Abstract
Genetic testing for BRCA1, a DNA repair protein, can identify carriers of pathogenic variants associated with a substantially increased risk for breast and ovarian cancers. However, an association with increased risk is unclear for a large fraction of BRCA1 variants present in the human population. Most of these variants of uncertain clinical significance lead to amino acid changes in the BRCA1 protein. Functional assays are valuable tools to assess the potential pathogenicity of these variants. Here, we systematically probed the effects of substitutions in the C terminus of BRCA1: the N- and C-terminal borders of its tandem BRCT domain, the BRCT-[N-C] linker region, and the α1 and α′1 helices in BRCT-[N] and -[C]. Using a validated transcriptional assay based on a fusion of the GAL4 DNA-binding domain to the BRCA1 C terminus (amino acids 1396–1863), we assessed the functional impact of 99 missense variants of BRCA1. We include the data obtained for these 99 missense variants in a joint analysis to generate the likelihood of pathogenicity for 347 missense variants in BRCA1 using VarCall, a Bayesian integrative statistical model. The results from this analysis increase our understanding of BRCA1 regions less tolerant to changes, identify functional borders of structural domains, and predict the likelihood of pathogenicity for 98% of all BRCA1 missense variants in this region recorded in the population. This knowledge will be critical for improving risk assessment and clinical treatment of carriers of BRCA1 variants.
Keywords: BRCA1, cancer prevention, human genetics, protein conformation, breast cancer, BRCT domains, breast cancer risk, clinical annotation, clinical risk assessment, VarCall
Introduction
Carriers of germline variants in BRCA1 that disrupt protein expression, stability, or function are at increased high risk for developing early-onset breast and ovarian cancer (1, 2). The identification of women at risk is now an important aspect to inform and manage preventive and therapeutic clinical decisions. It requires accurate discrimination of genetic variants associated with high risk (relative risk > 4) from those variants that are not associated with a clinically relevant increase in risk (3, 4).
Variants in BRCA1 whose effect can be inferred using the genetic code can be accurately classified according to their pathogenicity (5). However, the effect of missense variants, in-frame deletions or insertions, and intronic variants need to be assessed using multiple sources of data posing a challenge for clinical annotation. Currently, rigorous clinical annotation of BRCA1 variants regarding pathogenicity is obtained using a multifactorial likelihood model that incorporates family history, segregation data, personal history, and co-occurrence data (5–8). Importantly, clinical recommendations associated with each class (i.e. 5, pathogenic; 4, likely pathogenic; 3, uncertain; 2, likely not pathogenic; and 1, not pathogenic) are available (9). Variants for which the association with increased risk has not been established are called variants of uncertain clinical significance (VUS).3
Missense variants, in particular, constitute a challenge for clinical annotation. Most of these missense VUS alleles are observed in very low frequency (<1/10,000) in the population and, thus family and population-based data are often insufficient to determine pathogenicity. Functional assays that directly or indirectly test for a biological or biochemical function of BRCA1 in the laboratory can also provide information to clinically annotate missense variants (10–15). However, functional data are not currently considered for the calculation of the multifactorial likelihood but only as a confirmatory source of information. Although genetic and epidemiological models will remain the gold standard methods to associate variant and increased risk, several variants may only be understood through functional assays (16, 17).
BRCA1 is a multifunctional protein central to the maintenance of genome integrity through homologous recombination (18). BRCA1 has also been implicated in transcription (19–21), is associated with the RNA polymerase II holoenzyme (22, 23), and is enriched at transcription start sites (24). The transcriptional activation (TA) assay has been extensively used to evaluate BRCA1 VUS (25). In this validated assay, a fusion of a heterologous DNA-binding domain (yeast GAL4 DBD) and the C terminus of BRCA1 drives the expression of a reporter gene. Pathogenic variants have compromised transcription activation, whereas nonpathogenic variants display activity comparable with the reference “WT” allele.
To allow for integration of functional assay results in the multifactorial model a Bayesian two-component mixture model, called VarCall, was developed that generates probabilities of pathogenicity given the functional data (26). Recent analysis of 249 missense variants confirmed the importance of the BRCT domains for BRCA1 function, but also revealed regions within the BRCT domains that seemed tolerant to mutations (25). Thus, to refine and extend the coverage of missense VUS in the BRCA1 C-terminal we tested 102 variants (99 missense and 3 truncating) probing the functional borders of the BRCT domains, α helices in BRCT-[N] and BRCT-[C], and the linker region between BRCT units. Finally, we conducted a joint analysis including all 347 BRCA1 missense variants tested to date.
Results
BRCA1 variants in this study
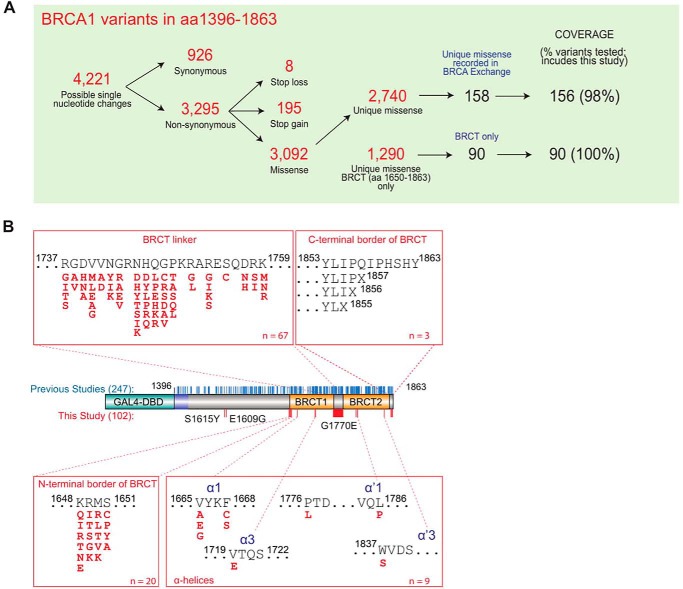
We tested 102 (99 missense and 3 truncating) VUS in the BRCA1 C-terminal region (aa 1396–1863) not previously analyzed using the TA assay, with a focus on structural features, divided in five groups: (a) 20 variants located at the border region between the disordered region and the BRCT-[N] at residues p.K1648 (to Gln, Glu, Thr, Arg, Ile, and Asn), p.R1649 (to Gly, Thr, Lys, Ile, and Ser), p.M1650 (to Leu, Val, Thr, Arg, and Lys), and p.S1651 (to Ala, Cys, Pro, and Tyr); (b) 67 variants located in the linker region connecting BRCT-[N] and BRCT-[C]; (c) 9 variants in α-helices α1 and α3 in BRCT-[N] and in α′1 and α′3 in BRCT-[C]; (d) 3 deletion mutants to probe the C-terminal border of BRCT domain; and (e) 3 assorted variants documented in the population (p.S1577Y, p.E1609G, and p.G1770E) (Fig. 1) (Table S1). We also examined two approaches for transcription data normalization using internal vector only and ectopic protein expression levels.
Figure 1.
BRCA1 variant functional annotation. A, overview of BRCA1 variants. B, diagram of BRCA1 C-terminal region (aa 1396–1863) and location of variants. Variants tested in previous studies and in the current study are shown as blue and red sticks, respectively. Missense VUS tested are grouped according to their structural feature.
N- and C-terminal border of BRCT domains
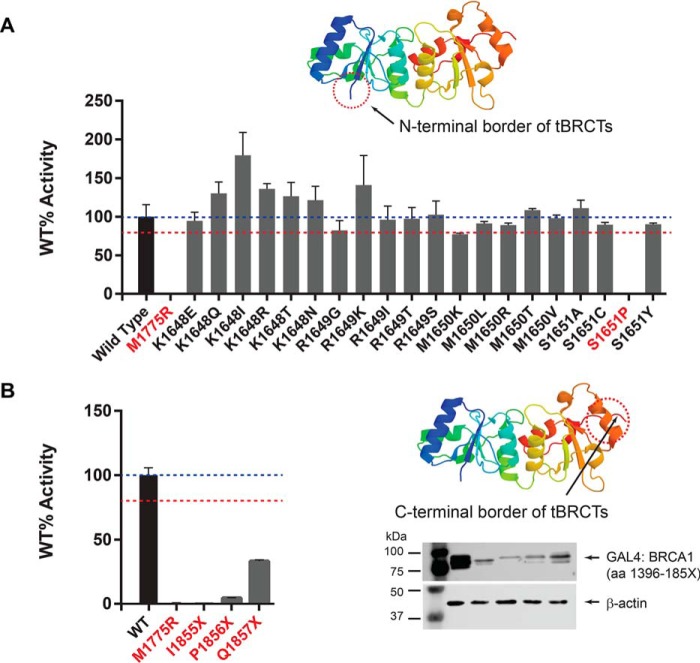
We interrogated 4 amino acid residue positions at the N-terminal border of BRCT domains (aa 1648–1651). When normalizing against an internal vector control for 20 variants in this group, 19 displayed transcription activity comparable with that of WT BRCA1 protein and variant p.S1651P was the only variant in this set with significantly compromised (<80%) function (Fig. 2A). These results suggest that residue p.S1651 is the first position of the tandem BRCT functional and structural unit sensitive to missense changes.
Figure 2.
Transcription activation assay for BRCA1 missense VUS at the N- and C-terminal borders of the BRCT domains. A, transcription activity of missense variants located at the transition between the disordered region and the BRCT domain. B, transcription activity of missense variants located at the C-terminal border of the BRCT domain. Western blots indicate steady state levels of expression of the fusion proteins containing C-terminal deletions. Regions of interest are indicated in the crystal structure diagram (Protein Data Bank 1Y98_A) by a dashed circle. Variants with significantly reduced activity (<80% WT) are denoted in red font.
Nonsense variants that result in an 11-amino acid truncation at the C-terminal region of BRCA1 lead to a dysfunctional protein (2). Deletion of eight final amino acid residues may also disrupt tBRCT function (27). To investigate the minimal region needed for BRCA1 activity we generated three nonsense variants: p.I1855X, p.P1856X, and p.Q1857X (Fig. 1B). All three deletions dramatically affected activity and expression levels of the protein carrying the deletion variants (Fig. 2B). This finding indicates that deletion of as few as 7 amino acid residues from the C terminus leads to protein instability, abrogation of BRCT function, and may therefore be pathogenic.
Linker region
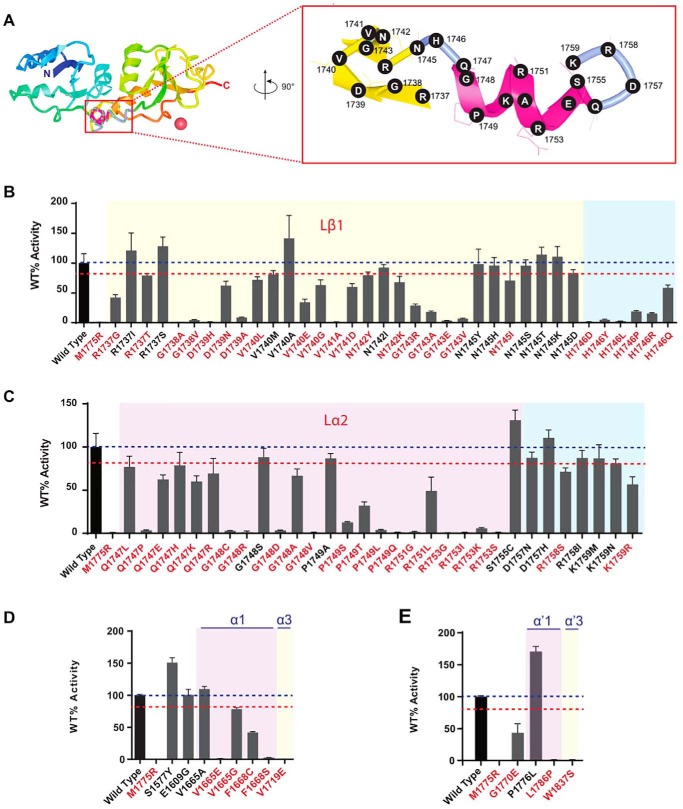
We tested 67 variants at 18 residue positions located across the BRCT linker region (Fig. 1B), and 49 variants displayed reduced activity (mean <80% WT activity). Although the majority of these low activity variants localize to secondary structures, every residue position probed (with the exception of 1755 and 1757) had at least one change that impacted function (<80% WT activity) (Fig. 3, A–C). Residue positions present in intervening sequences were more tolerant to changes than residues at the linker helices. In summary, missense changes in the linker region are likely to have an impact on function and most variants impact protein stability.
Figure 3.
Transcription activation assays for BRCA1 missense VUS at the BRCT linker region. A, structure of BRCA1 tandem BRCT domains (PDB 1JNX). The linker region is shown as a magenta structure perpendicular to the plane of the page (red box). The linker is rotated (90°) and enlarged to show amino acid residue positions tested. B and C, transcription activity of missense variants located at the linker region between BRCT domains. D, transcription activity of missense variants located at the disordered region, and intervening segments and α-helices of BRCT domains. Variants with significantly reduced activity (< 80% WT) are denoted in red font.
α Helices
We evaluated nine variants in four α-helices in both BRCT domains (α1, α3, α′1, and α′3) (Fig. 3, D and E). Seven variants had significantly reduced activity indicating the importance of these α-helices, despite the lack of functional impact of most variants previously analyzed (25).
Assorted variants
We tested two variants described in the population and located in the disordered region (p.S1577Y and p.E1609G) but they did not have an effect on activity (Fig. 3D), as expected. Variant p.G1770E, located in an intervening segment between β′1 and α′1 had a significant impact on function (Fig. 3E).
Protein expression levels
The previous analysis controlled for differences in transfection efficiency in each well by normalizing transcriptional activity, measured by firefly luciferase, by a Renilla luciferase control reporter (driven by HSV thymidine kinase promoter) (Fig. S1A). Protein expression levels of GAL4DBD:BRCA1 variants were assessed by Western blotting (Fig. S1B) to determine whether missense variants caused protein instability but were not used to normalize the results.
To test whether using protein levels for normalization would improve the assay accuracy we also assessed each variant's activity normalized by protein levels (GAL4DBD:BRCA1 and β-actin) (Fig. 4). Similar coefficient of variation distributions were obtained when normalizing by vector only (mean = 17.52, S.D. = 17.77) or by vector and protein levels (mean = 16.57, S.D. = 12.3) (Fig. S2A).
Figure 4.
Transcription activation assays for BRCA1 missense VUS normalized by internal vector and protein levels. A–E, transcription activity of missense variants located at the transition between the disordered region and the BRCT domain normalized against vector control and protein levels (Fig. S1). For comparison, squares denote whether the variant displayed activity similar to WT (open squares) or reduced (<80% WT; filled squares) activity when normalizing against vector control only.
Thirty-five variants, all located in the BRCT domains, showed discordant results between the two normalization methods (Fig. 4, A–D) when using the 80% threshold of activity. Of those, 33 variants were called defective when normalizing by vector only but similar to WT when normalizing to vector and protein levels due to low levels of expression (Fig. S1). However, although these variants were discordant using the arbitrary 80% activity threshold, after the generation of likelihood of pathogenicity by VarCall (see below), only five variants were discordant, indicating that this threshold may be too stringent.
Expression of these variants was consistently low in independent experiments suggesting these amino acid changes lead to protein instability rather than reflecting errors during transient transfections. Consistent with this, seven of 15 selected variants with compromised expression in mammalian cells also failed to express in stable yeast transformants (Fig. S2B). We selected variants p.V1740E and p.H1746D, which showed instability in mammalian cells but are stably expressed in yeast to assess reporter function. Both variants showed markedly reduced reporter transcription activation function, even when reaching steady state levels comparable with WT (Fig. S2C). Taken together, these results indicate that normalization using protein levels may lead to inflated activity levels when lower levels of expression are due to instability caused by the amino acid changes, which ultimately lead to defective function, and not due to errors during transient transfections.
Likelihood of pathogenicity
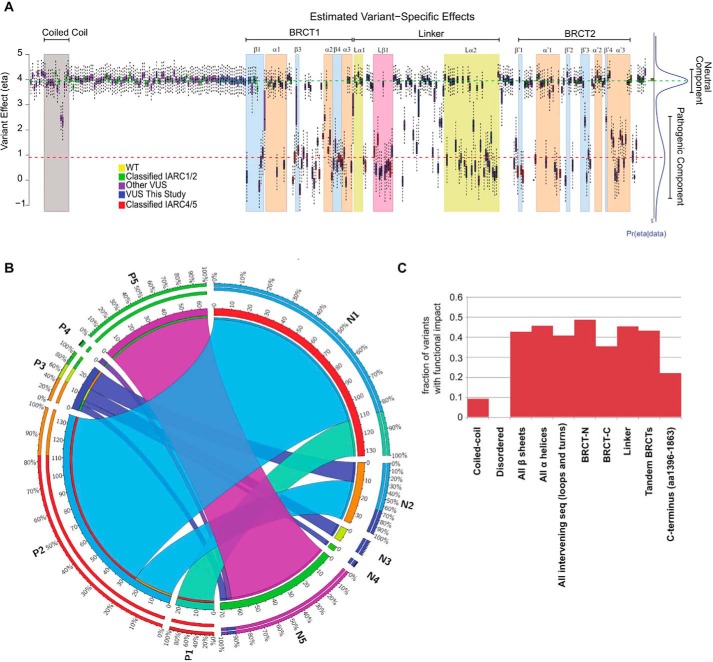
Next, we incorporated the results obtained for the 99 missense variants in the present study into the VarCall algorithm in a joint analysis with data from published variants to estimate the likelihood of pathogenicity of 347 variants (25, 26) (Table S2). The output from VarCall represents the likelihood of pathogenicity given the effects on the functional capacity of the variant as previously described (Fig. 5A) (Table S3).
Figure 5.
VarCall analysis of missense variants in the C-terminal region of the BRCA1 protein. A, in this study, transcriptional assays were performed for 99 missense variants (blue bars). Results of these transcriptional assays were combined with previous data (25) (combined dataset of 347 variants) and analyzed using VarCall to predict the likelihood of pathogenicity. See Fig. S3, a high resolution version, for additional details and variant labels. B, circos plot illustrates how the new series of variants improves classification for all variants. Variants assigned to fClasses (1–5) in Woods et al. (25) (denoted as Previous fClass 1–5) were re-assessed by VarCall (denoted as New fClass 1–5) after including the set of variants in the present study. C, fraction of variants tested that displayed impact on function grouped by structural features.
We previously developed a “functional class” (fClass) scoring scheme (25) using the posterior probability calculation of a variant being pathogenic in the TA assays (PrDel) output by VarCall to generate functional classifications (fClass): PrDel ≤ 0.001 as fClass 1 (nonpathogenic), 0.001 < PrDel ⩽ 0.05 as fClass 2 (likely not pathogenic), 0.05 > Pr-Del ⩽ 0.95 as fClass 3 (uncertain), 0.95 < PrDel ⩽ 0.99 as fClass 4 (likely pathogenic), and PrDel > 0.99 as fClass 5 (pathogenic). According to this scheme, 347 variants were assessed and only 13 variants remain as VUS (Table 1) (Fig. 5B) (for variant labels and details see Fig. S3). Of the 99 variants tested in this article only six remain as VUS (Table 1).
Table 1.
Improvement in classification of BRCA1 missense variants
To assess the performance of VarCall we used a reference panel of 49 known variants classified by the multifactorial model (6) (Table S4). For this assessment we combined IARC Class 1 with 2 (nonpathogenic and likely nonpathogenic) and IARC Class 4 with 5 classes (likely pathogenic and pathogenic). The assay displayed sensitivity of 0.95 (95% CI = 0.72–1.00) and specificity of 1.00 (95% CI = 0.84–1.00) when including a missense variant whose pathogenicity is due to splicing defects (p.R1495M). When using a panel of variants excluding this variant, with prior knowledge of splicing defects, sensitivity improves to 1 (95% CI = 0.78–1.0), confirming that the model can be used to classify VUS reliably given the functional data.
The contributions of the present data set (n = 99) are not limited to the proposed classification of these individual variants but also will provide refinement of previous classifications when the VarCall is run with the complete dataset (n = 347). Indeed, not only the fraction of VUS decreased significantly but also more variants are now classified as fClass 1 (Table 1) (Fig. 5B). The improvement of functional classifications with the present dataset can be visualized in Fig. 5B, which tracks how variants previously assigned fClasses (P1–5) are now assigned to fClasses after the addition of the present dataset (N1–N5). Note that 100% of variants assigned to P1 remained in the corresponding N1 fClass and also 100% of variants in P5 remain in N5. Thus, nonpathogenic (fClass 1) and pathogenic (fClass 5) assignments were stable (Fig. 5B). Importantly, all variants in P4 were further refined to N5 as did a large fraction (85%) of variants in P2, which were further refined to N1 (Fig. 5B).
All BRCT related features (β sheets, α helices, loops and turns, BRCT-[N], BRCT-[C], linker, and tandem BRCTs) were sensitive to missense changes with similar fraction of fClass 4 and 5 (mean = 0.43; median = 0.43; range = 0.35–0.48) (Fig. 5C, Table S5). Notably, no variant in the disordered region (n = 95) had a significant impact on function.
Data integration from multiple assays
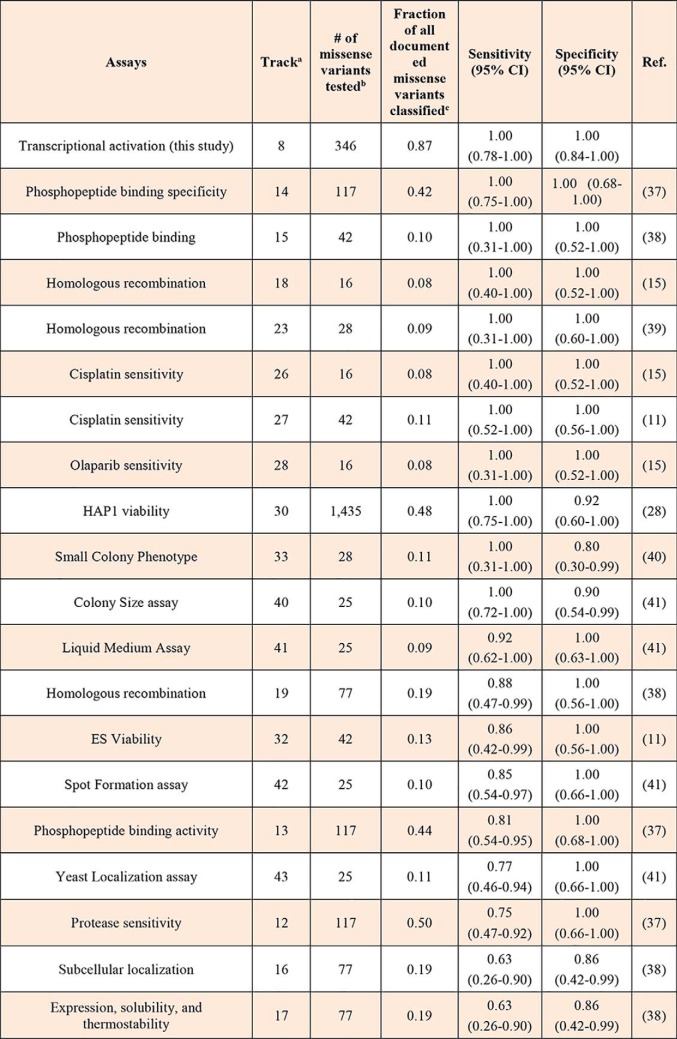
The data presented above corresponds to a single functional assay. Thus, it is conceivable that variants that have no functional impact on transcription might be defective in different assays. We compiled published data for missense variants from 35 functional assays that assess variants in the C terminus of BRCA1 (aa 1315–1863) (Table S6). Using the reference panel (Table S4) we calculated specificity and sensitivity for this region of the protein for 24 assays for which there was at least three pathogenic and 3 nonpathogenic reference variants tested (Table 2) (for full table, see Table S7).
Table 2.
Functional assays for BRCA1 variants (aa 1315–1863)
a Refers to tracks in Table S6.
b Variants tested in region aa 1315–1863.
c Documented variants (aa 1315–1863) are those reported in BRCA Exchange database (n = 206) (42) follows individual studies' cut-offs for classification.
All assays displayed high specificity (range 0.80–1.00) but a more variable sensitivity (range 0.63–1.00) with lower sensitivity found for yeast-based or protein stability assays, presumably due to a subset of pathogenic variants that are stabilized at lower temperatures and variants that do not affect protein stability, respectively (Table 2). Notably, the transcription assay described here achieves, for this region of the protein, the highest sensitivity and specificity of all assays, surpassing other functional assays assessing other canonical functions of BRCA1 such as homologous recombination (HR) (Table 2).
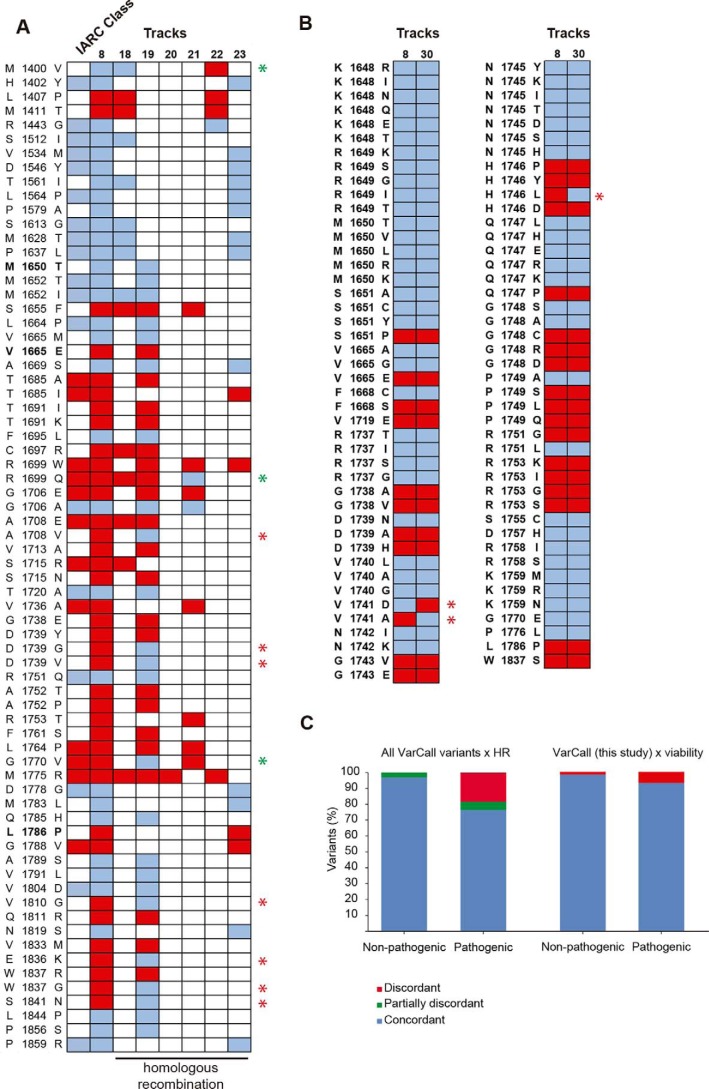
Next, we determined the concordance between VarCall classification for transcription activation and homologous recombination assays (Tracks 18–23; Table S6). There were 60/70 variants (86%) scored by VarCall with at least one additional homologous recombination assay result in complete agreement. Three additional variants had agreement between VarCall and at least one HR assay (Fig. 6, A and B).
Figure 6.
High concordance between VarCall classification and other functional assays. A, diagram showing classification of all missense variants in aa 1315–1863 as defective (red squares) or not (blue squares according to VarCall (Track 8) or to functional assays based on homologous recombination (Tracks 18–23 in Table S6). First row depicts IARC Class using the multifactorial model (Table S4) with red squares (Classes 4–5) and blue squares (Classes 1–2). Discordant results between VarCall and any HR assay are indicated by a red star. Green stars indicate discordant from any assay but in agreement with another HR assay. Variants are in bold font. B, diagram showing classification of variants tested in this study as defective (red squares) or not (blue squares) according to VarCall (Track 8) or to a saturation mutagenesis study (Track 30; Findlay et al. (28)) based on cell viability. Discordant results are indicated by a red star. C, fraction of missense variants with disagreements between assays.
However, due to a small sample size there was limited overlap with variants assessed in the present study (three, all concordant). To assess the variants in the present study we compared results from high throughput mutagenesis and cell viability for which there 87 variants classified by both methods. Concordance was also very high with 97% (84/87) agreement (Fig. 6C).
Discussion
VUS pose a challenge for genetic counseling. For BRCA1 and BRCA2 variants determination of pathogenicity is based on a multifactorial model incorporating data on segregation analysis, family and personal breast or ovarian cancer history, and co-occurrence (5, 7). Due to their low allele frequency many missense alleles of BRCA1 remain as VUS.
Functional assays have emerged as source of empirical data to aid in the determination of pathogenicity of missense VUS (16, 25). The transcriptional assay for variants in the BRCA1 C terminus, in combination with VarCall, was recently assessed for its performance and has been proposed to be used for clinical annotation (25). Recent progress in saturation genome editing for BRCA1 VUS functional analysis demonstrates the feasibility of achieving comprehensive functional assessment of VUS in cancer susceptibility genes for the purposes of clinical annotation of variants (28).
Here we compared the use of two normalization methods using internal vector control only and vector control plus GAL4 DBD BRCA1 fusion protein levels. Normalization using protein levels may lead to incorrect variant classification when loss-of-function variants are associated with protein instability. Although we do not recommend the use of protein expression levels to normalize reporter activity results, protein expression analysis provides important insights regarding the underlying reason for the loss of function. Interestingly, we estimate that ⅓ to ½ of variants that score as likely pathogenic or pathogenic in VarCall lead to unstable protein with a higher fraction of variants in secondary structures in the BRCT domains.
Inspection of VarCall results in Woods et al. (25) showed clearly that the likelihood of being pathogenic varies significantly depending on its location, reflecting the importance of secondary and tertiary structures for function, in particular the tandem BRCT domains. Thus, we focused on three areas of improvement on our ability to identify pathogenic variants using the transcription assay: first, the definition of the N- and C-terminal borders of functional domains; second, a more granular analysis of the linker region; and finally, the role of α-helices in the BRCT domains.
In the present study we analyzed 99 missense and 3 deletion variants and define the functional borders of the BRCT domains. Because digestion of a bacterially expressed BRCA1 C-terminal region (aa 1528–1863) resulted in a proteolytically stable fragment (aa 1646–1863) and further deletion of residues 1860–1863 yielded a soluble fragment used for structural determination, residue positions (1646–1859) have been loosely considered the functional borders of the BRCT domain (29). Structural determination further refined it to 1650–1859 as the first secondary structure in the BRCT-[N], a β-sheet (β1), starts at residue Met-1650 (29). Our data suggest that this position is also tolerant to changes, as all six variants tested at this position were fClass 1 or 2. Deletion analysis of the C-terminal end of BRCA1 in yeast indicated that 1856–1863 were dispensable for function (in the transcription assay), whereas aa p.I1855 was required for BRCT stability (30). p.Y1853, p.L1854, and p.I1855 represent a cluster of hydrophobic residues present at the end of Motif II in several BRCT domains (31). Our data showed that deletion starting at Gln-1857 significantly compromised the function in mammalian cells, consistent with a higher sensitivity of the transcriptional assay in mammalian cells than in yeast (32, 33). Thus, we propose that for clinical variant annotation the BRCT domains in BRCA1 should be conservatively considered (1650–1857). That is, a variant predicted to modify or truncate protein sequence at residues 1858–1863 is not considered clinically important.
Hayes et al. (30) identified several variants in the liker domain as sensitive to amino acid changes suggesting the existence of unidentified structural requirements for the linker region. A flexible linker with a helical central portion which connects both BRCT units was identified by X-ray crystallography (29). However, this region had not been systematically explored. Our dense mutagenesis of this region showed that Lβ1-intervening sequence-Lα2 is extremely sensitive to most changes resulting from single nucleotide changes in those residue positions. Previous data had shown that most variants in α1 and α′1 (two corresponding helices in the N- and C-BRCT domains) did not affect function, a case different from α3 and α′3 where most variants have a dramatic effect on function (25). Analysis of the current set shows that some variants in these structures lead to a functional defect. The lack of missense variants in the disordered region showing any impact on function (0 of 95 tested), confirms that this region is unlikely to harbor pathogenic variants. The significant fraction of pathogenic/likely pathogenic variants across all BRCT-related features (β sheets, α helices, loops and turns, BRCT-[N], BRCT-[C], linker, and tandem BRCTs) highlights their requirement for the integrity of the tandem BRCT domains.
We had previously identified a patch of highly conserved amino acid residues forming a groove on the opposite face from the known phosphopeptide-binding pocket formed by residues Thr-1684, Thr-1685, His-1686, Lys-1711, Trp-1712, and Arg-1753, specific to BRCA1 BRCT (34). Interestingly, the conservative change p.R1753K, which is predicted by in silico tools to have no impact on function, shows severely impaired activity, further supporting a role for this putative binding site. Further studies will be needed to investigate this groove on the BRCT surface.
A significant challenge in functional assessment of VUS is the concern that a variant scoring as nonpathogenic in one assay may confer risk of disease through a function other than the one being tested in the assay. Here, using the same reference panel we show that most functional assays achieve high sensitivity and specificity and there is a high level of concordance between assays. Importantly, there was no discordance in variants that scored nonpathogenic in VarCall and in homologous recombination assays. The reason for the excellent agreement across assays of distinct functions (HR, transcription, viability, and cisplatin sensitivity) may be partly due to the significant fraction of variants that lead to the absence of stable protein product, which would affect any readout. It is unlikely that any assay will be ideal and integration of results from different systems will be critical to improve clinical classification of variants.
In summary, data from this study confirms the high accuracy of the transcription assay for BRCA1 missense variants indicating that it can aid in the classification of VUS for clinical decisions. We performed a systematic analysis of several structural features and provide assessment for 98% of all variants recorded in the C-terminal region of BRCA1 with a small fraction (4%) of variants remaining as VUS. Although at this point these data should not be used as a sole source of information to classify variants, results are encouraging and warrant further exploration on how to use functional data for clinical annotation of VUS.
Experimental procedures
Plasmid constructs
We used the pcDNA3 (SG) BRCA1 13/24 (WT) plasmid containing a reference cDNA of the human BRCA1 gene (GenBankTM accession U14680) (OMIM 113705) coding for amino acid residues 1396–1863 (exons 13–24), as an in-frame fusion to GAL4 DNA-binding domain (DBD). Controls were pathogenic (IARC Class 5) variants (p.M1775R and p.Y1853X) and benign (IARC Class 1) variant (p.S1613G) as previously described (35).
VUS were introduced into pcDNA3 (SG) BRCA1 13/24 (WT) by site-directed mutagenesis using the QuikChange II Site-directed Mutagenesis Kit, following the manufacturer's protocol (Agilent, Santa Clara, CA). Primers used for site-directed mutagenesis are listed in Table S1. All plasmids containing variants were subjected to Sanger sequencing to confirm the correct reading frame with GAL4 DBD, the presence of the variant to be tested, and absence of additional mutations introduced during PCR amplification.
TA assay in human cells
Variants were assessed by the TA assay (27, 35, 36) using three technical replicates for each variant in at least two independent experiments for a total of at least six measurements per variant. Briefly, BRCA1 constructs containing missense VUS or controls were co-transfected in HEK293FT cells with the pG5Luc plasmid with a Photinus pyralis luciferase reporter gene driven by GAL4-binding sites, and the phGR-TK plasmid, which constitutively expresses the internal control Renilla luciferase. Cells were transfected using FuGENE HD (Roche Applied Science) or polyethylenimine (Sigma) and harvested 24 h after transfection. Transcriptional activity was assayed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). For use in VarCall luciferase activity was normalized using the internal constitutive expression of Renilla sp. luciferase and variant activity was depicted as % of the “WT” activity. For comparison purposes we also provide luciferase activity (as described above) normalized by expression levels measured by Western blotting. Note that Western blots for expression levels are obtained from parallel cultures and not directly from the wells in which luciferase activity is being measured.
TA assay in yeast cells
The TA assay was also conducted in EGY48 yeast cells using a plasmid reporter JK103, which has one LexA operator driving LacZ, and expressing a fusion of LexA:BRCA1 aa 1396–1864 as previously described (30).
Western blotting
Cells were harvested 24 h after transfection and lysed with RIPA buffer (NaCl 150 mm; Tris-Cl 10 mm (pH 7.4); EDTA 5 mm (pH 8.0); Triton X-100, 1%; sodium deoxycholate, 0.1%; and SDS, 0.1%). Whole cell extracts were evaluated by immunoblotting using mouse monoclonal α-GAL4 DBD (Santa Cruz Biotechnology, Dallas, TX; catalog number sc-510), rabbit polyclonal α-LexA DBD (Millipore Sigma, Burlington, MA; catalog number 06-719), or α-BRCA1 C terminus (Santa Cruz; catalog number sc-642) as a primary antibody and horseradish peroxidase-conjugated α-mouse or α-rabbit IgG (Santa Cruz Biotechnology, catalog numbers sc-2005 and sc-2030) as secondary antibodies.
VarCall
Results were analyzed using the computation model VarCall (26) with the input dataset as in Table S2, which merged the results from the Woods et al. (25) dataset with the missense variants in this study. The Woods et al. (25) dataset incorrectly included variant p.M1652W, which was not a natural occurrence and variant p.A1752R, which contained a typographical error. These are corrected in the present dataset. The input dataset contained (Table S2) assays on 367 variants and the WT reference (includes one frameshift, three nonsense, and four in-frame deletions, including the 99 missense variants in the present study, and 4,974 individual measurements (1,291 from the present study). For performance evaluation we compared results to a panel of 63 variants classified by the multifactorial model (IARC Class 1, 26 variants; IARC Class 2, 15 variants; IARC Class 4, variants 5; IARC Class 5, variants 17) (Table S4). Performance metrics for VarCall were calculated using Vassarstats Clinical Calculator (http://vassarstats.net/clin1.html).4
Author contributions
V. C. F., G. S.-K., F. C., E. S. I., A. N. A. M., and M. A. C. data curation; V. C. F. and V. A. G. formal analysis; V. C. F., G. S.-K., F. C., E. S. I., A. N. A. M., and M. A. C. supervision; V. C. F. and E. S. I. validation; V. C. F., V. A. G., G. D. P., C. S., K. A., T. C. N., G. d. G., R. B. V. A., C. H., T. T. G., and R. F. S. investigation; V. C. F., V. A. G., G. D. P., C. S., K. A., T. C. N., G. d. G., R. B. V. A., C. H., T. T. G., and R. F. S. methodology; V. C. F. writing-original draft; G. S.-K., F. C., E. S. I., A. N. A. M., and M. A. C. conceptualization; G. S.-K., F.C., A. N. A. M., and M. A. C. writing-review and editing; E. S. I. software; A. N. A. M. and M. A. C. resources; A. N. A. M. and M. A. C. funding acquisition.
Supplementary Material
Acknowledgments
We thank Bryony Thompson for helpful suggestions and Steven Hart and Xueli Li for technical assistance and all individuals and families who have generously donated their time, samples, and information to facilitate research on the predisposition factors of cancer.
This work was supported by National Institutes of Health Grant U01 CA116167, the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Conselho Nacional de Desenvolvimento Cientifico e Tecnológico, Fundação do Câncer (Programa de Oncobiologia), and in part by the Molecular Genomics Core Facilities at the Moffitt Cancer Center through National Institutes of Health NCI CCSG Grant P30-CA76292. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S7 and Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- VUS
- variants of uncertain clinical significance
- TA
- transcriptional activation
- aa
- amino acid(s)
- HR
- homologous recombination
- DBD
- DNA-binding domain
- CI
- confidence interval.
References
- 1. Mavaddat N., Peock S., Frost D., Ellis S., Platte R., Fineberg E., Evans D. G., Izatt L., Eeles R. A., Adlard J., Davidson R., Eccles D., Cole T., et al. (2013) Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 105, 812–822 10.1093/jnci/djt095 [DOI] [PubMed] [Google Scholar]
- 2. Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., and Ding W. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 3. Frank T. S., Deffenbaugh A. M., Reid J. E., Hulick M., Ward B. E., Lingenfelter B., Gumpper K. L., Scholl T., Tavtigian S. V., Pruss D. R., and Critchfield G. C. (2002) Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 20, 1480–1490 10.1200/JCO.2002.20.6.1480 [DOI] [PubMed] [Google Scholar]
- 4. Easton D. F., Pharoah P. D., Antoniou A. C., Tischkowitz M., Tavtigian S. V., Nathanson K. L., Devilee P., Meindl A., Couch F. J., Southery M., et al. (2015) Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 372, 2243–2257 10.1056/NEJMsr1501341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldgar D. E., Easton D. F., Deffenbaugh A. M., Monteiro A. N., Tavtigian S. V., and Couch F. J., Breast Cancer Information Core (BIC) Steering Committee (2004) Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am. J. Hum. Genet. 75, 535–544 10.1086/424388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindor N. M., Guidugli L., Wang X., Vallée M. P., Monteiro A. N., Tavtigian S., Goldgar D. E., and Couch F. J. (2012) A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 33, 8–21 10.1002/humu.21627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Easton D. F., Deffenbaugh A. M., Pruss D., Frye C., Wenstrup R. J., Allen-Brady K., Tavtigian S. V., Monteiro A. N., Iversen E. S., Couch F. J., and Goldgar D. E. (2007) A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 81, 873–883 10.1086/521032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldgar D. E., Easton D. F., Byrnes G. B., Spurdle A. B., Iversen E. S., Greenblatt M. S., and IARC Unclassified Genetic Variants Working Group (2008) Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 29, 1265–1272 10.1002/humu.20897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plon S. E., Eccles D. M., Easton D., Foulkes W. D., Genuardi M., Greenblatt M. S., Hogervorst F. B., Hoogerbrugge N., Spurdle A. B., Tavtigian S. V., IARC Unclassified Genetic Variants Working Group (2008) Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 29, 1282–1291 10.1002/humu.20880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Millot G. A., Carvalho M. A., Caputo S. M., Vreeswijk M. P., Brown m M. A., Webb M., Rouleau E., Neuhausen S. L., Hansen Tv., Galli A., Brandão R. D., Blok M. J., Velkova A., et al. (2012) A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 33, 1526–1537 10.1002/humu.22150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bouwman P., van der Gulden H., van der Heijden I., Drost R., Klijn C. N., Prasetyanti P., Pieterse M., Wientjens E., Seibler J., Hogervorst F. B., and Jonkers J. (2013) A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 3, 1142–1155 10.1158/2159-8290.CD-13-0094 [DOI] [PubMed] [Google Scholar]
- 12. Brzovic P. S., Meza J. E., King M. C., and Klevit R. E. (2001) BRCA1 RING domain cancer-predisposing mutations: structural consequences and effects on protein-protein interactions. J. Biol. Chem. 276, 41399–41406 10.1074/jbc.M106551200 [DOI] [PubMed] [Google Scholar]
- 13. Morris J. R., Pangon L., Boutell C., Katagiri T., Keep N. H., and Solomon E. (2006) Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 15, 599–606 10.1093/hmg/ddi476 [DOI] [PubMed] [Google Scholar]
- 14. Starita L. M., Young D. L., Islam M., Kitzman J. O., Gullingsrud J., Hause R. J., Fowler D. M., Parvin J. D., Shendure J., and Fields S. (2015) Massively parallel functional analysis of BRCA1 RING domain variants. Genetics 200, 413–422 10.1534/genetics.115.175802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anantha R. W., Simhadri S., Foo T. K., Miao S., Liu J., Shen Z., Ganesan S., and Xia B. (2017) Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife 6, e21350 10.7554/eLife.21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toland A. E., and Andreassen P. R. (2017) DNA repair-related functional assays for the classification of BRCA1 and BRCA2 variants: a critical review and needs assessment. J. Med. Genet. 54, 721–731 10.1136/jmedgenet-2017-104707 [DOI] [PubMed] [Google Scholar]
- 17. Starita L. M., Ahituv N., Dunham M. J., Kitzman J. O., Roth F. P., Seelig G., Shendure J., and Fowler D. M. (2017) Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 101, 315–325 10.1016/j.ajhg.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., and Livingston D. M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 10.1016/S0092-8674(00)81847-4 [DOI] [PubMed] [Google Scholar]
- 19. Monteiro A. N., August A., and Hanafusa H. (1996) Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc. Natl. Acad. Sci. U.S.A. 93, 13595–13599 10.1073/pnas.93.24.13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nadeau G., Boufaied N., Moisan A., Lemieux K. M., Cayanan C., Monteiro A. N., and Gaudreau L. (2000) BRCA1 can stimulate gene transcription by a unique mechanism. EMBO Rep. 1, 260–265 10.1093/embo-reports/kvd059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kleiman F. E., and Manley J. L. (1999) Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285, 1576–1579 10.1126/science.285.5433.1576 [DOI] [PubMed] [Google Scholar]
- 22. Scully R., Anderson S. F., Chao D. M., Wei W., Ye L., Young R. A., Livingston D. M., and Parvin J. D. (1997) BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 94, 5605–5610 10.1073/pnas.94.11.5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anderson S. F., Schlegel B. P., Nakajima T., Wolpin E. S., and Parvin J. D. (1998) BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 19, 254–256 10.1038/930 [DOI] [PubMed] [Google Scholar]
- 24. Dunham I., and ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woods N. T., Baskin R., Golubeva V., Jhuraney A., De-Gregoriis G., Vaclova T., Goldgar D. E., Couch F. J., Carvalho M. A., Iversen E. S., and Monteiro A. N. (2016) Functional assays provide a robust tool for the clinical annotation of genetic variants of uncertain significance. NPJ Genom. Med. 1, 16001 10.1038/npjgenmed.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iversen E. S., Couch F. J., Goldgar D. E., Tavtigian S. V., and Monteiro A. N. A. (2011) A computational method to classify variants of uncertain significance using functional assay data with application to BRCA1. Cancer Epidemiol. Biomarkers Prev. 20, 1078–1088 10.1158/1055-9965.EPI-10-1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carvalho M. A., Marsillac S. M., Karchin R., Manoukian S., Grist S., Swaby R. F., Urmenyi T. P., Rondinelli E., Silva R., Gayol L., et al. (2007) Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 67, 1494–1501 10.1158/0008-5472.CAN-06-3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Findlay G. M., Daza R. M., Martin B., Zhang M. D., Leith A. P., Gasperini M., Janizek J. D., Huang X., Starita L. M., and Shendure J. (2018) Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 10.1038/s41586-018-0461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Williams R. S., Green R., and Glover J. N. (2001) Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 8, 838–842 10.1038/nsb1001-838 [DOI] [PubMed] [Google Scholar]
- 30. Hayes F., Cayanan C., Barillà D., and Monteiro A. N. (2000) Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 60, 2411–2418 [PMC free article] [PubMed] [Google Scholar]
- 31. Bork P., Hofmann K., Bucher P., Neuwald A. F., Altschul S. F., and Koonin E. V. (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11, 68–76 10.1096/fasebj.11.1.9034168 [DOI] [PubMed] [Google Scholar]
- 32. Worley T., Vallon-Christersson J., Billack B., Borg A., and Monteiro A. N. (2002) A naturally occurring allele of BRCA1 coding for a temperature-sensitive mutant protein. Cancer Biol. Ther. 1, 497–501 10.4161/cbt.1.5.164 [DOI] [PubMed] [Google Scholar]
- 33. Carvalho M. A., Billack B., Chan E., Worley T., Cayanan C., and Monteiro A. N. (2002) Mutations in the BRCT domain confer temperature sensitivity to BRCA1 in transcription activation. Cancer Biol. Ther. 1, 502–508 10.4161/cbt.1.5.165 [DOI] [PubMed] [Google Scholar]
- 34. Karchin R., Monteiro A. N., Tavtigian S. V., Carvalho M. A., and Sali A. (2007) Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput. Biol. 3, e26 10.1371/journal.pcbi.0030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phelan C. M., Dapic V., Tice B., Favis R., Kwan E., Barany F., Manoukian S., Radice P., van der Luijt R. B., van Nesselrooij B. P., et al. (2005) Classification of BRCA1 missense variants of unknown clinical significance. J. Med. Genet. 42, 138–146 10.1136/jmg.2004.024711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vallon-Christersson J., Cayanan C., Haraldsson K., Loman N., Bergthorsson J. T., Brøndum-Nielsen K., Gerdes A. M., Møller P., Kristoffersson U., Olsson H., Borg A., and Monteiro A. N. (2001) Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum. Mol. Genet. 10, 353–360 10.1093/hmg/10.4.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee M. S., Green R., Marsillac S. M., Coquelle N., Williams R. S., Yeung T., Foo D., Hau D. D., Hui B., Monteiro A. N., and Glover J. N. (2010) Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 70, 4880–4890 10.1158/0008-5472.CAN-09-4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petitalot A., Dardillac E., Jacquet E., Nhiri N., Guirouilh-Barbat J., Julien P., Bouazzaoui I., Bonte D., Feunteun J., et al. (2018) Combining homologous recombination and phosphopeptide-binding data to predict the impact of BRCA1 BRCT variants on cancer risk. Mol. Cancer Res. 17, 54–69 10.1186/s12943-018-0790-4 [DOI] [PubMed] [Google Scholar]
- 39. Lu C., Xie M., Wendl M. C., Wang J., McLellan M. D., Leiserson M. D., Huang K. L., Wyczalkowski M. A., Jayasinghe R., Banerjee T., et al. (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 6, 10086 10.1038/ncomms10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coyne R. S., McDonald H. B., Edgemon K., and Brody L. C. (2004) Functional characterization of BRCA1 sequence variants using a yeast small colony phenotype assay. Cancer Biol. Ther. 3, 453–457 10.4161/cbt.3.5.809 [DOI] [PubMed] [Google Scholar]
- 41. Thouvenot P., Ben Yamin B., Fourrière L., Lescure A., Boudier T., Del Nery E., Chauchereau A., Goldgar D. E., Houdayer C., Stoppa-Lyonnet D., Nicolas A., and Millot G. A. (2016) Functional assessment of genetic variants with outcomes adapted to clinical decision-making. PLoS Genet. 12, e1006096 10.1371/journal.pgen.1006096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cline M. S., Liao R. G., Parsons M. T., Paten B., Alquaddoomi F., Antoniou A., Baxter S., Brody L., Cook-Deegan R., Coffin A., Couch F. J., et al. (2018) BRCA challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 14, e1007752 10.1371/journal.pgen.1007752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.