Abstract
The G-rich Pu39 region of the P1 promoter of the oncogene BCL-2, an apoptosis regulator, can fold into multiple G-quadruplex (G4) structures. Bcl2-2345 and Bcl2-1245 are two major G4 species forming with high thermal stability and distinct topologies in the Pu39 region, but their folding/unfolding kinetics have not yet been investigated. Here, we used magnetic tweezers to measure the mechanical stability and the folding/unfolding kinetics of the Bcl2-2345 and Bcl2-1245 G4 structures. We report that the hybrid-stranded Bcl2-2345 G4 had a lower mechanical stability than the parallel-stranded Bcl2-1245 G4. We observed that the Bcl2-2345 G4 is a kinetically favored structure, whereas the Bcl2-1245 G4, with a slow unfolding rate, may function as a kinetic barrier for transcription. We also determined that in addition to the Bcl2-2345 and Bcl2-1245 G4s, other stable DNA secondary structures, such as a hybrid-stranded Bcl2-1234 G4, can also form in the Pu39 sequence. The characterization of the folding/unfolding kinetics of specific G4s reported here sheds light on the participation of G4s during gene transcription and provides information for designing G4-targeting small molecules that could modulate BCL-2 gene expression.
Keywords: G-quadruplex, B-cell lymphoma 2 (Bcl-2), single-molecule biophysics, promoter, transcription regulation, DNA secondary structure, folding/unfolding kinetics, force spectroscopy, magnetic tweezers, force spectroscopy, oncogene promoter
Introduction
The human B-cell CLL/lymphoma 2 (BCL-2) gene encodes a mitochondrial membrane protein, which plays an essential role in the inhibition of apoptosis in cells. Overexpression of BCL-2 is closely related to a large number of cancers (1–3). Increased BCL-2 expression is also related to resistance to chemotherapy and γ-ray therapy (4). The human BCL-2 gene has two promoters, P1 and P2. P1 is a GC-rich promoter that is responsible for the majority of the BCL-2 gene expression. A 39-bp GC-rich sequence (Pu39), located 58–19 bp upstream of the P1 promoter, can form four-stranded DNA structures called G-quadruplexes (G4s)2 (5). G4s that are formed in the promoter region act as molecular switches that control BCL-2 transcription (5, 6). Deletion of the Pu39 region increases the promoter activity by 2.1-fold compared with the full-length WT promoter (7). Stabilization of the BCL-2 G4s by small molecules reduces the expression of BCL-2 and leads to apoptosis in HL-60 cells, which is promising for cancer therapeutics (8).
The Pu39 sequence of BCL-2 contains six guanine tracts (3–5 consecutive guanines) and has the potential to form multiple G4 structures with 15 possible combinations from these different G-tracts (Fig. 1A and Table S1). The first G4 solution structure solved for the Pu39 sequence was the Bcl2-2345 G4 (also named Bcl2-Mid4), which involved the middle four consecutive runs of G-tracts 2, 3, 4, and 5 (Fig. 1B) (5, 9). The Bcl2-2345 G4 is a hybrid-stranded G4 structure in which three strands are oriented in one direction and the fourth strand is oriented in the opposite direction (5, 9). Later, dimethyl sulfate (DMS) footprinting experiments indicated that the G4s from the full-length Pu39 sequence formed in K+ involved the G-tracts 1, 2, 4, and 5 and was named Bcl2-1245 G4 (10). Bcl2-1245 G4 is a parallel-stranded structure in which the four strands are oriented in the same direction (Fig. 1B) (10). DMS footprinting studies showed that G-tracts 1, 2, 3, 4, and 5 were protected from chemical cleavage due to the formation of G4 structures. Thus, the Bcl2-2345 and Bcl2-1245 G4s are likely to be two major G4 species formed in the Pu39 sequence (5, 10, 11). The presence of different DNA secondary structures in the BCL-2 promoter region may be necessary for the precise regulation of gene transcription, as each DNA secondary structure can be recognized by various transcription factors (7, 12). Different topologies, such as hybrid and parallel-stranded topologies of the Bcl2-2345 and Bcl2-1245 G4 structures, provided structural differences for being recognized by transcription factors and for designing drugs targeting a specific G4 structure (13). In addition, both the Bcl2-2345 and Bcl2-1245 G4s contain a long middle loop, 7 and 13 nt in length, respectively, which provides another region for specific recognition. A recent study showed that a 7-mer peptide nucleic acid (PNA) that binds to the 7-nt loop region of Bcl2-2345 increased the formation of Bcl2-2345 G4 and reduced the expression of the BCL-2 gene in cancer cells, which indicates that the Bcl2-2345 G4 structure is a potent target for modulating BCL-2 gene expression (14).
Figure 1.
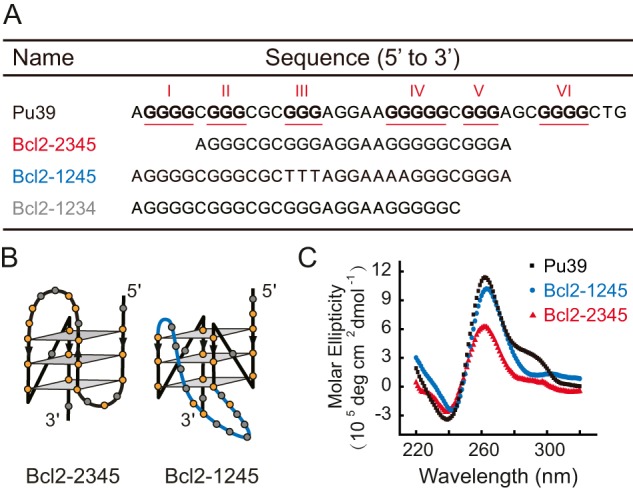
A, the sequences of the WT Pu39 region and the truncated mutants Bcl2-2345, Bcl2-1245, and Bcl2-1234. The six G-tracts with three or more guanines are underlined and numbered using Roman numerals. B, schematic structures of Bcl2-2345 and Bcl2-1245 G4s according to previous NMR studies (9, 10). The 13-nt middle loop of the Bcl2-1245 G4 is shown in blue. C, CD spectra of the Pu39, Bcl2-1245, and Bcl2-2345 sequence measured in 100 mm KCl at room temperature.
To understand the biological roles of promoter G4s, the characterization of the stability and folding/unfolding kinetics of different G4 species is necessary. Both Bcl2-2345 and Bcl2-1245 G4s demonstrate high thermodynamic stability in which the melting temperatures of Bcl2-2345 and Bcl2-1245 G4s in ∼100 mm K+ solution were determined to be ∼75 and ∼71 °C, respectively (9, 10). However, the kinetics of the Bcl2-2345 and Bcl2-1245 G4s remain unclear because G4 structures with distinct folding/unfolding kinetics may have similar melting temperatures. The folding/unfolding kinetics of G4s determine the possible participation of G4s during transcription and are also crucial for designing small molecules targeting Bcl2-2345 and Bcl2-1245 G4s.
Single-molecule force spectroscopy technologies, such as atomic force spectroscopy, optical tweezers, and magnetic tweezers, have emerged as a powerful tool to explore the mechanical stability and folding/unfolding kinetics of biomolecule structures, including G4s (15–17). A previous single-molecule force spectroscopy study reported multiple unfolding force peaks of the Pu39 sequence, which suggested the formation of multiple stable DNA secondary structures (18). It is unclear whether these structures correspond to Bcl2-2345 and Bcl2-1245 G4s or other stable G-rich secondary structures. The respective folding/unfolding kinetics of these two G4s have not yet been investigated, which impairs our understanding of how Bcl2-2345 and Bcl2-1245 G4 structures play roles in the transcriptional regulation of the BCL-2 gene. Here, we studied the folding/unfolding kinetics of these two major G4s in the Pu39 region based on the mechanical manipulation of a single-stranded DNA (ssDNA)2 that contains sequences corresponding to Bcl2-2345 and Bcl2-1245 G4s. By comparing with the data obtained from the Pu39 sequence, we also probed the potential existence of other stable G4 structures.
Results
The formation of G4s from DNA sequences used in our experiments was confirmed by CD experiments in 100 mm K+ solution. We first focused on Bcl2-2345 and Bcl2-1245 G4s, as previous DMS footprinting experiments suggested that these two G4s are the major species in the Pu39 sequence (5, 10). Fig. 1C shows that the CD spectrum of the Bcl2-1245 sequence has a minimum at ∼240 nm and a maximum at ∼260 nm, which indicates the formation of parallel-stranded G4 structures. The small positive peak at ∼290 nm of Bcl2-2345 indicates the formation of a mixed parallel/antiparallel-stranded G4 structure, which agrees with previous studies (9, 10). The CD spectrum of the Pu39 sequence is similar to the Bcl2-2345, suggesting that mixed secondary structures are formed in this sequence.
Fig. 2A shows the experimental design for the single-molecule magnetic tweezers study for BCL-2 G4s (19). A single-stranded G4-forming DNA sequence spanned between two duplex DNA handles is tethered between a coverslip and a streptavidin-coated paramagnetic bead. The extension change of the tethered DNA molecule is measured based on the change in the bead height, which was analyzed using the diffraction pattern of the bead recorded by a CCD camera (20, 21).
Figure 2.
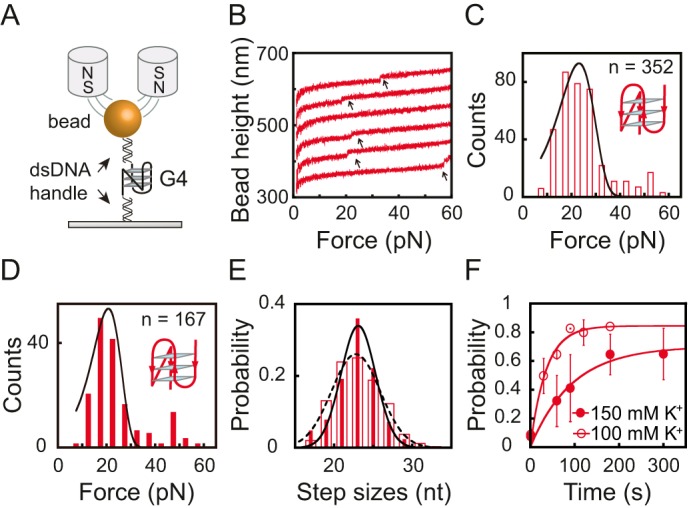
A, schematic of the G4 DNA and the magnetic tweezers measurement. The G4-forming ssDNA is sandwiched between two dsDNA handles (489 and 601 bp). B, typical force-bead height curves of the Bcl2-2345 sequence in repeating stretching cycles in 100 mm KCl. The curves are shifted along the bead height axis for visual clarity. The G4 unfolding results in a sudden bead height jump (black arrows). C and D, unfolding force histograms for the Bcl2-2345 G4 in 100 mm KCl (C) and 150 mm KCl (D). E, unfolding step size histogram of the Bcl2-2345 G4 in 100 mm KCl (dotted line) and 150 mm KCl (solid line). Data were fitted by a single Gaussian equation. F, the time evolution of the folding probability pfold(t) of the Bcl2-2345 sequence in 100 mm KCl (open circles) and in 150 mm KCl (solid circles).
To obtain the unfolding kinetics, we used the force-ramp measurements, where the force increased linearly with time (i.e. constant loading rate) by changing the magnet–bead distance through a programmed trajectory (19, 21). Due to the slow folding/unfolding transition of the Bcl2 G4s, constant force measurements were not used (17, 22). Fig. 2B shows typical force–bead height curves of the Bcl2-2345 sequence, and these curves were obtained by repeating the stretching cycles in the presence of 100 mm KCl. The DNA molecule was held at a low force of 1 pN for 90 s to allow refolding, and then the force was increased to ∼60 pN at a loading rate of 2 pN/s for the Bcl2-2345 sequence. The unfolding transition of the G4 was observed from a sudden extension jump in the bead height curve (indicated by an arrow in Fig. 2B), and the unfolding force was measured at the transition. After reaching ∼60 pN, the force was jumped to 1 pN to allow the refolding of the G4s. After refolding, the procedure was repeated.
Fig. 2 (C and D) shows the unfolding force distribution of the Bcl2-2345 G4 obtained by repeating the force-ramp cycles in 100 mm KCl (n = 352) and 150 mm KCl (n = 167), respectively. At each condition, the data were obtained from more than 10 independent tethers. The unfolding force distribution of the Bcl2-2345 G4 shows a single peak at 21 ± 7 pN (average ± S.D.) in 100 mm KCl and at 19 ± 5 pN in 150 mm KCl. The unfolding rate of the Bcl2-2345 G4 was obtained by fitting the unfolding force distribution (Fig. 2, C and D) with Bell's model as follows,
| (Eq. 1) |
where r represents the loading rate, ku is the zero-force unfolding rate, Δxu is the transition distance from the folded state to the transition state, kB is the Boltzmann constant, and T is the absolute temperature. The best-fitting parameters for the Bcl2-2345 G4 were ku = (1.2 ± 0.4) × 10−2 s−1 and Δxu = 0.6 ± 0.1 nm in 100 mm KCl and ku = (1.0 ± 0.4) × 10−2 s−1 and Δxu = 0.7 ± 0.1 nm in 150 mm KCl (average ± S.E.). The unfolding step size distribution of Bcl2-2345 revealed a single peak at 23 ± 3 nt in 100 mm KCl (dotted line) and 23 ± 2 nt in 150 mm KCl (solid line) (average ± S.D.), which is consistent with the number of nucleotides in the Bcl2-2345 G4 structure (Fig. 2E) (9, 10). Both unfolding forces and unfolding step sizes of Bcl2-2345 showed similar distributions in 100 and 150 mm KCl, suggesting that the potassium concentration has only minor effects on the unfolding kinetics of Bcl2-2345.
The refolding kinetics of the G4s were quantified based on measuring the time evolution of the refolding probability. More specifically, the G4s were first mechanically unfolded in the aforementioned force-ramping procedure. The force was subsequently jumped to 1 pN and was held at this force for various holding time intervals (t). If a G4 unfolding step was observed, it indicated that G4 was folded at 1 pN before the force was increased. Repeating the procedure N times, we obtained the probability of refolding of the G4 over the holding time t at 1 pN by pfold(t) = M/N, where M is the number of cycles in which the G4 refolded. As shown in Fig. 2F, Bcl2-2345 pfold(t) data were fitted with a single-exponential function as follows: pfold(t) = pst(1 − e−kt). Here, pst is the steady-state folding probability at 1 pN, and kf is the folding rate. Using this approach, the values of pst and kf were determined to be ∼84% and 0.028 ± 0.005 s−1 in 100 mm KCl and ∼70% and 0.011 ± 0.003 s−1 in 150 mm KCl. These results suggested that the folding rate and folding probability of the Bcl2-2345 G4 decreased when the potassium concentration increased.
We next analyzed the unfolding and refolding kinetics of Bcl2-1245 G4 using the same force-ramp methods (Fig. 3, A–D). Due to the low refolding probability in 100 mm KCl (pfold ∼10% at 300 s; see Fig. 3D, open circle), we used 150 mm KCl, which showed a higher folding probability (pfold ∼27% at 300 s) than in 100 mm KCl but was still at a physiologically relevant potassium concentration (23). A total of 170 unfolding events were measured from more than 10 independent DNA tethers. The unfolding force distribution of the Bcl2-1245 G4 shows a single peak at 55 ± 6 pN (average ± S.D.). The best-fitting parameters for the Bcl2-1245 G4 were ku = (1.7 ± 0.9) × 10−7 s−1 and Δxu = 0.93 ± 0.04 nm (average ± S.E.) (Fig. 3B). The unfolding step size distribution also revealed a single peak at 30 ± 2 nt in 150 mm KCl (average ± S.D.), which is consistent with the number of nucleotides in the Bcl2-1245 G4 sequence (Fig. 3C). We also measured the average unfolding force and unfolding step size of Bcl2-1245 in 100 mm KCl, and the values of 47 ± 7 pN and 27 ± 7 nt (n = 12 from more than 7 beads), respectively, were similar to those measured in 150 mm KCl. Based on the time evolution of the refolding probability of Bcl2-1245 G4, the steady-state folding probability pst and the folding rate kf were determined to be ∼33% and 0.005 ± 0.001 s−1 for Bcl2-1245 in 150 mm KCl (Fig. 3D).
Figure 3.
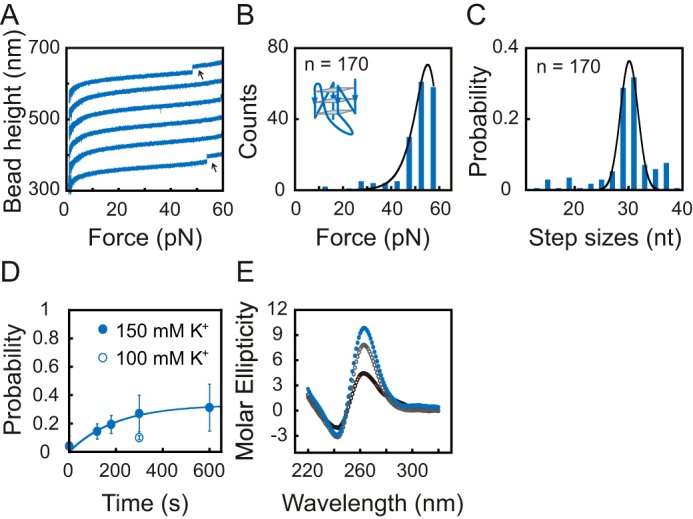
A, typical force–bead height curves of the Bcl2-1245 sequence in repeating stretching cycles in 150 mm KCl. The curves are shifted along the bead height axis for visual clarity. G4 unfolding results in a sudden bead height jump (black arrows). B, unfolding force histogram of the Bcl2-1245 G4. Data were fitted by Bell's model equation (solid line). C, unfolding step size histogram of the Bcl2-1245 G4. Data were fitted by a single-peak Gaussian equation (solid line) with a peak value of 30 ± 3 nt. D, the time evolution of the folding probability pfold(t) of Bcl2-1245 sequence in 100 mm KCl (open circle) and in 150 mm KCl (solid circles). E, CD spectra of Bcl2-1245 DNA prepared by thermal annealing in 150 mm KCl (solid blue circles), in situ refolding after adding 150 mm KCl for 1 min (open black circles), and 10 min (open gray circles) at room temperature.
The ∼33% of the steady-state folding probability of Bcl2-1245 G4 cannot be explained based on the reversible unfolding of Bcl2-1245 at 1 pN, as the unfolding rate was much lower than the folding rate. Based on the drastically different unfolding and refolding rates at 1 pN, one would expect ∼100% of the steady-state folding probability. To explain this unexpectedly low steady-state folding probability, we reasoned that after force-jumping to 1 pN, the unfolded ssDNA could refold into some metastable intermediate secondary structures, which prevents the formation of G4 over our experimental timescale for refolding (several minutes). To test this possibility, we compared the CD spectra of the Bcl2-1245 G4 prepared using fast in situ refolding induced by suddenly adding 150 mm KCl and thermal annealing by reducing the temperature gradually over several hours. The CD spectra (Fig. 3E) of the DNA sample prepared using a fast in situ refolding protocol at room temperature showed a lower peak at 260 nm than that prepared using a thermal annealing protocol, indicating that fewer G4 structures were formed by a fast in situ refolding protocol. Measuring the equilibrium folding probability of the Bcl2-1245 sequence may require a timescale longer than 10 min, which was difficult to achieve in repeating stretching cycles. This result supports our explanation of the low steady-state folding probability of the Bcl2-1245 G4.
It has been known that the WT Pu39 sequence can form multiple G4 structures, including Bcl2-2345 and Bcl2-1245 G4s, using different combinations of the G-tracts. To obtain new insights into the G-rich structures formed in the Pu39 sequence, we measured the distributions of unfolding forces and step sizes of the stable DNA secondary structures formed on the Pu39 sequence using the same force-ramp experiments at 2 pN/s in 100 mm KCl buffer (Fig. 4, A and B). The results show a broad distribution of unfolding forces and step sizes, which is consistent with a previous single-molecule force spectroscopy study (18). The broad distribution of unfolding forces and step sizes suggest that multiple stable DNA secondary structures form in the Pu39 sequence.
Figure 4.
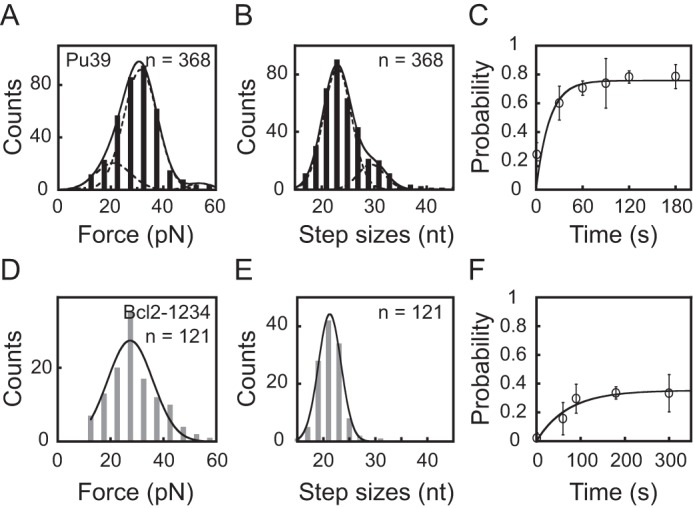
A, unfolding force histogram of the Pu39 sequence in 100 mm KCl. Data were fitted by a three-peak Gaussian equation (solid line) with peak values of 21 ± 7, 31 ± 6, and 54 ± 5 pN. B, unfolding step size histogram of the Pu39 sequence. Data were fitted by a two-peak Gaussian equation (solid line) with peak values of 23 ± 3 and 30 ± 2 nt. C, the time evolution of the folding probability pfold(t) of the Pu39 sequence. D, unfolding force histogram of Bcl2-1234 G4. Data were fitted by a single-peak Gaussian equation (solid line) with a peak value of 27 ± 9 pN. E, unfolding step size histogram of Bcl2-1234 G4. Data were fitted by a single-peak Gaussian equation (solid line) with a peak value of 21 ± 2 nt. F, the time evolution of the folding probability pfold(t) of the Bcl2-1234 sequence.
Assuming that the Bcl2-2345 and Bcl2-1245 G4s are the only two types of G4s that can be formed in the Pu39 sequence, we find that a two-species model with unfolding force peaks at 21 pN (Bcl2-2345) and 54 pN (Bcl2-1245) was unable to fit the experimental data obtained for the Pu39 sequence. The peak value of the Bcl2-1245 G4 was the average unfolding force of Bcl2-1245 G4 (54 ± 8 pN, n = 15) measured using the same conditions as the Pu39 sequence at 100 mm KCl and 2 pN/s loading rates. To explain the data, we used a three-species model that contained the contributions from Bcl2-2345, Bcl2-1245 G4s, and an unknown species to fit the data. Treating the relative abundance of the three species as well as the peak and variance of the unknown species as free parameters, the experimentally measured unfolding force distribution can be explained (black curve in Fig. 4A). Based on the areas occupied by these species, their fractions of occurrence were estimated to be 14% for Bcl2-2345 G4 unfolded at 21 ± 7 pN, 60% for the unknown species unfolded at 31 ± 6 pN, and 2% for Bcl2-1245 G4 unfolded at 54 ± 5 pN. Notably, the unknown species occupies more than half of the occurrence.
Fig. 4B shows the distribution of the number of released nucleotides during unfolding, which were fitted using a double-Gaussian model; one peaked at 23 ± 3 nt and the other peaked at 30 ± 2 nt, occupying 64 and 12%, respectively. It is expected that the unfolding of Bcl2-2345 G4 should release ∼23 nt; however, based on the unfolding force distribution, the occurrence probability for the Bcl2-2345 G4 is ∼14%. Therefore, 50% of the fraction that released ∼23 nt belongs to an unknown species. Based on the same calculation, 10% of the fraction that released ∼30 nt was from another unknown species. This result suggests that multiple unknown species formed in the Pu39 sequence. Fig. 4C shows that the pfold(t) of the Pu39 sequence reached a steady-state folding probability of 76% within 60 s with an apparent folding rate of 0.06 ± 0.03 s−1.
To identify the major unknown species occupying 50% of the fraction, we also measured the CD spectra of other truncated mutants, including all of the possible combinations of the four G-tracts in the Pu39 sequence in 100 mm KCl (Table S1 and Fig. S1). Our results show that, in addition to Bcl2-1356 and Bcl2-2356 (Table S1), 11 other truncated mutants also form G4 structures at room temperature (20–23 °C) (Fig. S1). By listing the sequences in the order of length (number of nucleotides) (Table S1), we found that within the 11 mutants, only Bcl2-1234 and Bcl2-3456 have lengths smaller than 30 nt and consist of the length of major unknown species (∼23 nt, ∼50% of the fraction). Our CD spectra show that both Bcl2-1234 and Bcl2-3456 sequences form hybrid-stranded G4 structures (Fig. S1). Using the same force-ramp experiments, we characterized the Bcl2-1234 and Bcl2-3456 G4s. The unfolding force distribution of Bcl2-1234 (Fig. 4D) showed a single peak at 27 ± 9 pN, which was similar to that of the unknown species (31 ± 6 pN). By fitting the data using Bell's model (Equation 1), we obtained ku = (0.9 ± 0.4) × 10−2 s−1 and Δxu = 0.5 ± 0.1 nm. The unfolding step size distribution of Bcl2-1234 (Fig. 4D) revealed a single peak at 21 ± 2 nt, which was also consistent with the unknown species in the Pu39 sequence. The involvement of G-tracts 1, 2, 3, and 4 also agrees well with previous DMS footprinting results (5, 10). Fig. 4F shows that the pfold(t) of Bcl2-1234 reached a steady-state folding probability of 35% within 100 s with an apparent folding rate of 0.014 ± 0.005 s−1. The refolding probability of Bcl2-3456 was extremely low (< 3% at 300 s, n = 97), suggesting that Bcl2-3456 is unlikely to be the major unknown species.
Discussion
In summary, we analyzed the folding/unfolding kinetics of Bcl2-2345, Bcl2-1245, and Bcl2-1234 G4s forming at the Pu39 region of the P1 promoter of the oncogene BCL-2. We showed that the hybrid-stranded Bcl2-2345 G4 is a kinetically favored structure with a faster folding and unfolding rates. In contrast, the parallel-stranded Bcl2-1245 G4 has much slower folding and unfolding rates. In addition to the previously reported Bcl2-2345 and Bcl2-1245 G4s, our results strongly suggested the existence of other stable secondary structures formed on the Pu39 sequence. We showed that a hybrid-stranded Bcl2-1234 G4 occupied 50% of the unfolding force and unfolding step size distribution of the Pu39 sequence. Together with previous DMS footprinting results (5, 10, 11), our results suggested that Bcl2-1234, Bcl2-2345, and Bcl2-1245 are likely to be three major G4 structures that co-exist and are interchangeable in the Pu39 region.
The unfolding rates of the hybrid-stranded Bcl2-2345 and Bcl2-1234 G4s are on the order of 10−2 s−1, which is similar to the hybrid-stranded telomeric G4 (22). The unfolding rate of Bcl2-1245 is on the order of 10−7 s−1, which is similar to the parallel-stranded Myc2345 G4 (24). The similar unfolding rates of different sequences with the same quadruplex topology suggest that the unfolding rates of G4s may be related to the quadruplex topology. To understand how the G4 topology determines the unfolding kinetics of intramolecular G4s, further single-molecule force spectroscopy and structural studies on various G4-forming sequences are still needed.
Despite the low occurrence probability, the parallel-stranded Bcl2-1245 G4 has the slowest unfolding rate compared with other structures formed in the Pu39 sequence. Such a slow unfolding rate is potentially important for suppressing transcription. Interestingly, a broad class of promoter G4-forming sequences containing two G3NG3 motifs (three guanines linked with a single nucleotide loop) and different lengths of the middle loop, such as the promoter of oncogenes VEGF (25, 26), HIF-1α (27), and c-KIT (28, 29), have been shown to prefer to form parallel-stranded G4 structures (30). These parallel-stranded G4 structures likely share a common feature of the ultra-slow unfolding rate for transcription suppression.
In addition, we also found that the unfolding force distribution of Bcl2-2345 and Bcl2-1245 G4s was insensitive to the potassium concentration in the range of 100–150 mm, but the folding rates and the folding probability of Bcl2-2345 and Bcl2-1245 sequences showed significant differences in 100 and 150 mm KCl. A lower folding probability was observed for the hybrid-stranded Bcl2-2345 G4 in 150 mm than in 100 mm KCl, whereas the parallel-stranded Bcl2-1245 G4 showed a higher folding probability in 150 mm than in 100 mm KCl. These results suggested that the potassium concentration may have different effects on the folding rates of G4s with different topologies.
Experimental procedures
Oligonucleotides and DNA preparation
Oligonucleotides for single-molecule experiments were purchased from Integrated DNA Technologies (Coralville, IA) and Sangon Biotech (Shanghai, China). The 5′-thiol–labeled 489-bp and 5′-biotin–labeled 601-bp dsDNA handles were prepared by PCR using TaKaRa Ex Taq Polymerase (Takara, Beijing, China) and λ-phage DNA as the template (Thermo Scientific). PCR products were purified by a Universal DNA Purification Kit (TIANGEN Biotech, Beijing, China) and digested with the restriction enzyme BstXI (Thermo Scientific). The G4-forming oligonucleotides were annealed with two flanking sequences and ligated with two dsDNA handles. The ligated DNA products were purified by gel extraction using a TIANgel Midi Purification Kit (TIANGEN Biotech).
CD spectroscopy
Oligonucleotides for CD experiments were purchased from Sangon Biotech. Oligonucleotides, at a final concentration of 10 μm, were suspended in 10 mm Tris-HCl, pH 8.0, buffer with 100 or 150 mm KCl. The DNA samples were heated at 95 °C for 5 min followed by a slow cooling process to room temperature over several hours. The CD spectra were collected on a Jasco-810 spectropolarimeter using five scans from 220 to 320 nm at room temperature of 20–23 °C. A quartz cell with a 1-mm optical path length was used. A buffer baseline was collected in the same quartz cell and was subtracted from the sample spectra.
Single-molecule magnetic tweezers experiments
A flow chamber was constructed on a (3-aminopropyl)triethoxysilane–modified coverslip (Sigma-Aldrich, Shanghai China). The thiol-labeled DNA construct was covalently attached to the amine group of the (3-aminopropyl)triethoxysilane–modified coverslip via a Sulfo-SMCC cross-linker (Thermo Scientific). The flow chamber was blocked with BSA solution (10 mg/ml BSA (Sigma-Aldrich), 1 mm 2-mercaptoethanol, 1× PBS buffer, pH 7.4). The 2.8-μm-diameter streptavidin-coated paramagnetic beads (Dynal M280, Thermo Fisher Scientific) were introduced to attach to the biotin-labeled 5′-end of the DNA construct. Magnetic tweezers experiments were carried out at room temperature of 20–23 °C in a 10 mm Tris-HCl, pH 8.0, buffer with 100 mm KCl. The 2 pN/s loading rate was used for Bcl2-2345, Pu39, and Bcl2-1234 sequences. Due to the low folding probability of Bcl2-1245 in the 100 mm KCl buffer (lower than 10% at 300 s), a higher 150 mm KCl concentration and 0.2 pN/s loading rate were used for the Bcl2-1245 unfolding force and folding probability measurements. A slow loading rate can ensure that unfolding would occur at forces below 60 pN, thus avoiding interference from the DNA overstretching transitions of the dsDNA handles that typically occur at >60 pN (31). The magnetic tweezers were controlled by the LabVIEW program (National Instruments).
Unfolding step size calculation
The unfolding step size determination was based on a noise-beating step detection algorithm as described in our previous study (22). The step sizes in nanometers were converted to the number of nucleotides released based on the force–extension curves for ssDNA described by Cocco et al. (32),
| (Eq. 2) |
with the following parameters: h = 0.34 nm, a1 = 0.21, a2 = 0.34, f1 = 0.0037 pN, f2 = 2.9 pN, f3 = 8000 pN, and a3 = 2.1 ln([K+]/0.0025)/ln(0.15/0.0025) − 0.1, where [K+] is the salt concentration.
Author contributions
Y. C., J. Y., and H. Y. conceptualization; Y. C., Y. L., C. Z., and H. Y. data curation; Y. C., Y. L., and H. Y. formal analysis; Y. C. and H. Y. validation; Y. C., Q. T., Y. L., Y. Z., C. Z., J. Y., and H. Y. investigation; Y. C., Q. T., Y. L., Y. Z., C. Z., J. Y., and H. Y. methodology; Y. C., J. Y., and H. Y. writing-original draft; Y. C. and H. Y. project administration; Y. C., Y. L., J. Y., and H. Y. writing-review and editing; J. Y. and H. Y. supervision; J. Y. and H. Y. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Danzhou Yang and Dr. Xinghua Zhang for advice and suggestions.
This work was supported by National Natural Science Foundation of China Grant 21708009 (to H. Y.); Fundamental Research Fund for the Central Universities Grant 2017KFYXJJ153 (to H. Y.); National Research Foundation, Prime Minister's Office, Singapore and the Ministry of Education under the Research Centres of Excellence programme (to J. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Fig. S1.
- G4
- G-quadruplex
- ssDNA
- single-stranded DNA
- nt
- nucleotides
- pN
- piconewtons
- DMS
- dimethyl sulfate.
References
- 1. McDonnell T. J., Troncoso P., Brisbay S. M., Logothetis C., Chung L. W., Hsieh J. T., Tu S. M., and Campbell M. L. (1992) Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res. 52, 6940–6944 [PubMed] [Google Scholar]
- 2. Pezzella F., Turley H., Kuzu I., Tungekar M. F., Dunnill M. S., Pierce C. B., Harris A., Gatter K. C., and Mason D. Y. (1993) bcl-2 protein in non-small-cell lung carcinoma. N. Engl. J. Med. 329, 690–694 10.1056/NEJM199309023291003 [DOI] [PubMed] [Google Scholar]
- 3. Joensuu H., Pylkkänen L., and Toikkanen S. (1994) Bcl-2 protein expression and long-term survival in breast cancer. Am. J. Pathol. 145, 1191–1198 [PMC free article] [PubMed] [Google Scholar]
- 4. Reed J. C., Kitada S., Takayama S., and Miyashita T. (1994) Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin's lymphoma and lymphocytic leukemia cell lines. Ann. Oncol. 5, Suppl. 1, 61–65 [DOI] [PubMed] [Google Scholar]
- 5. Dexheimer T. S., Sun D., and Hurley L. H. (2006) Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 128, 5404–5415 10.1021/ja0563861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Young R. L., and Korsmeyer S. J. (1993) A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol. Cell. Biol. 13, 3686–3697 10.1128/MCB.13.6.3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heckman C., Mochon E., Arcinas M., and Boxer L. M. (1997) The WT1 protein is a negative regulator of the normal bcl-2 allele in t(14;18) lymphomas. J. Biol. Chem. 272, 19609–19614 10.1074/jbc.272.31.19609 [DOI] [PubMed] [Google Scholar]
- 8. Wang X. D., Ou T. M., Lu Y. J., Li Z., Xu Z., Xi C., Tan J. H., Huang S. L., An L. K., Li D., Gu L. Q., and Huang Z. S. (2010) Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindoline derivatives. J. Med. Chem. 53, 4390–4398 10.1021/jm100445e [DOI] [PubMed] [Google Scholar]
- 9. Dai J., Dexheimer T. S., Chen D., Carver M., Ambrus A., Jones R. A., and Yang D. (2006) An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 128, 1096–1098 10.1021/ja055636a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agrawal P., Lin C., Mathad R. I., Carver M., and Yang D. (2014) The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J. Am. Chem. Soc. 136, 1750–1753 10.1021/ja4118945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Onel B., Carver M., Wu G., Timonina D., Kalarn S., Larriva M., and Yang D. (2016) A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc. 138, 2563–2570 10.1021/jacs.5b08596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui Y., Koirala D., Kang H., Dhakal S., Yangyuoru P., Hurley L. H., and Mao H. (2014) Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL. Nucleic Acids Res. 42, 5755–5764 10.1093/nar/gku185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amato J., Pagano A., Capasso D., Di Gaetano S., Giustiniano M., Novellino E., Randazzo A., and Pagano B. (2018) Targeting the BCL2 gene promoter G-quadruplex with a new class of furopyridazinone-based molecules. ChemMedChem 13, 406–410 10.1002/cmdc.201700749 [DOI] [PubMed] [Google Scholar]
- 14. Falanga A. P., Cerullo V., Marzano M., Feola S., Oliviero G., Piccialli G., and Borbone N. (2019) Peptide Nucleic Acid-functionalized adenoviral vectors targeting G-quadruplexes in the P1 promoter of Bcl-2 proto-oncogene: a new tool for gene modulation in anti-cancer therapy. Bioconj. Chem. 30, 572–582 10.1021/acs.bioconjchem.8b00674 [DOI] [PubMed] [Google Scholar]
- 15. Lynch S., Baker H., Byker S. G., Zhou D., and Sinniah K. (2009) Single molecule force spectroscopy on G-quadruplex DNA. Chemistry 15, 8113–8116 10.1002/chem.200901390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu Z., Schonhoft J. D., Dhakal S., Bajracharya R., Hegde R., Basu S., and Mao H. (2009) ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 131, 1876–1882 10.1021/ja806782s [DOI] [PubMed] [Google Scholar]
- 17. Long X., Parks J. W., Bagshaw C. R., and Stone M. D. (2013) Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 41, 2746–2755 10.1093/nar/gks1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui Y., Kong D., Ghimire C., Xu C., and Mao H. (2016) Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry 55, 2291–2299 10.1021/acs.biochem.6b00016 [DOI] [PubMed] [Google Scholar]
- 19. You H., Le S., Chen H., Qin L., and Yan J. (2017) Single-molecule manipulation of G-quadruplexes by magnetic tweezers. J. Vis. Exp. 127, e56328 10.3791/56328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H., Fu H., Zhu X., Cong P., Nakamura F., and Yan J. (2011) Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys. J. 100, 517–523 10.1016/j.bpj.2010.12.3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao X., Zeng X., Lu C., and Yan J. (2017) Studying the mechanical responses of proteins using magnetic tweezers. Nanotechnology 28, 414002 10.1088/1361-6528/aa837e [DOI] [PubMed] [Google Scholar]
- 22. You H., Zeng X., Xu Y., Lim C. J., Efremov A. K., Phan A. T., and Yan J. (2014) Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 42, 8789–8795 10.1093/nar/gku581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linzell J. L., and Peaker M. (1971) Intracellular concentrations of sodium, potassium and chloride in the lactating mammary gland and their relation to the secretory mechanism. J. Physiol. 216, 683–700 10.1113/jphysiol.1971.sp009547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. You H., Wu J., Shao F., and Yan J. (2015) Stability and kinetics of c-MYC promoter G-quadruplexes studied by single-molecule manipulation. J. Am. Chem. Soc. 137, 2424–2427 10.1021/ja511680u [DOI] [PubMed] [Google Scholar]
- 25. Sun D., Guo K., Rusche J. J., and Hurley L. H. (2005) Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 33, 6070–6080 10.1093/nar/gki917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agrawal P., Hatzakis E., Guo K., Carver M., and Yang D. (2013) Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 41, 10584–10592 10.1093/nar/gkt784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Armond R., Wood S., Sun D., Hurley L. H., and Ebbinghaus S. W. (2005) Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry 44, 16341–16350 10.1021/bi051618u [DOI] [PubMed] [Google Scholar]
- 28. Rankin S., Reszka A. P., Huppert J., Zloh M., Parkinson G. N., Todd A. K., Ladame S., Balasubramanian S., and Neidle S. (2005) Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 127, 10584–10589 10.1021/ja050823u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsu S. T., Varnai P., Bugaut A., Reszka A. P., Neidle S., and Balasubramanian S. (2009) A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 131, 13399–13409 10.1021/ja904007p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y., and Yang D. (2012) Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. Chapter 17, Unit 17.5 10.1002/0471142700.nc1705s50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X., Qu Y., Chen H., Rouzina I., Zhang S., Doyle P. S., and Yan J. (2014) Interconversion between three overstretched DNA structures. J. Am. Chem. Soc. 136, 16073–16080 10.1021/ja5090805 [DOI] [PubMed] [Google Scholar]
- 32. Cocco S., Yan J., Léger J. F., Chatenay D., and Marko J. F. (2004) Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70, 011910 10.1103/PhysRevE.70.011910 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


