Abstract
The American Diabetes Association convened a research symposium, “Epigenetics and Epigenomics: Implications for Diabetes and Obesity” on 17–19 November 2017. International experts in genetics, epigenetics, computational biology, and physiology discussed the current state of understanding of the relationships between genetics, epigenetics, and environment in diabetes and examined existing evidence for the role of epigenetic factors in regulating metabolism and the risk of diabetes and its complications. The authors summarize the presentations, which highlight how the complex interactions between genes and environment may in part be mediated through epigenetic changes and how information about nutritional and other environmental stimuli can be transmitted to the next generation. In addition, the authors present expert consensus on knowledge gaps and research recommendations for the field.
Overview
The last decade has brought an explosion of interest in the epigenome, generally defined as changes to chromatin structure and function that do not involve altering the sequence of DNA. Epigenetic control is critical to both normal homeostasis and disease. Given that risk for diabetes and its complications is linked to both genetic and environmental factors, it is not surprising that there are now more than 1,000 articles that address the intersection of diabetes and epigenetics or epigenomics. There are multiple layers of epigenetic regulation: direct methylation of cytosine or adenine residues, covalent modifications to histone proteins, higher-order chromatin structure, and noncoding RNA (1). Each of these has been implicated in cellular processes relevant to diabetes. In fact, there is a long history of associations between epigenetics and diabetes, obesity, and other metabolic disorders. The agouti Ay mouse, for example, is an inbred strain that exhibits severe obesity resulting from epigenomic modification of a gene that affects food intake (2). Similarly, obesity seen in human conditions like Prader-Willi syndrome occurs because of aberrant imprinting, a specialized form of epigenomic modification (3). Newer data demonstrate that modifying the epigenome via the manipulation of specific chromatin-modifying enzymes has important metabolic consequences, causing or ameliorating obesity and hyperglycemia, depending on the specific experimental paradigm (4). Finally, there is a wealth of data implicating epigenetic mechanisms in the risk of inherited diabetes and obesity, best documented in humans through unfortunate experiments of history like the Dutch Hunger Winter (5), in addition to a plethora of rodent studies. The purpose of this meeting was to discuss new data linking epigenomics to metabolism and diabetes. The wide range of results presented reflect the broad contribution of epigenomics to many areas of diabetes research and highlight knowledge gaps and research recommendations to move the field forward toward therapeutic intervention (Table 1).
Table 1.
Research knowledge gaps and recommendations
| Epigenomics to understand GWAS |
| • Include critical cell and tissue types at different developmental stages relevant to metabolism in data sets and ensure integration of data sets. |
| • Develop novel high-throughput methods and single-cell resolution techniques to define and validate epigenomic function, including validation of appropriate animal models for the study in question. |
| • Collect human tissues from longitudinal studies with healthy and disease comparator sets and consider sex-specific effects in human and animal studies. |
| • Examine epigenomic contributions to diabetes and obesity pathophysiology earlier in disease progression to identify targets for prevention and cure. |
| Using the epigenome to understand β-cells and periphery |
| • Examine how nutrients and other environmental cues affect chromatin state in cell types controlling energy homeostasis. |
| • Determine the degree to which nutrient excess–induced epigenetic changes are reversible and how epigenetic imprints affect cell function after the stressor is removed. |
| • Determine whether distinct stressors are associated with the same epigenetic marks or different ones (i.e., whether there are common effects on various organs). |
| • Identify which chromatin modifications have the most functional impact to enhance opportunities for therapeutic intervention. |
| • Explore cell-specific drug targeting strategies to avoid broad systemic effects of modifying epigenetic regulators. Targeting the pathway from an environmental cue to the nucleus may allow more specific effects. |
| Using the epigenome to understand metabolism and complications |
| • Determine how cells transduce signals from environmental exposures to induce epigenetic changes in the nucleus. Studying the vascular endothelium might be helpful because it lines blood vessels in all tissues and, along with blood cells, is an initial sensor of environmental changes. |
| • Examine epigenome modifications in specific blood/immune/inflammatory cell types, as well as target tissues where available, to determine their involvement in complications of diabetes, including in monozygotic twins discordant for diabetes or complications. |
| • Explore EWAS in clinical cohorts of diabetes complications and metabolic memory. |
| • Collect and compare different tissues and encourage clinical trials to collect DNA from sorted cells moving forward. |
| • Consider tissue cross talk and hormone action and explore unifying mechanisms of metabolic diseases and complications, such as immune modulation, microbiome, and the gut-brain axis. |
| Epigenetic signals and intergenerational risk of chronic disease |
| • Determine specific epigenetic mechanisms responsible for intergenerational, nongenetic transmission of disease risk and whether the mechanisms influence the embryo or the placenta to a greater extent. |
| • Examine whether epigenetic mechanisms operate to ensure optimal survival of the next generation or simply as means to transmit environmental information to the next generation and optimize subsequent responses. |
| • Define the role of early-life exposures in mediating disease risk in populations transitioning from undernutrition to overnutrition or in individuals conceived via artificial reproduction techniques. |
| • Determine whether epigenetic mechanisms responsible for intergenerational risk can be used to develop markers of high risk and whether risk can be modified by early-life postnatal interventions or interventions that improve the metabolic health of both parents prior to reproduction. |
| • Standardize experimental animal models to determine whether epigenetic responses to diverse nutrient/metabolic stressors are unique or common. |
| Developing epigenetic therapeutics for metabolic disease |
| • Profile individual cell types, such as β-cells, rather than heterogenous tissues in the exploration of epigenetic therapeutics. |
| • Explore viral or other delivery vectors with cell-type specificity to all diabetes relevant tissues to achieve targeted therapy activating or repressing a given gene. |
| • Consider evaluation of drug safety for potential epigenetic modification therapies that are long acting for chronic diseases such as diabetes and obesity. |
| • Examine established therapies with effects beyond glycemic control to identify potential epigenomic effects in particular cell types. |
| • Define health outcomes to offspring of parents undergoing therapies that alter the epigenetic profile. |
While the term “epigenetics” has been formally defined for decades as a change in the state of expression of a gene (or a trait) that does not involve a mutation but that is nevertheless inherited (at least through a mitotic division) in the absence of the signal or event that initiated the change, the newer term “epigenomics” has, to our knowledge, never been formally defined. “Epigenomics” has been widely adopted operationally by the research community to indicate studies, on the genome-wide level, that focus on the analysis of DNA methylation, histone modifications, and noncoding RNAs (6,7).
Epigenomics to Understand GWAS
Over the last decade, genome-wide association studies (GWAS) have identified thousands of loci associated with human traits and diseases. Most of these risk-associated variants are in the noncoding portion of the genome. Thus far, analysis of risk alleles has focused on determining the molecular mechanism by which a given variant confers diabetes risk one locus at a time—a slow and costly endeavor. More recently, the focus has shifted toward the systematic characterization of loci using high-throughput methods (Fig. 1).
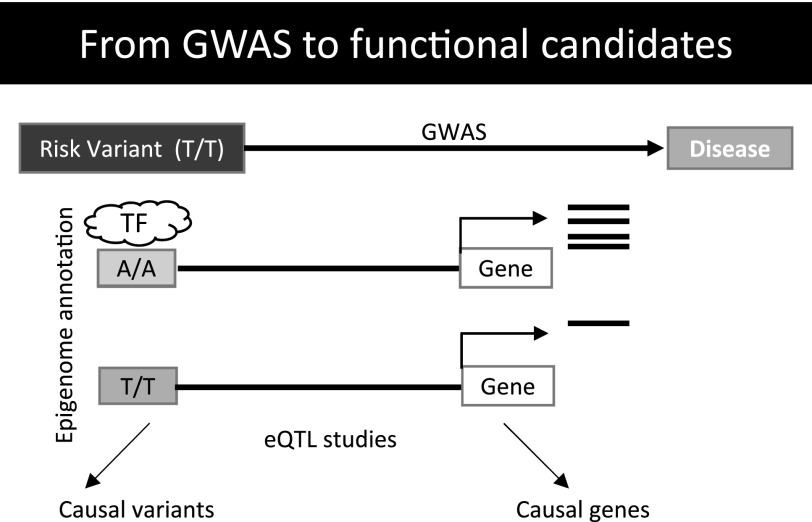
Figure 1.
From GWAS to functional candidates. Most complex trait variants are localized to the noncoding regions of the genome. Causal variants are enriched on cell-type–specific regulatory elements and alter transcription factor binding and influence the expression of causal genes. Given the small effect of each variant on gene regulation and disease phenotype, it is likely that networks of genes influence disease.
Thus far, more than 400 genetic loci have been associated with the risk for type 2 diabetes, many of which are thought to have their main function in β-cells (8). The recent determination of distinct chromatin states through global mapping of key histone marks has greatly facilitated the identification of risk-conferring single nucleotide polymorphisms (9,10). Anna L. Gloyn presented the first systematic survey of the molecular mechanism of type 2 diabetes–associated variants. By combining epigenomic annotations with genetic fine mapping results, they were able to identify the risk-conferring single nucleotide polymorphism for multiple risk loci, including variants that disrupt the melatonin receptor (MTNR1B), ADCY5 (11) and ZMIZ1 (12). Using islet-specific epigenome maps followed by functional screens of positional target genes, Gloyn and colleagues identified effects for 37 of 75 risk loci for type 2 diabetes, providing one of the first demonstrations of a truly scalable, high-throughput screen of cellular phenotypes for GWAS (13).
Identifying the disease-relevant cell types is challenging, even with tissue-specific epigenomes. Marcelo A. Nóbrega showcased a framework that makes this possible. The TCF7L2 locus, with the strongest known type 2 diabetes association, contains numerous enhancers that are active across a variety of cell types. Allele-specific effects on enhancer activity point to three tissues: pancreatic islets, enteroendocrine cells, and adipocytes, but it remains unclear in which the variant is causal. By adding an extra copy of the gene in all tissues of transgenic mice, which led to insulin resistance (14), Nóbrega and colleagues were then able to remove the extra copies from each of the three candidate tissues in order to reveal which one was involved in the disease mechanism. Indeed, only overexpression of TCF7L2 in adipocytes is sufficient to produce insulin resistance. They then narrowed down the causal variant within the haplotype using gene editing, showing how a single nucleotide change, the risk T allele of rs7903146, is sufficient to repress adipogenesis and produce hypertrophic cells.
The Claussnitzer laboratory described the functional characterization of the FTO locus, the genomic area with the strongest statistical association to obesity, which exemplifies the crucial role of epigenomics in identifying the causal variant, genes, and cell type of action (15). Chromatin state maps from the National Institutes of Health Roadmap Epigenomics Mapping Consortium and Hi-C links between enhancers and genes allowed them to jumpstart their search for allele-specific effects of variants on the epigenome and tissue-specific expression quantitative trait loci (eQTLs) with the neighboring genes. In a second study, epigenomic data helped to elucidate a pleiotropic effect of the ADCY5 locus in both fat and bone. The activation of this gene leads to increased lipolysis and secretion of fatty acids into the bloodstream by adipocytes, and increased bone formation through stimulated differentiation in osteoblasts—effects that were validated through a fast and scalable CRISPR method that allows for the cost-effective screening of entire credible sets.
Studies of the genetics of complications of diabetes, such as diabetic kidney disease, use similar approaches. The Susztak laboratory has generated whole-kidney and kidney cell-type–specific epigenome maps and eQTL maps to annotate kidney disease GWAS loci. Integration of GWAS and eQTL maps highlighted several important genes and pathways for diabetes complications (16,17).
Reference epigenomes are one of the first data sources used to illuminate risk-associated variants, but all available resources are static snapshots derived from a small number of individuals. The Snyder group is focused on better understanding the temporal and personal dimension of epigenomes in health and disease by pairing constant phenotyping through wearables and frequent collection of omics data, including methylomes, epigenomes, and transcriptomes, among others. These data were collected for 105 subjects as part of the Integrated Personal Omics Profiling (iPOP) study (18). In insulin-resistant individuals, they found two major pathways that tracked with body-weight changes: inflammation and hypertrophic cardiomyopathy (19). These studies highlighted the importance of establishing precise, personalized omics profiles, as their interpretation was highly dependent on each individual’s baseline.
Using the Epigenome to Understand β-cells and Periphery
The metabolic environment has major effects on the function of cells involved in the regulation of energy metabolism (20–23). Longer-term adaptive responses are largely mediated through regulation of gene transcription, through the interaction between transcription factors, coregulators, and the basal transcriptional machinery. Recent evidence shows that metabolic signals play critical roles in determining chromatin structure (24). Most chromatin-modifying enzymes require substrates or cofactors that are intermediates of cell metabolism. Therefore, fluctuation of metabolite levels could modulate activities of these enzymes and effect changes in gene transcription by influencing chromatin state. Defining the molecular connections between cellular metabolism, the epigenome, gene transcription, and cell function could provide an opportunity for therapeutic intervention via targeting of the epigenome. The speakers in this session presented findings addressing these questions in the context of β-cells, liver, adipose, and skeletal muscle.
Epigenetic mechanisms have been shown to contribute to the decline in β-cell proliferation with age (25,26). By establishing genome-wide maps of DNA methylation in β-cells from young and aged mice, the Kaestner laboratory showed that proproliferative gene loci are de novo methylated with age, while enhancers near genes involved in glucose metabolism undergo demethylation (27). Similar age-dependent DNA methylation patterns occur in human β-cells, suggesting that aging could affect β-cell function via modulation of the epigenome. Further illustrating the dynamic regulation of the epigenome in β-cells, Sander and coworkers reported changes in histone acetylation during feeding and fasting at key genes controlling metabolic processes (unpublished data). They further identified a chromatin-modifying enzyme that is recruited to sites of dynamic chromatin in β-cells and regulates fasting blood glucose levels. Together, these studies provide the first insights into chromatin dynamics in β-cells and suggest that modulation of the epigenome is a mechanism by which β-cells adapt their insulin secretory response to changing metabolic demands.
In addition to nutrient state, the circadian clock also has significant effects on cellular metabolism (28,29). Peripheral clocks are present in most tissues involved in the regulation of metabolic homeostasis, including β-cells, liver, adipose, and muscle. Since core clock transcription factors are partners of chromatin-modifying enzymes, disruption of circadian rhythms can affect cell function through regulation of the epigenome. The Lazar laboratory identified nuclear receptors, other transcription factors, and histone deacetylases (HDACs) as important components of the transcriptional complexes involved in circadian regulation of liver metabolism and fasting blood glucose levels (unpublished data). Their studies demonstrate highly dynamic regulation of liver chromatin state and architecture during fasting/feeding transitions and in response to circadian rhythm. The Rosen laboratory examined chromatin state in brown, beige, and white adipocytes in response to cold and heat exposure and observed that beige adipocytes have unique plasticity not shared by interscapular brown adipocytes (30). Further, beige cells that have been “whitened” after warming retain certain epigenetic features of the cold state. This mechanism allows for rapid reactivation of the thermogenic program upon re-exposure to cold, thereby protecting animals from hypothermia. These findings exemplify the importance of the epigenome for retaining a memory of prior environmental influences and imply that metabolic history cannot be easily erased and may determine future responses. Mechanisms of epigenetic memory also appear to be relevant in skeletal muscle, where high-fat feeding has been shown to leave an epigenetic imprint even after return to normal diet (31). Barres and coworkers showed that exercise can counteract negative effects of obesity on muscle cell metabolism, acting, at least in part, through the regulation of chromatin state (unpublished data). An overarching observation across all tissues is that the epigenome is highly plastic in response to environmental cues. We are also beginning to recognize that past metabolic experiences can be “memorized” at the level of the epigenome and can influence future responses (Fig. 2).
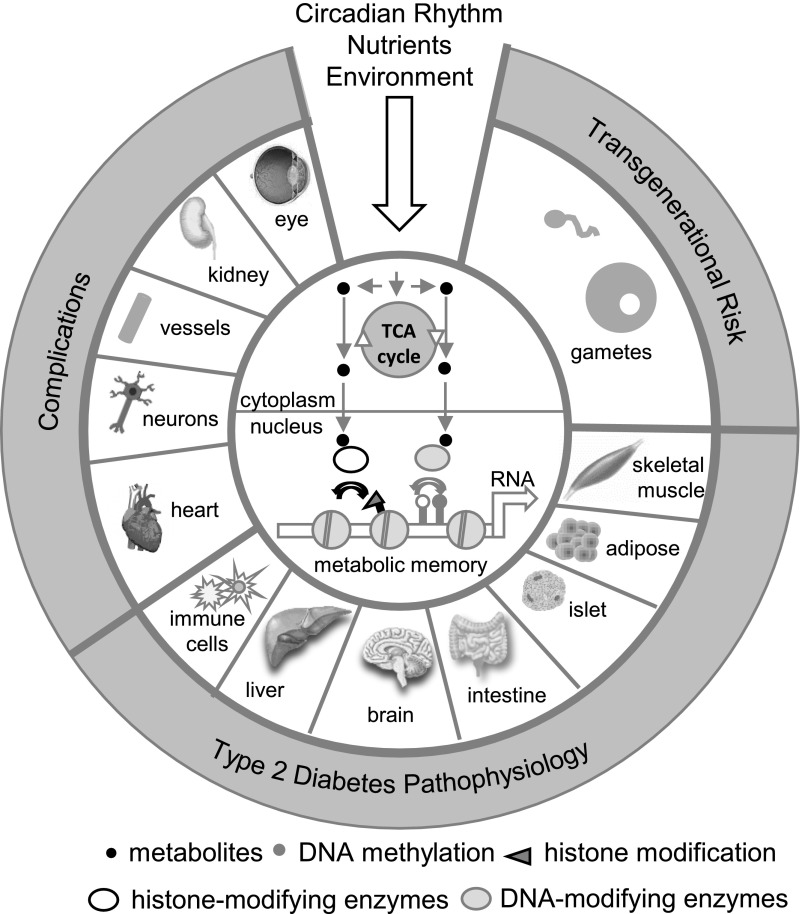
Figure 2.
Effects of nutrients, circadian rhythm, and other environmental cues on the epigenome and pathogenesis of type 2 diabetes. Systemic environmental factors affect cellular metabolism and concentrations of intermediate metabolites that are used as substrates and cofactors for enzymes that coordinate epigenetic status (i.e., histone- and DNA-modifying enzymes). Alterations to the epigenome can have lasting effects on cellular responses that persist independent of the environmental stimulus (“metabolic memory”). The epigenetic reprogramming of insulin target tissues, islet cells, and immune cells contributes to the development of type 2 diabetes, while reprogramming of gametes has transgenerational effects. Hyperglycemia leads to persisting epigenetic changes in tissues involved in diabetes complications.
Using the Epigenome to Understand Metabolism and Complications
Diabetes is associated with significantly accelerated rates of micro- and macrovascular complications. Clinical trials in people with diabetes have underscored the beneficial effects of intensive glycemic control for preventing the progression of complications. Interestingly, the rate of diabetes complications can be affected by glucose levels that were experienced years earlier, a phenomenon called metabolic memory (32). This is exemplified by results of the Diabetes Control and Complications Trial (DCCT), which demonstrated that patients with type 1 diabetes placed on intensive glycemic control had a lower incidence and severity of complications relative to those on conventional therapy (33,34). After the trial was complete, in the follow-up observational Epidemiology of Diabetes Intervention and Complications (EDIC) study, the HbA1c levels of both groups converged, and they remained similar for many years. Despite this, patients in the original intensive treatment group continued to have a significantly lower risk of developing complications compared with the original conventionally treated group, even after 30 years (33). Studies of patients with type 2 diabetes have found similar benefits of strict glycemic control (35).
Epigenetic changes might provide the biological explanation for the long-lasting impact of metabolic changes, as metabolite levels can influence the epigenome and such changes are maintained during cell division. It is therefore imperative to determine the contribution of epigenetic mechanisms and perform epigenome-wide studies (EWAS) to reveal epigenotypes and epimutations (36) that might be involved in the development or progression of diabetes complications. Furthermore, superimposing EWAS and GWAS results can yield more meaningful information about biology and causality. The speakers of this session highlighted the emerging role of epigenetics and epigenomics in vascular complications of diabetes, diet-induced metabolic disorders, and metabolic memory, as well as the intersection between GWAS and EWAS.
Studies of cells and tissues from patients with diabetes have revealed clear differences in epigenetic marks at key genes associated with complications, including fibrotic and inflammatory genes. Susztak and coworkers used kidney tissues from patients with chronic kidney disease and diabetes to show epigenetic changes at renal disease-related genes, demonstrating the importance of studying the correct target tissue (37,38). Epigenetic mechanisms have been shown to be involved in cellular models of metabolic memory (32). Natarajan and coworkers used genomic DNA from the white blood cells of patients with type 1 diabetes enrolled in DCCT/EDIC to demonstrate a direct association between epigenetic variations and human metabolic memory (39,40). They found a persistence of DNA methylation variations at key CpG sites in patient whole blood, monocytes, and lymphocytes over 16 years from the DCCT to EDIC. The association of DNA methylation with HbA1c at several CpG sites also persisted over this period. Together, these data provide strong connections between epigenetics and metabolic memory.
Prediabetes and obesity/insulin resistance are also associated with epigenomic changes. The Schones laboratory showed that an obesogenic diet leads to epigenomic modifications in the liver that alter gene regulatory networks (41,42). Moreover, epigenetic variation in the livers of several strains of mice with differential susceptibility to metabolic disease occurs at transposable elements (43). Evolutionarily younger transposons with binding sites for inflammatory transcription factors are a major component of epigenetic variation across these strains, and inflammatory stimuli appear to be triggers of dysregulation of these transposons, potentially contributing to long-term disease risk.
Changes in the circadian rhythm and dietary patterns can also affect metabolic state and systemic glucose regulation via epigenomic mechanisms. The Panda laboratory has shown that time-restricted feeding (TRF) without changing nutrient quality or quantity improves daily oscillations in metabolic pathways (44,45). TRF prevents excessive weight gain, adiposity, glucose intolerance, systemic inflammation, hepatosteatosis, and hypercholesterolemia, independent of diet type. TRF is also associated with increased endurance, motor coordination, and brown fat function. Similar therapeutic benefits of TRF are also observed in high-fat diet–induced obese mice and mice genetically predisposed to obesity. Panda and colleagues have begun to monitor daily eating patterns in humans using a novel, unbiased, and scalable method. They also recently examined the novel roles of specific RNA binding proteins like NONO and noncoding RNAs in circadian rhythm and metabolism research (46). These results suggest that restricting food intake to specific times might be a potential new intervention for diabetes and obesity, even without altering the type or quantity of the food consumed.
Epigenetic Signals and Intergenerational Risk of Chronic Disease
Both human and animal studies indicate that environmental exposures during early life can increase risk for chronic disease during adulthood. Diverse exposures such as under- or overnutrition, placental dysfunction, hypoxia, maternal diabetes or obesity, and alterations in early postnatal growth can impact the development of key metabolic organs, altering body composition, stem cell populations, and tissue function of offspring. Even transient exposures can induce persistent epigenomic modifications that may alter transcriptional responses to aging and other environmental stressors during adult life, further increasing disease risk. Furthermore, environmental or metabolic insults experienced during either development or adult life can affect germ cells, potentially contributing to phenotypes in subsequent generations.
The Simmons laboratory has made important contributions highlighting the role of maternal obesity in pregnancy as a mediator of developmental programming. Maternal obesity in rodents increases adiposity in offspring via effects on both the maternal intrauterine environment and oocyte (47–50). Maternal obesity has an even greater impact on the developing placenta, altering transcription and metabolism, particularly within lipid metabolic pathways, and increasing inflammation (51).
The Ferguson-Smith laboratory has identified epigenetic marks responsive to the metabolic environment that contribute to disease (unpublished data). Repeat elements present within the human genome and their regulated silencing by epigenetic modifications are important in this regard. New “metastable epialleles” identified in mice may be sites of epigenetic modulation by diet and other environmental stressors, which can also influence expression of endogenous genes. These loci are variably methylated between individuals but consistently methylated in different tissues in the same individual. Interestingly, these repeats are 100% methylated in sperm and variably methylated in offspring, indicating they can be reprogrammed postfertilization.
The Patti laboratory has focused on mechanisms responsible for intergenerational transmission of metabolic disease risk. Data from animal models indicate that health of offspring can be influenced by the metabolic health of either parent at the time of breeding or by environmental exposures previously experienced by their parents or grandparents. This paradigm suggests that epigenetic marks within parental germ cells or noncoding RNA transmitted during breeding may influence early postfertilization transcriptional responses. For example, maternal undernutrition during late pregnancy results in increased adiposity and glucose intolerance in first-generation (F1) offspring; both male and female F1 offspring produce F2 with increased adiposity and glucose intolerance, even in the absence of further nutritional perturbations (52).
While maternal contribution to disease risk is well recognized, disease risk transmission via F1 fathers suggests that male germ cells or other components of the sperm contribute epigenetic information capable of influencing postfertilization transcription in offspring. Sperm DNA methylation is altered in males subjected to undernutrition during intrauterine life, particularly at sites influencing development (53). Whether differential DNA methylation could impact early-life development is a key focus of current studies.
The Pospisilik laboratory has demonstrated that nutritional conditions in male flies can alter germ cell chromatin structure and increase adiposity in offspring (54). RNA sequence analysis of both sperm and embryos reveals that paternal high sugar intake impacts transcriptional pathways regulating energy metabolism, cell cycle, and patterning via polycomb-, SetDB1-, and Su(var)-dependent mechanisms. Interestingly, many of these genes are also enriched in differentially expressed genes in obese versus lean humans (55). Collectively, these data illustrate that chromatin structure may be an effector mechanism for intergenerational metabolic disease.
The Rando laboratory demonstrated that paternal diet alters levels of small noncoding RNA in sperm, such as tRNA fragments, which are derived from epididymosomes released from somatic cells and acquired by sperm as they mature in the epididymis (56). In turn, these tRNA fragments may influence early embryonic phenotypes via altered expression of MERVL transposons (56). Together, these data indicate that both the male germ cell and small RNA added during epididymal transit are essential components of sperm that can influence offspring development. More broadly, this process suggests a regulated mechanism by which somatic cells can alter the epigenetic information transmitted to offspring via sperm.
Collectively, these data indicate that current health or prior environmental exposures may promote a vicious cycle of intergenerational disease risk (Fig. 3). Unfortunately, data from human populations are limited due to the difficulty in isolating nongenetic from genetic effects when environmental factors cannot be carefully controlled. Nevertheless, observational studies in the Dutch Hunger Winter, Överkalix, and other human cohorts do support the idea that environmental exposures can influence health and metabolism of grandchildren. Such nongenetic intergenerational amplification of disease risk effects could contribute to the global increase in prevalence of obesity, diabetes, and related complications over a very short time frame.
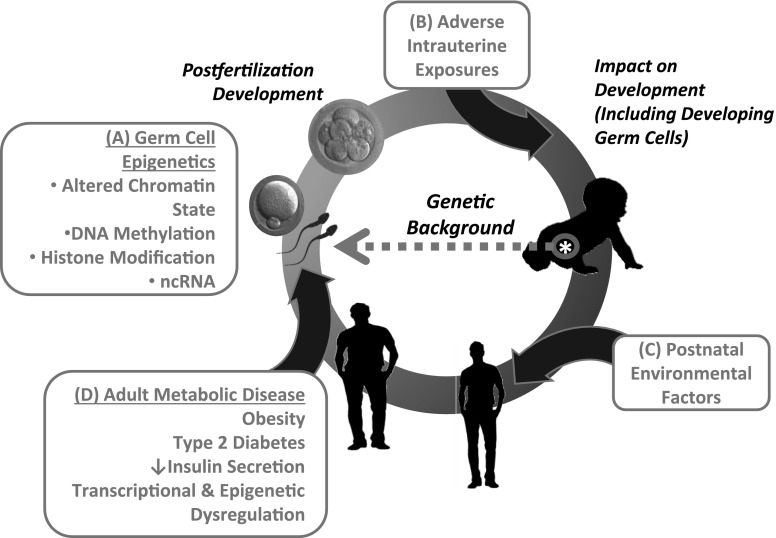
Figure 3.
Effects of adverse intrauterine exposures and postnatal environmental factors on pathogenesis of adult metabolic disease and transgenerational disease risk. A: Epigenetic marks within parental germ cells, such as DNA methylation, histone modification, or noncoding RNA (ncRNA) (e.g., in semen) may influence early postfertilization transcriptional responses, altering development and ultimate health of offspring. Adverse intrauterine exposures (B) and postnatal environmental factors (C) may also contribute to disease risk via epigenetic modifications. D: Transcriptional and epigenetic dysregulation associated with adult metabolic disease can also impact germ cells, promoting transgenerational disease risk. Note that these effects occur in the context of the individual’s genetic background, which can modulate responses to environmental exposures.
Developing Epigenetic Therapeutics for Metabolic Disease
Historically, cancer cells have served as a paradigm for epigenomic dysregulation, as chromatin and DNA methylation are widely abnormal, which was reviewed by Peter A. Jones. In fact, many tumor suppressor genes that are active in normal cells are turned off in cancer cells via methylation of their promoters (57). Therefore, it is not surprising that multiple systemically acting epigenomic drugs that target DNA methyltransferases have been developed and approved by the U.S. Food and Drug Administration for use in patients (58). More recently, agents that target chromatin-associated epigenomic writers such as EZH2, erasers such as LSD1, and readers such as BRD4 are being added as therapeutic agents (59). Jay Bradner highlighted the use of one such compound, JQ1, in the treatment of cancer (unpublished data). While useful for advanced malignancy, these types of globally acting drugs may not be applicable to the treatment of diabetes due to their sometimes severe side effects, their simultaneous action on multiple pathways, and their lack of cell-type specificity.
Recently, multiple approaches have been developed for locus-specific epigenetic targeting to alter the activity of selected genes. Thus, the Jaenisch laboratory used a modified CRISPR/Cas9 system to either methylate or demethylate key regulatory gene elements on cytosine residues to control their expression (60). While the native version of the CRISPR/Cas9 system has been used extensively to introduce mutations, in the version for epigenomic editing, the nuclease activity of Cas9 has been inactivated and the protein is fused to the catalytic domains of DNA methyltransferases, or Tet enzymes, which mediate hydroxymethylation and eventual demethylation at targeted CpG elements. Thus far, it appears that this type of manipulation is indeed locus specific; however, careful studies will need to be performed to ensure that there are no off-target effects, as they could alter the activity of other genes with unintended consequences. For example, a recent publication from the Meissner group demonstrated that dCas9-DNA methyltransferase fusion proteins have widespread off-target effects, suggesting that this technology warrants further refinement (61).
Related to β-cells, the Kaestner laboratory used a similar experimental paradigm to produce targeted “epimutations,” or changes to the epigenetic state of a gene of interest, by designing a modular DNA-binding protein (termed “TALE”) fused to the catalytic domain of the aforementioned Tet1 enzyme (unpublished data). In line with the results from the Jaenisch group, they could control the expression of a key cell cycle regulator by generating an epimutation and in this case stimulate the replication of human β-cells in the xenotransplant setting.
Conclusions
In summary, the symposium provided an excellent overview of the current status of epigenetic studies in diabetes research. Several new themes have emerged. It is clear that diabetes and its complications develop as a joint effect of DNA sequence variations and environmental effects leading to changes in the cellular phenotype. The field of epigenetics and epigenomics is developing rapidly, and multiple groups are working on the generation of a solid and clinically annotated cell-type–specific epigenome atlas. The epigenome maps will be used to interpret genetic variation and highlight the way variation leads to disease development. Furthermore, the integration of the epigenome maps with genomic and genetic data can improve our understanding of the mechanisms whereby environmental changes contribute to diabetes development. In addition to a better understanding of disease pathogenesis, epigenetic regulation provides unique therapeutic opportunities for the treatment of diabetes and its complications. In summary, epigenetic and epigenomic research will be a critical cornerstone for a better understanding of diabetes during the coming decade.
Article Information
Acknowledgments. The authors gratefully acknowledge the research symposium steering committee members and speakers for the excellent presentations, discussions, and contributions to the conference. In addition to the writing group members, Melina Claussnitzer, Beth Israel Deaconess Medical Center, Harvard Medical School, and Broad Institute, participated on the planning committee. Invited speakers included Romain Barrès, University of Copenhagen; Bradley E. Bernstein, Harvard Medical School and Broad Institute; Jay Bradner, Novartis Institutes for BioMedical Research; John M. Denu, University of Wisconsin; Anne Ferguson-Smith, University of Cambridge; Anna L. Gloyn, University of Oxford; Rudolf Jaenisch, Whitehead Institute; Peter A. Jones, Van Andel Research Institute; Mitchell A. Lazar, University of Pennsylvania Perelman School of Medicine; Marcelo A. Nóbrega, The University of Chicago; Satchidananda Panda, Salk Institute for Biological Studies; J. Andrew Pospisilik, Max Plank Institute for Immunobiology; Oliver Rando, University of Massachusetts Medical School; Dustin Schones, Beckman Research Institute of City of Hope; Rebecca Simmons, University of Pennsylvania Perelman School of Medicine; and Michael Snyder, Stanford University.
Funding and Duality of Interest. E.D.R. reports grants from Merck and personal fees from Novartis, outside the submitted work. R.N. reports grants from the National Institutes of Health and JDRF during the conduct of the study. M.-E.P. reports grants from Janssen Pharmaceuticals, Xeris Pharmaceuticals, Ethicon, MedImmune, Covidien, Dexcom, and AstraZeneca; supplies for clinical trials from Novo Nordisk, Insulet, and Nestle; a role as a clinical trial investigator for Xoma; and personal fees from Eiger BioPharmaceuticals, outside the submitted work. K.S. reports funding from the National Institutes of Health, JDRF, Lilly, Boehringer Ingelheim, GSK, Regeneron, Merck, ONO Pharma, and Celgene and honoraria from Bayer. No other potential conflicts of interest relevant to this article were reported.
References
- 1.Keating ST, El-Osta A. Epigenetics and metabolism. Circ Res 2015;116:715–736 [DOI] [PubMed] [Google Scholar]
- 2.Bouchard C. The genetics of obesity: from genetic epidemiology to molecular markers. Mol Med Today 1995;1:45–50 [DOI] [PubMed] [Google Scholar]
- 3.Ohta T, Gray TA, Rogan PK, et al. Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 1999;64:397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen ED. Epigenomic and transcriptional control of insulin resistance. J Intern Med 2016;280:443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobi EW, Slieker RC, Luijk R, et al.; Biobank-based Integrative Omics Studies Consortium . DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci Adv 2018;4:eaao4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deans C, Maggert KA. What do you mean, “epigenetic”? Genetics 2015;199:887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev 2009;23:781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan A, Taliun D, Thurner M, et al. Fine-mapping of an expanded set of type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps [preprint article online]. 9 January 2018. Available from https://www.biorxiv.org/content/early/2018/01/09/245506. Accessed 11 May 2018 [DOI] [PMC free article] [PubMed]
- 9.Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 2014;46:136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest 2013;123:1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurner M, van de Bunt M, Torres JM, et al. Integration of human pancreatic islet genomic data refines regulatory mechanisms at type 2 diabetes susceptibility loci. eLife 2018;7:e31977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Bunt M, Manning Fox JE, Dai X, et al. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet 2015;11:e1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomsen SK, Ceroni A, van de Bunt M, et al. Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants. Diabetes 2016;65:3805–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey KA, Savic D, Zielinski M, et al. Evidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice. Hum Mol Genet 2015;24:1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claussnitzer M, Dankel SN, Kim K-H, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 2015;373:895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko Y-A, Yi H, Qiu C, et al. Genetic-variation-driven gene-expression changes highlight genes with important functions for kidney disease. Am J Hum Genet 2017;100:940–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Susztak K. Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol 2014;25:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Dunn J, Salins D, et al. Digital health: tracking physiomes and activity using wearable biosensors reveals useful health-related information. PLoS Biol 2017;15:e2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piening BD, Zhou W, Contrepois K, et al. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst 2018;6:157–170.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsatsoulis A, Mantzaris MD, Bellou S, Andrikoula M. Insulin resistance: an adaptive mechanism becomes maladaptive in the current environment - an evolutionary perspective. Metabolism 2013;62:622–633 [DOI] [PubMed] [Google Scholar]
- 21.Rui L. Energy metabolism in the liver. Compr Physiol 2014;4:177–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wortham M, Sander M. Mechanisms of β-cell functional adaptation to changes in workload. Diabetes Obes Metab 2016;18(Suppl. 1):78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sylow L, Kleinert M, Richter EA, Jensen TE. Exercise-stimulated glucose uptake - regulation and implications for glycaemic control. Nat Rev Endocrinol 2017;13:133–148 [DOI] [PubMed] [Google Scholar]
- 24.Sharma U, Rando OJ. Metabolic inputs into the epigenome. Cell Metab 2017;25:544–558 [DOI] [PubMed] [Google Scholar]
- 25.Dhawan S, Tschen S-I, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev 2009;23:906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Gu X, Su IH, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 2009;23:975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avrahami D, Li C, Zhang J, et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab 2015;22:619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papazyan R, Zhang Y, Lazar MA. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 2016;23:1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science 2016;354:994–999 [DOI] [PubMed] [Google Scholar]
- 30.Roh HC, Tsai LTY, Shao M, et al. Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metab 2018;27:1121–1137.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsen SC, Brøns C, Bork-Jensen J, et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia 2012;55:3341–3349 [DOI] [PubMed] [Google Scholar]
- 32.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 2015;58:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathan DM; DCCT/EDIC Research Group . The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications study at 30 years: overview. Diabetes Care 2014;37:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA 2002;287:2563–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HAW. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 2008;359:1577–1589 [DOI] [PubMed] [Google Scholar]
- 36.Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 2017;18:441–451 [DOI] [PubMed] [Google Scholar]
- 37.Beckerman P, Ko Y-A, Susztak K. Epigenetics: a new way to look at kidney diseases. Nephrol Dial Transplant 2014;29:1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko Y-A, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol 2013;14:R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao F, Chen Z, Genuth S, et al.; DCCT/EDIC Research Group . Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014;63:1748–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Miao F, Paterson AD, et al.; DCCT/EDIC Research Group . Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A 2016;113:E3002–E3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung A, Parks BW, Du J, et al. Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem 2014;289:23557–23567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung A, Trac C, Du J, Natarajan R, Schones DE. Persistent chromatin modifications induced by high fat diet. J Biol Chem 2016;291:10446–10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du J, Leung A, Trac C, et al. Chromatin variation associated with liver metabolism is mediated by transposable elements. Epigenetics Chromatin 2016;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longo VD, Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab 2016;23:1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panda S. Circadian physiology of metabolism. Science 2016;354:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benegiamo G, Mure LS, Erikson G, et al. The RNA-binding protein NONO coordinates hepatic adaptation to feeding. Cell Metab 2018;27:404–418.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jungheim ES, Schoeller EL, Marquard KL, Louden ED, Schaffer JE, Moley KH. Diet-induced obesity model: abnormal oocytes and persistent growth abnormalities in the offspring. Endocrinology 2010;151:4039–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luzzo KM, Wang Q, Purcell SH, et al. High fat diet induced developmental defects in the mouse: oocyte meiotic aneuploidy and fetal growth retardation/brain defects. PLoS One 2012;7:e49217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasson IE, Vitins AP, Mainigi MA, Moley KH, Simmons RA. Pre-gestational vs gestational exposure to maternal obesity differentially programs the offspring in mice. Diabetologia 2015;58:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igosheva N, Abramov AY, Poston L, et al. Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLoS One 2010;5:e10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart TJ, O’Neill K, Condon D, et al. Diet-induced obesity alters the maternal metabolome and early placenta transcriptome and decreases placenta vascularity in the mouse. Biol Reprod 2018;98:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jimenez-Chillaron JC, Isganaitis E, Charalambous M, et al. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 2009;58:460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martínez D, Pentinat T, Ribó S, et al. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metab 2014;19:941–951 [DOI] [PubMed] [Google Scholar]
- 54.Öst A, Lempradl A, Casas E, et al. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 2014;159:1352–1364 [DOI] [PubMed] [Google Scholar]
- 55.Dalgaard K, Landgraf K, Heyne S, et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell 2016;164:353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol 2010;28:1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Costanzo A, Del Gaudio N, Migliaccio A, Altucci L. Epigenetic drugs against cancer: an evolving landscape. Arch Toxicol 2014;88:1651–1668 [DOI] [PubMed] [Google Scholar]
- 60.Liu XS, Wu H, Ji X, et al. Editing DNA methylation in the mammalian genome. Cell 2016;167:233–247.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galonska C, Charlton J, Mattei AL, et al. Genome-wide tracking of dCas9-methyltransferase footprints. Nat Commun 2018;9:597. [DOI] [PMC free article] [PubMed] [Google Scholar]