Abstract
Escherichia coli and Klebsiella pneumoniae are opportunistic pathogens that are commonly associated with infections at mucosal surfaces, such as the lung or the gut. The host response against these types of infections includes the release of epithelial-derived antimicrobial factors such as lipocalin-2 (LCN-2), a protein that specifically inhibits the iron acquisition of Enterobacteriaceae by binding and neutralizing the bacterial iron-scavenging molecule enterobactin. Regulation of epithelial antimicrobial responses, including the release of LCN-2, has previously been shown to depend on IL-22, a cytokine produced by innate lymphoid cells type 3 (ILC3) during Enterobacteriaceae infections. However, much remains unknown about the extent to which antimicrobial responses are regulated by IL-22 and how IL-22 regulates the expression and production of LCN-2 in intestinal epithelial cells (IECs). Our study demonstrates how IL-22–induced activation of STAT3 synergizes with NF-κB–activating cytokines to enhance LCN-2 expression in human IECs and elucidates how ILC3 are involved in LCN-2–mediated host defense against Enterobacteriaceae. Together, these results provide new insight into the role of ILC3 in regulating LCN-2 expression in human IECs and could prove useful in future studies aimed at understanding the host response against Enterobacteriaceae as well as for the development of antimicrobial therapies against Enterobacteriaceae-related infections.
Keywords: innate immunity, antimicrobial peptide (AMP), microbiology, mucosal immunology, signal transduction, interleukin-22, STAT3
Introduction
Enterobacteriaceae is a large family of Gram-negative bacteria that are commonly found as part of the human intestinal microbiota and include species such as Escherichia coli and Klebsiella pneumoniae (1, 2). Although intestinal colonization by these bacteria is often asymptomatic in healthy individuals, they pose a major threat to immunocompromised patients, where colonization can lead to intestinal infection, but also infections at other mucosal surfaces, such as the lung or the urinary tract (3–6). Intestinal epithelial cells (IECs)2 play a crucial role in the protection against such infections by producing a variety of antimicrobial factors (7). For instance, regenerating islet-derived protein 3γ (REG3γ), which targets both Gram-positive and Gram-negative bacteria, is produced by epithelial cells and prevents intestinal microbes from penetrating the sterile mucous layer separating the microbiota from host tissues (8). Alternatively, epithelial-derived lipocalin-2 (LCN-2) provides protection by binding and neutralizing enterobactin, an important iron-scavenging molecule expressed specifically by Enterobacteriaceae (9–12). Importantly, although some of these antimicrobial factors are constitutively expressed, others can be affected by a variety of host and/or microbial factors, which allows regulation of antimicrobial responses during homeostasis and infection (13–16).
One of the cell types that plays an important role in the regulation of antimicrobial responses in IECs are innate lymphoid cells type 3 (ILC3) (17, 18). These cells are tissue-resident under homeostatic conditions (19–21) and are marked by the expression of transcription factors Retinoic Acid-Related (RAR)-related orphan receptor-γ (22) and aryl hydrocarbon receptor (23). ILC3 are activated by IL-1β and IL-23, which are produced by macrophages and dendritic cells during Enterobacteriaceae infections (24). As a response, ILC3 produce a variety of cytokines, including IL-22, IL-17A, TNF, and GM-CSF (25, 26), which gives them a similar cytokine profile as TH17 and TH22 cells (27) and likely explains the partial redundancy of these cell types during infections (28–30). Of the cytokines produced by ILC3, IL-22 has received much attention due to its importance in the protection against Enterobacteriaceae infections (25, 31–37) and was found to be produced in large amounts by intestinal and tonsillar NKp44+ ILC3 (38). IL-22 signals through a dimer of the IL10R2 and IL-22R1 receptors, the latter of which is exclusively expressed on nonhematopoietic cells (39). Upon activation, this receptor complex has been described to activate STAT3 through classical JAK–STAT signaling events (39) and influences a variety of epithelial host–defense programs, such as the expression of tight-junction proteins (40) and the production of antimicrobial factors, including LCN-2, Reg3β, and Reg3γ (35–37). Importantly, clinical studies have shown that patients suffering from ulcerative colitis have increased expression of the IL-22–binding protein, a factor that neutralizes IL-22 activity (41), whereas perturbed IL-22 expression in the inflamed mucosa of Crohn's disease patients was linked to disturbed regulation of antimicrobial factors and tight junctions in epithelial cells (42). These findings demonstrate the importance of IL-22 in the regulation of antimicrobial responses in the gut and the promotion of intestinal health in humans.
Nevertheless, although it is clear that ILC3 and IL-22 play an important role in countering intestinal Enterobacteriaceae infections, much remains unclear about the pathways that connect the activated ILC3 with the actual antimicrobial responses against these opportunistic pathogens. In this study, we aimed to uncover how ILC3-derived IL-22 regulates the antimicrobial response in IECs and which other factors are required to provide optimal IL-22–mediated protection against infections by Enterobacteriaceae.
Results
Synergy between cytokines in the up-regulation of antimicrobial gene expression in human IECs
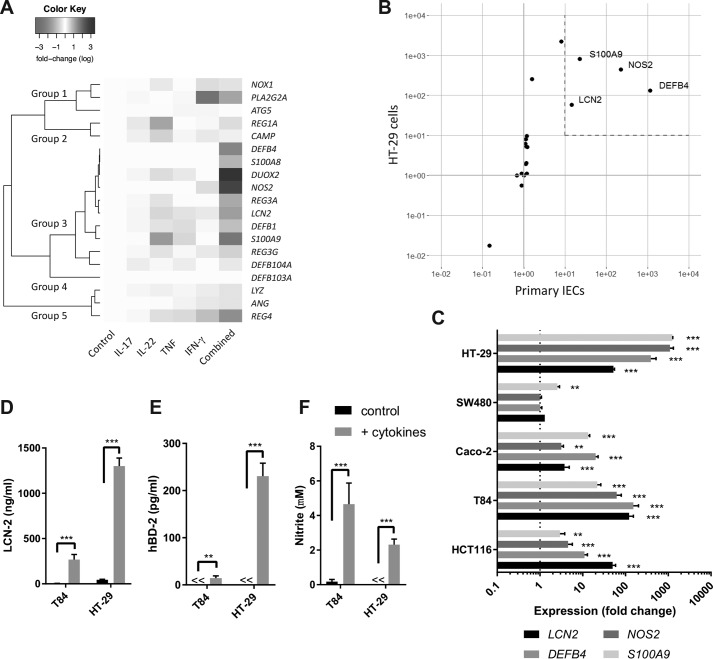
To better understand the role of IL-22 in the regulation of antimicrobial responses in human IECs, we aimed to identify how IL-22 affects antimicrobial gene expression in the context of other cytokines commonly expressed during Enterobacteriaceae infection, namely IL-17A, TNF, and IFN-γ (17, 24, 46–49). To this end, the human colonic epithelial cell line HT-29 was stimulated for 24 h with either IL-22, IL-17A, TNF, or IFN-γ alone, as well as in combination. Next, gene expression of 19 antimicrobial effector genes that have previously been associated with antimicrobial responses in the gut was assessed by qPCR (Tables S1 and S2). Expression patterns were grouped by hierarchical clustering, which yielded five distinct groups (Fig. 1A). Genes in group 1 showed the strongest up-regulation in response to IFN-γ, whereas genes in group 2 showed the strongest up-regulation in response to IL-22. Group 3 was the largest group and consisted of 10 genes that showed the strongest up-regulation in response to the combination of all four cytokines. Group 4 consisted of only DEFB103A, of which no expression was detectable in either stimulated or unstimulated samples. Finally, group 5 consisted of genes mostly down-regulated in response to stimulation, particularly in response to all four cytokines. Of all the single cytokine stimulations, IL-22 appeared most potent in up-regulating gene expression, with significant up-regulation of S100A9, REG1A, CAMP, and LCN2 (Fig. S1A). TNF and IFN-γ only significantly up-regulated S100A9 and PLA2G2A, respectively (Fig. S1, B and C), whereas IL-17A was unable to significantly up-regulate any of the tested genes (Fig. S1D). Combined cytokine treatment, however, was most potent and significantly up-regulated nine genes, which all clustered together in group 3 (Fig. S1E). Together, these results confirmed the importance of IL-22 in the regulation of antimicrobial responses in IECs and demonstrated that synergy is important in the cytokine-mediated up-regulation of the majority of antimicrobial genes tested in this study.
Figure 1.
Cytokine-mediated antimicrobial gene and protein expression in human IECs. A, HT-29 cells were stimulated for 24 h with IL-17A (50 ng/ml), IL-22 (50 ng/ml), TNF (20 ng/ml), IFN-γ (10 ng/ml), or a combination of these cytokines after which gene expression for the indicated genes was determined by qPCR. Subsequent hierarchical clustering was performed based on the calculated Pearson distance between genes. Colors represent average fold change on log scale compared with untreated controls based on three independent experiments. B, HT-29 cells and primary human CD45-depleted intestinal cells were stimulated for 24 h with IL-17A (50 ng/ml), IL-22 (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) after which gene expression for 19 antimicrobial genes was determined by qPCR. For HT-29 cells, values represent average fold change compared with no stimulation based on three independent experiments. For primary IECs, values indicate average fold change compared with no stimulation for three individual patient samples. C, HT-29, SW480, Caco-2, T84, and HCT116 cells were stimulated for 24 h with IL-17A (50 ng/ml), IL-22 (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) after which gene expression for the indicated genes was determined by qPCR. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Dunnett's post hoc test as compared with the unstimulated cells (expression value of 1). D–F, HT-29 and T84 cells were stimulated for 24 h with IL-17A (50 ng/ml), IL-22 (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) after which hBD-2 and LCN-2 concentrations in supernatant were determined by ELISA, and nitrite concentrations were determined by Griess assay. Values indicate the mean with S.E. of three independent experiments. Statistical analysis performed with two-way ANOVA followed by Šidák's post hoc test. p < 0.005 = **, and p < 0.0005 = ***.
To compare the transcriptional response of HT-29 cells with the transcriptional response of primary cells, human colonic IECs were isolated from nonaffected tissue of colonic cancer resection material (Fig. S2) and stimulated with a combination of IL-22, IL-17A, TNF, and IFN-γ, after which gene expression was compared with identically stimulated HT-29 cells (Fig. 1B and Fig. S1, E and F). This showed that LCN2, NOS2, DEFB4, and S100A9 were significantly up-regulated >10-fold in both primary and HT-29 cells. In addition, other human IEC cell lines responded in a similar fashion to stimulation with IL-22, IL-17A, TNF, and IFN-γ, with significant up-regulation of LCN2, DEFB4, and NOS2 (4/5 cell lines) as well as significant up-regulation of S100A9 (5/5 cell lines) (Fig. 1C). Next, we assessed whether the increased expression of these four genes also resulted in the production and release of the antimicrobial factors. Release of LCN-2 was strongly induced in both HT-29 and T84 cells upon stimulation, reaching high concentrations close to 1 μg/ml (Fig. 1D). In addition, both hBD-2 (encoded by DEFB4) and NO release were significantly increased upon cytokine stimulation in both cell lines (Fig. 1, E and F). Surprisingly, despite the strong up-regulation of S100A9 in both cell lines, neither S100A9 nor S100A8/A9 (calprotectin heterodimer) could be detected in cell supernatants (data not shown). Together, our results demonstrate that LCN2, DEFB4, NOS2, and S100A9 are consistently up-regulated on a transcriptional level in both human IEC cell lines and primary human IECs upon stimulation with IL22, IL-17A, TNF, and IFN-γ and that hBD-2, LCN-2, and NO concentrations are elevated in supernatants in response to these cytokines.
IL-22 and IFN-γ regulate distinct antimicrobial genes in the context of IL-17A and TNF
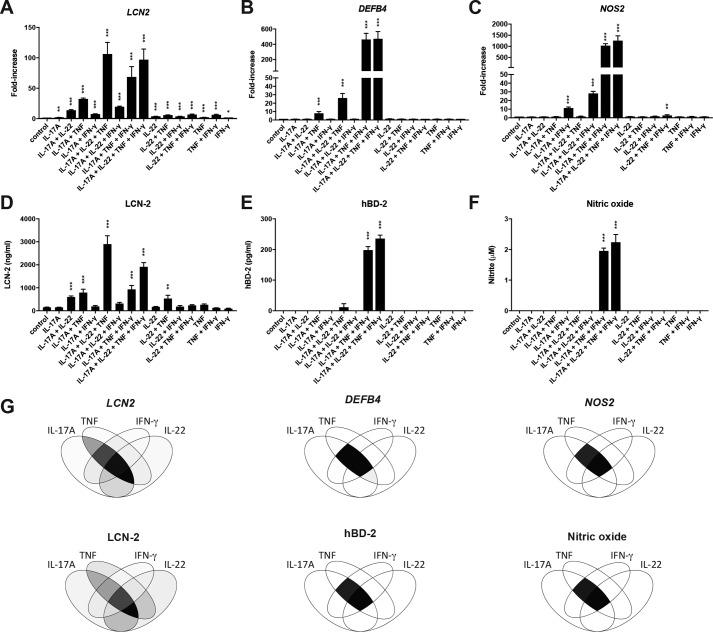
To better define which of the previously mentioned cytokines are specifically required for the optimal up-regulation of the different antimicrobial genes, HT-29 cells were stimulated with all possible cytokine combinations after which gene expression for LCN2, NOS2, and DEFB4 as well as release of LCN-2, NO, and hBD-2 were determined. LCN2 gene expression (Fig. 2A), as well as release of the LCN-2 protein (Fig. 2B), showed strong up-regulation in response to cytokine combinations containing both IL-17A and TNF. Interestingly, whereas IL-22 alone only had a minor impact on LCN2 expression and no effect on LCN-2 protein release, addition of IL-22 to the IL-17A/TNF combination resulted in the strongest up-regulation on both mRNA and protein levels. In addition, combining IL-22 plus TNF as well as IL-22 plus IL-17A also significantly increased both gene expression and protein release. In contrast, IFN-γ only enhanced IL-17A/TNF-induced LCN2 expression, albeit not as efficiently as IL-22, and did not have a positive effect on gene expression or protein release in any other combination. Expression of DEFB4 was significantly up-regulated in response to IL-17A/TNF (Fig. 2C). However, this stimulation did not lead to detectable hBD-2 protein levels in the cell supernatant (Fig. 2D). Addition of IFN-γ to IL-17A/TNF strongly enhanced the expression of DEFB4 and also led to a detectable increase in hBD-2 levels in the cell supernatant. In contrast, IL-22 appeared to only have a limited impact on both DEFB4 expression and hBD-2 release when combined with IL-17A/TNF. Finally, NOS2 expression was significantly increased in response to IL-17A plus IFN-γ and was strongly enhanced by further addition of TNF (Fig. 2E). IL-22 was able to slightly improve NOS2 expression in combination with IL-17A plus IFN-γ, but overall played a minor role in the regulation NOS2. Similar to hBD-2, NO production was only detectable when IL-17A, TNF, and IFN-γ were combined (Fig. 2F). Overall, these experiments point toward an important role for IL-17A and TNF in the regulation of all three antimicrobial factors, whereas IL-22 and IFN-γ appear to be more specific, with IL-22 strongly affecting LCN-2 expression and IFN-γ most strongly affecting expression of hBD-2 and NOS2 (Fig. 2G).
Figure 2.
Specific cytokine synergy regulates the expression and release of LCN-2, NO, and hBD-2. A–F, HT-29 cells were stimulated for 24 h with all possible combinations of IL-22 (50 ng/ml), IL-17A (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) after which qPCR was performed to determine LCN2 (A) DEFB4 (B), and NOS2 (C) expression. Protein levels for LCN-2 (D) and hBD-2 (E) were determined in supernatants by ELISA, whereas nitrite levels (F) were determined by Griess assay. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with one-way ANOVA followed by Dunnett's post hoc test as compared with unstimulated cells. G, Venn diagrams representing gene expression levels (left) and protein/nitrite levels (right) for different cytokine combinations. Color intensity corresponds to the values in A–F redistributed on a gray scale from 0 to 255. p < 0.05 = *, p < 0.005 = **, and p < 0.0005 = ***.
IEC-mediated inhibition of K. pneumoniae growth corresponds to LCN-2 production
Our results so far indicate that high concentrations of LCN-2 can be produced upon IEC activation with IL-22, IL-17A, and TNF. To determine whether this increased LCN-2 production could inhibit the growth of Enterobacteriaceae, we assessed the antimicrobial activity of HT-29 cell supernatant against two K. pneumoniae strains. The first strain expressed enterobactin (Ent), yersiniabactin (Yer), and salmochelin (Sal), whereas the second strain is an isogenic mutant only expressing Ent. Whereas these three factors are all iron-scavenging siderophores, only the activity of enterobactin is inhibited by LCN-2. Thus, expression of alternative siderophores such as Yer and Sal provides resistance against LCN-2 (45). Growth analysis of both strains in RPMI 1640 medium showed no significant difference in bacterial growth rates (Table 1). When grown in supernatant from unstimulated HT-29 cells, the Ent+ KP20 strain appeared to have a slight growth disadvantage compared with the Ent+/Yer+/Sal+ KP4 strain, albeit not significant. When grown in supernatant from IL-22, IL-17A, and TNF-stimulated cells however, the growth rate of the KP20 strain was significantly lower compared with the KP4 strain (p = 0.01), which is in line with the increased LCN-2 concentrations. Furthermore, the addition of an excess of iron, which renders LCN-2 ineffective, abolished the difference in growth rates between both strains (Table 1). Together, this indicates that LCN-2 produced by stimulated HT-29 cells is capable of inhibiting growth of LCN-2 susceptible K. pneumoniae.
Table 1.
Growth of K. pneumoniae in cell supernatants
Hill slope values ± S.E. were calculated from four-parameter fit models of bacterial growth curves. Hill slopes represent the estimated maximum growth rate during log phase, with higher values representing more rapid growth. Values indicate the mean with S.E. of three to four independent experiments.
| Media | Ent+/Yer+/Sal+ | Ent+ | p value |
|---|---|---|---|
| RPMI 1640 | 5.20 ± 0.28 | 5.19 ± 0.25 | 0.98 |
| Cell supernatant (control) | 4.97 ± 0.15 | 4.71 ± 0.08 | 0.22 |
| Cell supernatant + IL-22, IL-17A, TNF | 5.27 ± 0.10 | 4.69 ± 0.08 | 0.01a |
| Cell supernatant + IL-22, IL-17A, TNF + 5 μm Fe(III)SO4 | 9.13 ± 0.29 | 9.22 ± 0.57 | 0.90 |
a Statistical analysis was performed with two-tailed unpaired t tests. p < 0.05.
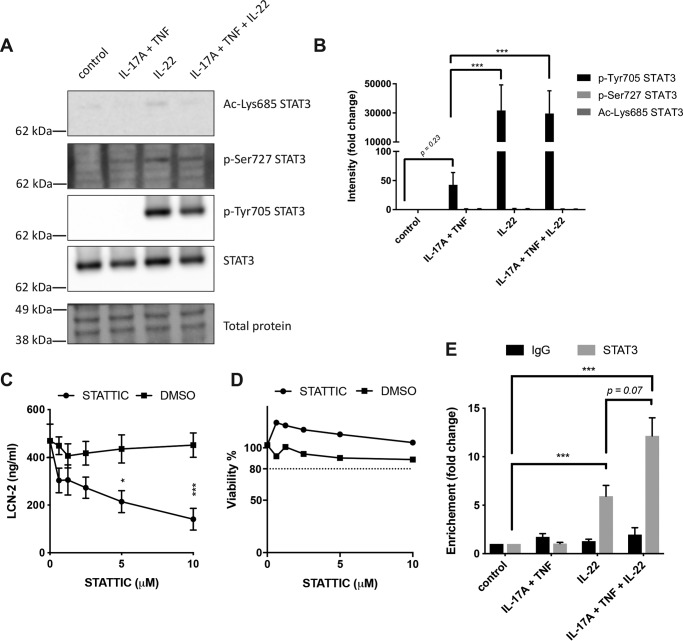
IL-22 promotes STAT3 activation and recruitment of STAT3 to the LCN2 promoter region
Activation of STAT3 has previously been described to be one of the main intracellular signaling events involved in IL-22–induced responses (39). To determine whether STAT3 signaling is involved in the up-regulation of LCN-2 in human IECs, Western blot analysis was performed. This showed that stimulation of HT-29 cells with IL-22 alone or in combination with IL-17A/TNF activated STAT3 through phosphorylation of Tyr-705 (Fig. 3, A and B), whereas stimulation with only IL-17A/TNF did not have a significant impact on Tyr-705 phosphorylation. In contrast, no significant changes were observed for phosphorylation of Ser-727 or acetylation of Lys-685. To determine the importance of STAT3 activation in LCN-2 production, we used STATTIC to inhibit STAT3 activation in HT-29 cells. This showed that the up-regulation of LCN-2 observed in IL-22, IL-17A, and TNF-stimulated cells was inhibited in a dose-dependent manner by STATTIC, albeit not completely (Fig. 3C). Importantly, no cytotoxic effects of the inhibitor were observed in a WST-1 assay (Fig. 3D). To determine whether activation of STAT3 results in the binding of STAT3 to the LCN2 promoter region, ChIP-qPCR analysis was performed. This showed that stimulation of HT-29 cells with IL-22 promoted binding of STAT3 to the LCN2 promotor region (Fig. 3E). In addition, whereas stimulation with IL-17A/TNF did not have an effect on STAT3 binding to the LCN2 promotor, stimulation with IL-22 in combination with IL-17A/TNF further enhanced STAT3 binding compared with IL-22 stimulation alone. Together, these results demonstrate that IL-22 induces Tyr-705 phosphorylation of STAT3 and promotes the binding of STAT3 to the LCN2 promotor in human IECs. In addition, this binding can be enhanced upon stimulation with a combination of IL-22 and IL-17A/TNF, and inhibition of STAT3 signaling prevents the IL-22–mediated enhancement of LCN-2 production in combination with IL-17A/TNF.
Figure 3.
Involvement of STAT3 in IL-22, TNF, and IL-17A–mediated LCN-2 production. A and B, HT-29 cells were stimulated for 30 min with the indicated cytokines after which protein lysate was used for Western blot analysis to determine levels of pSTAT3–Tyr-705, pSTAT3–Ser-727, ac-STAT3–Lys-685, STAT3, and total protein. A, images of Western blottings representative of three independent experiments. B, band intensity quantification. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. C and D, HT-29 cells were stimulated for 24 h with IL-22 (50 ng/ml), IL-17A (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) in combination with indicated concentrations of STATTIC or DMSO control after which LCN-2 concentrations were determined by ELISA (C) and HT-29 viability was determined by WST-1 assay (D). Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Dunnett's post hoc test. E, HT-29 cells were stimulated with indicated cytokine combinations for 30 min after which ChIP-qPCR was performed using anti-STAT3 or control IgG. Fold-increase in enrichment is shown compared with unstimulated cells. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. p < 0.05 = *, p < 0.005 = **, and p < 0.0005 = ***. Molecular masses indicate the location of the closest protein ladder marker on the blot.
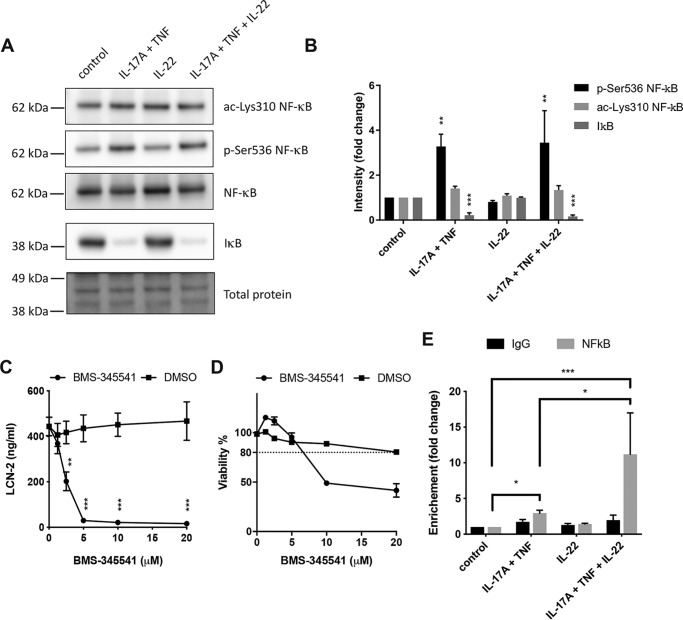
IL-17A and TNF induce NF-κB activation and NF-κB recruitment to the LCN2 promoter region
A previous study by Karlsen et al. (50) has shown that the combination of IL-17A and TNF induced LCN2 expression through activation of NF-κB in human lung epithelial cells. To determine the role of NF-κB in the regulation of LCN-2 in human IECs, HT-29 cells were stimulated with IL-22, IL-17A/TNF, or a combination of all three cytokines, after which NF-κB activation was assessed by Western blotting. Stimulation with IL-17A/TNF resulted in activation of the NF-κB pathway by both promoting IκB degradation as well as Ser-536 phosphorylation of NF-κB, while no changes in Lys-310 acetylation were observed (Fig. 4, A and B). Co-stimulation with IL-22 and IL-17A/TNF yielded a similar response that did not significantly differ from stimulation with IL-17A/TNF alone. In addition, IL-22 stimulation alone did not have any effect on NF-κB modifications nor IκB degradation. To verify that the activation of NF-κB was required for the release of LCN-2, the NF-κB inhibitor BMS-345541 was used to block NF-κB signaling during stimulation. BMS-345541 inhibited LCN-2 release in response to combined stimulation with IL-17A, TNF, and IL-22 and brought LCN-2 levels back to the basal expression levels of unstimulated cells (Fig. 4C). To verify that this reduction was not the result of toxic effects by the inhibitor, a WST-1 assay was performed, which showed that inhibition was reached at concentrations where no toxicity was observed (2.5–5 μm) (Fig. 4D). Finally, to assess whether NF-κB directly binds the LCN2 promotor and whether IL-22 might alter this binding, ChIP-qPCR analysis was performed. This showed that whereas stimulation with IL-17A/TNF induced the binding of NF-κB to the LCN2 promotor, more binding was observed upon stimulation with IL-17A, TNF, and IL-22 (Fig. 4E). In contrast, no significant NF-κB binding was observed upon stimulation with IL-22 alone. Together, these results show that IL-17A/TNF activated NF-κB signaling and promoted NF-κB binding to the LCN2 promotor. Moreover, co-stimulation with IL-22 and IL-17A/TNF further enhanced the binding of NF-κB to the LCN2 promotor region.
Figure 4.
Involvement of NF-κB in IL-22, TNF, and IL-17A-mediated LCN-2 production. A and B, HT-29 cells were stimulated for 30 min with the indicated cytokines after which protein lysate was used for Western blot analysis to determine levels of pNF-κB–Ser-536, ac-NF-κB–Lys-310, IκB, NF-κB, and total protein. A, images of Western blottings representative of three independent experiments. B, band intensity quantification. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. C and D, HT-29 cells were stimulated for 24 h with IL-22 (50 ng/ml), IL-17A (50 ng/ml), TNF (20 ng/ml), and IFN-γ (10 ng/ml) in combination with indicated concentrations of BMS-345541 or DMSO control after which LCN-2 concentrations were determined by ELISA (C) and HT-29 viability was determined by WST-1 assay (D). Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Dunnett's post hoc test. E, HT-29 cells were stimulated with indicated cytokine combinations for 30 min after which ChIP-qPCR was performed using anti-NF-κB or control IgG. Fold-increase in enrichment was compared with unstimulated cells. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. p < 0.05 = *, p < 0.005 = **, p < 0.0005 = ***. Molecular masses indicate the location of the closest protein ladder marker on the blot.
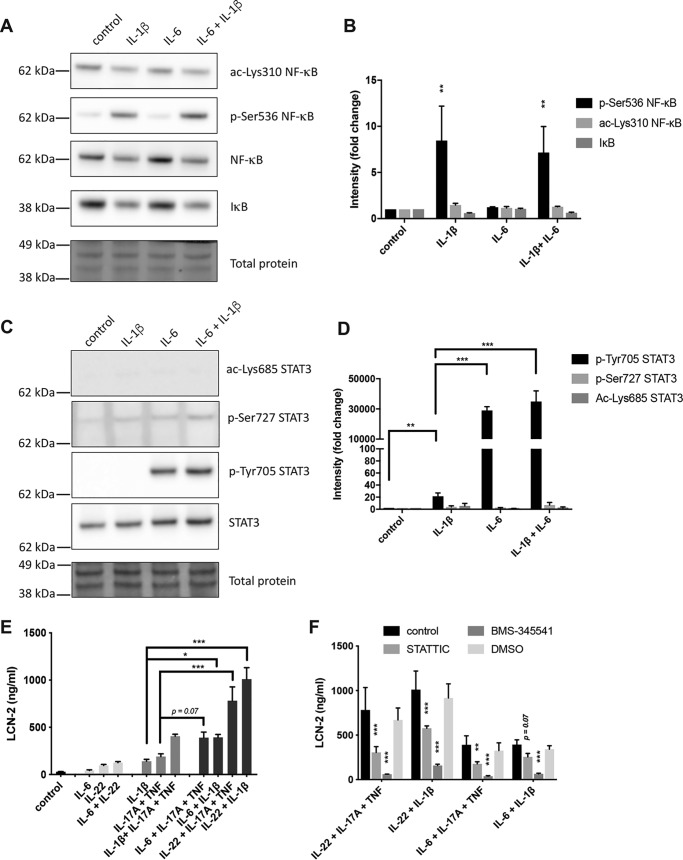
Alternative STAT3- and NF-κB–activating cytokines similarly enhanced LCN-2 expression
To determine whether the STAT3- and NF-κB–mediated LCN-2 production was a specific response to IL-22 and IL-17A/TNF, we sought to activate these pathways with two alternative cytokines, the NF-κB–activating cytokine IL-1β (51) and the STAT3-activating cytokine IL-6 (27). To confirm that these cytokines activate NF-κB and STAT3 in our model, Western blot analysis was performed on IL-1β, IL-6, and IL-1β/IL-6–stimulated HT-29 cells. This showed that IL-1β and IL-1β/IL-6 stimulation induced similar levels of Ser-536 NF-κB phosphorylation but were unable to significantly affect IκB degradation or Lys-310 acetylation (Fig. 5, A and B). In addition, IL-6 alone did not significantly affect IκB degradation nor any of the NF-κB modifications. Alternatively, IL-6 and IL-1β/IL-6 stimulation both increased Tyr-705 phosphorylation on STAT3 to a similar extent, while not affecting any of the other STAT3 modifications (Fig. 5, C and D). Notably, IL-1β alone was also capable of increasing STAT3 Tyr-705 phosphorylation, albeit to a much lower extent than IL-6 or IL-22. Next, HT-29 cells were stimulated with combinations of NF-κB inducers (IL-17A/TNF or IL-1β) and STAT3 inducers (IL-22 or IL-6) (Fig. 5E). Similar to stimulation with IL-17A/TNF, IL-1β was able to induce the release of LCN-2 protein, whereas IL-6 was unable to induce LCN-2 production, similar to IL-22. Stimulation with combinations of one NF-κB and one STAT3 activator led to increased LCN-2 production in all cases, especially in combinations containing IL-22. Interestingly, although a combination of IL-22 and IL-6 did not lead to strong LCN-2 production, combining IL-17A/TNF and IL-1β did increase LCN-2 production compared with IL-17A/TNF or IL-1β alone. Finally, to confirm that both STAT3 and NF-κB are involved in the synergistic activation of LCN-2, STATTIC and BMS-345541 were used to inhibit STAT3 and NF-κB, respectively. Inhibition with STATTIC resulted in partial inhibition of LCN-2 release induced by the different combinations of STAT3 and NF-κB inducers. In addition, inhibition with BMS-345541 completely inhibited LCN-2 release induced by these cytokine combinations (Fig. 5F). Together, these results indicated that alternative STAT3- and NF-κB–inducing cytokines appear to have a similar impact on LCN-2 production as IL-22 and IL-17A/TNF.
Figure 5.
Synergy between STAT3 and NF-κB inducers regulates LCN-2 production. A and B, HT-29 cells were stimulated for 30 min with the indicated cytokines after which protein lysate was used for Western blot analysis to determine levels of pNF-κB–Ser-536, ac-NF-κB–Lys-310, IκB, NF-κB, and total protein. A, images of Western blottings representative of three independent experiments. B, band intensity quantification. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. C and D, HT-29 cells were stimulated for 30 min with the indicated cytokines after which protein lysate was used for Western blot analysis to determine total levels of pSTAT3–Tyr-705, pSTAT3–Ser-727, ac-STAT3–Lys-685, STAT3, and total protein. C, images of Western blottings representative of two independent experiments. D, band intensity quantification. Values indicate the mean with S.E. of two independent experiments. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. E, HT-29 cells were stimulated for 24 h with the indicated combinations of IL-22 (50 ng/ml), IL-17A (50 ng/ml) + TNF (20 ng/ml), IL-6 (50 ng/ml), and IL-1β (50 ng/ml), after which LCN-2 concentrations were determined in the cell supernatant by ELISA. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with one-way ANOVA followed by Šidák's post hoc test. F, HT-29 cells were stimulated for 24 h with all indicated combinations of IL-22 (50 ng/ml), IL-17A (50 ng/ml) + TNF (20 ng/ml), IL-6 (50 ng/ml), and IL-1β (50 ng/ml) in combination with 5 μm BMS-345541, 2.5 μm STATTIC, or DMSO control, after which LCN-2 concentrations were determined in cell supernatant by ELISA. Values indicate the mean with S.E. of three independent experiments. Statistical analysis was performed with two-way ANOVA followed by Dunnett's post hoc test as compared with unstimulated cells. p < 0.05 = *, p < 0.005 = **, p < 0.0005 = ***. Molecular masses indicate the location of the closest protein ladder marker on the blot.
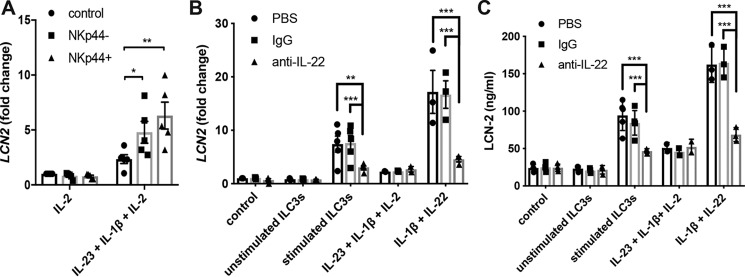
ILC3-derived IL-22 is involved in the regulation of LCN-2 in IECs
The stimulatory conditions used throughout this study relate to the epithelial cell microenvironment during infections. In this environment, ILC3 released IL-22, IL-17A, and TNF in response to the IL-23 and IL-1β produced by macrophages and dendritic cells, especially with NKp44+ ILC3 being marked as potent IL-22 producers (17, 25, 52). To mimic the interaction between IECs and ILC3 during infection, NKp44+ and NKp44− ILC3 were isolated from tonsil samples and stimulated with IL-2, IL-1β, and IL-23 to induce the release of IL-22. Subsequently, supernatants from these cells, containing both the IL-2, IL-1β, and IL-23 initially provided as well as IL-22 and other cytokines released from the ILC3 (25, 38), were used to stimulate HT-29 cells, after which LCN2 expression was determined. These results showed that whereas stimulation with IL-2, IL-1β, and IL-23 induced LCN-2 expression to some extent in HT-29 cells, both NKp44+ and NKp44− ILC3-derived supernatants further increased LCN-2 expression, with NKp44+ ILC3 appearing to be most potent (Fig. 6A). To determine whether this enhancement depended on the release of IL-22, a neutralizing IL-22 antibody was used to block IL-22–mediated signaling. Stimulation of HT-29 cells with ILC3 supernatant together with anti-IL-22 indeed reduced LCN2 expression (Fig. 6B) as well as LCN-2 protein release (Fig. 6C) compared with stimulation with IgG or PBS-supplemented ILC3 supernatant. In addition, the use of anti-IL-22 reduced LCN2 expression and LCN-2 protein release back to levels induced by IL-2, IL-1β, and IL-23 alone, which indicates that IL-22 is the main LCN-2–regulating factor released by the ILC3. Together, these results demonstrate the ability of ILC3 to regulate LCN-2 expression and production through IL-22 in human IECs.
Figure 6.
ILC3 regulates LCN-2 production in human IECs through IL-22 release. A, from patient samples (n = 5), isolated NKp44+ and NKp44− ILC3 were stimulated with 50 ng/ml IL-2, 50 ng/ml IL-1β, and 50 ng/ml IL-23 for 2 days, after which supernatant was collected separately for each patient sample to stimulate HT-29 cells for 24 h. RNA was isolated to determine LCN2 expression by qPCR. Graph shows the LCN2 response of HT-29 cells to individual supernatant samples as well as the mean with S.E. over all samples. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. B and C, from patient samples (n = 5), isolated ILC3 (combined NKp44+ and NKp44−) was stimulated with 50 ng/ml IL-2, 50 ng/ml IL-1β, and 50 ng/ml IL-23 for 2 days, after which supernatant was collected separately for each patient sample. HT-29 cells were stimulated for 24 h with indicated cytokines or ILC3 supernatant with added anti-IL-22 (25 μg/ml), control IgG (25 μg/ml), or PBS. B, RNA was isolated to determine LCN2 expression by qPCR, and C, supernatant was collected to determine LCN-2 concentrations in cell supernatant. Graphs show the LCN-2 response of HT-29 cells to individual supernatant samples as well as the mean with S.E. over all samples. Values indicate the mean with S.E. of three independent experiments. n = 3–5, except for IL-23 + IL-1β + IL-2, which is n = 2. Statistical analysis was performed with two-way ANOVA followed by Tukey's post hoc test. p < 0.05 = *, p < 0.005 = **, and p < 0.0005 = ***.
Discussion
IL-22 produced by ILC3 during intestinal infections is an important factor in the regulation of innate host defense mechanisms and has previously been reported for its protective function during Enterobacteriaceae infections (28, 31, 36, 53–57). This study provides a deeper understanding on how IL-22 in collaboration with other cytokines released during infection is involved in the synergistic regulation of a variety of antimicrobial genes in IECs, including LCN2, DEFB4, NOS2, and S100A9. In addition, our findings demonstrate how activation of ILC3 and the subsequent production of IL-22 influences LCN-2 production and how the STAT3 and NF-κB-dependent production of LCN-2 by IECs can provide protection against Enterobacteriaceae infections.
Screening the effects of IL-22, IL-17A, TNF, and IFN-γ on the expression of 19 antimicrobial genes showed that of these cytokines IL-22 was most effective in terms of the number of genes (4/19) that were significantly up-regulated upon stimulation with single cytokines. Combinational stimulation with IL-22, IL-17A, TNF, and IFN-γ, however, resulted in significant up-regulation of 9 of the 19 genes screened in HT-29 cells and induced significant up-regulation of LCN2, DEFB4, NOS2, and S100A9 in both HT-29 cells and primary human epithelial cells. Although the up-regulation of LCN2, DEFB4, and NOS2 corresponded to the increased release of LCN-2, hBD-2, and NO in HT-29 and T84 supernatants, no S100A9 or S100A8/A9 could be detected on a protein level, despite strong mRNA expression. This corresponds to a previous study that observed a lack of S100A9 protein release in murine colonocytes upon in vivo Salmonella typhimurium, despite high S100A9 gene expression (58). Notably, exclusive stimulation with IL-1β was found to induce significant LCN2 expression and LCN-2 protein release in HT-29 cells, which is in line with its previously described ability to up-regulate antimicrobial responses, such as increased LCN2 expression in human lung epithelial cells (51), as well as increased hBD-2 release from both epithelial cells and keratinocytes (59, 60).
Although a strong link between IL-22 and LCN-2 production has been observed in several in vivo studies (61–63), our results, as well as those from others (35, 64), show that IL-22 on its own appears to have a limited impact on LCN-2 production in human IECs. Upon stimulation of HT-29 cells, no significant LCN-2 release could be observed, despite the activation of STAT3 and the observed binding of STAT3 to the LCN-2 promoter region. In contrast, we observed that activation of NF-κB by e.g. IL-1β or the combination of IL-17A/TNF induced binding of NF-κB to the LCN2 promotor region and subsequently induced LCN-2 production, which corresponded to the previously reported NF-κB binding on the LCN2 promotor region in human lung epithelial cells (50, 51). Interestingly, once IL-22 was combined with one of these NF-κB inducers, it significantly increased LCN-2 production in a STAT3-dependent manner. Importantly, STAT3 inhibition only partially inhibited LCN-2 production in response to IL-22 and IL-17A/TNF, whereas NF-κB inhibition completely inhibited the LCN-2 response to these cytokines. Together, these results indicate a strong supportive role for IL-22 and STAT3 in the regulation of NF-κB–induced LCN-2 production.
Whereas the IL-22–mediated effects on LCN-2 production could have been the result of altered post-translational modifications on NF-κB, no changes on these modifications could be detected between stimulation with IL-22 plus IL-17A/TNF or IL-17A/TNF stimulation alone. Similarly, no changes in STAT3 activation were observed when comparing stimulation with IL-22 plus IL-17A/TNF to IL-22 stimulation alone. In contrast, binding of both STAT3 and NF-κB to the LCN-2 promoter was enhanced upon simultaneous activation of both factors. This enhanced binding could be the result of increased chromatin access, as both transcription factors bind in close proximity in the LCN-2 promotor region (50, 65), and thereby could assist each other in opening up the chromatin. In addition, opening of the chromatin by relatively small transcription factors, such as STAT3 and NF-κB, has recently been modeled to enhance the access to DNA by larger transcription factors, such as p300, a relatively large DNA-binding molecule, which has previously been shown to bind both STAT3 and NF-κB on the ICAM gene (66, 67). Interestingly, regardless of the exact mechanisms involved in the synergy between NF-κB and STAT3, other cell types appear to be affected by this synergy as well. ILC3 and TH17 cells, for instance, are strongly activated by simultaneous stimulation by IL-23 and IL-1β, which are cytokines activating STAT3 and NF-κB, respectively (68, 69). It will be interesting to investigate in more detail how these two transcription factors collaborate in the regulation of gene expression in other cell types and which additional transcription factors influence the transcription of genes involved in the antimicrobial responses of the gut.
Although our study has focused on a set of cytokines commonly found expressed during Enterobacteriaceae infections, other signals released during infection might play a similar role in the induction of LCN-2. E. coli-derived flagellin for instance can activate NF-κB, which can subsequently lead to LCN-2 production in epithelial cells (70–73). In addition, even in the absence of infection, commensals can potentially induce NF-κB signaling through activation of Toll-like receptors or the inflammasome, the latter of which results in the production of IL-18, a potent NF-κB inducer (74–77). Although it will be difficult to fully elucidate to what extent each potential signal contributes to LCN-2 production, especially due to the observed synergies, a better understanding of the factors involved will help to obtain a better grasp on the interactions between the immune system and our microbiota.
During steady state, ILC3 are thought to be the main source of IL-22, and its production can be activated by signals from the intestinal microbiota, such as aryl hydrocarbon receptor ligands (55, 78). Alternatively, adaptive lymphocytes, such as TH17 cells, TH22 cells, and mucosa-associated invariant T-cells (79–81), as well as the recently described IL-22–expressing neutrophils (82), can contribute to the production of IL-22 during infections. The plethora of cellular IL-22 sources highlights the importance of this particular cytokine and likely explains the redundancy of some of these cell types in providing protection against infections (28–30).
In conclusion, our results show how ILC3-derived IL-22 regulates LCN-2 production in a STAT3-dependent manner in human IECs in synergy with NF-κB activation signals. Together, these results fill a gap in the available knowledge on the regulation of antimicrobial responses in IECs and may prove useful in the general elucidation of our host defense mechanisms against infections. In addition, our results provide important insight that might be useful in the development of new therapies aimed to give specific protection against Enterobacteriaceae infection by up-regulating LCN-2 expression in the host.
Materials and methods
Reagents
The following cytokines were obtained from BioTechne (Minneapolis, MN) and used at the following concentrations unless stated otherwise: 50 ng/ml human IL-22 (782-IL-010); 50 ng/ml human IL-17A (317-ILB-050); 20 ng/ml human TNF (210-TA-005); 10 ng/ml human IFN-γ (285-IF-100); 50 ng/ml human IL-1β (201-LB-005); 50 ng/ml human IL-23; and 50 ng/ml human IL-6 (206-IL-010). Human IL-2 (PeproTech, Stockholm, Sweden) was used at 10 units/ml. The NF-κB inhibitor BMS-345541 (43) and STAT3 inhibitor STATTIC (44) were obtained from Sigma. The IL-22 neutralizing antibody (Monoclonal Mouse IgG1, MAB7821) was obtained from BioTechne.
Patient samples
Human ILCs were obtained from adult (n = 10, age 18–53 years) or pediatric (n = 1, age 5 years) patients undergoing tonsillectomy due to obstructive sleep apnea syndrome. Primary human epithelial cells were obtained from the nonaffected part of the colon from resection material of patients undergoing colorectal cancer surgery (n = 3, age 41–81). Informed consents were obtained from the patients or their parents if they were younger than 18 years.
Ethical statement
The sample collection was approved by the regional ethical committee at Karolinska Institutet, Stockholm, Sweden. All patients provided written informed consent before inclusion. The study was performed according to the Declaration of Helsinki.
Cell isolation and sorting
To isolate tonsil cells, whole tonsils were cut into small pieces and ground through a 100-μm cell strainer using a plunger of a plastic syringe. Mononuclear cells (MNCs) were isolated from the obtained cell suspension using Ficoll gradient density centrifugation. For the subsequent FACS purification of ILC3, MNCs were depleted of T cells, B cells, and monocytes using anti-CD3, anti-CD19, and anti-CD14 microbeads and LD columns (all Miltenyi Biotec, Bergisch Gladbach, Germany). CD3−CD19−CD14− MNCs were surface-stained for 30 min at room temperature. The following FITC-conjugated antibodies were used for defining lineage marker-positive cells: anti-CD14 (TÜK4; Agilent Technologies Sweden, Kista, Sweden); anti-FcϵRIα (AER-37), anti-CD34 (581), anti-CD123 (6H6), anti-CD1a (HI149), anti-TCRαβ (IP26), CD94 (DX22), and anti-TCRγδ (B1) (all from BioLegend, San Diego); anti-BDCA2 (AC144; Miltenyi Biotec); and anti-CD19 (4G7; BD Biosciences, Stockholm, Sweden). Additionally, cells were stained with anti-CD3 in BV785 (OKT3; Biolegend), anti-CD161 in APC-A750 (191B8), anti-CD127 in PC7 (R34.34), anti-CD117 in PC5.5 (104D2D1), and NKp44 in PC5 (Z231) (all from Beckman Coulter, Bromma, Sweden), as well as anti-CRTH2 in V450 (BM16; BD Biosciences). ILC3 were sort-purified as lineage−CD45+CD3−CD127+CD161+CD117+CRTH2− lymphocytes. Cells were sorted using BD FACSAriaTM fusion cell sorter.
To isolate primary human epithelial cells, muscle and adipose tissues were removed, and the remaining colon tissue was mechanically disrupted, followed by enzymatic digestion with 250 μg/ml DNase and collagenase II at 37 °C and magnetic stirring at 450 rpm for 45 min. Cell suspension was filtered using a 70-μm cell strainer. Subsequently, hematopoietic cells were removed by magnetic depletion of CD45-expressing cells using anti-CD45 microbeads and LD columns (both Miltenyi Biotec), according to the manufacturer's instructions. Cells were either seeded for cell culture or stained with anti-CD31-PB (WM59; Biolegend), anti-CD90-BV605 (5E10; Biolegend), anti-EpCAM-AlexaFluor647 (9C4; Biolegend), and LIVE/DEADTM Fixable Near-IR Dead Cell Stain (ThermoFisher Scientific, Waltham, MA).
Cell culture
Human colonic epithelial cell lines HT-29, T84, HCT116, and SW480 were cultured in RPMI 1640 medium (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS) (ThermoFisher Scientific) and 2 mm l-glutamine (ThermoFisher Scientific). Caco-2 cells were cultured in Dulbecco's modified Eagle's medium (ThermoFisher Scientific) supplemented with 10% FBS and 2 mm l-glutamine. Sorted tonsil ILCs were cultured in Yssel's-supplemented Iscove's modified Dulbecco's medium with 1% normal human serum (Sigma) and 10 units/ml IL-2. In case of stimulation, cells were additionally treated with 50 ng/ml IL-1β and 50 ng/ml IL-23. All cells mentioned above were cultured at 37 °C, 5.0% CO2. Primary human epithelial cells were cultured in SmGMTM-2 medium (Lonza, Basel, Switzerland) in rat-tail type 1 collagen (ThermoFisher Scientific)–coated plates at 33 °C and 5.0% CO2, according to the manufacturer's instructions. Stimulations were performed 24 h after plating the cells.
RNA isolation and qPCR
To determine gene expression, RNA was isolated with the Isolate II RNA mini kit (BIO-line, London, UK) according to the manufacturer's protocol. RNA concentrations were measured by Nanodrop (Saveen & Werner, Limhamn, Sweden), and 500 ng of RNA was used for cDNA synthesis using the cDNA synthesis kit (BIO-line). cDNA was subsequently used for qPCR in combination with Maxima SYBR Green (ThermoFisher Scientific) and forward/reverse primers (Table S1) (Sigma) in a CFX96 qPCR (Bio-Rad, Solna, Sweden). Cq values were corrected for 18S housekeeping gene expression. Cq values over 35 were given an arbitrary Cq value of 35. Fold-increase compared with unstimulated controls was calculated, and statistical analyses were performed on log-transformed fold-increase expression values.
Chromatin-immunoprecipitation (ChIP)
ChIP was performed with the SimpleChIP Plus Enzymatic Chromatin IP kit (catalog no. 9005S, Cell Signaling Technologies, Leiden, the Netherlands) together with the following antibodies from Cell Signaling Technologies: normal rabbit IgG (catalog no. 2729), STAT3 rabbit mAb (catalog no. 12640S), and NF-κB p65 (D14E12) XP rabbit mAb. For stimulation, HT-29 cells were stimulated for 30 min at 37 °C with the indicated cytokines in RPMI 1640 medium after which cells were fixed in 1% formaldehyde (methanol-free 16% stock, ThermoFisher Scientific) for 10 min at room temperature. All subsequent steps were performed according to the SimpleChIP Plus Enzymatic Chromatin IP kit protocol. DNA quantification was performed using a CFX96 qPCR (BIO-RAD) with primers listed in Table S1. Statistical analysis was performed on log-transformed data.
Enzyme-linked immunosorbent assay (ELISA)
Fresh or frozen (−20 °C) cell supernatants were diluted in 1% BSA Reagent Diluent (BioTechne) before analysis. DUOset ELISA kits were obtained for human LCN-2, human S100A9, and human S100A8/A9 (all BioTechne) and human β-defensin 2 (hBD-2) (PeproTech). ELISAs were performed in Maxisorp 96-well plates (ThermoFisher Scientific), according to the manufacturer's' instructions. The OD was determined at 450 nm and corrected for 570 nm background values with a TECAN Infinite m200 plate reader (TECAN Group Ltd., Männedorf, Switzerland). Standard curves were calculated by four-parameter fit modeling in PRISM 7.03 (Graphpad, La Jolla, CA), from which sample concentrations were determined. Statistical analysis was performed on square root-transformed data.
Griess assay
Nitric oxide (NO) production was measured by determining nitrite levels in cell supernatant using the Griess assay. Cell supernatants (50 μl) were mixed with 50 μl of 1% sulfanilamide (Sigma) in 5% phosphoric acid. After a 5-min incubation in the dark at room temperature, 50 μl of 0.1% N-(1-naphthyl)ethylenediamine dihydrochloride (Sigma) was added, followed by another 5-min incubation in the dark at room temperature. OD values at 550 nm were measured with a TECAN Infinite m200 plate reader. Standard curves were calculated by four-parameter fit modeling in PRISM 7.03, from which sample concentrations were determined. Statistical analysis was performed on square root-transformed data.
SDS-PAGE
HT-29 cells were washed with ice-cold PBS and lysed with RIPA buffer containing protease inhibitor mixture cOmplete ULTRA tablets (Roche Applied Science, Basel, Switzerland) and PHOSSTOP phosphatase inhibitors (Roche Applied Science). After scraping the lysed cells, lysate was sonicated and subsequently spun down at 14,000 × g to remove debris. Protein samples were stored at −20 °C until further use. Protein concentrations were determined with the Pierce BCA kit (ThermoFisher Scientific) according to the manufacturer's protocol. OD was measured at 562 nm with a TECAN Infinite m200 plate reader, and standard curves were calculated by four-parameter fit modeling in PRISM 7.03. Protein samples were mixed with 5× Pierce Lane Marker Reducing buffer (ThermoFisher Scientific) and heated to 96 °C for 10 min. Samples were loaded on precast NuPAGE 4% BisTris gels (ThermoFisher Scientific) and run in NuPAGE MES SDS Running buffer (ThermoFisher Scientific). SeeBlue Plus2 prestained protein marker (ThermoFisher Scientific) was used as a molecular weight ladder. Samples were run at 200 V.
Western blot analysis
SDS-PAGE samples were transferred to an Amersham Biosciences HyBond 0.2-μm polyvinylidene difluoride membrane (GE Healthcare) using NuPAGE Transfer Buffer (ThermoFisher Scientific). After the transfer, the membrane was washed in TBST (ThermoFisher Scientific) at room temperature for 5 min. Next, the membrane was blocked with 5% nonfat dry milk (Semper, Sundbyberg, Sweden) in TBST at room temperature for 1 h followed by three 10-min washes in TBST. Membranes were incubated overnight at 4 °C with the following primary antibodies from Cell Signaling Technologies: STAT3 rabbit mAb (12640S); phospho-STAT3 (Tyr-705) XP rabbit mAb (9145S); phospho-STAT3 (Ser-727) mouse mAb (9136S); acetyl-STAT3 (Lys-685) rabbit Ab (2523S); and IκB-α (N-terminal) mouse (4814S). Staining was performed in 5% BSA in TBST or 5% nonfat dry milk in TBST, according to the manufacturer's antibody-specific instructions. After incubation, the membrane was washed three times for 10 min with TBST, followed by incubation with secondary antibody for 1 h at room temperature in 5% nonfat dry milk in TBST. Secondary antibodies used were anti-rabbit IgG, HRP-linked Ab (Cell Signaling Technologies, 7076S) and anti-mouse IgG, HRP-linked Ab (Cell Signaling Technologies, 7074S). Finally, the membrane was washed three times for 10 min with TBST, incubated with Amersham Biosciences ECL Prime ECL reagent (GE Healthcare), and imaged using a SynGene G:Box-ChemiXX6 (SynGene, Cambridge, UK) obtaining 16-bit images. After imaging, the membranes were washed three times for 10 min in TBST and stripped with stripping buffer (2% SDS (Sigma), 16% Tris-HCl, pH 6.8 (Merck), and 0.1 m DTT (Sigma) in water) for 1 h at 37 °C. After stripping, membranes were washed six times for 5 min with TBST and blocked with 5% nonfat dry milk in TBST, after which a new primary staining could be performed. Alternatively, a total protein staining was performed with Amido Black (Sigma) by incubating the membrane in 1× Amido Black for 1 min and destaining the membrane with 25% isopropyl alcohol, 10% acetic acid in water for 30 min. Analysis of the band intensities on the imaged membrane was performed with ImageJ (National Institutes of Health, Bethesda), and statistics were performed on log-transformed data. Brightness and contrast adjustments on images were done equally over the entire images.
WST-1 viability assay
WST-1 reagent was used to assess mitochondrial activity as a marker for cell viability. Cells were incubated with 1:10-diluted WST1 reagent (Sigma) in RPMI 1640 medium at 37 °C for ∼10 min, after which supernatants were transferred to a new 96-well TC-culture plate, and the OD was measured with a TECAN Infinite m200 plate reader (TECAN Group Ltd.) at 450 nm and corrected for 620 nm background signal. WST1 reagent diluted 1:10 in the absence of cells was used as a negative control, whereas untreated cells were used as a positive control. Percentage of viability was calculated by subtracting the negative control from all samples and setting the signal from untreated cells to 100%.
Bacterial growth
Bacterial growth was assessed on two isogenic K. pneumoniae strains derived from K. pneumoniae KPRR1 (45). Strain KP4 expressed Ent, Yer, and Sal, whereas strains KP20 only expressed Ent. Bacteria were grown in RPMI 1640 medium + 200 μm 2,2-bipyridyl (Sigma) overnight at 37 °C with shaking. For growth analysis, bacteria were diluted to OD 0.001 in different media conditions. This included fresh RPMI 1640 medium, RPMI 1640 cell supernatants from 24-h cultures of unstimulated or stimulated (IL-22, IL-17A, and TNF) HT-29 cells, or 24-h cell supernatants from stimulated HT-29 cells with addition of 5 μm Fe(III)SO4 (Sigma). Bacterial growth was assessed by measuring OD over time with a BioScreen C device (Oy Growth Curves Ab Ltd., Helsinki, Finland). Growth curves were analyzed by fitting a four-parameter fit model and extracting Hill slopes representing the optimal growth rate in log phase.
Statistics and data analysis
Data transformation and statistical analysis was performed using PRISM 7.03 (Graphpad). Hierarchical clustering on qPCR data were performed by calculating the Pearson distance with the Pearson correlation coefficient followed by complete linkage clustering using R (R Core Team, Vienna, Austria) and the “hyperSpec,” “RColorBrewer,” and “gplots” packages. The graphs were made in Graphpad and R.
Author contributions
M. C., B. A., J. M., and P. B. conceptualization; M. C., S. K. G., and D. U. data curation; M. C. and A. R. formal analysis; M. C., A. R., S. K. G., and D. U. investigation; M. C., A. R., S. K. G., D. U., U. L., and J. M. methodology; M. C. and P. B. writing-original draft; M. C., B. A., J. M., and P. B. writing-review and editing; A. R., U. L., J. M., and P. B. resources; B. A., J. M., and P. B. supervision; B. A. and P. B. funding acquisition; B. A. and P. B. validation; P. B. project administration.
Supplementary Material
Acknowledgments
We acknowledge Dr. Michael Bachman, University of Michigan, for sharing the isogenic K. pneumoniae mutants containing specific siderophore deletions.
This work was supported by the Swedish Research Council (Vetenskapsrådet), the Scandinavian Society for Antimicrobial Chemotherapy (SSAC), The Groschinsky Foundation, the Karolinska Institutet (to P. B.), and German Research Foundation (Deutsche Forschungsgemeinschaft) Postdoctoral Fellowship RA 2986/1-1 (to A. R.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
- IEC
- intestinal epithelial cell
- ILC3
- innate lymphoid cells type 3
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR
- IFN
- interferon
- TNF
- tumor necrosis factor
- IL
- interleukin
- Ent
- enterobactin
- Yer
- yersiniabactin
- Sal
- salmochelin
- ILC
- innate lymphoid cell
- MNC
- mononuclear cell
- FBS
- fetal bovine serum
- Ab
- antibody
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Lloyd-Price J., Abu-Ali G., and Huttenhower C. (2016) The healthy human microbiome. Genome Med. 8, 51 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byndloss M. X., Pernitzsch S. R., and Bäumler A. J. (2018) Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol. 11, 1299–1305 10.1038/s41385-018-0010-y [DOI] [PubMed] [Google Scholar]
- 3. Goering R. V., Dockrell H., Zuckerman M., Roitt I., and Chiodini P. L. (2013) Mims' Medical Microbiology, 5th Ed., Elsevier/Saunders, Philadelphia [Google Scholar]
- 4. Kalanuria A. A., Ziai W., Zai W., and Mirski M. (2014) Ventilator-associated pneumonia in the ICU. Crit. Care 18, 208 10.1186/cc13775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fletcher S. M., McLaws M.-L., and Ellis J. T. (2013) Prevalence of gastrointestinal pathogens in developed and developing countries: systematic review and meta-analysis. J. Public Health Res. 2, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magill S. S., Edwards J. R., Bamberg W., Beldavs Z. G., Dumyati G., Kainer M. A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., Ray S. M., Thompson D. L., Wilson L. E., Fridkin S. K., and Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. (2014) Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208 10.1056/NEJMoa1306801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antoni L., Nuding S., Weller D., Gersemann M., Ott G., Wehkamp J., and Stange E. F. (2013) Human colonic mucus is a reservoir for antimicrobial peptides. J. Crohns Colitis 7, e652–64 10.1016/j.crohns.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 8. Loonen L. M., Stolte E. H., Jaklofsky M. T., Meijerink M., Dekker J., van Baarlen P., and Wells J. M. (2014) REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 7, 939–947 10.1038/mi.2013.109 [DOI] [PubMed] [Google Scholar]
- 9. Cramer E. P., Dahl S. L., Rozell B., Knudsen K. J., Thomsen K., Moser C., Cowland J. B., and Borregaard N. (2017) Lipocalin-2 from both myeloid cells and the epithelium combats Klebsiella pneumoniae lung infection in mice. Blood 129, 2813–2817 10.1182/blood-2016-11-753434 [DOI] [PubMed] [Google Scholar]
- 10. Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., and Aderem A. (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 10.1038/nature03104 [DOI] [PubMed] [Google Scholar]
- 11. Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., and Strong R. K. (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043 10.1016/S1097-2765(02)00708-6 [DOI] [PubMed] [Google Scholar]
- 12. Steigedal M., Marstad A., Haug M., Damås J. K., Strong R. K., Roberts P. L., Himpsl S. D., Stapleton A., Hooton T. M., Mobley H. L., Hawn T. R., and Flo T. H. (2014) Lipocalin 2 imparts selective pressure on bacterial growth in the bladder and is elevated in women with urinary tract infection. J. Immunol. 193, 6081–6089 10.4049/jimmunol.1401528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mukherjee S., and Hooper L. V. (2015) Antimicrobial defense of the intestine. Immunity 42, 28–39 10.1016/j.immuni.2014.12.028 [DOI] [PubMed] [Google Scholar]
- 14. Lewis S. B., Prior A., Ellis S. J., Cook V., Chan S. S., Gelson W., and Schüller S. (2016) Flagellin induces β-defensin 2 in human colonic ex vivo infection with enterohemorrhagic Escherichia coli. Front. Cell. Infect. Microbiol. 6, 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Neil D. A., Porter E. M., Elewaut D., Anderson G. M., Eckmann L., Ganz T., and Kagnoff M. F. (1999) Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163, 6718–6724 [PubMed] [Google Scholar]
- 16. Fischer N., Sechet E., Friedman R., Amiot A., Sobhani I., Nigro G., Sansonetti P. J., and Sperandio B. (2016) Histone deacetylase inhibition enhances antimicrobial peptide but not inflammatory cytokine expression upon bacterial challenge. Proc. Natl. Acad. Sci. U.S.A. 113, E2993–E3001 10.1073/pnas.1605997113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geremia A., and Arancibia-Cárcamo C. V. (2017) Innate lymphoid cells in intestinal inflammation. Front. Immunol. 8, 1296 10.3389/fimmu.2017.01296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okumura R., and Takeda K. (2017) Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 49, e338 10.1038/emm.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gasteiger G., Fan X., Dikiy S., Lee S. Y., and Rudensky A. Y. (2015) Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 10.1126/science.aac9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simoni Y., Fehlings M., Kløverpris H. N., McGovern N., Koo S.-L., Loh C. Y., Lim S., Kurioka A., Fergusson J. R., Tang C.-L., Kam M. H., Dennis K., Lim T. K. H., Fui A. C. Y., et al. (2017) Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity 46, 148–161 10.1016/j.immuni.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weiner J., Zuber J., Shonts B., Yang S., Fu J., Martinez M., Farber D. L., Kato T., and Sykes M. (2017) Long-term persistence of innate lymphoid cells in the gut after intestinal transplantation. Transplantation 101, 2449–2454 10.1097/TP.0000000000001593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spits H., Artis D., Colonna M., Diefenbach A., Di Santo J. P., Eberl G., Koyasu S., Locksley R. M., McKenzie A. N., Mebius R. E., Powrie F., and Vivier E. (2013) Innate lymphoid cells–a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- 23. Qiu J., Heller J. J., Guo X., Chen Z. M., Fish K., Fu Y.-X., and Zhou L. (2012) The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 10.1016/j.immuni.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins J. W., Keeney K. M., Crepin V. F., Rathinam V. A., Fitzgerald K. A., Finlay B. B., and Frankel G. (2014) Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 12, 612–623 10.1038/nrmicro3315 [DOI] [PubMed] [Google Scholar]
- 25. Björklund Å. K, Forkel M., Picelli S., Konya V., Theorell J., Friberg D., Sandberg R., and Mjösberg J. (2016) The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17, 451–460 10.1038/ni.3368 [DOI] [PubMed] [Google Scholar]
- 26. Hoorweg K., Peters C. P., Cornelissen F., Aparicio-Domingo P., Papazian N., Kazemier G., Mjösberg J. M., Spits H., and Cupedo T. (2012) Functional differences between human NKp44− and NKp44+ RORC+ innate lymphoid cells. Front. Immunol. 3, 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Simone V., Franzè E., Ronchetti G., Colantoni A., Fantini M. C., Di Fusco D., Sica G. S., Sileri P., MacDonald T. T., Pallone F., Monteleone G., and Stolfi C. (2015) Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-κB to promote colorectal cancer cell growth. Oncogene 34, 3493–3503 10.1038/onc.2014.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rankin L. C., Girard-Madoux M. J., Seillet C., Mielke L. A., Kerdiles Y., Fenis A., Wieduwild E., Putoczki T., Mondot S., Lantz O., Demon D., Papenfuss A. T., Smyth G. K., Lamkanfi M., Carotta S., et al. (2016) Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 17, 179–186 10.1038/ni.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song C., Lee J. S., Gilfillan S., Robinette M. L., Newberry R. D., Stappenbeck T. S., Mack M., Cella M., and Colonna M. (2015) Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212, 1869–1882 10.1084/jem.20151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vély F., Barlogis V., Vallentin B., Neven B., Piperoglou C., Ebbo M., Perchet T., Petit M., Yessaad N., Touzot F., Bruneau J., Mahlaoui N., Zucchini N., Farnarier C., Michel G., et al. (2016) Evidence of innate lymphoid cell redundancy in humans. Nat. Immunol. 17, 1291–1299 10.1038/ni.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo S.-U., Kuffa P., Kitamoto S., Nagao-Kitamoto H., Rousseau J., Kim Y.-G., Núñez G., and Kamada N. (2015) Intestinal macrophages arising from CCR2+ monocytes control pathogen infection by activating innate lymphoid cells. Nat. Commun. 6, 8010 10.1038/ncomms9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu J., Guo X., Chen Z. E., He L., Sonnenberg G. F., Artis D., Fu Y.-X., and Zhou L. (2013) Group 3 innate lymphoid cells inhibit T-cell–mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 39, 386–399 10.1016/j.immuni.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H. J., Langa F., Di Santo J. P., and Eberl G. (2010) Lineage relationship analysis of RORγt + innate lymphoid cells. Science 330, 665–669 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- 34. Sonnenberg G. F., Monticelli L. A., Alenghat T., Fung T. C., Hutnick N. A., Kunisawa J., Shibata N., Grunberg S., Sinha R., Zahm A. M., Tardif M. R., Sathaliyawala T., Kubota M., Farber D. L., Collman R. G., et al. (2012) Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 10.1126/science.1222551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raffatellu M., George M. D., Akiyama Y., Hornsby M. J., Nuccio S.-P., Paixao T. A., Butler B. P., Chu H., Santos R. L., Berger T., Mak T. W., Tsolis R. M., Bevins C. L., Solnick J. V., Dandekar S., and Bäumler A. J. (2009) Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 10.1016/j.chom.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng Y., Valdez P. A., Danilenko D. M., Hu Y., Sa S. M., Gong Q., Abbas A. R., Modrusan Z., Ghilardi N., de Sauvage F. J., and Ouyang W. (2008) Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- 37. Backert I., Koralov S. B., Wirtz S., Kitowski V., Billmeier U., Martini E., Hofmann K., Hildner K., Wittkopf N., Brecht K., Waldner M., Rajewsky K., Neurath M. F., Becker C., and Neufert C. (2014) STAT3 activation in Th17 and Th22 cells controls IL-22–mediated epithelial host defense during infectious colitis. J. Immunol. 193, 3779–3791 10.4049/jimmunol.1303076 [DOI] [PubMed] [Google Scholar]
- 38. Konya V., Czarnewski P., Forkel M., Rao A., Kokkinou E., Villablanca E. J., Almer S., Lindforss U., Friberg D., Höög C., Bergman P., and Mjösberg J. (2018) Vitamin D downregulates the IL-23 receptor pathway in human mucosal group 3 innate lymphoid cells. J. Allergy Clin. Immunol. 141, 279–292 10.1016/j.jaci.2017.01.045 [DOI] [PubMed] [Google Scholar]
- 39. Sabat R., Ouyang W., and Wolk K. (2014) Therapeutic opportunities of the IL-22–IL-22R1 system. Nat. Rev. Drug Discov. 13, 21–38 10.1038/nrd4176 [DOI] [PubMed] [Google Scholar]
- 40. Tsai P.-Y., Zhang B., He W.-Q., Zha J.-M., Odenwald M. A., Singh G., Tamura A., Shen L., Sailer A., Yeruva S., Kuo W.-T., Fu Y.-X., Tsukita S., and Turner J. R. (2017) IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe 21, 671–681.e4 10.1016/j.chom.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin J. C., Bériou G., Heslan M., Bossard C., Jarry A., Abidi A., Hulin P., Ménoret S., Thinard R., Anegon I., Jacqueline C., Lardeux B., Halary F., Renauld J.-C., Bourreille A., and Josien R. (2016) IL-22BP is produced by eosinophils in human gut and blocks IL-22 protective actions during colitis. Mucosal Immunol. 9, 539–549 10.1038/mi.2015.83 [DOI] [PubMed] [Google Scholar]
- 42. Mizuno S., Mikami Y., Kamada N., Handa T., Hayashi A., Sato T., Matsuoka K., Matano M., Ohta Y., Sugita A., Koganei K., Sahara R., Takazoe M., Hisamatsu T., and Kanai T. (2014) Cross-talk between RORγt+ innate lymphoid cells and intestinal macrophages induces mucosal IL-22 production in Crohn's disease. Inflamm. Bowel Dis. 20, 1426–1434 10.1097/MIB.0000000000000105 [DOI] [PubMed] [Google Scholar]
- 43. Burke J. R., Pattoli M. A., Gregor K. R., Brassil P. J., MacMaster J. F., McIntyre K. W., Yang X., Iotzova V. S., Clarke W., Strnad J., Qiu Y., and Zusi F. C. (2003) BMS-345541 is a highly selective inhibitor of IκB kinase that binds at an allosteric site of the enzyme and blocks NF-κB–dependent transcription in mice. J. Biol. Chem. 278, 1450–1456 10.1074/jbc.M209677200 [DOI] [PubMed] [Google Scholar]
- 44. Schust J., Sperl B., Hollis A., Mayer T. U., and Berg T. (2006) Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 13, 1235–1242 10.1016/j.chembiol.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 45. Bachman M. A., Oyler J. E., Burns S. H., Caza M., Lépine F., Dozois C. M., and Weiser J. N. (2011) Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316 10.1128/IAI.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Friedrich C., Mamareli P., Thiemann S., Kruse F., Wang Z., Holzmann B., Strowig T., Sparwasser T., and Lochner M. (2017) MyD88 signaling in dendritic cells and the intestinal epithelium controls immunity against intestinal infection with C. rodentium. PLOS Pathog. 13, e1006357 10.1371/journal.ppat.1006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Borenshtein D., Nambiar P. R., Groff E. B., Fox J. G., and Schauer D. B. (2007) Development of fatal colitis in FVB mice infected with Citrobacter rodentium. Infect. Immun. 75, 3271–3281 10.1128/IAI.01810-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geddes K., Rubino S. J., Magalhaes J. G., Streutker C., Le Bourhis L., Cho J. H., Robertson S. J., Kim C. J., Kaul R., Philpott D. J., and Girardin S. E. (2011) Identification of an innate T helper type 17 response to intestinal bacterial pathogens. Nat. Med. 17, 837–844 10.1038/nm.2391 [DOI] [PubMed] [Google Scholar]
- 49. Tourret J., Willing B. P., Croxen M. A., Dufour N., Dion S., Wachtel S., Denamur E., and Finlay B. B. (2016) Small intestine early innate immunity response during intestinal colonization by Escherichia coli depends on its extra-intestinal virulence status. PLoS ONE 11, e0153034 10.1371/journal.pone.0153034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karlsen J. R., Borregaard N., and Cowland J. B. (2010) Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-α is controlled by IκB-ζ but neither by C/EBP-β nor C/EBP-δ. J. Biol. Chem. 285, 14088–14100 10.1074/jbc.M109.017129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cowland J. B., Sørensen O. E., Sehested M., and Borregaard N. (2003) Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1β, but not by TNF-α. J. Immunol. 171, 6630–6639 10.4049/jimmunol.171.12.6630 [DOI] [PubMed] [Google Scholar]
- 52. Mjösberg J., and Spits H. (2016) Human innate lymphoid cells. J. Allergy Clin. Immunol. 138, 1265–1276 10.1016/j.jaci.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 53. Tumanov A. V., Koroleva E. P., Guo X., Wang Y., Kruglov A., Nedospasov S., and Fu Y.-X. (2011) Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe 10, 44–53 10.1016/j.chom.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Satoh-Takayama N., Serafini N., Verrier T., Rekiki A., Renauld J.-C., Frankel G., and Di Santo J. P. (2014) The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity 41, 776–788 10.1016/j.immuni.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 55. Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T., Ishikawa E., Shima T., Hara T., Kado S., Jinnohara T., et al. (2015) Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 10.1016/j.cell.2015.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dalli J., Colas R. A., Arnardottir H., and Serhan C. N. (2017) Vagal regulation of group 3 innate lymphoid cells and the immunoresolvent PCTR1 controls infection resolution. Immunity 46, 92–105 10.1016/j.immuni.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiong H., Keith J. W., Samilo D. W., Carter R. A., Leiner I. M., and Pamer E. G. (2016) Innate lymphocyte/Ly6C(hi) monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell 165, 679–689 10.1016/j.cell.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu J. Z., Jellbauer S., Poe A. J., Ton V., Pesciaroli M., Kehl-Fie T. E., Restrepo N. A., Hosking M. P., Edwards R. A., Battistoni A., Pasquali P., Lane T. E., Chazin W. J., Vogl T., Roth J., et al. (2012) Zinc Sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239 10.1016/j.chom.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pioli P. A., Weaver L. K., Schaefer T. M., Wright J. A., Wira C. R., and Guyre P. M. (2006) Lipopolysaccharide-induced IL-1β production by human uterine macrophages up-regulates uterine epithelial cell expression of human β-defensin 2. J. Immunol. 176, 6647–6655 10.4049/jimmunol.176.11.6647 [DOI] [PubMed] [Google Scholar]
- 60. Wehkamp K., Schwichtenberg L., Schröder J.-M., and Harder J. (2006) Pseudomonas aeruginosa- and IL-1β–mediated induction of human β-defensin-2 in keratinocytes is controlled by NF-κB and AP-1. J. Invest. Dermatol. 126, 121–127 10.1038/sj.jid.5700020 [DOI] [PubMed] [Google Scholar]
- 61. Zheng M., Horne W., McAleer J. P., Pociask D., Eddens T., Good M., Gao B., and Kolls J. K. (2016) Therapeutic role of interleukin 22 in experimental intra-abdominal Klebsiella pneumoniae infection in mice. Infect. Immun. 84, 782–789 10.1128/IAI.01268-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wozniak K. L., Hole C. R., Yano J., Fidel P. L. Jr., and Wormley F. L. Jr. (2014) Characterization of IL-22 and antimicrobial peptide production in mice protected against pulmonary Cryptococcus neoformans infection. Microbiology 160, 1440–1452 10.1099/mic.0.073445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Behnsen J., Jellbauer S., Wong C. P., Edwards R. A., George M. D., Ouyang W., and Raffatellu M. (2014) The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273 10.1016/j.immuni.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cayatte C., Joyce-Shaikh B., Vega F., Boniface K., Grein J., Murphy E., Blumenschein W. M., Chen S., Malinao M.-C., Basham B., Pierce R. H., Bowman E. P., McKenzie B. S., Elson C. O., Faubion W. A., et al. (2012) Biomarkers of therapeutic response in the IL-23 pathway in inflammatory bowel disease. Clin. Transl. Gastroenterol. 3, e10–e10 10.1038/ctg.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jung M., Weigert A., Tausendschön M., Mora J., Ören B., Sola A., Hotter G., Muta T., and Brüne B. (2012) Interleukin-10–induced neutrophil gelatinase-associated lipocalin production in macrophages with consequences for tumor growth. Mol. Cell. Biol. 32, 3938–3948 10.1128/MCB.00413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kesanakurti D., Chetty C., Rajasekhar Maddirela D., Gujrati M., and Rao J. S. (2013) Essential role of cooperative NF-κB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene 32, 5144–5155 10.1038/onc.2012.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maeshima K., Kaizu K., Tamura S., Nozaki T., Kokubo T., and Takahashi K. (2015) The physical size of transcription factors is key to transcriptional regulation in chromatin domains. J. Phys. Condens. Matter 27, 064116 10.1088/0953-8984/27/6/064116 [DOI] [PubMed] [Google Scholar]
- 68. Glatzer T., Killig M., Meisig J., Ommert I., Luetke-Eversloh M., Babic M., Paclik D., Blüthgen N., Seidl R., Seifarth C., Gröne J., Lenarz M., Stölzel K., Fugmann D., Porgador A., et al. (2013) RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity 38, 1223–1235 10.1016/j.immuni.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 69. Revu S., Wu J., Henkel M., Rittenhouse N., Menk A., Delgoffe G. M., Poholek A. C., and McGeachy M. J. (2018) IL-23 and IL-1β drive human Th17 cell differentiation and metabolic reprogramming in absence of CD28 costimulation. Cell Rep. 22, 2642–2653 10.1016/j.celrep.2018.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Singh V., Yeoh B. S., Chassaing B., Zhang B., Saha P., Xiao X., Awasthi D., Shashidharamurthy R., Dikshit M., Gewirtz A., and Vijay-Kumar M. (2016) Microbiota-inducible innate immune siderophore binding protein lipocalin 2 is critical for intestinal homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2, 482–498.e6 10.1016/j.jcmgh.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Giacomin P. R., Moy R. H., Noti M., Osborne L. C., Siracusa M. C., Alenghat T., Liu B., McCorkell K. A., Troy A. E., Rak G. D., Hu Y., May M. J., Ma H.-L., Fouser L. A., Sonnenberg G. F., and Artis D. (2015) Epithelial-intrinsic IKKα expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. J. Exp. Med. 212, 1513–1528 10.1084/jem.20141831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cañas M.-A., Fábrega M.-J., Giménez R., Badia J., and Baldomà L. (2018) Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front. Microbiol. 9, 498 10.3389/fmicb.2018.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bambou J.-C., Giraud A., Menard S., Begue B., Rakotobe S., Heyman M., Taddei F., Cerf-Bensussan N., and Gaboriau-Routhiau V. (2004) In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J. Biol. Chem. 279, 42984–42992 10.1074/jbc.M405410200 [DOI] [PubMed] [Google Scholar]
- 74. Levy M., Thaiss C. A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J. A., David E., Savidor A., Korem T., Herzig Y., Pevsner-Fischer M., Shapiro H., Christ A., Harmelin A., Halpern Z., et al. (2015) Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kanther M., Sun X., Mühlbauer M., Mackey L. C., Flynn E. J. 3rd., Bagnat M., Jobin C., and Rawls J. F. (2011) Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology 141, 197–207 10.1053/j.gastro.2011.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sansone C. L., Cohen J., Yasunaga A., Xu J., Osborn G., Subramanian H., Gold B., Buchon N., and Cherry S. (2015) Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe 18, 571–581 10.1016/j.chom.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., and Medzhitov R. (2004) Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 78. Cervantes-Barragan L., and Colonna M. (2018) Chemical sensing in development and function of intestinal lymphocytes. Curr. Opin. Immunol. 50, 112–116 10.1016/j.coi.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., and Lantz O. (2011) Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- 80. Kara E. E., Comerford I., Fenix K. A., Bastow C. R., Gregor C. E., McKenzie D. R., and McColl S. R. (2014) Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathog. 10, e1003905 10.1371/journal.ppat.1003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liang S. C., Tan X.-Y., Luxenberg D. P., Karim R., Dunussi-Joannopoulos K., Collins M., and Fouser L. A. (2006) Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 10.1084/jem.20061308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zindl C. L., Lai J.-F., Lee Y. K., Maynard C. L., Harbour S. N., Ouyang W., Chaplin D. D., and Weaver C. T. (2013) IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc. Natl. Acad. Sci. U.S.A. 110, 12768–12773 10.1073/pnas.1300318110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.