Abstract
Yin Yang 1 (YY1) is a zinc-finger protein that plays critical roles in various biological processes by interacting with DNA and numerous protein partners. YY1 has been reported to play dual biological functions as either an oncogene or tumor suppressor in the development and progression of multiple cancers, but its role in human nasopharyngeal carcinoma (NPC) has not yet been revealed. In this study, we found that YY1 overexpression significantly inhibits cell proliferation and cell-cycle progression from G1 to S and promotes apoptosis in NPC cells. Moreover, we identified YY1 as a component of the c-Myc complex and observed that ectopic expression of YY1 inhibits c-Myc transcriptional activity, as well as the promoter activity and expression of the c-Myc target gene microRNA-141 (miR-141). Furthermore, restoring miR-141 expression could at least partially reverse the inhibitory effect of YY1 on cell proliferation and tumor growth and on the expression of some critical c-Myc targets, such as PTEN/AKT pathway components both in vitro and in vivo. We also found that YY1 expression is reduced in NPC tissues, negatively correlates with miR-141 expression and clinical stages in NPC patients, and positively correlates with survival prognosis. Our results reveal a previously unappreciated mechanism in which the YY1/c-Myc/miR-141 axis plays a critical role in NPC progression and may provide some potential and valuable targets for the diagnosis and treatment of NPC.
Keywords: tumor, transcription regulation, proliferation, cell cycle, apoptosis, tumor suppressor gene, c-Myc, microRNA 141, nasopharyngeal carcinoma (NPC), Yin Yang 1 (YY1)
Introduction
Nasopharyngeal carcinoma (NPC)2 is a head-and-neck epithelial malignancy that occurs frequently in Southern China, which is a most peculiar cancer with a distinctly skewed geographic and ethnic distribution (1). A multifactorial etiology has been suggested for NPC, including a dynamic interplay between genetic predisposition, Epstein-Barr virus infection, and environmental carcinogens (2). However, the molecular mechanisms that regulate NPC initiation and promote its malignant progression remain obscure. Despite marked improvements in NPC patient prognosis in recent decades (3), most patients are still diagnosed with advanced stage NPC (4). Therefore, identifying sensitive biomarkers and drug targets is essential to enable early diagnosis and improve treatment outcomes for early-stage NPC patients.
c-Myc is an important oncoprotein whose abnormal expression contributes to 30–50% of human malignancies, being overexpressed in more than 70% of human malignancies, including nasopharyngeal carcinoma, lymphomas, neuroblastomas, melanomas, breast, ovarian, prostate, and liver cancers (5–7). In c-Myc–dependent cancer models, c-Myc inactivation cannot only regress tumor growth but also induce cell death, suggesting that c-Myc inactivation can impact a range of human cancers (8). As a critical transcription factor, c-Myc is widely involved in cell proliferation, apoptosis, differentiation, metabolism, somatic cell reprogramming, and other key processes under normal or pathological conditions via the regulation of numerous target genes (9). Therefore, strategies for down-regulating the c-Myc transcriptional activity are thus widely sought. We previously found that miR-141 functions as an oncogenic miRNA in NPC and plays critical roles in NPC development and progression (10). Moreover, c-Myc is known to specifically bind the miR-141 promoter region and thus regulate the transcriptional activation of miR-141, whereas c-Myc knockdown has been shown to activate PTEN/AKT signaling and suppress cell proliferation and tumor growth via transcriptional down-regulation of miR-141 in NPC cells (11–13).
c-Myc always exerts its functions through the transcriptional regulation of its downstream target genes, which depend on the formation of the Myc/Max/Mad complex. c-Myc binds Max through its basic helix-loop-helix zipper domain, and these heterodimers bind specifically to 5′-CACGTG-3′ E-box sequences to activate transcription (14, 15). Alternatively, transcriptional repression by Mad is mediated through its interaction with mSin3, which results in the recruitment of histone deacetylases and corepressor molecules and thus leads to the transcriptional repression of target genes (16). Further exploration of the molecular mechanism of c-Myc in NPC using bioinformatics analyses revealed Yin Yang-1 (YY1) as a potential c-Myc–interacting protein that might be involved in the regulation of c-Myc target genes (17, 18). YY1 is a ubiquitously expressed member of the GLI–Kruppel family of zinc-finger transcription factors that is abnormally expressed in most human tumors and exerts dual biological functions as a tumor suppressor or oncogene through the regulation of different target genes or signaling pathways (19, 20). These dual functions in different cancers are probably because YY1 can both positively and negatively regulate gene expression, as well as interact with a multitude of proteins with diverse functions (21). Crystal structures of YY1 with different binding partners reveal that YY1 is a key scaffold protein that functionally interfaces with various partners to regulate gene transcription and participate in multiple signaling pathways. However, the precise biological function of YY1 in NPC remains unclear.
In this study, we revealed that YY1 significantly inhibits cell proliferation and cell-cycle progression and induces apoptosis in NPC cells. Moreover, YY1 was identified as a component of the c-Myc complex, and ectopic expression of YY1 is able to inhibit c-Myc transcriptional activity, as well as the promoter activity and expression of the critical downstream target gene miR-141. Furthermore, restoring the expression of miR-141 at least partially reverses the inhibitory effects of YY1 on cell proliferation, cell-cycle progression, apoptosis and tumor growth, as well as the expression of some critical c-Myc targets, such as the PTEN/AKT pathway, both in vitro and in vivo. In addition, YY1 is also down-regulated in NPC tissues, and the YY1 protein level is negatively correlated with the clinical stage in NPC patients and miR-141 expression, while positively correlated with survival prognosis. Taken together, our results demonstrate that the YY1/c-Myc/miR-141 axis plays a critical role in the development and progression of NPC, thereby providing potential targets for the diagnosis and treatment of NPC.
Results
YY1 inhibits cell proliferation and promotes apoptosis in NPC cells
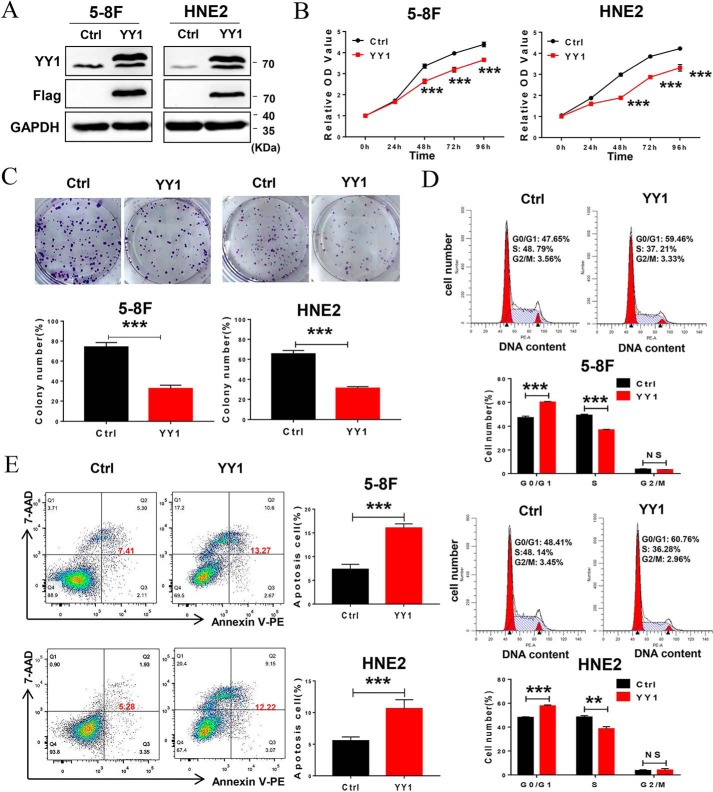
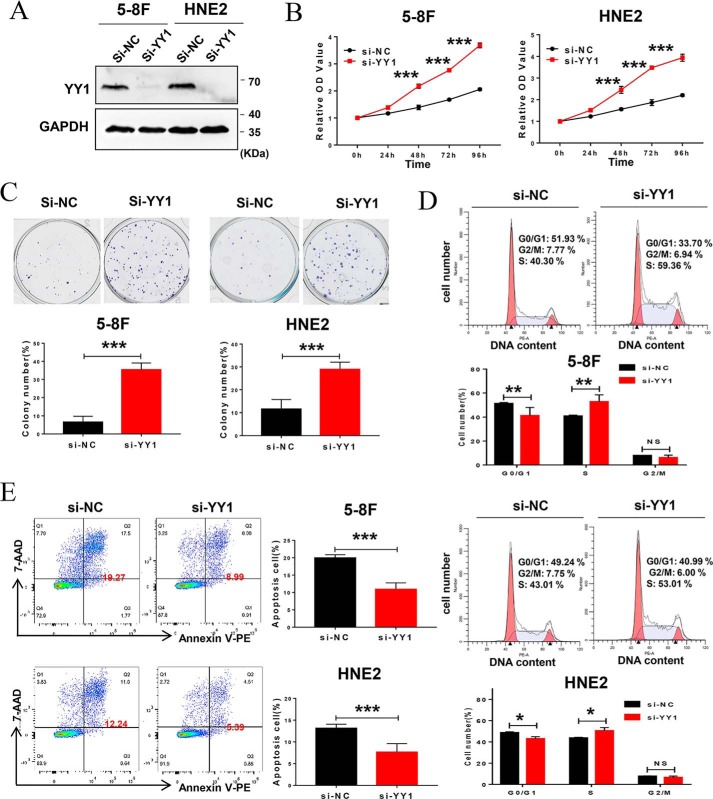
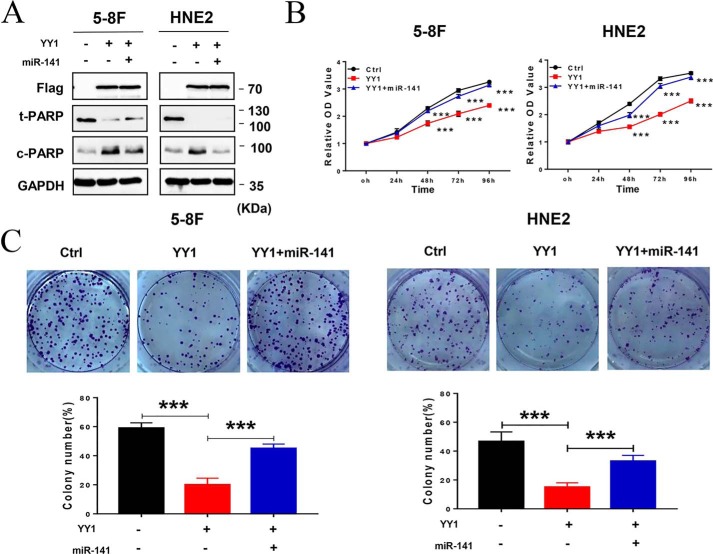
As an important transcription factor, YY1 plays dual biological roles as an oncogene or tumor suppressor in different tumors. However, its role in nasopharyngeal carcinoma has not been defined. To confirm the role of YY1 in NPC progression, 5-8F and HNE2 cell lines stably overexpressing YY1 were constructed, and the expression of exogenous YY1 was confirmed by Western blotting (Fig. 1A). Then, we investigated the effect of YY1 on cell proliferation. CCK-8 assays show that YY1 overexpression inhibits cell proliferation in both 5-8F and HNE2 cell lines (Fig. 1B), which was further confirmed by colony-forming assays. YY1-overexpressing 5-8F and HNE2 cells formed ∼40 and 50% fewer colonies, respectively, compared with their negative controls (Fig. 1C). To define the underlying biological mechanisms, we further analyzed the effect of YY1 overexpression on cell-cycle progression (Fig. 1D) and found that ectopic expression of YY1 significantly increases the G0/G1 population, followed by a decrease in the S-phase, in both 5-8F and HNE2 cells compared with control cells. Furthermore, we examined the effect of YY1 on cell apoptosis by conducting annexin V-PE/7-AAD double-staining and flow-cytometry analysis after the cells were serum-starved for 24 h, and the results show that both 5-8F and HNE2 cells overexpressing YY1 have significantly higher percentages of apoptotic cells compared with negative control (Fig. 1E). Furthermore, we investigated the effect of YY1 knockdown on cell biological functions by RNAi technology with si-YY1 (Fig. S1 and Fig. 2A), and the results show that knockdown of YY1 increases cell proliferation, colony formation, and cell-cycle G1/S transition (Fig. 2, B–D) and inhibits cell apoptosis (Fig. 2E). Taken together, these data suggest that YY1 inhibits cell proliferation and the G1–S transition and promotes apoptosis in both 5-8F and HNE2 cells and thus functions as a potential tumor suppressor in NPC cells.
Figure 1.
YY1 functions as a tumor suppressor in nasopharyngeal carcinoma. A, Western blotting using antibodies against YY1 and FLAG tag to confirm exogenous YY1 protein levels. GAPDH served as an internal control (Ctrl). B, CCK-8 assays of 5-8F and HNE2 stably overexpressing YY1 or negative control cells. C, colony-forming assays (upper panel) and quantification of colony 800/well (lower panel). D, cell-cycle analysis by flow cytometry. E, flow-cytometry analysis of cell apoptosis via annexin V-PE and 7-AAD double staining. Error bars represent the mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance. All experiments were performed in triplicate.
Figure 2.
Depletion of YY1 by siRNA promotes cell proliferation and inhibits apoptosis in NPC cell lines. A, 5-8F or HNE2 cells were transfected with scramble siRNA (negative control) and YY1 siRNA pool, respectively, and Western blotting was used to analyze the silence efficiency of YY1, and GAPDH served as an internal control. B, CCK-8 assays of 5-8F and HNE2 with transfection of YY1 siRNAs or negative control. C, colony-forming assays and quantification of colony 800/well (lower panel). D, cell-cycle analysis by flow cytometry. E, flow-cytometry analysis of cell apoptosis via annexin V-PE and 7-AAD double staining. Error bars represent the mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, no significance. All experiments were performed in triplicate.
YY1 negatively regulates c-Myc transcriptional activity via the c-Myc/Max/Mad network
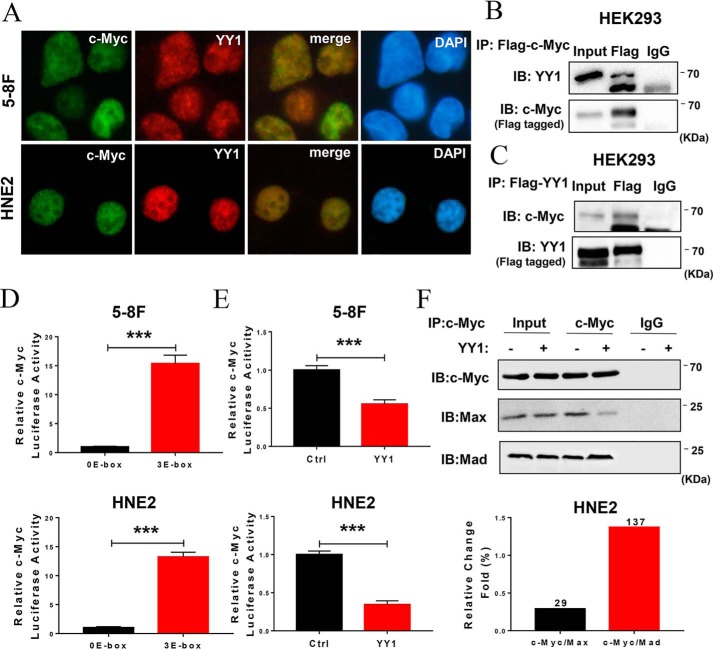
Our previous work showed that knockdown of c-Myc significantly inhibits cell proliferation and promotes apoptosis in NPC cells. Further bioinformatics analysis revealed YY1 as a potential c-Myc–interacting protein. To confirm the interaction between c-Myc and YY1, we analyzed the subcellular localization of YY1 and c-Myc in 5-8F and HNE2 cells by immunofluorescence, and the results show that YY1 is colocalized with c-Myc in the nucleus (Fig. 3A). YY1 was also found to interact with c-Myc, as performed by co-immunoprecipitation experiments with the lysates from HEK293 cells co-transfected with c-Myc and YY1 (Fig. 3, B and C). Because both YY1 and c-Myc are critical transcription factors, we then explored the effect of YY1 on the expression of c-Myc as well as the effect of c-Myc on the expression of YY1. Western blotting and qRT-PCR indicated that overexpression or knockdown of YY1 has no effect on c-Myc protein or mRNA levels (Fig. S2, A, B, and E). Meanwhile, knockdown of c-Myc similarly fails to alter YY1 protein or mRNA levels (Fig. S2, C and D). Therefore, these results suggest that YY1 is a component of the c-Myc transcriptional complex and a negative regulator of c-Myc transcriptional activity.
Figure 3.
YY1 regulates c-Myc transcriptional activity via the c-Myc/Max/Mad network. A, subcellular colocalization of endogenous YY1 and c-Myc detected by immunofluorescence in 5-8F and HNE2 cell lines. Images were obtained via confocal microscopy. B and C, co-IP assays and Western blotting confirmed the interaction between YY1 and c-Myc in HEK293 cells. D and E, dual-luciferase reporter assays confirmed the transcriptional activity of c-Myc toward the consensus E-box sequence (D) and the effect of YY1 overexpression on c-Myc activity (E) in HNE2 and 5-8F cells. Data are normalized to Renilla activity (pRL-TK). F, effect of YY1 on c-Myc binding to Max or Mad was investigated by co-IP and Western blotting, and the amount of c-Myc binding to Max or Mad reflected the transcriptional activity intensity of c-Myc. Error bars represent the mean ± S.D. ***, p < 0.001. All experiments were performed in triplicate. IB, immunoblot.
c-Myc is a helix-loop-helix leucine zipper protein that dimerizes with an obligate partner, Max, to bind DNA sites, 5′-CACGTG-3′, termed E-boxes. Therefore, we first confirmed the promoter activity of a c-Myc target consensus sequence containing three E-boxes in NPC cells (Fig. 3D), and we then investigated the effect of YY1 on c-Myc transcriptional activity using double-luciferase reporter assays. The results show that c-Myc activity was significantly down-regulated in YY1-overexpressing cells compared with control cells (Fig. 3E), suggesting that YY1 might directly play a negative role in c-Myc–mediated transcription. Furthermore, co-IP experiments were performed on total protein extracts to confirm that overexpression of YY1 reduces the formation of Max and c-Myc heterodimers ∼71% compared with the negative control. Conversely, the Mad and c-Myc protein complex increases ∼37% (Fig. 3F). These results indicate that YY1 inhibits c-Myc transcriptional activity toward the target gene by decreasing the binding of the c-Myc/Max/Mad complex to E-box sequences.
YY1 negatively regulates the transcription and expression of c-Myc oncogenic target miR-141
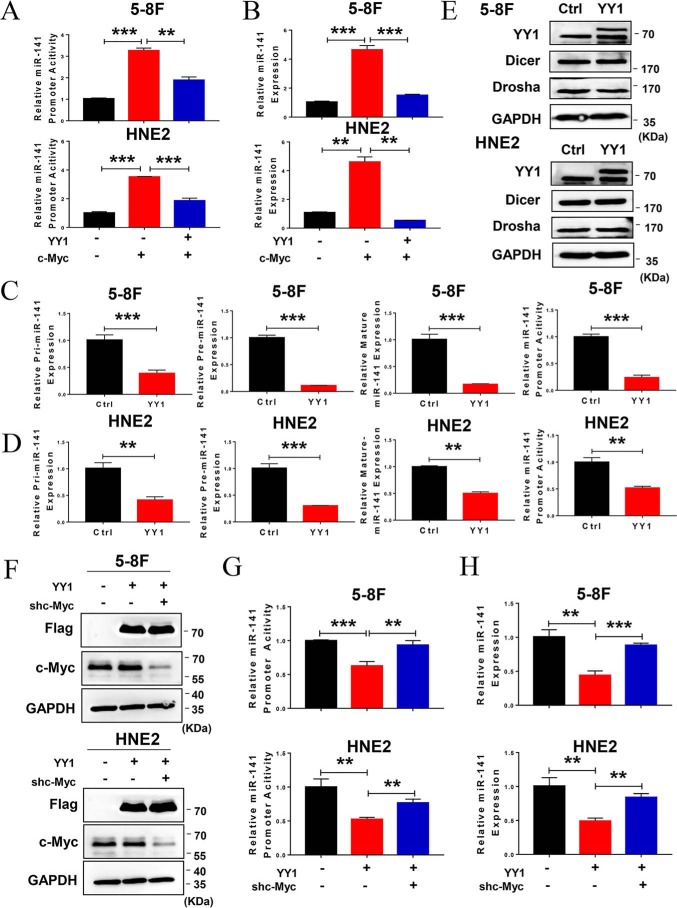
Because YY1 is a component of the c-Myc transcriptional complex and negatively regulates c-Myc transcriptional activity, miR-141 was previously identified as a critical oncogenic miRNA that is directly transcriptionally regulated by c-Myc. We further verified the functions of miR-141 in HNE2 cells used in this study and found that miR-141 promotes cell proliferation and colony formation (Fig. S3, A and B). Thus, we hypothesized that YY1 is a negative regulator of the c-Myc/miR-141 transcriptional axis and thus plays a critical role in the progression of NPC. Therefore, we first used dual-luciferase report assays and real-time PCR to detect the effect of YY1 on c-Myc–dependent miR-141 transcription and expression. As a result, ectopic expression of c-Myc led to the up-regulation of miR-141 promoter activity and expression, which is consistent with previous results (12), whereas YY1 overexpression decreases the effect of c-Myc on miR-141 promoter activity and expression in 5-8F and HNE2 cells (Fig. 4, A and B). Next, we investigated the effect of YY1 on the expression of pri-, pre-, and mature miR-141, as well as the promoter activity of miR-141 driven by endogenous c-Myc in NPC cells. The qRT-PCR results show that overexpressing YY1 down-regulates the expression of pri-, pre-, and mature miR-141, as well as miR-141 promoter activity (Fig. 4, C and D), but they present no effect on the expression levels of Drosha and Dicer, which are involved in miRNA processing (Fig. 4E). Inversely, knockdown of YY1 increases the expression of miR-141 (Fig. S2F). To verify whether the regulation of YY1 on miR-141 depends on c-Myc, we examined the effect of YY1 overexpression on the promoter activity and expression of miR-141 in both 5-8F and HNE2 cells with c-Myc knockdown, and the results show that c-Myc knockdown causes the deregulation of YY1 on the promoter activity and expression of miR-141 (Fig. 4, F–H). These findings demonstrate that YY1 transcriptionally down-regulates miR-141 expression by negatively regulating c-Myc transcriptional activity.
Figure 4.
YY1 negatively regulates c-Myc–mediated miR-141 transcription. A, dual-luciferase reporter assay shows the effect of YY1 overexpression on miR-141 promoter activity regulated by c-Myc in 5-8F and HNE2 cells. Data are normalized to Renilla activity (pRL-TK). B, qRT-PCR analysis shows the effect of YY1 overexpression on miR-141 levels regulated by c-Myc in 5-8F and HNE2 cells. U6 served as an internal control. C and D, dual-luciferase reporter assay and qRT-PCR analysis show miR-141 promoter activity and pri-miR-141, pre-miR-141, and mature miR-141 levels in YY1-overexpressing 5-8F (C) and HNE2 (D) cells. U6 served as an internal control. E, Western blotting of Dicer and Drosha protein levels in YY1-overexpressing 5-8F and HNE2 cells. GAPDH served as an internal control. F, Western blotting was performed to confirm the protein levels of exogenous YY1 and endogenous c-Myc by using antibodies against FLAG tag and c-Myc in 5-8F and HNE2 cells. GAPDH served as an internal control. G, dual-luciferase reporter assay shows the effect of YY1overexpression on miR-141 promoter activity in c-Myc–knockdown 5-8F and HNE2 cells. Data are normalized to Renilla activity (pRL-TK). H, qRT-PCR analysis shows the effect of YY1 overexpression on miR-141 levels in c-Myc–knockdown 5-8F and HNE2 cells. U6 served as an internal control. Error bars represent the mean ± S.D. **, p < 0.01; ***, p < 0.001. All experiments were performed in triplicate.
Restoring miR-141 levels reverses the effect of YY1 on cell proliferation and apoptosis in NPC cells
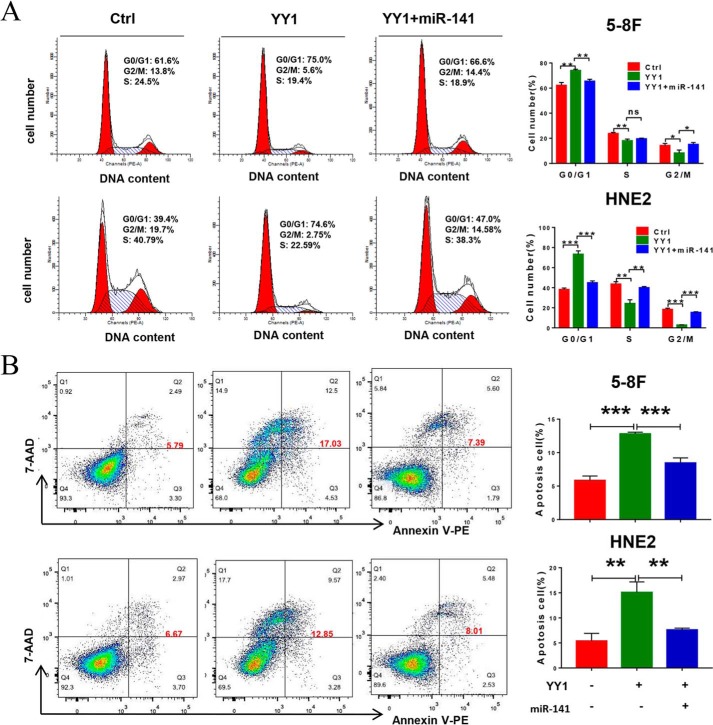
Given that YY1 functions as a tumor suppressor in NPC by inhibiting cell proliferation and inducing apoptosis and that YY1 down-regulates oncogenic miR-141 expression, we wondered whether the effect of YY1 on cell proliferation and apoptosis is mediated by its negative regulation of miR-141. Therefore, rescue assays were performed by cotransfecting 5-8F/YY1 and HNE2/YY1 cells with the miR-141 mimic. The expression of exogenous miR-141 was confirmed by qRT-PCR (Fig. S4). To determine the impact of miR-141 restoration on cell proliferation and colony formation by NPC cells, we performed CCK-8 and colony-forming assays. As expected, ectopic expression of YY1 obviously inhibits cell proliferation and colony formation in 5-8F and HNE2 cells (YY1), whereas restoring the expression of miR-141 in YY1-overexpressing cells (YY1 + miR-141) markedly rescues cell proliferation and colony formation compared with YY1-overexpressing cells (YY1) (Fig. 5, B and C). Furthermore, we analyzed the effect of miR-141 rescue on cell-cycle progression. The results show that overexpression of YY1 arrests cell cycle cells in the G0/G1 phase, but restoring the expression of miR-141 causes a significant decrease in the G0/G1 population, followed by an increase in S-phase cells compared with YY1 overexpression alone (Fig. 6A). Furthermore, overexpression of YY1 significantly increases the percentage of apoptotic cells in the NPC cells when compared with the negative controls, while restoring miR-141 expression dramatically rescues the effect of YY1 on apoptosis induction in NPC cells (Fig. 6B). Consistent with the results from flow-cytometry assays, Western blotting results show that overexpression of YY1 results in a significant increase of c-PARP, a critical apoptosis marker (11), while restoring the expression of miR-141 dramatically recovers the expression of c-PARP (Fig. 5A). Taken together, these findings demonstrate that rescuing miR-141 expression at least partially reverses the tumor-suppressive effect of YY1 on cell proliferation and apoptosis in vitro, suggesting that YY1 plays critical roles in cell-cycle arrest and initiation of apoptosis by negatively regulating miR-141 transcription during NPC progression.
Figure 5.
Restoring miR-141 levels reverses the effect of YY1 on cell proliferation and colony formation in NPC cells. A, Western blotting using antibodies against FLAG and the apoptosis marker c-PARP. GAPDH served as an internal control. B, CCK-8 assays of 5-8F and HNE2 stably overexpressing YY1 or negative control (Ctrl). C, colony-forming assays and quantification of colony 800/well. Error bars represent the mean ± S.D. ***, p < 0.001. All experiments were performed in triplicate.
Figure 6.
Restoring miR-141 levels reverses the effect of YY1 on cell cycle and apoptosis in NPC cells. A, cell-cycle analysis by flow cytometry. B, flow-cytometry analysis of cell apoptosis by annexin V-PE and 7-AAD double staining. Error bars represent the mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, no significance. All experiments were performed in triplicate.
miR-141 can rescue the tumor-suppressive effect of YY1 on tumor growth in vivo
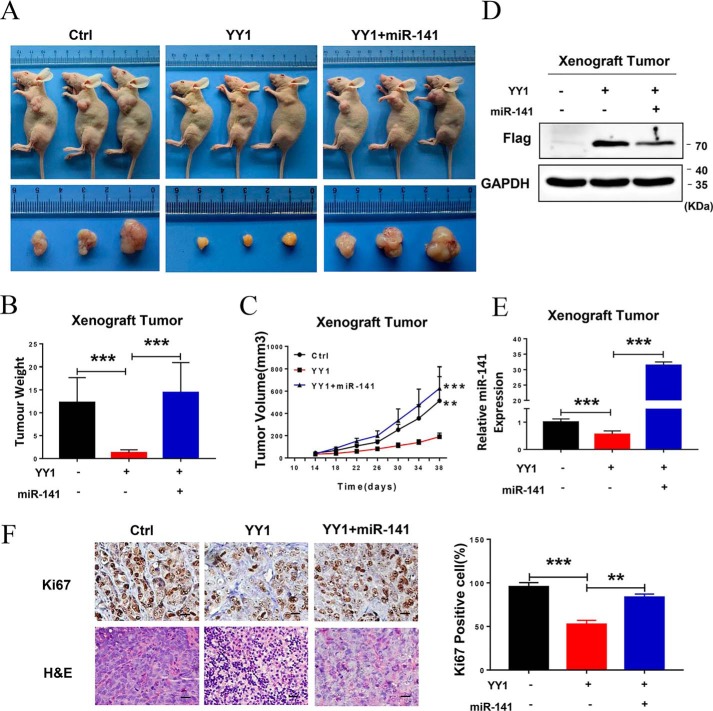
Because our in vitro results show that YY1 regulates miR-141 to affect NPC cell proliferation and apoptosis, we next investigated whether YY1 inhibits tumor growth through down-regulation of miR-141 in vivo. Therefore, we performed in vivo experiments using xenograft tumor models in nude mice. 5-8F/control, 5-8F/YY1, and 5-8F/YY1 with transfection of the miR-141 mimic were injected subcutaneously into the flanks of 5-week-old female nude mice, respectively. All xenograft model mice were killed on day 38 in order to examine the final tumor volume, and the results show that tumor size in nude mice injected with 5-8F/YY1 cells is obviously reduced compared with the negative control, whereas the size could be dramatically recovered when miR-141 expression was restored in 5-8F/YY1 cells (Fig. 7A). Meanwhile, xenograft tumor weight and growth rate with miR-141 restoration significantly increase compared with the 5–8 F/YY1 group (Fig. 7, B and C). In addition, the expression of YY1 and miR-141 in xenograft tumor tissues was confirmed by Western blotting and qRT-PCR (Fig. 7, D and E). To further confirm the effect of miR-141 restoration on tumor growth and apoptosis in vivo, we analyzed the percentage of cells expressing the proliferation marker Ki67, and the results show that ectopic expression of YY1 significantly reduces the number of Ki67-positive cells, whereas miR-141 restoration significantly recovers the number of Ki67-positive cells, which was also confirmed by H&E staining (Fig. 7F). These results indicate that YY1 inhibits tumor growth and presents anti-tumor effects in vivo, while restoring the expression of miR-141 significantly reverses these effects.
Figure 7.
YY1 inhibits tumor growth by down-regulating miR-141 expression in vivo. A, representative images (top panel) and tumor images (bottom panel) of the 5-8F xenograft model in nude mice. B, tumor weight quantification (n = 5). C, tumor growth curve (n = 5). D, Western blotting using anti-FLAG antibodies confirmed YY1 protein levels in xenograft tumor tissues. GAPDH served as an internal control. E, qRT-PCR assay confirming the expression of mature miR-141 in xenograft tumor tissues. U6 served as an internal control. F, representative IHC images of tumor samples that were stained with hematoxylin and eosin (H&E) and Ki67 (left). The percentages of Ki67-positive cells were measured (right). Three tumors were analyzed per group. Original magnification, ×200; scale bars, 50 μm. Error bars represent the mean ± S.E. **, p < 0.01; ***, p < 0.001.
YY1 inhibits tumor growth by repressing miR-141/PTEN/AKT signaling
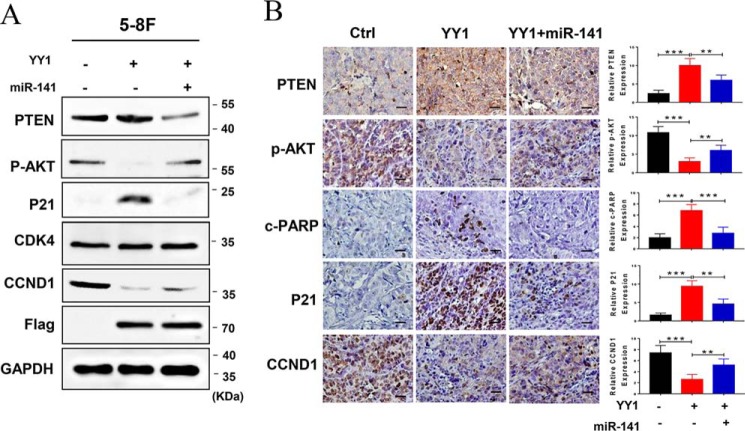
We previously reported that PTEN functions as a tumor suppressor by negatively regulating the AKT-signaling pathway. PTEN was identified to be a direct target of miR-141 and was post-transcriptionally down-regulated by miR-141 in NPC cells. Moreover, increasing evidence indicated that the aberrant PTEN/AKT pathway activation is essential for the development and progression of NPC. Therefore, we performed Western blotting and immunohistochemistry (IHC) to detect the expression of some critical components of the PTEN/AKT pathway in 5-8F cells and the corresponding xenograft tumor tissues. As a result, PTEN protein increases and p-AKT decreases in YY1-overexpressed NPC cells and their xenograft tumor tissues compared with the negative controls, while restoring miR-141 expression causes a significant reversion. In addition, we detected the expression of some molecules related to cell cycle and apoptosis, and the results reveal that CCND1 is largely decreased, while p21 and c-PARP are obviously increased. Moreover, restoring miR-141 expression in YY1-overexpressing cells results in a significant reversion, although CDK4 levels remain unaltered (Fig. 8, A and B). Consequently, these data show that YY1 inhibits cell proliferation and induces apoptosis by down-regulating the miR-141/PTEN/AKT pathway in vitro and in vivo.
Figure 8.
YY1 inhibits tumor growth by repressing miR-141/PTEN/AKT signaling. A, relative protein expression levels in miR-141 mimic-transfected 5-8F and HNE2 stable cell lines were analyzed by Western blotting. GAPDH served as an internal control. B, IHC (DAB staining) for PTEN, p-AKT, p21, CCND1, and c-PARP in the 5-8F xenograft model. Three tumors were analyzed per group. Original magnification, ×200; scale bars, 50 μm. Error bars represent the mean ± S.D. Control: vector + miR-NC; YY1: YY1 + miR-NC. Error bars represent the mean ± S.D. **, p < 0.01; ***, p < 0.001.
YY1 expression was negatively correlated with miR-141 and survival in human NPC patients
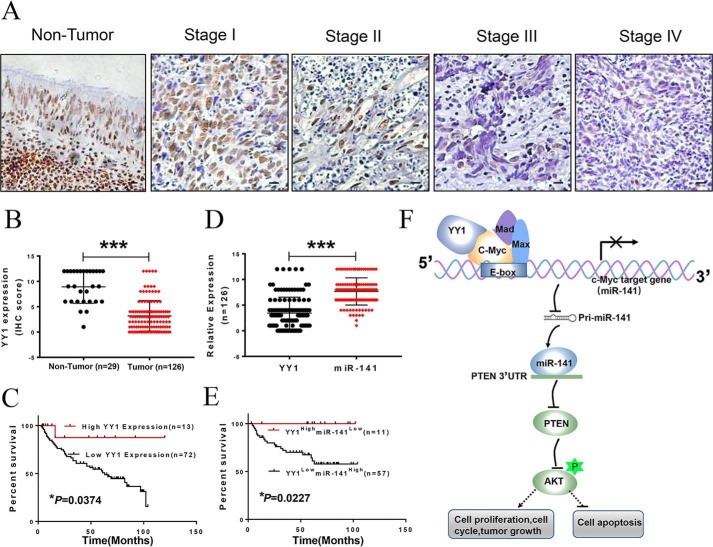
To confirm the role of YY1 and miR-141 in NPC, we detected their expression in NPC tissues derived from 29 healthy donors and 126 patients varying in age, gender, histological type, and the clinical stage using IHC staining and in situ hybridization (ISH). As a result, most patients exhibit significant reduction of the YY1 protein, and YY1 expression is decreasing with the increasing clinical TNM stage in NPC patients (Table 1 and Fig. 9, A and B). We also analyzed the association between YY1 expression and the survival of NPC patients with follow-up data selected from the total of NPC samples. The survival time ranged from 3 to 120 months, and the OS rate of patients with high YY1 expression is significantly higher compared with low YY1 expression (Fig. 9C). These results indicate that YY1 plays an essential role in NPC development. However, the average miR-141 levels in NPC tissues were higher than those in nontumor NPC control tissues (Fig. S5, A and B), and the OS rate of patients with low miR-141 expression is significantly higher than that in patients with high miR-141 expression (Fig. S5C), and the expression level of miR-141 is negatively correlated with the protein level of YY1 in NPC patients (r = −0.207, p < 0.001, Table 2) (Fig. 9D). We then assessed the relationship between the expression levels of YY1 and miR-141 and prognosis. The results show that low YY1 and high miR-141 expressions in NPC patients are both associated with poorer survival rates. Conversely, high YY1 and low miR-141 expression correlated with favorable survival in NPC patients (Fig. 9E). These findings indicate that both YY1 and miR-141 might be involved in NPC progression and that YY1 might be a negative regulator of miR-141 in NPC. In conclusion, YY1 is a component of the c-Myc transcriptional complex and negatively regulates c-Myc transcriptional activity by disrupting the c-Myc/Max complex; therefore, YY1 inhibits cell proliferation and tumor growth and initiates apoptosis through inactivation of the c-Myc/miR-141/PTEN/AKT axis in NPC (Fig. 9F).
Table 1.
Association between the expression of YY1, miR-141 and NPC clinical pathological features (n = 126)
The following abbreviations were used: DNC, differentiated nonkeratinized nasopharyngeal carcinoma; UDNC, undifferentiated non-eratinized nasopharyngeal carcinoma; H, high expression; L, low expression. Statistical analysis was performed using the Chi-squared test. *, p < 0.05; ***, p < 0.001.
| Characteristics (N) | YY1 |
miR-141 |
YY1/miR-141 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| H (%) | L (%) | p value | H (%) | L (%) | p value | H-L (%) | l-H (%) | p value | |
| Age (year) | |||||||||
| ≤40 (n = 52) | 29 | 71 | 0.7156 | 88 | 12 | 0.8325 | 6 | 67 | 0.6587 |
| >40 (n = 74) | 23 | 77 | 81 | 19 | 11 | 65 | |||
| Gender | |||||||||
| Female (n = 25) | 23 | 77 | 0.5013 | 84 | 16 | 0.9316 | 8 | 68 | 0.8474 |
| Male (n = 101) | 32 | 68 | 84 | 16 | 9 | 65 | |||
| Histological type | |||||||||
| DNC (n = 33) | 18 | 82 | 0.3481 | 82 | 18 | 0.664 | 9 | 61 | 0.5624 |
| UDNC (n = 93) | 28 | 72 | 85 | 15 | 9 | 68 | |||
| Clinical stages | |||||||||
| Stage I (n = 15) | 60 | 40 | 0.0247* | 67 | 33 | <0.001*** | 27 | 40 | 0.0158* |
| Stage II (n = 52) | 25 | 75 | 77 | 23 | 6 | 56 | |||
| Stage III (n = 34) | 24 | 76 | 97 | 3 | 6 | 74 | |||
| Stage IV (n = 25) | 8 | 92 | 92 | 8 | 0 | 92 | |||
Figure 9.
YY1 expression is negatively correlated with miR-141 and poor prognosis in NPC patients. A, representative images of YY1 expression in noncancerous nasopharyngeal tissues and different clinical TNM stages of NPC detected by IHC. B, box diagram of YY1 expression in noncancerous control (n = 29) and NPC tissues (n = 126). C and E, Kaplan-Meier overall survival analysis of NPC patients by expression of YY1 (C) or both of YY1 and miR-141 (E). Clinicopathological characteristics and statistical significance were assessed using the log-rank test. Kaplan-Meier curves show that worse survival rates in 64 NPC patients with low YY1 protein expression and high miR-141 expression compared with the related controls. D, YY1 and miR-141 expression in NPC. F, working model showing the mechanism of YY1 in negatively regulating cell proliferation and tumor growth in NPC through the inactivation of the c-Myc/miR-141/PTEN/AKT pathway. Error bars represent the mean ± S.D. *, p < 0.05; ***, p < 0.001.
Table 2.
The expression level of miR-141 was negatively correlated with YY1 in NPC patients (n = 126)
Statistical analysis was performed using the Pearson correlation analysis, ***, p < 0.001.
| miR-141 expression | YY1 expression |
||
|---|---|---|---|
| High (n = 28) | Low (n = 98) | ||
| High (n = 108) | 22 | 86 | r = −0.2078 |
| Low (n = 18) | 6 | 12 | ***, p < 0.001 |
Discussion
The YY1 transcription factor plays pivotal roles in normal biological processes such as development, differentiation, replication, and cell proliferation (22). The roles of YY1 in cancer have been explored recently. Interestingly, although YY1 functions as an oncogene in most cancers (23), it also acts as a tumor suppressor, such as in pancreatic cancer (24) and breast cancer (25), although its function in NPC has not been reported. In this study, we found that the expression of YY1 significantly decreased in NPC tissues compared with normal controls and that low YY1 levels inversely correlated with OS in NPC patients. Furthermore, overexpressing YY1 effectively inhibited cell proliferation and promoted apoptosis in NPC cell lines, and xenograft data in nude mice also confirmed that ectopic expression of YY1 can inhibit tumor growth in vivo. These data support a role for YY1 as a tumor suppressor in NPC and provide us with a new perspective on the function of YY1. In recent years, more and more genes with dual functions as oncogenes or tumor suppressors have been found in different of tumor types, as well as different stages of tumor development and progression, such as klf4 (26), atf2 (27), and tgf-β (28), indicating that the function of some genes is spatiotemporally dynamic and cannot be simply considered as inherently oncogenic or tumor-suppressive for functional mapping. To our knowledge, this is the first report to define the function of YY1 in NPC and to provide more exact data to support the dual roles of YY1 in different types of tumors.
YY1 was first discovered as a multifunctional transcription factor that can selectively initiate, activate, or repress transcription depending upon the context in which it binds to the recruited cofactors (20). Previously, Wu et al. (29) and Sankar et al. (30) reported that YY1 regulates the transcriptional activity of p53 by inhibiting its interaction with the coactivator p300 and by enhancing its interaction with the negative regulator Mdm2. More recently, Schlisio et al. (31) reported that YY1 and E2F2/E2F3 constitute a protein complex to regulate the Cdc6 promoter activity. In this study, YY1 was identified to be a c-Myc co-factor and to down-regulate c-Myc transcriptional activity. As we all know, c-Myc is a critical growth regulator that is commonly overexpressed in a wide range of cancers. Mechanistically, c-Myc is a helix-loop-helix leucine zipper protein that dimerizes with an obligate partner, Max, to bind DNA sites, 5′-CACGTG-3′, termed the E-box. Because binding to Max is necessary for all known c-Myc activities, the establishment of a link between YY1 and c-Myc in tumorigenesis is very interesting (32). In this study, YY1 was identified as a potent inhibitor of c-Myc transcriptional activity by reducing the formation of c-Myc and Max heterodimers, and overexpression of YY1 inhibits the activity of the promoters containing E-boxes. Therefore, YY1 acts as the most direct inhibitor for the interaction between c-Myc and Max, which suggests that YY1 reduces c-Myc–mediated transcription by disrupting the c-Myc/Max complex. It is expected to provide a new target for clinical treatment.
Impressively, accumulating evidence indicates that YY1 might be involved in the regulation of c-Myc–mediated target gene expression. Previous studies have shown that knocking down c-Myc significantly inhibits cell proliferation and tumor growth and down-regulates miR-141 in NPC cells by directly binding to its promoter (11). miR-141 is a member of the miR200 family that has been shown to be dysregulated in a wide variety of cancers and plays critical oncogenic roles in NPC (10). Meanwhile, c-Myc was identified as a negative regulator of the bromodomain-containing protein 7 (BRD7) gene, which is a tumor suppressor in multiple types of cancers, as well as a cofactor of c-Myc (33–35). Therefore, BRD7 expression and c-Myc activation form a double-negative feedback loop that controls cell proliferation and tumor growth in NPC by targeting the downstream gene miR-141 (12). In this study, we found that YY1 overexpression does not affect the mRNA and protein levels of c-Myc, but significantly reduces miR-141 expression, including pri-, pre-, and mature miR-141, as well as miR-141 promoter activity, suggesting that YY1 inhibits the activation of the c-Myc/miR-141 transcriptional axis. Additionally, the restoration of miR-141 significantly rescues the tumor-suppressive effect of YY1 overexpression in NPC cells. These results demonstrate that YY1 inhibits cell proliferation and tumor growth in NPC at least partially by inactivating the c-Myc/miR-141 axis. PTEN was reported as a direct target of miR-141, and it is post-transcriptionally down-regulated by miR-141 in NPC cells. Here, we confirmed that ectopic expression of YY1 in NPC cells results in increased PTEN protein level and decreased AKT phosphorylation, and restoring miR-141 rescues the alterations in PTEN expression and AKT phosphorylation, which demonstrates that the PTEN/AKT pathway might play critical roles in YY1-mediated tumor suppression.
Taken together, our results reveal a precise mechanism by which YY1 functions as a critical regulatory factor for c-Myc–mediated miR-141 transcription in NPC development and progression. As c-Myc is known to be deregulated in 30–50% of human malignancies with poor prognosis, finding more c-Myc targeting genes that are coregulated by YY1 and further investigating their function in NPC development and progression remain critical goals in the future.
Experimental procedures
Tissue samples
NPC samples (n = 126) and noncancerous nasopharyngeal tissues (n = 29) were all collected at the Second Xiangya Hospital of Central South University (Changsha, China); noncancerous nasopharyngeal tissues were collected from independent patients with chronic inflammation of the nasopharyngeal mucosa or polyps. The clinicopathological characteristics of the NPC patients are shown in Table 1. The NPC patients range in age from 23 to 74 years old, and 85 of the 126 patients have valid follow-up data. The longest survival time is 104 months. OS is defined as the time from diagnosis to the date of death or the date that the patient was last known to be alive. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C. Clinicopathological data are reviewed, and TNM staging classifications are based on the criteria of the American Joint Committee on Cancer (AJCC, 6th Ed.).
Cell lines
The human NPC cell lines 5-8F and HNE2 were preserved in our laboratory and were cultured in RPMI 1640 medium (Gibco-BRL, Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL, Invitrogen, Paisley, UK) in a humidified incubator with 5% CO2 at 37 °C. The embryonic kidney cell lines HEK293 and HEK293T were also preserved in our laboratory and maintained at 37 °C with a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL, Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum (Gibco-BRL, Invitrogen, Paisley, UK).
Cell transfections
Exponentially growing cells were trypsinized, counted, and seeded into 6-well plates to ensure 60–80% cell confluence on the following day for transfection. All of the constructs and oligonucleotides were transfected into the indicated cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. The hsa-miR-141 mimic (miR-141) and negative control (miR-NC) were purchased from RiBoBio (Guangdong, China), which was used to restore the expression of miR-141 in YY1-overexpressing cells according to the manufacturer's instructions. The specific small interfering RNAs (siRNAs) for YY1 were purchased from RiBoBio, and the siRNA sequences are as follows (5′ → 3′): siRNA#1, CGACGACTACATTGAACAA; siRNA#2, GCACAAAGATGTTCAGGGA. The two recombinant plasmids of c-Myc knockdown (pRNATU6.1/sh-c-Myc) and c-Myc overexpression (PIRES2-EGFP/2FLAG-c-Myc) were constructed by our laboratory as described previously (11).
Stable cell lines
As for the recombined lentiviral expression vector (pCDH-EF1-MCS-T2A-copGFP/YY1), the full-length ORF of human YY1 with three FLAG tags was linked to lentivirus expression vector pCDH-EF1-MCS-T2A-copGFP. For the construction of NPC stable cell lines with YY1 overexpression (HNE2/YY1 and 5-8F/YY1), the packaging vectors pMD2.G, psPAX2, and the recombined lentiviral expression vector were co-transfected into HEK293T packaging cells using Lipofectamine3000 reagent (Invitrogen). After transfection for 6 h, the medium was replaced with fresh medium with 10% FBS. After 30 h, virus particles in the medium were collected, filtered, and transduced into target cells. Then, flow-cytometry assay was further used to screen positive cells. Finally, the NPC stable cell lines with YY1 overexpression were obtained.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from tissue samples and cell lines using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized with 2 μg of total RNA using PrimeScriptTM first strand cDNA synthesis kit (TaKaRa, Dalian, China). Quantitative real-time PCR was performed using the miDETECT A Track miRNA qRT-PCR kit (RiBoBio) and the iCycler real-time PCR detection system (Bio-Rad) following the manufacturer's protocol. The U6 small nuclear RNA was used for normalization. The relative expression ratio of mature miR-141, pre-miR-141, and pri-miR-141 was calculated using the 2−ΔΔCt method. The primers for miR-141, pre-miR141, U6, pri-miR141, c-Myc, and GAPDH were described previously (11, 12). The relative amount of mRNA or gene to internal control was calculated using the equation 2−ΔΔCt method. Each sample was conducted in triplicate.
Western blot analysis
Cells and tissue samples were lysed in RIPA buffer in the presence of Protease Inhibitor Mixture and PhoSTOP (Roche Applied Science, Basel, Switzerland). Protein was quantified using a BCA protein assay kit (Pierce). Protein (30–80 μg) was separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA). The membranes were then blocked with 5% nonfat milk in TBS and then incubated with the following primary antibodies: anti-YY1 (dilution 1:500, Santa Cruz Biotechnology, SC-734); anti-c-Myc (dilution 1:1000, CST, D84C12); anti-FLAG (dilution 1:2000, Sigma, F-10084); anti-MAX (dilution 1:500, Santa Cruz Biotechnology, SC-8011); anti-Mad (dilution 1:500, Santa Cruz Biotechnology, SC-8012); anti-PARP (dilution 1:1000, CST, 46011); anti-c-PARP (dilution 1:1000, CST, ASP214); anti-CCND1 (dilution 1:500, Santa Cruz Biotechnology, SC-753); anti-CDK4 (dilution 1:500, Santa Cruz Biotechnology, SC-260); anti-PTEN (dilution 1:500, Bioworld Technology, BS1305); anti-pS473AKT (dilution 1:500, Bioworld Technology, BS4007); anti-AKT (dilution 1:500, Bioworld Technology, BS1810); anti-p21 (dilution 1:1000, CST, 12D1); anti-Drosha (dilution 1:1000, Proteintech, 55001-1-AP); anti-Dicer (dilution 1:500, Santa Cruz Biotechnology, SC-136979); or anti-GAPDH (dilution 1:1000, Proteintech, 10494-1-Ig) at 4 °C overnight. The membranes incubated with primary antibodies were then washed with PBST solution for 10 min in triplicate, then incubated with secondary antibodies, and washed. Signals were detected by an enhanced chemiluminescence detection system as per the manufacturer's protocol (Bio-Rad).
Immunofluorescence assay
Subcellular localization of endogenous c-Myc and exogenous YY1 was analyzed by immunofluorescence assay. 5-8F and HNE2 cells were seeded on glass coverslips, and then fixed with 4% paraformaldehyde for 15 min at room temperature and washed with PBS. Cells were then permeabilized with 0.3% Triton X-100 for 5 min at room temperature. Cells were then washed three times with PBS and incubated for 15 min in 5% goat serum and then incubated at 4 °C overnight with the primary antibody anti-c-Myc (dilution 1:800; CST) and anti-YY1 (dilution 1:100; Santa Cruz Biotechnology). After three washes for 5 min with PBS, cells were incubated for 1 h at 37 °C in the dark with the secondary antibody DyLight 488 AffiniPure goat anti-rabbit IgG (H+L) (dilution 1:1000; abbkine,A23220) and DyLight 594 AffiniPure goat anti-mouse IgG (H+L) (dilution 1:1000; Abbkine, A23410). Coverslips were washed with PBS and visualized using an epifluorescence microscope.
Co-IP
After transfecting for ∼36 h, the cells were lysed with lysis buffer for co-IP using the co-IP kit (Beyotime, Beijing, China). The cell lysates were then premixed with protease inhibitor mixture (Roche Applied Science) on ice for 20 min and then centrifuged at 12,000 × g for 15 min. Equal amounts of protein (1 mg) with 30 μl of protein A/G magnetic beads (Selleck, Houston, TX) precoated with anti-FLAG antibodies (Sigma) or anti-mouse IgG (Santa Cruz Biotechnology) were immunoprecipitated for 16–18 h at 4 °C. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and then followed by Western blotting assay probed with the corresponding antibody.
Luciferase reporter assays
The luciferase vector with E-box was a gift from Prof. Guoliang Qing (Wuhan University of Medical Research Institute, China) (36). The recombinant reporter vectors fused with the miR-141 promoter (pGL3 enhancer/miR-141P) were constructed by our laboratory as described previously (11). The proper insertions were further confirmed by sequencing. The cells were seeded in 24-well plates. After 12 h, pIRES2-EGFP/2FLAG-c-Myc and the luciferase vector with E-box or pGL3 enhancer/miR-141P were transiently co-transfected into 5-8F and HNE2-stable NPC cells with YY1 overexpression in triplicate using Lipofectamine 3000, respectively, and the pRL-TK vector (Promega) containing Renilla luciferase was used as an internal control. Cells were harvested 36 h post-transfection. Firefly and Renilla luciferase activities were measured using a dual-luciferase reporter kit (Promega) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
CCK-8 and colony-forming assays
Cell proliferation assays were performed using the cell counting kit-8 (CCK-8) (Selleck, Houston, TX) according to the manufacturer's instructions. Briefly, cells were seeded into 96-well plates (1000 cells per well). After the indicated time points (0, 24, 48, 72, and 96 h), 10 μl of CCK-8 reagent was added to each well containing 100 μl of growth medium. After incubation at 37 °C for 3 h, the number of viable cells was determined by measuring the optical density at 450 nm on a Beckman microplate reader (Beckman, Brea, CA).
For colony-forming assays, ∼800 cells per well were added to a 6-well plate, with three wells per sample. After incubation for 14 days, the cells were washed twice with PBS and stained with Crystal Violet Staining Solution (Beyotime, Beijing, China). Colonies containing more than 50 cells were counted as one positive colony. The plate clone formation efficiency was calculated as (number of colonies/number of cells inoculated) × 100%. All experiments were performed in triplicate.
IHC staining and ISH
For IHC, paraffin-embedded specimens were cut into 4-μm sections and baked for 1 h at 65 °C. After deparaffinization and rehydration, endogenous peroxidase was blocked using 0.3% hydrogen peroxide methanol for 20 min, and then heat-induced antigen retrieval was performed for 15 min in a microwave oven. Next, the sections were blocked with 5% goat serum for 30 min and then incubated overnight with the following: anti-YY1 (dilution 1:200, Abcam, EPR4652); anti-c-PARP (dilution 1:100, CST, ASP214); anti-CCND1 (dilution 1:100, Santa Cruz Biotechnology, SC-753); anti-PTEN (dilution 1:100, Bioworld Technology, BS1305); anti-pS473AKT (dilution 1:100, Bioworld Technology, BS4007); and anti-p21 (dilution 1:100, CST,12D1) primary antibodies using predetermined optimal dilutions and incubation times. Immune complexes were visualized using the MaxVision HRP-polymer IHC kit detection system, peroxidase/DAB, rabbit/mouse (MaxVision, Fuzhou, China), according to the manufacturer's protocol. Nuclei were counterstained with hematoxylin (Beyotime). Positive-control slides were included in every experiment in addition to the internal positive control. Antibody specificity was determined using a matched IgG isotype antibody as a negative control.
For ISH staining, the mature Hsa-miR-141–specific probe and negative control (Scramble) were purchased from Sangon (Sangon Biotech, Shanghai, China). An in situ hybridization kit (Boster, Wuhan, China) was used to perform probe hybridization according to the manufacturer's instructions.
The results of IHC and ISH were analyzed by examination and photography under microscopy with ×400 magnification. All the slices were evaluated by two pathologists without knowledge of the clinical outcome. The percentage of immune-positive cells and the staining intensities were evaluated in each sample. The percentage of immune-positive cells was graded on a scale of 0–4, where no staining was scored as 0; 1–10% of cells stained was scored as 1; 11–50% was scored as 2; 51–80% was scored as 3; and 81–100% was scored as 4. The staining intensities were graded from 0 to 3, where 0 was defined as negative; 1 as weak; 2 as moderate; and 3 as strong. The score of the molecule of interest in samples was calculated as the product of intensity and percentage scores, ranging from 0 to 12. The expression level was divided as low or high by the median total score (37, 38).
5-8F tumor xenograft model
All animal studies were approved by the Institutional Animal Care and Use Committee of Central South University (Changsha, China). For xenograft experiments, a total volume of 150 μl (8 × 106 cells in 0.9% saline solution with Matrigel) of 5-8F cells stably expressing YY1 or YY1 plus miR-141 mimic or negative control were injected subcutaneously into 5-week-old male BALB/C nude mice (n = 5 for each group). Mice were checked every 4 days, and tumor nodules were measured with a caliper. Tumor volume was evaluated using the following formula: volume = (length × width2)/2. Tumor growth curves were calculated. The three experimental groups were killed after 38 days. All tumor grafts were excised, weighed, harvested, fixed, and embedded. Anti-Ki67 (dilution 1:100, Bioworld), anti-c-PARP, anti-PTEN, anti-p-AKT, anti-P21, and anti-CCND1 antibodies were used to detect the expression of the corresponding molecules related to cell proliferation, apoptosis, or the PTEN/AKT pathway using immunohistochemistry. Samples were observed on an Olympus microscope (Olympus, Tokyo, Japan), and the results were calculated as the mean percentage of cells positive for Ki67 or other molecules in 10 different ×200 fields.
Statistical analysis
GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) and SPSS 18.0 (SPSS, Chicago, IL) were used to perform statistical analyses. The relationships between miR-141 and YY1 expression levels and clinical pathological characteristics in NPC were assessed using the χ2 test. Pearson correlation analysis was used to assess the significance of the correlation between YY1 and miR-141 expression in NPC. Kaplan-Meier analysis was performed for OS curves, and statistical significance was assessed using the log-rank test. Differences between groups were analyzed using Student's t test for pairs of groups or using one-way analysis of variance for more than two groups. A two-tailed value of p < 0.05 was considered statistically significant.
Author contributions
M. L. conceptualization; M. L., Y. L., Q. W., S. F., Xiayu Li, J. Z., K. C., and Y. Q. resources; M. L., W. N., Y. Z., and Z. L. software; M. L., Y. W., and H. W. formal analysis; M. L., C. W., and H. M. investigation; M. L., Y. L., H. W., and G. L. methodology; M. L. writing-original draft; M. L., G. L., and M. Z. writing-review and editing; Q. W. and S. F. visualization; W. X., Xiaoling Li, and M. Z. supervision; Z. Z. validation; M. Z. funding acquisition; M. Z. project administration.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 81772990 and 81572748, the “111” Project Grant 111-2-12, and the Central South University Graduate Research and Innovation Project Grants 2018zzts823 and 2018zzts233. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- NPC
- nasopharyngeal carcinoma
- BRD7
- bromodomain containing protein 7
- FBS
- fetal bovine serum
- IP
- immunoprecipitation
- IHC
- immunohistochemical staining
- ISH
- in situ hybridization
- DAB
- 3,3′-diaminobenzidine
- 7-AAD
- 7-aminoactinomycin D
- PE
- phycoerythrin
- PARP
- poly(ADP-ribose) polymerase
- c-PARP
- cleaved PARP
- YY1
- Yin Yang 1
- OS
- overall survival
- qRT-PCR
- quantitative RT-PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TNM
- tumor node metastasis.
References
- 1. Chua M. L. K., Wee J. T. S., Hui E. P., and Chan A. T. C. (2016) Nasopharyngeal carcinoma. Lancet 387, 1012–1024 10.1016/S0140-6736(15)00055-0 [DOI] [PubMed] [Google Scholar]
- 2. Masmoudi A., Toumi N., Khanfir A., Kallel-Slimi L., Daoud J., Karray H., and Frikha M. (2007) Epstein-Barr virus-targeted immunotherapy for nasopharyngeal carcinoma. Cancer Treat. Rev. 33, 499–505 10.1016/j.ctrv.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 3. Lee A. W., Ma B. B., Ng W. T., and Chan A. T. (2015) Management of nasopharyngeal carcinoma: current practice and future perspective. J. Clin. Oncol. 33, 3356–3364 10.1200/JCO.2015.60.9347 [DOI] [PubMed] [Google Scholar]
- 4. Pan J. J., Ng W. T., Zong J. F., Chan L. L., O'Sullivan B., Lin S. J., Sze H. C., Chen Y. B., Choi H. C., Guo Q. J., Kan W. K., Xiao Y. P., Wei X., Le Q. T., Glastonbury C. M., et al. (2016) Proposal for the 8th edition of the AJCC/UICC staging system for nasopharyngeal cancer in the era of intensity-modulated radiotherapy. Cancer 122, 546–558 10.1002/cncr.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyer N., and Penn L. Z. (2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- 6. Trop-Steinberg S., and Azar Y. (2018) Is Myc an important biomarker? Myc expression in immune disorders and cancer. Am. J. Med. Sci. 355, 67–75 10.1016/j.amjms.2017.06.007 [DOI] [PubMed] [Google Scholar]
- 7. Nesbit C. E., Tersak J. M., and Prochownik E. V. (1999) MYC oncogenes and human neoplastic disease. Oncogene 18, 3004–3016 10.1038/sj.onc.1202746 [DOI] [PubMed] [Google Scholar]
- 8. Adhikary S., and Eilers M. (2005) Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645 10.1038/nrm1703 [DOI] [PubMed] [Google Scholar]
- 9. Rahl P. B., and Young R. A. (2014) MYC and transcription elongation. Cold Spring Harb. Perspect. Med. 4, a020990 10.1101/cshperspect.a020990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L., Deng T., Li X., Liu H., Zhou H., Ma J., Wu M., Zhou M., Shen S., Li X., Niu Z., Zhang W., Shi L., Xiang B., Lu J., et al. (2010) microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis 31, 559–566 10.1093/carcin/bgp335 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Zhao R., Wang H., Luo Y., Wang X., Niu W., Zhou Y., Wen Q., Fan S., Li X., Xiong W., Ma J., Li X., Tan M., Li G., et al. (2016) miR-141 is involved in BRD7-mediated cell proliferation and tumor formation through suppression of the PTEN/AKT pathway in nasopharyngeal carcinoma. Cell Death Dis. 7, e2156 10.1038/cddis.2016.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Zhao R., Wei Y., Li M., Wang H., Niu W., Zhou Y., Qiu Y., Fan S., Zhan Y., Xiong W., Zhou Y., Li X., Li Z., Li G., et al. (2018) BRD7 expression and c-Myc activation forms a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141. J. Exp. Clin. Cancer Res. 37, 64 10.1186/s13046-018-0734-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niu Z., Liu H., Zhou M., Wang H., Liu Y., Li X., Xiong W., Ma J., Li X., and Li G. (2015) Knockdown of c-Myc inhibits cell proliferation by negatively regulating the Cdk/Rb/E2F pathway in nasopharyngeal carcinoma cells. Acta Biochim. Biophys. Sin. 47, 183–191 10.1093/abbs/gmu129 [DOI] [PubMed] [Google Scholar]
- 14. Ecevit O., Khan M. A., and Goss D. J. (2010) Kinetic analysis of the interaction of b/HLH/Z transcription factors Myc, Max, and Mad with cognate DNA. Biochemistry 49, 2627–2635 10.1021/bi901913a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lüscher B., and Larsson L. G. (1999) The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 18, 2955–2966 10.1038/sj.onc.1202750 [DOI] [PubMed] [Google Scholar]
- 16. Nair S. K., and Burley S. K. (2006) Structural aspects of interactions within the Myc/Max/Mad network. Curr. Top. Microbiol. Immunol. 302, 123–143 [DOI] [PubMed] [Google Scholar]
- 17. Shrivastava A., Yu J., Artandi S., and Calame K. (1996) YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc. Natl. Acad. Sci. U.S.A. 93, 10638–10641 10.1073/pnas.93.20.10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Austen M., Cerni C., Lüscher-Firzlaff J. M., and Lüscher B. (1998) YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 17, 511–520 10.1038/sj.onc.1201968 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Q., Stovall D. B., Inoue K., and Sui G. (2011) The oncogenic role of Yin Yang 1. Crit. Rev. Oncog. 16, 163–197 10.1615/CritRevOncog.v16.i3-4.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gordon S., Akopyan G., Garban H., and Bonavida B. (2006) Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25, 1125–1142 10.1038/sj.onc.1209080 [DOI] [PubMed] [Google Scholar]
- 21. Khachigian L. M. (2018) The Yin and Yang of YY1 in tumor growth and suppression. Int. J. Cancer 143, 460–465 10.1002/ijc.31255 [DOI] [PubMed] [Google Scholar]
- 22. Shi Y., Lee J. S., and Galvin K. M. (1997) Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332, F49–F66 [DOI] [PubMed] [Google Scholar]
- 23. Castellano G., Torrisi E., Ligresti G., Malaponte G., Militello L., Russo A. E., McCubrey J. A., Canevari S., and Libra M. (2009) The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle 8, 1367–1372 10.4161/cc.8.9.8314 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J. J., Zhu Y., Xie K. L., Peng Y. P., Tao J. Q., Tang J., Li Z., Xu Z. K., Dai C. C., Qian Z. Y., Jiang K. R., Wu J. L., Gao W. T., Du Q., and Miao Y. (2014) Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol. Cancer 13, 130 10.1186/1476-4598-13-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee M. H., Lahusen T., Wang R. H., Xiao C., Xu X., Hwang Y. S., He W. W., Shi Y., and Deng C. X. (2012) Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene 31, 116–127 10.1038/onc.2011.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen Y., Chen T. J., and Lacorazza H. D. (2017) Novel tumor-suppressor function of KLF4 in pediatric T-cell acute lymphoblastic leukemia. Exp. Hematol. 53, 16–25 10.1016/j.exphem.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhoumik A., Fichtman B., Derossi C., Breitwieser W., Kluger H. M., Davis S., Subtil A., Meltzer P., Krajewski S., Jones N., and Ronai Z. (2008) Suppressor role of activating transcription factor 2 (ATF2) in skin cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 1674–1679 10.1073/pnas.0706057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu J., Acharya S., Sahin O., Zhang Q., Saito Y., Yao J., Wang H., Li P., Zhang L., Lowery F. J., Kuo W. L., Xiao Y., Ensor J., Sahin A. A., Zhang X. H., et al. (2015) 14-3-3ζ turns TGF-β's function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell 27, 177–192 10.1016/j.ccell.2014.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu S., Kasim V., Kano M. R., Tanaka S., Ohba S., Miura Y., Miyata K., Liu X., Matsuhashi A., Chung U. I., Yang L., Kataoka K., Nishiyama N., and Miyagishi M. (2013) Transcription factor YY1 contributes to tumor growth by stabilizing hypoxia factor HIF-1α in a p53-independent manner. Cancer Res. 73, 1787–1799 10.1158/0008-5472.CAN-12-0366 [DOI] [PubMed] [Google Scholar]
- 30. Sankar N., Baluchamy S., Kadeppagari R. K., Singhal G., Weitzman S., and Thimmapaya B. (2008) p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene 27, 5717–5728 10.1038/onc.2008.181 [DOI] [PubMed] [Google Scholar]
- 31. Schlisio S., Halperin T., Vidal M., and Nevins J. R. (2002) Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21, 5775–5786 10.1093/emboj/cdf577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beaulieu M. E., McDuff F. O., Bédard M., Montagne M., and Lavigne P. (2013) Methods for the expression, purification, preparation, and biophysical characterization of constructs of the c-Myc and Max b-HLH-LZs. Methods Mol. Biol. 1012, 7–20 10.1007/978-1-62703-429-6_2 [DOI] [PubMed] [Google Scholar]
- 33. Liu H., Peng C., Zhou M., Zhou J., Shen S., Zhou H., Xiong W., Luo X., Peng S., Niu Z., Ouyang J., Li X., and Li G. (2006) Cloning and characterization of the BRD7 gene promoter. DNA Cell Biol. 25, 346–358 10.1089/dna.2006.25.346 [DOI] [PubMed] [Google Scholar]
- 34. Liu H., Zhou M., Luo X., Zhang L., Niu Z., Peng C., Ma J., Peng S., Zhou H., Xiang B., Li X., Li S., He J., Li X., and Li G. (2008) Transcriptional regulation of BRD7 expression by Sp1 and c-Myc. BMC Mol. Biol. 9, 111 10.1186/1471-2199-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu X., Li Z., and Shen J. (2016) BRD7: a novel tumor suppressor gene in different cancers. Am. J. Transl. Res. 8, 742–748 [PMC free article] [PubMed] [Google Scholar]
- 36. Yue M., Jiang J., Gao P., Liu H., and Qing G. (2017) Oncogenic MYC activates a feed forward regulatory loop promoting essential amino acid metabolism and tumorigenesis. Cell Rep. 21, 3819–3832 10.1016/j.celrep.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 37. Wen Q., Alnemah M. M., Luo J., Wang W., Chu S., Chen L., Li J., Xu L., Li M., Zhou J., and Fan S. (2015) FLOT-2 is an independent prognostic marker in oral squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 8, 8236–8243 [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao R., Liu Y., Wang H., Yang J., Niu W., Fan S., Xiong W., Ma J., Li X., Phillips J. B., Tan M., Qiu Y., Li G., and Zhou M. (2017) BRD7 plays an anti-inflammatory role during early acute inflammation by inhibiting activation of the NF-κB signaling pathway. Cell. Mol. Immunol. 14, 830–841 10.1038/cmi.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.