-3 was not saturated even when LAG-3 was expressed with the strongest promoter tested. In addition, changes in the amount of LAG-3 on the cell surface directly affected the inhibitory effect of LAG-3, indicating that regulation of its cell surface amount is effective for the regulation of T cell activation.
The expression level of LAG-3 has been reported to differ largely depending on the cell type. CD4+ T cells with exhaustion-like phenotypes have been reported to express a high amount of LAG-3, whereas a small subset of intestinal CD4+ T cells with regulatory function have been reported to weakly express LAG-3 (7, 22–25). The impact of LAG-3 on cell-autonomous function may thus differ among different types of cells. In addition, a stronger TCR signal induces higher LAG-3 expression, suggesting that the induction of LAG-3 likely serves as negative feedback to prevent excess activation (8).
By dissecting the IC region of LAG-3, we found that LAG-3 inhibits T cell activation by using two distinct mechanisms that are dependent on the FXXL motif in the PR and the EX repeat at the C terminus. Because LAG-3ΔEX showed an inhibitory capacity comparable with intact LAG-3, FXXL motif–dependent inhibition predominates over EX repeat–dependent inhibition, at least regarding inhibition of IL-2 production by DO11.10 T cells upon activation. On the other hand, EX repeat–dependent inhibition functions complementarily in the absence of FXXL motif–dependent inhibition. Intriguingly, the LAG-3 mutant that is defective in both FXXL motif– and EX repeat–dependent inhibitory mechanisms provided co-stimulation, suggesting that the PR of LAG-3 may contain a co-stimulatory motif(s) in addition to co-inhibitory motifs. Further analyses are required to reveal the physiological meaning of the co-stimulatory potential of LAG-3.
Because the FXXL motif resembles the YXXL sequence in the immunoreceptor tyrosine–based inhibitory motif, phosphotyrosine-independent signaling adaptor proteins may be involved in the inhibitory signal via the FXXL motif in LAG-3. For example, μ-adaptin, which is involved in endocytosis of membrane molecules, has been reported to recognize the FXXL motif in β-arrestin2 (26). The C-terminal tail of platelet-derived growth factor β receptor is also rich in Glu and Pro. Intriguingly, this C-terminal tail has been reported to inhibit its tyrosine kinase activity autonomously (27). Although LAG-3 itself does not have tyrosine kinase activity, the EX repeat of LAG-3 may also inhibit another tyrosine kinase that is activated by antigen stimulation. Currently, how the FXXL motif and EX repeat regulate TCR-proximal and downstream molecules is unknown. Further analyses are expected to delineate the precise molecular mechanisms of how these motifs independently or cooperatively regulate signaling pathways in T cell activation.
LAG-3 inhibits the proliferation of T cells and production of cytokines, such as IL-2 and IFNγ, from T cells (8, 28, 29). In addition, LAG-3 has been reported to play critical roles in regulatory T cells (24). In this study, we clearly demonstrate that LAG-3 inhibits IL-2 production from T cells upon antigen stimulation by using the FXXL motif and EX repeat. The requirement of both or either of these motifs in LAG-3–dependent regulation of other T cell functions remains to be verified.
We have reported recently that LAG-3 does not bind to MHCII universally but selectively binds to stable pMHCII and preferentially inhibits the activation of T cells reactive to stable pMHCII (14). Identification of the FXXL motif and EX repeat in this study further supports the idea that LAG-3 accumulates at the immunological synapse and transduces an inhibitory signal via its IC region to attenuate the TCR-proximal signal.
As mentioned above, we could not reproduce the former observation by Vignali et al. (15) that deletion of the KIEELE sequence abrogated the inhibitory function of LAG-3. Although the reason for this discrepancy is currently unknown, it should be noted that the PR was not analyzed in the study, and thus the relative effects of the PR and the KIEELE sequence have not been compared (15). Based on our observation, deletion of the PR might have shown greater effect compared with the deletion of KIEELE sequence under the former experimental condition as well. In addition, the expression levels of LAG-3 mutants substantially differed among different mutants. Because we observed a strong positive correlation between the cell surface amount and the inhibitory effect of LAG-3, we need to take into consideration the expression level of mutants. Otherwise, we may under- or overestimate the inhibitory capacity of mutants. By comparing the expression level and the inhibitory effect of mutants with the reference values obtained from the panel of DO11.10 T cells expressing LAG-3 to variable degrees, we could obtain unambiguous results.
While we were preparing this manuscript, Chen and co-workers (30) reported that LAG-3 inhibits T cell activation by associating with fibrinogen-like protein 1 (FGL1). Although the exact contribution of FGL1 and stable pMHCII to the inhibitory function of LAG-3 needs future verification, complete blockade of LAG-3 function by TKB58, which interferes with the binding between D1 of LAG-3 and pMHCII, strongly suggests that stable pMHCII is primarily involved in inhibition by LAG-3 (14).
Because of the success of tumor immunotherapy targeting CTLA-4 and PD-1, the therapeutic potencies of other inhibitory co-receptors are being extensively explored. However, their actual inhibitory mechanisms, functional differences, and collaboration are not well understood. Here we demonstrated that LAG-3 transduced two independent inhibitory signals through the FXXL motif in the PR and the C-terminal EX repeat. These motifs have not been reported previously for inhibitory co-receptors, suggesting that LAG-3 inhibits T cell activation using nonredundant inhibitory mechanisms with the other inhibitory co-receptors. Thus, combinatorial therapy of LAG-3 with the other inhibitory co-receptors is expected to provide synergistic effects.
Experimental procedures
Cell culture
DO11.10 T and IIA1.6 cells, which were kindly provided by Tasuku Honjo (Kyoto University) and Tomohiro Kurosaki (Osaka University), respectively, were cultured in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) FBS (Biowest), 0.5 mm monothioglycerol (Wako), 2 mm l-alanyl-l-glutamine dipeptide (Gibco), 100 units/ml penicillin (Nacalai Tesque), and 100 μg/ml streptomycin (Nacalai Tesque). Plat-E cells, which were kindly provided by Toshio Kitamura (University of Tokyo), were maintained in DMEM (Gibco) supplemented with 10% (v/v) FBS, 100 units/ml penicillin (Nacalai Tesque), and 100 μg/ml streptomycin (Nacalai Tesque).
Plasmids and retroviral gene transduction
Fragments of cDNA were amplified by PCR and cloned into retroviral expression plasmid vectors modified from pFB-ires-Neo (Agilent), with which cDNAs are transcribed under pLTR. Mutant cDNAs with deletions and amino acid substitutions and chimeric cDNAs of mouse and human LAG-3 were generated by overlap extension PCR. To control the expression levels, fragments of cDNA were cloned into retroviral expression plasmid vectors modified from pSUPER.retro.puro (OligoEngine), the promoter region of which was replaced with promoters of human elongation factor 1α (EF-1α), cytomegalovirus enhancer/chicken β-actin (CAG), β-actin, pCMV (cytomegalovirus), SV40 (simian virus 40), and MC1 (polyoma virus enhancer/herpes simplex virus thymidine kinase) coupled with or without a poly(A) signal. Plasmids were transfected using FuGENE HD (Promega) into Plat-E cells cultured in high-glucose DMEM (Gibco) supplemented with 20% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin, and supernatants containing viruses were used to transduce genes into target cells. Infected cells were selected with G418 (Wako) or puromycin (Sigma-Aldrich) or by cell sorting.
Soluble protein
The fragment of cDNA encoding D1 to D4 of mouse LAG-3 (LAG-3–EC) was amplified by PCR. The five-stranded coiled-coil domain of cartilage oligomeric matrix protein (COMP) (31) with a DYKDDDDK tag, tobacco etch virus protease cleavage site, and a PA tag (GVAMPGAEDDVV) was added to the C terminus of LAG-3–EC for the pentamer. The Fc region of human IgG1 (hIgG1Fc) was added to the C terminus of LAG-3–EC for the dimer. A strep-tag II (WSHPQFEK) was added to the C terminus of LAG-3–EC for the monomer. Chimeric cDNAs were cloned into expression vectors modified from pEBMulti-Neo (Wako). Plasmids were transfected into Plat-E cells using Avalanche-Omni transfection reagent (EZ Biosystems), and the culture supernatants were collected after 48 h. Pentameric LAG-3–EC was purified using anti-PA tag Ab beads (Wako), followed by cleavage by TurboTEV Protease (Accelagen). Dimeric LAG-3–EC was purified using protein G (GE Healthcare).
Mice
BALB/cCrSlc-Lag3−/− (LAG-3aida/aida) mice (8) were housed under specific pathogen-free conditions in environmentally controlled clean rooms. All mouse protocols were approved by the Animal Experimentation Committee of Tokushima University. All experimental procedures complied with institutional regulations in accordance with the Act on Welfare and Management of Animals and related guidelines in Japan.
Generation of anti-mouse LAG-3 monoclonal Abs
BALB/cCrSlc-Lag3−/− mice were immunized with dimeric mouse LAG-3–EC, and lymph node cells of immunized mice were fused with SP2/o cells using the envelope of hemagglutinating virus of Japan (GenomONE-CF, Ishihara Sangyo Kaisha). Culture supernatants of hybridoma clones were tested for their reactivity to DO11.10 T cells overexpressing mouse LAG-3 by flow cytometry.
Antibody and flow cytometry analysis
Cells were stained with the indicated Abs. Data were obtained with Gallios (Beckman Coulter) and analyzed using FlowJo (Tree Star). Anti-mouse LAG-3 Ab (C9B7W) and its isotype control (rat IgG1, eBRG1) were purchased from Thermo Fisher Scientific. The anti-DYKDDDDK tag (L5) Ab and isotype control for TKB58 (mouse IgG1, MOPC-21) were purchased from BioLegend. The FITC-labeled F(ab')2 fragment of the goat anti-mouse IgG (H+L) Ab was purchased from Jackson ImmunoResearch Laboratories. The FITC-labeled goat anti-rat IgG (H+L) Ab was purchased from Southern Biotech. TKB58 and TKB27 were purified from culture supernatants of hybridomas using protein G–Sepharose. Purified TKB58 was labeled with CF®633-Dye (Biotium).
Biolayer interferometry
Monomeric mouse LAG-3–EC with a streptavidin tag was immobilized on streptavidin-coated biosensor chips (Pall ForteBio), and the association of anti-mouse LAG-3 Abs at different concentration was monitored using BLItz (Pall ForteBio). Chips were washed with PBS to analyze the dissociation kinetics. The association rate constant (ka), dissociation rate constant (kd), and dissociation constant (kD) were calculated using BLItz Pro software.
Stimulation of DO11.10 T cells
DO11.10 T cells infected with or without retroviral vectors containing the indicated cDNAs (5 × 104 cells/well) were co-cultured with IIA1.6 cells (1 × 104 cells/well) with cognate OVA323–339 peptide (ISQAVHAAHAEINEAGR, >95% purity, Sigma-Aldrich Japan or eurofins Genomics) for 24 h. The concentration of IL-2 in the culture supernatant was quantified by ELISA (BioLegend). To block mouse LAG-3–mediated inhibition, anti-mouse LAG3 Ab or isotype matched control IgG was added throughout the co-culture assay. The percentage of LAG-3–dependent inhibition of IL-2 production was calculated as the ratio of IL-2 concentration in the presence or absence of anti-LAG-3–blocking Ab (unless specified, TKB58). To block CD4, anti-mouse CD4 Ab (5 μg/ml, GK1.5, Thermo Fisher Scientific) was added throughout the co-culture assay. The estimated inhibitory effect by intact LAG-3 at the same expression level of each LAG-3 mutant was deduced from the regression line obtained from the panel of DO11.10 T cells expressing LAG-3 to variable degrees. The expression-adjusted inhibitory function of each LAG-3 mutant was calculated by dividing the raw percentage of inhibition by each LAG-3 mutant by the estimated percentage of inhibition by intact LAG-3 at the same expression level.
Statistical analysis
Two-tailed unpaired Student's t test was used to evaluate statistical significance.
Author contributions
T. K. M., D. S., and T. O. conceptualization; T. K. M., D. S., I.-m. O., T. M., and T. O. resources; T. K. M., D. S., I.-m. O., T. M., and T. O. data curation; T. K. M., D. S., I.-m. O., T. M., and T. O. formal analysis; T. K. M., D. S., I.-m. O., T. M., and T. O. investigation; D. S., I.-m. O., and T. O. validation; D. S. and T. O. visualization; D. S., I.-m. O., T. M., and T. O. methodology; D. S. writing-original draft; D. S., I.-m. O., and T. O. writing-review and editing; I.-m. O. and T. O. project administration; T. O. supervision; T. O. funding acquisition.
Acknowledgments
We thank Drs. T. Honjo, T. Kurosaki, and T. Kitamura for kindly providing DO11.10 T, IIA1.6, and Plat-E cells, respectively; H. Tsuduki, A. Otsuka, Y. Okamoto, M. Aoki, and R. Matsumura for technical and secretarial assistance; and the other members of our laboratory for helpful discussions.
This work was supported in part by the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency, Basic Science and Platform Technology Program for Innovative Biological Medicine of the Japan Agency for Medical Research and Development JP18am0301007 (to T. O.), and Grant-in-Aid by the Japan Society for the Promotion of Science JP18H05417 (to T. O.). The authors declare that they have no conflicts of interest with the contents of this article.
- TCR
- T cell receptor
- EC
- extracellular
- D
- domain
- MHCII
- major histocompatibility complex class II
- IC
- intracellular
- Ab
- antibody
- PR
- membrane-proximal region
- cDNA
- complementary DNA
- MFI
- mean fluorescence intensity.
References
- 1. Esensten J. H., Helou Y. A., Chopra G., Weiss A., and Bluestone J. A. (2016) CD28 Costimulation: from mechanism to therapy. Immunity 44, 973–988 10.1016/j.immuni.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Okazaki T., Chikuma S., Iwai Y., Fagarasan S., and Honjo T. (2013) A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 14, 1212–1218 10.1038/ni.2762 [DOI] [PubMed] [Google Scholar]
- 3. Schildberg F. A., Klein S. R., Freeman G. J., and Sharpe A. H. (2016) Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44, 955–972 10.1016/j.immuni.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Topalian S. L., Taube J. M., Anders R. A., and Pardoll D. M. (2016) Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bettini M., Szymczak-Workman A. L., Forbes K., Castellaw A. H., Selby M., Pan X., Drake C. G., Korman A. J., and Vignali D. A. (2011) Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J. Immunol. 187, 3493–3498 10.4049/jimmunol.1100714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackburn S. D., Shin H., Haining W. N., Zou T., Workman C. J., Polley A., Betts M. R., Freeman G. J., Vignali D. A., and Wherry E. J. (2009) Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10, 29–37 10.1038/ni.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butler N. S., Moebius J., Pewe L. L., Traore B., Doumbo O. K., Tygrett L. T., Waldschmidt T. J., Crompton P. D., and Harty J. T. (2011) Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage Plasmodium infection. Nat. Immunol. 13, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okazaki T., Okazaki I. M., Wang J., Sugiura D., Nakaki F., Yoshida T., Kato Y., Fagarasan S., Muramatsu M., Eto T., Hioki K., and Honjo T. (2011) PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J. Exp. Med. 208, 395–407 10.1084/jem.20100466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Triebel F., Jitsukawa S., Baixeras E., Roman-Roman S., Genevee C., Viegas-Pequignot E., and Hercend T. (1990) LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 171, 1393–1405 10.1084/jem.171.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woo S. R., Turnis M. E., Goldberg M. V., Bankoti J., Selby M., Nirschl C. J., Bettini M. L., Gravano D. M., Vogel P., Liu C. L., Tangsombatvisit S., Grosso J. F., Netto G., Smeltzer M. P., Chaux A., et al. (2012) Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 10.1158/0008-5472.CAN-11-1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrews L. P., Marciscano A. E., Drake C. G., and Vignali D. A. (2017) LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 276, 80–96 10.1111/imr.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huard B., Prigent P., Tournier M., Bruniquel D., and Triebel F. (1995) CD4/major histocompatibility complex class II interaction analyzed with CD4- and lymphocyte activation gene-3 (LAG-3)-Ig fusion proteins. Eur. J. Immunol. 25, 2718–2721 10.1002/eji.1830250949 [DOI] [PubMed] [Google Scholar]
- 13. Workman C. J., Rice D. S., Dugger K. J., Kurschner C., and Vignali D. A. (2002) Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur. J. Immunol. 32, 2255–2263 [DOI] [PubMed] [Google Scholar]
- 14. Maruhashi T., Okazaki I. M., Sugiura D., Takahashi S., Maeda T. K., Shimizu K., and Okazaki T. (2018) LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat. Immunol. 19, 1415–1426 10.1038/s41590-018-0217-9 [DOI] [PubMed] [Google Scholar]
- 15. Workman C. J., Dugger K. J., and Vignali D. A. (2002) Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J. Immunol. 169, 5392–5395 10.4049/jimmunol.169.10.5392 [DOI] [PubMed] [Google Scholar]
- 16. Iouzalen N., Andreae S., Hannier S., and Triebel F. (2001) LAP, a lymphocyte activation gene-3 (LAG-3)-associated protein that binds to a repeated EP motif in the intracellular region of LAG-3, may participate in the down-regulation of the CD3/TCR activation pathway. Eur. J. Immunol. 31, 2885–2891 [DOI] [PubMed] [Google Scholar]
- 17. Baixeras E., Huard B., Miossec C., Jitsukawa S., Martin M., Hercend T., Auffray C., Triebel F., and Piatier-Tonneau D. (1992) Characterization of the lymphocyte activation gene 3-encoded protein: a new ligand for human leukocyte antigen class II antigens. J. Exp. Med. 176, 327–337 10.1084/jem.176.2.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Laethem F., Sarafova S. D., Park J. H., Tai X., Pobezinsky L., Guinter T. I., Adoro S., Adams A., Sharrow S. O., Feigenbaum L., and Singer A. (2007) Deletion of CD4 and CD8 coreceptors permits generation of αβT cells that recognize antigens independently of the MHC. Immunity 27, 735–750 10.1016/j.immuni.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 19. Van Laethem F., Tikhonova A. N., Pobezinsky L. A., Tai X., Kimura M. Y., Le Saout C., Guinter T. I., Adams A., Sharrow S. O., Bernhardt G., Feigenbaum L., and Singer A. (2013) Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 154, 1326–1341 10.1016/j.cell.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bae J., Lee S. J., Park C. G., Lee Y. S., and Chun T. (2014) Trafficking of LAG-3 to the surface on activated T cells via its cytoplasmic domain and protein kinase C signaling. J. Immunol. 193, 3101–3112 10.4049/jimmunol.1401025 [DOI] [PubMed] [Google Scholar]
- 21. Li N., Wang Y., Forbes K., Vignali K. M., Heale B. S., Saftig P., Hartmann D., Black R. A., Rossi J. J., Blobel C. P., Dempsey P. J., Workman C. J., and Vignali D. A. (2007) Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 26, 494–504 10.1038/sj.emboj.7601520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gagliani N., Magnani C. F., Huber S., Gianolini M. E., Pala M., Licona-Limon P., Guo B., Herbert D. R., Bulfone A., Trentini F., Di Serio C., Bacchetta R., Andreani M., Brockmann L., Gregori S., et al. (2013) Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 19, 739–746 10.1038/nm.3179 [DOI] [PubMed] [Google Scholar]
- 23. Okamura T., Fujio K., Shibuya M., Sumitomo S., Shoda H., Sakaguchi S., and Yamamoto K. (2009) CD4+CD25-LAG3+ regulatory T cells controlled by the transcription factor Egr-2. Proc. Natl. Acad. Sci. U.S.A. 106, 13974–13979 10.1073/pnas.0906872106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C. T., Workman C. J., Flies D., Pan X., Marson A. L., Zhou G., Hipkiss E. L., Ravi S., Kowalski J., Levitsky H. I., Powell J. D., Pardoll D. M., Drake C. G., and Vignali D. A. (2004) Role of LAG-3 in regulatory T cells. Immunity 21, 503–513 10.1016/j.immuni.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 25. Camisaschi C., Casati C., Rini F., Perego M., De Filippo A., Triebel F., Parmiani G., Belli F., Rivoltini L., and Castelli C. (2010) LAG-3 expression defines a subset of CD4+CD25(high)Foxp3+ regulatory T cells that are expanded at tumor sites. J. Immunol. 184, 6545–6551 10.4049/jimmunol.0903879 [DOI] [PubMed] [Google Scholar]
- 26. Marion S., Fralish G. B., Laporte S., Caron M. G., and Barak L. S. (2007) N-terminal tyrosine modulation of the endocytic adaptor function of the beta-arrestins. J. Biol. Chem. 282, 18937–18944 10.1074/jbc.M700090200 [DOI] [PubMed] [Google Scholar]
- 27. Chiara F., Bishayee S., Heldin C. H., and Demoulin J. B. (2004) Autoinhibition of the platelet-derived growth factor β-receptor tyrosine kinase by its C-terminal tail. J. Biol. Chem. 279, 19732–19738 10.1074/jbc.M314070200 [DOI] [PubMed] [Google Scholar]
- 28. Workman C. J., and Vignali D. A. (2003) The CD4-related molecule, LAG-3 (CD223), regulates the expansion of activated T cells. Eur. J. Immunol. 33, 970–979 10.1002/eji.200323382 [DOI] [PubMed] [Google Scholar]
- 29. Workman C. J., and Vignali D. A. (2005) Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223). J. Immunol. 174, 688–695 10.4049/jimmunol.174.2.688 [DOI] [PubMed] [Google Scholar]
- 30. Wang J., Sanmamed M. F., Datar I., Su T. T., Ji L., Sun J., Chen L., Chen Y., Zhu G., Yin W., Zheng L., Zhou T., Badri T., Yao S., Zhu S., et al. (2019) Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 176, 334–347.e12 10.1016/j.cell.2018.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terskikh A. V., Le Doussal J. M., Crameri R., Fisch I., Mach J. P., and Kajava A. V. (1997) “Peptabody”: a new type of high avidity binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 1663–1668 10.1073/pnas.94.5.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]