Abstract
Two smyd1 paralogues, smyd1a and smyd1b, have been identified in zebrafish. Although Smyd1b function has been reported in fast muscle, its function in slow muscle and the function of Smyd1a, in general, are uncertain. In this study, we generated 2 smyd1a mutant alleles and analyzed the muscle defects in smyd1a and smyd1b single and double mutants in zebrafish. We demonstrated that knockout of smyd1a alone had no visible effect on muscle development and fish survival. This was in contrast to the smyd1b mutant, which exhibited skeletal and cardiac muscle defects, leading to early embryonic lethality. The smyd1a and smyd1b double mutants, however, showed a stronger muscle defect compared with smyd1a or smyd1b mutation alone, namely, the complete disruption of sarcomere organization in slow and fast muscles. Immunostaining revealed that smyd1a; smyd1b double mutations had no effect on myosin gene expression but resulted in a dramatic reduction of myosin protein levels in muscle cells of zebrafish embryos. This was accompanied by the up-regulation of hsp40 and hsp90-α1 gene expression. Together, our studies indicate that both Smyd1a and Smyd1b partake in slow and fast muscle development although Smyd1b plays a dominant role compared with Smyd1a.—Cai, M., Han, L., Liu, L., He, F., Chu, W., Zhang, J., Tian, Z., Du, S. Defective sarcomere assembly in smyd1a and smyd1b zebrafish mutants.
Keywords: muscle development, myofibrillogenesis, CRISPR, chaperone
Skeletal muscle cell differentiation is a complex process that depends on the coordinated gene expression and assembly of myofibrillar proteins into highly organized sarcomeres, the fundamental unit of muscle contraction. The sarcomere is defined as the segment between 2 neighboring Z lines, covering 2 major compartments, namely I-band and A-band, with the M line in the center. The Z-line anchors the actin thin filaments of the I-band. The M-line anchors the myosin thick filaments of the A-band. The sarcomere assembly of these multiprotein complexes follow ordered pathways that are highly regulated at the transcriptional, translational, and posttranslational levels (1, 2). Genetic mutations that disrupt sarcomere assembly often result in embryonic lethality or congenital myopathies (3–5).
Recent studies have demonstrated that zebrafish Smyd1b and mouse Smyd1 play a vital role in muscle cell differentiation and sarcomere assembly in skeletal and cardiac muscles (6–15). Smyd1 is a SET domain containing lysine methyltransferase specifically expressed in skeletal and cardiac muscles (6, 7). The muscle-specific expression of Smyd1 is regulated by the myogenic transcriptional factors myogenic differentiation (MyoD), myocyte enhancer factor 2, and serum response factor (16–18). It appears that Smyd1 functions downstream of MyoD and Myogenin, because loss of Smyd1 had no effect on MyoD and Myogenin gene expression and early myoblast specification but completely disrupted the sarcomere organization during myofiber maturation (7–9, 13). The molecular mechanism by which Smyd1 regulates myofibril assembly is not clear. It has been suggested that Smyd1 acts as a transcriptional regulator controlling gene expression (6, 18). A recent study demonstrated that Smyd1 directly controls the expression of peroxisome proliferator-activated receptor γ coactivator 1α to regulate mitochondrial energetics in the heart (19). Intriguingly, several studies have shown that Smyd1 protein is translocated from the nucleus to the cytoplasm after myoblast differentiation into myotubes (20). Biochemical analysis revealed that Smyd1b associates with myosin and myosin chaperones, Unc45b and Hsp90-α1, which were required for myosin protein folding and myofibril assembly (9, 21–24), suggesting a role in the muscle cell cytoplasm.
Genetic studies demonstrated that Smyd1 is required for both early myogenesis and later myofiber maturation during muscle development in mice and zebrafish (7, 8, 12–14). Muscle-specific deletion of Smyd1 in mouse embryos using Myf5cre impaired myoblast differentiation and resulted in fewer myofibers and perinatal death (12). In contrast, deletion of Smyd1 specifically in skeletal myocytes after birth using Myf6cre produced a nondegenerative myopathy in mice (14). The mutant mice were viable but exhibited myofiber hypotrophy, myofibrillar disorganization, and a high percentage of immature myofibers with centralized nuclei (14). Loss of smyd1b, a smyd1 ortholog in zebrafish, resulted in defective myofibril assembly in skeletal and cardiac muscles of zebrafish embryos, leading to embryonic death around d 6 after fertilization (7–9, 13).
Zebrafish contain 2 smyd1 genes, smyd1a and smyd1b, located on chromosome 5 and 8, respectively (11, 25). Although the Smyd1b function is well characterized in fast muscle, its function in slow muscle remains uncertain. Two contradictory findings have been reported (7, 9). We demonstrated that knockdown of smyd1b disrupted muscle development in both slow and fast muscles in zebrafish embryos (7, 9). However, using the zebrafish smyd1b mutant, flatline (Flamvo47a), it was reported that Smyd1b function was restricted to fast muscles and that the loss of Smyd1b had no effect on slow muscle development (8). Smyd1a function also remains controversial based on 2 morpholino (MO) knockdown studies (11, 26). We have shown that knockdown of smyd1a had no visible effect on muscle development in zebrafish embryos (11). However, a recent report suggested that loss of smyd1a interfered with myofibrillar integrity in zebrafish skeletal and cardiac muscles (26).
To clarify these controversies regarding the function of Smyd1a and Smyd1b in myofibril assembly, we generated 2 novel smyd1a mutant alleles in zebrafish using the clustered regularly interspaced short palindromic repeat (CRISPR) technology and characterized the muscle phenotype in the smyd1a and smyd1bsa15678 single and double mutants. Our data revealed that homozygous smyd1a mutant embryos had normal muscle development, growth, and survival. In contrast, knockout of smyd1b resulted in defective sarcomere organization in both slow and fast muscles of early-stage zebrafish embryos. Moreover, smyd1a and smyd1b double mutations resulted in a stronger muscle defect with a complete disruption of myofibril organization and up-regulation of Hsp40 and hsp90 expression in embryonic skeletal muscles. Ectopic expression of zebrafish smyd1a, smyd1b or mouse Smyd1 transgene was able to rescue the muscle defects in the smyd1a; smyd1b double mutants. Together, these data indicate that Smyd1a and Smyd1b are both involved in myofibril assembly during muscle cell differentiation, although Smyd1b plays a more critical role.
MATERIALS AND METHODS
Ethics statement
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA). The protocol was approved by the Institutional Animal Care and Use Committee of University of Maryland Baltimore (Permit: 0516005). To ease pain and facilitate animal handling, fish embryos over 1 d old were anesthetized in 0.6 mM Tricaine (pH 7.0) before fixation in 4% paraformaldehyde for whole-mount in situ hybridization and immunostaining.
Zebrafish lines and maintenance
All adult zebrafish were kept at the zebrafish facility at the Institute of Marine and Environmental Technology, University of Maryland. The smyd1amb4 and smyd1amb5 mutant alleles were generated in our laboratory by using CRISPR/CRISPR-associated protein 9 (Cas9) system. The smyd1bsa15678 mutant was obtained from ZIRC (27). The Myom3mn0067gt zebrafish line expressing a Myom3–red florescent protein (RFP) fusion protein was obtained from Steve Ekker’s laboratory at Mayo Clinic (28). Zebrafish larvae and adults were maintained at 28.5°C in a recirculating aquatic system at a photoperiod of 14/10-h light/dark cycle.
DNA constructs
The following DNA constructs were used in this study. The pGEM-T-zfsmyd1a plasmid contained the full-length coding sequence of zebrafish smyd1a (NM_205540) (11). The Tg (smyd1b:zfsmyd1bmyc) transgene expressed an N-terminal myc-tagged zebrafish Smyd1b (7). The Tg (smyd1b:zfsmyd1amyc) transgene expressed a C-terminal myc-tagged zebrafish Smyd1a (11). The Tg(smyd1b:mSmyd1myc) transgene expressed a C-terminal myc-tagged mouse Smyd1 (11).
Design and synthesis of Smyd1a and Smyd1b single-guide RNAs
A smyd1a–single-guide RNA (sgRNA) target including the underlined PAM sequence (GGACCTGAAGGAGCTCAAAGCGG) was identified in exon 3 of the zebrafish smyd1a gene (http://zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx). Two oligonucleotides, Smyd1a-Cas-P1 and Smyd1a-Cas-P2, were synthesized with a complementary sequence covering the target site (Table 1). The Smyd1a-Cas-P1 and Smyd1a-Cas-P2 primers were denatured and annealed to form a duplex oligonucleotide. The oligo duplex was cloned into the BsaI site of pDR274 vector purchased from Addgene (Watertown, MA, USA) (29). The final DNA construct was named Smyd1a-sgRNA-pDR274 and confirmed by DNA sequencing. The Smyd1a-sgRNA-pDR274 DNA plasmid was digested with DraI, purified and used as the DNA template for Smyd1a-sgRNA synthesis.
TABLE 1.
Primers for sgRNA synthesis and PCR analyses
| Primer | Sequence, 5′–3′ |
|---|---|
| Smyd1a-Cas-P1 | TAGGACCTGAAGGAGCTCAAAG |
| Smyd1a-Cas-P2 | AAACCTTTGAGCTCCTTCAGGT |
| Smyd1b-sgRNA-F1 | GATCACTAATACGACTCACTATAGAATCAGCTGACGACTCTGGGTTTTAGAGCTAGAAAT |
| Cas9-P4 | AAAAGCACCGACTCGGTGCC |
| Smyd1a-F2 | AGTCTGGTTGCTCGTATCCTGTGGCG |
| Smyd1a-R2 | CTGCTGTCAAATTGTGTGCTGAAGC |
| Smyd1b-sg4/5-F1 | AGTCTGGCGGCCCGTATCCTGTGGC |
| Smyd1b-sg4/5-R1 | ACCACGCCGAGGATGTGCGAGACGC |
| Smyd1b-mut-F1 | CGTGTGAATGGGGCGTCATAGATGTG |
| Smyd1b-mut-R2 | GGCCAGCAGTCGTGGTTCACCAAAC |
| zfhsp40-P9-qPCR | ATCAGCACGCTGGACAACAGATCGC |
| Zfhsp40-P10-qPCR | CGGTCAGCATGATGTGCTCTCGTAC |
| zfhsp90-α1-P6-qPCR | AGCCAGACTTCGGTGAATCAA |
| zfhsp90-α1-P7-qPCR | TTCTCTCTGTTTCTCAATGTAAA |
| zfmyhz2-P4-qPCR | GCTCACCTACCAGACTGAGGA |
| zfmyhz2-P5-qPCR | ACTCAGCAATATCAGCACGCT |
| zfsmyhc1-P4 | GCTCACCTACCAGACTGAGGA |
| zfsmyhc1-P5-qPCR | CATCTTGTTGACCTGAGATTCA |
| zfef1a-P3-qPCR | CTTCAACGCTCAGGTCATCAT |
| zfef1a-P4-qPCR | ACAGCAAAGCGACCAAGAGGA |
| zfSmyd1b-Bopx-qPCR | AAAGCCTTCGCCATCCTGATGATCAC |
| zfSmyd1b-Bop4-qPCR | CGTTGACCTGAGCGTGTCTGTCACTGTGG |
| zfSmyd1a-P1-qPCR | AGCATGACCGTGGAGAAGACGGAC |
| zfSmyd1a-E2-R1-qPCR | GACACGTGCGGTCGCAGTAATGCGC |
The underlined sequences in Smyd1a-Cas-P1 and Smyd1b-sgRNA-F1 are the gene-specific target sequences within the respective sgRNAs. Mut, mutation.
A smyd1b-sgRNA target site was identified in the sense strand of exon 3 of the zebrafish smyd1b gene (Table 1). The DNA template for synthesizing smyd1b-sgRNA was generated by PCR using the pDR274 plasmid as a template as described (30). The PCR was carried out using the Phusion DNA polymerase (F530S; Thermo Fisher Scientific, Waltham, MA, USA) with a smyd1b target-specific forward primer (Smyd1b-sgRNA-F1) and a common reverse primer (Cas9-P4) (Table 1). The PCR product was purified and used as a template for synthesizing the smyd1b-sgRNA in vitro.
The smyd1a and smyd1b sgRNAs were synthesized in vitro using the MaxScript T7 Transcription Kit (1312; Thermo Fisher Scientific). The transcription was carried out in vitro for 4 h at 37°C using ∼1000 ng linearized Smyd1a-sgRNA-pDR274 plasmid or ∼200 ng purified PCR product (for smyd1b-sgRNA) as DNA a template, respectively. The sgRNA products were purified using MegaClear TM Kit (1908; Thermo Fisher Scientific).
Cas9 mRNA preparation and microinjection
Cas9 mRNA was prepared by in vitro transcription using XbaI linearized pT3TS-nCas9n DNA plasmid as a template (31). The Cas9 mRNA was transcribed with the T3 RNA Polymerase Kit (1348; Thermo Fisher Scientific) and purified using the MegaClear Kit. The recombinant Cas9 nuclease (GeneArt Platinum Cas9 Nuclease) was purchased from Thermo Fisher Scientific (B25640).
sgRNAs were mixed with equal volume of Cas9 mRNA (400 ng/μl) or Cas9 protein (500 ng/μl) and coinjected into zebrafish embryos at the 1-cell stage. The final concentration of sgRNA was 35 ng/µl for smyd1a-sgRNA and 32 ng/µl for smyd1b-sgRNA. The final concentration of Cas9 mRNA or Cas9 protein was 200 or 250 ng/µl, respectively. Each embryo was injected with ∼2 nl of the mixed sgRNA/Cas9 solution.
Genotyping using DNA from fish embryos and caudal fin clips
To genotype fish embryos, genomic DNA was isolated from the head region or a whole embryo at 48 h postfertilization (hpf) using the alkaline method (30). To genotype juvenile and adult zebrafish, caudal fin was cut from individual zebrafish. The fin clip was used for DNA extraction using the alkaline method (30). The PCR reaction was carried out in a 25-μl reaction using GoTaq DNA polymerase (Promega, Madison, WI, USA). To genotype CRISPR-induced mutations, smyd1a DNA fragment was amplified using Smyd1a-F2 and Smyd1a-R2 primers (Table 1), whereas smyd1b was amplified with Smyd1b-sg4/5-F1 and Smyd1b-sg4/5-R1 primers (Table 1). The PCR products of smyd1a were digested with SacI. The SacI site within the Smyd1a-sgRNA target sequence was abolished in the smyd1amb4 and smyd1amb5 mutant alleles containing a 7 bp or 10 deletion, respectively, thus simplifying the mutant screening and genotyping. The smyd1bsa15678 mutant genotyping was performed by PCR using Smyd1b-mut-F1 and Smyd1b-mut-R2 primers (Table 1). The PCR products of smyd1b were digested with DpnII. The DpnII site in the PCR product of smyd1bsa15678 mutant was destroyed by the C544 > T mutation.
To assay the efficiency of smyd1b-sgRNA/Cas9-induced mutagenesis in injected F0 embryos, 10 injected embryos were randomly selected at 48 hpf for DNA extraction and PCR analysis. The PCR products were digested with ExoSAP-IT [78200; U.S. Biochemical Corp. (USB), Cleveland, OH, USA] and sequenced directly using the Smyd1b-sg4/5-F1 primer.
Whole-mount in situ hybridization and immunostaining
Whole-mount in situ hybridization was carried out using digoxigenin-labeled RNA antisense probes as previously described (32). The NcoI linearized pGEM-T-zfsmyd1a plasmid was transcribed with Sp6 RNA polymerase. In situ hybridization for MyoD, slow and fast myosin heavy chains (MyHCs), was carried out using the respective digoxigenin-labeled probes as previously described (23). The images were acquired using a Leica dissecting microscope M12 equipped with a cool charge-coupled device digital camera (DX8; Olympus, Tokyo, Japan).
Immunostaining was carried out using whole-mount zebrafish embryos at 14, 28, 48, and 72 hpf as previously described (7). To increase permeability, embryos of 48 and 72 hpf were digested with 1 mg/ml collagenase for 45 and 75 min, respectively. Immunostaining was performed with the following primary antibodies: anti-α-actinin (clone EA-53, A7811; MilliporeSigma, Burlington, MA, USA), anti-MHC for slow muscles [F59, Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA, USA], anti-myomesin (mMaC myomesin B4, DSHB), anti-myosin light chain (MyLC) (F310, DSHB), anti-myosin (MF-20, DSHB), and anti-myc tag (71D10; Cell Signaling Technology, Danvers, MA, USA). Secondary antibodies were Alexa Fluor 488- or Alexa Fluor 555-conjugated anti-IgG secondary antibodies (A11001 and A31630; Thermo Fisher Scientific). After being washed with PBST 3 times (30 min each), trunk muscles of embryos were dissected and mounted in Vectashield (H-1000; Vector Laboratories, Burlingame, CA, USA). The images were acquired and photographed using a confocal microscope (SP8; Leica Microsystems, Buffalo Grove, IL, USA).
Phalloidin and nuclear staining
For nuclear staining, the embryos were incubated with 1 μg/ml Hoechst 33258 (B2883; MilliporeSigma) for 1 h at room temperature in the dark. For double staining with Hoechst and phalloidin, the fixed embryos were incubated with 1 μg/ml Hoechst 33258 (B2883; MilliporeSigma) and 50 ng/ml Phalloidin-TRITC (P1951; MilliporeSigma) for 1 h at room temperature in the dark. After being washed with PBST 3 times (30 min each), the trunk region of fish embryos was dissected and mounted in Vectashied (H-1000; Vector Laboratories). The embryos were photographed using a Leica SP8 confocal microscope.
Electrophoresis and Western blotting
Wild-type (WT) or smyd1a; smyd1b mutant zebrafish embryos were dechorinated manually at 24 and 48 hpf. The embryos (40 embryos each group) were washed with 1 ml of PBS and crushed gently to remove the yolk by pipetting with a glass pipet in 0.5 ml of PBS. The embryos were collected by a quick spin at 3000 rpm for 1 min. The embryo extract was washed once with 0.5 ml of PBS and solubilized in 80 µl of 2× SDS loading buffer (0.125 M Tris-Cl pH 6.8, 4% SDS, 20% Glycerol, 0.2 M DTT, 0.02% Bromophenol Blue) containing 1 mM PMSF and protease inhibitor (P8340; MilliporeSigma) by passing through the 21-gauge needle 10–15 times. The proteins were denatured by boiling for 3 min and analyzed on a 7.5% SDS-PAGE gel. Proteins from ∼5 embryos were loaded on each lane of the SDS-PAGE gel. Proteins from the gel were transferred onto a PVDF membrane (Immobilion-P; MilliporeSigma, Burlington, MA, USA) by electrophoresis. Immunodetection of MyHC or γ-tubulin was carried out with their primary antibodies (MF20; DSHB and T6557; MilliporeSigma) and followed by a peroxidase-conjugated secondary antibody. All blots were developed using the ECL Prime Western Blotting System (RPN2232; GE Healthcare, Waukesha, WI, USA).
Real-time quantitative PCR analysis
Total RNA was isolated from the WT and mutant embryos using Trizol (Thermo Fisher Scientific) according to the manufacturer’s protocol. Two to 3 replicates of 30 embryos for each replicate were used for both WT and mutant groups. Total RNA was transcribed using the RevertAid First Strand cDNA Synthesis Kit to produce cDNA according to the manufacturer’s protocol (K1621; Thermo Fisher Scientific). Real-time PCR was carried out using the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR reaction was carried out using the Powerup Sybr Green Master Mix (A25741; Thermo Fisher Scientific). Three technical replicates were performed for each PCR reaction. The quantitative RT-PCR (qRT-PCR) primers are listed in Table 1. Expression levels were analyzed using the Prism software (GraphPad Software, La Jolla, CA, USA) and normalized against elongation factor 1α (ef-1α). Fold change was calculated using the ΔΔCt method. All the results were expressed as means ± sem. Differences between WT and mutants were analyzed by a Student’s t test. A value of P < 0.05 was considered as the level of significance.
RESULTS
Smyd1b mutation disrupted sarcomere organization in slow muscle of early-stage zebrafish embryos
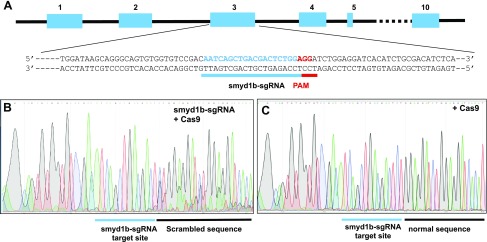
Previous studies using MO knockdown and zebrafish mutant reported contradictory findings regarding Smyd1b function in the slow muscle of zebrafish embryos (7–9, 13). To decipher these contradictory findings, we decided to knock out smyd1b using CRISPR. A smyd1b gene-specific sgRNA was targeted to a sequence in exon 3 (Fig. 1A). The smyd1b-sgRNA could not work on the smyd1a gene because the protospacer adjacent motif sequence was not found in the corresponding region in the smyd1a gene. Sequence analysis confirmed that the coinjection of smyd1b-sgRNA and Cas9 protein induced mutations at the smyd1b target site in the injected zebrafish embryos (Fig. 1B). The indel mutations resulted in scrambled sequence after the target site (−3 position from the protospacer adjacent motif sequence, PAM), leading to the appearance of multiple peaks on the sequence chromatograph. In contrast, injection of Cas9 protein alone failed to induce mutations at the smyd1b-sgRNA target site and all peaks appeared as single read on the chromatograph, which is identical to the WT smyd1b sequence (Fig. 1C).
Figure 1.
Knockout of smyd1b by CRISPR. A) An smyd1b specific sgRNA, Smyd1b-sgRNA was targeted to the exon 3. B, C) The Smyd1b-sgRNA was coinjected with Cas9 protein into Myom3mn0067gt zebrafish embryos at 1–2 cell stages. The injected embryos were analyzed by DNA sequencing and immunostaining at 48 hpf. DNA sequence analysis (sense strand) of Smyd1b-sgRNA/Cas9–injected (B) and Cas9-injected WT control (C) embryos shows the presence of scrambled multiple peaks after the Smyd1b-sgRNA target site in the Smyd1b-sgRNA/Cas9–injected embryos.
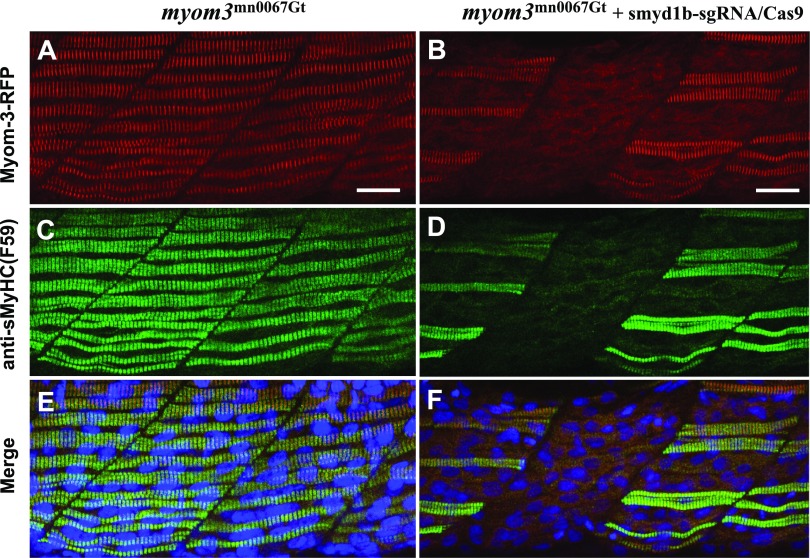
To determine whether knockout of smyd1b affected sarcomere organization in slow muscles, we took advantage of the Myom3mn0067gt zebrafish line generated by transposon-mediated gene trapping that resulted in the RFP gene integration into the Myom3 gene and the localization of a muscle-specific Myom3-RFP fusion protein at the sarcomere M lines (28, 33). The smyd1b gene was mutated in Myom3mn0067gt zebrafish embryos by coinjecting smyd1b-sgRNA with Cas9 protein into the Myom3mn0067gt embryos. Compared with the uninjected control (Fig. 2A), a mosaic pattern in loss of Myom3-RFP expression was clearly observed in ∼40–50% of the slow myofibers of the injected F0 zebrafish embryos (n = 60) (Fig. 2B). The loss of Myom3-RFP positive slow myofibers was unlikely due to a nonspecific injection effect because injection of Cas9 protein alone failed to induce indel mutations and loss of Myom3-RFP expression in slow muscles of the injected embryos (data not shown).
Figure 2.
Knockout of smyd1b by CRISPR disrupted sarcomere organization in slow myofibers. The Smyd1b-sgRNA was coinjected with Cas9 protein into Myom3mn0067gt zebrafish embryos at 1–2 cell stages. Myom3-RFP expression (A, B) and anti-sMyHC (F59) immunostaining (C, D) show the respective M line and thick filament organization in slow fibers of control (A, C) and Smyd1b-sgRNA/Cas9–injected (B, D) embryos at 24 hpf. E, F) Merged images of A, C or B, D, respectively. Scale bars, 20 μm (A, B).
To further validate the function of smyd1b in slow muscles, we analyzed the thick filament organization in slow muscles using a slow fiber-specific anti-MyHC antibody (F59) staining (Fig. 2C, D). A clear loss of myosin thick filament organization was observed in 40–50% of the slow muscles of the smyd1b-sgRNA/Cas9–injected F0 embryos (Fig. 2D). Moreover, compared with the control in Fig. 2E, there was a perfect correlation between the lack of Myom3-RFP organization and the loss of myosin thick filaments in slow fibers of the injected F0 embryos (Fig. 2F). Together, these data indicate that Smyd1b plays a critical role in sarcomere organization in slow muscle of zebrafish embryos.
The early slow muscle defect was gradually recovered in smyd1b mutant embryos
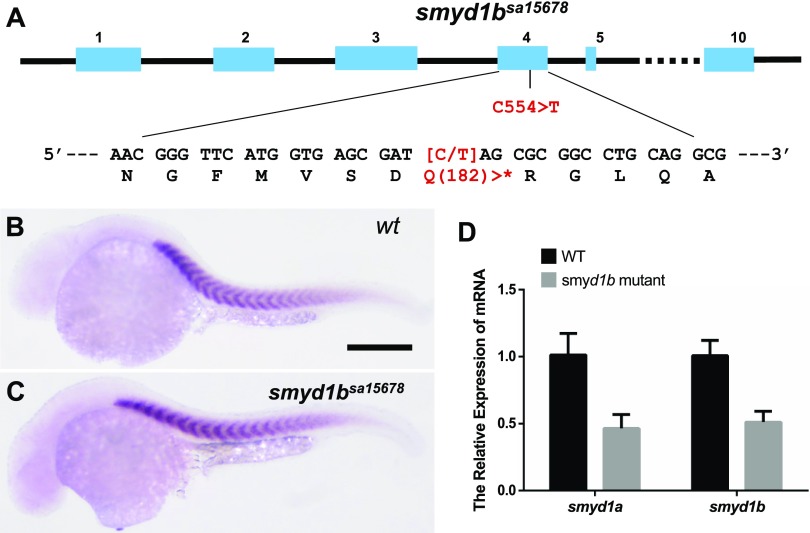
To determine the effect of smyd1b mutation on slow muscle development, we obtained a novel smyd1bsa15678/+ mutant allele that carries a C544 > T nonsense mutation in codon Q182 of smyd1b mRNA (Fig. 3A) (27). Homozygous smyd1bsa15678 mutant was generated by in-crossing of the smyd1bsa15678/+ mutants. Approximately 25% of the produced embryos (103/405) showed the lack of muscle contraction at 24 hpf, and these embryos were confirmed to be smyd1bsa15678 homozygous mutants. The rest of the embryos had normal muscle contraction and represented the WT and smyd1bsa15678/+ mutants, respectively. The locomotion defect was found to be 100% penetrant in all smyd1bsa15678 homozygous mutants produced by various spawning pairs in multiple generations.
Figure 3.
The effect of nonsense mutation in smyd1bsa15678 on smyd1a and smyd1b expression. A) Diagram shows the C554 > T mutation in the smyd1bsa15678 mutant. The C554 > T mutation created a premature in-frame stop codon (*) at codon Q182. B, C) In situ hybridization comparing smyd1b mRNA expression in WT control (B) and smyd1bsa15678 (C) mutant embryos at 24 hpf. Scale bar (B), 100 μm. D) qRT-PCR analysis compared the levels of smyd1a and smyd1b mRNA transcripts in WT control and smyd1bsa15678 mutant embryos at 24 hpf.
To test whether the C544 > T nonsense mutation in smyd1bsa15678 caused a nonsense-mediated RNA decay, we analyzed smyd1b mRNA expression in smyd1bsa15678 mutant and WT control embryos by whole-mount in situ hybridization and qRT-PCR. The data revealed a comparable signal from the in situ hybridization in WT control and smyd1b mutant (Fig. 3B, C). qRT-PCR results further confirmed that there was no significant change of smyd1b mRNA levels in the smyd1bsa15678 mutant embryos (Fig. 3D). The smyd1bsa15678 mutant mRNA transcripts, if translated, could only produce a truncated Smyd1b mutant protein with only 181 aa at the N terminus, missing 2/3 of the entire protein sequence containing the critical SET and C-terminal domains required for its biologic function.
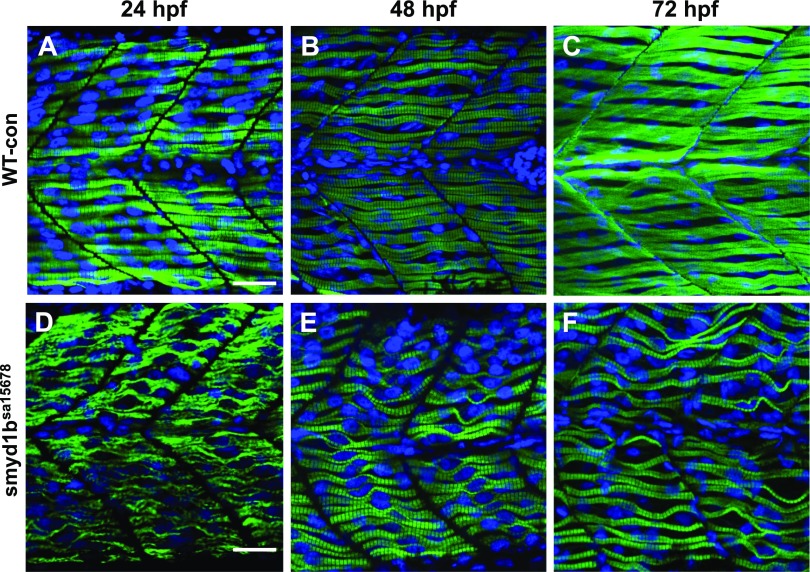
To determine whether loss of smyd1b affected myofibrillogenesis, sarcomere organization was characterized in slow muscles of the mutant and WT sibling at 24, 48, and 72 hpf by immunostaining with anti-myosin antibody (Fig. 4). Compared with the WT sibling (Fig. 4A–C), smyd1bsa15678 homozygous mutant embryos showed a poor sarcomere organization of myosin thick filaments with a 100% penetrance (n = 30) at 24 hpf (Fig. 4D). Interestingly, we noted that myosin thick filament organization gradually appeared in the slow fibers of smyd1bsa15678 homozygous mutant embryos at 48 and 72 hpf although myofibers in the mutant were not as straight as the control fibers (Fig. 4E, F).
Figure 4.
The effect of smyd1b mutation on thick filament organization in slow muscle fibers. Zebrafish embryos of WT control sibling (WT-con) and smyd1bsa15678 mutant were fixed at 24, 48, and 72 hpf. Myosin thick filament organization was analyzed by immunostaining with the anti-sMyHC (F59) antibody. The images represent side view of trunk muscles. A–C) Lateral view of myosin thick filament organization in slow muscle fibers of WT control sibling at 24 (A), 48 (B), and 72 (C) hpf. D–F) Lateral view of thick filament organization in slow muscle fibers of smyd1bsa15678 mutant embryos at 24 (D), 48 (E), and 72 (F) hpf. To better visualize the myofibril defect, the image (D) was taken using a higher gain compared with the gain used for taking the image (A). Scale bars, 25 µm (A, D).
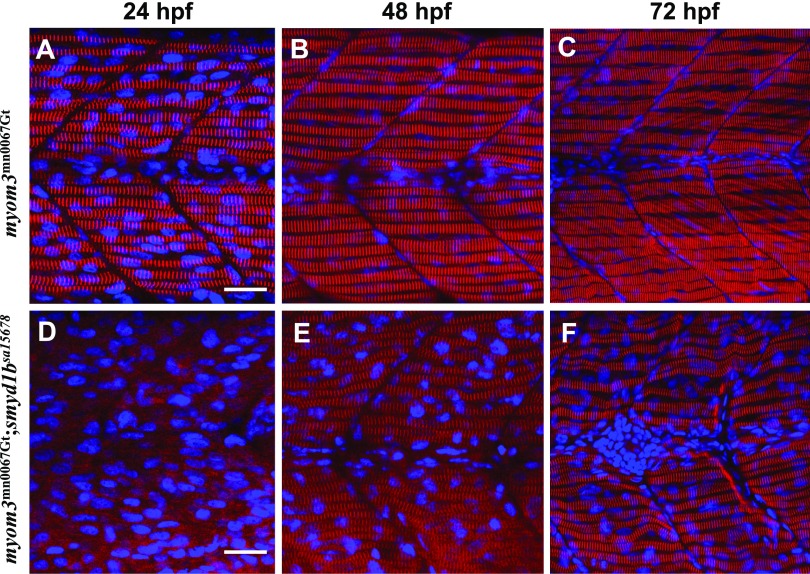
To determine whether smyd1bsa15678 mutation also disrupted other sarcomere structures in the slow muscle, we took advantage of the Myom3mn0067gt zebrafish line that expressed a Myom3-RFP localized specifically at the M line of slow muscles (30, 33). The Myom3mn0067gt zebrafish was crossed with the smyd1bsa15678/+ mutant line. The Myom3mn0067gt; smyd1bsa15678/+ mutants were identified by genotyping using fin clip DNA. The Myom3mn0067gt; smyd1bsa15678/+ mutants were in-crossed to generate Myom3mn0067gt; smyd1b+/+, Myom3mn0067gt; smyd1bsa15678/+, and Myom3mn0067gt; smyd1bsa15678 mutant embryos. Myom3-RFP localization was determined in the Myom3mn0067gt (Fig. 5A–C) and Myom3mn0067gt; smyd1bsa15678 mutant embryos (Fig. 5D–F). The data showed that loss of smyd1b completely abolished the M-line organization of Myom3-RFP in slow myofibers at 24 hpf (Fig. 5D). The M-line defect was, however, gradually reduced at 48 and 72 hpf (Fig. 5E, F). Collectively, these studies indicate that Smyd1b is initially required for slow muscle development in early-stage embryos at 24 hpf. However, the slow muscle defect was gradually reduced in smyd1bsa15678 homozygous mutant embryos at 48 and 72 hpf, suggesting that other redundant factors may be involved.
Figure 5.
The effect of smyd1b mutation on M-line organization in slow muscle fibers. Zebrafish embryos of WT Myom3mn0067gt and Myom3mn0067gt; smyd1bsa15678 mutants were fixed at 24, 48, and 72 hpf. M-line organization was analyzed by directly detecting Myom3–RFP localization. The images represent side view of trunk muscles. A–C) Lateral view of M-line localization of Mym3-RFP in slow muscle fibers of Myom3mn0067gt embryos at 24 (A), 48 (B), and 72 (C) hpf. D–F) Lateral view of M-line localization of Mym3-RFP in slow muscle fibers of Myom3mn0067gt; smyd1bsa15678 mutant embryos at 24 (D), 48 (E), and 72 (F) hpf. Scale bars, 25 µm (A, D).
smyd1a mutant embryos were viable and showed no defects in muscle development
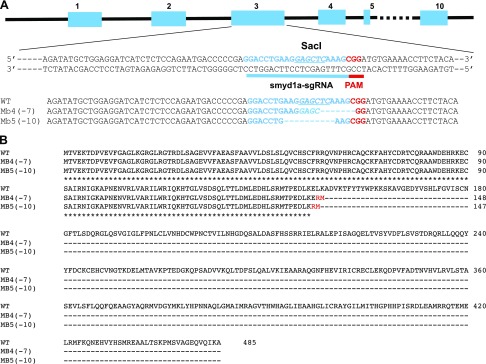
To determine whether Smyd1a plays a redundant role in muscle development, we decided to knock out the smyd1a gene in zebrafish using the CRISPR technology (Fig. 6A). Two mutant alleles, smyd1amb4 and smyd1amb5, were generated. The smyd1amb4 and smyd1amb5 mutant alleles contained a 7 bp (TCAAAGC) and a 10 bp (AAGGAGCTCA) deletion, respectively (Fig. 6A). The indel mutations in smyd1amb4 and smyd1amb5 caused reading frame shift mutations at codon 147 and 146, respectively. The frame shift mutations resulted in premature translational termination, removing the essential SET and C-terminal domains required for Smyd1 function (Fig. 6B).
Figure 6.
Smyd1a-sgRNA design and indel mutations generated by CRISPR in the zebrafish smyd1a gene. A) The Smyd1a-sgRNA was designed to target the exon 3 sequence in the smyd1a gene in zebrafish. The Smyd1a-sgRNA was coinjected with Cas9 mRNA into WT zebrafish embryos at 1–2 cell stages. Two mutant alleles (smyd1amb4 and smyd1amb5) were generated that contain a 7 or 10 bp deletion, respectively. B) The smyd1amb4 and smyd1amb5 mutant alleles contained a reading frame shift mutation at position 147 and 146 with respect to the ATG start codon, respectively, leading to premature termination and truncation of almost 2/3 of the Smyd1a protein containing the essential middle and C-terminal regions.
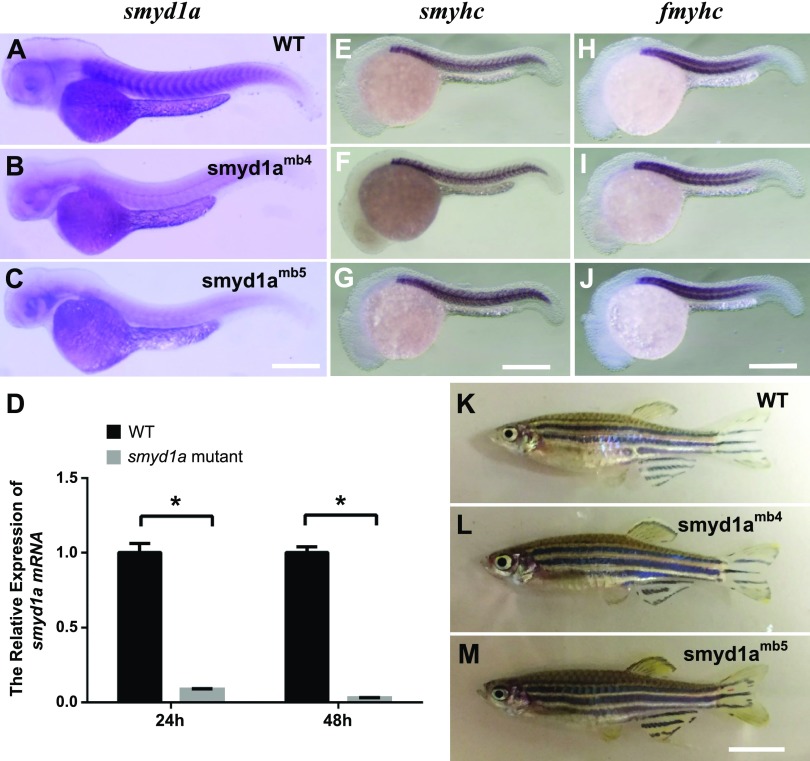
To assess whether these reading frame shift mutations could lead to nonsense-medicated RNA decay, we compared smyd1a mRNA expression in WT, smyd1amb4, and smyd1amb5 mutants by whole-mount in situ hybridization. The results revealed a dramatic reduction of smyd1a mRNA levels in the smyd1amb4 and smyd1amb5 mutant embryos compared with WT control (Fig. 7A–C). qRT-PCR revealed a 7–10-fold decrease of smyd1a transcript levels in smyd1amb5 mutant embryos at 24 and 48 hpf, indicating a nonsense-mediated RNA decay (Fig. 7D).
Figure 7.
Loss of smyd1a had no effect on myogenesis and fish growth. A–C) In situ hybridization comparing smyd1a mRNA expression in WT control (A), smyd1amb4 (B), and smyd1amb5 (C) mutant embryos at 48 hpf. D) qRT-PCR analysis compared the levels of smyd1a mRNA transcripts in WT control and smyd1amb5 mutant embryos at 24 and 48 hpf. *P < 0.0001. E–J) In situ hybridization comparing MyHC gene expression in slow (E–G) and fast (H–J) muscles in WT control (E, H) or smyd1amb4 (F, I) and smyd1amb5 (G, J) mutant embryos at 24 hpf. The Smyhc1 and Fmyhc probes are slow and fast muscle specific, respectively. K–M) Morphologic comparison of WT control (K), smyd1amb4 (L), and smyd1amb5mutant fish (M) at 90 d old. Scale bars: 150 μm (C); 120 μm (G, J); 700 μm (M).
To determine whether loss of smyd1a had a subtle effect on muscle development, we analyzed myosin gene expression during myogenesis by whole-mount in situ hybridization. Compared with WT control (Fig. 7E, H), myosin mRNA expression appeared normal in both slow and fast muscle cells in smyd1amb4 and smyd1amb5 mutant embryos (Fig. 7F, G, I, J). These data indicate that loss of smyd1a had no visible effect on early myogenesis in zebrafish embryos. Viable homozygous mutants were generated for both smyd1amb4 and smyd1amb5 mutant alleles. Unlike the smyd1bsa15678 homozygous mutants, the smyd1amb4 and smyd1amb5 homozygous mutants developed normally and grew into reproductive adult fish (Fig. 7L, M), suggesting that Smyd1a is not required for fish growth and survival.
smyd1a mutation alone had no visible effect on sarcomere organization in slow and fast muscles
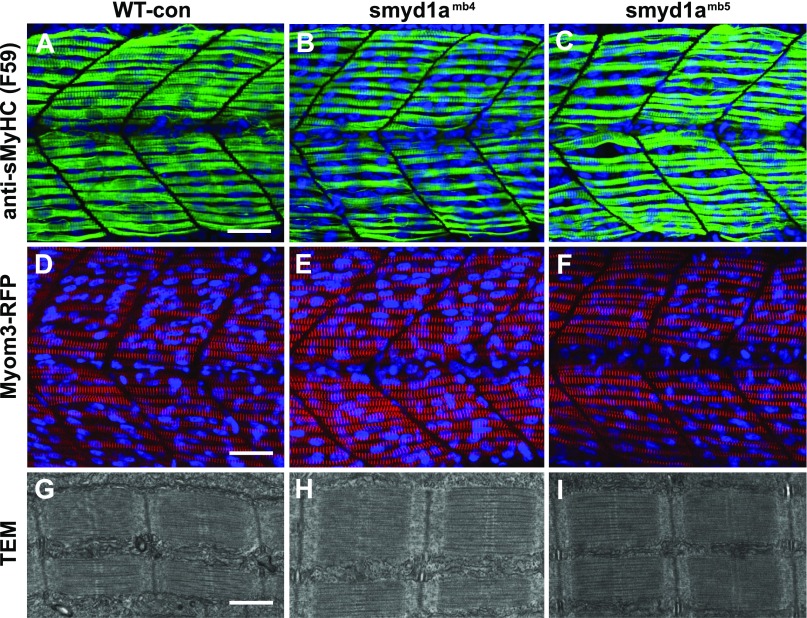
It is well known that Smyd1b, the homolog of Smyd1a, plays a vital role in muscle cell differentiation and sarcomere assembly in skeletal and cardiac muscles of zebrafish embryos (7–9, 11, 13). To determine whether loss of smyd1a disrupted sarcomere organization during muscle cell differentiation, we analyzed the thick filament organization in smyd1a mutant embryos by immunostaining with a slow muscle-specific anti-sMyHC antibody (F59) (Fig. 8A–C). In addition, we generated Myom3mn0067gt; smyd1amb4 and Myom3mn0067gt; smyd1amb5 mutants and characterized M-line organization of Myom3-RFP in slow myofibers (Fig. 8D–F). Compared with WT sibling, (Fig. 8A, D), no visible difference was detected in myosin thick filament or M-line organization in slow myofibers of smyd1amb4 or smyd1amb5 mutant embryos (Fig. 8B, C, E, F). Moreover, transmission electron microscopy (TEM) analysis revealed highly organized sarcomeres in slow fibers of smyd1amb4 or smyd1amb5 mutant embryos (Fig. 8H, I).
Figure 8.
The effect of smyd1a deficiency on sarcomere organization in slow muscle fibers. Zebrafish embryos of WT sibling, smyd1amb4, smyd1amb5, Myom3mn0067gt, Myom3mn0067gt; smyd1amb4, and Myom3mn0067gt; smyd1amb5 embryos were fixed at 24 hpf. The sarcomere organization was analyzed by immunostaining with the anti-MyHC (F59) antibody (A–C), direct observation of Myom-3-RFP localization (D–F), and TEM analysis (G–I). Lateral view of myosin thick filament organization in slow muscle fibers (A–C) of WT control (A), smyd1amb4 (B), and smyd1amb5 (C) mutant embryos by F59 antibody staining at 24 hpf. Lateral view of M-line organization of Myom-3-RFP in slow fibers of Myom3mn0067gt (D), Myom3mn0067gt; smyd1amb4 (E), and Myom3mn0067gt; smyd1amb5 (F) mutant embryos at 24 hpf. TEM analysis shows the sarcomere organization in slow myofibers of WT control (G), smyd1amb4 (H), and smyd1amb5 (I) mutant embryos at 48 hpf. Scale bars: 30 µm (A, D); 500 nm (G).
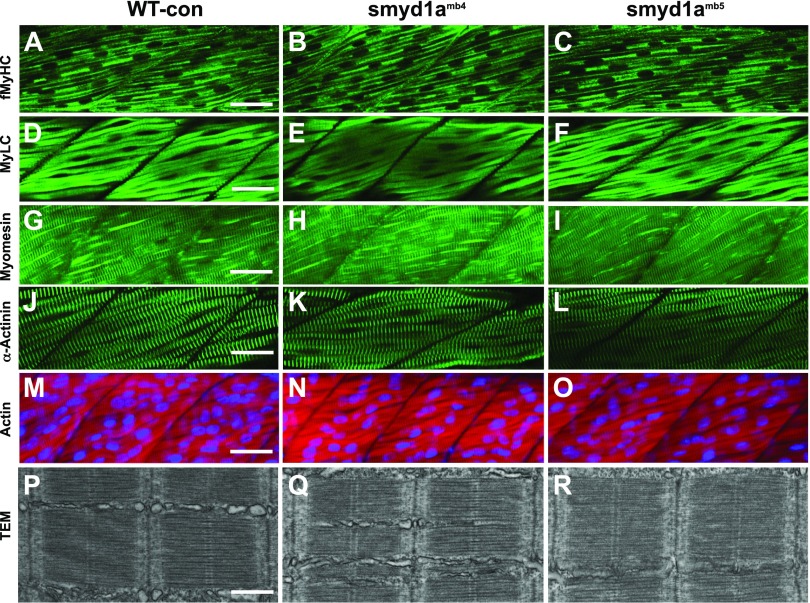
To determine whether Smyd1a mutation affected sarcomere organization in fast muscles, we characterized the sarcomere structures in fast myofibers of smyd1amb4 and smyd1amb5 mutant embryos by immunostaining and EM analyses (Fig. 9). The result showed that smyd1amb4 and smyd1amb5 mutant embryos contained highly organized thick and thin filaments as well as Z and M lines in their fast myofibers (Fig. 9A–O). Moreover, TEM analysis revealed no detectable difference in sarcomere organization between the WT and the smyd1a mutants (Fig. 9P–R). Collectively, these data indicate that loss of smyd1a alone had no detectable effect on sarcomere organization in both slow and fast muscles during muscle cell differentiation.
Figure 9.
The effect of smyd1a deficiency on sarcomere organization in fast muscle fibers. Zebrafish embryos of WT sibling, smyd1amb4, and smyd1amb5 mutant embryos were fixed at 48 or 72 hpf. The key sarcomere structures were analyzed by staining with Phalloidin-TRITC and various antibodies, as well as TEM. A–C) Lateral view of myosin light chain organization in fast muscle fibers of WT control (A), smyd1amb4 (B), and smyd1amb5 (C) mutant embryos shown by anti-MyHC (MF20) antibody staining at 72 hpf. D–F) Lateral view of myosin light chain organization in fast muscle fibers of WT control (D), smyd1amb4 (E), and smyd1amb5 (F) mutant embryos shown by anti-MyLC (F310) antibody staining at 48 hpf. G–I) Lateral view of M-line organization in fast muscle fibers of WT control (G), smyd1amb4 (H), and smyd1amb5 (I) mutant embryos by anti-Myomesin antibody staining at 48 hpf. J–L) Lateral view of Z-line organization in fast muscle fibers of WT control (J), smyd1amb4 (K), and smyd1amb5 (L) mutant embryos by anti-α-actinin antibody staining at 48 hpf. M–O) Lateral view of actin thin filament organization in fast muscle fibers of WT control (M), smyd1amb4 (N), and smyd1amb5 (O) mutant embryos analyzed by Phalloidin-TRITC staining at 48 hpf. P–R) TEM analysis shows the sarcomere organization in fast myofibers of WT control (P), smyd1amb4 (Q), and smyd1amb5 (R) mutant embryos at 48 hpf. Scale bars: 40 µm (A, D, G, J, M); 500 nm (P).
smyd1a and smyd1b double mutants showed a stronger muscle phenotype in slow muscles
To determine whether loss of both smyd1a and smyd1b together could result in a stronger muscle defect, we first generated smyd1amb5/+; smyd1b sa15678/+ double heterozygous mutant by crossing the smyd1amb5 homozygous mutant with smyd1bsa15678/+ heterozygous mutant. The smyd1amb5/+; smyd1b sa15678/+ heterozygous double mutants grew normally into reproductive adults. The smyd1amb5/+; smyd1b sa15678/+ heterozygous double mutants were in-crossed to generate fish embryos of 9 distinct genotypes, including smyd1a+/+; smyd1b+/+, smyd1a+/+; smyd1bsa15678/+, smyd1a+/+; smyd1bsa15678, smyd1amb5/+; smyd1b+/+, smyd1amb5/+; smyd1bsa15678/+, smyd1amb5/+; smyd1bsa15678, smyd1amb5; smyd1b+/+, smyd1amb5; smyd1bsa15678/+, and smyd1amb5; smyd1bsa15678. Except the smyd1a+/+; smyd1bsa15678 single, smyd1amb5/+; smyd1bsa15678, and smyd1amb5; smyd1bsa15678 double mutants, all other types of mutants were viable and grew into reproductive adults in a predicted Mendelian ratio. Collectively, these data indicate that Smyd1b is more critical than Smyd1a in muscle development and fish survival.
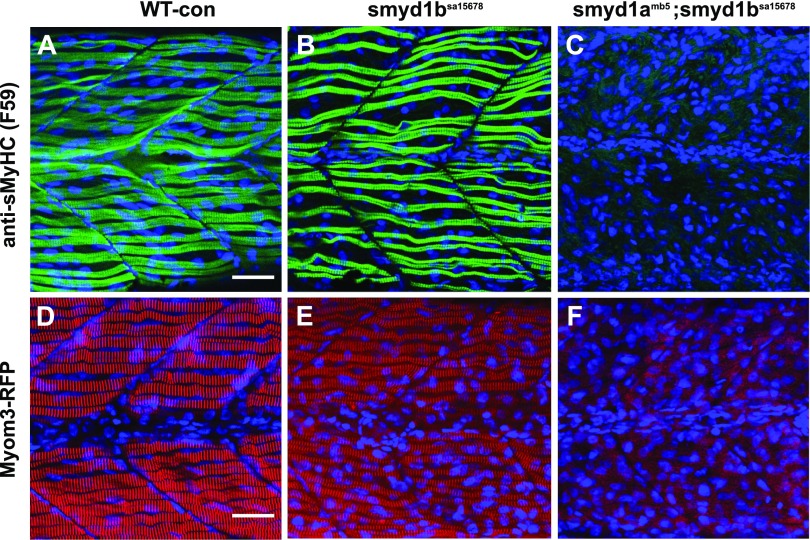
The smyd1amb5; smyd1bsa15678/+ mutants were identified and used for in-cross to generate smyd1amb5; smyd1bsa15678 double mutant embryos. As expected, ∼25% of the embryos (31/128) from above in-cross showed the lack of locomotion at 24, 48, and 72 hpf. The paralyzed embryos were genotyped by PCR and found to be exclusively smyd1amb5; smyd1bsa15678 double mutants. To assess whether loss of smyd1a and smyd1b together had a stronger effect on slow muscles, we compared the thick filament in slow fibers of smyd1bsa15678 single and smyd1amb5; smyd1bsa15678 double mutants (Fig. 10A–C). The results showed that in contrast to the smyd1bsa15678 single mutation, which had organized sarcomeres in slow muscle fibers at 72 hpf (Fig. 10B), smyd1amb5; smyd1bsa15678 double mutations resulted in a complete disruption of myosin thick filament organization in slow muscle fibers at 72 hpf (Fig. 10C).
Figure 10.
The effect of smyd1b single or smyd1a; smyd1b double mutation on myofibril organization in slow muscles. Zebrafish embryos of WT control, smyd1bsa15678, and smyd1amb5; smyd1bsa15678 mutant embryos were fixed at 72 hpf. Thick filaments in slow muscles were analyzed by immunostaining with anti-MyHC (F59) antibody (A–C), whereas the M-line organization in fast muscles was analyzed by Myom3-RFP localization in the slow myofibers of trunk muscles (D–F). Lateral view of myosin light chain organization in slow muscle fibers of WT control (A), smyd1bsa15678 (B), and smyd1amb5; smyd1bsa15678 (C) mutant embryos at 72 hpf. Lateral view of M-line organization of Myom3-RFP in slow muscle fibers of WT Myom3mn0067gt control (D), Myom3mn0067gt; smyd1bsa15678 (E), and Myom3mn0067gt; smyd1amb5; smyd1bsa15678 (F) mutant embryos at 72 hpf. Scale bar, 30 µm (A, D).
To investigate the effect of smyd1a; smyd1b double mutation on the M-line organization, smyd1amb5; smyd1bsa15678/+ double mutants were crossed with Smyd1amb5; Myom3mn0067gt. The smyd1amb5; smyd1bsa15678/+; Myom3mn0067gt were identified from the above cross at adult state by fin clip PCR. The smyd1amb5; smyd1bsa15678/+; Myom3mn0067gt were in-crossed to generate the smyd1amb5; smyd1bsa15678; Myom3mn0067gt fish embryos. Myom3-RFP localization was compared between the WT control, smyd1b sa15678 single, and smyd1amb5; smyd1bsa15678 double mutant Myom3mn0067gt embryos (Fig. 10D–F). The data showed that loss of smyd1a and smyd1b together resulted in a complete disruption of M-line organization in slow fibers at 72 hpf (Fig. 10F). These data indicate that both Smyd1a and Smyd1b are involved in slow muscle development because smyd1a and smyd1b double mutation gave a stronger muscle defect compared with smyd1a or smyd1b mutant alone. Collectively, these data indicate that Smyd1a and Smyd1b together play an important role in sarcomere assembly in slow muscles of zebrafish embryos.
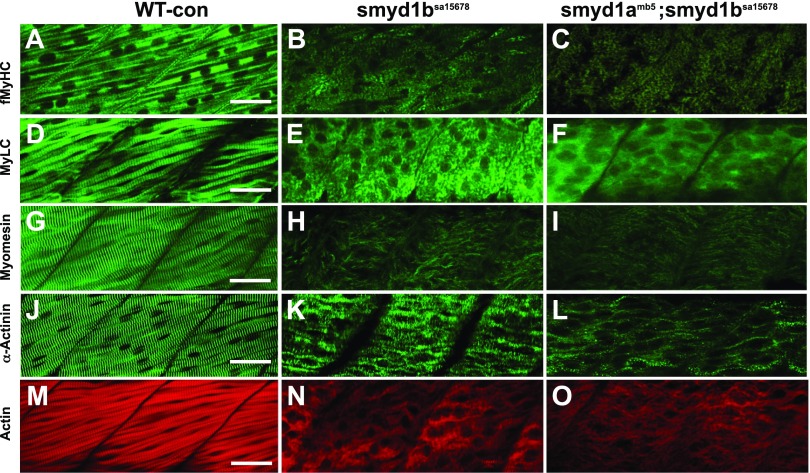
To determine whether the smyd1amb5 single and smyd1amb5; smyd1bsa15678 double mutants exhibited similar or distinct muscle defects in fast muscles, we compared the key sarcomere organization in smyd1bsa15678 single and smyd1amb5; smyd1bsa15678 double mutants (Fig. 11). The key sarcomere structures were examined by staining with phalloidin and various antibodies against MyLC, MyHC, α-actinin, and myomesin. The results showed that the smyd1b mutation alone resulted in a dramatic disruption of sarcomere organization in fast fibers. There was little or no sarcomeric localization of myomesin, MyHC, and MyLC that could be detected in the smyd1bsa15678 single (Fig. 11B, E, H) and smyd1amb5; smyd1bsa15678 double mutants (Fig. 11C, F, I). A weak sarcomeric organization of α-actinin and α-actin was observed in both smyd1bsa15678 single (Fig. 11K, N) and smyd1amb5; smyd1bsa15678 double mutants (Fig. 11L, O).
Figure 11.
The effect of smyd1b single or smyd1a; smyd1b double mutation on myofibril organization in fast muscles. A–C) Lateral view of myosin thick filament organization in fast muscle fibers of WT control (A), smyd1bsa15678 (B), and smyd1amb5;smyd1bsa15678 (C) mutant embryos by anti-MyHC (MF-20) antibody staining at 72 hpf. D–F) Lateral view of myosin light chain organization in fast muscle fibers of WT control (D), smyd1bsa15678 (E), and smyd1amb5;smyd1bsa15678 (F) mutant embryos by anti-MyLC (F310) antibody staining at 48 hpf. G–I) Lateral view of M-line organization in fast muscle fibers of WT control (G), smyd1bsa15678 (H), and smyd1amb5;smyd1bsa15678 (I) mutant embryos by anti-myomesin antibody staining at 48 hpf. J–L) Lateral view of Z-line organization in fast muscle fibers of WT control (J), smyd1bsa15678 (K), and smyd1amb5;smyd1bsa15678 (L) mutant embryos by anti-α-actinin antibody staining at 48 hpf. M–O) Lateral view of actin thin filament organization in fast muscle fibers of WT control (M), smyd1bsa15678 (N), and smyd1amb5;smyd1bsa15678 (O) mutant embryos by Phalloidin-TRITC staining at 48 hpf. Scale bars, 30 µm (A, D, G, J, M).
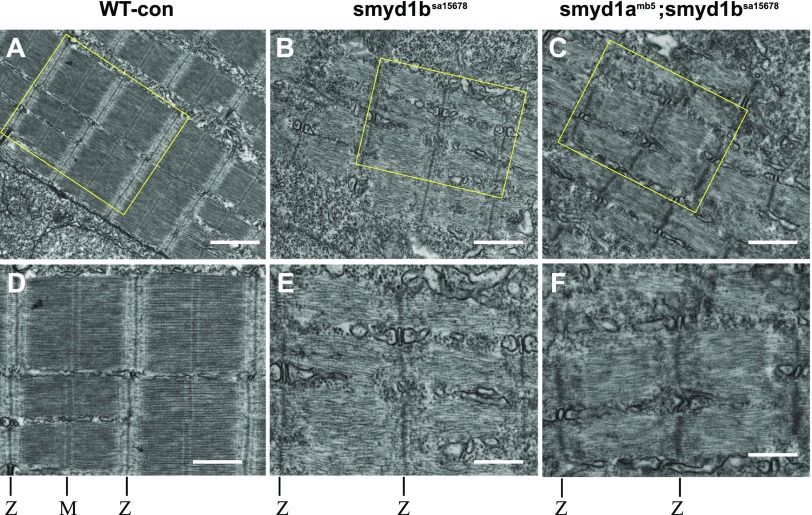
To validate these findings, we analyzed the subcellular structure in skeletal muscles of WT, smyd1bsa15678 single, and smyd1amb5; smyd1bsa15678 double mutant embryos by TEM analysis at 48 hpf (Fig. 12). Compared with the WT control (Fig. 12A, D), the results showed a dramatic reduction of myofibrils in the muscle cells of smyd1bsa15678 single or smyd1amb5; smyd1bsa15678 double mutant embryos (Fig. 12B, C). Close examination revealed sarcomere-like structures in the smyd1bsa15678 or smyd1amb5; smyd1bsa15678 mutant fibers that contained the Z lines and thin filaments (Fig. 12E, F). However, no M line and thick filaments could be detected in these sarcomere-like structures in the mutants (Fig. 12E, F). Given that loss of smyd1b or smyd1a and smyd1b together produced similar defects in fast muscles, these results suggest that Smyd1b is essential for fast muscle development in zebrafish embryos.
Figure 12.
The effect of smyd1b single or smyd1a; smyd1b double mutation on subcellular sarcomere organization in fast muscles. TEM analysis shows the sarcomere organization in fast myofibers of WT control (A, D), smyd1bsa15678 (B, E) and smyd1amb5; smyd1bsa15678 (C, F) mutant embryos at 48 hpf. Images (D, E, F) are larger images from part of A–C, respectively. The Z line and M line are indicated (D–F). Scale bars: 850 nm (A–C); 500 nm (D–F).
The effect smyd1a and smyd1b mutations on myosin and heat shock protein gene expression
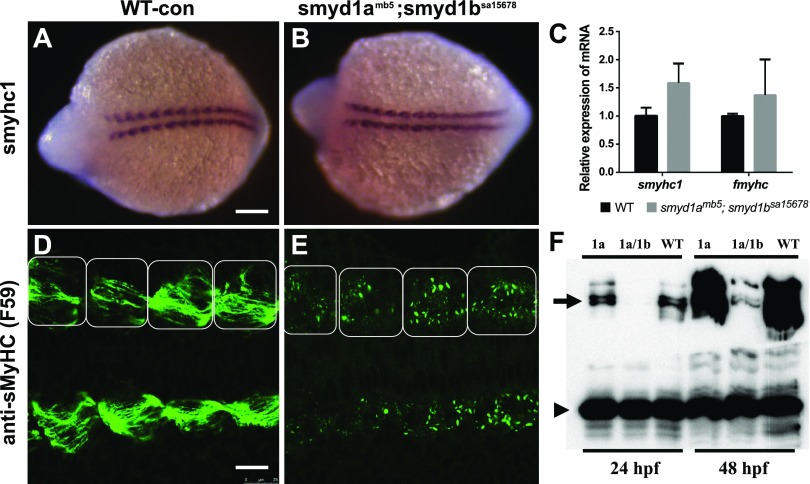
The molecular mechanism underlying the lack of sarcomere organization in smyd1a;smyd1b mutant is not clear. Previous studies have shown that knockdown of Smyd1b resulted in a significant reduction in MyHC protein levels in zebrafish embryos at 24 and 72 hpf (9). To determine whether loss of smyd1a and smyd1b had an early effect on MyHC expression in myocytes before the differentiation into myofibers, we analyzed MHC mRNA and protein expression by whole-mount in situ hybridization, qRT-PCR, and anti-MyHC immunostaining in zebrafish embryos at the 9 somite stage (Fig. 13). The in situ data showed that loss of smyd1a and smyd1b had no significant effect on MyHC mRNA expression in the slow muscle (Fig. 13B). This was further confirmed by qRT-PCR analysis (Fig. 13C). However, compared with WT control (Fig. 13D), signal from the anti-MyHC immunostaining was greatly reduced in adaxial cells (slow muscle precursors) of smyd1a; smyd1b mutant embryos (Fig. 13E). Unlike the control embryo, in which MyHC staining appeared as strings of premyofibrils (34) in adaxial cells (Fig. 13D), the fluorescent signals in the mutant embryos appeared much weaker as small aggregates (Fig. 13E).
Figure 13.
The effect of smyd1a and smyd1b deficiency on myosin expression in early-stage zebrafish embryos. Zebrafish embryos from WT or smyd1amb5;smyd1bsa15678 mutant were fixed at 14 hpf. A–F) Slow MyHC 1 mRNA and protein expression were analyzed by whole-mount in situ hybridization (A, B) and antibody staining (D, E), respectively. The images represent dorsal view of the embryos. In situ hybridization comparing MyoD gene expression in WT control (A) or smyd1amb5; smyd1bsa15678 mutant (B) embryos at 14 hpf. C) qRT-PCR analysis compared the levels of Smyhc and Fmyhc mRNA transcripts in WT control and smyd1amb5; smyd1bsa15678 mutant embryos at 24 hpf. Immunostaining with anti-MHC (F59) shows the MyHC protein expression in WT control (D) or smyd1amb5; smyd1bsa15678 mutant (E) embryos at 14 hpf. The muscle segments/somites are indicated by the boxes. F) Western blot analysis compared the levels of MyHC protein expression in smyd1amb5, smyd1amb5; smyd1bsa15678 mutants and WT control embryos at 24 and 48 hpf. Scale bars: 150 µm (A); 25 µm (D).
To test whether smyd1amb5; smyd1bsa15678 mutant embryos had low levels of myosin protein expression, we performed Western blot analysis. The data showed a dramatic reduction of MyHC protein levels in the smyd1amb5; smyd1bsa15678 mutant embryos compared with WT and smyd1a mutants (Fig. 13F). Collectively, these data indicated that smyd1amb5; smyd1bsa15678 double mutations had no significant effect on MyHC mRNA expression. However, loss of smyd1a and smyd1b together resulted in increased MyHC protein aggregation and degradation in muscle cells leading to diminished sarcomere organization.
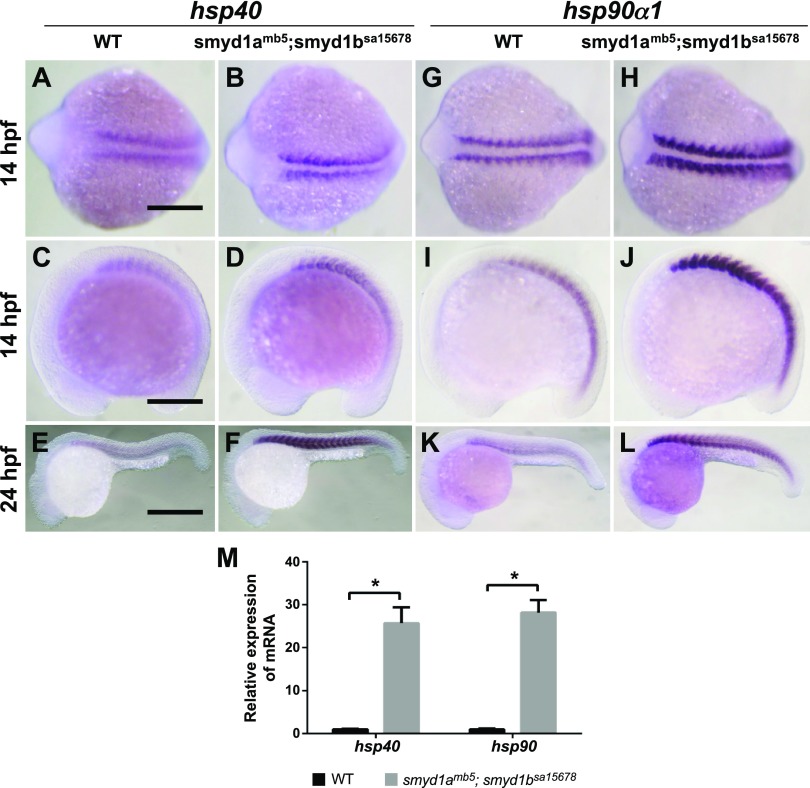
Up-regulation of heat shock protein gene expression is associated with stress response from protein misfolding and degradation. To determine the effect of smyd1a; smyd1b mutation on heat shock protein gene expression, we analyzed Hsp40 and Hsp90a1 mRNA expression in WT and smyd1amb5; smyd1bsa15678 mutant embryos at 14 and 24 hpf. A strong up-regulation of hsp40 and hsp90a1 expression was detected in smyd1amb5; smyd1bsa15678 double mutant embryos at 14 and 24 hpf (Fig. 14). This was further confirmed by qRT-PCR, which revealed a 25- and 28-fold increase for hsp40 and hsp90a1 expression in smyd1a; smyd1b double mutant embryos, respectively (Fig. 14M). In contrast, loss of smyd1a alone had no effect on heat shock protein gene expression (Data not shown). Together, these data indicate that loss of smyd1a and smyd1b triggered a muscle-specific stress response, suggesting that they may play a critical role in muscle protein stability and sarcomere assembly.
Figure 14.
Loss of smyd1a and smyd1b in zebrafish up-regulates hsp40 and hsp90-α1 gene expression. A–F) In situ hybridization comparing hsp40 gene expression in WT control (A, C, E) or smyd1amb5; smyd1asa15678 double mutant (B, D, F) embryos at 14 (A–D) and 24 (E, F) hpf, respectively. G–L) In situ hybridization comparing hsp90-α1 gene expression in WT control (G, I, K) or smyd1amb5; smyd1asa15678 mutant (H, J, L) embryos at 14 (G–J) and 24 (K, L) hpf, respectively. Scale bars: 200 µm (A, C); 500 µm (E). M) qRT-PCR analysis compared the levels of hsp40 and hsp90-α1 mRNA transcripts in WT control and smyd1amb5; smyd1asa15678 double mutant embryos at 24 hpf *P < 0.05.
Ectopic expression of zebrafish smyd1a, smyd1b or mouse Smyd1 rescued the myofibril defects in smyd1a; smyd1b mutant
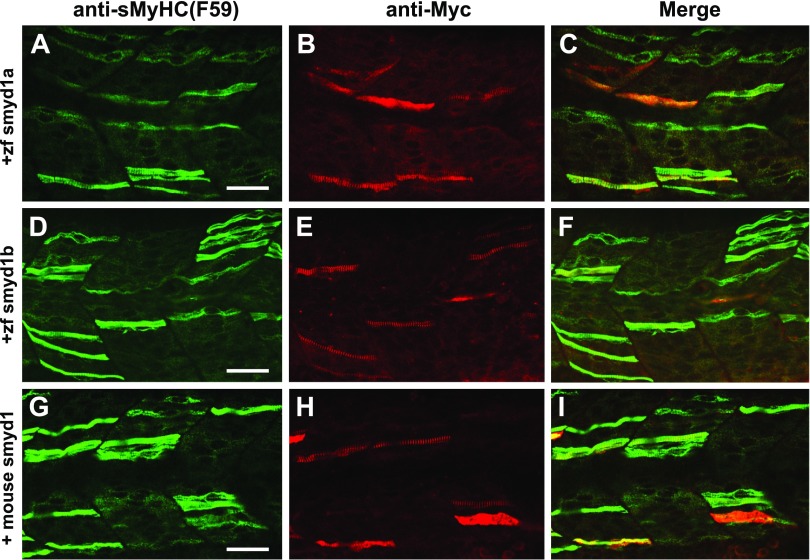
Sequence analysis revealed that Smyd1 proteins are highly conserved from zebrafish to mice and humans. To test whether Smyd1 function is conserved during evolution, we tested whether expression of the zebrafish smyd1a or smyd1b or mouse Smyd1 could rescue the muscle defects in the smyd1amb5; smyd1bsa15678 double mutant embryos. The Tg (smyd1b:zfsmyd1amyc) or Tg (smyd1b:zfsmyd1bmyc) transgene expressing a myc-tagged zebrafish Smyd1a or Smyd1b was microinjected into smyd1amb4; smyd1bsa15678 double mutant zebrafish embryos. Immunostaining revealed a mosaic pattern of slow myofibers with clear thick filaments in 60% (n = 30) and 67% (n = 30) of the Tg(smyd1b:zfsmyd1amyc)-(Fig. 15A–C) or Tg (smyd1b:zfsmyd1bmyc)-injected (Fig. 15D, F) mutant embryos. The mosaic pattern of slow fiber formation is consistent with the nature of ectopic gene expression via DNA plasmid injection. This was confirmed by the mosaic pattern of Smyd1bmyc and Smyd1bmyc expression from immunostaining with anti-myc antibody (Fig. 15B, C, E, F). Similarly, injection of the Tg (smyd1:mSmyd1myc) transgene expressing an myc-tagged mouse Smyd1 could rescue the myofibril defects in smyd1amb5; smyd1bsa15678 double mutant in a cell-autonomous manner (Fig. 15G–I). Collectively, these data indicate that Smyd1 function is conserved during evolution.
Figure 15.
The myofibril defects in smyd1amb5; smyd1asa15678 double mutant could be rescued by ectopic expression of zebrafish smyd1a, smyd1b, or mouse Smyd1. The smyd1amb5; smyd1asa15678 mutant embryos were injected with DNA constructs expressing the zebrafish smyd1amyc, smyd1bmyc, or mouse Smyd1myc at 1 or 2 cell stages. Myosin thick filament organization and transgene expression were analyzed by double immunostaining with anti-MHC (F59, green) and anti-myc (9E10, red) antibodies. A–C) Double immunostaining with anti-MHC (A) and anti-myc (B) antibodies shows the thick filament organization in myofibers expressing the myc-tagged zebrafish smyd1a transgene at 24 hpf. Merged picture (C) of A and B. D–F) Double immunostaining with anti-MHC (D) and anti-myc (E) antibodies shows the thick filament organization in myofibers expressing the myc-tagged zebrafish smyd1b transgene at 24 hpf. F) Merged picture of D and E. G–I) Double immunostaining with anti-MHC (G) and anti-myc (H) antibodies shows the thick filament organization in myofibers expressing the myc-tagged mouse Smyd1 transgene at 24 hpf. merged picture (I) of G with H. Scale bars, 40 µm (A, D, G).
DISCUSSION
In this study, we analyzed the effects of smyd1a and smyd1b mutations on muscle development in zebrafish embryos. We demonstrated that smyd1a mutation alone had no visible effect on muscle development, growth, and zebrafish survival. This is in contrast to the loss of smyd1b mutations that resulted in skeletal and cardiac muscle defects leading to early embryonic lethality. smyd1a and smyd1b double mutations, however, yield a stronger muscle defect compared with smyd1a or smyd1b mutation alone. Together, our studies indicate that both Smyd1a and Smyd1b partake in slow and fast muscle development, although Smyd1b plays a dominant role compared with Smyd1a.
Smyd1a mutation alone resulted in no visible muscle defect in slow and fast muscle
We demonstrated in this study that knockout of smyd1a alone had no visible effect on muscle development in slow and fast muscles. This is in contrast to a recent report that knockdown of smyd1a using MO or transient injection of CRISPR/Cas9 RNP complex interfered with heart and skeletal muscle function and sarcomere organization in fast muscles of zebrafish embryos (26). The reason underlying these opposite findings is not clear. It has been noted that there is a poor correlation between MO-induced and mutant phenotypes in zebrafish studies (35). The poor correlation has been partially explained by the genetic compensation induced by deleterious genetic mutations but not MO gene knockdown (36). In addition, it has been suggested that alternative splicing or exon skipping in the mutants might also contribute to the recovery of gene function (37, 38). However, these theories might not explain the different results from the smyd1a knockdown and knockout studies. We have reported previously that knockdown of smyd1a alone had no visible muscle defects, unless it was knocked down together with the smyd1b gene (11). In contrast, Paone and colleagues showed that knockdown of Smyd1a had a skeletal and cardiac muscle phenotypes in zebrafish embryos, although different MOs were used in these 2 knockdown studies (11, 26).
It should be noted that CRISPR/Cas9 was also used to mutate the smyd1a gene by Paone and colleagues, and they showed that coinjection of smyd1a-sgRNA/Cas9 RNP complex into zebrafish embryos resulted in CRISPR/Cas9-induced mutants that resemble the smyd1a MO knockdown phenotype (26). However, these studies were performed on the F0 injected embryos where the off-target effects and toxicity could not be ruled out (26). In our study reported here, we used 2 mutant alleles generated by CRISPR and carried out the phenotypical analysis at the F2, F3, and F4 generations, minimizing the off-target effect. Our data revealed that smyd1amb4 and smyd1amb5 mutants had no visible phenotype in fast and slow muscles when smyd1a is mutated alone. These mutant alleles are likely to be null mutants because they contained nonsense mutations and showed nonsense-medicated mRNA decade. In addition, the mutant data were in agreement with results from our previous MO knockdown studies (11). Moreover, other evidence also indicated that loss of smyd1a might not have a strong impact on muscle development. We showed that, in contrast to smyd1b mutation, loss of smyd1a did not up-regulate hsp90-α1 gene expression. This is consistent with previous findings showing that loss of smyd1a did not trigger misfolded myosin response in zebrafish (26).
Smyd1a and Smyd1b are required for slow muscle development
Previous studies using MO knockdown and zebrafish mutant showed contradictory findings regarding Smyd1 function in the slow muscle of zebrafish embryos (7–9, 13). Just and colleagues reported that smyd1b expression was restricted to fast muscles, and loss of smyd1b in flatline (Flamvo47a) mutant had no effect on slow muscle development (8). In contrast, we demonstrated that knockdown of smyd1b by MO abolished sarcomere assembly in both slow and fast myofibers of zebrafish embryos (7, 9, 11). The reason behind the discrepancy was not clear. One possible explanation is the use of different genetic approaches, zebrafish mutant vs. knockdown. However, data from our study here clearly showed a slow muscle defect in smyd1bsa15678 mutant allele and CRISPR-induced smyd1b mutant embryos at 24 hpf. The slow muscle defect in the smyd1bsa15678 mutant was consistent with the results from MO knockdown studies, arguing again the idea that the discrepancy was due to the use of different genetic approaches.
We could not completely rule out the possibility that the discrepancy could be due to the use of different mutant alleles. Just and colleagues used a flatline (Flamvo47a) zebrafish mutant, which carries a nonsense mutation in codon 73 of the zebrafish smyd1b gene (8). The smyd1bsa15678 mutant allele used in our studies carries a nonsense mutation in codon 182 (27). Interestingly, Prill and colleagues noted that slow myofibrils become disrupted in still hearttm123a (sth), another smyd1b mutant allele at 48 hpf (13). The still hearttm123a (sth) zebrafish mutant carries an insertion of 9 nt that contain an in-frame stop codon at position 45 (13). All these mutants contained premature stop codons in the smyd1b gene, thus unlikely producing a functional protein.
We speculate that different timing of phenotypic analyses might contribute to the data discrepancy between these studies. We noted that although the smyd1bsa15678 mutant embryos showed a sarcomere defect 24 hp, the slow muscle defect was partially recovered at 48 and 72 hpf. Just and colleagues characterized the slow muscle phenotype in smyd1b mutant embryos at 48 hpf when some of the slow muscle defect might have been recovered (8). In contrast, we characterized the muscle defects at multiple stages at 24, 48, and 72 hpf, enabling us to observe the muscle defects at the early stage of 24 hpf.
Our study here further indicates that the recovery of slow muscle defect in smyd1b mutant at 48 and 72 hpf was likely due to the expression of smyd1a. smyd1a expression started around 24 hpf and increased significantly at 48 and 72 hpf (11). The timing of smyd1a expression coincides nicely with the slow muscle recovery in smyd1b mutant. Consistent with this idea, we showed that the muscle recovery was completely eliminated in the smyd1a and smyd1b double mutant at 48 and 72 hpf. Collectively, these studies provide a possible explanation in clarifying the discrepancy of previous findings regarding Smyd1a and Smyd1b function in slow muscles.
Rescue of muscle defect in Smyd1a; Smyd1b double mutant by ectopic expression of zebrafish Smyd1a or Smyd1b or mouse Smyd1
We demonstrated in this study that the muscle defects in smyd1a; smyd1b double mutant could be rescued by the ectopic expression of zebrafish smyd1a or smyd1b under the control of the smyd1b muscle-specific promoter. Previous studies by us and others have shown that ectopic expression of smyd1a or smyd1b was able to rescue the muscle defect in smyd1b mutant or knockdown embryos (7, 8, 11). Our studies here further extended these findings that expression of Smyd1a or Smyd1b alone could rescue the muscle defects in the smyd1a; smyd1b double mutants. The rescue of the smyd1a; smyd1b double mutants by smyd1a alone was surprising because smyd1b mutant showed a clear muscle defect although possessing the normal smyd1a gene. We speculate that the phenotypic difference between smyd1a and smyd1b mutants might be caused by the distinct patterns of smyd1a and smyd1b gene expression during muscle development. Consistent with this idea, we have shown that smyd1a expressed later than smyd1b in developing zebrafish embryos (11). By expressing the smyd1a under the control of the smyd1b promoter, we showed that Smyd1a could functionally replace Smyd1b if expressed in similar temporal and spatial patterns as smyd1b.
Up-regulation of heat shock gene expression in Smyd1 mutant embryos
We demonstrated in this study that loss of smyd1a and smyd1b together up-regulated heat shock protein gene expression. This is consistent with previous finding with respect to hsp90a1 expression in smyd1b deficient embryos (8, 9). Previous studies by microarray showed that smyd1b deficiency resulted in a broad up-regulation of heat shock gene expression (8, 9). Consistent with these findings, we demonstrated in this study that expression of another muscle-specific chaperone DnaJ, also known as Hsp40, was also up-regulated in the smyd1b or smyd1a;smyd1b mutant embryos. DnaJ/Hsp40 determines the specificity of Hsp70s by stabilizing their interaction with substrate proteins. The up-regulation of heat shock protein gene expression in smyd1a;smyd1b double mutants likely represents a robust stress response because of the increased muscle protein misfolding and defective sarcomere assembly in muscle cells of Smyd1 mutant embryos.
Molecular mechanism of Smyd1 function
Currently, the molecular mechanism by which Smyd1 functions in myofibrillogenesis is not clear. It appears that Smyd1 plays a critical role in stimulating muscle cell differentiation and sarcomere organization as demonstrated by the loss of function studies in vitro and zebrafish and mouse embryos in vivo (7–9, 12–14), and forced expression of Smyd1 accelerated mouse C2C12 myoblast differentiation in vitro (18). Moreover, modulation of Smyd1 expression by microRNA206 has been implicated in human embryonal rhabdomyosarcoma formation (39). Biochemical analysis revealed that Smyd1 has a histone methyltransferase activity in vitro that has been implicated in transcriptional regulation via histone modification (6, 7). In addition, it has been reported that Smyd1 interacts with skeletal Naca (skNAC), a transcription factor involved in cardiac development and skeletal muscle growth and regeneration (40, 41). Several target genes have been identified in cardiac muscles. A recent study demonstrated that Smyd1 plays an important role in cardiac energetics through transcriptional control of peroxisome proliferator-activated receptor γ coactivator 1α, a key regulator of mitochondrial energetics (19). Moreover, Smyd1 functions as a transcriptional repressor that binds directly to the promoter regions of tgfβ3 and nppa/ANF and inhibits their expression (42). Interestingly, Franklin and colleagues showed that this repression was not associated with significant changes in H3K4 trimethylation at targeted loci, questioning the model of Smyd1 function by targeting histone modification in vivo (42).
Smyd1b has a chaperone-like function in myosin folding and assembly (13). In agreement with the chaperone function, we showed in this study that loss of smyd1 had no effect on myosin gene expression but resulted in a dramatic reduction of myosin protein levels in myocytes of developing zebrafish embryos. It has been shown that Smyd1 interacts with MyHC and myosin chaperones including Hsp90-α1 and Unc45b (8, 9). It is not surprising that muscle cells have a high demand for protein quality control given that they express abundant large size muscle proteins that are assembled into a highly organized protein complex, the sarcomeres (9). It is well known that molecular chaperones, such as Hsp90-α1 and Unc45b, are specifically expressed in muscle cells and play vital roles in myosin folding and sarcomere assembly in zebrafish (23, 24, 43). Genetic deficiency in any of these genes resulted in similar skeletal muscle defects in zebrafish embryos (21, 22).
Protein methylation plays critical roles in protein folding and stability (44–46). It has been demonstrated that sarcomere proteins, such as myosin, actin, and creatine kinase, are methylated at the lysine residues (47, 48). We have shown that knockdown of Smyd1b resulted in reduced protein methylation including myosin in zebrafish embryos (9). The significant reduction of myosin methylation might contribute to increased myosin protein misfolding and up-regulation of heat shock gene expression in smyd1a; myd1b mutant zebrafish embryos. However, it remains to be determined whether Smyd1a and Smyd1b are involved directly or indirectly in myosin protein methylation and stability.
ACKNOWLEDGMENTS
The authors thank Ming-Yuan Liu (University of Maryland, Baltimore) for fish care, Siping Li (University of Maryland, Baltimore) for statistical analysis of the qRT-PCR data, and Nick Du (University of Washington, Seattle, WA, USA) for proofreading the manuscript. This research was supported by funding from the U.S. National Institute of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR072703-01A1 to S.D.), and National Natural Science Foundation of China (Grants 31701039 to M.C. and 31820103016 to J.Z.). The authors also acknowledge the contribution of Electron Microscopy Core Imaging Facility of the University of Maryland. This work utilized electron microscopy sample preparation instruments purchased with funding from the NIH Shared Instrument Grant (1S10 RR26870). The authors declare no conflicts of interest.
Glossary
- Cas9
CRISPR-associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeat
- hpf
hour postfertilization
- MO
morpholino
- MyHC
myosin heavy chain
- MyLC
myosin light chain
- MyoD
myogenic differentiation
- qRT-PCR
quantitative RT-PCR
- RFP
red florescent protein
- sgRNA
single-guide RNA
- TEM
transmission electron microscopy
- WT
wild type
AUTHOR CONTRIBUTIONS
M. Cai, L. Han, and S. Du designed research; M. Cai, L. Han, L. Liu, F. He, and S. Du performed research; W. Chu, J. Zhang, Z. Tian, and S. Du analyzed data; and M. Cai and S. Du wrote the manuscript.
REFERENCES
- 1.Ehler E., Gautel M. (2008) The sarcomere and sarcomerogenesis. Adv. Exp. Med. Biol. 642, 1–14 [DOI] [PubMed] [Google Scholar]
- 2.Sanger J. W., Kang S., Siebrands C. C., Freeman N., Du A., Wang J., Stout A. L., Sanger J. M. (2005) How to build a myofibril. J. Muscle Res. Cell Motil. 26, 343–354 [DOI] [PubMed] [Google Scholar]
- 3.Laing N. G. (2008) The Sarcomere and Skeletal Muscle Disease. Advances in Experimental Medicine and Biology, Vol 642, Springer, New York: [Google Scholar]
- 4.Hwang P. M., Sykes B. D. (2015) Targeting the sarcomere to correct muscle function. Nat. Rev. Drug Discov. 14, 313–328 [DOI] [PubMed] [Google Scholar]
- 5.De Winter J. M., Ottenheijm C. A. C. (2017) Sarcomere dysfunction in nemaline myopathy. J. Neuromuscul. Dis. 4, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb P. D., Pierce S. A., Sims R. J., Yamagishi H., Weihe E. K., Harriss J. V., Maika S. D., Kuziel W. A., King H. L., Olson E. N., Nakagawa O., Srivastava D. (2002) Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 31, 25–32 [DOI] [PubMed] [Google Scholar]
- 7.Tan X., Rotllant J., Li H., De Deyne P., Du S. J. (2006) SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc. Natl. Acad. Sci. USA 103, 2713–2718; erratum: 7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Just S., Meder B., Berger I. M., Etard C., Trano N., Patzel E., Hassel D., Marquart S., Dahme T., Vogel B., Fishman M. C., Katus H. A., Strähle U., Rottbauer W. (2011) The myosin-interacting protein SMYD1 is essential for sarcomere organization. J. Cell Sci. 124, 3127–3136 [DOI] [PubMed] [Google Scholar]
- 9.Li H., Zhong Y., Wang Z., Gao J., Xu J., Chu W., Zhang J., Fang S., Du S. J. (2013) Smyd1b is required for skeletal and cardiac muscle function in zebrafish. Mol. Biol. Cell 24, 3511–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du S. J., Tan X., Zhang J. (2014) SMYD proteins: key regulators in skeletal and cardiac muscle development and function. Anat. Rec. (Hoboken) 297, 1650–1662 [DOI] [PubMed] [Google Scholar]
- 11.Gao J., Li J., Li B. J., Yagil E., Zhang J., Du S. J. (2014) Expression and functional characterization of Smyd1a in myofibril organization of skeletal muscles. PLoS One 9, e86808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagandla H., Lopez S., Yu W., Rasmussen T. L., Tucker H. O., Schwartz R. J., Stewart M. D. (2016) Defective myogenesis in the absence of the muscle-specific lysine methyltransferase SMYD1. Dev. Biol. 410, 86–97 [DOI] [PubMed] [Google Scholar]
- 13.Prill K., Windsor Reid P., Wohlgemuth S. L., Pilgrim D. B. (2015) Still heart encodes a structural HMT, SMYD1b, with chaperone-like function during fast muscle sarcomere assembly. PLoS One 10, e0142528; erratum: 11, e0148027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart M. D., Lopez S., Nagandla H., Soibam B., Benham A., Nguyen J., Valenzuela N., Wu H. J., Burns A. R., Rasmussen T. L., Tucker H. O., Schwartz R. J. (2016) Mouse myofibers lacking the SMYD1 methyltransferase are susceptible to atrophy, internalization of nuclei and myofibrillar disarray. Dis. Model. Mech. 9, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkholz J., Eberle R., Boller K., Munz B. (2018) siRNA-mediated inhibition of skNAC and Smyd1 expression disrupts myofibril organization: immunofluorescence and electron microscopy study in C2C12 cells. Micron 108, 6–10 [DOI] [PubMed] [Google Scholar]
- 16.Phan D., Rasmussen T. L., Nakagawa O., McAnally J., Gottlieb P. D., Tucker P. W., Richardson J. A., Bassel-Duby R., Olson E. N. (2005) BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development 132, 2669–2678 [DOI] [PubMed] [Google Scholar]
- 17.Du S. J., Rotllant J., Tan X. (2006) Muscle-specific expression of the smyd1 gene is controlled by its 5.3-kb promoter and 5′-flanking sequence in zebrafish embryos. Dev. Dyn. 235, 3306–3315 [DOI] [PubMed] [Google Scholar]
- 18.Li D., Niu Z., Yu W., Qian Y., Wang Q., Li Q., Yi Z., Luo J., Wu X., Wang Y., Schwartz R. J., Liu M. (2009) SMYD1, the myogenic activator, is a direct target of serum response factor and myogenin. Nucleic Acids Res. 37, 7059–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren J. S., Tracy C. M., Miller M. R., Makaju A., Szulik M. W., Oka S. I., Yuzyuk T. N., Cox J. E., Kumar A., Lozier B. K., Wang L., Llana J. G., Sabry A. D., Cawley K. M., Barton D. W., Han Y. H., Boudina S., Fiehn O., Tucker H. O., Zaitsev A. V., Franklin S. (2018) Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart. Proc. Natl. Acad. Sci. USA 115, E7871–E7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims R. J., III, Weihe E. K., Zhu L., O’Malley S., Harriss J. V., Gottlieb P. D. (2002) m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J. Biol. Chem. 277, 26524–26529 [DOI] [PubMed] [Google Scholar]
- 21.Wohlgemuth S. L., Crawford B. D., Pilgrim D. B. (2007) The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev. Biol. 303, 483–492 [DOI] [PubMed] [Google Scholar]
- 22.Etard C., Behra M., Fischer N., Hutcheson D., Geisler R., Strähle U. (2007) The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev. Biol. 308, 133–143 [DOI] [PubMed] [Google Scholar]
- 23.Du S. J., Li H., Bian Y., Zhong Y. (2008) Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc. Natl. Acad. Sci. USA 105, 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins T. A., Haramis A. P., Etard C., Prodromou C., Vaughan C. K., Ashworth R., Ray S., Behra M., Holder N., Talbot W. S., Pearl L. H., Strähle U., Wilson S. W. (2008) The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development 135, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X. J., Xu P. F., Zhou T., Hu M., Fu C. T., Zhang Y., Jin Y., Chen Y., Chen S. J., Huang Q. H., Liu T. X., Chen Z. (2008) Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One 3, e1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paone C., Rudeck S., Etard C., Strähle U., Rottbauer W., Just S. (2018) Loss of zebrafish Smyd1a interferes with myofibrillar integrity without triggering the misfolded myosin response. Biochem. Biophys. Res. Commun. 496, 339–345 [DOI] [PubMed] [Google Scholar]
- 27. Busch-Nentwich, E., Kettleborough, R., Dooley, C. M., Scahill, C., Sealy, I., White, R., Herd, C., Mehroke, S., Wali, N., Carruthers, S., Hall, A., Collins, J., Gibbons, R., Pusztai, Z., Clark, R., Stemple, D. L. (2013) Sanger Institute zebrafish mutation project mutant data submission. ZFIN Direct Data Submission. Accessed January 13, 2016, at: http://zfin.org.
- 28.Clark K. J., Balciunas D., Pogoda H. M., Ding Y., Westcot S. E., Bedell V. M., Greenwood T. M., Urban M. D., Skuster K. J., Petzold A. M., Ni J., Nielsen A. L., Patowary A., Scaria V., Sivasubbu S., Xu X., Hammerschmidt M., Ekker S. C. (2011) In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat. Methods 8, 506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J. R., Joung J. K. (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai M., Si Y., Zhang J., Tian Z., Du S. (2018) Zebrafish embryonic slow muscle is a rapid system for genetic analysis of sarcomere organization by CRISPR/Cas9, but not NgAgo. Mar. Biotechnol. (NY) 20, 168–181 [DOI] [PubMed] [Google Scholar]
- 31.Jao L. E., Wente S. R., Chen W. (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thisse C., Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 33.Xu J., Gao J., Li J., Xue L., Clark K. J., Ekker S. C., Du S. J. (2012) Functional analysis of slow myosin heavy chain 1 and myomesin-3 in sarcomere organization in zebrafish embryonic slow muscles. J. Genet. Genomics 39, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanger J. W., Wang J., Holloway B., Du A., Sanger J. M. (2009) Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motil. Cytoskeleton 66, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kok F. O., Shin M., Ni C. W., Gupta A., Grosse A. S., van Impel A., Kirchmaier B. C., Peterson-Maduro J., Kourkoulis G., Male I., DeSantis D. F., Sheppard-Tindell S., Ebarasi L., Betsholtz C., Schulte-Merker S., Wolfe S. A., Lawson N. D. (2015) Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., Stainier D. Y. (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 [DOI] [PubMed] [Google Scholar]
- 37.Anderson J. L., Mulligan T. S., Shen M. C., Wang H., Scahill C. M., Tan F. J., Du S. J., Busch-Nentwich E. M., Farber S. A. (2017) mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 13, e1007105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lalonde S., Stone O. A., Lessard S., Lavertu A., Desjardins J., Beaudoin M., Rivas M., Stainier D. Y. R., Lettre G. (2017) Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS One 12, e0178700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coda D. M., Lingua M. F., Morena D., Foglizzo V., Bersani F., Ala U., Ponzetto C., Taulli R. (2015) SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle 14, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkholz J., Orgeur M., Stricker S., Munz B. (2015) skNAC and Smyd1 in transcriptional control. Exp. Cell Res. 336, 182–191 [DOI] [PubMed] [Google Scholar]
- 41.Park C. Y., Pierce S. A., von Drehle M., Ivey K. N., Morgan J. A., Blau H. M., Srivastava D. (2010) skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc. Natl. Acad. Sci. USA 107, 20750–20755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franklin S., Kimball T., Rasmussen T. L., Rosa-Garrido M., Chen H., Tran T., Miller M. R., Gray R., Jiang S., Ren S., Wang Y., Tucker H. O., Vondriska T. M. (2016) The chromatin-binding protein Smyd1 restricts adult mammalian heart growth. Am. J. Physiol. Heart Circ. Physiol. 311, H1234–H1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernick E. P., Zhang P. J., Du S. (2010) Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol. 11, 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X. D., Lamb A., Chen L. F. (2009) Methylation, a new epigenetic mark for protein stability. Epigenetics 4, 429–433 [DOI] [PubMed] [Google Scholar]
- 45.Egorova K. S., Olenkina O. M., Olenina L. V. (2010) Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc.) 75, 535–548 [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Wen H., Shi X. (2012) Lysine methylation: beyond histones. Acta Biochim. Biophys. Sin. (Shanghai) 44, 14–27 [DOI] [PubMed] [Google Scholar]
- 47.Tong S. W., Elzinga M. (1983) The sequence of the NH2-terminal 204-residue fragment of the heavy chain of rabbit skeletal muscle myosin. J. Biol. Chem. 258, 13100–13110 [PubMed] [Google Scholar]
- 48.Iwabata H., Yoshida M., Komatsu Y. (2005) Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics 5, 4653–4664 [DOI] [PubMed] [Google Scholar]