Abstract
The first version of the Standard PREanalytical Code (SPREC) was developed in 2009 by the International Society for Biological and Environmental Repositories (ISBER) Biospecimen Science Working Group to facilitate documentation and communication of the most important preanalytical quality parameters of different types of biospecimens used for research. This same Working Group has now updated the SPREC to version 2.0, presented here, so that it contains more options to allow for recent technological developments. Existing elements have been fine tuned. An interface to the Biospecimen Reporting for Improved Study Quality (BRISQ) has been defined, and informatics solutions for SPREC implementation have been developed. A glossary with SPREC-related definitions has also been added.
Introduction
The SPREC (Standard PREanalytical Code) was developed in 2009 to provide a comprehensive and easy-to-implement tool to document the in vitro preanalytical (collection, processing and storage) details of biospecimens.1 The objective of the SPREC is to facilitate annotation of biospecimens with preanalytical factors that fulfill two criteria: (a) their variation is known or highly suspected to impact the results of downstream analyses, and (b) they are within the control of the biobank and thus can be anticipated and standardized in standard operating procedures (SOPs). The original SPREC is applicable to animal biospecimens, and more specifically to mammals. SPREC development in botanical collections and storage has also been explored.2
Since then and as originally anticipated, new technologies have been developed for specimen collection (e.g., new anticoagulants), processing (e.g., new tissue stabilization methods), and storage (e.g., new dry, room temperature storage media) necessitating an update of the original codes. Furthermore, with the implementation of SPREC version 1.0 in different biobanks, feedback has been received and suggestions on possible improvements considered. We are aware of at least thirteen biobanks or biobank networks in the United States, Europe, Korea, and Australia,3 and of at least three commercial biobank LIMS who have already implemented the SPREC (personal communications). The SPREC has been incorporated in the “Minimum data set for sharing biobank samples, information, and data” (MIABIS), developed by the Biobanking and Biomolecular Resources Research Infrastructure Sweden (BBMRI.se)4 and is also being implemented as a national standard both for healthcare and research in BBMRI.se (personal communication). An application programming interface (API) module has been developed that allows SPREC to be defined from sample collection and processing protocols, submitted to the Molecular Methods database (www.molmeth.org) that is supported by BBMRI.5 Furthermore, SPREC is mentioned in the College of American Pathologists (CAP) biobank accreditation checklist, which is being tested in pilot audits (personal communication). Currently, the application of SPREC to stem cell biobanks is being evaluated by Demiroglu and colleagues at the University Medical Center in Göttingen, Germany (manuscript in preparation). Thus, this new version 2.0 has been developed, and is expected to be more comprehensive and easier to implement.
SPREC Version 2.0
In updating SPREC 1.0, and for the sake of continuity, no significant changes to codes were made, while new options have been added as detailed in Tables 1 and 2. The new options in SPREC 2.0 include the following:
Table 1.
Preanalytical Variables Included in SPREC (7-element long SPREC), Version SPREC 2.0, Applied to Fluid Samples
| Type of sample | |
|---|---|
| Ascites fluid | ASC |
| Amniotic fluid | AMN |
| Bronchoalveolar lavage | BAL |
| Blood (whole) | BLD |
| Bone marrow aspirate | BMA |
| Breast milk | BMK |
| Buccal cells | BUC |
| Unficolled buffy coat, viable | BUF |
| Unficolled buffy coat, nonviable | BFF |
| Ficoll mononuclear cells, viable | CEL |
| Fresh cells from nonblood specimen type | CEN |
| Cells from nonblood specimen type(e.g., ascites, amniotic), viable | CLN |
| Cord blood | CRD |
| Cerebrospinal fluid | CSF |
| Dried whole blood (e.g., Guthrie cards) | DWB |
| Nasal washing | NAS |
| Ficoll mononuclear cells, nonviable | PEL |
| Cells from non blood specimen type (e.g., ascites, amniotic), nonviable | PEN |
| Pleural fluid | PFL |
| Plasma, single spun | PL1 |
| Plasma, double spun | PL2 |
| Red blood cells | RBC |
| Saliva | SAL |
| Semen | SEM |
| Serum | SER |
| Sputum | SPT |
| Stool | STL |
| Synovial fluid | SYN |
| Tears | TER |
| 24 h urine | U24 |
| Urine, random (“spot”) | URN |
| Urine, first morning | URM |
| Urine, timed | URT |
| Other | ZZZ |
| Type of primary container | |
|---|---|
| Acid citrate dextrose | ACD |
| Additives | ADD |
| Serum tube without clot activator | CAT |
| Citrate phosphate dextrose | CPD |
| Cell Preparation Tube® | CPT |
| EDTA and gel | EDG |
| Lithium heparin | HEP |
| Hirudin | HIR |
| Lithium heparin and gel | LHG |
| Oragene collection container or equivalent | ORG |
| PAXgene® blood RNA+ | PAX |
| Potassium EDTA | PED |
| Polyethylene tube sterile | PET |
| S8820 protease inhibitor tablets or equivalent | PI1 |
| Protease inhibitors | PIX |
| Polypropylene tube sterile | PPS |
| PAXgene® blood DNA | PXD |
| PAXgene® bone marrow RNA | PXR |
| Sodium citrate | SCI |
| Sodium EDTA | SED |
| Sodium heparin | SHP |
| Sodium fluoride/potassium oxalate | SPO |
| Serum separator tube with clot activator | SST |
| Tempus® tube | TEM |
| Trace elements tube | TRC |
| Unknown | XXX |
| Other | ZZZ |
| Pre-centrifugation (delay between collection and processing) | ||
|---|---|---|
| RT* | <2 h | A |
| 2°C–10°C | <2 h | B |
| RT | 2–4 h | C |
| 2°C–10°C | 2–4 h | D |
| RT | 4–8 h | E |
| 2°C–10°C | 4–8 h | F |
| RT | 8–12 h | G |
| 2°C–10°C | 8–12 h | H |
| RT | 12–24 h | I |
| 2°C–10°C | 12–24 h | J |
| RT | 24–48 h | K |
| 2°C–10°C | 24–48 h | L |
| RT | >48 h | M |
| 2°C–10°C | >48 h | N |
| 35°C–38°C | <2 h | O |
| Unknown | X | |
| Other | Z | |
| Centrifugation | ||
|---|---|---|
| RT 10–15 min | <3000 g no braking | A |
| RT 10–15 min | <3000 g with braking | B |
| 2°C–10 °C 10–15 min | <3000 g no braking | C |
| 2°C–10 °C 10–15 min | <3000 g with braking | D |
| RT 10–15 min | 3000–6000 g with braking | E |
| 2°C–10°C 10–15 min | 3000 g to 6000 g with braking | F |
| RT 10–15 min | 6000 g to 10000 g with braking | G |
| 2°C–10 °C 10–15 min | 6000 g to 10000 g with braking | H |
| RT 10–15 min | >10000 g with braking | I |
| 2°C–10 °C 10–15 min | >10000 g with braking | J |
| RT 30 min | <1000 g no braking | M |
| No centrifugation | N | |
| Unknown | X | |
| Other | Z |
| Second centrifugation | ||
|---|---|---|
| RT 10–15 min | <3000 g no braking | A |
| RT 10–15 min | <3000 g with braking | B |
| 2°C–10°C 10–15 min | <3000 g no braking | C |
| 2°C–10°C 10–15 min | <3000 g with braking | D |
| RT 10–15 min | 3000–6000 g with braking | E |
| 2°C–10°C 10–15 min | 3000–6000 g with braking | F |
| RT 10–15 min | 6000–10,000 g with braking | G |
| 2°C–10°C 10–15 min | 6000–10,000 g with braking | H |
| RT 10–15 min | >10,000 g with braking | I |
| 2°C–10°C 10–15 min | >10,000 g with braking | J |
| No centrifugation | N | |
| Unknown | X | |
| Other | Z |
| Post-centrifugation delay | |
|---|---|
| <1 h 2°C–10°C | A |
| <1 h RT | B |
| 1 to 2 h 2°C–10°C | C |
| 1 to 2 h RT | D |
| 2 to 8 h 2°C–10°C | E |
| 2 to 8 h RT | F |
| 8 to 24 h 2°C–10°C | G |
| 8 to 24 h RT | H |
| >24 h 2°C–10°C | I |
| >24 h RT | J |
| Not applicable | N |
| Unknown | X |
| Other | Z |
| Long-term storage | ||
|---|---|---|
| PP tube 0.5- to 2 mL** | (−85)°C–(−60)°C | A |
| PP tube 0.5- to 2 mL | (−35)°C–(−18)°C | B |
| PP tube 0.5- to 2 mL | <-135°C | V |
| Cryotube 1- to 2 mL | LN*** | C |
| Cryotube 1- to 2 mL | (−85)°C–(−60) °C | D |
| Cryotube 1- to 2-mL | Programmable freezing to <-135°C | E |
| Plastic cryo straw | LN*** | F |
| Straw | (−85)°C–(−60)°C | G |
| Straw | (−35)°C–(−18)°C | H |
| Straw | Programmable freezing to <-135°C | I |
| PP tube ≥5 mL | (−85)°C–(−60)°C | J |
| PP tube ≥5 mL | (−35)°C–(−18)°C | K |
| Microplate | (−85)°C–(−60)°C | L |
| Microplate | (−35)°C–(−18)°C | M |
| Cryotube 1- to 2 mL | LN*** after temporary (−85)°C–(−60)°C | N |
| Plastic cryo straw | LN*** after temporary (−85)°C–(−60)°C | O |
| Paraffin block | RT or 2°C–10°C | P |
| Bag | LN*** | Q |
| Dry technology medium | RT | R |
| PP tube 40- to 500 μL | (−85)°C–(−60)°C | S |
| PP tube 40- to 500 μL | (−35)°C–(−18)°C | T |
| PP tube 40- to 500 μL | <-135°C | W |
| Original primary container | (−35)°C–(−18)°C or (−85)°C–(−60)°C | Y |
| Unknown | X | |
| Other | Z |
New elements are screened. Codes in bold come from the Laboratory Data Management System (LDMS).
RT, room temperature: 18°C–28°C; **PP, polypropylene; ***LN, liquid nitrogen, referring to either vapor- or liquid-phase (this information being documented in the biobank's SOPs)
Volumes refer to container size.
Table 2.
Preanalytical Variables Included in the SPREC (7-element long SPREC), Version SPREC 2.0, Applied to Solid Samples
| Type of sample | |
|---|---|
| Fresh cells from non blood specimen type (e.g., biopsy) | CEN |
| Cells from non blood specimen type (e.g., dissociated tissue), viable | CLN |
| Cells from fine needle aspirate | FNA |
| Hair | HAR |
| Cells from laser capture microdissected tissue | LCM |
| Cells from non blood specimen type (e.g., dissociated tissue), nonviable | PEN |
| Placenta | PLC |
| Solid tissue | TIS |
| Disrupted tissue, non-viable | TCM |
| Other | ZZZ |
| Type of collection | |
|---|---|
| Autopsy < 6 h postmortem | A06 |
| Autopsy 6–12 h postmortem | A12 |
| Autopsy 12–24 h postmortem | A24 |
| Autopsy 24–48 h postmortem | A48 |
| Autopsy 48–72 h postmortem | A72 |
| Biopsy in culture media | BCM |
| Biopsy | BPS |
| Biopsy in normal saline or phosphate buffered saline | BSL |
| Biopsy in tissue low temperature transport media | BTM |
| Fine needle aspirate | FNA |
| Punction | PUN |
| Surgical excision in culture media | SCM |
| Surgical excision | SRG |
| Surgical excision in normal saline or phosphate buffered saline | SSL |
| Surgical excision in tissue low temperature transport media | STM |
| Surgical excision in vacuum container | VAC |
| Swab | SWB |
| Other | ZZZ |
| Warm ischemia time | |
|---|---|
| < 2 min | A |
| 2–10 min | B |
| 10–20 min | C |
| 20–30 min | D |
| 30–60 min | E |
| >60 min | F |
| Unknown | X |
| Not applicable (e.g., biopsy) | N |
| Other | Z |
| Cold ischemia time | |
|---|---|
| < 2 min | A |
| 2–10 min | B |
| 10–20 min | C |
| 20–30 min | D |
| 30–60 min | E |
| >60 min | F |
| Unknown | X |
| Not applicable (e.g., autopsy) | N |
| Other | Z |
| Fixation/stabilization type | |
| Non-aldehyde with acetic acid | ACA |
| Aldehyde-based | ALD |
| Allprotect® tissue reagent | ALL |
| Alcohol-based | ETH |
| Nonbuffered formalin | FOR |
| Heat stabilization | HST |
| Snap freezing | SNP |
| Non-aldehyde based without acetic acid | NAA |
| Neutral buffered formalin | NBF |
| Optimum cutting temperature medium | OCT |
| PAXgene® tissue | PXT |
| RNA Later® | RNL |
| Unknown | XXX |
| Other | ZZZ |
| Fixation time | |
|---|---|
| <15 min | A |
| 15 min–1 h | B |
| 1–4 h | C |
| 4–8 h | D |
| 8–24 h | E |
| 24–48 h | F |
| 48–72 h | G |
| Not applicable | N |
| Unknown | X |
| Other | Z |
| Long-term storage | ||
|---|---|---|
| PP tube 0.5–2 mL** | (−85)°C–(−60)°C | A |
| PP tube 0.5–2 mL | (−35)°C–(−18)°C | B |
| PP tube 0.5–2 mL | <-135°C | V |
| Cryotube 1–2 mL | Liquid nitrogen*** | C |
| Cryotube 1–2 mL | (−85)°C–(−60)°C | D |
| Cryotube 1–2 mL | Programmable freezing to <-135°C | E |
| Plastic cryostraw | Liquid nitrogen | F |
| Straw | (−85)°C–(−60)°C | G |
| Straw | (−35)°C–(−18)°C | H |
| Straw | Programmable freezing to <-135°C | I |
| PP tube ≥5 mL | (−85)°C–(−60)°C | J |
| PP tube ≥5 mL | (−35)°C–(−18)°C | K |
| Microplate | (−85)°C–(−60)°C | L |
| Microplate | (−35)°C–(−18)°C | M |
| Cryotube 1–2 mL | LN*** after temporary (−85)°C–(−60)°C | N |
| Straw | LN*** after temporary (−85)°C–(−60)°C | O |
| Paraffin block | RT or 2°C–10°C | P |
| Bag | LN*** | Q |
| Dry technology medium | RT | R |
| PP tube 40–500 μL | (−85)°C–(−60)°C | S |
| PP tube 40–500 μL | (−35)°C–(−18)°C | T |
| PP tube 40–500 μL | <-135 ° C | W |
| Original primary container | (−35)-(−18)°C or (−85)-(−60) °C | Y |
| Unknown | X | |
| Other | Z |
New elements are screened. Codes in bold come from the Laboratory Data Management System (LDMS).
RT, room temperature: 18°C–28°C; **PP, polypropylene; ***Liquid nitrogen refers to either vapor or liquid phase (this information being documented in the biobank's SOPs)
Volumes refer to container size.
Fluid samples
Sample types
Dried whole blood (e.g., Guthrie cards) and red blood cell fraction were added. A distinction was made between random, timed, and first morning urine specimen.
Types of primary containers
Sodium-heparin collection tube, EDTA, and heparin collection tubes with a gel separation plug and cell preparation tubes (CPT®) were added. A distinction was made between serum separation tubes with silica clot activator and tubes without additive. No distinction is now made between evacuated (Vacutainer®-type) and nonevacuated (Monovette®-type) collection tubes. Polyethylene containers (often used for urine collection) and additives (often used in urine collection) were added.
Centrifugation conditions
Initial centrifugation conditions of 30 min at room temperature (RT) with a relative G-force <1000 g and without braking used (most appropriate for density centrifugation and isolation of mononuclear cells) were added. For all the centrifugation options, a 10-min centrifugation time was replaced by 10–15 min to provide more flexibility. A “not applicable” option in the post-centrifugation element was added.
Storage conditions
Long-term storage options were included for liquid nitrogen preceded by temporary −80°C storage (in either cryotubes or straws). The temperature options for paraffin blocs were revised from “RT” only to “RT or 2°C–10°C”. Other new storage options include a dry technology medium at RT, bag storage, original primary container, and tubes from 40–500 μL sizes. Storage at refrigerated temperatures has been harmonized throughout the SPREC and corresponds to 2°C–10°C.
Solid samples
Sample types
Placenta was added as a new sample type. The abbreviation LCM appeared twice in the sample type element in version 1.0. In version 2.0, LCM corresponds to cells from laser capture microdissected tissue, while TCM corresponds to cells from mechanically disrupted tissue.
For biopsies and surgical excisions, options for collection (and subsequent transport) in either saline, culture media, or low-temperature transport media (e.g., AQIX, Hyperthermosol, Unisol, Thermo-ROS) or in vacuum containers were added.
“Fixation” type became “fixation/stabilization” type and now includes heat stabilization, PAXgene® tissue fixation, and Allprotect® tissue fixation/stabilization options.
A “not applicable” option was added in the Fixation time element.
Support Tools for Implementation of SPREC Version 2.0
The integration of SPREC into biobank databases generally requires the inclusion of SPREC drop-lists among the “tables” in the software. This is relatively easy with customized software, but the supplier would need to make a customized development for off-the-shelf software. Two tools have been developed in order to facilitate SPREC implementation and interfacing with either customized or off-the-shelf biobank software.
A default SPREC can be set per sample type according to the SOPs of the collection project. Thus, only deviations from the SOP SPREC need to be documented. These can be highlighted, making any deviation from pre-defined sample quality or any nonconforming in processing directly visible. However, active generation of the SPREC for each individual sample is to be preferred as it enforces traceability.
Although the use of the SPREC requires an additional initial investment of time, it is expected that this will be more than repaid by the ease and accuracy of subsequent sample search and selection.
SPRECware
The first tool (SPRECware) requires entry of each samples' SPREC elements. SPRECware6 is an IT architecture, that is, a collection of software and communication tools that have been developed in order to foster the adoption of SPREC by biobanks and biolaboratories. Basic information concerning preanalytical processing of a given sample is selected from drop-down menus. The resulting SPREC is generated together with the corresponding barcode, and all the preanalytical data with the SPREC are stored in a local database.
One component, SPRECbase, has been recently released and is freely downloadable from http://www.sprecware.org. This software supports both coding/printing barcode labels and decoding in text of a sample's SPREC, then storing all the preanalytical data in a local database. These barcodes simplify storing and exchanging corresponding information about the samples.
In a working environment, SPRECware is intended to be connected to the laboratory/biobank information system (LIS/BIS). The required data can then be loaded from the originating biobank's LIS/BIS database and translated into the SPREC. Samples are thus labeled with both the human-readable and barcoded format. The barcode provides the receiver organization with the data documenting the specimen and can be stored in the LIS/BIS. This approach provides fully automated data transfer without manual writing or reading when the organizations exchanging specimens both use SPREC. In the case that the receiving organization has not adopted the SPREC, it is still possible to read the SPREC (e.g., by a barcode scanner) and download the explicit preanalytical data by means of an internet connection.
Since the SPREC does not encapsulate any personal data, it can be used for searches on sample availability. This possibility of a preliminary check would allow a researcher to select material with the most appropriate preanalytical characteristics for a planned experiment.6 (Even in the case of derivatives (e.g., DNA, RNA), the SPREC information on the collected samples, from which derivatives were produced, is highly valuable.
SPRECalc
Unlike the SPRECware which requires entry of the SPREC options (A, B, X…), the second tool (SPRECalc) requires entry of the corresponding preanalytical variables themselves (temperature, time…); this tool then performs automatic calculation of the time-associated elements and generation of SPREC.
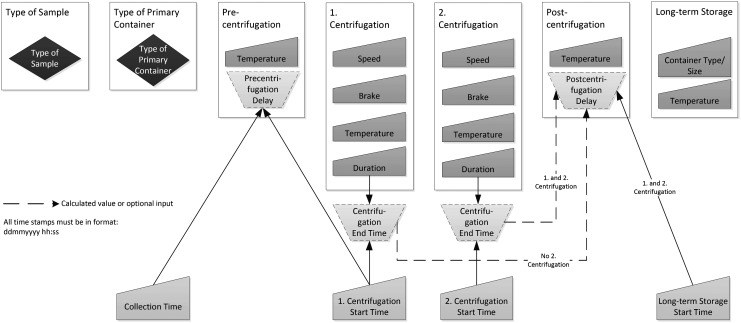
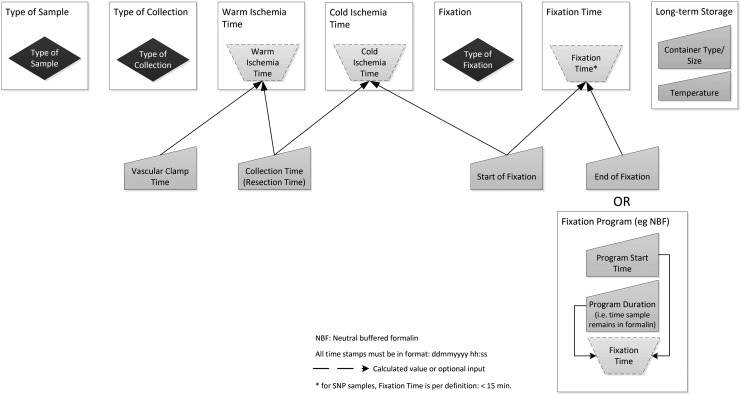
The Excel-based SPRECalc automatically calculates SPREC time-associated elements for solid and fluid samples, using the data logic depicted in Figures 1 and 2. Data entry is supported by pull-down lists (blue-colored fields in the Excel tool) to minimize inconsistent data entry. The dropdowns are based upon SPREC version 2.0 as specified in this article and are provided in the “Lists” worksheet. Time stamps have to be entered in the format dd/mm/yyyy hh:mm:ss (pink-colored fields in the Excel tool) to allow correct calculation of time spans. The format of the related columns is pre-defined.
FIG. 1.
The elements and their relationships needed for automated SPREC calculation for fluid samples.
FIG. 2.
The elements and their relationships needed for automated SPREC calculation for solid samples.
To ensure that automatic SPREC calculation and generation can be performed, the data items in Table 3 must be either captured manually or retrieved automatically (i.e., via barcode scanning or RFID tagging).
Table 3.
Date Items for Automatic SPREC Calculation
| Fluid Samples | Solid Samples |
|---|---|
| Type of sample | Type of sample |
| Type of primary sample container | Type of collection |
| Collection time | Vascular clamping time at surgery (denotes beginning of warm ischemia) |
| Time of first centrifugation start | Time of collection/Time of excision (denotes end of warm ischemia time and beginning of cold ischemia time) |
| Sample temperature between collection and first centrifugation | Fixation/stabilization type |
| First centrifugation speed | Fixation start time (denotes end of cold ischemia time for fixed tissue) |
| First centrifugation brake | Fixation end time (alternatively for automated processes: program start time, program type with related duration) |
| First centrifugation temperature | Fixation end time (alternatively for automated processes: program start time, program type with related duration) |
| Time of first centrifugation start1 | Temperature of long term storage |
| Time of first centrifugation end (or duration of first centrifugation) | Type of container of long term storage |
| Second centrifugation speed | |
| Second centrifugation brake | |
| Second centrifugation temperature | |
| Time of second centrifugation start | |
| Time of second centrifugation end2 (or duration of second centrifugation) | |
| Time of putting into temporary storage | |
| Temperature of temporary storage | |
| Time of putting into long term storage | |
| Sample Temperature between end of last centrifugation and putting into long term storage | |
| Temperature of long term storage | |
| Type of container of long term storage |
Time of freezing in the case of PAXgene®.
Time of last washing centrifugation in the case of viable cells.
Gray fields contain automatic calculations/translations, displaying either the time period (e.g., warm/cold ischemia time, fixation time), or “help” columns for identification of the applicable SPREC.
For solid samples, the fixation time can be determined in two different ways. Option 1 is designed for use with automated fixation systems (fixation start time, program type, and program start time need to be recorded). Option 2 is targeted for nonautomated fixation steps (fixation start and end time need to be entered).
With respect to option 1, the “Fixation Programs” worksheet allows the user to predefine fixation programs and their respective duration. Selection of the applicable program during data entry automatically uses the related duration to calculate the correct fixation time, provided fixation start time and program start time have been entered as well. For each sample, SPREC is then automatically displayed following the coding rules specified in this article. All of these fields are protected against changes.
The tool also contains the following quality-related features. The data entry worksheet provides its own “quality control” features, since Excel-specific error messages in the calculated columns (e.g., ###### or #REF) indicate data inconsistency (e.g., the fixation start time entered is before the collection time). Systematic investigation of such errors supports quality checks to improve data quality and consistency.
Default SPRECs can be defined for each collection according to established SOPs for sample collection. Nonconformities can be detected automatically by the system and flagged in dedicated columns, with a flexible way of setting target parameters (e.g., fixation time shall be <= 24 h). Alternatively the same nonconformities can be specifically custom-programmed directly in the final SPREC columns, using the “conditional formatting” functionalities of Excel. These nonconformities should then be investigated as required by the organization's Quality Management System (QMS) as part of biobank accreditation.
In addition to columns with a “Z” option (“Other”), text columns are provided, allowing recording of more detailed information in cases where other specifications than those listed in the SPREC were applied. If a value is unknown, then “unknown” should be entered.
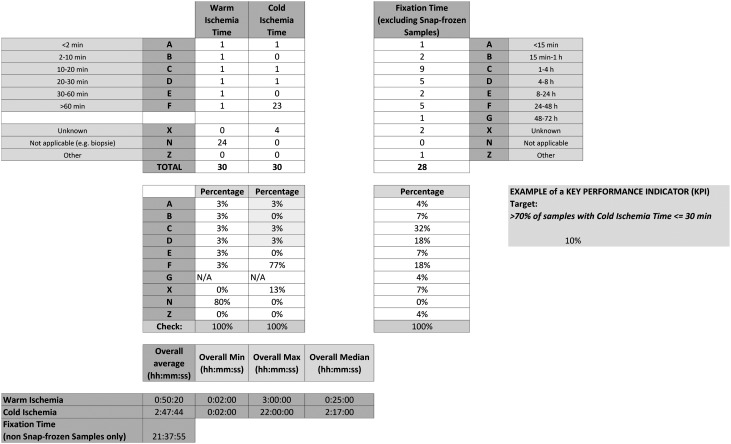
The “Statistics” worksheet displays summary statistics about the sample collection (e.g., number and percentage of samples per code type, average, minimum, maximum and median for selected time spans) related to selected SPREC items (e.g., warm/cold ischemia time and fixation time for solid samples). This summary sheet also allows the continuous automated calculation of key performance indicators (KPI), as required by quality norms. An example is shown for the percentage of solid samples in the collection with warm ischemia times below 30 min (Fig. 3). For example, a report can be generated stating the percentage of solid samples in the collection with warm ischemia times below 30 minutes. Reports of variables of interest can be programmed as required by the user. The Excel tool is available on http://www.isber.org/SPRECtools.cfm.
FIG. 3.
Screenshot of SPRECalc statistics function.
Although SPRECalc is a stand-alone tool, the final SPREC can be imported into a biobank database at regular intervals. This solution provides a simple way of integrating the SPREC into custom software, since no data logic or drop-down lists need to be created. It is also possible to combine the SPREC tools described here, the SPRECalc and SPRECware, for generating and encoding SPRECs, respectively.
SPREC Interface with Reporting Recommendations
A recent publication7 proposed reporting recommendations : Biospecimen Reporting for Improved Study Quality (BRISQ). These recommendations include the type of information that should be reported in scientific publications and regulatory submissions based on work with biospecimens. Whereas these recommendations present the different data items that should be reported, they do not provide a standardized way of reporting them. The SPREC provides this standardized format and can be efficiently used to fulfill all of the BRISQ requirements that relate to in vitro variables: covering the workflow between the time of specimen collection and long-term storage. Table 4 presents the interface between the BRISQ and the seven basic SPREC elements (Table 4).
Table 4.
Comparison of the BRISQ and SPREC Data Elements
| BRISQ data element | SPREC data element |
|---|---|
| Biospecimen type | Sample type |
| Anatomical site | NI [not included] |
| Disease status of patients | OS [out of scope] |
| Clinical characteristics of patients | OS |
| Vital state of patients | Type of collection |
| Clinical diagnosis of patients | OS |
| Pathology diagnosis | OS |
| Collection mechanism | Type of primary container |
| Pre-centrifugation delay | |
| Centrifugation | |
| Second centrifugation | |
| Post-centrifugation delay | |
| Type of collection | |
| Warm ischemia time | |
| Cold ischemia time | |
| Type of stabilization | Type of primary container |
| Type of collection | |
| Type of long-term preservation | Fixation/stabilization type |
| Fixation time | |
| Long-term storage | |
| Constitution of preservative | Fixation/stabilization type |
| Storage temperature | Long-term storage |
| Storage duration | NI |
| Shipping temperature | NI |
| Composition assessment and selection | OS |
BRISQ data elements that are out of scope (OS) are in vivo- or pathology-related elements: These are not considered to be SPREC elements. BRISQ data elements that are not included (NI) are those that relate to events during long term storage. The SPREC covers processes until the specimens are placed in long term storage.
Conclusion
SPREC version 2.0 includes several new options, mainly new sample types and additional storage conditions. Development of the SPREC version 2.0 was based on valuable input from many different biobanks worldwide that had previously implemented SPREC version 1.0. Furthermore, we describe readily accessible tools that facilitate implementation of SPREC in biobank databases and fulfill requirements of the BRISQ.7
Glossary
Additives, urine: The most common urine preservatives are boric acid, tartaric acid, or ascorbic acid. They reduce the risk of bacterial overgrowth and specimen decomposition. Preservation at room temperature ranges from 24 to 72 hours.
Buffy coat: The layer of white cells that forms between the layer of red cells and the plasma when unclotted blood is centrifuged or allowed to stand.
Cell preparation tube: Blood collection tube containing cell separation media in the form of gel and/or filter.
Cryotubes: High density polypropylene tubes manufactured to withstand ultra-low temperature conditions.
Dissociated tissue: Tissue that has undergone mechanical (e.g., scalpel) or chemical (e.g., protein digestion) separation, allowing whole cells to be retrieved.
Disrupted tissue: Tissue that has undergone mechanical (e.g., sonication, freeze-thaw) or chemical (e.g., lysis buffer) cellular disintegration.
Long-term storage: Storage of samples at their final destination temperature after possible temporary storage at conditions, imposed by technical (e.g., overnight storage of viable cells at −80°C before transfer into liquid nitrogen) or logistical (e.g., overnight storage of serum at −20°C before transfer to −80°C in central storage facility) reasons.
Pre-centrifugation delay: Time between collection and first centrifugation of the samples. In the case of PAXgene® tubes, this corresponds to the delay between blood collection and freezing of the tube (in this case, equivalent of “pre-processing” delay). In the case of no centrifugation at all, the “pre-centrifugation delay” corresponds to the delay between collection time and start of long-term storage.
Post-centrifugation delay: Time between the last centrifugation and storage of the samples. In the case of viable cells, this corresponds to the delay between the last washing centrifugation and the time of cryopreservation. In the case of PAXgene® tubes, the “post-centrifugation delay” is not applicable. In the case of no centrifugation at all, the “post-centrifugation delay” is not applicable.
Programmable freezing: Freezing in either a programmable rate freezer or alternative solutions such as Mr Frosty.
Puncture: Surgical piercing of an anatomical tissue to obtain material for clinicopathological examination. Includes cisternal puncture, lumbar puncture, spinal puncture, sternal puncture.
Room Temperature: Defined as 18°C–28°C.
Temporary Storage: Initial-term storage in conditions different from the long-term storage conditions.
Contributor Information
Collaborators: [International Society for Biological and Environmental Repositories (ISBER) Working Group on Biospecimen Science]
Author Disclosure Statement
No competing financial interests exist.
References
- 1.ISBER Working Group on Biospecimen Science. Standard Pre-analytical coding for biospecimens: Defining the sample PREanalytical Code (SPREC) Cancer Epidemiol Biomarkers Prevention. 2010;19:1004–1011. doi: 10.1158/1055-9965.EPI-09-1268. [DOI] [PubMed] [Google Scholar]
- 2.Benson EE. Betsou F. Amaral R. Santos LMA. Harding K. Standard PREanalytical Codes (SPREC): A new paradigm for environmental biobanking sectors explored in algal culture collections. Biopreserv Biobank. 2011;9:1–12. doi: 10.1089/bio.2011.0035. [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui R. Ploetz C. Drepper J, et al. Promoting the use of high-quality biospecimens from medical research: Integrating quality coding standards for pre-analytical treatment in the German Biobank Registry. Biopreserv Biobank. 2012;10:187. [Google Scholar]
- 4.Norlin L. Fransson MN. Eriksson M, et al. A minimum data set for sharing biobank samples, information, and data—MIABIS. Biopreserv Biobank. 2012;10:343–348. doi: 10.1089/bio.2012.0003. [DOI] [PubMed] [Google Scholar]
- 5.Klingström T. Soldatova L. Stevens R, et al. A workshop on laboratory protocol standards for the molecular methods database. N Biotechnol. 2012 Jun 2; doi: 10.1016/j.nbt.2012.05.019. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Nanni U. Betsou F. Riondino S, et al. SPRECware: Software tools for Standard PREanalytical Code (SPREC) labeling: Effective exchange and search of stored biospecimens. Int J Biol Markers. 2012 doi: 10.5301/JBM.2012.9718. in press. [DOI] [PubMed] [Google Scholar]
- 7.Moore HM. Kelly A. Jewell SD, et al. Biospecimen reporting for improved study quality. Biopreserv Biobank. 2011;9:57–70. doi: 10.1089/bio.2010.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]